Abstract
Phase variation is frequently utilized by bacterial species to affect gene expression such that phenotypic variants are maintained within populations, ensuring survival as environmental or host conditions change. Unusual among Helicobacter pylori phase variable or contingency genes is arsS, encoding a sensory histidine kinase involved in the acid acclimation of the organism. The presence of a 3′ homopolymeric cytosine tract of variable length in arsS among Helicobacter pylori strains allows for the expression of various functional ArsS isoforms, differing in carboxy-terminal protein domains. In this study, we analyzed this 3′ arsS region via amplified fragment length polymorphism (AFLP) and sequencing analyses for H. pylori populations from 3 different gastric sites of 12 patients. Our data indicate the presence of multiple arsS alleles within each population of H. pylori derived from the gastric antrum, cardia, or corpus of these patients. We also show that H. pylori, derived from the same anatomical site and patient, are predicted to express multiple ArsS isoforms in each population investigated. Furthermore, we identify a polymorphic deletion within arsS that generates another alternate ArsS C-terminal end. These findings suggest that four C-terminal variations of ArsS adds to the complexity of the ArsRS acid adaptation mechanism as a whole and may influence the ability of H. pylori to persist in the gastric niche for decades.
Keywords: Helicobacter pylori, arsS, polymorphism, homopolymeric tract
1. INTRODUCTION
Helicobacter pylori is a spiral shaped, Gram-negative, microaerophilic, highly motile bacterium [1, 2, 3, 4]. It is a neutralophile that is ecologically restricted to the mucus layer overlaying the human gastric epithelium and an etiologic agent of peptic ulcer disease, chronic active gastritis, and non-ulcer dyspepsia [1, 2, 3, 4, 5]. When untreated, infection may persist for decades, increasing the risk for the development of gastric malignancies such as mucosal-associated lymphoid tissue (MALT) lymphoma and gastric adenocarcinoma [5, 6, 7]. H. pylori encounters marked pH fluctuations during its decades long persistence within the gastric environment as pH levels in the stomach lumen can vary from 5 to 1 depending if the host is in a fed or fasting state [7, 8].
Many studies show that H. pylori possesses unique acid acclimation mechanisms to maintain its periplasmic and cytoplasmic pH levels near neutrality [4, 7, 9, 10, 11]. These buffering mechanisms allow H. pylori to withstand severe acid shock and to grow at moderately low pH levels [2, 7]. One important acid acclimation mechanism is the two-component signal transduction system ArsRS, which is composed of a sensory histidine kinase, ArsS, and its cognate response regulator, ArsR [2, 7, 12]. In response to acidic conditions, ArsS dimerizes to promote autophosphorylation at a conserved histidine residue within its C-terminal transmitter domain [2, 13]. Phosphorylated ArsS can serve as a phosphoryl donor for ArsR, which subsequently regulates gene expression [2, 12]. Interestingly, arsS mutants are viable while arsR mutants are not, suggesting an essential function for non-phosphorylated ArsR [2, 12, 14].
H. pylori strains exhibit remarkable genetic variation and one mechanism promoting this genetic heterogeneity is slipped-strand mispairing during DNA replication [15, 16]. Slipped-strand mispairing occurs more frequently at repetitive sequences, resulting in misalignment of template and nascent DNA strands [15, 16]. Thus, insertion and deletion mutations (indels) are more frequent in sequence repeats of single nucleotides (homopolymeric tracts) or multiple nucleotides (heteropolymeric tracts). Short sequence repeats and the associated indels result in phase variation, a mechanism utilized by bacteria for transcriptional or translational regulation in response to alterations in environmental or host conditions [17].
arsS exhibits unusual sequence variation due to a 3′ homopolymeric tract that differs in the number of repeated cytosine nucleotides among H. pylori strains [12, 18, 19]. Different arsS alleles possessing cytosine repeat length polymorphisms are predicted to be expressed as alternate, yet functional, ArsS isoforms in vitro and, thus far, these ArsS variants have been considered to be strain specific [12]. In this study, we hypothesized that slipped-strand mispairing at the arsS cytosine tract would lead to the generation of alternate alleles within populations of H. pylori. Here, we demonstrate substantial polymorphic variation in the 3′ arsS homopolymeric cytosine tract of all H. pylori populations examined. We also determined that a predominant ArsS isoform is predicted in H. pylori populations from different patients and, in some cases, different populations of the same patient. We additionally document a previously unidentified deletion polymorphism in the 3′ arsS region, leading to a novel alternate ArsS C-terminal domain.
2. MATERIALS AND METHODS
2.1 Patients, bacterial populations, and growth conditions
Populations of clinical H. pylori strains used in this study were cultured from gastric biopsies harvested from the antrum, cardia, and corpus (A, Ca, and Cs) of 12 patients (patients 1 – 12) who were scheduled for endoscopy at the Veterans Administration Hospital in Nashville, Tennessee. All but one patient was male, and patient age ranged from 50 to 74 years with a median age of 62.5 years. H. pylori populations were collected by sweep culture of all resulting H. pylori colonies. To minimize selection of mutants more fit to in vitro conditions, H. pylori populations were generated from cryopreserved, low passage stocks for each DNA extraction. All populations were cultured on Trypticase Soy Agar II plates with 5% sheep blood (BD) and incubated in a humidified environment at 37°C and 5% CO2. Specific information on patients’ age, gender, and clinical findings as well as documented virulence factor genes cagA, vacA, and iceA status of the H. pylori isolates is located in Supplemental Table 1.
