Abstract
The SMK box (SAM-III) translational riboswitches were identified in S-adenosyl-L-methionine (SAM) synthetase metK genes in members of the Lactobacillales. This riboswitch switches between two alternative conformations in response to the intracellular SAM concentration and controls metK expression at the level of translation initiation. We previously reported the crystal structure of the SAM-bound SMK box riboswitch. In this study we combined SHAPE chemical probing with mutagenesis to probe the ligand-induced conformational switching mechanism. We revealed that while the majority of the apo SMK box RNA molecules exist in an alternatively base paired (ON) conformation, a subset of them pre-organize into a SAM-bound-like (READY) conformation, which upon SAM exposure is selectively stabilized into the SAM-bound (OFF) conformation through an induced-fit mechanism. Mutagenesis showed that the ON state is only slightly more stable than the READY state, as several single-nucleotide substitutions in a hypervariable region outside the SAM-binding core can alter the folding landscape to favor the READY state. Such SMK variants display a “constitutively-OFF” behavior both in vitro and in vivo. Time-resolved and temperature-dependent SHAPE analyses revealed adaptation of the SMK box RNA to its mesothermal working environment. The latter analysis revealed that the SAM-bound SMK box RNA follows a two-step folding/unfolding process.
Introduction
Riboswitches are cis-acting non-coding RNAs that regulate gene expression by switching their conformation in response to changes in metal ion or metabolite concentration, environmental temperature, or a specific uncharged tRNA 1; 2; 3. Riboswitches regulate a significant number of gene families involved in metabolism, and are especially prevalent in Gram-positive bacteria including a number of important pathogens 2; 4; 5. Canonical riboswitches contain a ligand-sensing aptamer domain and a downstream expression platform. Upon binding to its cognitive ligand, the metabolite-sensing aptamer domain of the riboswitch switches its conformation and induces a structural change at the expression platform, which affects expression of the gene encoded downstream of the riboswitch, usually at the level of either transcription attenuation or translation initiation.
The S-adenosyl-L-methionine (S AM)-responsive riboswitches are arguably the most prevalent families of metabolite-binding riboswitches 6; 7; 8; 9; 10; 11; 12; 13l ; 14; 15; 16. Crystal structures are available for the S box (SAM-I) 17; 18, SAM-II 19, and SMK box (SAM-III) 20 elements. These riboswitches adopt completely different structures and recognize the SAM molecule in distinct manners. They, however, share a common mechanism to enforce the requirement of a positive charge in the sulfonium moiety of SAM through electrostatic interactions.
The SMK box riboswitch inhibits the translation initiation of the SAM synthetase (metK) gene in bacteria in the Lactobacillales through sequestration of the Shine-Dalgarno (SD) sequence 12; 16; 20. We previously determined the crystal structure of the Enterococcus faecalis SMK box riboswitch and revealed that this RNA folds into a “Y” shaped arrangement that organizes an array of conserved nucleotides around the SAM-binding site at a three-way junction 20. The SD sequence is an integral component of the riboswitch structure and is directly involved in SAM recognition 20. We further determined the crystal structure of the SMK RNA bound to S-adenosyl-L-homocysteine (SAH), a natural SAM analog, to reveal the mechanism used by the SMK to distinguish cognate from near-cognate ligands 20.
In this study, we use chemical probing methods to reveal the ligand-induced conformational switching mechanism in the E. faecalis SMK box riboswitch. We show that the SMK RNA adopts alternatively base-paired “ON” and “OFF” conformations in the apo and SAM-bound states, respectively. Through mutagenesis, we further reveal the presence of an important conformational intermediate (referred to as the “READY” state), where the apo SMK RNA folds into a SAM-bound-like conformation pre-organized to accept the ligand. The subtle but reproducible differences between the READY and OFF states suggest that further conformational changes occur upon SAM entry through an induced-fit mechanism. The energetic difference between the ON and READY conformations is a matter of making or breaking of two to three hydrogen bonds, as a single point mutation in a hypervariable region outside the SAM-binding core selectively stabilized the READY conformation in the apo SMK and resulted in a constitutive-OFF riboswitch behavior in vitro and in vivo. Our results suggest that naturally occurring riboswitches maintain an intricate energetic balance between alternative conformational states in order to fulfill their gene regulatory function, and that variable sequences outside the ligand-sensing core of riboswitches may play important roles in maintaining the conformational balance. We further use temperature-dependent SHAPE analysis to show that the E. faecalis SMK box riboswitch is adapted to the mesothermal environment. Lower temperatures markedly reduce the kinetics of the SAM-induced conformational transition, whereas increasing the temperature causes the disruption of tertiary interactions. These results suggest that the thermodynamic and kinetic behavior of the SMK box riboswitch is fine-tuned towards the preferred host environment of E. faecalis.
Results
Chemical probing analysis of SAM-induced conformational dynamics in SMK RNA
Our previous structural studies provided high-resolution insights into the ligand-recognition mechanism in the SMK box riboswitch 20. Here we employed chemical probing methods to further characterize the ligand-induced conformational dynamics within the SMK RNA. The RNA used in this study contained the entire SMK RNA regulatory element including the aptamer domain (U21-A94), the upstream alternative base pairing elements (A11-G20) and the downstream sequences up to the AUG start codon. Two different chemical probing methods were used in the initial characterizations to cross-validate the results.
