Abstract
In both metazoan development and metastatic cancer, migrating cells must carry out a detailed, complex program of sensing cues, binding substrates, and moving their cytoskeletons. The linker cell in Caenorhabditis elegans males undergoes a stereotyped migration that guides gonad organogenesis, occurs with precise timing, and requires the nuclear hormone receptor NHR-67. To better understand how this occurs, we performed RNA-seq of individually staged and dissected linker cells, comparing transcriptomes from linker cells of third-stage (L3) larvae, fourth-stage (L4) larvae, and nhr-67-RNAi–treated L4 larvae. We observed expression of 8,000–10,000 genes in the linker cell, 22–25% of which were up- or down-regulated 20-fold during development by NHR-67. Of genes that we tested by RNAi, 22% (45 of 204) were required for normal shape and migration, suggesting that many NHR-67–dependent, linker cell-enriched genes play roles in this migration. One unexpected class of genes up-regulated by NHR-67 was tandem pore potassium channels, which are required for normal linker-cell migration. We also found phenotypes for genes with human orthologs but no previously described migratory function. Our results provide an extensive catalog of genes that act in a migrating cell, identify unique molecular functions involved in nematode cell migration, and suggest similar functions in humans.
Keywords: major sperm protein, nematode development, transcriptional profiling, twin pore potassium channels, conserved uncharacterized proteins
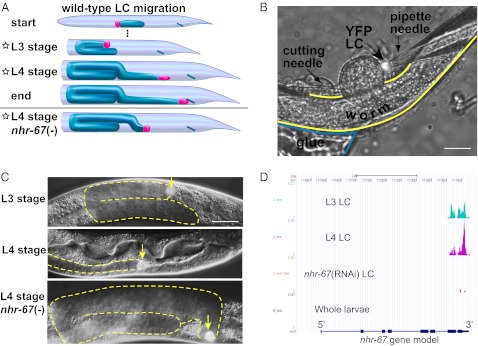
Cell migration is pervasive in animal development and is also an unwanted part of human cancer. Several different varieties of cell migration, ranging from social protozoa to the human immune system, have been extensively studied (1), but there remains a need for detailed analysis of the different components of migration in a simple cell type that can be exhaustively probed through anatomy, genetics, and genomics. We have undertaken to develop the linker cell (LC) of Caenorhabditis elegans as such a model. The LC is a single cell, attached to the front of a proliferating male gonad, which carries out a stereotypic, long-range migration to generate the shape of the mature gonad (Fig. 1A). During 25 h of the second to fourth larval stages (L2 through L4), the LC moves anteriorly along the ventral body wall, turns dorsally, proceeds posteriorly, switches sides once more to the ventral bodywall, continues posteriorward, reaches the cloaca, and eventually dies (2–4). Throughout this process, the proliferating and lengthening male gonad follows the LC’s path to the cloaca. Stage-specific changes occur during the middle to late stages of migration (L3 and L4 larval stages): the LC changes shape from a spheroid to a pointed ellipsoid; it increases its speed of migration; and the netrin receptor gene unc-5 is down-regulated within the LC, which causes it to switch body walls ventrally (Fig. 1A; ref. 3). These changes between the L3 and L4 stages require the orphan nuclear receptor NHR-67, whose orthologs TAILLESS and TLX are required for embryonic and neural development in Drosophila and mice (5, 6). Unlike most known regulators of migrating cells, NHR-67 acts temporally rather than spatially; in the absence of NHR-67, the LC of the L4 stage has the characteristics of the earlier L3 stage (Fig. 1; ref. 3).
Fig. 1.
LC biology, dissection, and transcriptional profiling. (A) The LC position and gonad shape during LC migration. The LC (red), located at the tip of the gonad (blue), begins migrating toward the head, performs a U-turn while switching to the dorsal bodywall, migrates posteriorly and turns downward, and continues to migrate toward the tail until stopping at the cloaca (blue line). The bottom panel shows LC migration in an nhr-67(RNAi) animal at the same L4 stage timepoint as the wild-type animal. Stars indicate the types of LCs collected for profiling. (B) A YFP-labeled LC is dissected from an animal glued to an agar pad by extruding the gonad with a cutting needle and collecting the LC into a patch pipet. (Scale bar, 20 μm.) (C) Three classes of LCs microdissected for RT-PCR and RNA-seq. Trailing gonads are outlined in yellow. The nhr-67(RNAi) L4-stage LC is morphologically retarded, still showing the spheroid shape normally seen in L3-stage LCs. (Scale bar, 20 μm.) (D) The RNA-seq profile for nhr-67 showed tissue-specificity of transcripts and efficacy of RNAi. Our RT-PCR protocol (13) deliberately limited amplification to the 3′ exons of genes.
