SUMMARY
Clustered regularly interspaced short palindromic repeats (CRISPRs), together with an operon of CRISPR-associated (Cas) proteins, form an RNA-based prokaryotic immune system against exogenous genetic elements. Cas5 family proteins are found in several Type I CRISPR-Cas systems. Here we report the molecular function of Subtype I-C/Dvulg Cas5d from B. halodurans. We show that Cas5d cleaves pre-crRNA into unit length by recognizing both the hairpin structure and the 3′ single stranded sequence in the CRISPR repeat region. Cas5d structure reveals a ferredoxin domain-based architecture and a catalytic triad formed by Y46, K116 and H117 residues. We further show that after pre-crRNA processing, Cas5d assembles with crRNA, Csd1, and Csd2 proteins to form a multi-subunit interference complex similar to E. coli Cascade (CRISPR-associated complex for antiviral defense) in architecture. Our results suggest that formation of a crRNA-presenting Cascade-like complex is likely a common theme among Type I CRISPR subtypes.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPRs) are found in about ~45% of sequenced bacteria and ~83% of archaea genomes and participate in RNA-based adaptive immunity against exogenous genetic elements such as viruses (phages), invading conjugative plasmids, and transposable elements (Deveau et al., 2010; Horvath and Barrangou, 2010; Karginov and Hannon, 2010; Marraffini and Sontheimer, 2010; Sorek et al., 2008; van der Oost et al., 2009; Waters and Storz, 2009; Wiedenheft et al., 2012). CRISPR loci are composed of invariable repeat sequences about 21-48 base pairs (bp) in length, interspaced by variable spacer sequences (26-72 bp) derived from epichromosomal origins (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005). Adjacent to CRISPR loci is a cas (CRISPR associated) operon, encoding a cluster of Cas proteins involved in CRISPR interference and adaptation processes. The cas genes can be further classified into a set of ‘core’ genes and subtype-specific genes based on phylogenetic analysis (Haft et al., 2005; Makarova et al., 2011a; Makarova et al., 2006; Makarova et al., 2011b). CRISPR-Cas systems are recently classified into three major types (Type I, II, and III) based on the mutually exclusive presence of cas3, cas9/csn1, and cas10/cmr2 genes, respectively. Type I system is the most wide-spread, and can be further classified into six different subtypes (I-A to I-F) based on the presence of distinct subtype-specific genes (Makarova et al., 2011b).
At the molecular level, CRISPR interference can be divided into three stages: (i) CRISPR adaptation upon exposure to foreign genetic elements through the insertion of a new spacer into the 5′-end of the genomic CRISPR array; (ii) expression of the nascent CRISPR RNA transcript (pre-crRNA) and enzymatic digestion into the mature form (crRNA); and (iii) crRNA-mediated interference to degrade foreign genetic elements (Barrangou et al., 2007; Brouns et al., 2008; Carte et al., 2008; Deltcheva et al., 2011; Haft et al., 2005; Hale et al., 2009; Haurwitz et al., 2010; Makarova et al., 2006). The pre-crRNA processing in some CRISPR subtypes has been shown to be mediated by ferredoxin domain containing endoribonucleases encoded in the cas operon. These include Cse3 protein from Subtype I-E (previously named Ecoli subtype), Csy4 from Subtype I-F (Ypest), and Cas6 from Subtype I-A (Apern), I-B (Tneap), and III-B (Cmr) have been shown to process the pre-crRNA in their corresponding CRISPR subtype (Brouns et al., 2008; Carte et al., 2008; Deltcheva et al., 2011; Haurwitz et al., 2010). Structure and mechanistic diversity were found among these pre-crRNA processors. Cse3 and Csy4 recognize the palindromic RNA stemloop in pre-crRNAs in a site- and structure-specific manner (Gesner et al., 2011; Haurwitz et al., 2010; Sashital et al., 2011). Endoribonucleolytic cleavage proceeds through a general acid-base mechanism to generate a 5′ hydroxyl and a 2′,3′-cyclic phosphate. The stemloop portion of the mature crRNA was shown to remain associated with the endoribonuclease to participate in the formation of the bigger Cascade (CRISPR-associated complex for antiviral defense) complex. This complex was thought to display the crRNA for subsequent destruction of invading nucleic-acid sequences (Jore et al., 2011; Wiedenheft et al., 2011). Although using a similar enzymatic mechanism, Cas6 preferentially recognizes the non-palindromic single stranded (ss-) RNA repeats and processes them using a ‘wrapping-mechanism’ (Wang et al., 2011). By contrast, the pre-crRNAs processing in the Subtype II CRISPR-cas systems uses a completely different mechanism involving Csn1 recognition and RNase III cleavage guided by the trans-encoded small RNAs (Deltcheva et al., 2011).
CRISPR interference has not been well studied in the Subtype I-C CRISPR organisms. The molecular function of the three Subtype I-C specific proteins Cas5d, Csd1, and Csd2 has not been reported. In this study, we show that pre-crRNA processing, the key molecular event that initiates the CRISPR interference, is carried out by the Cas5d protein in the Subtype I-C organism B. halodurans C-125. Such enzymatic activity has never been demonstrated for the Cas5 family of proteins, which are widely present in several CRISPR subtypes, therefore warrants more careful investigation. Our results show that Cas5d recognizes both the base of the pre-crRNA stemloop and the 3′ single stranded (ss)-RNA sequence, and cleaves the substrate in a metal-independent manner. The crystal structure of Cas5d reveals a ferredoxin-based architecture, with structural features and a cleavage site distinct from other known pre-crRNA processing factors. Mutagenesis indicates that residues Y46, K116 and H117 in Cas5d play critical roles in endoribonuclease activity, likely forming a catalytic triad. Furthermore, we show that after pre-crRNA cleavage, Cas5d assembles into a ~400-kDa Cascade-like complex together with crRNA and the other two subtype-specific proteins Csd1 and Csd2, suggesting that Cas5d further participates in the crRNA-mediated DNA silencing step. This Subtype I-C Cascade complex has much tighter affinity and greater specificity for the crRNA, a reflection of additional RNA contacts from other components of the Cascade complex.