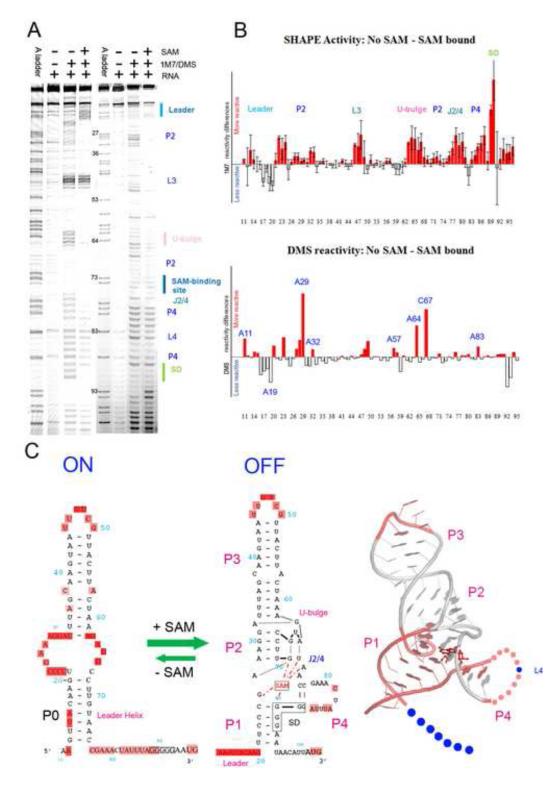
In selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) experiments, the extent of nucleotide 2′-hydroxyl alkylation by 1-methyl-7-nitroisatoic anhydride (1M7) was quantified to detect local conformational flexibility in the RNA structure. A side-by-side comparison of the SHAPE probing pattern of the apo and SAM-bound SMK RNA is shown in Figure 1A. The probing signal for each nucleotide was then quantified and normalized against the background (Figure 1A, lane 2) to generate SHAPE reactivity profiles (Figure 1B). Both the apo and SAM-bound SMK displayed distinctive protection patterns, suggesting that SMK adopts distinct structures in these two functional states (Figure 1A, Supplemental Figure 1). The SHAPE reactivity profile of the SAM-bound OFF state correlated very well with the reported crystal structure (Supplemental Figure 1, Figure 1C) 20. Specifically, SHAPE reactivity was markedly lower for residues inside the SAM-binding aptamer domain defined by the crystal structure. Within the aptamer domain, residues revealed by the crystal structure to participate in secondary structure formation (P1 – P4) and SAM-induced tertiary structures (G25, A26, G70-A72, and the AUG tri-nucleotide bulge) showed reduced SHAPE reactivity. By contrast, the two hypervariable loops (L3 and L4) in the aptamer domain, which were replaced by stable GAAA tetraloops as a necessary step to obtain high-resolution crystal structure, displayed markedly higher SHAPE reactivity. Interestingly, although the hypervariable 11-nucleotide (nt) L4 loop in the E. faecalis SMK box RNA has the potential to form an AU-rich helical structure, its elevated SHAPE reactivity suggested that such a structure does not form stably under physiological conditions, as illustrated by the elevated SHAPE activities in the L4 region in Figure 1A. This is consistent with the phylogenetic analysis showing that there is no defined structure beyond the short P4 helix, and that the length and sequence content in L4 vary widely among SMK box riboswitches 12.
Figure 1.
SHAPE and DMS probing of the apo and SAM-bound SMK RNA. (A). SHAPE (lanes 2-4) and DMS (lanes 6-8) chemical probing reactions separated by sequencing gel electrophoresis. Structural motifs were marked on the right. Lanes 1and 5, ddTTP sequencing revealing the adenosine positions, which are one-nucleotide higher than the chemical probing lanes. Adjusted base register was indicated next to lane 5. (B). Quantified reactivity profile revealed structural changes upon SAM binding. Upper panel: SHAPE analysis; lower panel: DMS probing analysis. Upward red bars represent protected residues due to SAM-induced structure formation. Grey downward bars represent exposed bases upon SAM-binding. Residue numbers are indicated on the X-axis. Standard deviations were generated from five sets of independent measurements. (C). Left: Secondary structure models of the ON and OFF states based on the chemical probing analysis. Flexible regions in each state are highlighted in pink ovals. The five consecutive Gs in solid black boxes indicate the ribosome binding sites (SD sequence). SAM is shown in red. SAM-mediated tertiary interactions are shown as red dotted lines. Right: SHAPE reactivity differences mapped onto the crystal structure of the SAM-bound SMK box riboswitch 20. Residues in pink are protected upon SAM-binding in SHAPE, in blue are exposed. Leader sequence is presented by dots because it is not included in the crystal structure. The 11-nt L4 loop is also shown in dots because it was converted to a GAAA tetraloop in the crystal structure. It was slightly protected upon SAM-binding because it was constrained from ss-RNA to a loop. The extremely reactive L3 loop also showed slight protection upon SAM-binding presumably due to scaling artifact. In the crystal structure P3 was shortened and L3 was replaced with a GAAA tetraloop.
The SHAPE profile of the apo SMK revealed a predominant ON conformation (Supplemental Figure 1, Figure 1C), which agreed well with the alternatively base paired secondary structure model proposed based on the enzymatic probing assay 12 and confirmed by NMR spectroscopy 21. All major helical structures except P3 in the OFF state underwent reorganization. The 6-bp P1 in the OFF state was replaced by a more stable 8-bp leader duplex (designated P0) between sequences upstream of the 5′ region of P1 and the J3/4 sequences in the ON state; P2 and P4 were completely disrupted, and bases in these regions displayed significantly higher SHAPE reactivity (Figure 1A, B). As a result, the SAM-binding site is completely disrupted in the ON state, and the sequestered SD sequence was accessible to allow translation initiation by the 30S ribosome. The protection of the upstream sequence and the de-protection of the anti-Shine-Dalgarno sequence (ASD) presented a signature SHAPE pattern that distinguishes the ON and OFF conformations.
