Abstract
The astonishing variation in the shape and size of bird beaks reflects a wide range of dietary specializations that played an important role in avian diversification. Among Darwin’s finches, ground finches (Geospiza spp.) have beaks that represent scaling variations of the same shape, which are generated by alterations in the signaling pathways that regulate growth of the two skeletal components of the beak: the prenasal cartilage (pnc) and the premaxillary bone (pmx). Whether this developmental mechanism is responsible for variation within groups of other closely related bird species, however, has remained unknown. Here, we report that the Caribbean bullfinches (Loxigilla spp.), which are closely related to Darwin’s finches, have independently evolved beaks of a novel shape, different from Geospiza, but also varying from each other only in scaling. However, despite sharing the same beak shape, the signaling pathways and tissues patterning Loxigilla beaks differ among the three species. In Loxigilla noctis, as in Geospiza, the pnc develops first, shaped by Bmp4 and CaM signaling, followed by the development of the pmx, regulated by TGFβIIr, β-catenin, and Dkk3 signaling. In contrast, beak morphogenesis in Loxigilla violacea and Loxigilla portoricensis is generated almost exclusively by the pmx through a mechanism in which Ihh and Bmp4 synergize to promote expansion of bone tissue. Together, our results demonstrate high flexibility in the relationship between morphology and underlying developmental causes, where different developmental programs can generate identical shapes, and similar developmental programs can pattern different shapes.
Keywords: convergent evolution, craniofacial, morphogenesis
The role that genes play during development is key to understanding evolutionary processes that generate morphological diversity (1–3). Comprising 30 orders, 193 families, 2,099 genera, and close to 10,000 species, birds are the most diverse group of land vertebrates, and much of their success can be attributed to adaptive variation in beak morphology, a trait closely associated with feeding habits and ecological niche (4). The adaptive significance and diversity of bird beaks offers an excellent opportunity for evolutionary developmental studies probing the mechanisms underlying morphological diversification.
The shape of the beak determines its functional properties (5). We previously showed that in Darwin’s finches—a group of 14 closely related species representing a classic example of an adaptive radiation (6, 7)—beak shapes can be classified into three unique morphological groups based on mathematical similarity (8). Within each group (termed A, B, and C), beak shapes differ only in scale along specific dimensions (i.e., depth and/or length) and can be shown using scaling transformations to be expressions of a single common shape. By definition, variation between groups cannot be accounted for by changes in beak scales alone, implying that species in different morphological groups have fundamentally different beak shapes (characterized by a different upper-beak curvature profile) beyond changes in scale (8). Hereafter, we refer to birds as having the same beak shape if they differ only in scale along the depth and/or length dimensions (and thus are in the same morphological group).
In Darwin’s finches of the monophyletic genus Geospiza, which all belong to morphological group A, beaks are patterned by a common underlying molecular and developmental mechanism (9–11). At early embryonic stages (stages 26 and 27), Bmp4 and calmodulin (CaM) regulate the growth of the prenasal cartilage (pnc) skeleton (9), (10). Subsequently, the pnc ceases its expansion (12), and beak morphogenesis is completed by the developing premaxillary bone (pmx), which forms from a separate condensation and is patterned by a network of unrelated yet interacting regulatory genes, TGFβIIr, β-catenin, and Dkk3 (11) at the later stages 28–31. Differences in scaling between species arise through changes in the signaling pathways that alter the pnc and the pmx, the two separate developmental modules that form the beak, along different axes of growth (9–11). However, it is unknown whether this mechanism is unique to Geospiza or is also responsible for generating scaling variations and novel beak shapes in other bird species. We hypothesized that the previously discovered mechanisms controlling beak diversity in Darwin’s finches would explain similar beak shapes in other more distantly related bird species. To address this hypothesis, we capitalize on the remarkable beak shape variation in the 13 species most closely related to Darwin’s finches (6, 13, 14). Together with Darwin’s finches, these birds, which are mainly endemic to the Caribbean islands, form a monophyletic and recently diverged clade known as the Tholospiza, the “dome finches,” because its members build dome-shaped nests with side entrances (14). Despite high genetic similarity, the Tholospiza have extraordinary levels of beak diversity that are comparable to those seen among members of disparate bird families on mainland (14, 15). The marked beak diversity of Tholospiza could be explained by ecological factors, such as strong selection pressures upon colonization of specific island niches, by unique aspects about the beak developmental genetic architecture of its ancestor, or by a combination of both (14). Here, we report that the three members of the genus Loxigilla, which form part of the Tholospiza, have evolved beaks of the same shape, different from that of Geospiza, varying among each other only in scaling. However, in contrast to Geospiza, Loxigilla species achieve identical beak shapes through distinct signaling pathways and tissues. In one species, beaks are patterned by the same mechanisms as in Geospiza, whereas the other two species use different signaling pathways and tissues. Overall, these results demonstrate flexibility between developmental mechanisms and morphology among the closely related members of Tholospiza.