2.2 arsS region AFLP
For each gastric population, 3 separate genomic DNA isolations were performed via CTAB extraction as previously described [20, 21]. Each DNA extraction was used in triplicate to amplify a ~300 base pair region encoding the 3′ end of arsS, including the homopolymeric cytosine tract, and part of the 5′ end of downstream gene hemB. Amplicons were generated according to manufacturer’s protocol with Expand High Fidelity PCR kit (Roche) with 6-carboxymethyl fluorescein (FAM)-labeled primer arsS F-1 FAM (CTTCTAACCCCAGCCAAGCCCATGG) and unlabeled hemB R-1 (CGCTGCTTCGTAATCTTCTCAATCG). Amplification conditions consisted of a hot start at 94°C/2 minutes, 30 cycles of 94°C/30 seconds, 60°C/30 seconds, and 72°C/30 seconds, and a final extension at 72°C/7 minutes. PCR was performed 3 times for each DNA extraction for a total of 9 PCR reactions for each population.
One μL of 1:100 diluted PCR samples was added to 12μL Hi-Di Formamide (Applied Biosystems, ABI) plus 0.25μL GeneScan ROX500 Size Standard (ABI) in 96-well plates. Samples were denatured at 95°C for 3 minutes and analyzed with an ABI3130 Genetic Analyzer. Amplicon frequency was quantified in Microsoft Excel with data generated from GeneMapper version 4.0 (ABI). For each PCR run, individual area under the curve for each amplicon was summed to determine total area under the curve for all amplicons. Individual amplicon frequency was calculated by dividing individual area by total area. To integrate AFLP data from the 9 PCR runs for each gastric population, frequencies of corresponding amplicons were used to calculate average and standard error of the mean (SEM).
Each amplicon length was considered to be representative of an individual arsS allele. Amplicons that differed in length by multiples of 3bp were considered to be alleles encoding in the same arsS open reading frame (ORF). Thus, allele (amplicon) frequency data was used to calculate ORF frequency by taking the sum of the frequencies of amplicons that differed in length by multiples of 3bp. ORF SEM was also calculated with corresponding allele (amplicon) SEM values.
2.3 Statistics
To determine the significance of different amplicon lengths within individual populations (i.e. antral, cardia, or corpus populations), one-way ANOVAs with Dunnett post-tests were performed (GraphPad Prism 5.0). The significance of amplicon length between these individual gastric regional populations of the same patient was determined with one-way ANOVAs with Bonferroni post-tests (GraphPad Prism 5.0). One-way ANOVAs with Bonferroni post-tests were also used to calculate significance for the predicted ORFs within and among populations of an individual patient. Thus, comparisons were made with H. pylori antrum, cardia, and corpus populations of individual patients. Comparisons among H. pylori populations of different patients were not considered.
2.4 arsS region sequencing
Unlabeled arsS region amplicons were generated with primers arsS F-1 and hemB R-1 as described above. Amplicons were cloned using pCR-Blunt II-TOPO (Invitrogen) according to manufacturer’s protocols. Plasmid DNA was purified with QIAprep Spin Miniprep Kit or IBI High-Speed Plasmid Mini Kit and 10 cloned 3′ arsS amplicons from each H. pylori gastric population were sequenced.
Both strands of each cloned amplicon were sequenced using BigDye Terminator version 3.1 (ABI). In general, 20 uL sequencing reactions were prepared with 1X Big Dye Terminator v1.1/v3.1 Sequencing Buffer, 0.25X Big Dye Terminator v3.1 Sequencing Reaction Mix, 10μM of T7 (20mer; Promega) or Sp6 (19mer; Promega) promoter primer, 10μL of purified plasmid DNA, and sdH2O. Reactions were achieved with thermal cycling conditions of 94°C/5 minutes and 26 cycles of 94°C/45 seconds, 50°C/30 seconds, and 60°C/4 minutes. Reactions were purified with Performa Dye Terminator Removal (DTR) Gel Filtration Cartridges (Edge Biosystems) according to manufacturer’s protocol. Purified sequencing reactions were vacuum-dried and resuspended in 12μL Hi-Di Formamide, denatured for 3 minutes at 95°C, and analyzed with an ABI3130 Genetic Analyzer. SequencingAnalysis version 5.2 (ABI) software was used for base calling and sequencing analyses were performed with MacVector version 9.0 (Accelrys, Inc.), 4Peaks version 1.7.2 (Mek&Tosj.com), and WebLogo version 3.0 (Threeplusone.com).
Using nine complete H. pylori genomes accessible from National Center for Biotechnology Information (NCBI) databases as of July 2010, we determined that no two completely sequenced H. pylori strains were identical in nucleotide sequence within the 3′ arsS region amplified in our analyses [18, 19, 22, 23, 24, 25, 26, 27, 28]. Based on sequence alignments for the amplified arsS region, the two most highly related arsS sequences are H. pylori strains 26695 and HPAG1. These strains differed by only two nucleotide substitutions when disregarding length differences in their arsS cytosine tracts [19, 27]. To make our analyses more stringent, we considered one base pair differences, disregarding polycytosine tract length polymorphisms, as a metric to denote individual H. pylori sequence types within each gastric population.
3. RESULTS
3.1 arsS polymorphisms
Amplified fragment length polymorphism (AFLP) PCR was utilized to determine whether amplicons of variable base pair (bp) length could be generated from the 3′ arsS region of H. pylori populations isolated from various sites within the stomachs of 12 patients. Resulting AFLP data show six to nine amplicons of variable length from each of the populations investigated in this study (Table 1). The 3′ arsS region of interest was predicted to be approximately 300 bp in length and AFLP data showed that amplicons of 296 bp through 307 bp were generated from the various populations. We quantified each amplicon as a proportion of the total amplicon yield. This indicated that 33 of 36 H. pylori populations produced an amplicon whose quantity was significantly greater than other amplicons from the same gastric population from the same patient (p<0.01) (Table 1). Amplicons of 302 bp, 301 bp, and 300 bp predominated in 33%, 25%, and 19.4% of the populations, respectively.
Table 1.