Dimethylsulfate (DMS) probing was used to validate the alternative base pairing structure model. DMS probes the solvent accessibility of the Watson-Crick face of adenosine (via N1 modification) and cytosine (via N3 modification). Consistent with the SHAPE analysis, DMS revealed that adenosine and cytosine nucleotides involved in Watson-Crick pairing in the ON state (A16-A19 in the leader sequence) were protected only in the absence of SAM, whereas residues such as A27-A29, which only base-pair in the SAM-bound OFF state, were protected only when SAM was present (Figure 1A, lanes 7 and 8, Figure 1B, lower panel). Similarly, A64 and C67 were protected only in the presence of SAM, consistent with their involvement in SAM-induced tertiary structure formation near the “tri-nucleotide bulge” 20.
Overall, the SHAPE and DMS analyses converged in support of the previous functional and structural studies showing that the SMK box riboswitch regulates translation initiation of downstream metK coding sequence through a SAM-dependent conformational switching mechanism 12; 16; 20. We focused on the SHAPE method henceforth for its ability to provide single-nucleotide resolution probing information as well as time-resolved and temperature-dependent details of conformation dynamics 22; 23; 24; 25.
Destabilization of the ON conformation unveils a READY conformation in apo SMK RNA
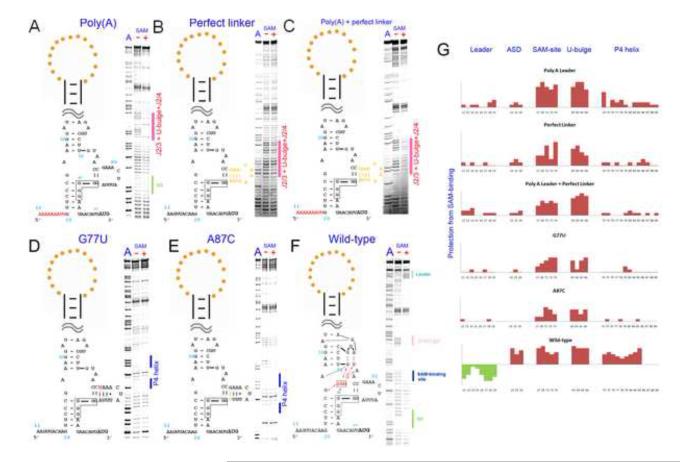
SHAPE analysis revealed that the SAM-binding site was completely disrupted when the RNA was in the ON state in the absence of SAM. An obvious questions then follows: how does the ON state SMK RNA sense the presence of SAM without a pre-organized binding pocket? We hypothesized that while the apo SMK RNA (“ON” state) molecules exist predominantly in the ON state, a fraction of them may occasionally visit a less stable intermediate conformation that contains a pre-organized SAM-binding pocket. SAM then selectively stabilizes this intermediate conformation through ligand-mediated tertiary interactions and drives the conformational equilibrium towards the OFF state. If an intermediate conformation physically exists, our hypothesis predicts that we would be able to reveal it by selectively destabilizing the ON conformation. The most obvious way to destabilize the ON conformation is a poly(A) substitution of the leader sequence (Figure 2A), which selectively disrupts the P0 base helix in the ON state while leaving the OFF state intact. Indeed, SHAPE reactivity of this poly(A) mutant in the absence of SAM clearly resembled that of the OFF state, but not the ON state (Figure 2A). Quantitative comparison of this intermediate conformation (which we termed the “READY” state) with the SAM-bound conformation revealed localized reactivity changes at regions around the SAM-binding pocket, including J2/4, the tri-nucleotide bulge (J3/2), and the SD residues (pink highlights in Figure 2A, and 2G). This suggested that certain SAM-induced tertiary structure features were induced by ligand-binding and therefore were only present in the OFF state, but not in the READY state.
Figure 2.
Mutations outside the SAM-binding domain can shift the conformational equilibrium to favor the READY state. SHAPE was carried out for the SMK mutants containing a poly(A) leader sequence (A), a perfect base-paired linker helix (B) both mutations poly(A)+perfect_linker (C), G77U point mutation (D), A87C point mutation (E) in the absence (−) or presence (+) of SAM. A wild type SMK control was done side-by-side in (F). Quantified reactivity profile revealed structural changes upon SAM binding are shown in (G). Upward red bars represent protected residues due to SAM-induced structure formation. Green downward bars represent exposed bases upon SAM-binding. Residue numbers and corresponding structural motifs are indicated. Note the SD sequence protection and the leader sequence de-protection in the (−) SAM lanes of these mutants as evidence for these mutants adopting READY instead of ON conformation in the absence of SAM. Reproducible minor SHAPE reactivity differences were found for these mutants in (−) and (+) SAM conditions (highlighted by pink bars in A-C) suggesting that the READY conformation is similar, but not identical to the OFF conformation due to the lack of SAM-induced tertiary interactions. L4 protection in (D, and E) due to additional base pairing formation was marked by the blue bars besides the SHAPE pattern.
