Abstract
Insulin activates sterol regulatory element-binding protein-1c (SREBP-1c) in liver, thereby increasing fatty acid and triglyceride synthesis. We created a line of transgenic rats that produce epitope-tagged human SREBP-1c in liver under control of the constitutive apolipoprotein E promoter/enhancer. This system allows us to dissect the pathway by which insulin stimulates SREBP-1c processing without interference by the insulin-mediated increase in SREBP-1c mRNA. Rats are used because freshly isolated rat hepatocytes respond much more robustly to insulin than do mouse hepatocytes. The data reveal that insulin-mediated stimulation of SREBP-1c processing requires the mechanistic target of rapamycin complex 1 (mTORC1), which also is required for insulin-mediated SREBP-1c mRNA induction. However, in contrast to mRNA induction, insulin stimulation of SREBP-1c processing is blocked by an inhibitor of p70 S6-kinase. The data indicate that the pathways for insulin enhancement of SREBP-1c mRNA and proteolytic processing diverge after mTORC1. Stimulation of processing requires the mTORC1 target p70 S6-kinase, whereas induction of mRNA bypasses this enzyme. Insulin stimulation of both processes is blocked by glucagon. The transgenic rat system will be useful in further defining the molecular mechanism for insulin stimulation of lipid synthesis in liver in normal and diabetic states.
Keywords: mTORC1 kinase, lipid metabolism
The liver plays a unique role in lipid metabolism because it is the only organ that synthesizes fatty acids (FAs) and triglycerides (TGs) for export to other tissues. These synthetic processes are controlled reciprocally by insulin and glucagon, which are secreted by the pancreas and delivered directly to the liver via the portal vein. Precise control is important because excess FA synthesis leads to elevated FAs in muscle, thereby contributing to the peripheral insulin resistance and lipotoxicity seen in type 2 diabetes. Excess FA synthesis also causes fatty liver, which sometimes leads to cirrhosis and liver failure (1, 2).
Insulin stimulates FA synthesis in liver by increasing the mRNA and the processed nuclear form of sterol regulatory element-binding protein-1c (SREBP-1c), a transcription factor that activates all the genes needed to produce FAs and TGs in liver (3). Of the three SREBP isoforms, SREBP-1c is the one whose expression is highest in liver, and it is the only one that is controlled primarily by insulin. For this reason, definitive studies of insulin-mediated activation of SREBP-1c must be performed with liver cells.
Studies of insulin action on liver cells are difficult because none of the established hepatocyte cell lines responds to insulin with the robustness observed in the livers of living animals. Moreover, freshly isolated hepatocytes lose their insulin responsiveness within 48–72 h after isolation. Therefore, studies must be performed in living animals or with freshly isolated hepatocytes that are less than 72 h old. Even more perplexing are species differences. Although mouse and rat livers manifest robust elevations in SREBP-1c mRNA when exposed to insulin in vivo, freshly isolated mouse hepatocytes show much less sensitivity to insulin than rat hepatocytes. Although numerous studies have dealt with the regulation of SREBP mRNA and processing in various tissues (reviewed in ref. 4), only a few have been carried out in freshly isolated rat hepatocytes (5–11).
SREBP-1c, like the other two SREBP isoforms, is synthesized as a membrane-bound protein embedded in endoplasmic reticulum (ER) membranes (12). Immediately after synthesis, SREBP forms a complex with Scap, a polytopic membrane protein that facilitates incorporation of the SREBP into COPII-coated vesicles that bud from the ER and fuse with the Golgi apparatus (13). There SREBP is processed by two proteases to liberate a soluble fragment that travels to the nucleus and activates transcription. Movement of the Scap/SREBP complex is inhibited by Insig proteins, intrinsic ER proteins that bind the Scap/SREBP complex and prevent its movement to the Golgi (13). The liver produces two Insig isoforms, Insig-1 and Insig-2, both of which retard the movement of Scap/SREBP complexes (14).
Insulin activates SREBP-1c in liver at two levels. It increases SREBP-1c processing to liberate the nuclear form, and it increases transcription of the SREBP-1c gene, leading to increased SREBP-1c mRNA and precursor protein (12, 15). Under optimal conditions with freshly isolated rat hepatocytes in cell culture, insulin can induce the SREBP-1c mRNA by as much as 40-fold within 6 h (8, 11). This increase is blocked by wortmannin, an inhibitor of phosphatidylinositol 3-kinase, an early enzyme in the insulin-signaling cascade. The increase also is blocked by low concentrations of rapamycin, indicating that the mechanistic target of rapamycin complex 1 (mTORC1) is required also. Importantly, the increase was not blocked by an inhibitor of p70 S6-kinase (S6K), one of the major downstream targets of mTORC1, suggesting that another target of mTOR is involved (11). Remarkably, rapamycin did not block the other major metabolic effect of insulin in liver, namely, the reduction in mRNAs encoding enzymes of gluconeogenesis. This finding indicated that the insulin-signaling pathway diverges upstream of mTORC1, with one pathway leading through mTORC1 to increase SREBP-1c mRNA and the other bypassing mTORC1 and downregulating the gluconeogenic mRNAs (11).
The other action of insulin on SREBP-1c, the increase in proteolytic processing, has been difficult to study because the increase in SREBP-1c mRNA and precursor protein levels makes an increase in nuclear SREBP-1c difficult to interpret. Hegarty et al. (9) attempted to circumvent this problem by first treating freshly isolated rat hepatocytes with an activator of liver X receptor (LXR), which increased SREBP-1c mRNA without stimulating proteolytic processing, and then adding insulin to stimulate processing. They found that the insulin stimulation of processing was blocked by wortmannin. However, further advances with this approach may be limited because of the requirement for an LXR activator, which might contribute to insulin’s action in stimulating SREBP-1c processing (8).