Summary of AFLP and sequencing data for 3′ arsS region amplicons generated from each H. pylori population of this study.
| arsS Amplicon AFLP Data | arsS Amplicon Sequence Data | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Patient Designation | Strain Designation | Gastric Site | Total # of Amplicon Lengths Detected | Amplicon Length Range (bp) | Predominant Amplicon Length (bp) | Significance of Predominant Amplicona | Common Predominant Ampliconb | Total # of Cytosine Tract Lengths Detected | Lengths of Cytosine Tracts (bp) Detected | Presence of Thymine Deletionc |
| Patient 1 | B215 | Antrum | 7 | 298 – 304 | 301 | *** | Yes | 2 | 12 – 13 | Yes |
| Cardia | 7 | 297 – 303 | 301 | *** | 2 | 12 – 13 | ||||
| Corpus | 6 | 298 – 303 | 301 | *** | 3 | 12 – 14 | ||||
|
| ||||||||||
| Patient 2 | B221 | Antrum | 6 | 299 – 304 | 302 | *** | Yes | 3 | 11 – 13 | Nod |
| Cardia | 6 | 299 – 304 | 302 | *** | 5 | 10 – 14 | ||||
| Corpus | 6 | 299 – 304 | 302 | *** | 4 | 11 – 14 | ||||
|
| ||||||||||
| Patient 3 | B253 | Antrum | 7 | 297 – 303 | 301 | *** | Yes | 4 | 11 – 14 | Yes |
| Cardia | 7 | 297 – 303 | 301 | *** | 3 | 11 – 13 | ||||
| Corpus | 6 | 298 – 303 | 301 | *** | 4 | 11 – 14 | ||||
|
| ||||||||||
| Patient 4 | B256 | Antrum | 7 | 299 – 305 | 303 | *** | No | 4 | 12 – 15 | No |
| Cardia | 7 | 299 – 305 | 303 | *** | 3 | 12 – 14 | ||||
| Corpus | 7 | 299 – 305 | 302 | *** | 4 | 11 – 14 | ||||
|
| ||||||||||
| Patient 5 | B266 | Antrum | 6 | 297 – 302 | 300 | *** | No | 4 | 10 – 13 | Yes |
| Cardia | 6 | 297 – 302 | 300 | *** | 4 | 11 – 13, 15 | ||||
| Corpus | 8 | 296 –303 | 301 | ns | 4 | 11 – 14 | ||||
|
| ||||||||||
| Patient 6 | B268 | Antrum | 6 | 299 – 304 | 302 | *** | No | 2 | 12 – 13 | No |
| Cardia | 7 | 299 – 305 | 303 | *** | 3 | 11, 13 – 14 | ||||
| Corpus | 6 | 299 – 304 | 302 | *** | 3 | 11 – 13 | ||||
|
| ||||||||||
| Patient 7 | B284 | Antrum | 8 | 299 – 306 | 304 | ** | No | 5 | 13 – 17 | Yes |
| Cardia | 9 | 298 – 306 | 303 | *** | 6 | 12 – 17 | ||||
| Corpus | 9 | 299 – 307 | 304 | *** | 5 | 12 – 16 | ||||
|
| ||||||||||
| Patient 8 | B292 | Antrum | 7 | 298 – 304 | 302 | *** | Yes | 5 | 10 – 14 | Yes |
| Cardia | 8 | 298 – 305 | 302 | *** | 4 | 12 – 15 | ||||
| Corpus | 7 | 298 – 304 | 302 | *** | 4 | 12 – 15 | ||||
|
| ||||||||||
| Patient 9 | B294 | Antrum | 7 | 296 – 302 | 299 | *** | No | 5 | 9, 11 – 14 | Yes |
| Cardia | 6 | 297 – 302 | 300 | *** | 3 | 11 – 13 | ||||
| Corpus | 6 | 297 – 302 | 300 | *** | 3 | 11 – 13 | ||||
|
| ||||||||||
| Patient 10 | B295 | Antrum | 9 | 298 – 306 | 301 | ns | No | 6 | 8, 11 – 15 | Yes |
| Cardia | 7 | 300 – 306 | 304 | *** | 5 | 10, 11, 14 – 16 | ||||
| Corpus | 6 | 298 – 303 | 301 | *** | 2 | 12 – 13 | ||||
|
| ||||||||||
| Patient 11 | B300 | Antrum | 6 | 297 – 302 | 300 | *** | Yes | 3 | 11 – 13 | Yes |
| Cardia | 7 | 297 – 303 | 300 | ns | 3 | 12 – 14 | ||||
| Corpus | 6 | 297 – 302 | 300 | *** | 3 | 11 – 13 | ||||
|
| ||||||||||
| Patient 12 | B301 | Antrum | 6 | 299 – 304 | 302 | *** | Yes | 5 | 9 – 13 | No |
| Cardia | 6 | 299 – 304 | 302 | *** | 3 | 9, 12 – 13 | ||||
| Corpus | 7 | 298 – 304 | 302 | *** | 4 | 10 – 13 | ||||
The statistical significance of the predominant amplicon versus other amplicon lengths was tested for individual H. pylori populations via one-way ANOVAs with Dunnett post-tests;
p<0.05,
p<0.01,
p<0.001,
p<0.0001.
H. pylori populations that shared the same predominant amplicon within a single patient were designated as having a common predominant amplicon. Commonality is only valid for populations within patients.
Sequenced amplicons from each H. pylori population of individual patients either encoded for a functional arsS stop codon (no deletion) or had a deletion of a thymine nucleotide within the arsS stop codon (deletion).
The thymine deletion was detected in a single sequence derived from the cardia H. pylori population of patient 2. The other 29 sequences from the populations of this patient were negative for the deletion.
H. pylori populations from different gastric sites of patients 1, 2, 3, 8, 11, and 12 produced a common quantitatively predominant amplicon from all gastric regions that was significantly different than any other amplicon length (p<0.0001) for five of these six patients (Table 1). However, one of the three gastric H. pylori populations of patients 4, 5, 6, 7, 9, and 10 produced a quantitatively predominant amplicon of different length when compared to the predominant amplicon length for the other two H. pylori gastric populations of the individual patients. Difference in predominant amplicon length among populations of the same patient was significant (p<0.01) in 5 of these 6 patients (Table 1).