The ON state is only marginally more stable than the READY state in SMK RNA
Having proven that the READY state could stably exist, we next sought to quantify the energetic differences between the ON, READY, and OFF states. A standard free energy change (ΔGo, 298 K) of 8.6 kcal/mol between the ON and OFF states could be derived from the SAM dissociation constant of 0.5 μM 20. We further used mutagenesis to estimate the free energy difference between the ON and READY states. Here we sought to: (i) demonstrate that the READY state can be selectively stabilized to become the predominant conformation in the apo SMK RNA; and (ii) gauge the energetic difference between the ON and READY states. We focused our attention on the 11-nt hypervariable L4 loop outside the SAM-binding core. This loop was predicted to be poorly structured because it is poorly conserved in length and sequence, and we showed that in the E. faecalis SMK, it was indeed poorly structured and displayed high SHAPE reactivity, even though the L4 sequence has the potential to fold into an AU-rich stemloop (Figure 1A). Mutants promoting base pairing in L4 should not negatively affect the stability of the ON state as L4 is downstream of the ON structure. These mutations may, however, selectively stabilize the READY conformation by reducing its overall conformational entropy. This idea was first tested by a perfect linker mutant, in which L4 was replaced with a perfectly base paired stem capped with a GAAA tetraloop. SHAPE analysis confirmed that similar to the poly(A) mutant, this perfect linker construct folded predominantly into the READY instead of ON conformation in the absence of SAM (Figures 2B, 2G). Not surprisingly, a mutant containing both the poly(A) leader and a stabilized L4 folds into the READY conformation in the absence of SAM as well (Figures 2C, 2G).
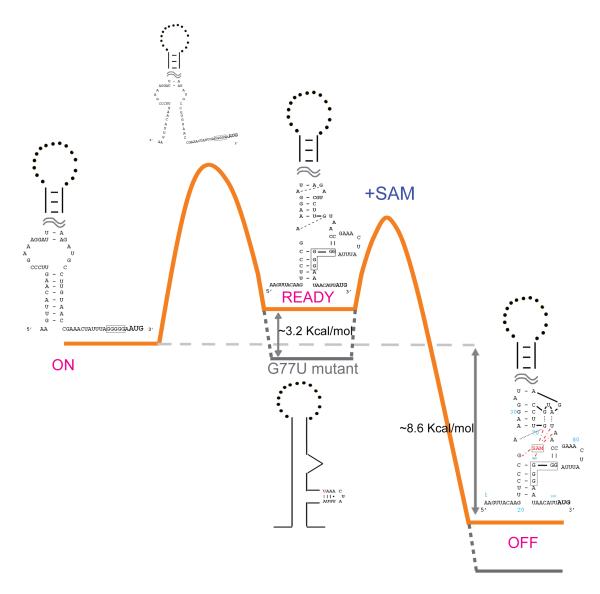
Next, to gauge the energetic difference between ON and READY states, we sought to identify a minimum perturbation that flipped the folding landscape of the apo SMK RNA. The G77•A87 mismatch in L4 was systematically varied in a total of five single-nucleotide substitutions. Among them, G77U and A87C (Figure 2D and 2E, respectively) converted the G•A mismatch to a U-A or G-C pair, respectively. Interestingly, SHAPE analysis revealed not only the intended base pair formation between positions 77 and 87, but also a drastic stabilization of the entire L4 region (Figures 2D, 2E, and 2G). At least three additional base pairs (A78-85, A79-U84, and A80-U83) were evident. In each case, stabilization of L4 was coupled with a predominant READY conformation in the apo SMK RNA. The quantitative comparison of SHAPE reactivity data suggested that the READY conformation was similar to the OFF conformation, with the exception that nucleotides surrounding the SAM-binding site and the U-bulge were more reactive, hence more flexible (Figure 2G). Interestingly, replacing G77•A87 with a G77•U87 wobble pair did not lead to complete stabilization of the L4 loop, and the SMK RNA displayed a mixture of ON and READY conformations in SHAPE analysis (data not shown), suggesting that the free energy difference between these two states is close to zero in this mutant. As a negative control, a G77•G87 mismatch generated by an A87G mutation did not show perturbation of the conformational equilibrium (data not shown). All together, our data are consistent with a free energy diagram (Figure 3) in which the energetic difference between the ON and READY states is quite small. A secondary structure analysis using prediction programs such as RNAfold 26; 27; 28, Kinefold 29; 30, and Mfold 27 suggested that replacement of the G77•A87 mismatch with a U-A or G•U pair stabilizes the P4/L4 stemloop by ~3.2 kcal/mol and ~2.5 kcal/mol, respectively. Such a small gain of free energy is apparently sufficient to make the READY state the most stable conformation in apo SMK.
Figure 3.
Proposed free energy landscape of the SMK riboswitch associating with SAM. Orange trajectory: folding path of wild-type SMK RNA with SAM; Grey dotted trajectory: folding path of the G77U mutant. Proposed ON, READY, and OFF secondary structures and the first transition state are shown. Energetic differences calculated from SAM-binding KD or estimated from mutagenesis are marked.
Selective stabilization of the READY state improves the affinity of SMK for SAM
Based on the free energy diagram, more free energy can be gained when the SMK RNA folds into the OFF state from a pre-organized READY state, instead of the ON state. This should be reflected as tighter SAM binding constants. To test this, we measured the SAM dissociation constants of the wild type SMK RNA as well as four READY mutants, G77U, A87C, poly(A) leader, and poly(A) leader + perfect_linker, using an equilibrium dialysis assay. Results showed that all four mutants displayed tighter SAM-binding KD values than the wild type SMK (Supplemental Figure 2, Table 1). Poly(A)+perfect_linker SMK, which is expected to adopt the most stable READY state, offers the most significant improvement in SAM binding, lowering the KD value by as much as five-fold (from 0.9 μM to 0.2 μM).
Table 1.