To discover what genes are specifically expressed in the LC, how they change during LC development, and how NHR-67 controls them, we sought to characterize the transcriptome of the developing LC. Previous analyses of the migratory transcriptome have tended to rely on microarrays of cell populations, such as populations of border cells in Drosophila (7, 8) and heart cells in Ciona (9). More recently, RNA-seq (10) and functional genomics (11) have been used to identify mammalian genes involved in the transition to invasive mesenchymal cell types. In C. elegans, it should be possible to combine such RNA-seq with single-cell analysis to achieve a comprehensive inventory of genes involved in migration, with precise temporal resolution. Because the LC is a solitary and highly dynamic postembryonic cell, neither embryonic cell cultures nor cell-specific pulldown from mass worm cultures (12) were promising approaches to obtaining LCs in bulk. We thus sequenced single-cell RT-PCRs (13) from individually dissected wild-type LCs from the mid-L3 and mid-L4 larval stages, along with mid-L4–stage LCs undergoing RNAi of nhr-67 (Fig. 1 B and C; ref. 3).
Results
RNA-seq of Individual Migrating Linker Cells.
We harvested individual yellow fluorescent protein (YFP)-labeled LCs for RNA-seq by cutting them free of the gonad with a laser microbeam, cutting open the midbody (14), and pipetting individual LCs (Fig. 1B). We ran single-cell RT-PCRs (13) on wild-type LCs from the mid-L3 and mid-L4 larval stages, as well as mid-L4 LCs with RNAi-inactivated nhr-67 (3). As a control for housekeeping genes (15) and genes expressed in other differentiated cell types (16), we also amplified RNA from whole mixed-stage hermaphrodites (primarily larvae). To minimize variable amplification of transcripts from five individual cells, we initially pooled aliquots of RT-PCRs from each cell type before sequencing. To determine cell-to-cell variability, we later sequenced RT-PCRs of each isolated cell individually and to greater depth. Gene expression was measured as reads per kilobase of exon model per million mapped reads (RPKM; ref. 17).
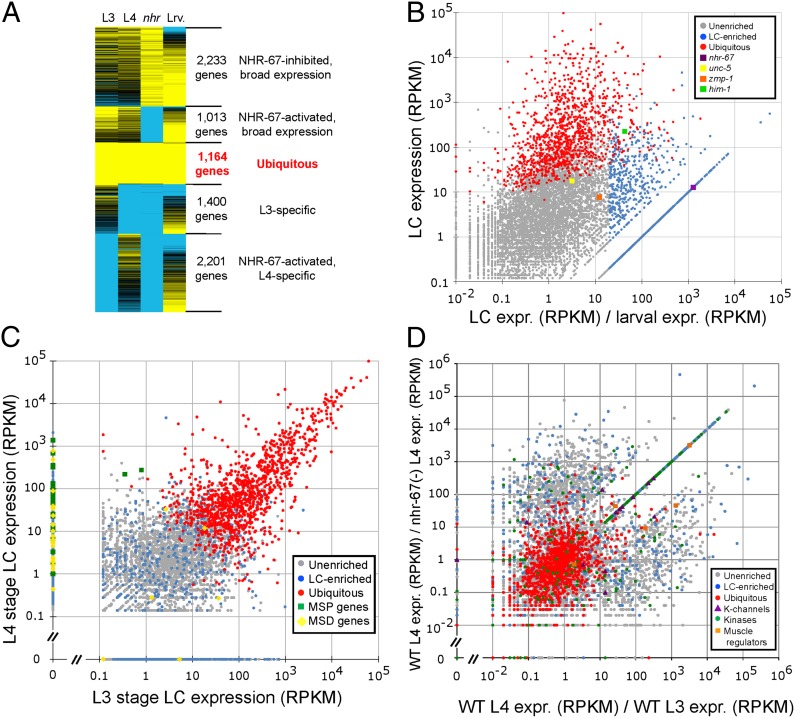
We detected expression for 8,000–10,000 C. elegans genes in wild-type L3- and L4-stage LCs (Fig. 2, and SI Appendix, Fig. S1 and Tables S1–S5). In particular, 8,011 genes showed expression in RNA-seq of pooled L3- and L4-stage LCs (SI Appendix, Table S3). This dataset, which was acquired first, largely overlapped with our later data from individual LCs (with 7,915 genes redetected in the latter dataset) and was used in most analyses below. A total of 10,064 genes were detected in all wild-type L3- or L4-stage LCs, pooled and individual. In any individual L3- or L4-stage LC, we generally detected fewer genes on average (mean, 3,531 and 5,083) than in the L3- and L4-stage pools (5,740 and 6,603). Conversely, we detected more expressed genes in the aggregated, deeper RNA-seq data from 10 individual LCs (6,528 in five L3s; 9,153 in five L4s; 9,968 in 10 L3s and L4s) than in RNA-seq of their pools. RNA-seq of any individual cell was thus less sensitive than that of pools, which in turn was less sensitive than aggregating data from several individual cells. Gene-expression values for the mean individual L3, L4, and nhr-67(RNAi) LCs correlated well with expression values from pooled LCs of the same type (r2 = 0.56–0.93; SI Appendix, Table S6). Correlation for mean individual LCs vs. pooled whole larvae was lower (r2 = 0.15–0.20), as expected.
Fig. 2.