RESULTS
Subtype I-C/Dvulg Cas5d endoribonucleolyticly cleaves pre-crRNA
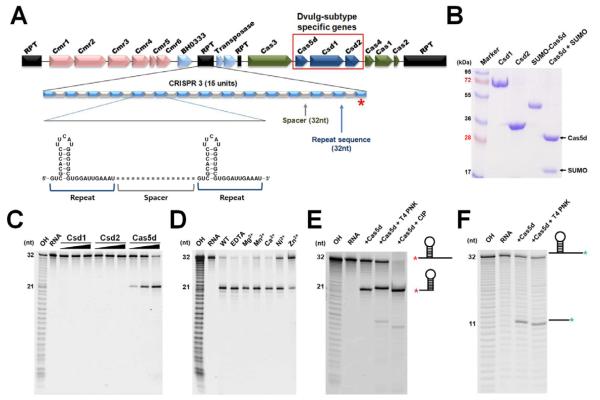
The B. halodurans strain C-125 encodes a Subtype III-B (cmr1-6) and a Subtype I-C CRISPR-Cas system. The latter system contains five CRISPR loci and seven cas genes (cas1-4, csd1-2, and cas5d) (Figure 1A). The highly conserved repeats in the CRISPR loci encode a 7-bp stemloop flanked by the 2- and 11-nucleotide (nt) overhangs at the 5′ and 3′ ends, respectively. To identify the pre-crRNA processing factor in the Subtype I-C CRISPR-Cas system, subtype-specific Cas proteins Csd1, Csd2 and Cas5d from B. halodurans C-125 were purified and incubated with a 32-nt RNA containing the conserved repeat sequence from B. halodurans C-125 CRISPR loci 3 (Figures 1B and 1C). Cas5d, but not Csd1 or Csd2, specifically cleaved the crRNA repeat at the 3′ base of the stemloop structure, between residues G21 and U22. crRNA repeat containing a 2′-deoxy substitution at G21 abolished Cas5d cleavage, confirming the cleavage site (data not shown). No further processing of the crRNA was observed when Csd1 and Csd2 were mixed with Cas5 (Figure 1C). We further verified that Cas5d was capable of processing a pre-crRNA containing multiple repeat-spacer-repeat sequences into the mature crRNAs (Figure S1). The endoribonucleolytic cleavage activity by Cas5d was metal independent, as addition of divalent cations (Mg2+, Mn2+, Ca2+, Ni2+, or Zn2+) or the metal chelator EDTA had no effect on cleavage rate and pattern (Figure 1D). Cas5d Cleavage produced a 2′,3′-cyclic phosphate that could be removed by T4 polynucleotide kinase (PNK) but not calf intestine phosphatase (CIP), and a 5′-OH detected by phosphorylation, in the 5′ and 3′ halves of the crRNA products, respectively (Figure 1E and 1F). This product feature and metal dependency study suggest that, similar to other characterized pre-crRNA processing factors, Cas5d most likely uses a general acid-base catalysis strategy to cleave the RNA.
Figure 1. Cas5d is the pre-crRNA processor in the Subtype I-C/Dvulg CRISPR-Cas system.
(A) Diagram of the CRISPR/Cas system in B. halodurans C-125, which consists of three CRISPR loci, four core cas genes (cas1, cas2, cas3, and cas4), three Subtype I-C subtype specific cas genes (csd1, csd2 and cas5d), and a Subtype III-B cas genes. The organization and representative sequence of the CRISPR 3 locus is illustrated. (B) SDS-PAGE of the purified subtype-specific B. halodurans Csd1, Csd2 and Cas5d proteins. (C) Pre-crRNA processing assay identified processing activity in Cas5d, but not in Csd1 or Csd2. 5′-HEX-labeled CRISPR repeat RNA (0.2 μM) was used as the substrate with a titration of purified Csd1, Csd2, Cas5d, or equimolar amount of three proteins together (0.04, 0.2 and 1.0 μM). Cas5 cleaved between G21 and U22 in the 32-nt repeat sequence as indicated by the alkaline hydrolysis (OH) sequence ladder. (D) Metal ion-independent endoribonuclease activity of Cas5d. Cleavage reaction was carried out in the presence of 10 mM Mg2+, Mn2+, Ca2+, Ni2+, Zn2+, or EDTA. (E) To verify the presence of a 2′,3′-cyclic phosphate in the 5′-half of the crRNA product, 0.2 μM 5′-HEX were incubated with 1 μM Cas5d at 37 °C for 20 min, the reactions were then further incubated with either T4 polynucleotide kinase (PNK) or Calf Intestine Phosphotase (CIP) for an additional 30 min, separated on 18% sequencing gel, and scanned using typhoon 2900. T4 PNK, but not CIP, is capable of removing the 2′,3′-cyclic phosphate, causing the 5′ product to migrate slower. (F) To verify the presence of a 5′-OH in the 3′-half of the cleavage product, the 3′-Fluoresein labeled pre-crRNA repeats were incubated with Cas5d in similar reaction condition. Further incubation with T4 PNK in the presence of ATP added a phosphate to the 5′-OH and caused the 3′ product to migrate slightly faster. See also Figure S1.
Cas5d processes the pre-crRNA in a sequence- and site-specific manner
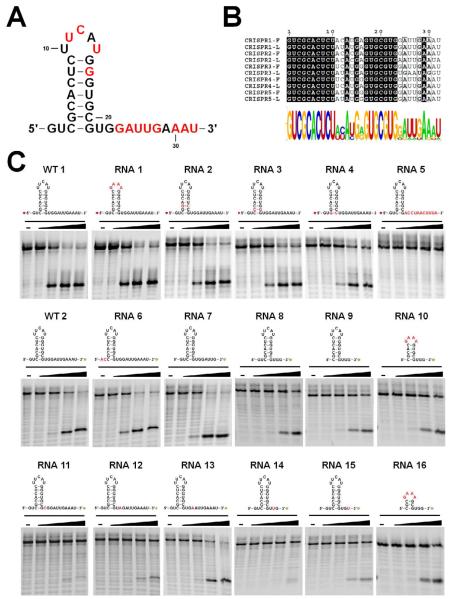
The substrate specificity of Cas5d was deduced by varying the pre-crRNA repeat sequence in a systematic fashion. The five CRISPR loci in B. halodurans C-125 converge into a highly conserved repeat sequence, with minor variations at the top of the stemloop and at the distal 3′ end (5′-GUCGCACUCUUCAUGGGUGCGUGGAUUGAAAU-3′; bold letters indicate variable sequence). This suggests that Cas5d likely processes all pre-crRNA transcripts from the five CRISPR loci (Figures 2A and 2B). A total of sixteen 5′ or 3′ fluorescently labeled RNA substrates were subjected to the cleavage assay by the Cas5d protein (Figures 2C and Table S1). Four titrations of Cas5d were used for each substrate to quantify the relative activity of Cas5d. Our results showed that recognition was weak to the distal end of the pre-crRNA stemloop, as replacing the UUCAU pentaloop with a stable GAAA tetraloop (RNA1 in Figure 2C). The base pairs at the base of the stemloop were systematically swapped (C3-G21, G4-C20, and C5-G19 to G3-C21, C4-G20 and G5-C19, respectively in RNA2, 3 and 4, Figure 2C), and a moderate inhibitory effect was observed as the swapping approached closer toward the cleavage site (RNA4>RNA3>RNA2). This suggests that there is stronger sequence-specific recognition towards the base of the stemloop. Interestingly, our data also revealed that while deleting the 5′ GU overhang had little effect on Cas5d activity (RNA 6 in Figure 2C), there existed strong recognition to the 3′ overhang. Flipping it (from U22 to U32) to its complementary sequence completely abolished the Cas5d processing (RNA 5 in Figure 2C). Further deletion (RNA 7, 8, 9, 10) and point mutation (RNA 11-13) mapping revealed that the recognition focused mostly on the tri-nucleotide sequence U22G23G24 immediately following the cleavage site (Figure 2C). U22C, G23A, and G23U substitutions almost completely abolished Cas5d’s activity (RNA 11, 12, and 14 in Figure 2C), and G24A and G24U had a moderate effect (RNA 13 and 15 in Figure 2C). These results are consistent with the CRISPR repeat conservation (Figure 2B). Taken together, Cas5d was able to efficiently process the minimum crRNA repeat substrate containing a 3-bp GAAA stemloop followed by a 3′ overhang as short as 3-nt (RNA 16 in Figure 2C). Since Cas5d is expected to tolerate the sequence variations among all B. halodurans CRISPR loci, we consider it to be the only pre-crRNA processor in B. halodurans. The strong recognition to the 3′ overhang region of the CRISPR repeats of Cas5d is similar to the behavior of Subtype I-E Cse3, but different from that of Subtype I-F Csy4.
