Abstract
Hypothesis directed proteomics offers higher throughput over global analyses. We show that immunoprecipitation (IP)–tandem mass spectrometry (LC-MS/MS) in H929 multiple myeloma (MM) cancer cells led to the discovery of a rare and unexpected BCR–ABL fusion, informing a therapeutic intervention using imatinib (Gleevec). BCR–ABL is the driving mutation in chronic myeloid leukemia (CML) and is uncommon to other cancers. Three different IP–MS experiments central to cell signaling pathways were sufficient to discover a BCR–ABL fusion in H929 cells: phosphotyrosine (pY) peptide IP, p85 regulatory subunit of phosphoinositide-3-kinase (PI3K) IP, and the GRB2 adaptor IP. The pY peptides inform tyrosine kinase activity, p85 IP informs the activating adaptors and receptor tyrosine kinases (RTKs) involved in AKT activation and GRB2 IP identifies RTKs and adaptors leading to ERK activation. Integration of the bait–prey data from the three separate experiments identified the BCR–ABL protein complex, which was confirmed by biochemistry, cytogenetic methods, and DNA sequencing revealed the e14a2 fusion transcript. The tyrosine phosphatase SHP2 and the GAB2 adaptor protein, important for MAPK signaling, were common to all three IP–MS experiments. The comparative treatment of tyrosine kinase inhibitor (TKI) drugs revealed only imatinib, the standard of care in CML, was inhibitory to BCR–ABL leading to down-regulation of pERK and pS6K and inhibiting cell proliferation. These data suggest a model for directed proteomics from patient tumor samples for selecting the appropriate TKI drug(s) based on IP and LC-MS/MS. The data also suggest that MM patients, in addition to CML patients, may benefit from BCR–ABL diagnostic screening.
Keywords: targeted therapies, personalized medicine, protein–protein interactions
Proteomics studies over the last decade have provided significant advancements in our understanding of the functional landscape of proteins, their posttranslational modifications and the protein–protein interactions (PPI) (1–3). This is especially important for diseases such as cancer because proteins and their transient posttranslational modifications (PTMs) reflect the changes cells undergo during pathogenic development. Modern high-resolution mass spectrometers are well suited for these analyses and have led the efforts in this field of proteomics, allowing researchers to analyze dynamic changes in cell lines or tissue and to distinguish between normal and unhealthy states (4, 5). PPI networks and phosphoproteomics are important for understanding the function and regulation of pathways leading to cell growth and proliferation in diseases such as cancer. The advantage of analyzing purified protein complexes is the ability to identify specific interacting proteins and posttranslational modifications that may otherwise go undetected in large-scale global analyses. This can be achieved by performing an immunoprecipitation (IP) with a bait protein and analyzing the prey proteins using microcapillary–tandem mass spectrometry (LC-MS/MS) (6–13). These experiments have included various types of affinity tags and have been overlaid with RNAi genetic screens (14, 15). Undoubtedly, PPI networks provide a valuable framework for a better understanding of the functional organization of the proteome (16).
Here, we focused on a hypothesis-directed approach to identify the active signaling pathways that drive cancers. To this end, we immunoprecipitated proteins that have clinical significance in cell proliferation such as the central nodes in the AKT and ERK signaling pathways. The p85 regulatory subunit of phosphoinositide-3-kinase (PI3K) binds pYXXM motif-containing proteins to the SRC homology 2 (SH2) domains of p85, thus recruiting the p110 catalytic subunit to the plasma membrane for activation (5, 17). Activated p110 phosphorylates its lipid substrate phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3) and binds to the pleckstrin homology (PH) domain of the serine/threonine kinase AKT, which is frequently elevated in cancers (18). We recently showed that we can immunopurify p85 and quantify the activating PI3K adaptors and RTKs by targeted LC-MS/MS (5). In addition, we show here that we can IP the growth-factor-receptor-bound protein 2 (GRB2) adaptor to identify associated activators of the extracellular signal-regulated kinase (ERK), which is also frequently elevated in cancers (19). GRB2 is an important SH2/SH3 adaptor protein linking receptor tyrosine kinases (RTK) and downstream targets and has implications in malignancies (20). The SH2 domain of GRB2 binds to the phosphorylated tyrosine residue of an adaptor protein or RTK and the SH3 domain binds proteins such as SOS leading to RAS/RAF binding and ERK activation (20, 21). Tyrosine phosphorylation (pY) is one of the most important and most studied posttranslational modification (PTM) of proteins because it regulates signaling pathway activity and important cellular functions including cell proliferation, growth, survival, etc. (22). Abnormalities in tyrosine kinases play a role in the pathogenesis of numerous inherited or acquired human diseases from cancer to immune deficiencies (23, 24). Enrichment of low-abundance tyrosine phosphorylated proteins is necessary for a successful comprehensive LC-MS/MS analysis. One common method for gaining insight into a broad range of active tyrosine signaling pathways in a cell can be achieved by an IP–MS analysis of phosphotyrosine containing peptides from tryptic digested cell lysates (25, 26).
Here we show a clinically relevant example using our three-pronged IP–MS technology (p85 IP, GRB2 IP, and pY IP) in multiple myeloma (MM) cells and how we identified a rare breakpoint cluster region-Abl tyrosine kinase (BCR-ABL) fusion protein by converging protein complexes and activated tyrosine kinases. BCR–ABL is unexpected in MM because, unlike the leukemias and lymphomas, MM typically has no specific chromosomal abnormalities and is characterized by a genomic instability that involves both ploidy and structural translocation (27). There have only been two reports of MM patients with a Philadelphia chromosome t(9;22)(q34;q11), a 64-y-old woman presenting a P190 BCR–ABL hybrid transcript by PCR analysis (28) and a 61-y-old man showing BCR–ABL-positive bone marrow after autologous bone marrow transplantation (29).