SAM-binding KD of the Wild-type and READY mutant SMK RNAs.
| Constructs | KD (μM) |
|---|---|
| Wild-type | 0.9±0.2 |
| G77U | 0.3±0.1 |
| A87C | 0.3±0.2 |
| Poly(A) leader | 0.5±0.1 |
| Poly(A) leader + perfect_linker | 0.2±0.1 |
READY mutants display constitutive-OFF regulatory properties to a various extent in vivo
Since ligand-induced conformational change is coupled with the gene regulatory function of the SMK box riboswitches, mutants predominantly adopting the READY conformation in the absence of SAM are not expected to function as a switch, and therefore may display a “constitutive-OFF” gene regulatory phenotype in vivo. We previously showed that the E. faecalis metK-lacZ translational fusion, expressed in the heterologous organism B. subtilis, was repressed ~5-fold in response to cellular SAM levels that were modulated through the addition or depletion of its precursor methionine to the media of the methionine auxotrophic strain BR15112; 31; 32. Here, we constructed metK-lacZ translational fusions containing point mutations at residues 77 and 87 to determine the effect of these mutations on β-galactosidase expression in vivo (Table 2).
Table 2.
Effect of SMK box riboswitch mutagenesis on the expression of E. faecalis metK-lacZ reporter gene expression in vivo.
β-Galactosidase activities are expressed in Miller units 40.
Ratio of expression during growth in the absence of methionine to expression during growth in the presence of methionine.
β-Galactosidase activity was elevated (110 Miller units, MU) in wild-type cells grown in low [SAM] conditions, and high [SAM] growth condition resulted in 4.2-fold repression (down to 26 MU), consistent with previous results 12. One of the READY mutants, A87C, resulted in significantly tighter repression of theβ-galactosidase activity in both low and high [SAM] conditions (0.10 and 0.059 MU, respectively), and the dynamic response range to SAM was only 1.7-fold. This SMK variant essentially behaved as a “constitutive-OFF” riboswitch presumably because the 30S ribosome was unable to gain access to the SD sequence in either high or low [SAM] conditions. The other READY mutant, G77U that converts the G77•A87 mismatch to a U–A pair, also greatly reduced the β-galactosidase reporter gene activity in cells grown in either low or high [SAM] conditions (8.4 and 0.29 MU, respectively), consistent with a “constitutive-OFF” behavior. A wider dynamic gene regulatory range (~30-fold, 6-times wider than the wild-type), however, was observed for this mutant, revealing an additional level of complexity as the result of the riboswitch-ribosome interactions. Conversely, the G77C substitution that maintained the mismatch in P4 (converting G77•A87 to C77•A87) appeared to largely maintain the wild type response level to SAM since the β-galactosidase activities were 210 and 85 MU for low and high [SAM] growth conditions, respectively; however, the mutation had a small destabilization effect for both states. Taken together, our data confirmed the prediction that the READY SMK mutants display constitutive-OFF regulatory properties in vivo.
SMK conformational transition is steeply temperature-dependent
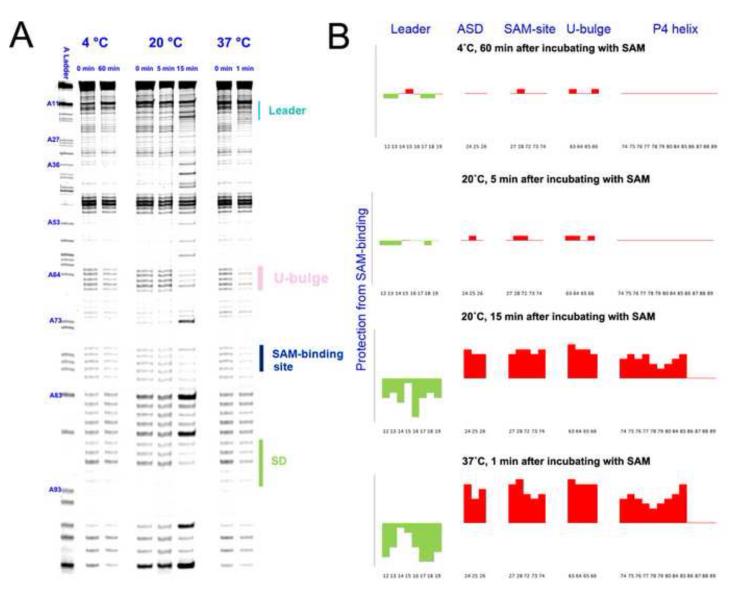
We next investigated the temperature-dependency of the conformational switching kinetics in the SMK box riboswitch using time-resolved SHAPE analysis. The first set of experiments was performed at 37 °C, the physiological temperature inside the gastrointestinal tracts of humans and other mammals where E. faecalis normally grows. At 37 °C, the SMK RNA switched from ON to OFF conformation within one minute upon SAM introduction (Figure 4A right panel, 4B). Since the temporal resolution of SHAPE by 1M7 is around 1 minute, the actual conformational switching kinetics may be much faster 24; 33. Our result suggested that the SMK box riboswitch was capable of down-regulating the translation of its host gene very quickly in response to intracellular SAM level.
Figure 4.
Time-resolved SHAPE analysis revealed a steep temperature dependency in the conformational transition of the SMK box riboswitch. (A) Time-dependent SHAPE resolved on the sequencing gel. (Left) At 4 °C, SMK remained in the ON conformation after 60 minutes of SAM introduction. (Middle) At 20 °C, SMK mostly remained in the ON conformation after 5 minutes of SAM introduction, but switched to mostly OFF conformation after 15 minutes of incubation. (Right) At 37 °C, SMK completed the ON-to-OFF transition within a minute. Residue numbers were labeled next to the sequencing lane on the left. Important structural motifs were marked on the right. Temperatures and time points are marked on the top. (B) Quantified reactivity profile revealed structural changes upon SAM binding. Upward red bars represent protected residues due to SAM-induced structure formation. Green downward bars represent exposed bases upon SAM-binding. Residue numbers and corresponding structural motifs are indicated.
