Abstract
The rate of meiotic crossing over (CO) varies considerably along chromosomes, leading to marked distortions between physical and genetic distances. The causes underlying this variation are being unraveled, and DNA sequence and chromatin states have emerged as key factors. However, the extent to which the suppression of COs within the repeat-rich pericentromeric regions of plant and mammalian chromosomes results from their high level of DNA polymorphisms and from their heterochromatic state, notably their dense DNA methylation, remains unknown. Here, we test the combined effect of removing sequence polymorphisms and repeat-associated DNA methylation on the meiotic recombination landscape of an Arabidopsis mapping population. To do so, we use genome-wide DNA methylation data from a large panel of isogenic epigenetic recombinant inbred lines (epiRILs) to derive a recombination map based on 126 meiotically stable, differentially methylated regions covering 81.9% of the genome. We demonstrate that the suppression of COs within pericentromeric regions of chromosomes persists in this experimental setting. Moreover, suppression is reinforced within 3-Mb regions flanking pericentromeric boundaries, and this effect appears to be compensated by increased recombination activity in chromosome arms. A direct comparison with 17 classical Arabidopsis crosses shows that these recombination changes place the epiRILs at the boundary of the range of natural variation but are not severe enough to transgress that boundary significantly. This level of robustness is remarkable, considering that this population represents an extreme with key recombination barriers having been forced to a minimum.
Keywords: decrease in DNA methylation 1, epigenetic inheritance, DNA methylome, epi-haplotype
Meiotic recombination is a fundamental process in genetics whereby maternally and paternally inherited homologous chromosomes exchange material, either nonreciprocally by gene conversion or reciprocally by crossing over (CO). COs are not distributed uniformly along the genome but occur more often in chromosome arms and are strongly suppressed in pericentromeric regions (1–3), partly as a result of sequence and chromatin determinants (1, 4–8). It is commonly believed that in plants and mammals high levels of DNA sequence polymorphisms as well as heterochromatic features associated with repeats, notably dense DNA methylation and transcriptional silencing, play a central role in this suppression (1, 4).
Suppression of COs by dense DNA methylation has been demonstrated experimentally in the fungus Ascobolus (7). Specifically, COs were reduced when the recombination interval was methylated on one homolog and were abolished almost completely when methylated on both homologs. In Arabidopsis, two recent mapping studies analyzed F2 progeny derived from crosses between Columbia ddm1 and met1 [Col(ddm1),Col(met1)] DNA methylation mutants and wild-type Landsberg [Ler(WT)] accessions and showed that loss of DNA methylation could not alleviate the suppression of COs in pericentromeric regions of chromosomes (9, 10). However, as pointed out by the authors, this experimental design could not rule out an inhibitory effect of sequence divergence between Col and Ler on COs.
An ideal design would use crosses between isogenic individuals, with one of the crossing partners having decreased DNA methylation levels throughout the genome (9). Melamed-Bessudo and Levy (9) implemented such an approach by crossing Col(ddm1) mutant to Col(WT). Using two fluorescent markers spanning a 16-centimorgan (cM) interval on the arm of chromosome 3, they detected increased CO rates in F2 plants derived from these parents relative to plants derived from a Col(WT)×Col(WT) control cross and concluded that COs in euchromatic regions can be up-regulated by loss of DNA methylation. A similar approach at a genome-wide scale and with high mapping resolution, particularly in pericentromeric regions, has not been attempted because of a lack of appropriate molecular and genetic tools. Hence, the combined effect of DNA methylation and sequence variation on COs has not been tested comprehensively in Arabidopsis or in any other higher eukaryote.
We previously reported the construction of a large population of epigenetic recombinant inbred lines (epiRILs) in Arabidopsis (11, 12), which provides a powerful experimental system to conduct such a test. These epiRILs were obtained by first crossing a fourth-generation plant homozygous for the recessive ddm1-2 mutation with a near-isogenic WT individual. The ddm1-2 mutation mostly affects transposable elements (TEs) and other repeats, which lose DNA methylation and become transcriptionally reactivated in a transmissible manner in many instances (11–14). However, transposition events are relatively rare (15).
Thus, F1 individuals can be considered homozygous throughout the genome, except at the DDM1 locus and at the few loci affected by TE mobilization, but have chromosome pairs that differ markedly in their DNA methylation levels and transcriptional activity over TEs and other repeats (11, 16). A single F1 DDM1/ddm1 individual was backcrossed to the WT parental line, and after the progeny homozygous for the WT DDM1 allele were selected, the epiRILs were propagated through seven rounds of selfing. In this design, more than 85% of all informative recombination events occur in the first two inbreeding generations (F1 and backcross), with fewer informative events being contributed by each subsequent generation (17).
Previous targeted analysis indicated that many of the parental differences in DNA methylation and transcriptional activity of repeats are inherited stably in the epiRILs (11, 12). Regions with segregating DNA methylation states therefore can serve as physical markers to detect the frequency and distribution of recombination events along chromosomes even though the two homologs have nearly identical DNA sequences.