For each of the 36 H. pylori gastric populations examined in this study, ten cloned amplicons containing 3′ arsS region amplicons were generated to confirm that AFLP results were due to differences in homopolymeric cytosine tract length. Our data show that at least two to as many as six different cytosine tract lengths could be documented from the amplicons generated from each gastric H. pylori population (Table 1). Amplicons from 88.9% of the populations possessed three or more different cytosine tract lengths. Only two different cytosine tract lengths were detected in four of the 36 gastric H. pylori populations. In total, sequence data indicated that homopolymeric tract lengths ranged from 8 to 17 tandem cytosine nucleotides, depending on the source population (Table 1). Cytosine tract lengths of 13, 12, and 14 represented 40%, 24.7%, and 15.8% of all sequenced amplicons, respectively. Due to the relatively small number of sequences analyzed, relationships between AFLP amplicon length and sequenced cytosine tract length were not considered. In sum, AFLP and sequencing analyses suggest that variant 3′ arsS homopolymeric tract lengths may be generated in vivo and suggested that predominant amplicon lengths could differ among H. pylori populations colonizing different regions of the stomach (Table 1).
Our sequence analysis of 360 cloned amplicons generated from the 36 H. pylori populations revealed a thymine deletion in 241 of the cloned sequences. This thymine deletion is not associated with a repeated sequence (Figure 1). This deletion eliminates the stop codon of one predicted arsS reading frame. Interestingly, 240 of these 241 sequences were found in the H. pylori populations of 8 patients (Table 1). Only one of the remaining 120 sequenced amplicons from populations of the other 4 patients possessed this stop codon thymine deletion. This sequence was generated from the cardia population of patient 2 and the deletion was not observed in sequences from the other 29 amplicons generated from populations from that patient. Outside this study, the NCBI sequence database indicates that H. pylori strains B38 and J99 are the only sequenced strains to possess this thymine deletion. Thus, differences in homopolymeric cytosine tract length as well as the presence or absence of this deletion appear to be common polymorphisms for this region of arsS.
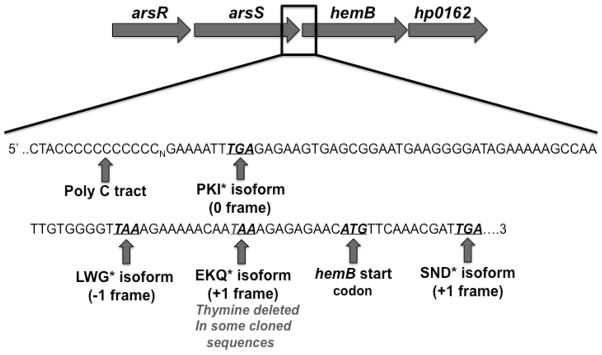
Figure 1. arsS polymorphisms and the arsRS operon.
The arsRS operon consists of arsR, arsS, hemB, and hp0162, a gene encoding a hypothetical protein. DNA sequence of the 3′ terminus of arsS and the 5′ region of hemB is shown. The arsS homopolymeric cytosine tract and associated stop codons of the different arsS ORFs are indicated. As the homopolymeric cytosine tract varies in length (Cn), different C-terminal regions of ArsS (PKI*, LWG*, EKQ*, and SND*) are encoded due to different ORFs. Many of the H. pylori populations in this study possessed a thymine deletion in the third stop codon (in italics), allowing for the alternate SND* C-terminal region to be predicted. The predicted stop codon of this C-terminal end is located within hemB gene.
3.2 Variant ArsS C-terminal regions
AFLP and sequence data indicated that the arsS amplicons could be considered as different alleles distinct in their 3′ regions due to differences in homopolymeric cytosine tract length and the presence/absence of a thymine deletion. Cytosine tract length differences and presence/absence of the thymine deletion can shift the arsS open reading frame (ORF) to alter the ArsS C-terminal amino acid sequence. Thus, AFLP amplicon lengths that varied by multiples of 3bp were presumed to be of the same arsS ORF and used to calculate total ORF frequencies. For each population, arsS amplicons of 301 bp, 302 bp, and 303 bp were designated to be of ORFs 1, 2, and 3, respectively. These variant alleles are predicted to express ArsS isoforms differing in their carboxy-terminal domains. The ORFs of all other amplicons were determined with the multiple of three bp parameter. A predominant ArsS ORF was apparent for each H. pylori population and ORFs 1, 2, and 3 were predominant in 33.3%, 41.7%, and 25% of all populations.
A predominant ArsS isoform was significant (p<0.05) for 29 of 36 populations, which may suggest a potential selective pressure on ArsS isoform expression dependent on the gastric environment. Overall, five of the 12 patients had H. pylori populations predicted to express a uniform predominant ORF regardless of the gastric site of origin. However, seven patients had H. pylori populations encoding for different predominant ORFs among their gastric regions, and the predominance difference amongst regions was significant (p<0.05) for the populations of five of these seven patients (Table 2). We speculate that local conditions in the cardia, corpus, or antrum of these five patients may differ sufficiently such that selection has allowed populations with differing ArsS isoforms to attain numeric predominance within the stomach of individual patients (Table 2).
Table 2.