In the current study, we have taken an alternative approach. To eliminate transcriptional effects of insulin on the SREBP-1c gene, we generated a line of transgenic rats that produce epitope-tagged SREBP-1c driven by a human apoE promoter/enhancer expression cassette, which is not regulated by insulin. Because rat hepatocytes give a dramatically more robust response to insulin than do mouse hepatocytes (6, 7), we injected our transgene into fertilized eggs of rats instead of mice. When treated with insulin, freshly isolated hepatocytes from these transgenic rats exhibited a consistent increase in the amount of the transgene-encoded nuclear SREBP-1c, allowing further dissection of the responsible signaling mechanism.
Results
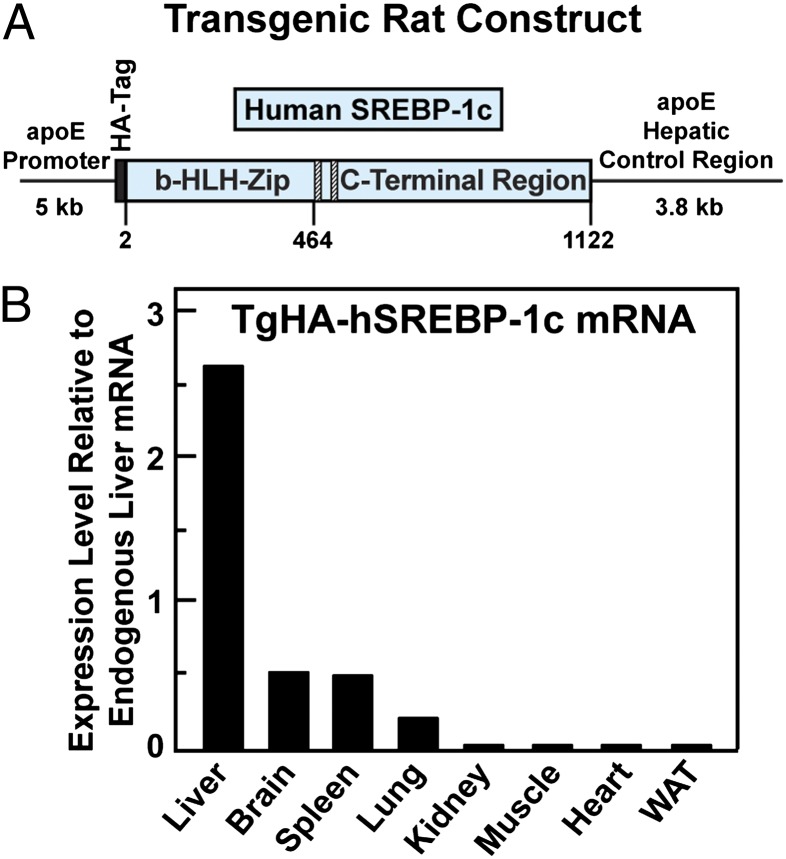
Fig. 1A shows the HA-tagged human SREBP-1c transgene that was used to generate the transgenic rats. Expression was mediated by the apoE promoter and its hepatic control region, which maximizes expression in hepatocytes. The transgenic rat line, hereafter designated “TgHA-hSREBP-1c,” was maintained as hemizygotes by breeding with WT Sprague–Dawley rats. In the liver, the amount of human SREBP-1c mRNA from the transgene was ∼2.5 times that of the endogenous rat SREBP-1c mRNA when the ad libitum-fed animals were killed 6 h into the dark cycle (Fig. 1B). Lower levels of transgene expression were seen in brain, spleen, and lung.
Fig. 1.
Generation of transgenic rats expressing HA-tagged full-length human SREBP-1c in liver. (A) The transgene construct contains a cDNA fragment encoding HA-tagged full-length human SREBP-1c (amino acids 2–1122) under control of human apoE promoter and its hepatic control region. (B) Tissue distribution of human HA-hSREBP-1c transgene. Male TgHA-hSREBP-1c rats (2–3 mo old) that were fed a chow diet ad libitum were killed 6 h into the dark cycle. Equal amounts of total RNA from the indicated tissues of four transgenic rats were pooled and subjected to real-time PCR. Each value represents the amount of transgenic human SREBP-1c mRNA in the indicated tissue relative to that of endogenous rat SREBP-1c mRNA in the liver, which is arbitrarily defined as 1. The cycle threshold (Ct) values for endogenous and transgenic SREBP-1c in the liver were 24.4 and 23.0, respectively.
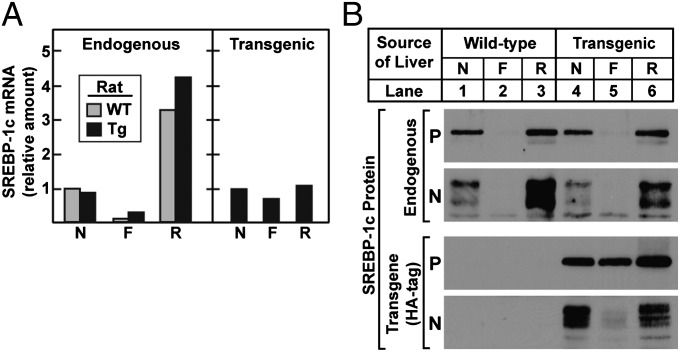
In WT rats, as observed previously (11), the amount of SREBP-1c mRNA declined dramatically after a 48-h fast and increased markedly after the animals had been re-fed with a high-carbohydrate diet for 6 h (Fig. 2A, Left). Nearly identical changes were observed when the endogenous SREBP-1c mRNA was measured in the transgenic rats. In marked contrast, the transgene mRNA did not decline significantly when the animals were fasted, nor did it rise when they were re-fed (Fig. 2A, Right). SREBP-1c protein levels were measured by immunoblots (Fig. 2B). The precursor and nuclear forms of endogenous SREBP-1c declined dramatically during fasting and were restored by refeeding in transgenic rats as well as in WT animals (lanes 1–6). Immunoblots of the SREBP-1c precursor generated by the transgene revealed little decrease on fasting (lanes 4–6). Nevertheless, the nuclear form was reduced dramatically, suggesting a decrease in proteolytic processing. Refeeding produced an increase in the nuclear form, again without a change in the precursor, suggesting that proteolytic processing was activated.