In this study we report the construction of a recombination map using genome-wide DNA methylation data from 123 epiRILs. This map was derived from 126 meiotically stable differentially methylated regions (DMRs) covering 81.9% of the total genome. Estimates of the genetic length for each chromosome revealed that global recombination rates are comparable with those of classical Arabidopsis crosses. On a local scale, we demonstrate that suppressed recombination activity within repeat-rich, pericentromeric regions of chromosomes is maintained robustly even after the removal of sequence polymorphisms and repeat-associated DNA methylation. Furthermore, we were able to identify 3-Mb regions flanking pericentromeric boundaries that appear to be subject to additional suppression and show that this effect is accompanied by increased recombination activity in chromosome arms. A direct comparison with 17 classical Arabidopsis crosses reveals that these recombination changes place the epiRILs at the boundary of the range of natural variation but appear not to be severe enough to transgress that boundary significantly.
Results
Construction of a Recombination Map Using Transgenerationally Stable DMRs.
To demonstrate that transgenerationally stable DMRs can be used for the construction of a recombination map in an isogenic population, we carried out methylated DNA immunoprecipitation followed by hybridization to a whole genome DNA tiling array (MeDIP-chip) on 123 epiRILs and on the two parental lines (256 array experiments including replicates). The 123 epiRILs originally were chosen using a selective (epi)genotyping strategy for two uncorrelated complex traits, flowering time and root length. We used a three-state Hidden Markov Model (HMM) to classify tiling array signals into three underlying DNA methylation states (18): unmethylated (U), intermediate methylation (I), or methylated (M). Benchmarking of these HMM calls against whole-genome bisulphite sequencing data (∼30×) for six epiRILs confirmed that both the MeDIP protocol and the analysis method performed well (SI Appendix, Fig. S1 and Table S1). Comparison of the two parental DNA methylomes revealed 2,611 DMRs representing clear instances of methylation loss in ddm1 (transitions from M to U). These DMRs (median length: 1,211 bp; range: 318–24,624 bp) were distributed throughout the genome but, as expected, were more abundant in pericentromeric regions (Fig. 1A and SI Appendix, Table S2) (19).
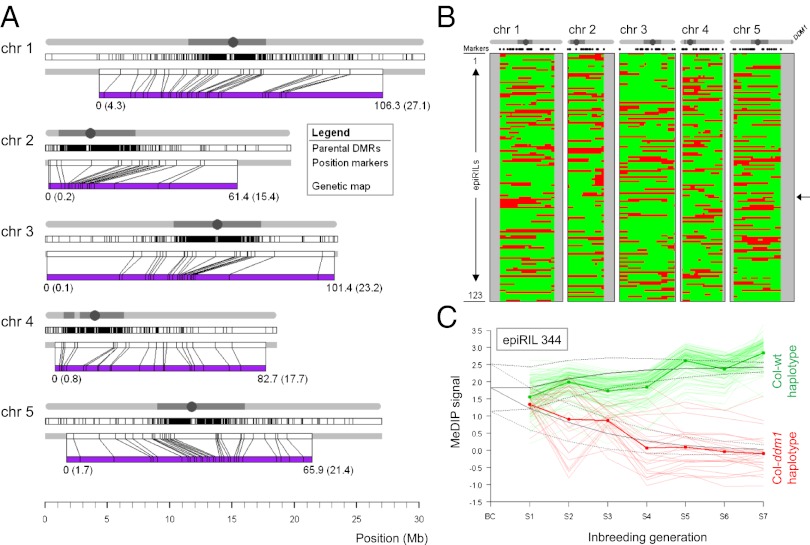
Fig. 1.
Recombination map construction. (A) Genome-wide distribution of the 2,611 parental DMRs (Top) and the 126 DMRs (i.e., markers; Middle) retained for construction of the recombination map (purple, Bottom) for each of the five Arabidopsis chromosomes. The mapping between physical and genetic positions of markers is shown. (B) Inference of inherited WT (green) and ddm1 (red) haplotypes along the genome (x-axis) as inferred from the recombination map for each of the 123 epiRILs (y-axis) (SI Appendix, Table S5). Chromosome extremities not covered by the genetic map are indicated in gray. The genome of epiRIL 344 is indicated by an arrow. A schematic representation of each chromosome is plotted above the map with the physical location of the DDM1 gene shown at the end of chromosome 5. (C) Transgenerational methylation data for epiRIL 344. Shown are the average methylation signals for the 126 markers, with regions that are predicted to become fixed for the ddm1 haplotypes (thin red lines) and the WT haplotypes (thin green lines) after seven selfing generations. The average signals (red and green thick solid lines) are in agreement with Mendelian inbreeding theory (black solid lines).