The conformational switching kinetics decreased sharply at lower temperatures. At 20 °C, SHAPE detected very little conformational changes 5 minutes after SAM addition. After 15 minutes, quantitative analysis showed that approximately 80% of the SMK RNAs underwent conformational changes (Figure 4A middle panel, 4B). Here the data suggested that although the kinetics was much slower, most of the SMK RNAs were able to overcome the activation energy barrier at 20 °C, a typical environment temperature outside the parasitic environment of E. faecalis. The significantly faster folding of the SMK RNA seen after 5 minutes also suggested that folding may not follow simple first-order kinetics, although additional data points are required to reveal the details. At 4 °C, the SMK RNA was kinetically trapped in the ON state and failed to overcome the activation energy barrier after 60 minutes of exposure to SAM (Figure 4A left panel, 4B). SMK was similarly trapped kinetically at 10 °C and 15 °C for the experimental duration of 30 minutes (data not shown). The ceiling of the working environment of SMK was later defined by the temperature–dependent SHAPE (shown below), which showed that the SAM-binding pocket melted away at above 47 °C. In summary, there is a strong temperature dependency in the conformational transition of the SMK RNA. The riboswitch appears to have been optimized kinetically and thermodynamically to function between 20 and 47 °C, with an optimum working range at around 37 °C.
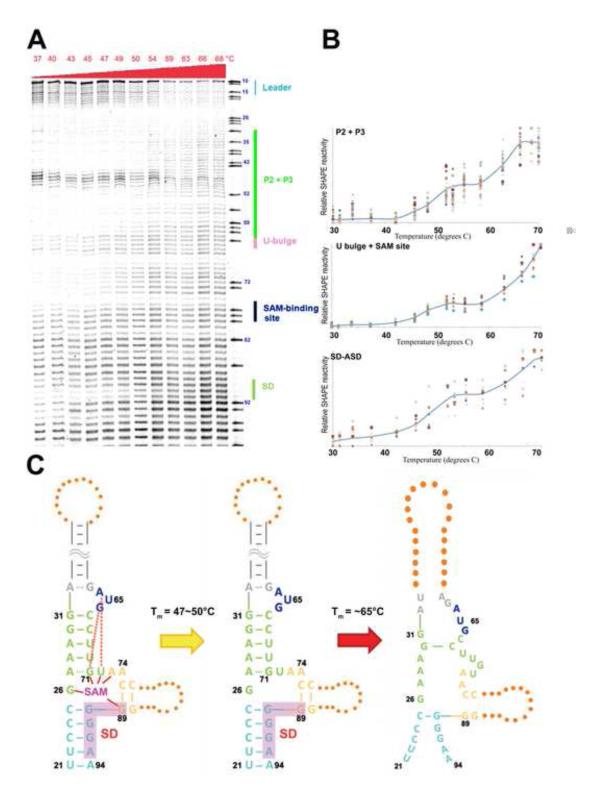
The SMK RNA unfolds in a non-hierarchical, two-step process with increasing temperatures
Recent studies revealed that the E. faecalis SMK box riboswitch is able to switch between ON and OFF conformations multiple times in response to changes in SAM concentration 34. The study suggests that the conformational dynamics of SMK is controlled thermodynamically, as depicted in our free energy diagram (Figure 3). To characterize the SMK unfolding process at higher resolution, we carried out temperature-dependent SHAPE experiments from 37 °C to 70 °C (Figure 5A). Quantitative analysis revealed that residues in the same structural elements follow a similar unfolding behavior (Figure 5B). Interestingly, structural elements such as the P2, P3 U-bulge, SAM-binding pocket, and the SD/ASD helix, followed a double-sigmoidal unfolding curve (Figure 5B). The first structural transition occurred at Tm=47~50 °C. Here the slight increase in SHAPE reactivity in structural elements surrounding the SAM-binding site was consistent with lose of SAM-mediated tertiary interactions (Figure 5C); even though the overall three-way junction architecture remained intact, the SMK RNA was no longer expected to be capable of SAM binding. The second structure transition followed at 65 °C, where the three-way junction architecture collapsed as the secondary structures melted away (Figure 5C).
Figure 5.
Two-step unfolding pathway in SMK revealed by the temperature-dependent SHAPE. (A) SHAPE profile of the SAM-bound SMK aptamer domain incubated at increasing temperature. The adenosine sequencing ladder and RNA structure are shown to the right. (B) Unfolding profile of local RNA structures after quantification. Data are plotted on a linear scale; solid blue line indicates average melting behavior of individual residues in a given local RNA structure. (C) Proposed model of the thermo-unfolding pathway of the OFF state SMK box riboswitch. Ligand-induced tertiary interactions are lost during the first melting transition. Nearly all secondary structures are lost during the second melting transition.
Discussion
Many riboswitches, especially those functioning at the transcription level, appear to have been evolved to switch only once. For example, the Bacillus subtilis yitJ S-box(SAM-I) riboswitch is transcribed into a pre-organized aptamer domain, ready to accept a SAM molecule 35; 36. If SAM is absent for an extended time, however, transcription of the downstream sequence by RNA polymerase (RNAP) triggers the collapse of the aptamer domain to form alternative structures. The presence of SAM, on the other hand, stabilizes the aptamer domain and triggers the downstream terminator hairpin formation, causing the transcribing RNAP to terminate prematurely 6; 7; 8; 9. Since the action of the RNAP is rate-limiting and irreversible, such riboswitches are not under selection to switch multiple times in both directions, and their folding may not have reached the thermodynamic equilibrium in the switching process due to transcription-coupling.