Overall gene-expression patterns in LCs. (A) k-partitioning of the expression data from LC genes. Expression data from 8,011 genes expressed in pooled wild-type LCs (“LC genes”) were compared between LC types [L3, L4, nhr-67(RNAi) L4], and mixed-stage larvae (SI Appendix, Table S1 and Dataset S1). This process distinguished four sets of variably expressed genes from a fifth set of 1,164 ubiquitously expressed genes. Ubiquitous genes disproportionately encoded such housekeeping functions as protein translation and electron transport (SI Appendix, Table S7). Yellow and blue denote high and low expression levels, respectively; ubiquitous genes are labeled in red. Genes belonging to each partition are listed in Dataset S1. An alternative partitioning is shown in SI Appendix, Fig. S1. (B) LC/larval expression ratio versus maximum LC gene expression for LC genes. A set of 1,097 LC-enriched genes, including nhr-67, showed ≥20× more expression in wild-type LCs than in larvae, and are marked in blue. Two genes previously shown to be active in the LC (3), the netrin receptor unc-5 and the metalloprotease zmp-1, show more moderate LC-enrichment; conversely, the cohesin subunit gene him-1 (the activity of which in LCs we confirmed with both antibody staining and RNAi; SI Appendix, Table S9) is an instance of the LC-enriched class. Ubiquitous genes from A are shown in red; other, unenriched LC genes are in gray. (C) L3- vs. L4-stage expression for LC genes. Ubiquitous genes are easily distinguished from LC-enriched genes, which are more likely to show moderate and stage-specific expression. MSP and MSD genes (green and yellow) are strongly up-regulated at the L4 stage. (D) A comparison, for LC genes, of the ratio of L4-stage to L3-stage gene expression to the ratio of L4-stage to nhr-67(RNAi) L4-stage expression. Of LC-expressed genes, 25% showed ≥20-fold expression changes for which development and NHR-67 activity were correlated (SI Appendix, Table S3). Subsets of LC genes encoding potassium channels, protein kinases, and regulators of muscle contraction are shown. All three subsets are visibly overrepresented among genes up-regulated by NHR-67 in L4-stage LCs. The genes along the ascending diagonal have significant expression in wild-type L4-stage LCs, but no detectable expression in either wild-type L3-stage LCs or nhr-67(RNAi) L4-stage LCs; genes with no expression were given a nominal expression level of 0.01 RPKM.
Although we detected expression levels from 1.2 × 10−1 to 9.8 × 104 RPKM in pooled LCs, only 208 genes (2.6%) had ≥103 RPKM, and almost all of these were ubiquitously expressed housekeeping genes (Fig. 2 A and B, SI Appendix, Table S7, and Dataset S1). Of pooled LC genes, 3,963 (50%) showed ≥20-fold differences in RPKM between the L3 and L4 stages, which are 10 h apart (Fig. 2C and SI Appendix, Table S3). nhr-67 itself was clearly silenced by RNAi (Fig. 1D), and its silencing had large effects on LC gene expression, which correlated strongly with the development of wild-type LCs from L3 to L4 stages. In particular, 1,977 genes (25%) expressed in L4-stage LCs showed parallel ≥20-fold changes of expression (up- or down-regulated) versus both L3-stage and nhr-67(RNAi) L4-stage LCs (Fig. 2D and SI Appendix, Table S3). If mean expression in individual LCs was considered, these numbers were similar; the fraction of genes with parallel ≥20-fold changes of expression for development and NHR-67 activity was 22% (2,206 of 9,968). LCs expressed 426 genes encoding known or predicted transcription factors; of these, 180 (42%) had ≥20-fold differences of expression between wild-type vs. nhr-67(RNAi) L4-stage LCs (SI Appendix, Table S3). Much of the effect of NHR-67 on gene activity in the LC may thus result indirectly from changed expression of other, intermediate transcription factors.
Functional Validation by RNAi.
To validate the LC genes and find new genes required for LC migration, we inactivated 204 LC-enriched genes with postembryonic RNAi (Fig. 3 and SI Appendix, Table S8), and looked for defects in the L4 stage near the end of LC migration. We defined the overall LC-enrichment of a gene as being the ratio of its highest wild-type LC expression (at either L3 or L4 stages) to its expression in whole larvae; 1,097 (14%) of LC genes had a LC/larval ratio of ≥20 (Fig. 2B and SI Appendix, Table S3). For 405 of these genes, preferential LC expression was highly significant (false-discovery rate = 0.01; Dataset S1; ref. 18).
Fig. 3.
