Abstract
Glandular secreting trichomes on the surface of tomato plants and many of its relatives in the Solanaceae produce a mixture of O-acyl sugars that contribute to insect resistance. The majority of acyl sucroses produced by the cultivated tomato (Solanum lycopersicum) contain three or four short chain aliphatic acyl esters, and tetra-acyl sucroses have an acetyl group as one of the acyl chains. We previously reported overlapping S. lycopersicum × Solanum pennellii introgression lines (ILs) that fail to accumulate high levels of acetylated tetra-acyl sucroses. A survey of the annotated genes in this region of cultivated tomato chromosome 1 revealed three candidate acyltransferases that were tested for function using virus-induced gene silencing. A member of the BAHD family of acyltransferases (Solyc01g105580, SlAT2) was shown to encode an acetyl-CoA–dependent acyltransferase enzyme capable of acyl sucrose acetylation in vitro. RNAi suppression of SlAT2 in transgenic S. lycopersicum cv. M82 resulted in reduced acyl sugar acetylation, whereas expression of the functional S. lycopersicum allele of SlAT2 in the triacyl sucrose producing IL1-3 restored the ability of the IL to synthesize acetylated tetra-acyl sugars. Transgenic plants with the SlAT2 promoter driving GFP expression showed fluorescence in tips cells of long, slender trichomes that is consistent with acyl sugar acetylation occurring in these cells.
Glandular trichomes are epidermal hairs on the surface of many plants that synthesize and store or secrete a diverse set of specialized metabolites (1). Trichomes of some plants, especially within the Solanaceae, secrete compounds called acyl sugars that can account for up to 20% of the leaf dry weight (2). The role of acyl sugars in defense against insect herbivores (3–5) inspired several breeding programs aimed at increasing the levels of acyl sugars in the agronomically important crops tomato and potato (6, 7). In addition to functioning in insect defense, acyl sugars also have commercial uses as food additives and in cosmetic products (8).
Acyl sugars typically consist of aliphatic acyl groups of varying chain length esterified to hydroxyl groups of glucose or sucrose. Several previous studies showed that the acyl chains are primarily short- to medium-chain length branched and straight-chain aliphatic acids (9–11). For tomato acyl sucroses, the primary short acyl chains are acetate (C2), 2-methyl-propanoic acid (iso-C4; iC4), 3-methyl-butanoic acid (iso-C5; iC5), and 2-methyl-butanoic acid (anteiso-C5; aiC5); the four- and five-carbon groups are derivatives of branched-chain amino acids (12). Longer acyl groups are produced through a process of chain elongation that has been studied in a variety of species. Evidence for two-carbon elongation was presented for Solanum pennellii LA0716 and Datura metel (13, 14). In contrast, one-carbon elongation was demonstrated for several tobacco species and Petunia (13). This pathway is also used for production of other specialized metabolites, including the well-characterized methionine-derived glucosinolate pathway (15).
Despite the wide distribution and importance of acyl sugars, our understanding of how acyl chains are attached to sugars is rudimentary. The synthesis of acyl sugars having branched-chain acyl groups involves the branched-chain keto acid dehydrogenase complex that generates activated acyl-CoA esters (16). How these acyl-CoAs are used for synthesis of acyl sugars is unclear. Work using the wild tomato species S. pennellii suggests that a glucosyl transferase activity could form 1-O-acyl glucose using free acids and UDP-glucose (17). Activated 1-O-acyl glucose molecules were shown to be substrates for a serinecarboxypeptidase-like (SCPL) acyltransferase that catalyzes a disproportionation reaction to give free glucose and 1,2-di-O-acyl glucose (18). However, the relationship between this proposed pathway and acyl sucrose synthesis is unclear.
Aside from the SCPL gene from S. pennellii, no other genes are implicated in playing a role in assembly of the acyl glucoses, and no enzymes are known that are directly involved in assembly of acyl sucroses. We report identification of a tomato enzyme, ACYLTRANSFERASE2 (AT2), that is required for adding the acetyl (C2) group found in the major detectable tetra-acyl sucroses of the cultivated tomato Solanum lycopersicum cv. M82. This gene encodes an enzyme belonging to the BAHD class of acyltransferases (named based on the first four characterized enzymes of the family: BEAT, AHCT, HCBT, and DAT) (19). Enzyme assays, transient in vivo silencing, and testing in transgenic plants all provide evidence for the role of SlAT2 enzyme in acyl sucrose biosynthesis. As expected for an enzyme of acyl sucrose biosynthesis, the SlAT2 promoter drives reporter gene expression in the single tip cell of the long glandular trichomes that are proposed to produce and secrete acyl sugars. The AT2 gene is nonfunctional in the acyl glucose producing S. pennellii LA0716, consistent with the lack of acetylated acyl sugars in glands of this accession.
Results
Identification of Acyltransferase Candidates Within the IL1-3/IL1-4 Overlap Region.
We are using a combination of genetics, genomics, and biochemistry to identify enzymes that produce the specialized metabolites found in tomato glandular trichomes (20). Cultivated tomato M82 trichomes produce a variety of acyl sugars, most of which are tetra-acyl sucroses containing a single acetyl (C2) group (21). Screening for altered trichome chemistry in nearly isogenic introgression lines (ILs) derived by crossing the cultivated tomato S. lycopersicum cv. M82 and the wild tomato S. pennellii LA0716 (22) revealed changes in leaf trichome chemistry, including acyl sugars (21). Of particular interest, introduction of two overlapping regions of chromosome 1 from the wild relative causes loss of acetylation typically found on tetra-acyl sugars of M82 (Fig. 1A; i.e., S4:17, which is an acyl sucrose with four acyl groups of carbon chain lengths 2, 5, 5, and 5 for a total of 17 carbons). As a result, IL1-3 and IL1-4 accumulate predominantly triacyl sucroses (Fig. 1A; i.e., S3:15, which is an acyl sucrose with three acyl groups of carbon chain lengths 5, 5, and 5 for a total of 15 carbons). Because S. pennellii LA0716 does not produce acetylated acyl sugars (Fig. 1A), we hypothesized that introgressions 1-3 and 1-4 cover a region of chromosome 1 harboring an acetyltransferase that is functional in S. lycopersicum but either absent or nonfunctional in S. pennellii LA0716 (note the majority of acyl sugars are acyl glucoses, for example G3:19).