Results and Discussion
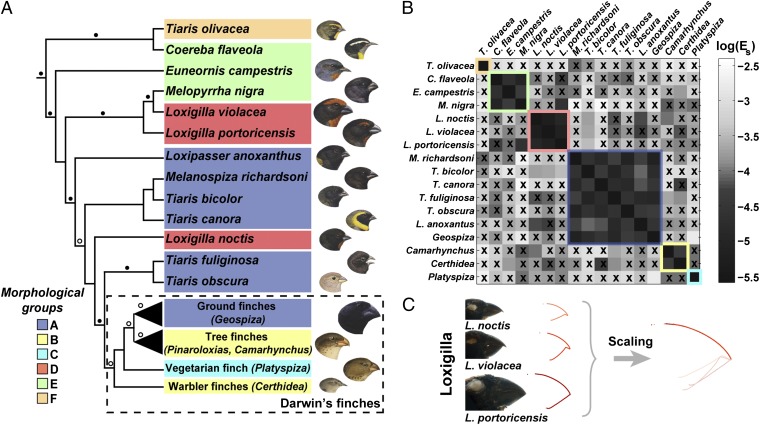
To provide a robust phylogenetic framework for our comparative study, analysis of beak shapes in Tholospiza species was undertaken with reference to a phylogenetic analysis reported here based on six genes (Fig. 1A and Materials and Methods), rather than the single genetic marker used previously (14, 16). Upper-beak profiles obtained from images of museum specimens of all Tholospiza members (Materials and Methods) (8) showed that although six of the 13 species (not including Darwin’s finches) belonged to the previously identified group A (8), there were three additional morphological groups, termed D, E, and F (Fig. 1 A and B and Fig. S1). To study in detail the developmental mechanisms that generate novel shapes and the variation within them, we chose to focus on beak morphogenesis in the Caribbean bullfinches of the genus Loxigilla (Loxigilla noctis, Loxigilla violacea, and Loxigilla portoricensis) for three reasons: (i) Loxigilla species (group D) have deep and wide conical seed-eating beaks that resemble those of Geospiza (group A) and thus the comparison of the developmental mechanisms of both groups has a relevant ecological context; (ii) distribution of their beak morphology (L. noctis has proportionally the least deep/wide beak, L. violacea an intermediately scaled beak, and L. portoricensis has the largest and deepest beak) (13) allows for analyzing the mechanisms originating scaling variation within this morphological group (Fig. 1C); and (iii) our beak shape analysis and the phylogenetic evidence from this and a previous study (14) shows that, although L. noctis, L. violacea, and L. portoricensis have been traditionally grouped under the same genus based on similarities in plumage coloration and beak characters (13), their beak shape has evolved convergently (Fig. 1A and Fig. S2), with L. noctis more closely related to Darwin’s finches than to the other two species of Loxigilla. Therefore, these birds are ideal to further investigate the principles of beak evolution, such as presence of possible developmental constraints in shape patterning.
Fig. 1.
Tholospiza phylogeny and classification of beak shapes. (A) ML phylogeny of Tholospiza based on six genes. Closed circles represent branches supported by Bayesian posterior probabilities higher than 0.95 and ML bootstrap support values higher than 70%; open circles represent branches with this level of support in one analysis but not the other. For Darwin’s finches, we show the summarized results from the detailed beak shape analysis that was done previously (8). Bird illustrations were reproduced with permission from refs. 33–36. (B) Heat map of pairwise comparisons between different beak shapes of species in Tholospiza. Crosses (x) indicate pairs where no minimum in the defined measures of shape difference, 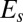 and
and 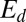 , as a function of the scaling factors could be found. Conversely, comparisons not marked with an x indicate that a minimum exists. The plotted color represents the residual of the shape difference measure,
, as a function of the scaling factors could be found. Conversely, comparisons not marked with an x indicate that a minimum exists. The plotted color represents the residual of the shape difference measure, 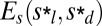 . The same results were obtained for the residual,
. The same results were obtained for the residual, 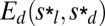 . For those pairs marked with an x, the plotted color indicates the minimal value of
. For those pairs marked with an x, the plotted color indicates the minimal value of 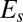 in the range of scaling factors that the experimental error allows to search for. In our analysis, two beak shapes collapse under scaling transformation if a minimum in both measures describing the difference between the shapes exists (no x) and the associated residual is low (black color; Materials and Methods) (8). Morphological groups, defined as groups of species where the beaks of all its members collapse onto each other under scaling transformations (and differ thus only by their scales, such as depth and length), are outlined in the phylogeny and in the heat map with colors (color legend for morphological groups is shown in the Lower Left). (C) Beak profiles of the three Loxigilla species as obtained from digitization of the beak profile (Left) and after being collapsed onto a common shape by nonuniform (anisotropic) scaling transformations (Right).
in the range of scaling factors that the experimental error allows to search for. In our analysis, two beak shapes collapse under scaling transformation if a minimum in both measures describing the difference between the shapes exists (no x) and the associated residual is low (black color; Materials and Methods) (8). Morphological groups, defined as groups of species where the beaks of all its members collapse onto each other under scaling transformations (and differ thus only by their scales, such as depth and length), are outlined in the phylogeny and in the heat map with colors (color legend for morphological groups is shown in the Lower Left). (C) Beak profiles of the three Loxigilla species as obtained from digitization of the beak profile (Left) and after being collapsed onto a common shape by nonuniform (anisotropic) scaling transformations (Right).