The mass spectrometry results were verified using biochemistry and histological analyses as well as DNA sequencing and compared with an array of MM cell lines and the K562 chronic myeloid leukemia (CML) cell line known to contain a BCR–ABL fusion. We show that imatinib (trade name Gleevec) selectively inhibits signaling in H929 cells and halts cell proliferation. This report shows the molecular investigation of BCR–ABL fusion in MM, and the results should serve as a template for the use of hypothesis directed proteomics on human tumor samples or cell lines derived from tumors for aiding the appropriate therapeutic selection of single or combinations of TKI drugs in patients.
Results
Analysis of Activating Signaling Pathways Using a Multifaceted Hypothesis-Directed Proteomics Approach.
Because cell proliferation and the growth of cancers are typically associated with signal transduction cascades from surface receptors to adaptors and downstream targets, we investigated a targeted proteomics strategy to enrich and identify the upstream activators of PI3K/AKT, GRB2/ERK, and complementary activated phosphotyrosine containing proteins. The strategy consists of performing three different IPs from a biological source such as cells or tumor tissue using antibodies against the endogenous protein. To investigate the PI3K-associated activators of AKT, the p85 regulatory subunit of PI3K is used as a bait protein; to investigate activators of ERK pathway, GRB2 is used as a bait protein. In addition, phosphotyrosine-containing peptides are immunoprecipitated from a trypsin-digested cell lysate to identify activated phosphotyrosine-containing proteins. The samples are analyzed by label-free LC-MS/MS (30). The resulting PPI data from the p85 and GRB2 IP–MS experiments and the protein products from identified phosphopeptides from the pY IP are then correlated in a simple network to clarify which upstream RTK and adaptor proteins may be involved in promoting cell proliferation in cancers.
Case Study Using H929 MM Cells.
H929 cells as well as other MM cells were investigated to explore which kinase signaling pathways may be responsible for driving proliferation and to explore whether any recently developed TKI drugs would be useful in inhibiting these cell lines. H929 cells are known to have a rearrangement of the c-Myc proto-oncogene and express c-Myc RNA and an activated N-RAS allele. They are positive for plasma cell antigen 1 (PCA-1), transferrin receptor and CD38 (T10), but are negative for HLA DR, CALLA, and markers of early B-cell development (31, 32).
p85 IP–MS PPI landscape.
The PPI landscape of p85 (PI3K) was investigated as the first phase in identifying drivers of cell proliferation in MM cells. H929 cells were lysed, and a polyclonal antibody recognizing the p85 regulatory subunit of PI3K was used to immunoprecipitate its associated proteins according to previously published studies (5, 33, 34). Immunoprecipitated p85 protein complexes were eluted and run by mini SDS/PAGE ∼1/6 of the gel distance and stopped. After staining, three gel sections were excised, one below and one above 55 kDa and the band for the IgG heavy chain to both capture the full molecular weight (MW) range and avoid excessive antibody contamination (5). Gel sections were digested with trypsin and run using data-dependent (shotgun) LC-MS/MS with a hybrid linear ion trap–orbitrap mass spectrometer to discover both canonical and unknown interacting proteins, and data were organized and validated using Scaffold protein identification software with three biological replicates for each experiment. After known contaminants such as human keratins, caseins from milk, trypsin, and BSA were removed from the analysis, 83 total proteins were associated with p85 from three independent biological replicate analyses. Sixteen proteins were common to all three replicates, including known p85-binding proteins like the p110 catalytic subunits PK3CA (p110α), PK3CB (p110β), and PK3CD (p110δ) and the known upstream activating adaptor GAB2 (35) and its associated protein SHP2 (5, 36). Incorporating two of three replicate runs yields 36 p85-interacting proteins, including the known activating adaptor IRS2 (37).
GRB2 IP–MS PPI landscape.
Because the p85 IP mainly captures kinases and adaptor proteins associated with AKT activation, we also immunopurified the GRB2 adaptor as a bait to capture proteins associated with the RAS/ERK signaling pathway in H929 cells (21, 38). We associated a total of 299 proteins with GRB2 from all three different IP–MS experiments. Twenty-seven proteins were common to all three biological replicates and, among these, several canonical GRB2-binding proteins were identified, including BCR, ABL1, SYK, and SHP2. From combining at least two biological replicates, we can identify 190 proteins including GAB2, p85α, p85β, SOS1, SOS2, CBL, and SHIP2. It is important to note that it is common to capture nonspecific binding proteins from IP–MS experiments, so most of the identified proteins are not intimately involved in a GRB2 complex. Nonspecific proteins can effectively be eliminated by performing multiple IP–MS experiments using the same bait or by intercalating different bait IP–MS studies, as we show here. A similar analysis was reported recently where FLAG-GRB2 was used as bait in HEK293T cells in combination with multiple reaction monitoring (MRM) to assess the intracellular GRB2 PPI landscape (19). In our data, SOS1, SOS2, GAB2, and SHP2 can be assigned to the MAP kinase pathway (39); however, it is known that the GRB2 adaptor also plays a role in cancer by binding to the cytoplasmic fusion tyrosine kinase BCR–ABL in CML cells (40) and, in addition, the proteins ABL1, BCR, and CBL can be assigned to the BCR–ABL pathway (41).
pY peptide landscape.
In contrast to pS and pT, pY has been successfully targeted by antibodies (26, 42, 43) and signaling pathway cascades are frequently initiated by pY sites on well defined signaling pathways leading to AKT and ERK activation, we decided to use this approach to help identify active pY peptides independent of p85 and Grb2 and integrate the data from all three IPs. We digested H929 cell lysate with trypsin and incubated the resulting purified peptides with either the monoclonal pTyr-100 antibody from Cell Signaling Technologies and monoclonal pTyr antibody from Sigma-Aldrich separately or with both antibodies combined. Phosphotyrosine-containing peptides were then identified using LC-MS/MS and, after excluding peptides that did not cotain pY and retaining pY peptides above a 1% false discovery rate (FDR) with a site localization probability above 75% according to Scaffold PTM software, we detected 74 total phosphopeptides representing 76 unique pY sites from 64 proteins (Table S1). Some of the phosphorylated proteins included p85, BCR, ABL1, BTK, CBL, SHC1, SHP2, SHIP2, FYN, YES, DOK1, and DOK2, which have been previously described as part of the BCR–ABL pathway (41). Given the relatively low number of pY-containing proteins identified, these data were significant.