Fig. 2.
Effect of fasting and refeeding on the hepatic levels of endogenous and transgenic SREBP-1c mRNAs and proteins in WT and TgHA-hSREBP-1c rats. Male WT and transgenic TgHA-hSREBP-1c (Tg) littermates (2–3 mo old, four rats per group) were subjected to fasting and refeeding as described in Materials and Methods. The nonfasted group (N) was fed a chow diet ad libitum, the fasted group (F) was fasted for 48 h, and the re-fed group (R) was fasted for 48 h and re-fed a high-carbohydrate diet for 6 h before study. (A) Equal amounts of total RNA from livers of four rats were pooled and subjected to real-time PCR. For the endogenous rat SREBP-1c mRNA, each value represents the amount relative to the amount in the nonfasted WT rats, which was arbitrarily defined as 1 (Ct value, 24.2). For the transgenic human SREBP-1c mRNA, the level in the nonfasted transgenic rats was arbitrarily defined as 1 (Ct value, 23.2). (B) Liver membrane and nuclear extract fractions were prepared individually and pooled from four rats per group. The mean values for the total amount of protein in the 24 membrane and nuclear extract fractions were 2,459 and 458 μg, respectively. Aliquots of membrane (30 μg) and nuclear extract (30 μg) fractions were subjected to 8% SDS/PAGE and immunoblot analysis with both anti-rat SREBP-1 and anti-HA antibodies to detect membrane-bound precursor (P) and cleaved nuclear (N) forms of endogenous rat SREBP-1 and HA-tagged transgenic human SREBP-1c, respectively. The exposure times for the autoradiograms were 1 s for all precursors, 10 s for transgenic nuclear extract, and 60 s for endogenous nuclear extracts.
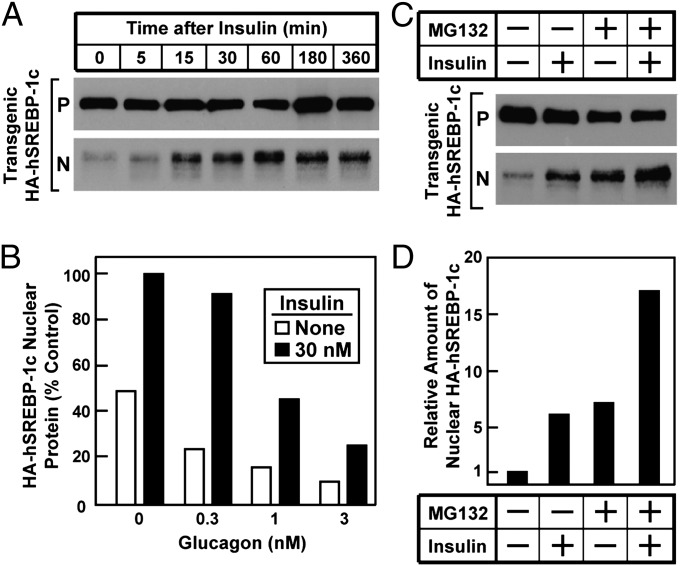
To determine whether insulin mediates the increase in SREBP-1c processing, we used fresh hepatocytes from the transgenic rats (Fig. 3). When the cells were exposed to 100 nM insulin, the increase in nuclear SREBP-1c was maximal within 15 min, and the nuclear protein remained elevated through 6 h (Fig. 3A). Similar results were obtained with insulin concentrations as low as 10 nM. The addition of glucagon decreased the amount of basal and insulin-induced nuclear SREBP-1c, as determined by densitometric staining of immunoblots (Fig. 3B). In the presence of 3 nM glucagon, basal nuclear SREBP-1c was reduced by 80%, and the absolute amount of insulin stimulation was reduced by 70%. Glucagon did not significantly reduce the amount of the precursor encoded by the SREBP-1c transgene.
Fig. 3.
Insulin-mediated stimulation of proteolytic processing of transgenic SREBP-1c in primary rat hepatocytes. Hepatocytes from nonfasted TgHA-hSREBP-1c rats were prepared and plated on day 0 as described in Materials and Methods. (A) Time course of insulin effect. On day 1, the cells were left untreated or treated with 100 nM insulin for the indicated time, harvested, and then pooled (three dishes of cells per sample) for immunoblot analysis of precursor (P) and nuclear (N) forms of transgenic HA-hSREBP-1c protein. The gels were exposed to film for 5 s. (B) Effect of glucagon. On day 1, cells were left untreated or were pretreated for 2 h with the indicated concentration of glucagon, after which the cells were left untreated or were treated with 30 nM insulin for 1 h before harvest as above. Immunoblot analysis of nuclear transgenic HA-hSREBP-1c was carried out and then was scanned and quantified by densitometry as described in SI Materials and Methods. The amount of the cleaved nuclear HA-hSREBP-1c protein in the cells without any treatment was arbitrarily set at 1. (C and D) Effect of proteasomal inhibitor. (C) On day 1, hepatocytes were left untreated or were treated 10 nM insulin for 1 h in the absence or presence of 10 μM MG132 (proteasomal inhibitor) and then were harvested for immunoblot analysis as in A. The gels were exposed to film for 5–10 s. (D) The gel of nuclear extract fractions (N) in C was scanned and quantified by densitometry as in B, with the amount of nuclear HA-hSREBP-1c protein in cells without any treatment arbitrarily set at 1.