Fig. 1.
A region of S. lycopersicum chromosome 1 controls acyl sucrose acetylation. (A) LC-time of flight MS total ion chromatograms from trichome and leaf surface extracts of M82, IL1-3, and LA0716 plants showing the major acyl sugar peaks. Peak annotation is defined as S for acyl sucrose or G for acyl glucose, followed by the number of acyl chains and the total number of carbons in the acyl chains (for example, S4:17 = acyl sucrose with four acyl chains of length 2, 5, 5, and 5 for a total of 17 carbons). The structure shown for M82 is S4:17 (2, 5, 5, 5), with an acetyl group in the 2 position (21), and the structure shown for IL1-3 is S3:15 (5, 5, 5), which lacks an acetyl group. (B) Diagram of introgressions covering chromosome 1, with markers defining the ends of IL1-3 and IL1-4 listed, and region of overlap shaded gray. Expanded region shows the organization of the three predicted BAHD acyltransferase-encoding genes, labeled SlAT1, SlAT2, and SlAT3.
Genetic markers that define ends of the IL1-3 and IL1-4 introgressions on chromosome 1 (Fig. 1B) were used to identify the genomic sequence of the overlap region present in the assembly of the tomato genome available at the Sol genomics network (SGN, assembly ITAG2.3; http://solgenomics.net) (23). This region was predicted to contain 352 genes, including three annotated to produce functional BAHD class acyltransferases (Solyc01g105550, SlAT1; Solyc01g105580, SlAT2; and Solyc01g105590, SlAT3) (Fig. 1B). In contrast to the cultivated tomato, DNA sequence alignments with the S. pennellii LA0716 alleles of each gene indicate that both SpAT1 (JQ899260) and SpAT2 (JQ899261) are mutated and predicted not to encode functional proteins (Fig. S1). SpAT1 has a 10-bp deletion compared with SlAT1, located starting at 665 bp of the predicted ORF, which would cause a frame shift leading to an early stop codon. SpAT2 has a more complex change that causes 172 bp at the beginning of the SlAT2 ORF to no longer align with SpAT2 (yellow highlighted sequence in Fig. S1). In contrast, the SpAT3 (JQ899262) sequence seems to encode a functional ORF.
A search of the EST data at SGN gave no evidence for expression of the three AT genes in any publically available S. lycopersicum cDNA library. Similarly, S. lycopersicum trichome RNAseq data (24) showed no contigs with sequence matching any of the candidates. The only publically available evidence of expression for any of the ATs came from a total leaf trichome RNAseq dataset from the wild relative of tomato Solanum habrochaites LA1777; a small number of reads have >95% identity to SlAT2 (25). To analyze directly whether any of the three AT genes are expressed in S. lycopersicum M82 trichomes, gene-specific PCR primers were used to test for expression using cDNA prepared from intact stems and petioles, isolated stem and petiole trichomes, and stem and petioles from which trichomes have been removed. SlAT2 transcript was detected in the intact stems and petioles (Fig. S2); however, no RT-PCR product was seen when using SlAT1 and SlAT3 primers that could produce a product from a genomic DNA-positive control. Furthermore, the expression of SlAT2 was specific to the isolated trichomes and was not detected in the underlying stem and petiole tissue after the trichomes were removed (Fig. S2).
SlAT2 Is Responsible for Tetra-Acyl Sugar Acetylation in Vivo.
Differences in AT2 sequence between S. lycopersicum and S. pennellii together with mRNA expression in trichomes suggested that this gene product functions in acyl sugar acetylation. To pursue this hypothesis, virus-induced gene silencing (VIGS) constructs were prepared to test for an in vivo role for SlAT1–SlAT3 acyltransferases in acyl sugar production. The sequences were designed to suppress expression of each individual gene (sequences highlighted in blue, Fig. S1) or all three genes simultaneously (sequence highlighted in green, Fig. S1). Analysis of acyl sugars in plants 14 d after infiltration showed a reduction in acyl sugar acetylation only upon suppression of SlAT2 alone or when all three SlATs were targeted together (SlAT*, Fig. S3A). In contrast, no statistically significant change in acyl sugars was found when only SlAT1 or SlAT3 were targeted for suppression. Although the change in acyl sugar acetylation, as shown by the ratio of S3:15/S4:17, was significantly altered by VIGS, the ratio is still ∼1,000 times lower than that seen for IL1-3 or 1-4 plants. The relatively small reduction in acyl sugar acetylation is presumably due to the nature of VIGS in tomato, where silencing is variable and incomplete (Fig. S3B) (26), and the fact that VIGS of SlAT genes did not cause morphological changes that allowed identification of silenced regions.