Integration of p85, GRB2, and pY IP–MS data.
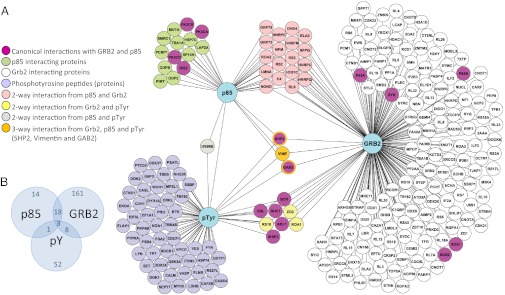
We then correlated the results of the p85, GRB2, and pY IP–MS studies in a simple PPI network based on common proteins using Cytoscape software (Fig. 1). To become part of the network, interacting proteins from the p85 and GRB2 IPs must have been present in at least two of three biological replicate runs passing a 1% FDR threshold. In total, 30 proteins overlapped in at least two of the three IP–MS experiments, including proteins like BCR, ABL1, SHIP2, CBL, p85, GAB2, and SHP2 (Fig. 1). Because GAB2 and SHP2 were present in all three IPs, it suggested that signaling in H929 cells proceeds via the GAB2 adaptor and the tyrosine phosphatase SHP2. GAB2 is a common adaptor protein that binds directly to RTKs and interacts with GRB2, SHP2, p85, and SHC1 (35). GAB2 is also known to be tyrosine phosphorylated by oncoproteins including SRC and BCR–ABL (44). In contrast, SHP2 has been shown to regulate PI3K and c-Kit activity (39, 45) and is essential for maintaining ERK signaling (46).
Fig. 1.
The network of the three-pronged IP–MS analysis. (A) The PPI network integrating data from three separate IP-LC-MS/MS experiments using endogenous antibodies to p85 (protein capture) and GRB2 (protein capture) as bait proteins and pTyr (peptide capture) to identify the active tyrosine kinase signaling pathways in H929 MM plasma cells. (B) Venn diagram showing the overlap of proteins/peptides from each IP–MS experiment.
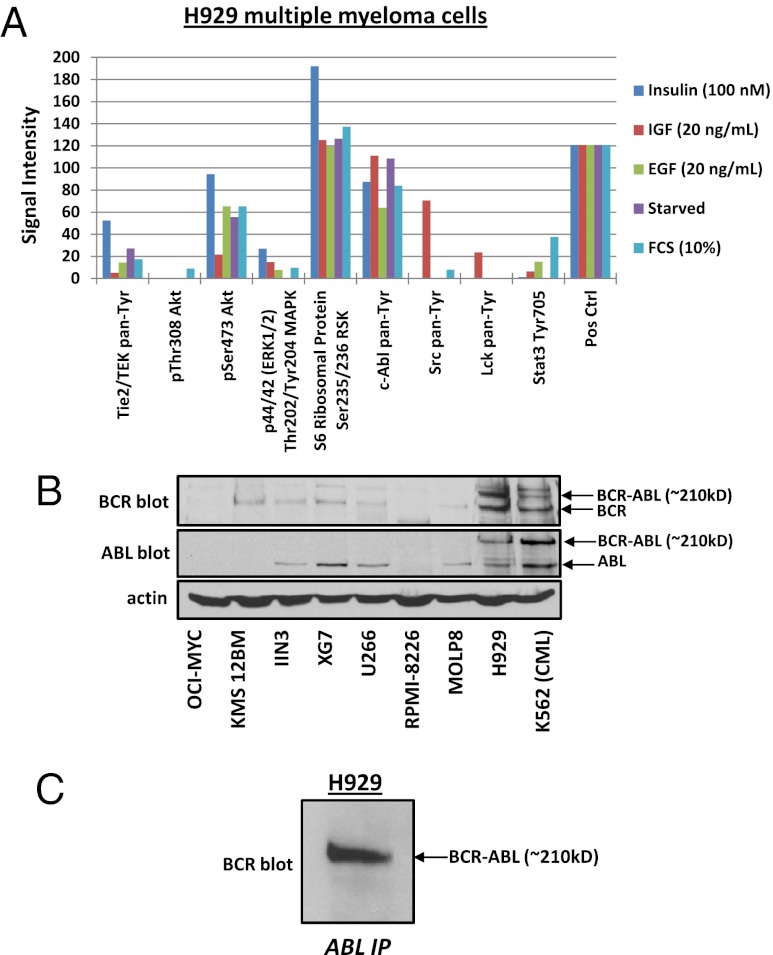
To correlate the identified pY peptides to protein level signaling, we performed an activity assay using the commercially available PathScan RTK Signaling antibody array kit that includes 39 different kinases and key signaling nodes (Fig. S1). H929 cells were stimulated with either 100 nM insulin, 20 ng/mL IGF, 20 ng/mL EGF, 10% (vol/vol) FCS, or were serum starved. Fig. 2A shows the few kinases for which we obtained an antibody signal after normalization to the negative control signal. The highest phosphorylation signal obtained relative to background was from the ABL tyrosine kinase and ribosomal S6 kinase; however, phosphorylation signals were also obtained for ERK1/2, Tie, AKT pS473, SRC, and STAT3. These tyrosine antibody data support the IP–MS results and suggest a possible signaling pathway regulated through the BCR–ABL fusion pathway.
Fig. 2.
Evidence for BCR–ABL signaling in H929 MM cells. (A) PathScan RTK signaling antibody array kit tests the phosphorylated signal level of kinases and key signaling nodes. After serum starvation, H929 MM cells were stimulated for 90 min with insulin, insulin growth factor (IGF), epidermal growth factor (EGF), or 10% FCS or were serum starved overnight. (B) Immunoblots of the MM cell lines OCI-MYC, KMS 12BM, IIN3, XG7, U266, RPMI-8226, MOLP8, H929, and K562 CML to test for the BCR–ABL fusion protein from whole cell lysates (WCL). (C) ABL IP is immunoblotted for BCR to check for the presence of the BCR–ABL fusion in H929 cells.