Figure 2. Substrate specificity in Cas5d probed using synthetic pre-crRNA constructs.
(A) Secondary structure of the CRISPR repeats in the pre-crRNA. Consensus and variable sequences are in black and red, respectively. (B) Conservation among the B. halodurans CRISPR RNA repeats shown in the form of sequence alignment [top, a tabulation of the first (F) and last (L) repeat in each CRISPR locus] and sequence logo (bottom). (C) Cas5d activity on pre-crRNAs containing sequence substitutions. 0.2 μM chemically synthesized RNAs with either 5′-HEX (red asterisk) or 3′-fluorescein (green asterisk) label were incubated at 25 °C for 20 minutes with increasing concentration of Cas5d (left to right: 0, 1, 2, 3 and 5 μM). Cleavage efficiency was analyzed using a 15% urea-PAGE gel, annotated with the sequence and secondary structure of each RNA. Base substitutions are highlighted in red. Detailed pre-crRNA processing activities are documented in Table S1.
Cas5d exhibits ferredoxin-based architecture
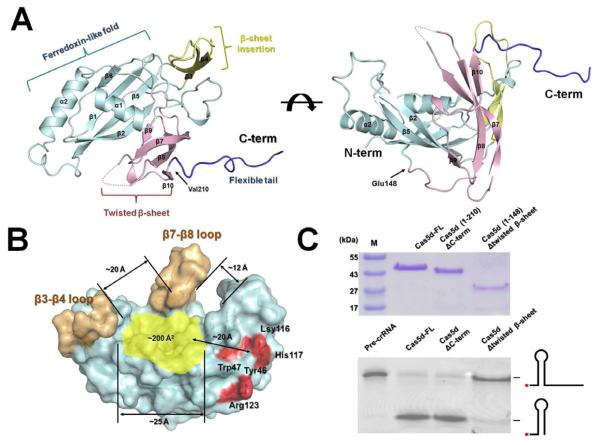
To gain deeper insight into the pre-crRNA cleavage mechanism, the crystal structure of the B. halodurans Cas5d protein was determined. Since the poor solubility of this protein hampered crystallization efforts, we purified Cas5d as an N-terminal SUMO fusion to high concentration, and carried out in situ proteolysis in the crystallization drops by including the SUMO-protease at a 1:100 molar ratio. The resulting Cas5d crystals diffracted X-ray to 1.7 Å, and allowed structure determination by single-wavelength anomalous dispersion (SAD) method from Se-methionine derivatized proteins (Figure 3A and Table 1). Two Cas5d molecules were observed in the asymmetric unit of the C2 space group (Figure S2); they agreed with each other with an r.m.s. deviation of 0.23 Å for Cα atoms. Since Cas5d behaved as a monomer as judged by size exclusion chromatography and dynamic light scattering measurement (data not shown), we hereby only describe the structure of one Cas5d molecule in the asymmetric unit. The electron density map of the Cas5d structure was well defined for most of the residues except the β7-β8 loop (Asn172-Leu182), and the C-terminus (Val209-Glu236).
Figure 3. Crystal structure of Csd5d.
(A) Cas5d consists of an N-terminal ferredoxin domain (β1- β6; cyan) with an additional β-sheet insertion (β3-β4; yellow). This insertion and a C-terminal twisted β-sheet (β7-β10; pink) form a “wall” fencing the ferredoxin domain (see the side view to the right). An unstructured C-terminal tail interacts with the ferredoxin fold of an adjacent molecule. (B) Surface representation of Cas5d highlighting the putative pre-crRNA binding pocket (in yellow). The “walls” by the β-sheet insertions were colored in gold, and the catalytic triad in red. (C) Top: purity of the Cas5d deletion mutants after purification. Bottom: Deletion mapping showing that the twisted β-sheet is required for the pre-crRNA cleavage activity of Cas5d, whereas the C-terminal tail is not. 0.2 μM of proteins and 0.2 μM of 5′-Hex labeled pre-crRNA were incubated with Cas5d proteins for 20 min at 25°C. See also Figure S2.
Table 1.
Data collection and refinement statistics of B. halodurans Cas5d
| Cas5d | Native | Se-Met |
|---|---|---|
| Data collection statistics | ||
| Beamline | CHESS A1 | CHESS A1 |
| Wavelength | 0.9770 | 0.9770 |
| Space group | C2 | C2 |
| Unit cell parameters (Å) | a = 86.50, b = 46.68 and c = 129.03 β =104.00 |
a = 86.61, b = 46.64 and c =129.20 β = 104.70 |
| Resolution (Å) | 20.0-1.70 (1.73-1.70) | 50.0-2.00 (2.03-2.00) |
| Completeness (%) | 91.8 (76.7) | 96.6 (89.9) |
| Redundancy | 3.1 (1.9) | 4.3 (2.7) |
| I/α(I) | 23.85 (2.79) | 20.40 (3.55) |
| Rmerge (%)a | 6.4 (20.5) | 11.6 (25.9) |
| Refinement statistics | ||
| Resolution (Å) | 20.0-1.70 | |
| Rwork/Rfree(%)b | 15.65/22.01 | |
| B-factor (Averaged) | ||
| Protein | 24.18 | |
| Water | 35.61 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.019 | |
| Bond angles (°) | 1.685 | |
| Ramachandran plot (%)c | ||
| Most favored | 100 |
Highest resolution shell is shown in parentheses.
Rmerge = ∑h ∑i∣ I(h,ii) − < I (h)> ∣ /∑h ∑iI (h,ii) ,where I (h,i) is the intensity of the i th measurement of reflection h and < I (h) is the mean value of I(h,i) for all i measurements.