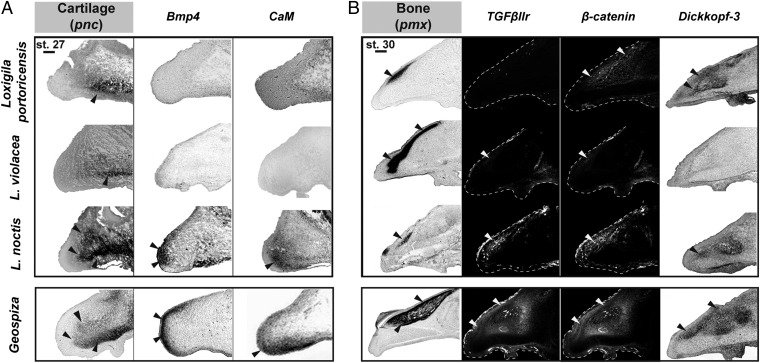
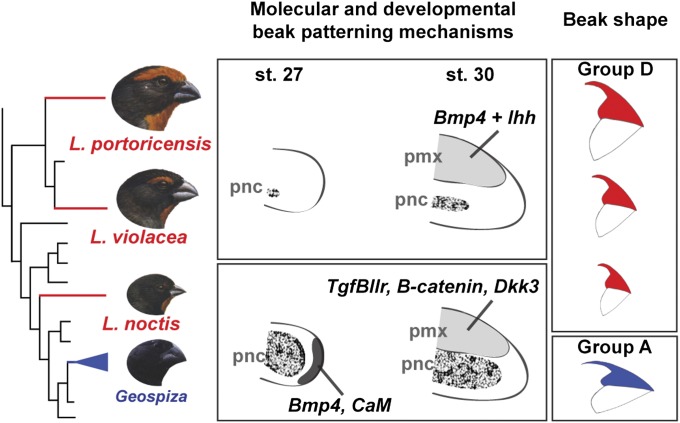
Because the pnc tissue plays an important role in patterning the beaks of Geospiza at early stages of development, through the action of Bmp4 and CaM (9, 10), we first wanted to examine the extent to which this tissue and signaling mechanisms were also contributing to beak morphogenesis in the three Loxigilla species. The expression of Col2a1, a cartilage marker, in stage 27 embryos reveals that the pnc occupies the majority of the developing beak in L. noctis, similar to what is seen in Geospiza, whereas the pnc represents proportionally a much smaller part of the developing beaks of L. violacea and L. portoricensis (Fig. 2A and Fig. S3). Bmp4 and CaM are specifically expressed around the rostral portion of the pnc in L. noctis, but are not expressed in L. violacea and in L. portoricensis at this stage (Fig. 2A and Figs. S3, S4A, and S5). In Geospiza, expression levels of Bmp4 and CaM correlate positively with the size of the pnc, and functional tests in chicken embryos show that these two genes drive outgrowth of this tissue at this stage of beak development (9, 10). Importantly, only mesenchymal Bmp4 controls beak skeleton morphology at these developmental stages (9). These results suggest that the pnc contributes to beak shape patterning in L. noctis through the action of Bmp4 and CaM, similar to what is observed in Geospiza (9, 10), but is not involved in shaping the beaks of L. violacea and L. portoricensis. Importantly, the pnc does not expand later in embryonic development, ruling out the possibility that this tissue plays a role in beak patterning at subsequent stages (Fig. S6).
Fig. 2.
Comparative analysis of gene expression patterns in the developing beaks of Loxigilla. (A) In L. noctis and in Darwin’s finches from the genus Geospiza (9, 10), the pnc, labeled with Col2a1, plays a marked role in beak patterning and occupies a larger portion of the developing beak than in L. violacea and L. portoricensis. At stage 27, Bmp4 and CaM are strongly expressed in L. noctis, similar to what is seen in Geospiza (9, 10), whereas they are not expressed in L. violacea or in L. portoricensis. (Scale bar: 0.1 mm.) (B) The pmx condensation, labeled with alkaline phosphatase, is shown for the three Loxigilla species. In stage 30 L. noctis and Geospiza (11) embryos, the size and location of the pmx condensation correlate with expression of TGFβIIr, β-catenin, and Dkk3. In L. violacea, these genes were expressed at small, almost undetectable levels that do not correlate with the large pmx condensation of this species. In L. portoricensis, TGFβIIr expression was not detected, and although β-catenin and Dkk3 were expressed at high levels, it is unlikely that these two latter genes are involved in patterning the pmx in this species (see text for details). (Scale bar: 0.2 mm.) Together, expression patterns in A and B indicate that the beak developmental program of L. noctis is similar to that of Geospiza (9–11), whereas it differs from that of L. violacea and L. portoricensis. Arrowheads in A and B show the specific regions where we detected expression of the skeletal markers and genes examined. Beak profiles for TGFβIIr and β-catenin stains are outlined with a white dashed line. Geospiza images were reproduced with permission from refs. 9–11 and are shown for comparison.