Verification of BCR–ABL Activity via Fusion in H929 Cells.
Because the proteomics and PathScan results led to an unexpected but recognizable signaling pathway that could be responsible for driving cell proliferation, it became clear that verification of a possible BCR–ABL fusion in H929 MM cells was necessary.
Western blotting and tandem mass spectrometry.
To test for the BCR–ABL fusion protein, whole cell lysates of H929 cells and control cell lines including the MM cell lines OCI-MYC, KMS 12BM, IIN3, XG7, U266, RPMI-8226, MOLP8, and the BCR–ABL-positive CML cell line K562 were immunoblotted for the proteins BCR and ABL1. Fig. 2B shows that H929 has not only the expected band for endogenous BCR at 140 kDa, but also an additional band with a higher MW of ∼210 kDa. The same result is observed for the immunoblot against the endogenous ABL1 protein, with an expected band at 120 kDa and an additional band at ∼210 kDa at the same position as BCR. The other MM cell lines are lacking the high MW bands at ∼210 kDa in the BCR and ABL blots. Only the Philadelphia-chromosome-positive CML cell line K562 shows a similar high MW band at ∼210 kDa for both BCR and ABL, which demonstrates a positive fusion event of BCR–ABL in the H929 cells (Fig. 2B). Additionally we performed an IP of the ABL protein on H929 cell lysate and immunoblotted for BCR. The Western blot in Fig. 2C shows the fusion with the molecular band at ∼210 kDa. To verify the Western blots, we used LC-MS/MS to sequence the high MW region of the ABL IP by excising the Coomassie-stained ∼210 kDa section from an SDS/PAGE gel and digesting separately with both trypsin and chymotrypsin (Fig. S2A). The amino acid coverage results (Fig. S2B) show peptides at a 1% FDR that were identified for both BCR and ABL from the ∼210-kDa gel region from replicate analyses. However, a peptide sequence containing both BCR and ABL components was not identified likely due to a small tryptic (QSSK) and chymotryptic (KQSSKAL) fusion peptide. Interestingly, Fig. S2C shows a singly charged MS precursor ion that is consistent with the chymotryptic peptide KQSSKAL with high mass accuracy, but fragmentation of the ion peak was poor and database searching did not yield statistically significant scores. Nevertheless, we confidently sequenced the peptides immediately surrounding the fusion site from the shifted gel band, consistent with the e14a2 form found from RT-PCR and gene sequencing.
DNA sequencing analysis of BCR–ABL fusion region.
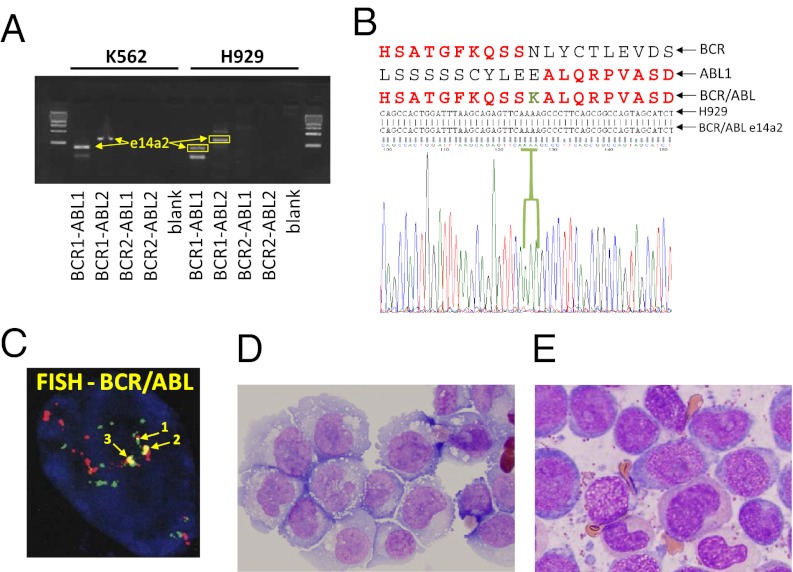
Additionally we were interested in identifying and confirming the BCR–ABL isoform present in H929 cells. RT-PCR was performed on H929 and K562 cells and used primers specific for the most frequent BCR–ABL isoforms including e13a2 (b2a2), e14a2 (b3a2), e13a3 (b2a3), e14a3 (b3a3), and e1a2 (47-49). As shown in Fig. 3A, the PCR showed the same DNA band pattern in H929 and K562 and the sequencing results for the extracted DNA of H929 showed clearly the existence of the isoform e14a2 (Fig. 3B) the same form described in K562 cells (50). This result in combination with the Western blotting and LC-MS/MS data proves the presence of a BCR–ABL fusion event in H929 cells.
Fig. 3.
Genomics and histopathology analyses. (A) Picture of agarose gel from H929 MM cells and CML K562 cells showing the prominent cDNA bands obtained from RT-PCR. (B) The obtained cDNA were sequenced with the Sanger method, and the translated protein sequence was aligned with the common and imatinib sensitive e14a2 form of BCR–ABL protein. (C) FISH analysis with a dual-color BCR–ABL fusion probe against the Philadelphia chromosome t(9, 22)(q34;q11) was performed to detect the BCR–ABL translocation in H929 cells. (D) H929 MM plasma cells visualized by hematoxylin and eosin (H&E) staining and microscopy. (E) Stable phase in vivo CML leukocytes visualized by Wright–Giemsa (WG) staining and microscopy.
Histopathology of H929 MM cells.