We examined the DNA methylation state at all parental DMRs in each of the 123 epiRILs and inferred their parent of origin (i.e., epigenotypes). Segregation was not compatible with stable inheritance of ddm1-induced DNA hypomethylation for 1,744 (66.8%) of the parental DMRs, and in most of these cases our data pointed to fully or partially penetrant reversion to WT DNA methylation. In contrast, 867 (33.2%) of the parental DMRs segregated in the expected 3:1 Mendelian ratio (SI Appendix, Fig. S2 and Table S3). Stable DMRs were associated with a comparatively lower abundance of siRNAs in the WT and ddm1 parental lines (SI Appendix, Fig. S3). These findings are in agreement with previous analyses (11, 12) and indicated that the 867 stable DMRs are not efficient targets of siRNA-mediated DNA remethylation, even after eight rounds of meiosis. These stable DMRs therefore could serve as physical markers in an extension of the Lander–Green algorithm (20) to derive a genetic map. After application of the algorithm and removal of mainly genetically redundant markers (i.e., markers located less than 0.0001 cM apart), 126 of the original 867 markers were retained (Fig. 1 A and B and SI Appendix, Fig. S2 and Table S4). These 126 markers covered ∼81.9% of the total genome (74.7, 77.0, 98.4, 91.1, and 73.0% of chromosomes 1, 2, 3, 4, and 5, respectively).
Many of the 126 markers contained TE sequences, consistent with the targeted effect of ddm1 on these and other repeats (SI Appendix, Fig. S4). However, in a vast majority of cases, markers included only TE relics, which likely have lost their capacity to be mobilized (SI Appendix, Table S6). Indeed, both comparative genomic hybridization (SI Appendix, Fig. S5) and preliminary whole-genome resequencing suggested that none of the 126 DMRs contain sequences that were mobilized in the parental ddm1 line or the epiRILs (SI Appendix, Table S6). Consistent with this finding, pair-wise recombination fractions between the 126 markers indicated a well-behaved and robust genetic map, reminiscent of those typically seen in classical crosses involving DNA sequence markers, with high correlation among linked loci and virtually no correlations among loci in different chromosomes (SI Appendix, Fig. S6). Moreover, all inferred ddm1-inherited nonrecombinant pericentromeric haplotypes contained significantly less DNA methylation and were more actively transcribed than their WT counterparts (SI Appendix, Figs. S7 and S8).
To test further the transgenerational stability of the 126 markers as well as our inference of the parental epigenotypes at these marker locations, we performed genome-wide DNA methylation analysis for one selected line (epiRIL 344) for each of its seven selfing generations (7 × 2 replicates = 14 array experiments). Fixation occurred for the predicted parental epigenotype in each case, and the rate of approach toward fixation was consistent with Mendelian inbreeding theory for a backcross-derived RIL (19) (Fig. 1C). Taken together, these results rule out any ambiguity in the actual location or DNA methylation state of the stable DMRs used for constructing the genetic map.
Total Genetic Length in the epiRILs Does Not Diverge Significantly from the Natural Range.
One approach for evaluating the epiRILs recombination map is by comparison with a Col(WT)×Col(WT)-derived reference cross. In this set-up, changes in recombination patterns can be attributed directly to DNA methylation loss. However, tracking recombination events in such a reference is experimentally challenging. It requires a system akin to the fluorescent marker reporters used by Melamed-Bessudo and Levy (9), which does not easily scale genome-wide. An alternative approach is to evaluate the epiRILs in the context of natural variation. In terms of DNA sequence and DNA methylome divergence of its founder parental lines, the epiRILs can be viewed as representing an extreme situation with key barriers to recombination having been forced to a minimum. An important question therefore is how genome-wide recombination patterns in this population compare with those seen in crosses derived from different pairs of natural accessions.
We estimated the genetic length for each of the five epiRIL chromosomes using Haldane’s map function. The lengths were 106.3, 61.4, 101.4, 82.7, and 65.9 cM for chromosomes 1–5, respectively, and correlated positively with physical chromosome length (SI Appendix, Fig. S9). The total length of the genetic map was 417.7 cM, yielding an average marker spacing of ∼0.804 Mb (3.45 cM). These estimates are similar to those previously reported for genetic maps based on classical Arabidopsis crosses (21–24). The use of other map functions that account for CO interference, such as the Kosambi or Carter and Falconer functions, yielded very similar results (SI Appendix, Fig. S10). To perform a more direct comparison between the epiRIL map and those of classical Arabidopsis crosses, we reanalyzed recombination data obtained for 17 F2 populations (24) that were derived from pairs of 18 distinct natural accessions. In total, these populations consisted of 7,045 plants (∼410 plants per cross; range: 235–462 plants), which were genotyped at 235 markers on average (range: 215–257 markers) (24). To facilitate a meaningful comparison, we constructed a consensus map using 83 markers that were shared across populations (SI Appendix, Fig. S11 and Table S7). Thorough testing showed that the reduction to 83 markers in the epiRIL and F2 maps led to no significant loss of information in capturing the linkage structure along chromosomes (SI Appendix, Figs. S12 and S13), and the 83 markers therefore were deemed appropriate for this comparative analysis.