Nuclear SREBPs are known to be degraded rapidly by proteasomal proteolysis (16). The rapidity of the insulin effect (Fig. 3A) suggests that insulin acts by increasing the production of the nuclear form rather than by blocking its degradation. To test this conclusion in another way, we made use of the proteasome inhibitor MG132 (Fig. 3 C and D). In the absence of MG132, the addition of insulin to the transgenic hepatocytes produced a sixfold increase in nuclear SREBP-1c as determined by densitometric scans. Addition of MG132 also caused an increase in nuclear SREBP-1c, presumably by preventing degradation. However, the amount of the nuclear protein rose further when insulin was added, even when the nuclear protein was stabilized by MG132, indicating that insulin acts by increasing production of the nuclear form rather than by blocking degradation.
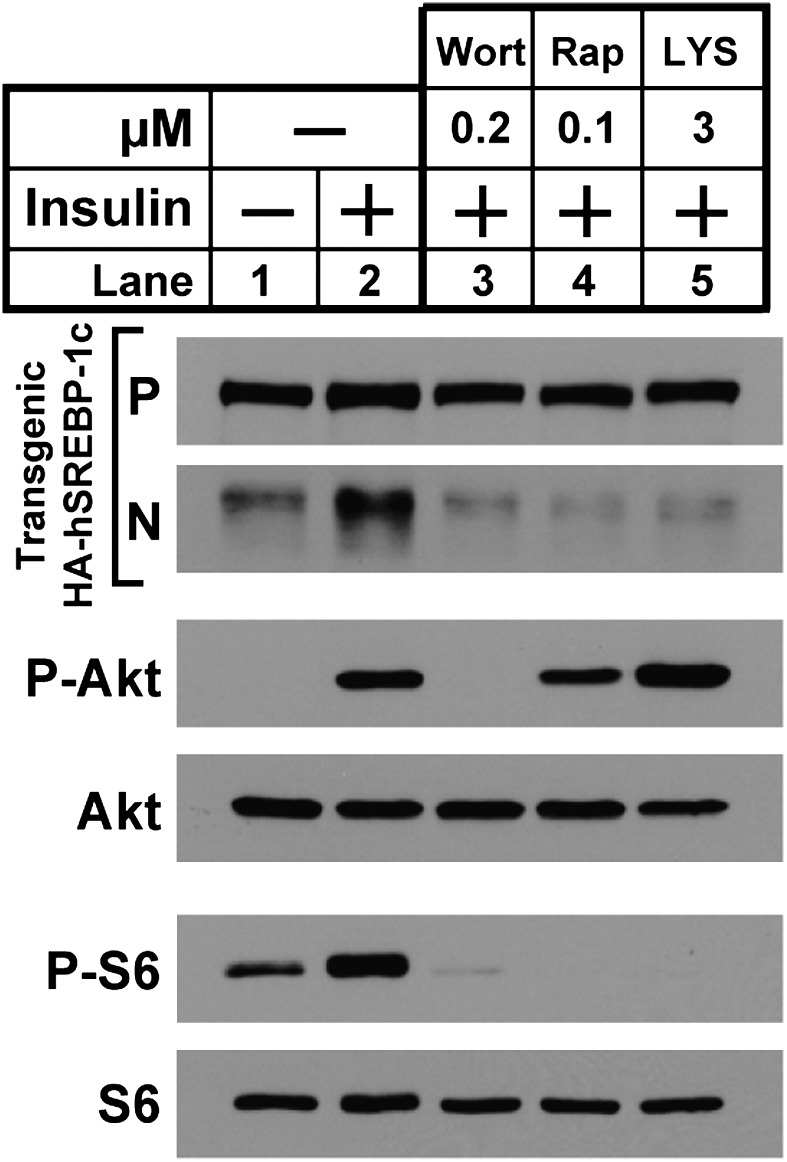
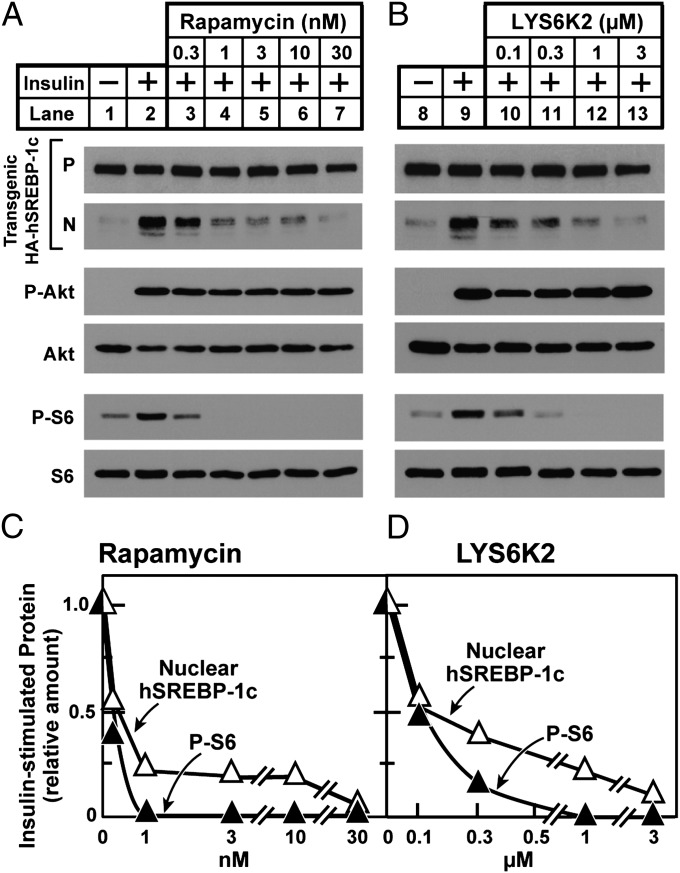
To dissect the insulin-signaling pathway that mediates the increase in SREBP-1c processing in transgenic hepatocytes, we used the three inhibitors shown in Fig. 4. In the absence of inhibitors, insulin increased the amount of nuclear SREBP-1c (lane 2). As expected, this increase was blocked by wortmannin (lane 3). It also was blocked by rapamycin (lane 4). The increase in SREBP-1c processing was inhibited by LYS6K2, an inhibitor of S6K that blocked the phosphorylation of S6 (lane 5). The result with LYS6K2 was unexpected, because we reported previously that LYS6K2 did not block the insulin-induced increase of SREBP-1c mRNA (11). As a control, neither rapamycin nor LYS6K2 blocked the insulin-induced phosphorylation of Akt.