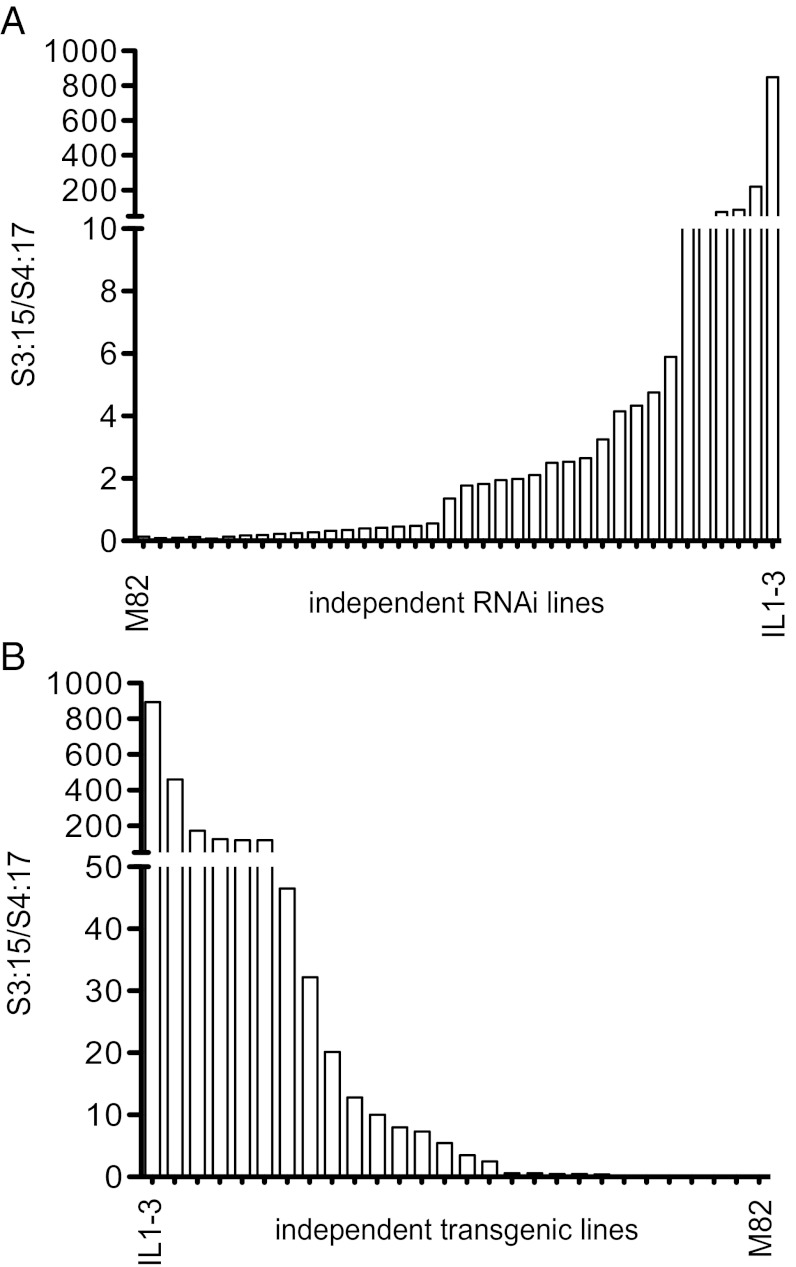
The unique ability of VIGS-mediated SlAT2 suppression to shift the acyl sugar composition toward increased triacyl sugars lacking the C2 chain is consistent with an in vivo role for this BAHD acyltransferase in acetylation. To test the impact of more-complete suppression of SlAT2 on production of tetra-acyl sucroses containing the C2 chain, stably transformed AT2 RNAi lines were generated in the tetra-acyl sucrose accumulating S. lycopersicum M82 background. The same 443-bp fragment corresponding to the 5′-UTR and the first 168 bp of the ORF used for VIGS was used to create an RNAi construct in the pHELLSGATE8 vector (27). Thirty-six independent transgenic tomato lines were generated and their acyl sugar composition analyzed (Fig. 2A). Wild-type M82 plants have an average S3:15/S4:17 ratio of 0.14 ± 0.04. Nineteen of the primary RNAi transformants had at least a 10-fold higher S3:15/S4:17 ratio than the WT parent, with some lines having very low levels of acetylated acyl sugars similar to that of IL1-3.
Fig. 2.
Transgenic analysis of AT2. Acyl sugar analysis showing level of acetylation represented by S3:15/S4:17 after RNAi suppression of SlAT2 in S. lycopersicum cv. M82 (A), or expression of the functional SlAT2 in IL1-3 (B). Data for 36 independent RNAi lines (A) and 26 independent transgenic IL1-3 lines (B) are from single measurements of each primary transformant. Data for M82 and IL1-3 are the average of three biological replicates.
Because of the introgression on chromosome 1, IL1-3 and IL1-4 plants harbor an apparently nonfunctional S. pennellii allele of AT2. We tested the hypothesis that this defect is responsible for reduced accumulation of C2-containing tetra-acyl sucroses in these ILs by generating transgenic IL1-3 lines expressing SlAT2 under the control of its own promoter. Of the 26 independent transgenic lines, 16 transformants had a S3:15/S4:17 ratio 100 times lower than the IL1-3 control plants (Fig. 2B). Eleven of these had a ratio >1,000 times lower than IL1-3, with acyl sugar acetylation levels nearly the same as M82. Together, the RNAi and IL1-3 complementation results indicate that SlAT2 is responsible for the majority of trichome acyl sugar acetylation in S. lycopersicum.
AT2 Acetylates Triacyl Sucroses in Vitro.