Estimates of the genetic length of each of the five chromosomes revealed substantial natural variation among the F2 populations (Figs. 2B and 3A). However, the genetic lengths of the epiRIL chromosomes did not diverge significantly from the natural range (Figs. 2B and 3A). The exception was chromosome 1, where we observed a significant increase relative to five of the F2 crosses. Overall, therefore, our data indicate that the global recombination rate in the epiRILs is not altered drastically. Nonetheless, we noted a clear, but nonsignificant, trend toward longer genetic lengths for chromosomes 1–4 as compared with the F2 populations (Fig. 3A); this trend is at least partly consistent with DNA methylation and DNA sequence polymorphisms acting as barriers to the global recombination rate in Arabidopsis.
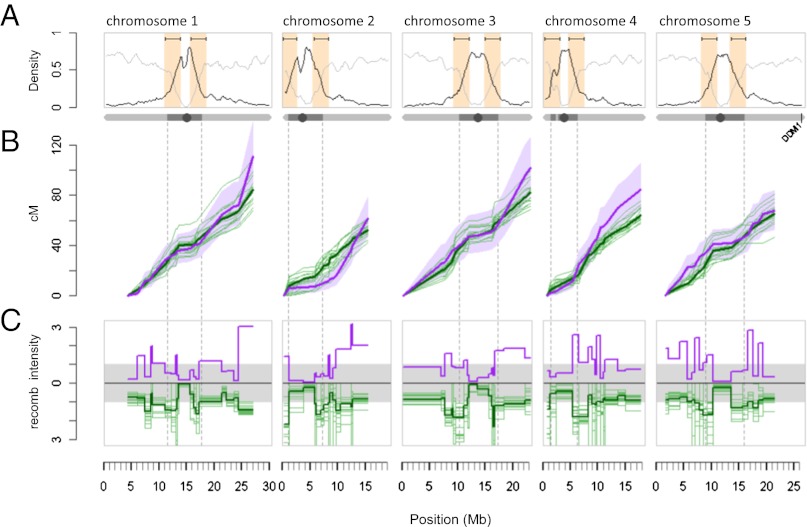
Fig. 2.
Comparison of global and local recombination patterns in the epiRILs and the 17 F2 populations (24). (A) Chromosome-wide gene (light gray line) and transposon (dark line) density distribution. The 3-Mb windows bracketing the intersection points between transposon- and gene-dense regions are indicated in orange. (B) Cumulative cM lengths of the epiRILs (thick purple line) and each of the F2 populations (thin green lines) using the consensus map. Purple shading shows the ±95% confidence interval (CI).The thick green line denotes the average F2 cumulative length (in cM). The dotted vertical lines define the pericentromeric regions of each chromosome. (C) The distribution of normalized recombination intensities (cM/Mb of a given marker interval divided by the cM/Mb chromosome average) shows suppression of recombination within pericentromeric regions and elevation at its boundaries. Color coding is as in B.
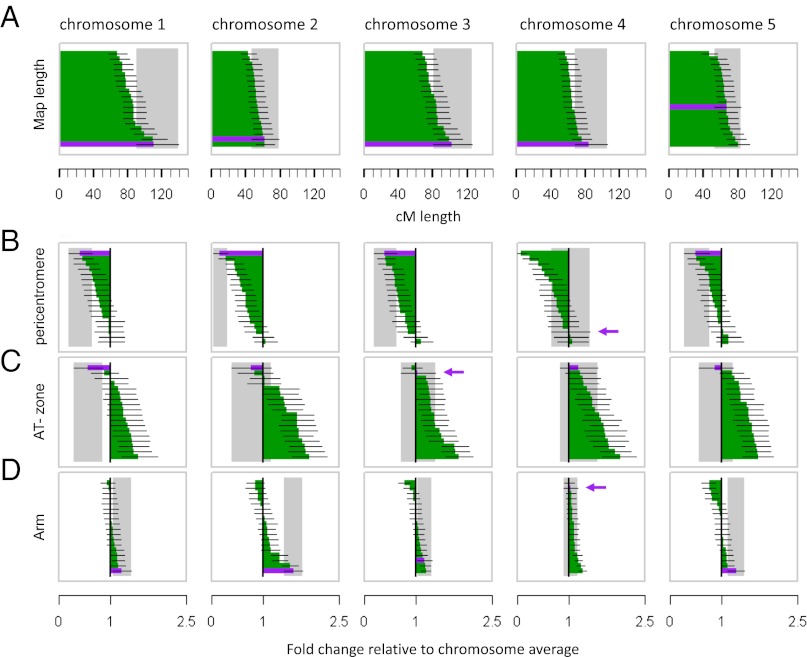
Fig. 3.
Estimated genetic lengths and fold-change recombination intensities. (A) Estimated genetic lengths (±95% CI) of the epiRILs (purple) and each of the 17 F2 populations (green) (24). (B–D) Fold change in recombination intensity [(cM/Mb) region/(cM/Mb) chromosome average] ±95% CI in pericentromeric regions (B), AT zones defined by a 3-Mb window bracketing the intersection point between transposon- and gene-dense regions at pericentromeric boundaries (C), and chromosome arms (D). Purple arrows indicate the location of the epiRILs when applicable. The values presented in each panel are ordered to highlight trends in the epiRILs recombination landscape. The identifiers of individual F2 crosses corresponding to this ordering can be found in SI Appendix, Table S11.