Because the discovery of BCR–ABL fusion in H929 cells using our three IP–MS proteomics approach represents an unexpected event in MM (28, 29), a FISH analysis with a dual-color BCR–ABL fusion probe against the Philadelphia chromosome t (9, 22)(q34;q11) was performed to detect the BCR–ABL translocation in H929 cells (Fig. 3C). In addition, we performed experiments to prove the MM plasma cell origin of our H929 and to exclude contamination with a CML cell line. By immunoperoxidase (IMPOX), H929 were decorated with CD138, a universal plasma cell marker routinely used in the clinical diagnosis of myeloma because of its plasma cell origin (Table 1). Also, the absence of reactivity with antibodies to MPO, CD33, and CD68 exclude the possibility of a contamination with CML cells. In contrast to H929, most MM plasma cells express and secrete Ig. However, the corresponding gene for Ig secretion, IFN regulatory factor 4 (IRF4), is required during an immune response for lymphocyte activation and the generation of plasma cells (51, 52). The lack of IRF4 expression in H929 cells may explain the absence of detectable Ig light chain signal by IHC. Fig. 3D shows photomicrographs of H929 MM plasma cells. The cells resemble typical plasmablasts, and Wright–Giemsa staining reveals an abundant blue cytoplasm more deeply stained toward the periphery, prominent Golgi complex, eccentrically located nuclei, and a large and prominent central nucleolus, all features characteristic of plasma cells but not of myeloid cells. In contrast, Fig. 3E shows typical CML leukocytes from a CML patient sample stained with Wright–Giemsa. The CML cells show features of neutrophilic differentiation, including cytoplasmic azurophilic granules, the appearance of secondary (pink) granules as nuclei begins to segment (cells with nonround nuclei in Fig. 3E). Even the more immature “blast” component (blue cells in Fig. 3E) differs significantly from H929 cells in that CML blasts have centrally placed nuclei with eccentrically placed nucleoli, far less cytoplasm, and rather inconspicuous Golgi apparatus.
Table 1.
Histopathology results of H929 cell line
| Marker | Plasma cells | H929 (MM) | CML |
| FISH [t(9;22)] = Philadelphia chromosome | − | + | + |
| MPO (myeloperoxidase) | − | − | + |
| CD68 | − | − | + |
| CD34 | − | − | + |
| CD33 | + or − | − | + |
| CD138 | + | + | − |
| CD45 | + or − | + | + |
| CD43 | + | + | + |
| CD79A | + | − | − |
| IgG | + | − | − |
| IgM | + | − | − |
| IgA | + | − | − |
| Kappa light chain | + | − | − |
| Lambda light chain | + | − | − |
| MUM1 (IRF4) | + | − | − |
Histopathology results performed by IHC and FISH in H929 MM cells compared with the typical phenotypes from neoplastic plasma cells and CML cells.
Effect of tyrosine kinase inhibitors on H929 signaling.
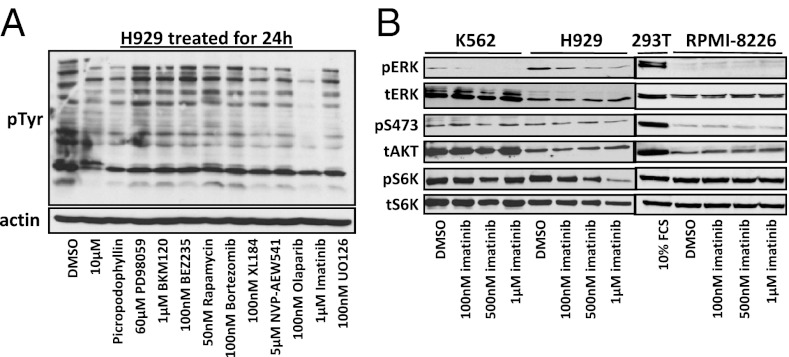
Because H929 cells showed a BCR–ABL fusion, we tested a variety of tyrosine kinase inhibitor drugs on H929 cells and probed the lysates using a pY antibody to test for a reduction in tyrosine signaling after 24 h of treatment, a common indicator that the TKIs are having an effect on the cell’s signaling network. We tried a variety of TKIs that target different signaling oncoproteins such as PI3K only (BKM-120 and LY294002), dual PI3K and mTOR (BEZ-235), MEK (U0126), mTOR (Rapamycin), dual ABL, and PDGFR (imatinib), the proteasome inhibitor bortezomib, and the tyrosine phosphatase inhibitor sodium orthovanadate (Na3VO4) as well as DMSO drug vehicle control (Fig. 4A). The results clearly show only a dramatic and selective reduction in tyrosine signaling in H929 cells with imatinib treatment due to ABL inhibition. Imatinib binds to the ATP-binding pocket of the c-ABL protein, prevents the kinase to become active and inhibits the ability to phosphorylate the downstream signaling pathways (53). We investigated various concentrations of imatinib and its effect on phosphorylated signaling proteins in H929 cells versus K562 BCR–ABL-positive cells and RPMI-8226 MM BCR–ABL-negative cells. We treated H929, K562, and RPMI-8226 with different concentrations of imatinib (100 nM, 500 nM, and 1 μM) for 1 h and immunoblotted for phospho-ERK, phospho-AKT (S473), and phospho-S6K (Fig. 4B). The comparison of K562 and H929 shows only H929 is sensitive to imatinib and decrease phosphorylation on S6K at a concentration of 500 nM. Imatinib can inhibit the phosphorylation of ERK in both cell lines; although K562 cells have a low level of p42 and no p44, ERK is completely inhibited by 500 nM. In H929, the level of ERK phosphorylation is higher than K562 and only the treatment with 500 μM imatinib significantly inhibited pERK. In both cell lines, pAKT (S473) levels are low and unaffected by imatinib. As expected, treatment of RPMI-8226 MM cells with imatinib had no effect on pERK, pAKT, or pS6K (Fig. 4B).
Fig. 4.
Phosphorylated signaling after drug treatment. (A) pY immunoblot of H929 lysate after 24-h incubation with various tyrosine kinase inhibitor drugs including imatinib and the proteasome inhibitor bortezomib. (B) Immunoblots showing the effect of 1-h treatment of increasing concentrations of imatinib on key signaling proteins pERK, pAKT, and pS6K from WCL of BCR–ABL fusion containing cells H929 (MM) and K562 (CML) as well as RPMI-8226 MM cells containing no BCR–ABL fusion and 293T cells for a control.
Cell proliferation assays.