The translational SMK box riboswitch, on the other hand, is capable of regulating the translation initiation of its host gene multiple times by switching back and forth between its ON and OFF conformations in response to changing cellular SAM concentrations 46. Therefore, the functional status of SMK at any given time is governed by the thermodynamic principles. This study established the thermodynamic framework for the SMK box riboswitch. First, we identified a folding intermediate, the READY state, along with the ON-to-OFF conformational transition. The presence of a pre-organized SAM-binding site in the READY state rationalizes the outstanding question about how the apo SMK RNA senses the presence of SAM, and points to an induced-fit mechanism in the READY-to-OFF conformational transition. Equivalent of the READY state was identified independently by NMR spectroscopy recently by deleting the leader sequence to destabilize the ON state 21. The secondary structure of the READY state defined by the NMR study agrees quite well with our model from SHAPE analysis 21. Second, our minimum perturbation experiments showed that the free energy difference between the ON and READY states was less than ~3.2 kcal/mol, estimated from secondary structure prediction programs26; 27; 28 (Figure 3). This converts to a maximum equilibrium constant of 4.5×10−3 between the READY and ON conformations. In other words, at least 0.5% of the apo SMK RNA molecules populate the READY state at any given time, mediating the ON-to-OFF conformational transition. Third, our results showed that maintaining a correct energetic balance between conformational states in SMK is as essential as keeping the required set of ligand-recognitions. Whereas losing key ligand-interactions lead to constitutive-ON phenotypes 20, here we show that SMK variants favoring the READY instead of ON state in the absence of SAM behaved in a constitutive-OFF fashion in vitro and in vivo. Moreover, we showed that such drastic behavior changes in SMK could be caused by single nucleotide substitutions in a hypervariable region outside the ligand-binding core, underlining the importance of such frequently overlooked variable sequence elements in maintaining the overall energetic balance between conformational states.
We further showed that not only is the energetic barrier low between the ON and READY states, but the transition kinetics is also fast. At 37 °C, the ON-to-OFF transition is complete within one minute, suggesting that the SMK RNA has been fine-tuned for rapid translational response. The switching behavior is, however, steeply sensitive to temperature. It becomes too slow to be physiological at temperatures below 15 °C, and the riboswitch starts to lose structural integrity above 47 °C. In a sense, the SMK RNA is not only a ligand-sensing switch, but also a temperature-sensor with different behaviors at different temperatures. It would be interesting to further investigate whether such differential behaviors in SMK may be part of the physiological adaptation to heat and cold stresses in E. faecalis.
Experimental procedures
RNA preparation
The Enterococcus faecalis SMK box RNA and all mutations were constructed to be flanked by a 5′-T7 promoter and 3′-reverse transcription start site using overlapping PCR and inserted into pJET cloning vectors (Invitrogen) for sequence verification. The DNA template for in vitro transcription was amplified in a 2 mL PCR reaction with terminal primers and was directly used in T7 RNAP run-off transcription as described 20; 37. The RNA product was purified and recovered by denaturing polyacrylamide gel electrophoresis (PAGE, 8% polyacrylamide, 8 M urea) as described 37. Eluted RNA was concentrated and buffer exchanged into 0.5X TE using Millipore centrifuge filters with 10 kDa cutoff. The purified RNAs have a final concentration around 50 μM.
DMS/SHAPE probing analysis
RNA was diluted into a final annealing mix containing 300 nM RNA, 111 mM NaCl, 111 mM Hepes pH 8.0, 11.1 mM MgCl2 (or no MgCl2 for -Mg samples). The annealing mix was heated at 65 °C for 5 min, then mixed with SAM to 500 μM final concentration (or appropriate amount of SAH), incubated at 37 °C for three more min, and cooled on ice. SHAPE was carried out as previously described with minor modifications 33; 38. 8 μL of RNA solution was mixed with 1 μL of 1M7 (from a stock solution of 100 mM in anhydrous DMSO) and allowed to react for 10 min at 30 °C or until the solution reaches a deep orange color. The reaction was quenched by the addition of 500 μL precipitation buffer [80% EtOH, 45μM NaCl, 0.45μM EDTA, 2 μl glycoblue (15mg/mL, Ambion, Inc)].
Dimethyl Sulfate (DMS) modification was performed as previously described 19. 10 pmol of annealed RNA mix prepared as described above were added to a total volume of 200 μL of DMS modification buffer (70 mM HEPES-KOH, pH 8.0, 10 mM MgCl2, 270 mM KCl, and an additional 100 μM SAM ) on ice. 1 μL of DMS stock, made from mixing one volume of pure DMS from Sigma with two volumes of ethanol, was added to every 5 pmol (100 μL) of RNA at 30 °C for 5 min, followed by the immediate addition of 50 μl of DMS quenching buffer (1 M Tris-acetate, pH 7.5, 1 M β-mercaptoethanol, 1.5 M NaOAc, 0.1 mM EDTA, and 1 μL glycoblue).
After SHAPE and DMS probing reactions, the modified RNA mix was mixed with 750 μl of −20 °C cold ethanol, stored at −80 °C for 30 min, and spun at 4 °C at 16000 g for 30 min. The resulting blue pellet was recovered and resuspended in 10 μL of 0.5X TE.