Summary of AFLP and sequencing data for predicted arsS ORFs from each H. pylori population of this study.
| AFLP arsS Amplicon Data | Sequenced arsS Amplicon Data | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Patient Designation | Strain Designation | Gastric Site | ORF Range | Predominant ORF | Significance of Predominant ORFa | Common Predominant ORFb | C-terminal Regions Detected | Thymine Deletionc |
| Patient 1 | B215 | Antrum | 1 – 3 | 1 | **** | Yes | LWG* SND* | Yes |
| Cardia | 1 – 3 | 1 | **** | LWG* SND* | ||||
| Corpus | 1 – 3 | 1 | **** | PKI* LWG* SND* | ||||
|
| ||||||||
| Patient 2 | B221 | Antrum | 1 – 3 | 2 | **** | Yes | PKI* LWG* EKQ* | Nod |
| Cardia | 1 – 3 | 2 | **** | PKI* LWG* EKQ* | ||||
| Corpus | 1 – 3 | 2 | **** | PKI* LWG* EKQ* | ||||
|
| ||||||||
| Patient 3 | B253 | Antrum | 1 – 3 | 1 | **** | Yes | PKI* LWG* SND* | Yes |
| Cardia | 1 – 3 | 1 | **** | PKI* LWG* SND* | ||||
| Corpus | 1 – 3 | 1 | **** | PKI* LWG* SND* | ||||
|
| ||||||||
| Patient 4 | B256 | Antrum | 1 – 3 | 3 | **** | No | PKI* LWG* EKQ* | No |
| Cardia | 1 – 3 | 3 | **** | PKI* LWG* EKQ* | ||||
| Corpus | 1 – 3 | 2 | ns | PKI* LWG* EKQ* | ||||
|
| ||||||||
| Patient 5 | B266 | Antrum | 1 – 3 | 3 | * | No | PKI* LWG* SND* | Yes |
| Cardia | 1 – 3 | 3 | ** | PKI* LWG* SND* | ||||
| Corpus | 1 – 3 | 1 | ns | PKI* LWG* SND* | ||||
|
| ||||||||
| Patient 6 | B268 | Antrum | 1 – 3 | 2 | **** | No | PKI* LWG* EKQ* | No |
| Cardia | 1 – 3 | 3 | **** | PKI* LWG* | ||||
| Corpus | 1 – 3 | 2 | **** | PKI* LWG* EKQ* | ||||
|
| ||||||||
| Patient 7 | B284 | Antrum | 1 – 3 | 1 | *** | No | PKI* LWG* SND* | Yes |
| Cardia | 1 – 3 | 2 | ns | PKI* LWG* SND* | ||||
| Corpus | 1 – 3 | 1 | **** | PKI* LWG* SND* | ||||
|
| ||||||||
| Patient 8 | B292 | Antrum | 1 – 3 | 2 | **** | Yes | PKI* LWG* SND* | Yes |
| Cardia | 1 – 3 | 2 | **** | PKI* LWG* SND* | ||||
| Corpus | 1 – 3 | 2 | **** | PKI* LWG* SND* | ||||
|
| ||||||||
| Patient 9 | B294 | Antrum | 1 – 3 | 2 | ns | No | PKI* LWG* SND* | Yes |
| Cardia | 1 – 3 | 3 | **** | PKI* LWG* SND* | ||||
| Corpus | 1 – 3 | 3 | **** | PKI* LWG* SND* | ||||
|
| ||||||||
| Patient 10 | B295 | Antrum | 1 – 3 | 2 | ns | No | PKI* LWG* SND* | Yes |
| Cardia | 1 – 3 | 1 | ns | PKI* LWG* SND* | ||||
| Corpus | 1 – 3 | 1 | *** | LWG* SND* | ||||
|
| ||||||||
| Patient 11 | B300 | Antrum | 1 – 3 | 3 | **** | No | PKI* LWG* SND* | Yes |
| Cardia | 1 – 3 | 1 | ns | PKI* LWG* SND* | ||||
| Corpus | 1 – 3 | 3 | **** | PKI* LWG* SND* | ||||
|
| ||||||||
| Patient 12 | B301 | Antrum | 1 – 3 | 2 | **** | Yes | PKI* LWG* EKQ* | No |
| Cardia | 1 – 3 | 2 | **** | LWG* EKQ* | ||||
| Corpus | 1 – 3 | 2 | **** | PKI* LWG* EKQ* | ||||
The statistical significance of the predominant amplicon versus other amplicon lengths was tested for individual H. pylori populations via one-way ANOVAs with Dunnett post-tests;
p<0.05,
p<0.01,
p<0.001,
p<0.0001.
H. pylori populations that shared the same predominant ORF within a single patient were designated as having a common predominant ORF. Commonality is only valid for populations within patients.
Sequenced amplicons from each H. pylori population of individual patients either encoded for a functional arsS stop codon (no deletion) or had a deletion of a thymine nucleotide within the arsS stop codon (deletion).
The thymine deletion was detected in a single sequence derived from the cardia H. pylori population of patient 2. The other 29 sequences from the populations of this patient were negative for the deletion.
As the ORF of arsS is shifted due to differences in homopolymeric cytosine tract length and the presence/absence of the thymine deletion, the translated ArsS amino acid sequence will be affected. arsS ORFs that differ by three bp multiples of cytosines in the homopolymeric tract should only differ by the number of proline amino acids translated in ArsS. However, differences in cytosine tract length paired with the presence/absence of the thymine deletion can considerably alter the translated ArsS C-terminal region. With the 3′ arsS polymorphisms now documented for each H. pylori population of this study, we performed in silico translation of the 360 cloned amplicon sequences to demonstrate that the polymorphic differences lead to the translation of variant ArsS C-terminal ends. Again, because of the relatively small number of sequenced amplicons, comparisons between AFLP-predicted arsS ORFs and sequence-predicted ArsS C-terminal ends were not considered. Although ArsS C-terminal domains vary from two to 31 amino acids past the poly proline regions, for simplicity, alternate C-terminal domains are abbreviated as the last 3 predicted amino acids prior to their respective stop codons, symbolized by *.