We then sought to determine the relative contribution of the pmx tissue to the developing beaks of Loxigilla by examining the expression of an osteoblast marker (alkaline phosphatase) in stage 30 embryos (Fig. 2B). Because our previous studies suggested that TGFβIIr, β-catenin, and Dkk3 pattern the pmx in stage 30 Geospiza (11) embryos, we examined the expression of these genes in Loxigilla embryos to test whether they were involved in regulating this tissue. In L. noctis, all three genes were expressed in the pmx at levels that correlated with the size of this tissue (Fig. 2B and Fig. S4B). In contrast, in L. violacea, the expression of these three molecules was restricted to a very small, almost undetectable domain that did not correlate with the large pmx condensation of this species (Fig. 2B and Figs. S4B and S5). Similarly, L. portoricensis showed undetectable levels of expression of TGFβIIr (Fig. 2B and Fig. S4B). In this species, however, β-catenin and Dkk3 were broadly expressed in the pmx relative to the other two Loxigilla species (Fig. 2B and Figs. S4B and S5). Elevated levels of β-catenin can promote osteogenesis when this molecule is mobilized into the nucleus (17), but contrary to what is seen in Geospiza, L. portoricensis has no accumulation of nuclear β-catenin in its pmx (Fig. S7), suggesting that this gene is not contributing to pmx expansion in this species at this embryonic stage. Alternatively, because up-regulation of Dkk3 drives pmx osteogenesis (11), the high expression levels of Dkk3 observed in L. portoricensis could explain the expansion of the pmx in this species (Fig. 2B and Fig. S4B). However, because Dkk3 up-regulation is known to increase beak depth and length (11), this molecule does not explain the larger beak depth and width seen in L. portoricensis relative to the other species of Tholospiza (14), indicating that other factors must be involved (11). Together, our results suggest that the TGFβIIr, β-catenin, and Dkk3 combined signaling network is involved in patterning the pmx of L. noctis, similar to what is seen in Geospiza (11), but does not pattern this tissue in L. violacea or in L. portoricensis.
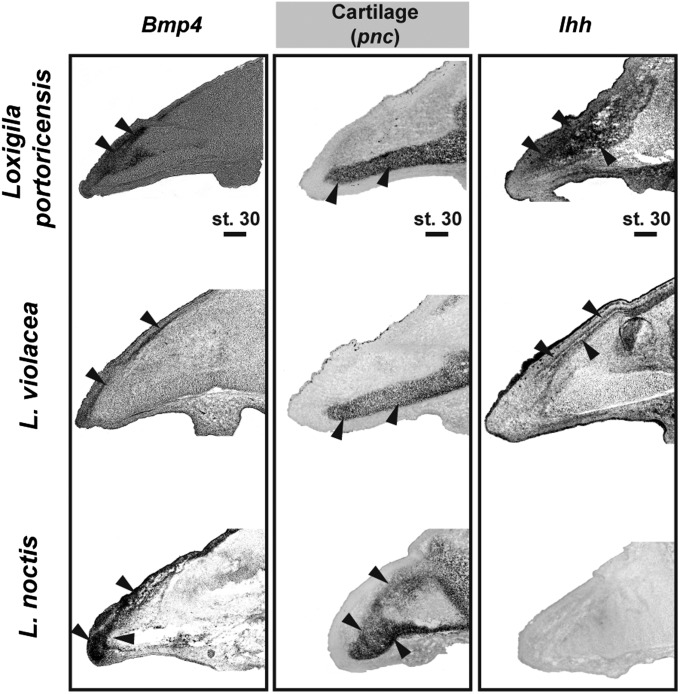
To establish which molecules regulate the pmx in L. violacea and L. portoricensis, we examined the expression of other skeletogenic factors known to control avian craniofacial development (18–21). We found that in these two species, early in development Bmp4 is primarily expressed in domains coinciding with their developing pmx (compare the bone condensation in Fig. 2B with expression of Bmp4 in Fig. 3). In contrast, in stage-matched L. noctis (Fig. 3) and in Geospiza (9) embryos, Bmp4 expression extends to areas around the pnc. Bmp4 expression around the pnc is known to cause a marked expansion of this skeletal tissue, as exemplified in L. noctis (Fig. 3) and in previous studies (9, 11, 21). However, Bmp4 is primarily expressed in the pmx of stage 30 L. violacea and L. portoricensis embryos, and therefore the pnc of these two species does not expand (compare their pnc with that of L. noctis in Fig. 3). Because up-regulation of Bmp4 in the developing pmx by itself does not cause bone expansion (see below) (11), we examined whether additional skeletogenic regulators expressed in the pmx of L. violacea and L. portoricensis could be amending the function of this gene. We found that in L. violacea and L. portoricensis expression of Indian hedgehog (Ihh) correlated spatially and temporally with the expression of Bmp4 in regions where pmx was formed (compare the bone condensation in Fig. 2B with expression of Bmp4 and Ihh in Fig. 3). Importantly, Ihh expression was not detected in L. noctis (Fig. 3 and Fig. S4C at the same stages and locations, showing that this pattern is particular to L. violacea and L. portoricensis.
Fig. 3.
Expression of Bmp4 and Ihh in Loxigilla. In L. violacea and L. portoricensis, Bmp4 is primarily expressed in areas that coincide with the pmx condensation (compare with location of bone condensation in Fig. 2B). In L. noctis, in contrast, Bmp4 expression surrounds the pnc (cartilage), where it causes a marked expansion of this tissue. In L. violacea and L. portoricensis, Ihh is expressed in coinciding domains with Bmp4 and the pmx condensation (compare with location of bone condensation in Fig. 2B), whereas this gene is not expressed in the beaks of L. noctis. (Scale bar: 0.2 mm.)