Fig. 4.
Effects of protein kinase inhibitors on insulin-stimulated proteolytic processing of transgenic SREBP-1c in rat hepatocytes. Hepatocytes from nonfasted TgHA-hSREBP-1c rats were prepared and plated on day 0. On day 1, cells were left untreated or were pretreated for 3 h with the indicated protein kinase inhibitor: 0.2 μM wortmannin (lane 3), 0.1 μM rapamycin (lane 4), or 3 μM LYS6K2 (lane 5). After this preincubation, the cells were left untreated or were treated with 10 nM insulin for 30 min, harvested, and then pooled (three dishes of cells per sample). Each sample was subjected to immunoblot analyses of the following proteins: precursor (P) and nuclear (N) forms of transgenic HA-hSREBP-1c, phosphorylated Akt (P-Akt), total Akt, phosphorylated ribosomal protein S6 (P-S6), and total S6. Gels were exposed to film for 3–25 s.
Fig. S1 shows a compilation of results from 10 independent experiments with rapamycin and eight experiments with LYS6K2, all of which found that both inhibitors blocked the insulin-induced processing of transgene-encoded SREBP-1c. To confirm the failure of LYS6K2 to block the mRNA induction of SREBP-1c, we used the conditions of Li et al. (11). Hepatocytes from the transgenic rats were pretreated for 30 min with vehicle, rapamycin, or LYS6K2. The cells then were incubated for 6 h in the absence or presence of 30 nM insulin. Cells were harvested, and the amount of endogenous SREBP-1c mRNA was quantified by real-time PCR. Nuclear extracts were prepared from identically treated cells and used for the measurement of nuclear SREBP-1c derived from the transgene as determined by SDS/PAGE, immunoblotting, and densitometric scanning (Fig. S2). After 6 h, insulin increased the amount of endogenous SREBP-1c mRNA by 14-fold. Rapamycin reduced this increase by 85%, but LYS6K2 had no effect. In similarly treated cells, insulin increased transgene-encoded nuclear SREBP-1c by 12-fold over the course of 6 h. Rapamycin blocked this increase by 64%, and LYS6K2 reduced it by 74%. Thus, the LYS6K2 inhibition was specific for protein processing and not for mRNA induction.
To provide further evidence that rapamycin and LYS6K2 block processing by inhibiting their respective enzymes, we performed dose–response curves with the inhibitors and measured phospho-S6 and nuclear SREBP-1c in the same cells (Fig. 5). Within the range of precision of these immunoblotting assays, the degree of inhibition of SREBP-1c processing by rapamycin and LYS6K2 was well correlated with the degree of reduction of phospho-S6, supporting the notion that the two inhibitors work by blocking the activity of S6K (Fig. 5 C and D). Again, insulin-induced phosphorylation of Akt was not blocked by either of the inhibitors.
Fig. 5.
mTORC1 and S6K are required for insulin to stimulate proteolytic processing of transgenic SREBP-1c in rat hepatocytes. Hepatocytes from nonfasted TgHA-hSREBP-1c rats were prepared and plated on day 0. On day 1, cells were left untreated or were pretreated for 2 h with the indicated concentration of rapamycin or LYS6K2, after which the cells were left untreated or were treated with 30 nM insulin for 1 h, harvested, and then pooled (three dishes of cells per sample) for immunoblot analysis. (A and B) Samples were subjected to immunoblot analysis as in Fig. 4. (C and D) Quantification of immunoblots. The gels of nuclear extract fractions (N) of transgenic HA-hSREBP-1c protein and cytosol fractions of phosphorylated ribosomal protein S6 (P-S6) in A and B were scanned and quantified by densitometry. The amount of nuclear HA-hSREBP-1c or cytosol P-S6 protein in cells treated with insulin alone (lanes 2 and 9) was arbitrarily set at 1.