Predicted protein sequence comparisons for the cultivated tomato SlAT genes compared with characterized BAHD proteins from other plant species showed that amino acid identities range from ∼16% to 34%, with the highest identity to a Clarkia breweri benzylalcohol acetyltransferase (Fig. S4). Because of the low similarity to other BAHD enzymes and the wide variety of substrates used by members of this family, protein sequence cannot be used to predict what reaction these candidates catalyze (28). However, VIGS and transgenic plant results led us to hypothesize that SlAT2 enzyme has triacyl sucrose acetyl transferase activity. Expression in Pichia pastoris yeast cells yielded soluble protein suitable for enzyme assays. Recombinant SlAT2 was engineered to include a C-terminal combined 6X His-cMyc tag to detect expression using anti-cMyc antibodies and to purify protein by Ni-affinity chromatography (Fig. S5). This SlAT2 enzyme preparation was used to test activity with acetyl-CoA as the acyl donor and three different acceptor substrates: glucose, sucrose, and a mixture of the two major and several minor unacetylated triacyl sucroses from IL1-3 trichomes. Reaction products were analyzed by liquid chromatography-mass spectrometry (LC-MS). No activity was seen when using glucose or sucrose as an acceptor. In contrast, the acyl sugars isolated from IL1-3, primarily S3:15 and S3:22 triacyl sucroses (Fig. 3A), were acetylated in vitro to produce the tetra-acyl sucroses S4:17 and S4:24, respectively (Fig. 3C; Fig. S6 shows the mass spectra of peaks labeled in Fig. 3). In addition to these two major acyl sugars, other triacyl sucroses of lower abundance were also acetylated (Table S1). SlAT2 and the triacyl sucrose substrate mixture were also tested for activity using the commercially available substrates isobutyryl-CoA (iC4) and isovaleryl-CoA (iC5), because these acyl chains are commonly found on S. lycopersicum acyl sugars. No activity was detected with either of these longer chain length acyl-CoAs. During Ni-affinity purification of SlAT2 expressed in Pichia, a yeast mitochondrial alcohol dehydrogenase copurified with SlAT2 (Fig. S5). However, no acyl sugar acetylation was observed when using protein from untransformed Pichia as the enzyme source (Fig. 3B). These results demonstrate that SlAT2 is capable of in vitro acetylation of a variety of triacyl sucroses.
Fig. 3.
Results of in vitro enzyme activity assay using recombinant SlAT2 fusion protein. The mixture of triacyl sucroses lacking an acetyl group collected from IL1-3 leaves (SI Materials and Methods, Acyl Sugar Substrate Preparation) was used as substrate for enzyme assays with recombinant SlAT2 and acetyl-CoA. (A) Acyl sugar substrate, with the two major nonacetylated compounds indicated. (B) Products from protein purified from untransformed Pichia incubated with IL1-3 acyl sugars and acetyl-CoA. (C) Products from incubation of SlAT2 with IL1-3 acyl sugars and acetyl-CoA. Peak labels indicate that these are acyl sucroses (S) and total number of carbons in the acyl chains, and the lengths of individual acyl chains are in parentheses.
SlAT2 Is Expressed in the Tip Cells of Type I/IV Trichomes.
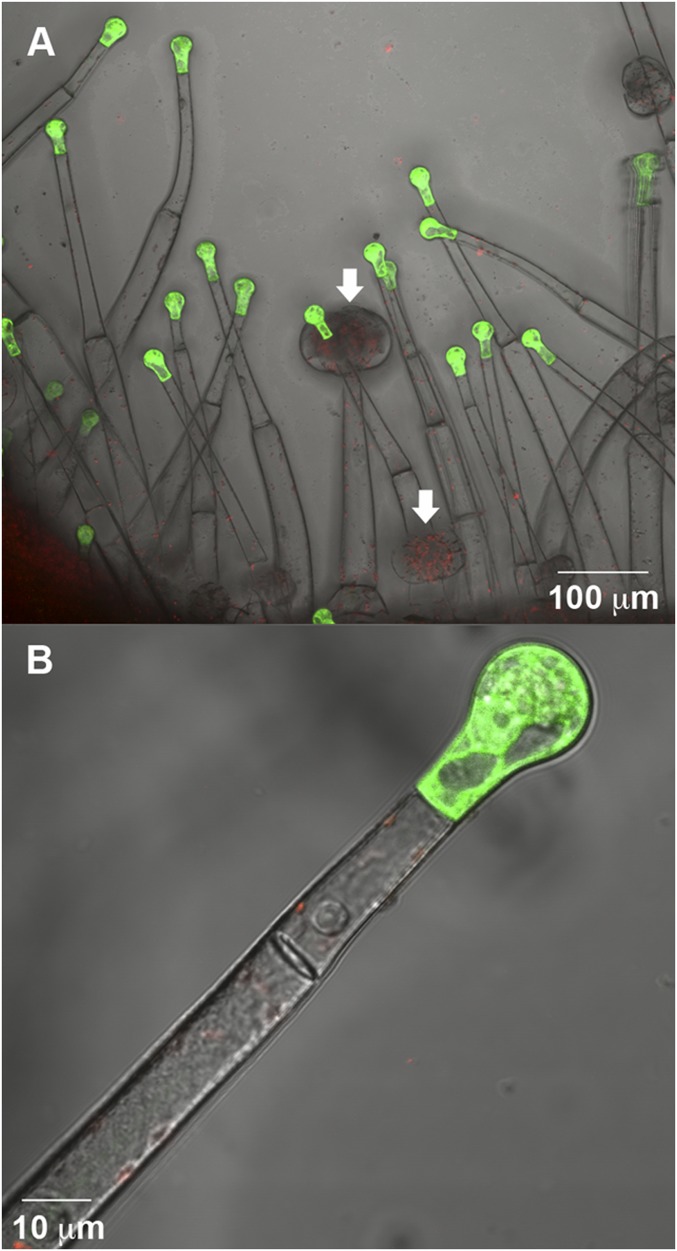
To identify the site of acyl sugar acetylation among the different trichome types of tomato, the SlAT2 promoter was used to drive expression of a GFP-beta-glucuronidase (GUS) fusion protein in M82 plants. Nine independent transgenic lines analyzed showed GFP fluorescence in the tip cell of the long and slender type I/IV trichomes (Fig. 4 A and B). No fluorescence was seen in other parts of the type I/IV trichome or in other trichome types including type VI trichomes (Fig. 4A). GFP fluorescence was seen in all type I/IV trichomes, regardless of what type of tissue they were located on (i.e., hypocotyl, cotyledon, stem, leaf, etc.) and whether from young growth chamber-grown seedlings or older greenhouse-grown plants.