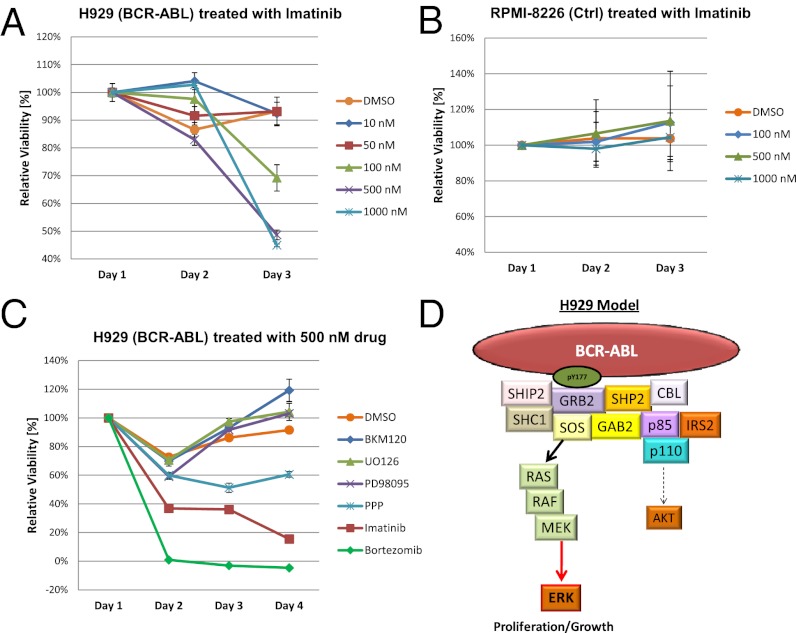
Because imatinib was effective in abrogating tyrosine signaling in H929 cells containing the BCR–ABL fusion, we performed a MTT cell proliferation assay in biological triplicates to test whether imatinib could stop the growth of H929 MM plasma cells. Fig. 5A shows the proliferation assay in H929 cells using different concentrations of imatinib over a 72-h period. Concentrations of 500 nM or more completely halt proliferation, and the effect of imatinib becomes apparent with as little as 100 nM. Fig. 5B shows that RPMI-8226 cell proliferation is not effected by imatinib even at high concentrations. To test whether other drugs have an effect on cell proliferation in H929 cells, we treated the cells with PD98095 (MEK-1 inhibitor), BKM-120 (PI3K inhibitor), Picropodophillin (PPP, IGF-1R inhibitor), bortezomib (proteasome inhibitor), or imatinib and monitored the proliferation for 96 h (Fig. 5C). BKM-120 and PD98095 with the concentration of 500 nM had no effect on the viability of the cells and the proliferation rate was similar to the control group. The drug PPP showed a slight effect in lowering the proliferation on H929 at the concentration of 500 nM, but only imatinib and bortezomib caused a dramatic loss in cell viability (Fig. 5C). The proteasome inhibitor bortezomib is involved in the first line of treatment for MM, so the dramatic effect on H929 cells is expected (54–56); however, the proliferation assay proves that imatinib could be as effective as bortezomib for treating MM when the BCR–ABL mutation is present.
Fig. 5.
MTT proliferation assays averaged over three experiments including SD. (A) H929 cells treated with increasing concentrations of imatinib (100 nM, 500 nM, and 1 μM) over 72 h. (B) RPMI-8226 cells treated with imatinib under the same conditions as H929. (C) H929 cells treated with 500 nM of various tyrosine kinase inhibitor drugs and the proteasome inhibitor bortezomib over a 96-h period. (D) The biological model for BCR–ABL signaling in H929 MM cells driving proliferation primarily through ERK via binding of the Grb2 adaptor to BCR-ABL and to a lesser degree through AKT via PI3K.
Discussion and Conclusions
Our results show that one can systematically use a hypothesis-directed proteomics approach to assess the activated RTK signaling pathways in cancers through a series of three separate IP-LC-MS/MS experiments without the use of chemical labels. The results also demonstrate that activated signaling pathways that drive the particular cancer can be discovered without prior knowledge of genetic information. These results provide evidence for the usefulness of proteomics approaches to probe individual patient tumors to gather information to help select the most appropriate single or combination of TKI drug candidate(s). Currently, no validated method exists for selecting TKI drugs, which are being developed at a rapid pace to block the activity of signaling pathways that drive cell proliferation and tumor growth (57). Although global proteomics still remains a challenge, targeted and/or directed applications that can be standardized across laboratories can be advantageous for propelling proteomics into the clinic.
MM is unique within the blood cell cancers with no specific chromosomal abnormalities (58), and Philadelphia-positive MM is a very rare and unexpected event. Until this report, only two reports of patient BCR–ABL fusions were detected using PCR amplification and gene sequencing; however, no molecular mechanistic data regarding expressed BCR–ABL protein was previously reported in MM (28, 29). The literature supports the existence of a relationship between MM and CML as well as a relationship between MM and the myeloproliferative disorders (59, 60). It is important to mention that the BCR–ABL fusion was most prominent in one lot of H929 cells, but an additional high molecular band for both BCR and ABL could be detected in other lots of H929 cells from different vendors and separate academic laboratories (Fig. S3). It only takes three IPs (p85, GRB2, and pY) to identify the major pathogenic abnormalities in kinase regulation or tyrosine phosphorylation, which are the major contributors for developing cancers (23, 61). We showed that imatinib was the most effective kinase inhibitor drug for inhibiting the growth of H929 cells due to the imatinib sensitive isoform e14a2 of BCR–ABL (47) and knowledge of the BCR–ABL proteomic identified defect was critical to these results. Cancer genomics can reveal a multitude of abnormalities in the form of mutations, deletions, and amplifications and some of these can be in the tens of thousands for a single cancer (62). However, directed IP–MS reveals less complex data and provides information about protein function through PTMs and PPIs that must be surmised from genetic information. The results also make a case for future testing of MM patients for not only BCR–ABL translocation but other common genetic mutations as part of the routine diagnosis.