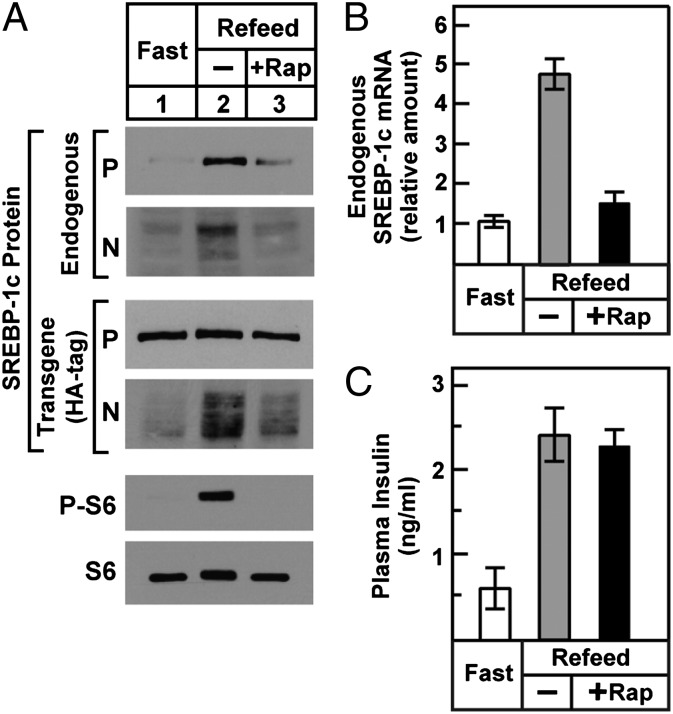
Fig. 6 shows an in vivo experiment designed to determine whether rapamycin blocks the increase in SREBP-1c processing in livers of rats after refeeding. Transgenic rats were fasted for 48 h and then re-fed for 3 h. One hour before refeeding, they were injected i.p. with vehicle or with rapamycin. Refeeding increased the amount of nuclear SREBP-1c derived from the transgene as well as the endogenous gene (Fig. 6A). Both of these feeding-induced increases were decreased by rapamycin. As previously reported (11), rapamycin also prevented the refeeding-induced increase in endogenous SREBP-1c mRNA (Fig. 6B). As a positive control, rapamycin prevented the increase in phospho-S6 that was induced by refeeding (Fig. 6A). After refeeding, plasma insulin rose equally in the vehicle- and rapamycin-treated rats (Fig. 6C), indicating that the rapamycin-mediated inhibition of SREBP-1c processing and transcription was not the result of decreased insulin availability.
Fig. 6.
Effect of mTORC1 inhibition on SREBP-1c processing (A) and SREBP-1c transcription (B) in livers of TgHA-hSREBP-1c rats subjected to fasting and refeeding. Male transgenic rats (2–3 mo old, four rats per group) were fasted for 48 h or were fasted for 48 h and then re-fed a high-carbohydrate diet for 3 h before study. Animals in the re-fed group were injected i.p. with vehicle or 20 mg/kg rapamycin (+Rap) 1 h before refeeding. (A) Liver membrane, nuclear extract, and cytosol fractions were prepared individually and pooled (four rats per group). Membrane and nuclear extract fractions were subjected to immunoblot analysis to detect precursor (P) and nuclear (N) forms of HA-tagged transgenic human SREBP-1c and endogenous rat SREBP-1 as in Fig. 2. Immunoblot analysis of cytosolic phosphorylated ribosomal protein S6 (P-S6) and total S6 was carried out as described in SI Materials and Methods. Gels were exposed to film for 3–60 s. (B) Total liver RNA was prepared individually and subjected to real-time PCR. Each bar (mean ± SEM of data from four rats) represents the amount of endogenous SREBP-1c mRNA relative to that in the fasted group, which was arbitrarily defined as 1. (C) Plasma insulin levels. Insulin was measured in plasma obtained immediately after administration of anesthesia as described in Materials and Methods. Each bar represents mean ± SEM of data from four rats.
Discussion
The pathways for insulin signaling in liver are complex. mTORC1 plays a central role, but it is embedded in an intricate web of feed-forward loops, feedback loops, bypass loops, and negative regulators of negative regulators whose interactions render definitive study difficult [for an illustration of this complexity, see figure 2 in Laplante and Sabatini (17)]. Moreover, insulin-signaling pathways differ in different cell types, and they are affected by the actions of other hormones. For this reason, our laboratory (6, 8, 11) and others (5, 9, 10) have focused on rat hepatocytes, which show robust responses to insulin both in living animals and immediately after their isolation from cell culture. Here we introduce a system for studying insulin stimulation of SREBP-1c processing, namely, a transgenic rat expressing human SREBP-1c mRNA in liver via a promoter/enhancer that is not regulated by insulin.
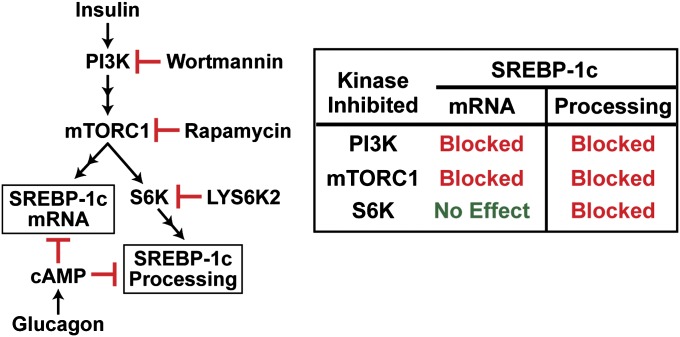
Earlier studies of mRNA expression in rat hepatocytes revealed a bifurcation in the insulin pathway in liver (11). One branch leads through mTORC1 and activates lipid synthesis through an increase in SREBP-1c; the other branch bypasses mTORC1 and leads to a reduction in gluconeogenesis mediated by a reduction in the activity of Foxo1 (18). Here we show that the branch leading to increased SREBP-1c itself bifurcates distal to mTORC1. This latter bifurcation is illustrated in Fig. 7. One pathway leads to an increase in SREBP-1c mRNA and SREBP-1c precursor protein. This action is not inhibited by the S6K inhibitor and therefore is independent of S6K. The other pathway leads to an increase in the proteolytic processing of the SREBP-1c precursor protein. This pathway is inhibited in parallel with the decline in phospho-S6 when S6K is blocked by LYS6K2 (Figs. 4 and 5). Both the SREBP-1c mRNA induction and the increase in SREBP-1c processing are blocked by low concentrations of rapamycin, indicating that mTORC1 is essential for both processes. Both pathways also are blocked by glucagon, acting through cAMP (Fig. 3B) (7, 15).