Rwork = ∑∥Fobs∣ − ∣Fcalc∥ / ∑∣Fobs∣, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively. Rfree was calculated as Rwork using a randomly selected subset (5.1%) of unique reflections not used for structure refinement.
Categories were defined by Molprobity.
Crystal structure of Cas5d revealed the presence of an N-terminal ferredoxin-like domain (Met1-Cys147) despite the lack of detectable homology at the primary sequence level. The topology of this domain (βαββββαβ) differs from other known ferredoxin folds (βαββαβ) by the insertion of a small lateral β-sheet of β3-β4 (Figures 3A and S2). The α1 helix (Tyr35-Ile45) packs against the β-sheet in the ferredoxin-like domain through extensive hydrophobic interactions. This helix connects to β1 and β2 through two flexible loops (Asp12-Tyr35 and Ile45-Thr50) (Figure S2). The α2 helix packs at the hydrophobic edge of β1 through hydrophobic interactions (Val5, Phe7, Ile45 and Val144 in ferredoxin fold; Leu121 and Leu125 in α2 helix) (Figure S2). A twisted β-sheet (β7-β10, Lys160-Pro208) packs at the opposite edge of the ferredoxin-like domain. The continuous antiparellel β-sheet in the ferredoxin domain generates a positively charged shallow groove of 13 × 15 Å2, forming a putative RNA binding pocket barricaded by the twisted β-sheet and other upward flexible loops (see discussion) (Figure 3B). Deletion of the twisted β-sheet (Lys160-Pro208) significantly reduced but did not completely eliminate the endoribonuclease activity (Figure 3C, lane 4), suggesting that this region facilitates the binding of the pre-crRNA, but probably does not contribute directly to the catalysis. The C-terminus of Cas5d (Val209-Glu236), required for the solubility of the SUMO-Cas5d protein, adopts an extended conformation to contact the neighboring molecule (Figure S2). This unstructured loop is not conserved among the Cas5d proteins, and deletion of this region did not affect cleavage activity (Figure 3C, lane 3). The twisted β-sheet (β7-β10) and the β3-β4 loop display more conformation dynamics, with significantly higher temperature B factors (~58 Å2) than the core ferredoxin domain (~21 Å2) (Figure S2).
Identification of the catalytic center of Cas5d
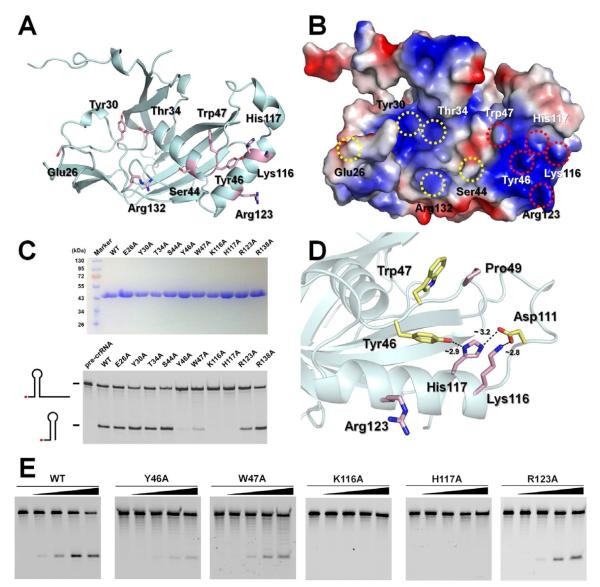
The shallow positively charged groove inside the ferredoxin domain of Cas5d is suggestive of the pre-crRNA binding site. Structure-guided mutagenesis was carried out to identify key RNA-binding and catalysis residues in Cas5d. Sequence alignment of the Cas5d family (sequence identity typically varied between 30 to 50%) from 10 different organisms identified 23 highly conserved residues (Figures S3). An alanine scan was carried out for ten surface residues (E26, Y30, T34, S44, Y46, W47, K116, H117, R123 and R138) located in or near the putative pre-crRNA binding groove (Figure 4A). Five of them (Y46A, W47A, K116A, H117A and R123A) greatly reduced the cleavage activity of Cas5d (Figure 4C). Further quantification revealed that Y46A, W47A, and R123A mutants reduced the cleavage activity to ~5, ~3 and ~10% of the wild-type level, respectively, and K116A and H117A mutations almost completely inactivated the enzyme (Figures 4D and 4E). These catalytically critical residues are clustered at a positive patch immediately outside the putative RNA-binding groove (Figure 4B). The close arrangement of Y46, K116, and H117, and the network of hydrogen bonds among them and D111, are suggestive of a catalytic triad that cleave the pre-crRNA through a general acid-base mechanism (Figure 4D). This is consistent with the metal-dependency study and the chemical structure of the cleavage products (Figure 1). Adjacent to the catalytic center, the vertically positioned W47 side chain stacks in front of the absolutely conserved P49 residue. We speculate that this residue stacks near the base of the pre-crRNA stemloop, serving to position the 3′ overhang near the catalytic center (see discussion). The R123 residue in the α2-helix is separated from the Y46-K116A-H117 center by ~9 Å, and may be involved in recognizing the sugar-phosphate backbone or the nucleotide base directly.
Figure 4. Identification of catalytic center in Cas5d via alanine scanning.
(A) Cartoon and (B) electrostatic surface representation of Cas5d highlighting the conserved surface residues targeted for alanine substitutions. (C) Top: purity of the Cas5d mutants after purification. Bottom: Cleavage activity of the Cas5d mutants. 0.2 μM wild-type or mutant Cas5d protein was incubated with 0.2 μM 5′-HEX labeled pre-crRNA repeat sequence for 20 min at 25°C. Reaction products were phenol-extracted and a nalyzed on urea-PAGE. Y46A, W47A, K116A, H117A and R123A mutants showed reduced pre-crRNA processing activity to various extent. (D) Putative catalytic triad in Cas5d. Tyr46, Lys116 and His117 residues form a charge relay network mediated by the hydrogen bonding network. The nearby Trp47 forms conserved Π-Π stacking with P49 residue. It is conceivable that Trp47 further stacks near the bottom of the pre-crRNA stemloop, positioning the 3′-tail to the catalytic triad. The conserved Arg123 residue is ~9 Å from the triad, it may move closer towards the catalytic center upon RNA binding. (E) Quantification of the mutational effect using Cas5d titrations. 5′-HEX-labeled CRISPR repeat RNA (0.2 μM) was used as the substrate with a titration of the purified mutant Cas5d proteins (0, 0.1, 0.2, 0.4 and 1.0 μM). See also Figure S3.