Materials and Methods
Detailed materials and methods including all reagents, drugs, cell lines, kits, assays, Western blots, histopathology, immunoprecipitations, mass spectrometry and gene sequencing used for this study can be found in SI Materials and Methods. The complete protein and peptide identification results from the p85, GRB2, and pY IP–MS data and PPI network can be found in SI Datasets S1, S2, S3, and S4. Figs. S1, S2, and S3 and Table S1 can also be found following the SI Materials and Methods section.
Supplementary Material
Acknowledgments
The authors thank Constantine Mitsiades (Dana–Farber Cancer Institute, DFCI) and Kenneth Anderson (DFCI) for helpful discussions and donating H929 cells; Alex Almasan (Taussig Cancer Institute) for the donation of H929 cells; Pier Paolo Pandolfi (Beth Israel Deaconess Medical Center, BIDMC) for the donation of the K562 cells; and Wenyi Wei (BIDMC) for the donation of OCI-MYC, KMS 12BM, IIN3, XG7, and U266 cells. We also thank Lewis Cantley and laboratory members for helpful discussions. This work was supported in part by National Institutes of Health Grant 5P01CA120964-05 and Dana–Farber/Harvard Cancer Center Support Grant 5P30CA006516-46 (both to J.M.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212759109/-/DCSupplemental.
References
- 1.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat Rev Genet. 2009;10:617–627. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 2.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 3.Schreiber TB, Mäusbacher N, Breitkopf SB, Grundner-Culemann K, Daub H. Quantitative phosphoproteomics—an emerging key technology in signal-transduction research. Proteomics. 2008;8:4416–4432. doi: 10.1002/pmic.200800132. [DOI] [PubMed] [Google Scholar]
- 4.Anderson NL, et al. A human proteome detection and quantitation project. Mol Cell Proteomics. 2009;8:883–886. doi: 10.1074/mcp.R800015-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X, et al. Using tandem mass spectrometry in targeted mode to identify activators of class IA PI3K in cancer. Cancer Res. 2011;71:5965–5975. doi: 10.1158/0008-5472.CAN-11-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia S, et al. Relative quantification of protein-protein interactions using a dual luciferase reporter pull-down assay system. PLoS ONE. 2011;6:e26414. doi: 10.1371/journal.pone.0026414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosley AL, et al. Highly reproducible label free quantitative proteomic analysis of RNA polymerase complexes. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.000687. M110 000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, et al. Mutant proteins as cancer-specific biomarkers. Proc Natl Acad Sci USA. 2011;108:2444–2449. doi: 10.1073/pnas.1019203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang G, et al. Mass spectrometry mapping of epidermal growth factor receptor phosphorylation related to oncogenic mutations and tyrosine kinase inhibitor sensitivity. J Proteome Res. 2011;10:305–319. doi: 10.1021/pr1006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gavin AC, Maeda K, Kühner S. Recent advances in charting protein-protein interaction: mass spectrometry-based approaches. Curr Opin Biotechnol. 2011;22:42–49. doi: 10.1016/j.copbio.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Charbonnier S, Gallego O, Gavin AC. The social network of a cell: Recent advances in interactome mapping. Biotechnol Annu Rev. 2008;14:1–28. doi: 10.1016/S1387-2656(08)00001-X. [DOI] [PubMed] [Google Scholar]
- 12.Aloy P, et al. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- 13.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 14.Friedman AA, et al. Proteomic and functional genomic landscape of receptor tyrosine kinase and ras to extracellular signal-regulated kinase signaling. Sci Signal. 2011;4:rs10. doi: 10.1126/scisignal.2002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stelzl U, et al. A human protein-protein interaction network: A resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, et al. Structure of lipid kinase p110β/p85β elucidates an unusual SH2-domain-mediated inhibitory mechanism. Mol Cell. 2011;41:567–578. doi: 10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bisson N, et al. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat Biotechnol. 2011;29:653–658. doi: 10.1038/nbt.1905. [DOI] [PubMed] [Google Scholar]
- 20.Cheng AM, et al. Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell. 1998;95:793–803. doi: 10.1016/s0092-8674(00)81702-x. [DOI] [PubMed] [Google Scholar]
- 21.Avruch J, et al. Ras activation of the Raf kinase: Tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog Horm Res. 2001;56:127–155. doi: 10.1210/rp.56.1.127. [DOI] [PubMed] [Google Scholar]
- 22.Bergström Lind S, et al. Toward a comprehensive characterization of the phosphotyrosine proteome. Cell Signal. 2011;23:1387–1395. doi: 10.1016/j.cellsig.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 23.Alonso A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi K, Ito F. Receptor tyrosine kinases and targeted cancer therapeutics. Biol Pharm Bull. 2011;34:1774–1780. doi: 10.1248/bpb.34.1774. [DOI] [PubMed] [Google Scholar]
- 25.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Rush J, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 27.Shaughnessy JD, Jr, Barlogie B. Integrating cytogenetics and gene expression profiling in the molecular analysis of multiple myeloma. Int J Hematol. 2002;76(Suppl 2):59–64. doi: 10.1007/BF03165089. [DOI] [PubMed] [Google Scholar]
- 28.Martiat P, et al. P190 BCR/ABL transcript in a case of Philadelphia-positive multiple myeloma. Leukemia. 1990;4:751–754. [PubMed] [Google Scholar]
- 29.Roper N, et al. An asymptomatic 61-year-old man with BCR-ABL-positive bone marrow following autologous transplantation for multiple myeloma. Am J Hematol. 2010;85:944–946. doi: 10.1002/ajh.21809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asara JM, Christofk HR, Freimark LM, Cantley LC. A label-free quantification method by MS/MS TIC compared to SILAC and spectral counting in a proteomics screen. Proteomics. 2008;8:994–999. doi: 10.1002/pmic.200700426. [DOI] [PubMed] [Google Scholar]