Fig. 4.
SlAT2::GFP is expressed in tip cells of long, slender trichomes of tomato. Images obtained using a confocal laser-scanning microscope using a 20× (A) or a 40× (B) objective and are overlays of GFP fluorescence (green), chlorophyll autofluorescence (red), and differential interference contrast images. (A) Confocal image showing GFP fluorescence in tip cells of long, slender trichomes only and no expression in type VI trichomes (white arrows). (B) Single optical section of a long trichome at higher magnification.
Discussion
In this study, we show that glandular trichomes of tomato express a BAHD-type acyltransferase (SlAT2) that acetylates triacyl sucroses. SlAT2 was identified through phenotypic screening of ILs (21) derived from an interspecific cross between S. lycopersicum M82 and S. pennellii LA0716 (22). These two parental species have distinct differences in their acyl sugar profiles, including accumulation of acetylated acyl sucroses in the cultivated species, and nonacetylated acyl glucoses in S. pennellii. Analysis of the tomato genome sequence covered by the specific chromosome substitution regions led to the discovery of a cluster of three BAHD acyltransferases in cultivated tomato. The allelic differences between the cultivated tomato M82 and wild species accession LA0716, combined with RT-PCR analysis of transcripts in M82, led us to test the hypothesis that SlAT2 is responsible for acyl sucrose acetylation. Functional analysis of SlAT2 by a combination of VIGS, RNAi, and transgenic complementation of IL1-3 confirmed the in vivo role of this gene in acyl sucrose acetylation.
We demonstrated that the SlAT2 gene is expressed in the single tip cell of the long glandular trichomes using the endogenous promoter to drive expression of GFP in transgenic tomato plants (Fig. 4). Previous studies using S. pennellii implicated type IV trichomes (tall trichomes with a single cell at the tip) in acyl sugar production (2). The expression of SlAT2 in the tip cells of type I/IV trichomes is consistent with acetylated acyl sugars being produced in these glands.
Although next-generation sequencing of cDNA was done using isolated trichomes from tomato (24), no ESTs were found for SlAT2. This is in contrast to ESTs of terpene biosynthetic enzymes, which represent ∼0.3% of the sequences from both total trichome and type VI gland cell preparations (20). We consider two nonmutually excluding hypotheses for the paucity of SlAT2 mRNA reads. First, the mRNAs for these enzymes may not accumulate to high levels in the tip cells. Second, the cDNA library consisted of total trichomes shaved from stems and petioles, which included the multicellular stalks along with the gland cells from these and other trichomes. The cell of the single tip cells represents a small percentage of the total cells isolated (Fig. 4), which could cause transcripts from these small apical gland cells to be underrepresented. In S. habrochaites, where acyl sugar production is higher than in S. lycopersicum M82, ESTs of the putative ShAT2 ortholog are found in a cDNA library from mixed leaf trichomes (25). If the remaining unknown steps of acyl sugar biosynthesis also take place in the gland cells, discovery of the other biosynthetic enzymes would be facilitated by RNAseq data from isolated tip cells.
Only one other qualitative difference in acyl sucrose composition was identified in our screening of the M82 × LA0716 ILs (19), limiting the utility of this population in further pathway elucidation. However, advances in phenotyping and genotyping techniques make it possible to work with a wider variety of genetically fixed or segregating populations from crosses between individuals with differences in metabolites of interest. These approaches, combined with genomic sequence analysis, should enable discovery of new biosynthetic genes for acyl sugars and other underexplored specialized metabolites in much the same way that SlAT2 was identified.
The SlAT2 enzyme belongs to the BAHD family of acyltransferases. These catalyze acyl-CoA–dependent transacylation reactions that transfer the acyl moiety from a CoA ester to an acceptor molecule, forming an acyl ester or an acyl amide linkage (25). Characterized members of this family catalyze diverse acyl transfer reactions using a range of acyl-CoA donors to modify a variety of metabolites, including terpenes, phenylpropanoids, polyamines, alkaloids, and aliphatic alcohols (19, 28). Assays with SlAT2 showed that this enzyme uses acetyl-CoA as acyl donor and triacyl sucroses as acceptor molecules (Fig. 3). SlAT2 did not show activity with glucose or sucrose as the acceptor, or when other commercially available acyl-CoAs (isobutyryl-CoA and isovaleryl-CoA) were tested as acyl donors. These assay results are consistent with the phenotype of IL1-3 and IL1-4, which accumulate nonacetylated triacyl sucroses. Other plants within the Solanaceae produce acetylated acyl glucoses (29); it remains to be seen whether SlAT2 or its orthologs are responsible for acetylating acyl glucose substrates.