In agreement with NCBI arsS ORF designations, we designated the 0 and −1 arsS ORFs as ArsS C-terminal ends PKI* and LWG* (Figure 1). Since the polymorphic thymine deletion was located within the stop codon of the +1 arsS ORF, two alternative C-terminal ends are predicted (Figure 1). These ends are designated as SND* or EKQ* depending on the thymine deletion presence or absence, respectively. The stop codon for the SND* C-terminal end overlaps the sequence encoding the downstream gene hemB (Figure 1). The peptide sequences of the alternative C-terminal ends vary in length by two to as many as 31 amino acids beyond the final proline encoded by the homopolymeric cytosine tract. These four alternate ArsS C-terminal ends also differ in the composition of their amino acid sequences (Figure 2). However, the predicted EKQ* and SND* ends appear to be homologous until the SND* sequence is extended due to the thymine deletion in the EKQ* stop codon (Figure 2).
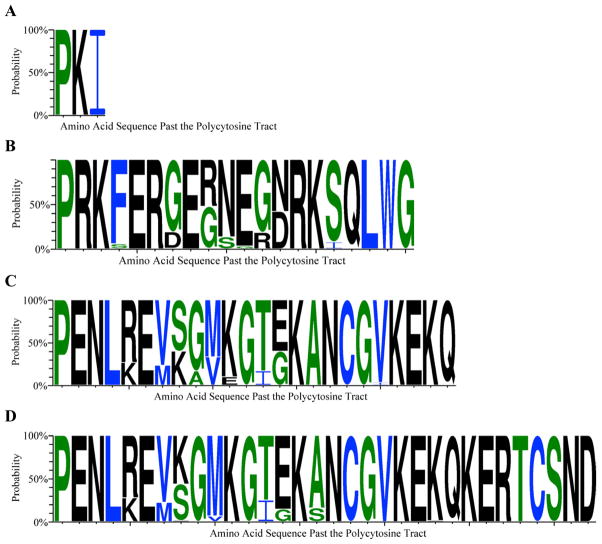
Figure 2. Variable ArsS C-terminal regions.
Sequence logos were generated based on the 360 cloned 3′ arsS region sequences and depict the 4 variable ArsS C-terminal regions past the homopolymeric cytosine tract. Amino acid sequence differences and similarities are apparent. The x-axis represents the amino acid sequence distal to the final proline (P) encoded by the homopolymeric cytosine tract. The y-axis represents the probability of the amino acid present. Amino acids are also colored due to their hydropathy index values, where hydrophobic amino acids are black, hydrophilic are blue, and neutral are green. (A) Sequence logo for PKI* was generated with 93 cloned 3′ arsS region sequences from 31 H. pylori populations. (B) Sequence logo for LWG* was derived from 158 sequences from 36 H. pylori populations. (C) Sequence logo EKQ* was generated with 30 sequences from 11 populations. (D) Sequence logo for SND* was derived from 76 3′ arsS region sequences from 24 populations.
From amplicon sequence data, the PKI* ArsS C-terminal end was predicted to be expressed by 31 of the 36 gastric H. pylori populations, but was not detected in 5 individual gastric regional populations from four patients (Table 2). The LWG* C-terminal end was predicted in all H. pylori populations investigated. The EKQ* C-terminal end was predicted in 11 populations of four patients and the SND* alternative +1 end was detected in 24 populations from the remaining eight patients (Table 2). PKI* and LWG* were predicted to be expressed by 93 (25.8%) and 158 (43.9%) of the 360 cloned 3′ arsS amplicons, respectively. Collectively, the alternative EKQ* and SND* ends, represented 107 (29.7%) of these sequences. Individually, the EKQ* and SND* C-terminal ends represented 30 (8.3%) and 77 (21.4%) of all sequenced amplicons. The remaining two (0.6%) cloned sequences possessed deletions within the arsS coding sequence, which may represent naturally occurring arsS null mutants or deletion mutations.
3.3 arsS point mutations
Our analyses of cloned arsS amplicon sequences generated from individual H. pylori populations from various gastric anatomical regions indicated that sequence variation due to point mutations outside the homopolymeric cytosine was present. Disregarding differences in hypermutable cytosine tract length, we considered single nucleotide mutations as a determinant of variant 3′ arsS sequence types. Thus, nucleotide sequences that were identical within a gastric population were considered to be of the same sequence type and sequences that possessed at least one nucleotide mutation were considered to be of an alternate sequence type. We frequently observed sequenced amplicons that possessed no nucleotide substitutions. At least 5 to as many as 10 sequenced amplicons had identical sequence types within each population. We also observed amplicons that differed in nucleotide sequence from 1 to as many as 11 nucleotides within some H. pylori gastric populations. In all, 29 of the 36 H. pylori gastric regional populations examined in the current study contained distinct 3′ arsS region sequence types, while only seven of 36 H. pylori populations showed identical arsS sequence types. In total, eight of twelve patients in the study showed evidence of polyclonal H. pylori infection.
4. DISCUSSION
Helicobacter pylori genome sequences deposited in NCBI databases suggest that individual strains possess a homopolymeric cytosine tract of specific length near the 3′ terminus of arsS [18, 19, 22, 23, 24, 25, 26, 27, 28]. In the present study, AFLP and sequence data generated with 3′ arsS amplicons indicate that individual H. pylori populations encode multiple 3′ arsS regions that can differ greatly in their cytosine tract lengths. AFLP data suggested that 6 to as many as 9 different 3′ arsS amplicons of variable length were present in each population investigated. Furthermore, sequencing data indicated that 8 to 17 repeated cytosine nucleotides could be detected in this 3′ arsS tract, depending on the population. The large number of cytosine tract lengths may be due in part to the fact that we queried entire H. pylori populations and not isolated, clonal strains as in previous studies [18, 19, 22, 23, 24, 25, 26, 27, 28]. However, Alm et al. observed 3 different 3′ arsS homopolymeric cytosine tract lengths among J99 sequencing reads, and suggested that slipped-strand mispairing or in vitro conditions could influence the generation of multiple tract lengths. Hence in the current study, H. pylori populations were harvested from gastric biopsy specimens and passed minimally in vitro to limit nonselective mutations and maintain original arsS homopolymeric cytosine tract length frequencies.