Suppression of Pericentromeric Recombination Persists in the epiRILs and Shows a Trend Toward Additional Reinforcement.
To explore the relationship between the epiRILs map and those of the different F2 crosses at a subchromosomal scale, we examined in more detail the distribution of recombination intensities, expressed as cM/Mb, for each marker interval along the genome (Fig. 2C). All populations, including the epiRILs, had clearly suppressed recombination activity across pericentromeric regions relative to the chromosome averages (Figs. 2C and 3B). The exception to this trend was chromosome 4, for which the epiRILs showed a slight increase of recombination intensity (Fig. 3B). However, the presence of the heterochromatic knob on chromosome 4 in the Columbia accession, but not in other accessions, makes this result difficult to interpret (10).
Specifically, recombination intensities in pericentromeric regions of epiRIL chromosomes 1, 2, 3, and 5 were, respectively, 2.50, 6.88, 2.53, and 2.01 times lower than the chromosome average, which compares to 1.27 (range: 0.97–2.15), 1.51 (range: 0.95–3.68), 1.52 (range: 0.90–2.48), and 1.20 (range: 0.87–1.98) in the F2 populations (Fig. 3B and SI Appendix, Table S8). This persistent suppression effect in the epiRIL agrees with the results of Melamed-Bessudo and Levy (9) and Mirouze et al. (10), who examined mapping populations derived from a Col(ddm1)×Ler(WT) and a Col(met1)×Ler(WT) cross, respectively. Hence, loss of DNA methylation appears to be insufficient to release pericentromeric suppression of recombination, even in the absence of DNA sequence polymorphisms. On the contrary, we found a clear trend toward enhanced suppression in the epiRILs: Recombination intensities in this population were consistently at the bottom of the natural range compared with the F2 populations, even though chromosome-wide recombination rates were comparatively large. Enhanced suppressive effects also were reported by Melamed-Bessudo and Levy (9) and Mirouze et al. (10), thus highlighting an unexpected and complex relationship between DNA methylation and the suppression of recombination in pericentromeric regions of Arabidopsis chromosomes.
Reinforced Suppression of Recombination Extends to Pericentromeric Boundaries in the epiRILs and Appears to Be Compensated by Increased Recombination in Chromosome Arms.
In contrast to core pericentromeric regions, recombination intensities in the F2 populations increase rapidly at pericentromeric boundaries with chromosome arms (Figs. 2C and 3C). An important property of these regions is that they correspond to major transitions in genome content from TE-rich to gene-rich sequences (Fig. 2A) and also have been described recently as hotspots of historical recombination activity at the species level (SI Appendix, Fig. S14) (25). We found that nearly 40% of all detected recombination breakpoints in the F2 populations mapped within a 3-Mb window bracketing the intersection point in these transition zones (henceforth referred to as “annotation transition zones”; AT zones), yielding local recombination intensities that were consistently above the chromosome averages (Fig. 3C and SI Appendix, Fig. S15 and Table S8).
This finding differs strongly from the situation seen in the epiRILs: AT zones accounted for only 25.31% of all detected recombinants in this population, and recombination intensities were close to the chromosome average (in chromosomes 3 and 4) or even below it (in chromosomes 1, 2, and 5) (Fig. 3C and SI Appendix, Table S8). These results suggest that the enhanced suppression of recombination seen in the epiRIL pericentromeric regions (see above) is driven at least in part by the more localized reduction of recombination within AT zones, which cover (on average) only 63.4% of the pericentromeric regions on either side of the centromeres. The two previous studies using mapping populations derived from crosses between Col(ddm1) and Ler(WT) (9) and between Col(met1)and Ler(WT) (10) were not able to delineate these local effects, most likely because of the sparsity of their genetic markers (two to three markers per pericentromeric region). Marker density in the epiRIL map, in contrast, was relatively high within AT zones and even permitted fine mapping of shared and nonshared recombination breakpoints to a resolution as low as 4 kb (SI Appendix, Figs. S16 and S17 and Tables S9 and S10).
Furthermore, our data indicate that suppression of recombination within AT zones in the epiRILs is accompanied by increased recombination in chromosome arms (Fig. 3D and SI Appendix, Fig. S18). This apparent compensatory effect reconciles the enhanced local suppression seen in the epiRILs with the earlier observation that chromosome-wide recombination rates are relatively large compared with the F2 populations. This effect was most pronounced on epiRIL chromosomes 1, 2, and 5 (the chromosomes with the strongest suppression in the AT zone), with recombination intensities being 1.23, 1.6, and 1.3 times above the chromosomes’ average (SI Appendix, Table S8). We failed to identify a similar trend in the F2 populations (SI Appendix, Fig. S18 and Table S8), suggesting that this effect is a specific feature of the epiRIL recombination landscape.