Cas5d assembles into a Cascade-like complex after pre-crRNA processing
A series of experiments were carried out to investigate the function of the Subtype I-C Cas5d protein after pre-crRNA processing reaction. Specifically, whether Cas5d, together with other Subtype I-C specific proteins Csd1 and Csd2, form in a multi-subunit protein complex as found in Subtype I-E (Ecoli) and Subtype I-F (Ypest) (denoted as Cascade in the Subtype I-E) (Brouns et al., 2008; Jore et al., 2011; Wiedenheft et al., 2011). Cas5d alone had rather weak affinity with the 5′ and 3′ halves of the pre-crRNA cleavage products as shown by the electrophoretic mobility shift assay (EMSA; Figure S4). This behavior is similar to that of Cas6, but differs from other processing factors such as Cse3 and Csy4 (Gesner et al., 2011; Jore et al., 2011; Wang et al., 2011; Wiedenheft et al., 2011). Mixing of B. halodurans crRNA, Cas5d, Csd1, and Csd2 proteins only led to small-sized and heterogeneous RNA-protein complexes as shown by size-exclusion chromatography (SEC, Figure S4). However, coexpression of CRISPR RNA together with the Cas5d, Csd1, and Csd2 proteins in E. coli BL21(DE3) cells led to the formation of a stable ~400-kDa complex as shown on SEC (Figures 5A and S4), which contained roughly two copies of Cas5d, six copies of Csd2, one copy of Csd1, and the processed crRNA (Figures 5B and 5C). Interestingly, besides much higher affinity for the crRNA, this Cascade-like complex appeared to have higher specificity for the repeat region in the crRNA. While changing the loop sequence in the crRNA repeat region had little effect on pre-crRNA processing by Cas5d (Figure 2C), the same change completely disrupted the formation of the Cascade-like complex (data not shown). This increased specificity is likely due to the presence of additional contacts to the repeat region of the crRNA from either Csd1 or Csd2 protein, which enables the Cascade-like complex to interact with the crRNA in the cell lysate with high affinity and specificity.
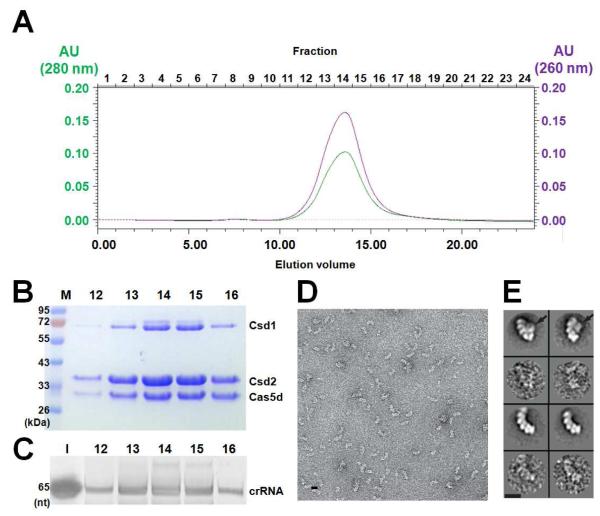
Figure 5. Reconstruction of Subtype I-C/Dvulg Cascade complex.
(A) Superose 6 SEC elution profile of the B. halodurans Cascade complex reconstituted from coexpressing Csd1, Csd2, Cas5d, and crRNA in E. coli. The molecular weight is ~400 kDa as compared to the MW standards. Analysis of the fractions in panel (A) was done using (B) Coomassie blue-stained SDS-PAGE gel or (C) SYBR-GOLD-stained denaturing gel. Fraction numbers are consistent with those shown in panel (A). (D) Negative staining electron micrograph of the B. halodurans Cascade. (E) Two major species were identified from the 2D classification (odd rows, corresponding raw images were shown underneath in even rows). The smaller complex (bottom two rows) likely lacked the Csd1 subunit (indicated by the arrows on the top two rows). Scale bars correspond to 10 nm. See also Figure S4.
Preliminary negative staining EM analysis confirmed the presence of a large macromolecular assembly in the SEC-purified Cascade sample (Figure 5D). Subsequent two-dimensional classification of the single particles revealed the presence of two major species in the electron micrograph, both adopting an elongated arched shape and containing multiple subunits (Figure 5E). The smaller species presumably lacked the Csd1 protein, as this protein had a tendency to dissociate from the complex during purification.
Cas5d, together with Csd1 and Csd2, complements Cas-deficient E. coli to silence plasmid DNA in vivo
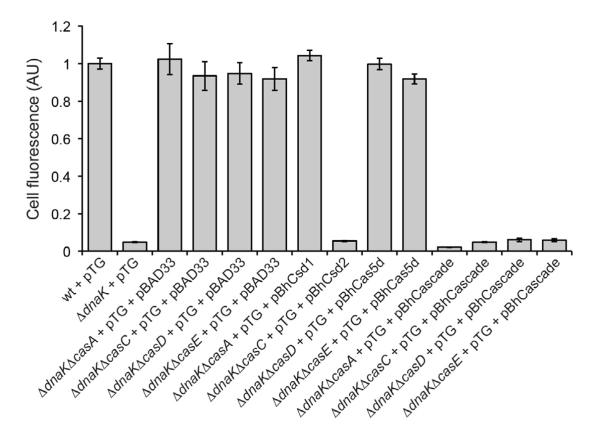
To test the in vivo activity of Cas5d and the newly identified Dvulg-Cascade complex, we utilized a previously established fluorescence-based genetic reporter of CRISPR-Cas interference in E. coli (Perez-Rodriguez et al., 2011). In this earlier study, we observed that GFP targeted to the twin-arginine translocation (Tat) pathway induced the CRISPR-Cas system in E. coli cells lacking the molecular chaperone DnaK. Activation of this CRIPSR-Cas response was dependent on the BaeRS two-component signaling pathway, which is typically involved in cell envelope stress responses. As a result, plasmid DNA encoding the Tat-targeted GFP reporter, namely ssTorA-GFP, was silenced and GFP fluorescence was abolished. This silencing phenotype required each Cas protein (i.e., Cascade and Cas1-3) and the genomic CRISPR region adjacent to the Cas operon; deletion of any of these components abolished the plasmid silencing. For example, silencing-competent DdnaK cells expressing ssTorA-GFP were virtually non-fluorescent but when the gene encoding one of the Cas proteins, such as CasA (Cse1), CasB (Cse2), CasC (Cse4), CasD (Cas5e), or CasE (Cse3) was deleted, silencing activity was abolished and cells became highly fluorescent (Figure 6). Using this genetic system, we attempted to complement the different E. coli Cas mutants with the functional or sequence homolog from B. halodurans, namely Csd1 for CasA, Csd2 for CasC, and Cas5d for CseD and CasE. Only the B. halodurans Csd2 enzyme was able to restore silencing activity to the corresponding CasC mutant E. coli strain (Figure 6). Interestingly, simultaneous coexpression of Cas5d, Csd1, and Csd2 resulted in a strong silencing phenotype in all of the mutant strain backgrounds (Figure 6), consistent with our biochemical reconstitution data showing that these proteins form a Cascade-like assembly. The E. coli pre-crRNA repeats bear certain degree of similarity with the B. halodurans pre-crRNA, especially the critical first three nucleotides in the 3′ overhang region, which may allow the B. halodurans Cas5d to process the E. coli pre-crRNA to a low degree. Taken together, these results suggest that the Subtype I-C Cascade complex formed in vivo is functionally active, using extrachromosomal DNA as the target.