Although SlAT2 uses acetyl-CoA as a substrate for acetylation of acyl sucroses, previous studies using S. pennellii LA0716 suggested that acyl-CoAs might not be the precursors for the initial steps of acyl glucose biosynthesis (17). Rather than using acyl-CoAs as the activated acyl donor, a glucosyltransferase (GT) activity was identified that could generate 1-O-acyl glucose from a free acid (isobutyrate) and UDP-glucose in vitro (17, 30). The 1-O-acyl glucose product could then serve as the activated donor in an acyl-transfer reaction catalyzed by an SCPL-like acyltransferase. In this reaction, the acyl moiety of one molecule of 1-O-acyl glucose is transferred to another molecule of 1-O-acyl glucose, giving a di-acyl glucose product (18, 31).
Although the disproportionation activity was shown for acyl glucose production, it was not demonstrated that this mechanism is also involved in the synthesis of acyl sucroses. Early labeling studies in tobacco led to the proposed pathway whereby α-keto acid intermediates of branched-chain amino acid metabolism are converted to acyl-CoAs (11). More recently, evidence for the in vivo involvement of acyl-CoAs in acyl sucrose production was reported (16): suppression of the branched-chain keto-acid dehydrogenase complex, which generates C4 and C5 branched-chain acyl-CoAs, caused a reduction in acyl sugars in tobacco and tomato. In contrast to our demonstration of acyl sucrose acetylation, we are not aware of genetic evidence for the role of the GT and SCPL steps in acyl sugar biosynthesis.
A variety of other questions remain regarding acyl sucrose biosynthesis in secretory glandular trichomes of S. lycopersicum. Where is the acetyl-CoA substrate for SlAT2 synthesized? The absence of apparent subcellular targeting signals suggests that SlAT2 resides in the cytoplasm. Therefore, a cytosolic acetyl-CoA pool seems the most likely source of the acetyl donor. This is consistent with prior results indicating that photosynthesis is not required to generate acetyl-CoA for acyl sucrose acetylation in tobacco (32). Does SlAT2 require a triacyl sucrose substrate, or could a mono- or di-acyl sucrose molecule serve as an acceptor substrate? Answering this question will require access to mono- and di-acyl sucroses. If the GT–SCPL route is not involved in acyl sucrose biosynthesis, do other BAHD enzymes play a role? In addition to SlAT2, two other acyltransferase genes were found on chromosome 1 in the introgression region. These are predicted to encode proteins with 68–74% identity to SlAT2. Similar to SlAT2, there is no evidence of SlAT1 or SlAT3 expression in any cDNA libraries from tomato trichome or any other tissue. However, unlike SlAT2, expression of SlAT1 and SlAT3 was not detected by RT-PCR in the tissues examined. It remains to be seen whether SlAT1 or SlAT3 play any role in acyl sugar biosynthesis.
Recently, C5 acyl chains released from N. attenuata acyl sucroses after insect feeding and metabolism were shown to act as attractants of herbivore predators (3). The wide variety of sugar backbones, chain length, and structural isomer combinations suggests the possibility of other functions for the array of naturally occurring acyl sugars. In addition to elucidation of the pathway, identification of more acyl sugar biosynthetic genes will allow transgenic manipulation of specific acyl sugars to further test their roles in stress adaptation and potentially to engineer resistance.
Materials and Methods
Candidate Gene Identification.
A map of the S. lycopersicum × S. pennellii ILs (http://tgrc.ucdavis.edu/pennellii-ILs.pdf) was used to identify genetic markers defining ends of the introgressions on chromosome 1. The sequence of markers TG237 and TG17 was used in a BLAST search of the tomato genome, and the sequence between represents the region covered by the overlap of ILs 1-3 and 1-4. Sequence of the S. pennellii alleles was kindly provided by Alisdair Fernie and Bjoern Usadel (Max Planck Institute of Molecular Plant Physiology, Potsdam-Golm, Germany). DNA sequences were aligned using MultAlin (http://multalin.toulouse.inra.fr/multalin/multalin.html).
Virus-Induced Gene Silencing.
VIGS was performed using the tobacco rattle virus vector system (33). Sequences targeting specific Chr. 1 acyltransferases (Fig. S1) were amplified using the primers listed in SI Materials and Methods, followed by cloning into pTRV2-LIC and transformation into Agrobacterium tumefaciens GV3101. Agrobacterium growth, inoculation, and plant growth conditions for VIGS are described in ref. 34. Acyl sugars were analyzed 14 d after inoculation of plants, as previously described (21).
Tomato Transformation.
Transformation into S. lycopersicum cv. M82 was performed using A. tumefaciens strain AGL0 according to published methods (35). The same SlAT2-specific sequence used for VIGS was also used to create a construct for RNAi suppression of SlAT2 expression in M82 plants. Using primers AT2F1-attB and AT2R1-attB (sequence in SI Materials and Methods), a 443-bp fragment of SlAT2 was amplified by PCR. This product was then used as template for another PCR using the attB1 and attB2 adapter primers followed by recombination into pDONR221 using BP Clonase II (Invitrogen). The RNAi transformation construct was made by recombination with LR Clonase II of AT2-pDONR221 and pHELLSGATE8 (27). To express a functional AT2 under the control of its native promoter, the full-length SlAT2 ORF with 1,494 bp upstream of the translation start site was amplified from an S. lycopersicum BAC clone (LeHBa0204G10; http://solgenomics.net) using primers pAT2-F1 and AT2seqR1. The resulting product was cloned into pCR8/GW/TOPO (Invitrogen), then moved into the plant transformation vector pK7WG (36) and transformed into IL1-3. Acyl sugars were analyzed in primary transformants after transfer to soil, as described previously (21). For in planta reporter gene analysis of SlAT2 expression, a 1,486-bp fragment (JQ975012) was amplified using the primers pAT2-F1 and pAT2-R and cloned into pCR8/GW/TOPO followed by recombination into pKGWFS7 (36). The resulting construct expressed a GFP-GUS fusion protein under the control of the SlAT2 promoter in stably transformed M82 plants. Plant growth conditions were as previously described (21).