Discussion
In this study we demonstrate that stable DNA methylation differences can be used as physical markers to derive genome-wide recombination patterns in a near isogenic population of epiRILs. We find that recombination suppression is maintained robustly in pericentromeric regions of the epiRILs, despite the extensive loss of sequence variation and of DNA methylation and transcriptional silencing over repeats. This observation indicates that these factors do not play a major role in the suppression of pericentromeric COs. This finding is contrary to common belief and is particularly intriguing given the interplay between recombination and transcription observed in yeast and the mouse (26, 27). Whether mechanisms exist in Arabidopsis that actively sequester the recombination machinery away from gene-promoter regions or other genomic elements, as in the mouse (27), remains to be determined.
Nonetheless, our results indicate that loss of DNA methylation over repeat sequences can lead to a local reinforcement of recombination suppression in pericentromeric regions and to increased recombination activity along chromosome arms. Similar results were reported by Melamed-Bessudo and Levy (9) and Mirouze et al. (10) using genetically divergent populations. Therefore we conclude that the absence of sequence polymorphisms is insufficient to counteract the enhanced suppressive effects induced by the loss of DNA methylation in pericentromeric regions. On the other hand, the lack of sequence polymorphisms still may be partly responsible for the increased recombination rates observed in chromosome extremities (9).
Melamed-Bessudo and Levy (9) demonstrated that ddm1-induced demethylation of only one homolog produces the same recombination changes seen when both homologs are demethylated. Our results and conclusions therefore should be generalizable to the two-homolog situation. However, it has been shown in Ascobolus that DNA methylation of a known recombination hotspot inhibits COs more severely when both homologs are methylated (7). Similar localized dosage effects may therefore also be present in Arabidopsis.
Our study and those of Melamed-Bessudo and Levy (9) and Mirouze et al. (10) have used well-characterized ddm1 and met1 DNA methylation mutants as a tool to perturb genome-wide methylation levels experimentally. Both ddm1 and met1 experience a nearly 70% reduction in DNA methylation levels genome-wide. This drastic loss probably sets an upper limit to the amount of demethylation that can be incurred in nature. Indeed, it is difficult to conceive of mechanisms that would elicit similar or more severe changes under natural settings, unless they involve spontaneous mutations in genes important for DNA methylation control, such as ddm1 or met1. Interestingly, a recent analysis of Arabidopsis mutation accumulation lines showed that drastic alterations in the methylome of one outlier line were likely caused by a spontaneous mutation in a methyl-transferase gene (28, 29), which must have arisen during just 30 generations of selfing. This observation suggests that similar events are certainly plausible under natural conditions.
An assessment of whether strong methylation loss can elicit recombination changes at magnitudes that are sufficient to drive genome evolution in this species has been lacking. Our study is an initial step in providing such an assessment. Our analysis of the 17 F2 populations derived from 18 natural accessions (24) allowed us to quantify the magnitude of the recombination changes observed in the epiRILs in the context of natural variation. Although we find that the epiRILs nearly always are situated at the boundary of the natural range, there is no strong evidence that local and global recombination patterns in this population markedly transgress the natural range. Indeed, in many cases, several of the F2 populations displayed even more extreme divergence from the F2 population average than did the epiRILs. These findings lead us to conclude that severe losses of DNA methylation along Arabidopsis chromosomes have no drastic implications for recombination-mediated genome evolution. This high level of robustness raises questions concerning the precise mechanisms that have shaped the recombination landscape in this species in the first place.
Of course, severe depletion of DNA methylation can drive other important events, such as large-scale structural rearrangements and polyploidization, which may impact the course of genome evolution. In addition, natural epigenetic variation, such as that associated with differential DNA methylation, can act on complex traits that are under natural selection (30), thereby changing linkage disequilibrium relations within and across chromosomes. However, understanding and documenting the impact of epigenetic variants on complex traits is challenging, mainly because of the technical difficulties in ruling out the confounding effect of DNA sequence polymorphisms (31). Because of this limitation, it has been argued that the epiRILs constitute an ideal system for the study of epigenetic inheritance in Arabidopsis (17, 32–34). We and others have shown recently that many adaptive phenotypes, such as plant height, flowering time, and growth rate, are highly heritable in this population (12, 35, 36). Segregating phenotypic effects also have been observed in another epiRIL population which was obtained from a cross between Col(met1) and Col(WT) (37).
A logical next step in the analysis of these populations is to map and characterize the epigenetic basis of these complex traits. The linkage map reported here (Fig. 1B) can be used in conjunction with classical quantitative trait-locus mapping methods to achieve this characterization in the ddm1-derived epiRILs. Ultimately, such efforts should contribute significantly to our understanding of epigenetics in adaptive evolution.
Materials and Methods
Methylome Analysis.