Fig. 6. Cas-dependent silencing of ssTorA-GFP in E. coli.
Cellular fluorescence of wild-type (wt) E. coli BW25113, BW25113 ΔdnaK, and isogenic cas mutant strains (e.g., BW25113 ΔdnaKΔcasE) expressing ssTorA-GFP from plasmid pTG. Complementation of silencing activity was assayed using pBhCsd1, pBhCsd2, pBhCas5d or pBhCascade (Cas5d/Csd1/Csd2) and compared to the empty vector (pBAD33) control. Data is the average of three replicate experiments and error is reported as the standard error of the mean.
DISCUSSION
Among the classified subtype-specific Cas proteins, the Cas5 family are found in Subtypes I-A (also known as Apern subtype: Cas5a), Subtype I-B (Tneap-Hmari: Cas5t-Cas5h), Subtype I-C (Dvulg: Cas5d) and Subtype I-E (Ecoli; Cas5e or CasD) (Haft et al., 2005; Makarova et al., 2011b). Proteins in this family contain a highly homologous N-terminal region and a divergent C-terminal sequence (Haft et al., 2005). The E. coli Cas5e (CasD) is a component of the crRNA-presenting Cascade complex, positioned near the 5′-handle of the crRNA (Brouns et al., 2008; Jore et al., 2011; Perez-Rodriguez et al., 2011; Wiedenheft et al., 2011). The Subtype I-A cas5a (from Sulfolobus solfataricus) was suggested to be responsible for nucleation and/or stabilization of the helical oligomeric structure of Csa2 (Lintner et al., 2011). In contrast, we show in this study that the Subtype I-C Cas5d protein from B. halodurans possesses endoribonuclease activity to process the pre-crRNA into unit length, a function never reported for Cas5 proteins. Moreover, we show that Cas5d recognizes both the stemloop structure and the 3′ overhang in the CRISPR repeat. Recognition of the latter part, which becomes the 5′ handle after the cleavage reaction, appears to be a hallmark of the Subtype I-C CRISPR-Cas system that distinguishes Cas5d from other pre-crRNA processing factors. Taken together, Cas5d appears to be a bifunctional enzyme that performs the function of both Cse3 (CasE) in processing the pre-crRNA and Cas5e (CasD) in binding to the crRNA 5′ handle.
Despite the lack of sequence homology, structure comparison reveals similarity between Cas5d and other pre-crRNA endoribonucleases (Carte et al., 2008; Ebihara et al., 2006; Gesner et al., 2011; Haurwitz et al., 2010; Sashital et al., 2011; Wang et al., 2011). Superimposition of the core ferredoxin domain in Cas5d with those in Cse3 (CasE) (PDB code: 2Y8W), Cas6 (3I4H) and Csy4 (2XLK) resulted in fairly large Cα r.m.s. deviations of 2.7, 3.4, and 4.2 Å, respectively (Figure S2). Unique structural features, however, exist peripheral to the core ferredoxin domain. For example, Cse3 and Cas6 contain a duplicated ferredoxin domain, and Csy4 has a distal Arg-rich loop that locks into place upon RNA binding. Similarly, Cas5d contains a small twisted β-sheet (β7-β10) following the ferredoxin domain, which we show is important for pre-crRNA binding (Figure 3).
Interaction between pre-crRNA and Cas5d is confined to a small region and appears to be transient. The concentrated Cas5d-RNA complex dissociates on the SEC chromatography and on EMSA (data not shown). This character hampered our effort to obtain a pre-crRNA-Cas5d complex structure from extensive co-crystallization and soaking experiments using chemically modified pre-crRNAs (or fragments) and/or catalytic mutant Cas5d proteins. Only fragmented extra densities suggestive of short ss-RNA backbones were observed near the catalytic triad, and at a cleft between the twisted β-sheet and the conserved W46 residues were sometimes observed from co-crystallization experiments (Figure S2). These density features did not allow reliable reconstruction of a pre-crRNA-Cas5d model. In contrast, we show that the Cas5d-containing Subtype I-C Cascade has much higher affinity for crRNA and there is hint of additional contacts towards the distal loop region of the crRNA repeat that is ignored by Cas5d alone, suggesting that other components of the Cascade may contribute to the selective pre-crRNA processing inside the cell. This also points to the possibility that processing of pre-crRNA by Cas5d and formation of the Subtype I-C Cascade may be spatially and temporally coupled.
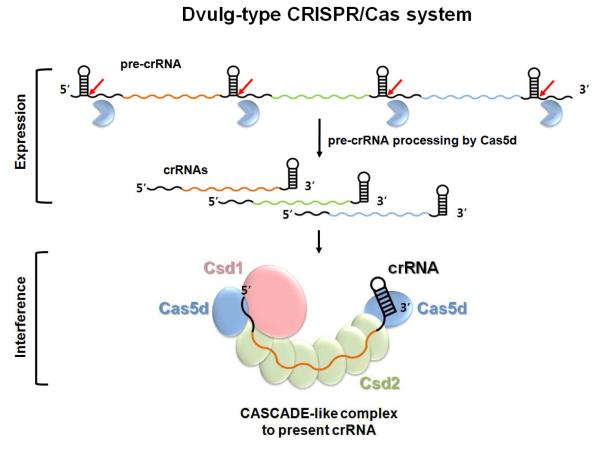
One interesting observation is the existence of several different subtypes among CRISPR-Cas systems (Haft et al., 2005; Makarova et al., 2011b). Each subtype encodes a group of subtype-specific cas genes. To what extent are the CRISPR interference mechanisms conserved among different subtypes? A recent study clustered all Cas3 containing CRISPR subtypes into a large Type I CRISPR-Cas system (Makarova et al., 2011b). Evidence suggests that the subtype-specific genes in two Type I CRISPR subtypes assemble around the mature crRNA into a multi-subunit protein complex (exemplified by the Cascade in the Subtype I-E/ Ecoli) (Brouns et al., 2008; Jore et al., 2011; Wiedenheft et al., 2011). This complex is speculated to promote the pairing of the crRNA with the target strand in a DNA-duplex and present the resulting R-loop structure for ss-DNA degradation by Cas3 (Jore et al., 2011; Wiedenheft et al., 2011). Here we demonstrated that in Subtype I-C/Dvulg, Cas5d assembles into a stable crRNA-presenting complex when coexpressed with the pre-crRNA, and the other two subtype-specific proteins Csd1 and Csd2. Our reconstitution experiment and the preliminary EM analysis pointed to both similarity and differences between the Subtype I-C/Dvulg Cascade and Subtype I-E/Ecoli Cascade. The apparent multiple copies of Csd2 in the B. halodurans Cascade suggest that Csd2 likely serves the equivalent function of Cse4 (CasC) in binding along the crRNA to display it in an extended conformation. The apparent single copy of Csd1 in B. halodurans Cascade, which was speculated to be a function homolog of E. coli Cse1 (CasA) (Makarova et al., 2011a; Makarova et al., 2006), may serve the equivalent function of CasA in assisting the binding of the crRNA 5′ handle. The fact that Csd1 was unable to complement the E. coli CasA mutant likely stems from the low sequence and structure similarities between these proteins, which may prevent hetero-assembly of Csd1 in E. coli Cascade. The B. halodurans Cascade appears to contain two copies of the Cas5d protein, and we speculate that they play the equivalent functions of CasE and CasD in E. coli Cascade by assembling at the opposite ends of the B. halodurans Cascade complex, where one binds to the crRNA 5′ handle and the other to the 3′ handle (Figure 7). Further mechanistic and structural details of the Subtype I-C B. halodurans Cascade complex await more detailed examination. Assuming that this model stands (Figure 7), our work supports the notion that a common molecular pathway is utilized by all Type I CRISPR-Cas systems to target double-stranded (ds)-DNA for degradation. In Subtype I-C/Dvulg, CRISPR interference involves three molecular events, formation of the crRNA-presenting Cascade complex, scanning the ds-DNA substrate and R-loop formation by Cascade, and Cas3-assisted degradation of ss-DNA around the R-loop region (Jore et al., 2011; Wiedenheft et al., 2011).