Fluorescence Microscopy.
GFP expression driven by the SlAT2 promoter was visualized using an Olympus FluoView 1000 laser scanning confocal microscope. Fluorescence from GFP was detected by excitation at 488 nm and a 505- to 525-nm emission filter. Chlorophyll autofluorescence was detected using an excitation wavelength of 543 nm and 650 nm long-pass emission filter.
Protein Expression and Enzyme Assay.
Initial attempts to express SlAT2 in Escherichia coli resulted in insoluble protein; however, expression in the methylotrophic yeast P. pastoris gave soluble protein suitable for enzyme activity assays. The full-length SlAT2 ORF was amplified using the forward primer 5′-GGTACCATGAATTGTTATATTGAAAT and the reverse primer 5′-GCGGCCGCGCTCCTAAGGAATGGAATGCTA that introduced KpnI and NotI restriction sites, respectively, for cloning into the same sites of the pPICZC vector (Invitrogen). The resulting plasmid creates an ORF with a C-terminal myc-6XHis tag. Transformation of P. pastoris strain X-33 by electroporation and induction of protein expression was performed according to the manufacturer’s instructions. Fifty-milliliter cultures of induced Pichia cells were washed once with ice-cold breaking buffer [50 mM sodium phosphate (pH 7.3), 150 mM NaCl, 5% (vol/vol) glycerol, and 5 mM β-mercaptoethanol] and lysed in 5 mL breaking buffer by vortexing with 0.5-mm glass beads (seven times, 30 s each with 30 s on ice between each vortexing). Recombinant myc-His–tagged SlAT2 was purified by Ni-affinity chromatography. Imidazole used to elute protein from the Ni-resin was removed by combining fractions containing SlAT2 and diluting 10-fold with breaking buffer, followed by concentrating protein using a centrifugal filter with 10,000 Da cutoff (Amicon). Protein measurement was performed using the bicinchoninic assay (BCA) assay (Pierce) with BSA as quantification standard.
SlAT2 enzyme activity assays were performed by incubating 1 μg of the partially purified recombinant protein in 30 μL of 50 mM sodium phosphate (pH 7.5) buffer containing 5 mM β-mercaptoethanol with 100 μM acetyl-CoA and a mixture of acyl sugar acceptor substrates collected from IL1-3 or IL1-4 plants. After incubating for 30 min at 30 °C, reactions were terminated by adding 60 μL of hot (60 °C) acetonitrile/isopropanol/formic acid (1:1:0.001). Ten microliters of the reaction mixture were analyzed by LC-MS as previously described (21). Acyl sugar acceptor substrate was collected from IL1-3 plants, as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the Solanum Trichome Project for their contributions during the project, especially Claire Taylor and Karin Hanisch for help with tomato transformation; Alisdair Fernie and Bjoern Usadel for providing S. pennellii genome sequence; and Shin-Han Shiu for help preparing the BAHD phylogeny. This work was supported by National Science Foundation Grants DBI-0604336 and IOS-1025636.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. JQ899257–JQ899262 (SlAT1, SlAT2, SlAT3, SpAT1, SpAT2, and SpAT3, respectively) and JQ975012 (SlAT2 promoter region)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207906109/-/DCSupplemental.
References
- 1.Schilmiller AL, Last RL, Pichersky E. Harnessing plant trichome biochemistry for the production of useful compounds. Plant J. 2008;54:702–711. doi: 10.1111/j.1365-313X.2008.03432.x. [DOI] [PubMed] [Google Scholar]
- 2.Fobes JF, Mudd JB, Marsden MP. Epicuticular lipid accumulation on the leaves of Lycopersicon pennellii (Corr.) D'Arcy and Lycopersicon esculentum Mill. Plant Physiol. 1985;77:567–570. doi: 10.1104/pp.77.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinhold A, Baldwin IT. Trichome-derived O-acyl sugars are a first meal for caterpillars that tags them for predation. Proc Natl Acad Sci USA. 2011;108:7855–7859. doi: 10.1073/pnas.1101306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez AE, Tingey WM, Mutschler MA. Acylsugars of Lycopersicon pennellii deter settling and feeding of the green peach aphid. J Econ Ent. 1993;86:34–39. [Google Scholar]
- 5.Chortyk OT, Kays SJ, Teng Q. Characterization of insecticidal sugar esters of petunia. J Agric Food Chem. 1997;45:270–275. [Google Scholar]
- 6.Lawson DM, Lunde CF, Mutschler MA. Marker-assisted transfer of acylsugar-mediated pest resistance from the wild tomato, Lycopersicon pennellii, to the cultivated tomato, Lycopersicon esculentum. Mol Breed. 1997;3:307–317. [Google Scholar]
- 7.Bonierbale MW, Plaisted RL, Pineda O, Tanksley SD. QTL analysis of trichome-mediated insect resistance in potato. Theor Appl Genet. 1994;87:973–987. doi: 10.1007/BF00225792. [DOI] [PubMed] [Google Scholar]
- 8.Hill K, Rhode O. Sugar-based surfactants for consumer products and technical applications. Fett Lipid. 1999;101:25–33. [Google Scholar]
- 9.Severson RF, et al. Isolation and characterization of the sucrose esters of the cuticular waxes of green tobacco leaf. J Agric Food Chem. 1985;33:870–875. [Google Scholar]
- 10.King RR, Calhoun LA, Singh RP, Boucher A. Sucrose esters associated with glandular trichomes of wild lycopersicon species. Phytochemistry. 1990;29:2115–2118. [Google Scholar]
- 11.Kandra G, Severson R, Wagner GJ. Modified branched-chain amino acid pathways give rise to acyl acids of sucrose esters exuded from tobacco leaf trichomes. Eur J Biochem. 1990;188:385–391. doi: 10.1111/j.1432-1033.1990.tb15415.x. [DOI] [PubMed] [Google Scholar]