Fig. 7.
Pathways for insulin stimulation of SREBP-1c mRNA and proteolytic processing diverge after mTORC1. Both processes are inhibited by glucagon acting through cAMP.
Although mTORC1 is essential for SREBP-1c activation in liver, it is not sufficient. This finding was reported initially by Yecies et al. (19), who found that SREBP-1c mRNA is not elevated in livers of mice with genetic ablation of TSC1, a negative regulator of mTORC1, even though these animals have markedly increased mTORC1 activity. Thus, insulin must regulate another process in addition to activation of mTORC1, and activation of both processes is required for induction of SREBP-1c mRNA.
The advantage of the current transgenic rat model is illustrated by the data in Fig. 2B. When WT rats were fasted, their SREBP-1c mRNA declined dramatically, and as a result the SREBP-1c precursor protein was not detectable. When the animals were re-fed, the SREBP-1c mRNA increased, and so did the precursor and nuclear forms of the protein (Fig. 2). From such an experiment, it is impossible to tell whether insulin increases SREBP-1c processing or whether it acts only to increase the amount of the SREBP-1c precursor, whereupon processing takes place through some constitutive mechanism. A similar problem was encountered by Bae et al. (20), who demonstrated clearly that knockdown of S6K by RNA interference in mouse liver reduced nuclear SREBP-1c after refeeding, but they could not distinguish an effect on processing versus an effect on mRNA induction. When encoded in rats by the transgene, SREBP-1c mRNA was present even in fasted animals, as was the precursor protein. However, there was very little nuclear form, indicating that processing was blocked. Refeeding increased the nuclear form without increasing the amount of precursor, indicating that the processing of SREBP-1c was enhanced (Fig. 2).
Although the SREBP-1c produced by the transgene provides a good model for studying proteolytic processing, it is not a model for SREBP-1c transcriptional activity. Insertion of an epitope tag at the N terminus of an SREBP markedly reduces its transcriptional activity. Indeed, in our transgenic rats, the amount of SREBP-1c target mRNAs, including acetyl CoA carboxylase and fatty acid synthase, was not elevated despite the 2.5-fold increase in SREBP-1c mRNA.
Because nuclear SREBPs are known to be degraded rapidly by proteasomes, we considered the possibility that insulin stabilized the nuclear form rather than increasing its production. Two observations argued against this possibility. First, insulin increased the steady-state level of nuclear SREBP-1c by four- to fivefold within 15 min (Fig. 3A), a rate that is much too fast to be attributed to stabilization. Second, insulin increased the amount of nuclear SREBP-1c even when degradation was blocked by a proteasome inhibitor (Fig. 3C).
In addition to causing a spike in plasma insulin, refeeding also causes a decline in glucagon (21). We showed earlier that glucagon blocks the insulin induction of SREBP-1c mRNA in rat hepatocytes (7). Here we show that glucagon also reduces the insulin-mediated increase of SREBP-1c processing in transgenic hepatocytes (Fig. 3B). Thus, in re-fed WT animals the dramatic increase in nuclear SREBP-1c represents the combined effects of insulin increase and glucagon reduction.
The transgenic rat model offers the promise of a system that should allow more careful dissection of the mechanism by which insulin activates SREBP-1c processing. In particular, it will be necessary to define the presumptive target of S6K that leads to the liberation of the Scap/SREBP-1c complex from the ER.
Materials and Methods
Materials.
We obtained powdered bovine insulin from Sigma (catalog no. I6634) and protein kinase inhibitors from sources as previously described (11). LYS6K2, obtained from Eli Lilly and Company, is a highly selective ATP inhibitor of P70 S6K, having been tested against 83 other kinases and 45 cell-surface receptors. A stock solution of 0.1 mM insulin was prepared by dissolving the powder in 1% (vol/vol) acetic acid, after which the solution was stored in multiple aliquots at −80 °C. All kinase inhibitors were prepared in dimethyl sulfoxide and stored at −20 °C. Culture media, FCS, and collagen-coated dishes were obtained from sources as described (11).
Generation of Transgenic Rats.
To generate transgenic rats expressing full-length human SREBP-1c in the liver, we used a pLiv-11 vector that contains the constitutive human apoE promoter/enhancer and its hepatic control region (22). The transgenic plasmid (pLiv-11-HA-hSREBP-1c) was generated by cloning a cDNA fragment encoding the ORF of human SREBP-1c with an N-terminal 3xHA tag into Mlu1-Cla1 sites of pLiv-11. The 11-kb SalI-SpeI fragment of pLiv-11-HA-hSREBP-1c then was isolated and injected into the pronucleus of Sprague–Dawley rat eggs as described (23). Transgenic founders were identified by dot blot analysis and mated with WT Sprague–Dawley rats. To genotype transgenic rats, ear-punch DNA was prepared with a direct lysis kit (Viagen Biotech Inc.) and used for PCR with the primers 5′-GTGCTGGGATTAGGCTGTTGCAGATAATGC-3′ and 5′-GGTACATCTTCAATGGAGTGGGTGCAGGCT-3′. Ear-punch DNA of transgenic rats produced a PCR product of 527 bp. The transgenic rats, hereafter designated TgHA-hSREBP-1c, were maintained as hemizygotes by breeding with WT Sprague–Dawley rats. Two independent lines were established, both exhibiting a two- to threefold overexpression of hepatic HA-hSREBP-1c mRNA relative to the endogenous SREBP-1c mRNA. All rats were housed in colony cages with a 12-h light/12-h dark cycle and were fed Teklad Rodent Diet 2016 (Harlan Teklad). Before blood and liver were obtained, rats were anesthetized in a bell-jar atmosphere containing isoflurane. All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee at University of Texas Southwestern Medical Center.