Using AFLP and sequence analyses to probe cytosine tract length differences and the presence/absence of the thymine deletion, we found that each H. pylori population of this study encoded various ArsS C-terminal regions. Beier et al. showed previously that different H. pylori strains with presumably strain-dependent ArsS C-terminal ends are each capable of ArsS autophosphorylation and phosphotransfer to the cognate response regulator, ArsR [12]. However, their study assumed that ArsS with a single C-terminal end was expressed for each of the H. pylori strains investigated. Our data indicate that multiple ArsS isoforms, variant at their C-terminal end, are expressed within each population investigated. Our study makes clear that H. pylori populations possess multiple arsS alleles leading to the expression of multiple ArsS isoforms with alternate C-termini. Thus, our data indicate that arsS is a contingency gene. It is interesting to speculate that ArsS isoforms, variant in their C-terminal end and still functional, may differ in regards to phosphotransfer capacity or efficiency; or perhaps each isoform or its encoding mRNA may have variable longevity in H. pylori. To test such hypotheses, we attempted to produce H. pylori strains capable of expressing ArsS with a single C-terminal end. However, these studies have been unsuccessful perhaps due to the importance of the hemB gene present in the arsRS operon.
The complexity of ArsS isoform functionality is increased with the discovery of a fourth (the SND*) C-terminal end. Interestingly, the thymine deletion that facilitates SND* translation is present in the sequenced H. pylori strains J99 and B38 [18, 19, 22, 23, 24, 25, 26, 27, 28]. In this study, we found that 8 of the 12 patients were infected with H. pylori that possessed this thymine deletion. We entertained the possibility that this was due to a bias in geographic locale as all patients in this study were evaluated in Nashville, Tennessee and strain J99 was also isolated from a patient in this geographic region. However, strain B38 was isolated from a patient in France [28]. Yet, the published sequences of H. pylori J99 and B38 do not suggest that the SND* C-terminal end of ArsS would be translated due to the apparent length of the homopolymeric cytosine tract [18, 28]. Thus, the acknowledgement of the SND* C-terminal end or its frequency has not been considered in prior studies.
We also detected nucleotide substitutions within regions of arsS outside the homopolymeric cytosine tract and the thymine deletion and considered each variant to be evidence of additional sequence types. Interestingly, Alm et al. also noted nucleotide substitutions in sequence reads of J99, but at low frequency [18]. Comparison of the 9 H. pylori genome sequences within the NCBI database at the time of this study demonstrate that at least two, and frequently more than two, nucleotide substitutions exist within the 3′ arsS region examined in our study [18, 19, 22, 23, 24, 25, 26, 27, 28]. At least 5 to as many as 10 arsS amplicons generated from each population had sequence types that possessed no nucleotide substitutions. However, there were also 3′ arsS amplicons that had between 1 and 11 nucleotide substitutions within certain populations. While polyclonal H. pylori infections are frequent in developing nations, our data showed that identical sequence types were generated from only 7 gastric H. pylori populations in 4 of the 12 patients examined [29]. Due to the somewhat limited scope of our study, we cannot contend that the remaining 29 populations are truly polyclonal. However, the notion that 8 of the 12 patients in our study may have multiple sequence types of H. pylori concurrently is interesting to consider, especially considering the well documented proclivity of H. pylori for horizontal gene transfer.
Supplementary Material
Highlights.
arsS is polymorphic at its homopolymeric cytosine tract within H. pylori populations
H. pylori populations are predicted to express multiple variant ArsS C-terminal domains.
A previously unrecognized ArsS C-terminal domain was documented in this study
Polyclonal infections of H. pylori may be common this patient group.
Acknowledgments
This work was supported by NIH grant R15 AI053062 to MHF and P01 CA116087, R01 DK58587, R01 CA77955 to RMP Jr. This research was also supported in part by a Howard Hughes Medical Institute grant through the Undergraduate Biological Sciences Education Program to the College of William and Mary.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pattison CP, Combs MJ, Marshall BJ. Helicobacter pylori and Peptic Ulcer Disease: Evolution to Revolution to Resolution. Am J Roentgenol. 1996;168:1415– 1420. doi: 10.2214/ajr.168.6.9168699. [DOI] [PubMed] [Google Scholar]
- 2.Pflock M, Finsterer N, Joseph B, Mollenkopf H, Meyer TF, Beier D. Characterization of the ArsRS regulon of Helicobacter pylori, Involved in Acid Adaptation. J Bacteriol. 2006;188:3449–3462. doi: 10.1128/JB.188.10.3449-3462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schär J, Sickmann A, Beier D. Phosphorylation-Independent Activity of Atypical Response Regulators of Helicobacter pylori. J Bacteriol. 2005;187:3100– 3109. doi: 10.1128/JB.187.9.3100-3109.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. Involvement of the HP0165-HP0166 two-component system in expression of some acidic-pH-upregulated genes of Helicobacter pylori. J Bacteriol. 2006;188:1750– 1761. doi: 10.1128/JB.188.5.1750-1761.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pflock M, Kennard S, Delany I, Scarlato V, Beier D. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect Immun. 2005;73:6437– 6445. doi: 10.1128/IAI.73.10.6437-6445.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsonnet J, Vandersteen D, Goates J, Sibley RK, Pritikin J, Chang Y. Helicobacter pylori Infection in Intestinal- and Diffuse-Type Gastric Adenocarcinomas. J Natl Cancer I. 1991;83:640– 643. doi: 10.1093/jnci/83.9.640. [DOI] [PubMed] [Google Scholar]