- 12.Walters DS, Steffens JC. Branched-chain amino acid metabolism in the biosynthesis of Lycoperiscon pennellii glucose esters. Plant Physiol. 1990;93:1544–1551. doi: 10.1104/pp.93.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroumova AB, Wagner GJ. Different elongation pathways in the biosynthesis of acyl groups of trichome exudate sugar esters from various solanaceous plants. Planta. 2003;216:1013–1021. doi: 10.1007/s00425-002-0954-7. [DOI] [PubMed] [Google Scholar]
- 14.van der Hoeven RS, Steffens JC. Biosynthesis and elongation of short- and medium-chain-length fatty acids. Plant Physiol. 2000;122:275–282. doi: 10.1104/pp.122.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Textor S, et al. Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: Recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta. 2004;218:1026–1035. doi: 10.1007/s00425-003-1184-3. [DOI] [PubMed] [Google Scholar]
- 16.Slocombe SP, et al. Transcriptomic and reverse genetic analyses of branched-chain fatty acid and acyl sugar production in Solanum pennellii and Nicotiana benthamiana. Plant Physiol. 2008;148:1830–1846. doi: 10.1104/pp.108.129510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghangas GS, Steffens JC. UDPglucose: Fatty acid transglucosylation and transacylation in triacylglucose biosynthesis. Proc Natl Acad Sci USA. 1993;90:9911–9915. doi: 10.1073/pnas.90.21.9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li AX, Steffens JC. An acyltransferase catalyzing the formation of diacylglucose is a serine carboxypeptidase-like protein. Proc Natl Acad Sci USA. 2000;97:6902–6907. doi: 10.1073/pnas.110154197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.St-Pierre B, Luca VD. Evolution of acyltransferase genes: Origin and diversification fo the BAHD superfamily of acyltransferases involved in secondary metabolism. Recent Adv Phytochem. 2000;34:285–315. [Google Scholar]
- 20.Schilmiller AL, et al. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci USA. 2009;106:10865–10870. doi: 10.1073/pnas.0904113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schilmiller A, et al. Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J. 2010;62:391–403. doi: 10.1111/j.1365-313X.2010.04154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eshed Y, Zamir D. A genomic library of Lycopersicon pennellii in L. esculentum: A tool for fine mapping of genes. Euphytica. 1994;79:175–179. [Google Scholar]
- 23.Bombarely A, et al. The Sol Genomics Network (solgenomics.net): Growing tomatoes using Perl. Nucleic Acids Res. 2011;39(Database issue) Suppl 1:D1149–D1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schilmiller AL, et al. Studies of a biochemical factory: Tomato trichome deep expressed sequence tag sequencing and proteomics. Plant Physiol. 2010;153:1212–1223. doi: 10.1104/pp.110.157214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDowell ET, et al. Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiol. 2011;155:524–539. doi: 10.1104/pp.110.167114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orzaez D, et al. A visual reporter system for virus-induced gene silencing in tomato fruit based on anthocyanin accumulation. Plant Physiol. 2009;150:1122–1134. doi: 10.1104/pp.109.139006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helliwell C, Waterhouse P. Constructs and methods for high-throughput gene silencing in plants. Methods. 2003;30:289–295. doi: 10.1016/s1046-2023(03)00036-7. [DOI] [PubMed] [Google Scholar]
- 28.D’Auria JC. Acyltransferases in plants: A good time to be BAHD. Curr Opin Plant Biol. 2006;9:331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 29.King RR, Calhoun LA, Singh RP. 3,4-Di-O- and 2,3,4-tri-O-acylated glucose esters from the glandular trichomes of nontuberous Solanum species. Phytochemistry. 1988;27:3765–3768. [Google Scholar]
- 30.Ghangas GS, Steffens JC. 1-O-acyl-beta-D-glucoses as fatty acid donors in transacylation reactions. Arch Biochem Biophys. 1995;316:370–377. doi: 10.1006/abbi.1995.1049. [DOI] [PubMed] [Google Scholar]
- 31.Li AX, Eannetta N, Ghangas GS, Steffens JC. Glucose polyester biosynthesis. Purification and characterization of a glucose acyltransferase. Plant Physiol. 1999;121:453–460. doi: 10.1104/pp.121.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kandra L, Wagner GJ. Studies of the site and mode of biosynthesis of tobacco trichome exudate components. Arch Biochem Biophys. 1988;265:425–432. doi: 10.1016/0003-9861(88)90145-2. [DOI] [PubMed] [Google Scholar]
- 33.Dong Y, Burch-Smith TM, Liu Y, Mamillapalli P, Dinesh-Kumar SP. A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol. 2007;145:1161–1170. doi: 10.1104/pp.107.107391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velásquez AC, Chakravarthy S, Martin GB. Virus-induced gene silencing (VIGS) in Nicotiana benthamiana and tomato. J Vis Exp. 2009;(28):e1292. doi: 10.3791/1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCormick S. In: Transformation of Tomato with Agrobacterium tumefaciens. Plant Tissue Culture Manual. Lindsey K, editor. Dordrecht, The Netherlands: Kluwer Academic; 1991. pp. 1–9. [Google Scholar]
- 36.Karimi M, Depicker A, Hilson P. Recombinational cloning with plant gateway vectors. Plant Physiol. 2007;145:1144–1154. doi: 10.1104/pp.107.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.