- 31.Gazdar AF, Oie HK, Kirsch IR, Hollis GF. Establishment and characterization of a human plasma cell myeloma culture having a rearranged cellular myc proto-oncogene. Blood. 1986;67:1542–1549. [PubMed] [Google Scholar]
- 32.Ernst TJ, Gazdar A, Ritz J, Shipp MA. Identification of a second transforming gene, rasn, in a human multiple myeloma line with a rearranged c-myc allele. Blood. 1988;72:1163–1167. [PubMed] [Google Scholar]
- 33.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 34.Guix M, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wickrema A, et al. Engagement of Gab1 and Gab2 in erythropoietin signaling. J Biol Chem. 1999;274:24469–24474. doi: 10.1074/jbc.274.35.24469. [DOI] [PubMed] [Google Scholar]
- 36.Gu H, Pratt JC, Burakoff SJ, Neel BG. Cloning of p97/Gab2, the major SHP2-binding protein in hematopoietic cells, reveals a novel pathway for cytokine-induced gene activation. Mol Cell. 1998;2:729–740. doi: 10.1016/s1097-2765(00)80288-9. [DOI] [PubMed] [Google Scholar]
- 37.Yonezawa K, et al. Insulin-dependent formation of a complex containing an 85-kDa subunit of phosphatidylinositol 3-kinase and tyrosine-phosphorylated insulin receptor substrate 1. J Biol Chem. 1992;267:25958–25965. [PubMed] [Google Scholar]
- 38.Aronheim A, et al. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell. 1994;78:949–961. doi: 10.1016/0092-8674(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 39.Yu M, et al. The scaffolding adapter Gab2, via Shp-2, regulates kit-evoked mast cell proliferation by activating the Rac/JNK pathway. J Biol Chem. 2006;281:28615–28626. doi: 10.1074/jbc.M603742200. [DOI] [PubMed] [Google Scholar]
- 40.Holgado-Madruga M, Emlet DR, Moscatello DK, Godwin AK, Wong AJ. A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature. 1996;379:560–564. doi: 10.1038/379560a0. [DOI] [PubMed] [Google Scholar]
- 41.Breitkopf SB, Oppermann FS, Keri G, Grammel M, Daub H. Proteomics analysis of cellular imatinib targets and their candidate downstream effectors. J Proteome Res. 2010;9:6033–6043. doi: 10.1021/pr1008527. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Heibeck TH, et al. An extensive survey of tyrosine phosphorylation revealing new sites in human mammary epithelial cells. J Proteome Res. 2009;8:3852–3861. doi: 10.1021/pr900044c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishida K, Hirano T. The role of Gab family scaffolding adapter proteins in the signal transduction of cytokine and growth factor receptors. Cancer Sci. 2003;94:1029–1033. doi: 10.1111/j.1349-7006.2003.tb01396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang SQ, et al. Receptor-specific regulation of phosphatidylinositol 3′-kinase activation by the protein tyrosine phosphatase Shp2. Mol Cell Biol. 2002;22:4062–4072. doi: 10.1128/MCB.22.12.4062-4072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maroun CR, Naujokas MA, Holgado-Madruga M, Wong AJ, Park M. The tyrosine phosphatase SHP-2 is required for sustained activation of extracellular signal-regulated kinase and epithelial morphogenesis downstream from the met receptor tyrosine kinase. Mol Cell Biol. 2000;20:8513–8525. doi: 10.1128/mcb.20.22.8513-8525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lucas CM, et al. Chronic myeloid leukemia patients with the e13a2 BCR-ABL fusion transcript have inferior responses to imatinib compared to patients with the e14a2 transcript. Haematologica. 2009;94:1362–1367. doi: 10.3324/haematol.2009.009134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sidorova JYu , et al. A rapid RT-PCR based method for the detection of BCR-ABL translocation. Mol Pathol. 1997;50:266–268. doi: 10.1136/mp.50.5.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emig M, et al. Accurate and rapid analysis of residual disease in patients with CML using specific fluorescent hybridization probes for real time quantitative RT-PCR. Leukemia. 1999;13:1825–1832. doi: 10.1038/sj.leu.2401566. [DOI] [PubMed] [Google Scholar]
- 50.Foroni L, et al. Guidelines for the measurement of BCR-ABL1 transcripts in chronic myeloid leukaemia. Br J Haematol. 2011;153:179–190. doi: 10.1111/j.1365-2141.2011.08603.x. [DOI] [PubMed] [Google Scholar]
- 51.Shaffer AL, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 53.Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 54.Dong H, et al. Dysregulation of unfolded protein response partially underlies proapoptotic activity of bortezomib in multiple myeloma cells. Leuk Lymphoma. 2009;50:974–984. doi: 10.1080/10428190902895780. [DOI] [PubMed] [Google Scholar]
- 55.Laubach JP, Richardson PG, Anderson KC. The evolution and impact of therapy in multiple myeloma. Med Oncol. 2010;27(Suppl 1):S1–S6. doi: 10.1007/s12032-010-9442-2. [DOI] [PubMed] [Google Scholar]
- 56.Hayden PJ, Mitsiades CS, Anderson KC, Richardson PG. Novel therapies in myeloma. Curr Opin Hematol. 2007;14:609–615. doi: 10.1097/MOH.0b013e3282f0e948. [DOI] [PubMed] [Google Scholar]
- 57.BCC Research 2012. Kinase Inhibitors: Global Markets. Available at http://www.bccresearch.com/report/kinase-inhibitors-global-markets-bio053c.html. Accessed August 31, 2012.
- 58.Sawyer JR. The prognostic significance of cytogenetics and molecular profiling in multiple myeloma. Cancer Genet. 2011;204:3–12. doi: 10.1016/j.cancergencyto.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Klenn PJ, Hyun BH, Lee YH, Zheng WY. Multiple myeloma and chronic myelogenous leukemia—a case report with literature review. Yonsei Med J. 1993;34:293–300. doi: 10.3349/ymj.1993.34.3.293. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka M, et al. Coexistence of chronic myelogenous leukemia and multiple myeloma. Case report and review of the literature. Acta Haematol. 1998;99:221–223. doi: 10.1159/000040843. [DOI] [PubMed] [Google Scholar]
- 61.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.