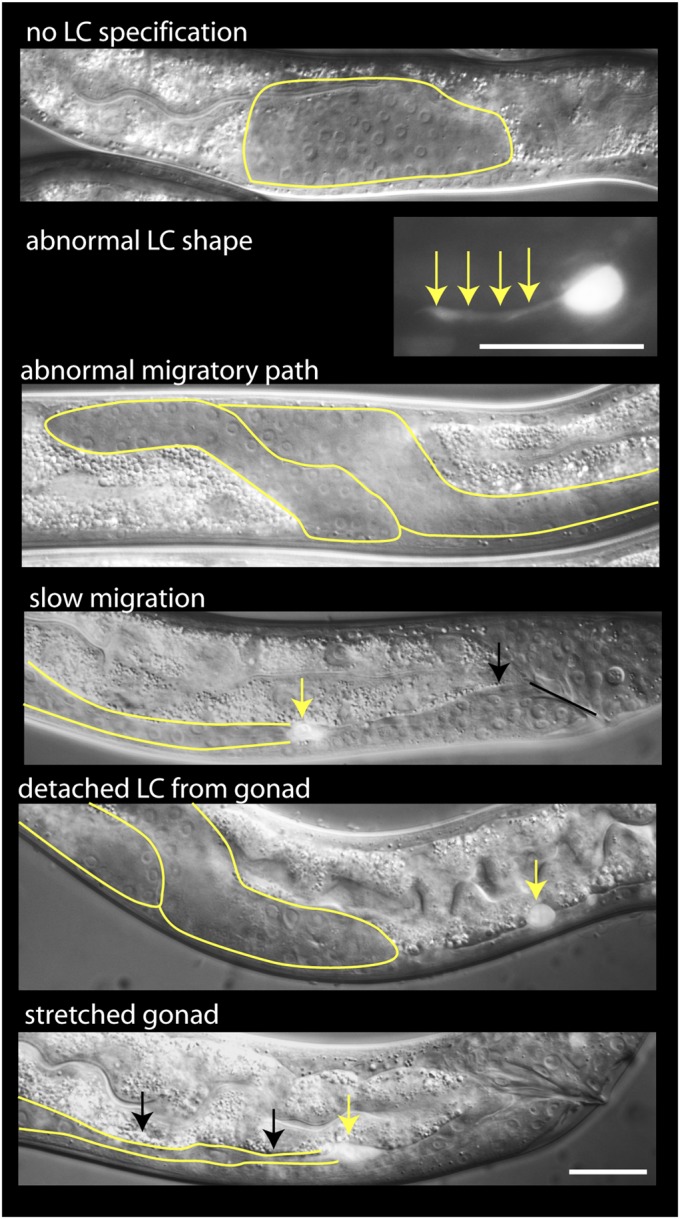
Types of LC migration phenotypes induced by RNAi. (Scale bar, 20 μm: applies to all panels except abnormal shapes panel, which is 10 μm.)
For RNAi, we selected LC-enriched genes, the products of which might be biologically interesting: transcription factors; unambiguous orthologs of human genes, particularly disease-associated genes; highly conserved genes of unknown function; and otherwise nondescript genes whose LC-enriched expression was strikingly high. RNAi gave phenotypes for 45 genes (22% of genes assayed; Fig. 3 and SI Appendix, Table S9). The frequency of phenotypes was only moderately dependent on which genes were chosen as targets: it was 17% (19 of 111) when transcription factors and genes with strict human orthologs were removed from the list, and 17% (2 of 12) among genes lacking known homologies or protein domains. Phenotypes included complete failure of the LC to develop, abnormal LC shapes, abnormal migratory paths, delayed migration, LCs detaching from their gonad, and stretched-out gonads (Fig. 3 and SI Appendix, Table S9). Delayed migration and abnormal shapes were the most prevalent phenotypes, seen for 44 and 39 genes respectively; RNAi of single genes usually caused multiple phenotypes, with only three genes having a single phenotype. Because 16 genes with LC defects had no phenotypes in previous genome-wide RNAi screens (Dataset S1), their defects were likely to be LC-specific rather than pleiotropic.
Some genes with RNAi phenotypes had homologs required for cell migration in mammals or Drosophila. Among these were two transcription factors: hlh-8, encoding a helix–loop–helix (HLH) protein orthologous to TWIST, which regulates cellular movements in Drosophila and mammals (19, 20); and crh-2, paralogous to SLBO, a C/EBPγ protein required for Drosophila border cell migration (7, 8). Others genes were regulators of the actin cytoskeleton, such as toca-1 and inft-1/inverted formin (21, 22). RNAi against inft-1 caused abnormal cell shapes, which paralleled the mutant phenotype of inft-1’s mammalian ortholog INF2 (23). Finally, sphk-1/sphingosine kinase and lim-9 were required for normal LC migration. Sphingosine kinase participates in several instances of vertebrate cell migration (24). lim-9’s mammalian ortholog FHL2 inhibits sphingosine kinase (25) and is involved in colon cancer invasion (26) and dendritic cell migration (27).
However, we also found migration phenotypes for highly conserved genes with no previously known role in cell migration, such as tpra-1, a seven-transmembrane receptor whose mammalian ortholog TPRA40 inhibits embryonic cell division (28), and maea-1, whose mammalian ortholog MAEA/EMP is required for developing erythroblasts to bind macrophages and mature into erythrocytes (29). Other unexpected migration phenotypes arose from structural maintenance of chromosome (SMC) genes, the products of which mediate both chromosome segregation and long-distance gene regulation (30). RNAi against him-1/SMC1, smc-3/SMC3, and smc-4/SMC4 induced the LC to separate from the gonad and migrate without it. Because these three genes are expressed both in the LC and in somatic gonadal cells (SI Appendix, Table S9), their products are likely to be required in either the LC or the somatic gonad for them to bind one another.
Cis-Regulatory Gene Assays.