Fasting and Refeeding Studies.
The rats were divided into three groups: nonfasted, fasted, and re-fed. The nonfasted group was fed a chow diet ad libitum. The fasted group was fasted for 48 h, and the re-fed group was fasted for 48 h and then re-fed a high-carbohydrate, fat-free diet (960238; MP Biomedicals) for 3 or 6 h before study. Feeding regimens were carried out in a staggered fashion so that all rats were euthanized at the same time. In one experiment (Fig. 6), rats were injected i.p. with either vehicle alone [14% ethanol, 5% (vol/vol) Tween 80, and 5% (vol/vol) polyethylene glycol 400] or vehicle containing 20 mg/kg rapamycin. Plasma insulin levels were measured with a commercial ELISA kit (Crystal Chem). The plasma was obtained from blood drawn from the inferior vena cava, which was collected on ice in EDTA-coated tubes, separated by centrifugation, and stored at −20 °C.
Primary Rat Hepatocytes.
Primary hepatocytes were isolated from nonfasted rats (male, 2–3 mo old) and cultured as previously described (11) with the following minor modifications. On day 0, isolated hepatocytes were plated onto collagen I-coated dishes (1 × 107 cells per 10-cm dish) in 10 mL medium A [Medium 199 (Life Technologies) supplemented with 5% (vol/vol) FCS, 100 nM dexamethasone, 100 units/mL penicillin, and 100 μg/mL streptomycin sulfate]. After incubation for 3–4 h at 37 °C in 5% CO2, the attached cells were washed once with PBS and then incubated at 37 °C overnight in medium B (medium A without FCS). Unless noted otherwise, on day 1 cells were pretreated for 2 h with a direct addition of 10 μL dimethyl sulfoxide with or without the indicated protein kinase inhibitor, after which the cells were treated with a direct addition of 10 μL acidified water with or without insulin. After incubation for the indicated time at 37 °C in 5% CO2, the cells were harvested for immunoblot and RNA analysis.
Other Methods.
Additional information about materials and methods is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jay Horton for a critical review of the manuscript; Monica Mendoza, Isis Soto, and Jeff Cormier for excellent technical assistance; and Eli Lilly and Company for the gift of LYS6K2. This work was supported by Grant HL20948 from the National Institutes of Health and by the Moss Heart Foundation. J.L.O., Y.Z., and M.S.F. are supported by Medical Scientist Training Program Grant ST32GM08014.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213343109/-/DCSupplemental.
See Commentary on page 15974.
References
- 1.Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114:147–152. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leavens KF, Birnbaum MJ. Insulin signaling to hepatic lipid metabolism in health and disease. Crit Rev Biochem Mol Biol. 2011;46:200–215. doi: 10.3109/10409238.2011.562481. [DOI] [PubMed] [Google Scholar]
- 3.Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krycer JR, Sharpe LJ, Luu W, Brown AJ. The Akt-SREBP nexus: Cell signaling meets lipid metabolism. Trends Endocrinol Metab. 2010;21:268–276. doi: 10.1016/j.tem.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Foretz M, Guichard C, Ferré P, Foufelle F. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc Natl Acad Sci USA. 1999;96:12737–12742. doi: 10.1073/pnas.96.22.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimomura I, et al. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci USA. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimomura I, et al. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol Cell. 2000;6:77–86. [PubMed] [Google Scholar]
- 8.Chen G, Liang G, Ou J, Goldstein JL, Brown MS. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci USA. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hegarty BD, et al. Distinct roles of insulin and liver X receptor in the induction and cleavage of sterol regulatory element-binding protein-1c. Proc Natl Acad Sci USA. 2005;102:791–796. doi: 10.1073/pnas.0405067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yellaturu CR, et al. Posttranslational processing of SREBP-1 in rat hepatocytes is regulated by insulin and cAMP. Biochem Biophys Res Commun. 2005;332:174–180. doi: 10.1016/j.bbrc.2005.04.112. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci USA. 2010;107:3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 13.Sun L-P, Seemann J, Goldstein JL, Brown MS. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci USA. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 15.Ferré P, Foufelle F. Hepatic steatosis: A role for de novo lipogenesis and the transcription factor SREBP-1c. Diabetes Obes Metab. 2010;12(Suppl 2):83–92. doi: 10.1111/j.1463-1326.2010.01275.x. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 17.Laplante M, Sabatini DM. mTOR Signaling. Cold Spring Harb Perspect Biol. 2012;4:a011593. doi: 10.1101/cshperspect.a011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu M, et al. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat Med. 2012;18:388–395. doi: 10.1038/nm.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yecies JL, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae EJ, et al. Liver-specific p70 S6 kinase depletion protects against hepatic steatosis and systemic insulin resistance. J Biol Chem. 2012;287:18769–18780. doi: 10.1074/jbc.M112.365544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unger RH, Cherrington AD. Glucagonocentric restructuring of diabetes: A pathophysiologic and therapeutic makeover. J Clin Invest. 2012;122:4–12. doi: 10.1172/JCI60016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelking LJ, et al. Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J Clin Invest. 2004;113:1168–1175. doi: 10.1172/JCI20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young SG, Lusis AJ, Hammer RE. In: Molecular Basis of Cardiovascular Disease. Chien KR, editor. Philadelphia: W.B. Saunders Company; 1999. pp. 37–85. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.