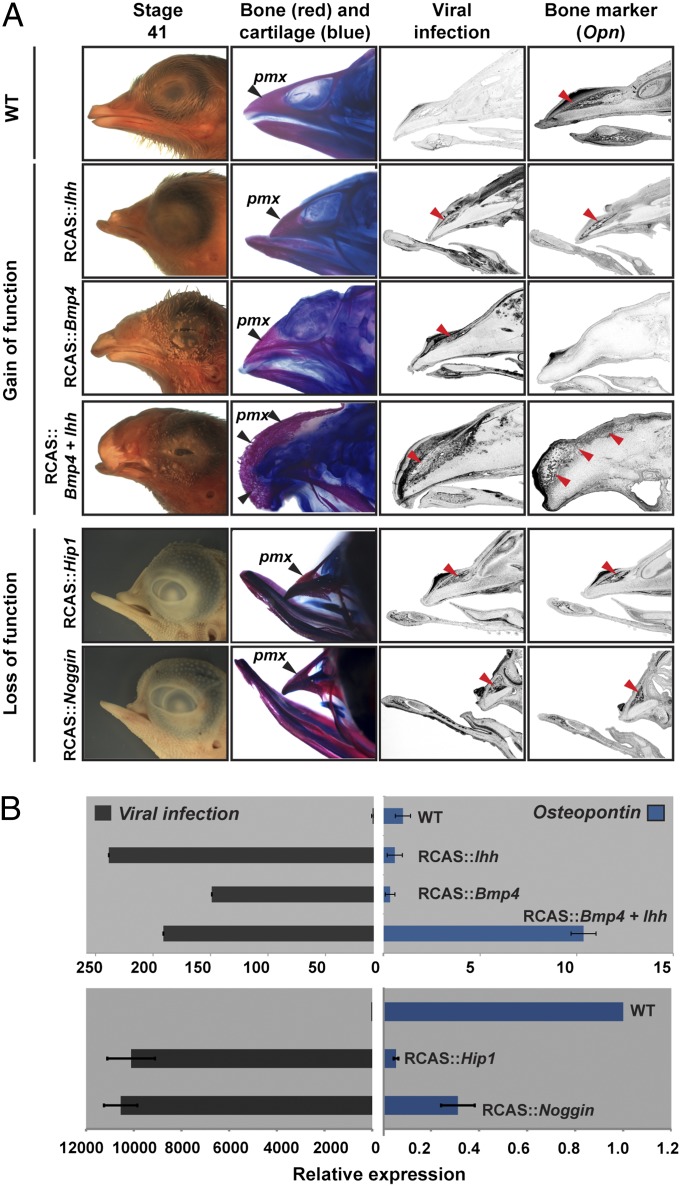
The coinciding pmx expression of Bmp4 and Ihh in L violacea and L. portoricensis led us to hypothesize that these two molecules were driving the pmx expansion in these species. To test this, we used avian retroviral vectors (RCAS) to overexpress these genes in the developing upper-beak prominence of chicken embryos, a useful model for avian functional experiments (9, 22). Increasing levels of Bmp4 alone (RCAS::Bmp4) yielded embryos with drastic expansions in the cranial cartilage (9, 21) (Fig. 4 and Fig. S8A), because this molecule positively regulates chondrogenic tissue (9, 21), but pmx was reduced. Increasing levels of Ihh (RCAS::Ihh) produced embryos with shorter beaks and less bone, because this molecule negatively regulates differentiation of dermal bone osteoblasts (21, 23) (Fig. 4 and Fig. S8A). However, simultaneous overexpression of both molecules (RCAS::Bmp4 + RCAS::Ihh) produced beaks with a markedly increased pmx (Fig. 4 and Fig. S8A). Because our injection techniques do not allow us to restrict infection to the developing pmx tissue, and some diffusion of signaling molecules occurs to other surrounding tissues, we also observed an increase in the pnc tissue (Fig. 3), an effect that is likely due exclusively to Bmp4 exposure, because Ihh alone does not influence cartilage growth in the developing beak (21). To verify that the excess bone seen in RCAS::Bmp4 + RCAS::Ihh-infected embryos is due to intramembranous ossification rather than to other processes, such as induction of endochondral ossification from ectopic cartilage, we infected the frontal bone of developing chicken embryos with RCAS::Bmp4 + RCAS::Ihh. Unlike the developing beak, the frontal bone does not have any physically associated cartilage tissue, and thus this experiment allowed us to restrict the infection exclusively to a bone of intramembranous origin. Similar to what is seen in the beak, we found that there was a marked increase in ossification and associated molecular markers (Fig. S8B).
Fig. 4.
Gain- and loss-of-function experiments demonstrate that Bmp4 and Ihh promote expansion of the pmx condensation. (A and B) Functional experiments in chicken embryos show that Ihh and Bmp4, two regulatory molecules that interfere with dermal bone development when up-regulated individually, can expand the pmx when overexpressed simultaneously. In contrast, down-regulation of either Bmp4 or Ihh pathways (RCAS::Noggin and RCAS::Hip1, respectively) causes a marked decrease in the pmx. (A) Lateral head views and alizarin red (bone)/alcian blue (cartilage) stains from stage 41 (embryonic day 15) embryos. We used RSCH (a viral specific construct) and Osteopontin (Opn) probes on sagittal sections of stage 39 (embryonic day 13) chicken embryos to reveal RCAS infection and late osteoblasts, respectively. Note that infection of all constructs is restricted to the pmx, as revealed by RCSH probe, therefore mimicking the expression domains seen in Loxigilla. Arrowheads in A indicate the location of the pmx and the signal from the mRNA probes used. (B) qPCR assays of stage 39 (embryonic day 13) embryos infected with the different constructs show the extent of the viral infection and expression levels of Opn, a bone marker. Embryos infected simultaneously with Bmp4 and Ihh had a 10-fold increase in Opn expression, relative to wild-type embryos or to embryos infected with only Bmp4 or Ihh. Expression levels are shown relative to wild-type uninfected controls. Two-tailed t tests were performed for each treatment against wild-type uninfected controls. Viral infections: RCAS::Bmp4 (n = 7; P = 2 × 10−5); RCAS::Ihh (n = 9; P = 9 × 10−5); RCAS::Bmp4 + RCAS::Ihh (n = 9; P = 5 × 10−6). Opn: RCAS::Bmp4 (n = 7; P = 0.03); RCAS::Ihh (n = 9; P = 0.004); RCAS::Bmp4 + RCAS::Ihh (n = 9; P = 1.2 × 10−6); RCAS::Noggin (n = 5; P = 0.002); RCAS::Hip1 (n = 5; P = 0.007). Bars represent SE measurements.