We made YFP reporters for the proximal promoter regions of 21 genes, and observed LC expression for 11 (SI Appendix, Tables S9 and S10). Because the transgenic reporters were driven solely by 5′-ward flanking regulatory sequences and not by intronic or 3′-ward ones, the expression frequency of 11 of 21 represents a lower limit on LC expression of these genes in vivo. Four genes showed YFP expression in LCs but not RNAi phenotypes, and five showed the reverse; however, six genes showed both LC expression (via transgene or antibody staining) and RNAi phenotypes, suggesting that the RNA-seq data reflect in vivo function in LCs. These six genes included arx-7, him-1, msp-3, smc-4, sphk-1, and srsx-18.
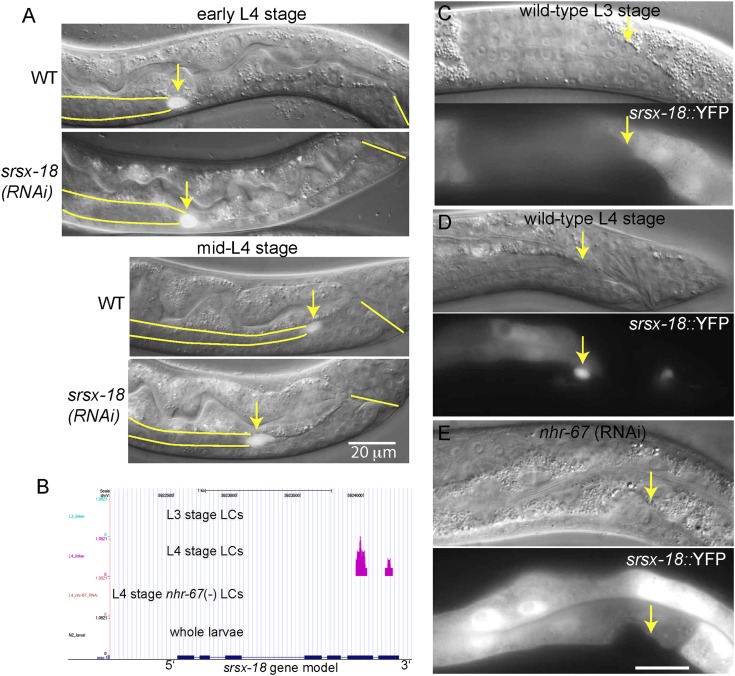
One example of an NHR-67–regulated gene is srsx-18, which encodes a putative G protein-coupled receptor (Fig. 4). In our RNA-seq data, srsx-18 is transcriptionally active solely in L4-stage LCs; in fact, it was detected in only one of our five individual L4 LCs (Dataset S1). A YFP transgene of srsx-18 recapitulated the results of the RNA-seq, with reporter activity in L4 LCs that was abolished by nhr-67(RNAi). srsx-18(RNAi) causes delayed migration, which is one of nhr-67’s RNAi phenotypes (3). srsx-18 may therefore be both a direct target of NHR-67 and a biologically important effector of NHR-67 in LCs.
Fig. 4.
srsx-18, a seven-transmembrane receptor, is a likely effector of NHR-67. (A) The LC migrates normally until near the end of the migration (early L4 stage, Upper two panels), but then slows down in srsx-18(RNAi) males. By the time the wild-type males have completed migration in the mid-L4 stage, srsx-18(RNAi) males are still migrating (Lower two panels). Note that delayed migration is one of nhr-67’s RNAi phenotypes (3). Arrow, LC; parallel lines, outline of gonad; single line, cloaca. (B) RNA-seq shows srsx-18 is transcriptionally active solely in wild-type L4-stage LCs, not in L3-stage LCs or in nhr-67(RNAi) L4-stage LCs. This L4-stage expression is regulated by nhr-67. (C and D) srsx-18::YFP is absent in the L3-stage but expressed in the L4-stage LC. (E) srsx-18::YFP expression in the L4-stage LC is abolished by nhr-67(RNAi). Other gene expression reporters are described in SI Appendix, Table S10.
Biological Functions Encoded by the LC Transcriptome.
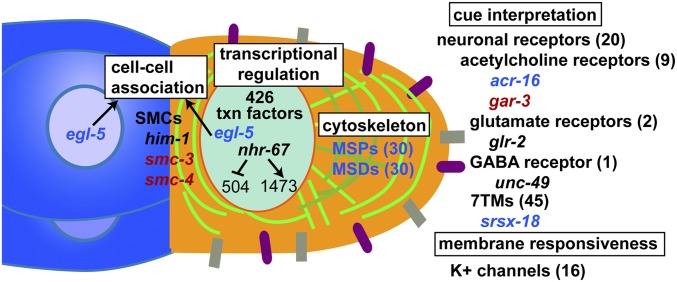
To find biological functions preferentially expressed by the LC transcriptome, we searched for Gene Ontology (GO) terms statistically overrepresented among genes with high LC-enrichment or among genes strongly up-regulated in the L4 stage (31). Functions enriched in highly LC-enriched genes included transcriptional regulation, cytoskeletal protein binding, intracellular protein transport, cell adhesion, G protein-coupled receptor signaling, cholinergic synaptic transmission, and axon components (SI Appendix, Table S11 and Dataset S1). Transcription factors included the E/daughterless ortholog hlh-2, hlh-8, and hlh-19. HLH-8 and HLH-19 both bind HLH-2 in two-hybrid assays (32); because we observed expression of hlh-19 and hlh-8 at the L3 and L4 stages respectively, their protein products could be successive heterodimeric partners of HLH-2 in the developing LC. One LC gene associated with G protein-coupled receptor signaling and required for normal migration was srsx-18 (Fig. 4 and SI Appendix, Table S9). LC genes associated with synaptic transmission included neurotransmitter receptors, which are generally considered neuronal; however, YFP reporters for the acetylcholine receptor genes acr-16 and gar-3 and the glutamate receptor glr-2 were expressed in the LC (SI Appendix, Table S9). Among the 45 LC genes associated with axon components were protein kinases, such as cam-1, pak-1, sax-1, unc-51, and vab-1, which are required for normal migration or morphology of axons and neurons (33–37).