MeDIP was carried out as previously described (18) followed by hybridization to a custom NimbleGen tiling array covering the Arabidopsis genome at 165 nt resolution (38). Including dye-swaps, we performed a total of 256 array experiments (SI Appendix, section 1). For each array, probe signals were classified into three underlying methylation states, methylated (M), intermediate (I), or unmethylated (U), using the HMM presented previously (SI Appendix, section 2) (18). These inferred methylation states were cross-validated against whole-genome bisulphite-sequencing data of six epiRILs (SI Appendix, section 3, Fig. S1, and Table S1).
Definition of Parental DMRs.
We conducted a probe-level comparison of the HMM calls between the ddm1 and WT parents (SI Appendix, section 4). Probe-level methylation calls were denoted as polymorphic when the parents differed (e.g., I in ddm1 and M in WT) and as nonpolymorphic when they were identical (e.g., U in ddm1 and U in WT). Neighboring probes reporting the same polymorphic state were collapsed into single regions. Hence, parental DMRs were defined as regions of at least three consecutive probes that reported the same extreme polymorphic state (i.e., transitions from M in WT to U in ddm1 or vice versa). We found 2,611 DMRs, all of which were U in the ddm1. Detailed summary statistics are given in SI Appendix, Table S2.
Calling of Parental Origin of DMRs in the epiRILs.
For any given epiRIL the parental origin of each DMR (i.e., epigenotype) was determined using an HMM-based inference method (SI Appendix, section 4).
Mendelian Segregation Criterion.
Under the assumption that DMRs were stable for eight generations of breeding, both WT- and ddm1-like parental states should appear according to Mendelian segregation ratios in the epiRILs. The sampling variation around these ratios was calculated from a binomial distribution taking into account the sample size (n = 123), the cross design, and the 8% F2 contamination previously reported (12). DMRs in the epiRILs showing a percentage of WT-like states between 62.7% and 83.3% (the expected value being 73%) were taken as putative transgenerationally stable markers. In total 867 parental DMRs fulfilled this criterion and were used subsequently as a starting point for map construction (SI Appendix, section 5, Fig. S2, and Table S3).
Extension of Lander–Green Algorithm.
Derivation of a genetic map using DMRs was carried out through an extension of the Lander-Green algorithm (20), which was designed to accommodate marker and individual specific error rates. Our implementation of this algorithm is detailed in SI Appendix, section 6.
Transcriptome Analysis of epiRILs and ddm1 Seedlings.
Whole-genome expression profiling was performed using a custom NimbleGen tiling array (37). For experimental details, see SI Appendix, section 7.
Transgenerational Analysis of DMRs.
MeDIP-chip was carried out for epiRIL 344 for seven generations of selfing after the backcross, following the protocol described above. At each generation, DNA from five siblings was pooled. The expected signal behavior was derived using a Markov Chain strategy, considering the Mendelian inheritance of the marker probes (SI Appendix, section 8).
Construction of the Consensus Map.
To facilitate a meaningful comparison of the epiRILs map with those of the 17 different F2 populations, we constructed a consensus map (SI Appendix, section 9 and Fig. S11) by using the epiRILs map as a reference and selecting from each of the F2 maps the SNPs closest to the reference, allowing a maximum distance of ±1.39 Mb. The average distance from reference was ±0.17 Mb, which led to little loss of information in capturing the recombination structure along the genome (SI Appendix, Figs. S12 and S13). Markers deemed too distant were not included in the consensus map. This process resulted in 83 markers (SI Appendix, Table S7).
Recombination Intensities at Major Annotation Transitions.
Fig. 2 A and C shows that the recombination intensity increases rapidly at the pericentromeric boundaries, which also coincide with major transitions in genome content from genes to transposons. To find the area where the recombination intensity is maximal, we implemented a sliding window approach (SI Appendix, section 10 and Fig. S15).
Note Added in Proof.
During the reviewing process, Yelina et al (Yelina NE, Choi K, Chelysheva L, Macaulay M, de Snoo B, et al. (2012) Epigenetic Remodeling of Meiotic Crossover Frequency in Arabidopsis thaliana DNA Methyltransferase Mutants. PLoS Genet 8(8): e1002844. doi:10.1371/journal.pgen.1002844) reported elevated centromere-proximal COs, coincident with pericentromeric decreases and distal increases in met1 mutants. However, total numbers of CO events were found to be similar between wild type and met1. These results support the trends observed in the epiRIL population.
Supplementary Material
Acknowledgments
We thank Tony Heitkam for help with the TE analysis. This work was funded by grants from the Ministère de la Recherche et de l’Enseignement Supérieur (to S.C., B.L., and M.E.); Agence Nationale de La Recherche TAG and MEIOMETH projects (to V.C.) and EPIMOBILE project (to V.C. and P.W.); European Union Seventh Framework Programme Network of Excellence EpiGeneSys (Award 257082; to VC); Netherlands Organisation for Scientific Research (NWO) (to F.J., M.L., and M.C.-T.); Consortium for Improving Plant Yield (CIPY) (to M.C.-T.); Netherlands Bioinformatics Centre (NBIC) (to R.W.); and EURATRANS (to R.C.J.). Work in the S.E.J. laboratory is supported by National Institutes of Health Grant GM60398. S.F. is a Special Fellow of the Leukemia & Lymphoma Society. S.E.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genome-wide data reported in this paper have been deposited in the Gene Expression Omnibus public functional genomics data repository, http://www.ncbi.nlm.nih.gov/geo/ [GEO accession nos. GSE37284 (methylomes), GSE37106 (transcriptomes), and GSE37285 (methylomes of intermediate generations)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212955109/-/DCSupplemental.