To complement our gain-of-function experiments, we performed reciprocal loss-of- function experiments using viruses carrying Noggin and Hip1, two known negative regulators of Bmp4 and Ihh signaling (24, 25), respectively. Contrary to the effect found when we simultaneously increased Bmp4 and Ihh signaling, down-regulation of these pathways led to a marked decrease of the pmx, as revealed by histological stains and osteogenic markers (Fig. 4 and Fig. S8A). Thus, our complementary gain- and loss-of-function experiments show that though Ihh and Bmp4 signaling is required for beak bone development, only the combined Ihh and Bmp4 synergy is sufficient and required to expand the pmx tissue. Moreover, beaks injected with RCAS::Bmp4 + RCAS::Ihh were significantly deeper and wider than those of wild-type embryos, but their length did not change (Fig. S9). In a good correlation, L. violacea and L. portoricensis show a relative increase in beak depth and width with respect to L. noctis, and the length remains unchanged (Fig. S9), suggesting that Ihh and Bmp4 can account for the scaling differences observed among the beaks of Loxigilla (Fig. S9).
We have shown that a set of different signaling pathways and developmental mechanisms, involving different tissues (cartilage and bone), can be associated with identical beak shapes varying only in scaling dimensions. Specifically, the beak developmental program in L. noctis is similar to that of Darwin’s finches of the genus Geospiza (9–11), with a marked contribution from two developmental modules: the pnc, shaped by Bmp4 and CaM signaling, followed by the pmx, regulated by TGFβIIr, β-catenin, and Dkk3 signaling. In contrast, in L. violacea and L. portoricensis, the contribution of the pnc to beak shape is negligible. Instead, beak patterning in these species is established by a single developmental module through a mechanism in which Ihh and Bmp4, two regulatory molecules that interfere with normal dermal bone development when up-regulated individually, synergize to promote expansion of pmx. It is worth pointing out that the gene expression differences seen in Loxigilla demonstrate formally that underlying developmental programs are different, whereas our functional experiments in chicken embryos serve to reinforce the conclusion that such differences, when mimicked in another bird system, can lead to variation in beak patterning and morphogenesis. The use of chicken embryos for functional tests assumes that the developmental gene toolkit for craniofacial morphogenesis and skeletogenesis is largely conserved in all birds, and indeed all vertebrates. For example, most of the known functions for molecules such as Ihh and Bmp4 come from studies on both chick and mouse embryos (23, 26–28). In fact, similar craniofacial mechanisms have been observed in groups as disparate as fishes and birds (e.g., Bmp4 plays a role in deep/strong jaw morphology in cichlids) (29). However, only functional experiments performed in Geospiza and Loxigilla will determine with certainty whether the genes examined here cause the species-specific morphologies.
Our results are in agreement with the evolutionary relations among the Tholospiza, which show that L. noctis is more closely related to Geospiza than to L. violacea or L. portoricensis (Fig. 1A). As suggested previously (14), it is possible that the ancestor of the Tholospiza possessed a unique developmental genetic architecture that facilitated the evolution of high levels of beak diversity and, as the various lineages colonized different environments and occupied specialized niches, they acquired specific developmental programs by either de novo evolution or by inheritance from the ancestor. Thus, the mechanisms that pattern the beaks of L. noctis and Geospiza could have arisen after a lineage leading up to these species diverged from the L. violacea and L. portoricensis lineage, or could represent a condition present in the ancestor of Tholospiza (Fig. 1A). Regardless of these two alternative scenarios, we have shown that different developmental pathways can be involved in development of the same beak shape.
Together, our results in Loxigilla and in Geospiza (9–11) show that the signaling pathways we uncovered so far regulate scaling variation within the beak shapes analyzed, and that there is a rather flexible association between both scaling and group shape variation and the underlying modular developmental mechanisms (Fig. 5). This finding is revelant, because scaling variation has significant biomechanical consequences that have been shown to play a critical role in a bird’s survival (5, 7). It still remains unknown how the group beak shapes are established and maintained throughout development. One possible explanation is that the exact level of activation of each signaling system might determine if the axes shift proportionally, therefore producing the same shape, or nonproportionally, therefore altering the shape. Alternatively, a distinct set of molecules might exist, which would be more directly involved in generating the differences in beak curvatures that are characteristic of the group beak shapes.
Fig. 5.
Different developmental mechanisms (signaling pathways and tissues) can generate beaks of the same shape, whereas beaks of different shapes can be generated by the same developmental mechanisms. In L. noctis and in Geospiza (9–11), beak morphology is formed by two developmental modules: first by the pnc, through the action of Bmp4 and CaM signaling, and then by the pmx, through the action of TGFβIIr, β-catenin, and Dkk3 signaling. In L. violacea and L. portoricensis, Bmp4 and Ihh signaling promote expansion of the pmx, which is the main developmental module responsible for shaping beak morphology. Despite these differences in signaling pathways and tissues, the three Loxigilla species have independently evolved a common beak shape (group D), which varies only in scaling and is different from that of Geospiza (group A). Branch lengths have been altered to highlight the species analyzed.