Figure 7. Mechanistic model for crRNA-mediated DNA silencing in Subtype I-C/Dvulg CRISPR-Cas system.
The pre-crRNA transcribed from the CRISPR loci is processed by Cas5d into mature crRNAs, each containing a spacer sequence between the 5′ and 3′ handles. Subsequently, Subtype I-C/Dvulg Cascade is formed from Cas5d, Csd1, and Csd2 proteins to present the crRNA in an extended conformation, initiating the process to invade and pair with the complementary ds-DNA, followed by ds-DNA degradation by Cas3. The stoichiometry and the location of each component were envisioned based on data generated here combined with knowledge from the E. coli Cascade (Brouns et al., 2008; Jore et al., 2011; Wiedenheft et al., 2011).
EXPERIMENTAL PROCEDURES
Protein expression and purification
Csd1 (UniProt Accession No.: Q9KFY2), Csd2 (Q9KFY1) and Cas5d (Q9KFY3) from B. halodurans were cloned into a modified pSUMO vector and expressed in E. coli BL21 star cells. Protein expression was induced by the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 12 h at 18°C. The cells were harvested by centrifugation, and lysed by sonication in buffer A (50 mM Tris-HCl, pH 7.5, 200 mM NaCl and 0.2 mM phenylmethylsulfonyl fluoride (PMSF). In the case of Csd1 and Csd2, soluble proteins were first purified using Ni-NTA resin as an N-terminal SUMO-fusion (Qiagen), followed by the incubation with SUMO protease to remove the SUMO tag overnight during dialysis to a low salt buffer. After a second pass through the Ni-NTA resin to remove the cleaved SUMO tag, the flow-through was further purified by Mono Q column (GE Healthcare), followed by Superdex 200 10/300 (GE Healthcare) into a buffer containing 10 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol and 2 mM DTT. Cas5d precipitated heavily after removing the SUMO tag, therefore the tag was not removed until crystallization using the in situ proteolysis method. Purification of the Se-methionine derivatized -Cas5d and Cas5d mutants were essentially the same as for the wild-type Cas5d. The purified proteins were concentrated to 20-30 mg/ml and stored at −80°C.
Preparation of the CRISPR RNAs
Pre-crRNAs containing the repeat sequence, with or without residue substitutions and deletions, were chemically synthesized with a HEX or fluorescein label at either the 5′- or 3′-end (from Sigma). They were dissolved in 10 mM HEPES, pH 7.5 and 50 mM NaCl, refolded by heating to 75°C for 5 min, and flash-cooled on ice. The pre-cr RNA containing multiple repeat-spacer units were transcribed from a pUC19 vector containing a T7 transcription cassette encoding the B. halodurans CRISPR3 locus. The run-off transcription and RNA purification were carried out as described (Ke and Doudna, 2004). Secondary structures of pre-crRNA were calculated using the MFOLD program (Zuker, 2003).
Pre-crRNA cleavage of Cas5d
All pre-crRNA processing reactions were performed at 25°C for 20 min. The 5′-HEX-labeled or 3′-fluorescein-labeled pre-crRNA repeats at 0.2-1.0 μM were incubated with Cas5d (0.2-10μM) in 20 mM HEPES pH 7.5, 100 mM KCl, 0.02% (v/v) Triton-X100, 10% (v/v) glycerol and 2 mM DTT. Metal-dependent nuclease activity was measured in the presence of 2.5 mM divalent cations (Mg2+, Mn2+, Ca2+, Zn2+, or Cu2+) or EDTA. Cleavage products were analyzed on 15% (w/v) sequencing gels after phenol extraction. The fluorescent signals were recorded and analyzed using a Typhoon 9400 scanner (GE Healthcare).
Crystallization, data collection and structure determination
Both native and Se-methionine derivatized Cas5d proteins were crystallized using the in situ proteolysis method, by mixing the SUMO-Cas5d fusion protein with the SUMO protease at a 100:1 ratio, in a buffer containing 0.1 M MES pH 6.0 and 0.8 M ammonium sulfate. The crystals were equilibrated in a cryo-protectant buffer containing reservoir buffer plus 30% (v/v) ethylene glycol. The Se-Single Anomalous Dispersion (SAD) data sets were collected at the A1 beamline in MacCHESS (Cornell High Energy Synchrotron Source) and 24ID beamlines at Advanced Photon Source. The datasets were indexed and processed using the program HKL2000 (Otwinowski and Minor, 1997). Phasing of Cas5d was carried out using the programs SOLVE (Terwilliger and Berendzen, 1999) and PHENIX AutoBuild (Terwilliger et al., 2008). The model building and refinement was initially done using the CNS program (Brunger et al., 1998), then using the COOT program (Emsley and Cowtan, 2004). Structure refinement was conducted using the Refmac5 program from the CCP4 suite (Winn et al., 2003). Final model structure was refined using the phenix.refine (Adams et al., 2010). All figures were generated using PyMOL (www.pymol.org).