- 7.Pflock M, Kennard S, Finsterer N, Beier D. Acid-responsive gene regulation in the human pathogen Helicobacter pylori. J Biotechnol. 2006;126:52– 60. doi: 10.1016/j.jbiotec.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 8.Pflock M, Müller S, Beier D. The CrdRS (HP1365-HP1364) Two-Component System Is Not Involved in pH-Responsive Gene Regulation in the Helicobacter pylori Strains 26695 and G27. Curr Microbiol. 2006;54:320– 324. doi: 10.1007/s00284-006-0520-9. [DOI] [PubMed] [Google Scholar]
- 9.Sachs G, Weeks DL, Wen Y, Marcus EA, Scott DR. Acid Acclimation by Helicobacter pylori. Physiology. 2005;20:429– 438. doi: 10.1152/physiol.00032.2005. [DOI] [PubMed] [Google Scholar]
- 10.Scott DR, Marcus EA, Wen Y, Oh J, Sachs G. Gene Expression in vivo Shows that Helicobacter pylori Colonizes an Acidic Niche on the Gastric Surface. P Natl Acad Sci USA. 2007;104:7235– 7240. doi: 10.1073/pnas.0702300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott DR, Marcus EA, Wen Y, Singh S, Feng J, Sachs G. Cytoplasmic Histidine Kinase (HP0244)-Regulated Assembly of Urease with UreI, a Channel for Urea and Its Metabolites, CO2, NH3, and NH4+ Is Necessary for Acid Survival of Helicobacter pylori. J Bacteriol. 2010;192:94– 103. doi: 10.1128/JB.00848-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beier D, Frank R. Molecular characterization of two-component systems of Helicobacter pylori. J Bacteriol. 2000;182:2068– 2076. doi: 10.1128/jb.182.8.2068-2076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stock AM, V, Robinson L, Goudreau PN. Two-Component Signal Transduction. Annu Rev Biochem. 2000;69:183– 215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 14.Dietz P, Gerlach G, Beier D. Identification of Target Genes Regulated by the Two-Component System HP166-HP165 of Helicobacter pylori. J Bacteriol. 2002;184:350– 362. doi: 10.1128/JB.184.2.350-362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Achtman M, Azuma T, Berg DE, Ito Y, Morelli G, Pan Z, Suerbaum S, Thompson SA, van der Ende A, van Doorn L. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459– 470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 16.Han S, Zschausch HE, Meyer HW, Schneider T, Loos M, Bhakdi S, Maeurer MJ. Helicobacter pylori: Clonal Population Structure and Restricted Transmission within Families Revealed by Molecular Typing. J Clin Microbiol. 2000;38:3646– 3651. doi: 10.1128/jcm.38.10.3646-3651.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson IR, Owen P, Nataro JP. Molecular switches - the ON and OFF of bacterial phase variation. Mol Microbiol. 1999;33:919– 932. doi: 10.1046/j.1365-2958.1999.01555.x. [DOI] [PubMed] [Google Scholar]
- 18.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176– 180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 19.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 20.McNulty SL, Mole BM, Dailidiene D, Segal I, Ally R, Mistry R, Secka O, Adegbola RA, Thomas JE, Lenarcic EM, Peek RM, Jr, Berg DE, Forsyth MH. Novel 180- and 480-Base-Pair Insertions in African and African-American Strains of Helicobacter pylori. J Clin Microbiol. 2004;42:5658– 5663. doi: 10.1128/JCM.42.12.5658-5663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orsi R, Bowen BM, Wiedmann M. Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC Genomics. 2010;11:1471– 2164. doi: 10.1186/1471-2164-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baltus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. The Complete Genome Sequence of Helicobacter pylori Strain G27. J Bacteriol. 2009;191:447– 448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer W, Windhager L, Karnholz A, Zeiller M, Zimmer R, Haas R. The complete genome sequence of Helicobacter pylori strain P12. 2008. Unpublished. [Google Scholar]
- 24.Kersulyte D, Kalia A, Gilman RH, Berg DE. Genome Sequence of Helicobacter pylori from the Remote Amazon: traces of Asian ancestry of the First Americans. 2010. Unpublished. [Google Scholar]
- 25.Kim S, Lee WK, Choi SH, Kang S, Park HS, Kim YS, Lee SG, Byun EY, Jeong JE, Park YH, Lee EJ, Kim JS, Ryu BD, Lee YS, Hahn Y, Yeom YI, Park SG, Youn HS, Ko GH, Choi MB, Park CH, Lim JY, Bae DW, Song JY, Park JU, Kang HL, Baik SC, Cho MJ, Yoo HS, Rhee KH. Helicobacter pylori. 2004:51. http://www.ncbi.nlm.nih.gov/nuccore/CP000012.1.
- 26.Kim S, Lee WK, Choi SH, Kang S, Park HS, Kim YS, Lee SG, Byun EY, Jeong JE, Park YH, Lee EJ, Kim JS, Ryu BD, Lee YS, Hahn Y, Yeom YI, Park SG, Youn HS, Ko GH, Choi MB, Park CH, Lim JY, Bae DW, Song JY, Park JU, Kang HL, Baik SC, Cho MJ, Yoo HS, Rhee KH. Helicobacter pylori. 2009:52. http://www.ncbi.nlm.nih.gov/nuccore/261838873.
- 27.Oh JD, Kling-Bäckhed H, Giannakis M, Xu J, Fulton RS, Fulton LA, Cordum HS, Wang C, Elliott G, Edwards J, Mardis ER, Engstrand LG, Gordon JI. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: Evolution during disease progression. P Natl Acad Sci USA. 2006;103:9999– 10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiberge J, Boursaux-Eude C, Lehours P, Dillies M, Creno S, Coppee J, Rouy Z, Lajus A, Ma L, Burucoa C, Ruskone-Foumestraux A, Courillon-Mallet A, De Reuse H, Boneca IG, Lamarque D, Megraud F, Delchier J, Medigue C, Bouchier C, Labigne A, Raymond J. Array-based hybridization of Helicobacter pylori isolates to the complete genome sequence of an isolate associated with MALT lymphoma. BMC Genomics. 2010;11:368. doi: 10.1186/1471-2164-11-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Logan Robert PH, Walker MM. Epidemiology and diagnosis of Helicobacter pylori infection. Br Med J. 2001;323:920– 922. doi: 10.1136/bmj.323.7318.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.