Genes up-regulated by NHR-67 from L3- to L4-stage LCs disproportionately encoded protein kinases and phosphatases, regulators of muscle contraction, and potassium channels (Fig. 2D, SI Appendix, Table S12, and Dataset S1). Genes associated with regulating muscle contraction included the myosin regulatory light-chain genes mlc-1 and mlc-2 and the troponin mup-2 (SI Appendix, Dataset S1); their up-regulation by NHR-67 is consistent with the observed changes of LC shape and size in L4 larvae (Fig. 1 A and C).
Among the genes preferentially up-regulated at the L4 stage were a set of 389 sperm-enriched genes, identified by Reinke et al. (38), which we also detected in wild-type LCs and found were disproportionately common among strongly up-regulated genes (SI Appendix, Fig. S2 and Table S13). A subset of these genes (Fig. 2C and SI Appendix, Fig. S2) encoded major sperm proteins (MSPs), motor proteins required for amoeboid movement of nematode sperm (39). LCs also expressed a broader class of MSP-domain (MSD) genes (Fig. 2C), the Ascaris suum homologs of which include regulators of MSP polymerization (39). Both sets of genes were much more strongly up-regulated in wild-type L4-stage LCs than in nhr-67(RNAi) ones (Fig. 2C and SI Appendix, Fig. S2 and Table S13). Although we cannot fully exclude the possibility that sperm-enriched genes detected in LCs arose from contamination during dissection of the male gonad, we can rule out sporadic contamination (because we detected them in five individual wild-type L4-stage LCs), and we reproducibly observed much lower expression levels in individual nhr-67(RNAi) L4-stage LCs (Dataset S1). We observed msp-3::YFP expression in the LC and distal tip cells, but not in other gonadal cells (SI Appendix, Fig. S3 and Table S9). RNAi against the most strongly expressed MSP gene, msp-3, produced abnormally round and delayed L4-stage LCs (SI Appendix, Fig. S3 and Table S9).
We tested five tandem pore potassium channel (twk) genes with RNAi, because 13 of 16 potassium channels expressed in the LC were from this class (40). Two twk genes yielded migration defects, in which the LC was abnormally slow to turn back from the dorsal to the ventral body wall (SI Appendix, Table S9). This finding was striking because C. elegans has 46 paralogous twk genes, which we had expected to be individually redundant. It is also notable that delayed ventral return is one of several defects seen for nhr-67(RNAi) LCs (3). These RNA-seq and RNAi data suggest that twk channels are significant effectors of NHR-67 in the migrating LC.
Discussion
We have used single-cell transcriptional profiling to capture the gene-expression profile of a single migrating cell required for organogenesis in C. elegans, both at two time points during migration and in a time point deprived of the transcription factor NHR-67. We detected the expression of 5,000 to 7,000 genes for each assay of pooled LCs, and a total of 8,000 to 10,000 wild-type LC genes. All of these disparate genes can be genetically perturbed, and their interactions studied, within the LC (Fig. 5). We found gene regulation to be highly dynamic during the course of cell migration, with half of the genes showing 20-fold changes of expression between two larval stages that are 10 h apart. Using both RNAi and GO-term enrichment analysis on the LCs assayed, we were able to identify several different classes of genes required for migration, including tandem pore potassium channels.
Fig. 5.
An overview of recently identified genes functioning in the LC, with possible functions in migration. Up- and down-regulation of genes during LC development by NHR-67 is shown, along with many genes up-regulated by NHR-67 and discussed in the main text. Genes marked in blue have ≥20-fold higher expression in L4-stage than in L3-stage LCs; genes marked in red are higher in L3-stage than L4; genes in black have a <20-fold difference. Further LC gene functions are given in SI Appendix, Tables S11 and S12.
MSPs are best known for acting in nematode sperm cells, which use MSPs rather than actin for amoeboid mobility (39). We observed expression of msp-3 in L4-stage LCs both with RNA-seq and with YFP transgenes, along with msp-3(RNAi) phenotypes in migrating LCs. A previous screen for genes required in axonal migration found one MSP domain-encoding gene (41). These data are consistent with the hypothesis that MSPs are not restricted to sperm cells, but also function in somatic cell and axonal migration. Such a function might explain why MSPs are conserved in parthenogenetic nematodes that lack sperm (42).