Preparation of deletion and mutant Cas5d
Deletion (M1-E148 and M1-V210) and point mutants (E26A, Y30A, T34A, S44A, Y46A, W47A, K116A, H117A, R123A and R138A) of Cas5d were constructed using a modified version of the Phusion site-directed mutagenesis method (New England Biolabs). The expected mutations were confirmed by DNA sequencing. A similar purification procedure was followed to obtain pure Cas5d mutant proteins.
Reconstruction of Subtype I-C/Dvulg Cascade-like complex
The B. halodurans genomic fragment containing cas5d, csd1, and csd2 genes was PCR-amplified and cloned into a Kan-resistant pET expression vectors with a His6-tag on the N-terminus of Cas5d and a Strep-II tag on the C-terminus of Csd2. An artificial CRISPR locus containing eight identical B. halodurans CRISPR-repeat-spacer-repeat units were cloned into an Amp-resistant pUC19 vector, under the control of the T7 transcription promoter and terminator (Figure S10). These two plasmids were co-transformed into BL21(DE3) star cells for coexpression of pre-crRNA with Cas proteins. The harvested cells were lysed by sonication in buffer A (50mM Tris-HCl, pH 7.5, 200 mM NaCl, 2 mM β-mercaptoethanol and 0.2 mM PMSF). The complex was purified using Ni-NTA affinity, and eluted using buffer A plus 300 mM imidazole. Eluted complex was immediately dialyzed into a buffer containing 10 mM Tris-HCl, pH 7.5, 50 mM NaCl and 2 mM β-mercaptoethanol at 4°C for 2 hr, concentrated and further purified on Superose 6 10/300 (GE Healthcare) equilibrated in a buffer containing 10 mM HEPES, pH 7.5, 50 mM KCl and 2 mM DTT. The molecular weight of the complex was estimated by comparing with SEC sizing standards. The stoichiometry of the eluted complex was analyzed by quantifying the intensity of the Coomassie stained protein bands on 12% SDS-PAGE gels. CRISPR RNA from the eluted complex was analyzed using a 10% denaturing urea gel, after phenol extraction. The CRISPR RNA was stained and visualized using SYBR-GOLD and Typhoon 9400 scanner (GE Healthcare).
Negative staining electron microscopy and image processing
Aliquots of the pooled SEC peak fractions containing the B. halodurans Cascade was thawed on ice and diluted to ~50 nM in a buffer containing 10mM HEPES pH 7.5, 20mM KCl and 2mM DTT. Immediately after dilution, 5 μl of the protein solution was negatively stained with 2% uranyl-formate solution on a holy carbon grid covered with a thin layer of carbon (Liu and Wang, 2011). The grid was examined under an FEI Tecnai-12 electron microscope operated at 120kV acceleration voltage. 30 micrographs were taken at a nominal magnification of 52,000 on Gatan Ultrascan4000 camera at a defocus of -0.6 micrometer. About 10,000 particle images were boxed out from these micrographs with a pixel size of 4.28 Å. Reference-free two dimensional alignment and classification were performed in IMAGIC-5 program (van Heel et al., 1996) through ten iterations of multi-variant statistical analysis and multi-reference alignment.
Genetic complementation of Cas-mediated silencing activity in E. coli
The gene encoding B. halodurans Cas5d was cloned between SacI and XmaI of pBAD33 (Guzman et al., 1995), creating plasmid pBhCas5d. A similar strategy was used to create pBhCsd1 and pBhCsd2. The operon encoding Cas5d, Csd1, and Csd2 was amplified from the B. halodurans genome using Phusion DNA polymerase (New England Biolabs) and cloned in pBAD33 between restriction sites SacI and XmaI, creating plasmid pBhCascade. Plasmid sequences were confirmed by sequencing. Plasmids were transformed in E. coli BW25113 ΔdnaK or isogenic mutants strains lacking cse1 (BW25113 ΔdnaKΔcse1), cse2 (BW25113 ΔdnaKΔcse2), cse3 (BW25113 ΔdnaKΔcse3) and cas5e (BW25113 ΔdnaKΔcas5e), all of which harbored plasmid pTG, which encodes the ssTorA-GFP reporter in vector pTrc99A (Perez-Rodriguez et al., 2011). Overnight cultures were subcultured 1:100 in fresh medium and induced at mid-log phase with 1 mM IPTG and 0.2% arabinose. Following incubation at 37°C for 4-6 h, fluorescent measurements were taken using a FACSCalibur flow cytometer (Becton Dickinson). All experiments were performed in triplicate.
Supplementary Material
Highlights.
▶ Identification of Cas5d as the pre-crRNA processor in Subtype I-C/Dvulg CRISPR-Cas system.
▶ Revealing the substrate specificity of in Cas5d endoribonuclease.
▶ Determination of Cas5d crystal structure, the first for Cas5 family of proteins.
▶ Identification of the catalytic triad in Cas5d through mutagenesis.
▶ Cas5d assembles into a Cascade-like complex with Csd1, Csd2 and crRNA.
ACKNOWLEDGMENTS
We thank the beamline staff at Macromolecular Diffraction at the beamline A1 at MACCHESS at the Cornell High Energy Synchrotron Source for assistance in data collection. This work is supported in part by the National Institutes of Health Grants GM-086766 and GM-059604 (to A.K), and by the National Research Foundation of Korea Grant from the Korean Ministry of Education, Science and Technology NRF-2010-357-C00106 (to K. H. N).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSSION NUMBERS Coordinates and structure factor have been deposited in the Protein Data Bank with accession number 4F3M.
SUPPLEMENTAL INFORMATION Supplemental information includes ten figures and one table can be found with this article online at doi:XXXX.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas System and Its Role in Phage-Bacteria Interactions. Annual Review of Microbiology, Vol. 2010;64:2010–64. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- Ebihara A, Yao M, Masui R, Tanaka I, Yokoyama S, Kuramitsu S. Crystal structure of hypothetical protein TTHB192 from Thermus thermophilus HB8 reveals a new protein family with an RNA recognition motif-like domain. Protein Science. 2006;15:1494–1499. doi: 10.1110/ps.062131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the Immune System of Bacteria and Archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ. The CRISPR System: Small RNA-Guided Defense in Bacteria and Archaea. Molecular Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke A, Doudna JA. Crystallization of RNA and RNA-protein complexes. Methods. 2004;34:408–414. doi: 10.1016/j.ymeth.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copie V, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang HW. Single particle electron microscopy reconstruction of the exosome complex using the random conical tilt method. Journal of visualized experiments : JoVE. 2011 doi: 10.3791/2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011a;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011b;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nature Reviews Genetics. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Perez-Rodriguez R, Haitjema C, Huang Q, Nam KH, Bernardis S, Ke A, DeLisa MP. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol Microbiol. 2011;79:584–599. doi: 10.1111/j.1365-2958.2010.07482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–687. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. Journal of structural biology. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.