This work has expanded our previous studies on Darwin’s finches by examining a group of related birds featuring similar levels of phenotypic diversity in a different geographical region, and in doing so we have demonstrated flexibility in the relationship between morphology and underlying developmental causes. Future efforts should be aimed at determining whether functional ecological demands can explain the ultimate causes driving the maintenance of the same beak shape within a lineage, the emergence of novel shapes as well as beak shape convergence among paraphyletic taxa, such as that found in Loxigilla. In addition to the proposed biomechanical studies, sampling more birds with drastically different beak shapes, such as the needle-shaped beaks of hummingbirds or the wide and shallow beaks of flycatchers, will help to determine whether other developmental mechanisms exist by which the entire beak diversity in avians is more comprehensively explained.
Materials and Methods
Phylogenetic Reconstruction.
For phylogenetic analyses we included representative Darwin’s finches and all 13 non-Darwin’s finches belonging to Tholospiza. We used sequences from two mitochondrial (cytochrome b and nicotinamide dehydrogenase subunit 2) and four nuclear markers (β-fibrinogen intron 5, myoglobin intron 2, recombination-activating gene, and aconitase 1 intron 10). Phylogenetic analyses were performed using maximum-likelihood and Bayesian inference methods. See SI Materials and Methods for additional details and sequence accession numbers.
Beak Shape Analysis.
The birds used for this analysis were obtained from the Museum of Comparative Zoology at Harvard University, and the beak shape analysis followed procedures outlined previously (8). See SI Materials and Methods for additional details.
Embryo Collection and Preparation.
Embryos of the three Loxigilla species were collected according to regulations established by the Secretaría de Estado de Medio Ambiente y Recursos Naturales (Dominican Republic), Departamento de Recursos Naturales y Ambientales (Puerto Rico), and the Natural Heritage Department, Ministry of Energy and the Environment (Barbados) using methods described in detail elsewhere (11). Embryos of L. noctis were collected in Holetown, Barbados (stage 27, n = 5; stage 30, n = 5); L. violacea embryos were collected in the Sierra de Bahoruco National Park, Dominican Republic (stage 27, n = 5; stage 30, n = 5); L. portoricensis embryos were collected in the surroundings of Guánica, Puerto Rico (stage 27, n = 5; stage 30, n = 5).
In Situ Hybridization and Immunohistochemistry.
In situ hybridizations, antibody stains, alkaline phosphatase assays, and quantifications of gene expression were performed as described previously (11). For immunostaining, we used anti-TGFβIIr (sc-400; Santa Cruz) and anti–β-catenin (610153; BD Transduction Laboratories) antibodies using methods described previously (11). In situ hybridizations were carried using chicken mRNA probes as described previously (9–11). See SI Materials and Methods for a detailed description on our methods for assessing gene expression in beaks and our use of controls.
Chicken Embryo Manipulations.
Fertilized eggs were obtained from SPAFAS, incubated at 37 °C, and staged according to Hamburger and Hamilton (30). Frontal nasal processes were infected at stage 24 with RCAS::Bmp4, RCAS::Ihh, RCAS::Noggin, and RCAS::Hip1 constructs, which have been described previously (9, 25, 31, 32). Embryos were collected at stage 39 (embryonic day 13) for in situ hybridizations and quantitative PCR (qPCR) and at stage 41 (embryonic day 15) for cartilage and bone staining. RCAS::Bmp4 and RCAS::Ihh were made using two different viral coats (“A” for RCAS::Bmp4 and “E” for RCAS::Ihh) (31, 32), allowing us to superinfect cells and test the simultaneous effect of overexpressing both genes. Stage 41 embryos were dehydrated in 95% (vol/vol) ethanol for 5 d and stained with alcian blue to reveal cartilage and with alizarin red to reveal bone. See SI Materials and Methods for qPCR methods. All animal experiments have been approved by the Institutional Biosafety Committee of the Faculty of Arts and Sciences and Committee on Microbiological Safety (COMS) of Harvard University.
Supplementary Material
Acknowledgments
We thank all the field assistants and participants of the field collecting trips for their help and advice, including J. Brocca, N. Corona, C. Kozak, F. Moscoso, and C. Perdomo. P. Grant, H. Hoekstra, M. Manceau, J. Losos, F. Jenkins, C. Tabin, and three anonymous reviewers provided comments on earlier versions of the manuscript. Environmental authorities in the Dominican Republic, Puerto Rico, and Barbados provided logistical support and help with permits. We thank Sigmar Stricker for providing the RCAS(B)::Hip1 construct. Support for this work was provided by National Science Foundation (NSF) Grants 10B-0616127 (to R.M. and A.A.), IBN-0217817 (to K.J.B.), and DEB-0315416 (to K.J.B.); and NSF Doctoral Dissertation Improvement Grant 0909695 (to R.M.). Support was also provided by the Kavli Institute for Bionano Science and Technology (Harvard University), the NSF’s Division of Mathematical Sciences, and National Institutes of Health Grant P50GM068763 (to O.C., J.A.F., and M.P.B.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HQ153049–HQ153089).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206205109/-/DCSupplemental.