For many genes required in LC migration, their roles may be conserved in other organisms. In the case of SMCs, mutation of cohesin accessory factors can impair neuronal and axonal migration in C. elegans and mice (30). Non-twk potassium channels are required for mammalian cell migration (43), and mammalian TWK channels are activated by both chemical and mechanical stimuli (44); mammalian TWK channels, like their C. elegans orthologs, could also have a migratory function. Similarly to NHR-67 in the LC, regulators of the actin and microtubule cytoskeleton are transcriptionally up-regulated by SLBO in migrating Drosophila border cells (7, 8). Although mammals do not have an exact equivalent to the MSP family, they have two genes encoding MSP domains (45), MOSPD1 and MOSPD3, with one isoform of MOSPD1 predicted to be cytoplasmic. The LC allows migration genes and their interplay to be studied within a single, precisely timed migrating cell, and may reveal novel effectors of cell migration in metazoa.
Materials and Methods
The strain PS4730 (syIs128[unc-119(+)(100 μg/mL) + lag-2::YFP(50 μg/mL)]II; unc-119(ed4)III; him-5(e1490)V) was used for LC dissections and for RNAi assays of LC migration. Feeding RNAi was carried out as in ref. 3. Worms were scored for LC defects by Nomarski microscopy, allowing more detailed screening than dissection microscopy previously used (3). Dissected cells were subjected to 3′-tailed RT-PCR by the method of ref. 13. RNA-seq was performed as in ref. 17; reads were mapped to the WS190 sequence of the C. elegans genome with bowtie (46); and RPKM counts were computed with ERANGE 3.1 (17). GO terms associated with LC-enriched genes were identified with FUNC (31). Further details are given in the SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank M. Goodman, M. Chalfie, and A. Mortazavi for early support in developing single-cell RT-PCR; B. Williams for providing control spike poly(A)+ RNAs; L. Schaeffer, D. Trout, and I. Antoshechkin of the Jacobs Genome Center for Illumina sequencing; A. Narayan for help setting up the dissection rig; J. Downes for scoring linker cell migration phenotypes; J. Thomas for his list of transcription factors; WormBase for gene annotations; and T. Brown, Y.-P. Hsueh, A. Roeder, K. Yook, and A. Zaslaver for comments. This work was supported by National Institutes of Health Grant GM084389 and by the Howard Hughes Medical Institute, with which P.W.S. is an Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank SRA database (accession no. SRA058596).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203045109/-/DCSupplemental.
References
- 1.Wedlich D. Cell Migration in Development and Disease. Wenheim, Germany: Wiley; 2005. [Google Scholar]
- 2.Abraham MC, Lu Y, Shaham S. A morphologically conserved nonapoptotic program promotes linker cell death in Caenorhabditis elegans. Dev Cell. 2007;12:73–86. doi: 10.1016/j.devcel.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Kato M, Sternberg PW. The C. elegans tailless/Tlx homolog nhr-67 regulates a stage-specific program of linker cell migration in male gonadogenesis. Development. 2009;136:3907–3915. doi: 10.1242/dev.035477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- 5.Morán E, Jiménez G. The tailless nuclear receptor acts as a dedicated repressor in the early Drosophila embryo. Mol Cell Biol. 2006;26:3446–3454. doi: 10.1128/MCB.26.9.3446-3454.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu Q, et al. Orphan nuclear receptor TLX activates Wnt/β-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat Cell Biol. 2010;12(1) doi: 10.1038/ncb2001. 31–40; supp pp 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borghese L, et al. Systematic analysis of the transcriptional switch inducing migration of border cells. Dev Cell. 2006;10:497–508. doi: 10.1016/j.devcel.2006.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, et al. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev Cell. 2006;10:483–495. doi: 10.1016/j.devcel.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Christiaen L, et al. The transcription/migration interface in heart precursors of Ciona intestinalis. Science. 2008;320:1349–1352. doi: 10.1126/science.1158170. [DOI] [PubMed] [Google Scholar]
- 10.Shapiro IM, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7:e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu MZ, et al. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchymal transition. Mol Cell. 2011;43:811–822. doi: 10.1016/j.molcel.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Spencer WC, et al. A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21:325–341. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 14.Lockery SR, Goodman MB. Tight-seal whole-cell patch clamping of Caenorhabditis elegans neurons. Methods Enzymol. 1998;293:201–217. doi: 10.1016/s0076-6879(98)93016-6. [DOI] [PubMed] [Google Scholar]
- 15.Ramsköld D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLOS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall D, Altun ZF. C. elegans Atlas. Cold Spring Harbor, New York: Cold Spring Harbor Lab Press; 2008. [Google Scholar]
- 17.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 18.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kölsch V, Seher T, Fernandez-Ballester GJ, Serrano L, Leptin M. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 2007;315:384–386. doi: 10.1126/science.1134833. [DOI] [PubMed] [Google Scholar]