References
- 1.Lichten M, de Massy B. The impressionistic landscape of meiotic recombination. Cell. 2011;147:267–270. doi: 10.1016/j.cell.2011.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mézard C, Vignard J, Drouaud J, Mercier R. The road to crossovers: Plants have their say. Trends Genet. 2007;23:91–99. doi: 10.1016/j.tig.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Muyt AD, Mercier R, Mézard C, Grelon M. Meiotic recombination and crossovers in plants. Genome Dyn. 2009;5:14–25. doi: 10.1159/000166616. [DOI] [PubMed] [Google Scholar]
- 4.Edlinger B, Schlögelhofer P. Have a break: Determinants of meiotic DNA double strand break (DSB) formation and processing in plants. J Exp Bot. 2011;62:1545–1563. doi: 10.1093/jxb/erq421. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Jinks-Robertson S. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics. 1999;151:1299–1313. doi: 10.1093/genetics/151.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emmanuel E, Yehuda E, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA. The role of AtMSH2 in homologous recombination in Arabidopsis thaliana. EMBO Rep. 2006;7:100–105. doi: 10.1038/sj.embor.7400577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maloisel L, Rossignol JL. Suppression of crossing-over by DNA methylation in Ascobolus. Genes Dev. 1998;12:1381–1389. doi: 10.1101/gad.12.9.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J, et al. Widespread gene conversion in centromere cores. PLoS Biol. 2010;8:e1000327. doi: 10.1371/journal.pbio.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melamed-Bessudo C, Levy AA. Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:E981–E988. doi: 10.1073/pnas.1120742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirouze M, et al. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:5880–5885. doi: 10.1073/pnas.1120841109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teixeira FK, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 12.Johannes F, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009;5:e1000530. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 14.Kakutani T, Munakata K, Richards EJ, Hirochika H. Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics. 1999;151:831–838. doi: 10.1093/genetics/151.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsukahara S, et al. Bursts of retrotransposition reproduced in Arabidopsis. Nature. 2009;461:423–426. doi: 10.1038/nature08351. [DOI] [PubMed] [Google Scholar]
- 16.Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 17.Johannes F, Colomé-Tatché M. Quantitative epigenetics through epigenomic perturbation of isogenic lines. Genetics. 2011;188:215–227. doi: 10.1534/genetics.111.127118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cortijo S, Wardenaar R, Colomé-Tatché M, Johannes F, Colot V. Genome-wide analysis of DNA methylation in Arabidopsis using MeDIP-chip. In: McKeown P, Spillane C, editors. Plant Epigenome: Understanding and Analysis. New York: Humana Press/Springer; 2012. [Google Scholar]
- 19.Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giraut L, et al. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 2011;7:e1002354. doi: 10.1371/journal.pgen.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drouaud J, et al. Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet. 2007;3:e106. doi: 10.1371/journal.pgen.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drouaud J, et al. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res. 2006;16:106–114. doi: 10.1101/gr.4319006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salomé PA, et al. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity (Edinb) 2012;108:447–455. doi: 10.1038/hdy.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horton MW, et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet. 2012;44:212–216. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan J, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitz RJ, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker C, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 30.Richards EJ. Population epigenetics. Curr Opin Genet Dev. 2008;18:221–226. doi: 10.1016/j.gde.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Johannes F, Colot V, Jansen RC. Epigenome dynamics: A quantitative genetics perspective. Nat Rev Genet. 2008;9:883–890. doi: 10.1038/nrg2467. [DOI] [PubMed] [Google Scholar]
- 32.Richards EJ. (2009) Quantitative epigenetics: DNA sequence variation need not apply. Genes Dev 15;23(14):1601–1605. [DOI] [PMC free article] [PubMed]
- 33.Weigel D. Natural variation in Arabidopsis thaliana: From molecular genetics to ecological genomics. Plant Physiol. 2012;158:2–22. doi: 10.1104/pp.111.189845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitz RJ, Ecker JR. Epigenetic and epigenomic variation in Arabidopsis thaliana. Trends Plant Sci. 2012;17:149–154. doi: 10.1016/j.tplants.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roux F, et al. Genome-wide epigenetic perturbation jump-starts patterns of heritable variation found in nature. Genetics. 2011;188:1015–1017. doi: 10.1534/genetics.111.128744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Latzel V, Zhang Y, Karlsson Moritz K, Fischer M, Bossdorf O. Epigenetic variation in plant responses to defense hormones. Ann Bot (Lond) 2012 doi: 10.1093/aob/mcs088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinders J, et al. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009;23:939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roudier F, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.