References
- 1.Darwin C. On the Origin of the Species by Means of Natural Selection. London: John Murray; 1859. [Google Scholar]
- 2.Huxley J. 1942. Evolution: The Modern Synthesis (Allen & Unwin, London)
- 3.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 4.Gill FB. Ornithology. New York: Freeman; 2007. [Google Scholar]
- 5.Herrel A, Speck T, Rowe NP. 2006. Ecology and Biomechanics: A Mechanical Approach to the Ecology of Animals and Plants (Taylor and Francis CRC, Boca Raton, FL)
- 6.Lack DL. Darwin's Finches. Cambridge: Cambridge Univ Press; 1947. [Google Scholar]
- 7.Grant PR. Ecology and Evolution of Darwin's Finches. Princeton, NJ: Princeton Univ Press; 1999. [Google Scholar]
- 8.Campàs O, Mallarino R, Herrel A, Abzhanov A, Brenner MP. Scaling and shear transformations capture beak shape variation in Darwin’s finches. Proc Natl Acad Sci USA. 2010;107:3356–3360. doi: 10.1073/pnas.0911575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 10.Abzhanov A, et al. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 11.Mallarino R, et al. Two developmental modules establish 3D beak-shape variation in Darwin’s finches. Proc Natl Acad Sci USA. 2011;108:4057–4062. doi: 10.1073/pnas.1011480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanken J, Hall BK. The Skull. Chicago: Univ of Chicago Press; 1993. [Google Scholar]
- 13.Bond J. Birds of the West Indies. Boston: Houghton Mifflin; 1993. [Google Scholar]
- 14.Burns KJ, Hackett SJ, Klein NK. Phylogenetic relationships and morphological diversity in Darwin’s finches and their relatives. Evolution. 2002;56:1240–1252. doi: 10.1111/j.0014-3820.2002.tb01435.x. [DOI] [PubMed] [Google Scholar]
- 15.Bowman RI. Morphological Differentiation and Adaptation in the Galápagos Finches. Berkeley: Univ of California Press; 1961. [Google Scholar]
- 16.Sato A, et al. On the origin of Darwin’s finches. Mol Biol Evol. 2001;18:299–311. doi: 10.1093/oxfordjournals.molbev.a003806. [DOI] [PubMed] [Google Scholar]
- 17.Hartmann C. A Wnt canon orchestrating osteoblastogenesis. Trends Cell Biol. 2006;16:151–158. doi: 10.1016/j.tcb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Schneider RA, Hu D, Rubenstein JL, Maden M, Helms JA. Local retinoid signaling coordinates forebrain and facial morphogenesis by maintaining FGF8 and SHH. Development. 2001;128:2755–2767. doi: 10.1242/dev.128.14.2755. [DOI] [PubMed] [Google Scholar]
- 19.Trainor PA, Melton KR, Manzanares M. Origins and plasticity of neural crest cells and their roles in jaw and craniofacial evolution. Int J Dev Biol. 2003;47:541–553. [PubMed] [Google Scholar]
- 20.Kulesa P, Ellies DL, Trainor PA. Comparative analysis of neural crest cell death, migration, and function during vertebrate embryogenesis. Dev Dyn. 2004;229:14–29. doi: 10.1002/dvdy.10485. [DOI] [PubMed] [Google Scholar]
- 21.Abzhanov A, Rodda SJ, McMahon AP, Tabin CJ. Regulation of skeletogenic differentiation in cranial dermal bone. Development. 2007;134:3133–3144. doi: 10.1242/dev.002709. [DOI] [PubMed] [Google Scholar]
- 22.Wu P, Jiang TX, Shen JY, Widelitz RB, Chuong CM. Morphoregulation of avian beaks: Comparative mapping of growth zone activities and morphological evolution. Dev Dyn. 2006;235:1400–1412. doi: 10.1002/dvdy.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmerman LB, De Jesús-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 25.Gao B, et al. A mutation in Ihh that causes digit abnormalities alters its signalling capacity and range. Nature. 2009;458:1196–1200. doi: 10.1038/nature07862. [DOI] [PubMed] [Google Scholar]
- 26.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 27.Shum L, Wang X, Kane AA, Nuckolls GH. BMP4 promotes chondrocyte proliferation and hypertrophy in the endochondral cranial base. Int J Dev Biol. 2003;47:423–431. [PubMed] [Google Scholar]
- 28.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 29.Albertson RC, Kocher TD. Genetic and developmental basis of cichlid trophic diversity. Heredity (Edinb) 2006;97:211–221. doi: 10.1038/sj.hdy.6800864. [DOI] [PubMed] [Google Scholar]
- 30.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 31.Kengaku M, et al. Distinct WNT pathways regulating AER formation and dorsoventral polarity in the chick limb bud. Science. 1998;280:1274–1277. doi: 10.1126/science.280.5367.1274. [DOI] [PubMed] [Google Scholar]
- 32.Vortkamp A, et al. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 33.Ridgely RS, Tudor G, Brown WL. The Birds of South America. Austin, TX: Univ of Texas Press; 1989. [Google Scholar]
- 34.Swash A, Still R, Lewington I. Birds, Mammals, and Reptiles of the Galápagos Islands: An Identification Guide. New Haven, CT: Yale Univ Press; 2000. [Google Scholar]
- 35.Raffaele HA, Wiley J, Garrido O, Keith A, Raffaele J. Birds of the West Indies. Princeton, NJ: Princeton Univ Press; 2003. [Google Scholar]
- 36.van Wyhe J, editor. The Complete Work of Charles Darwin Online. 2002. Available at http://darwin-online.org.uk/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.