- 20.Sahlin P, et al. Women with Saethre-Chotzen syndrome are at increased risk of breast cancer. Genes Chromosomes Cancer. 2007;46:656–660. doi: 10.1002/gcc.20449. [DOI] [PubMed] [Google Scholar]
- 21.Ho H-YH, et al. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 22.Madrid R, et al. The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev Cell. 2010;18:814–827. doi: 10.1016/j.devcel.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Brown EJ, et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010;42:72–76. doi: 10.1038/ng.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fyrst H, Saba JD. An update on sphingosine-1-phosphate and other sphingolipid mediators. Nat Chem Biol. 2010;6:489–497. doi: 10.1038/nchembio.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi H, et al. FHL-2 suppresses VEGF-induced phosphatidylinositol 3-kinase/Akt activation via interaction with sphingosine kinase-1. Arterioscler Thromb Vasc Biol. 2009;29:909–914. doi: 10.1161/ATVBAHA.108.178541. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W, et al. Four-and-a-half LIM protein 2 promotes invasive potential and epithelial-mesenchymal transition in colon cancer. Carcinogenesis. 2010;31:1220–1229. doi: 10.1093/carcin/bgq094. [DOI] [PubMed] [Google Scholar]
- 27.König K, et al. Four-and-a-half LIM domain protein 2 is a novel regulator of sphingosine 1-phosphate receptor 1 in CCL19-induced dendritic cell migration. J Immunol. 2010;185:1466–1475. doi: 10.4049/jimmunol.0903449. [DOI] [PubMed] [Google Scholar]
- 28.Aki T, Funakoshi T, Nishida-Kitayama J, Mizukami Y. TPRA40/GPR175 regulates early mouse embryogenesis through functional membrane transport by Sjögren’s syndrome-associated protein NA14. J Cell Physiol. 2008;217:194–206. doi: 10.1002/jcp.21492. [DOI] [PubMed] [Google Scholar]
- 29.Chasis JA, Mohandas N. Erythroblastic islands: Niches for erythropoiesis. Blood. 2008;112:470–478. doi: 10.1182/blood-2008-03-077883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bose T, Gerton JL. Cohesinopathies, gene expression, and chromatin organization. J Cell Biol. 2010;189:201–210. doi: 10.1083/jcb.200912129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prüfer K, et al. FUNC: A package for detecting significant associations between gene sets and ontological annotations. BMC Bioinformatics. 2007;8:41. doi: 10.1186/1471-2105-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grove CA, et al. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallegos ME, Bargmann CI. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron. 2004;44:239–249. doi: 10.1016/j.neuron.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 34.Hayashi Y, et al. A trophic role for Wnt-Ror kinase signaling during developmental pruning in Caenorhabditis elegans. Nat Neurosci. 2009;12:981–987. doi: 10.1038/nn.2347. [DOI] [PubMed] [Google Scholar]
- 35.Lucanic M, Kiley M, Ashcroft N, L’etoile N, Cheng HJ. The Caenorhabditis elegans P21-activated kinases are differentially required for UNC-6/netrin-mediated commissural motor axon guidance. Development. 2006;133:4549–4559. doi: 10.1242/dev.02648. [DOI] [PubMed] [Google Scholar]
- 36.Mohamed AM, Chin-Sang ID. Characterization of loss-of-function and gain-of-function Eph receptor tyrosine kinase signaling in C. elegans axon targeting and cell migration. Dev Biol. 2006;290:164–176. doi: 10.1016/j.ydbio.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 37.Ogura K, et al. Protein phosphatase 2A cooperates with the autophagy-related kinase UNC-51 to regulate axon guidance in Caenorhabditis elegans. Development. 2010;137:1657–1667. doi: 10.1242/dev.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 39.Smith H. Sperm motility and MSP. WormBook. 2006;Feb. 1:1–8. doi: 10.1895/wormbook.1.68.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salkoff L, et al. Potassium channels in C. elegans. WormBook. 2005;Dec. 30:1–15. doi: 10.1895/wormbook.1.42.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitz C, Kinge P, Hutter H. Axon guidance genes identified in a large-scale RNAi screen using the RNAi-hypersensitive Caenorhabditis elegans strain nre-1(hd20) lin-15b(hd126) Proc Natl Acad Sci USA. 2007;104:834–839. doi: 10.1073/pnas.0510527104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heger P, Kroiher M, Ndifon N, Schierenberg E. Conservation of MAP kinase activity and MSP genes in parthenogenetic nematodes. BMC Dev Biol. 2010;10:51. doi: 10.1186/1471-213X-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwab A, Hanley P, Fabian A, Stock C. Potassium channels keep mobile cells on the go. Physiology (Bethesda) 2008;23:212–220. doi: 10.1152/physiol.00003.2008. [DOI] [PubMed] [Google Scholar]
- 44.Mathie A, Al-Moubarak E, Veale EL. Gating of two pore domain potassium channels. J Physiol. 2010;588:3149–3156. doi: 10.1113/jphysiol.2010.192344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pall GS, et al. A novel transmembrane MSP-containing protein that plays a role in right ventricle development. Genomics. 2004;84:1051–1059. doi: 10.1016/j.ygeno.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 46.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.