The temperature gradient SHAPE procedure was performed in a similar manner as described previously 38; 39; 40. Briefly, 15 μL of the refolded SAM-bound SMK RNA was pre-equilibrated using a gradient PCR machine to the desired temperatures (20 °C, 30.0 °C, 33.8 °C, 43.9 °C, 48.4 °C, 50.0 °C, 52.0 °C, 53.3 °C, 58.7 °C, 58.9 °C, 63.0 °C, 66.4 °C, or 68.6 °C), and subsequently mixed with 1.5 μL 100mM 1M7 to initiate 2′-ribose hydroxyl alkylation. After the reactions had proceeded to completion, which varies between 3 and 15 minutes depending on the half life of 1M7 at different temperatures 22; 39, the reaction was then quenched, and RNA was recovered as described above.
Primer Extension
The reverse transcription and sequencing procedure after SHAPE or DMS probing were completed as described with minor modifications 38. 1 μL of fluorescently labeled DNA primer (HEX or 6-FAM-labelled GAA CCG GAC CGA AGC CCG at 30 μM) was annealed to the 10 μL recovered RNA by heating to 65 °C for 2 min, followed by incubation at 35 °C for 5 min. The sample was mixed with 6 μL reverse transcription buffer [167 mM Tris (pH 8.3), 250 mM KCl, 10 mM MgCl2, 1.67 mM each dNTP], pre-heated to 52 °C for 45 sec, then reverse transcribed by the addition of 1 μL Superscript III enzyme (Invitrogen, 200 units) at 52 °C for 20 min. The reactions were then quenched and the RNA template was digested away by the addition of 0.8 μL of 5M NaOH, followed by heating at 90 °C for 4 min. 29 μL of acid stop mix (4:25 (v/v) mixture of 1 M unbuffered Tris-HCl), and a stop solution (85% formamide, 0.5X TBE, 50 mM EDTA, pH 8.0)] was added to each reaction; the reaction was then incubated at 90 °C for 4 more minutes before being cooled to −20 °C. No tracking dyes were added to the reaction to avoid interference with the fragment analysis.
The same SHAPE and DMS samples were analyzed by PAGE (12% 29:1 acrylamide:bis-acrylamide, 0.5X TBE, 8 M urea). 5 to 10 μL of each sample was loaded per lane for 5 hours at constant power of 55 watts.
SHAPE Data Analysis
Sequencing of the band intensities read from a Typhoon phosphorimager (Molecular Dynamics) was quantified using program SAFA 41. Data analysis was performed in Microsoft Excel. Data points were averaged across three or four separate experiments on separate sequencing gels. A total of 11 bands including base 36-44 (P2 stem), 83 and 87 (flexible linker) that were similarly modified throughout all experiments were used for normalization and scaling. For temperature melting data, intensities were rescaled to a range of 0 to 1. Individual experiments at different temperatures were processed and graphed separately before being compared with each other and averaged to produce the final result to ensure reducibility and consistency.
Equilibrium dialysis
Equilibrium dialysis assays were conducted using a DispoEquilibrium Dialyzer (Harvard Biosciences) in which chambers A and B are separated by a 5,000 MWCO membrane. Each chamber contained 20 μl of either RNA or [3H]SAM. Chamber A contained [3H]SAM (50 μCi) at a concentration of 100 nM in a buffer containing 25 mM Tris-HCl (pH 7.5), 20 mM MgCl2 and 100 mM NaCl. Chamber B contained a variable amount of SMK RNA (from 0.01 to 100 μM) in the same buffer. Equilibrations were performed for at least 2 h at 20 °C while agitated on a shaker. Subsequently, 10 μl was withdrawn from each chamber and quantified using a liquid scintillation counter. Each data point was performed in triplicate. Initial fractional occupancy (Y) vs. [RNA] (X) was fitted to the equation Y=(Bmax*X)/(KD+X) by the program Origin, where Bmax is the maximum binding efficiency. Data was analyzed by the program Origin in order to obtain values for the dissociation constants (KDs). Binding energy was obtained from the equation G =-RTlnKD, where the gas constant R is 1.987 cal/mol K, and the absolute temperature T is 298 K.
E. faecalis metK-lacZ translational fusions in B. subtilis
A pFG328 plasmid encoding an E. faecalis metK-lacZ translational fusion downstream of the B. subtilis GlyQ promoter 12 was used as template for site-directed mutagenesis to generate the G77U, G77C, and A87C mutant constructs following the Stratagene protocol. Transformants were selected on LB agar 42 containing 5 μg/ml chloramphenicol and used for purification of plasmid DNA (Wizard Plus SV Miniprep DNA Purification System, Promega). Mutagenized plasmids were verified by DNA sequencing (GENEWIZ, Inc.) and used to transform B. subtilis strain 307A as previously described 6; 43; 44. Transducing phage was purified by passage through strain ZB449 and subsequently incorporated into strain BR151 for testing of β-galactosidase activity. B. subtilis transductants were plated on tryptose blood agar base medium (TBAB, Difco) containing 5 μg/ml chloramphenicol.
For β-galactosidase assays, cells were grown in Spizizen minimal media 45 supplemented with 50 μg/ml L-methionine (Sigma-Aldrich) to early exponential phase, harvested, and resuspended in Spizizen minimal media with or without methionine. Samples were collected after four hours of growth (shaking at 37 °C) and cell density was determined by measuring the optical density at 595 nm. β-Galactosidase assays were performed after permeabilization of the cells with toluene as previously described 42. Experiments were performed in triplicate.
Supplementary Material
Figure S1. Quantified SHAPE reactivity profile of the SMK RNA in the presence or absence of SAM. Blue bars, profile of the apo SMK RNA; yellow bars, profile of the SAM-bound form. Residue numbers and structure motifs are indicated. Standard deviations were generated from five sets of independent measurements.
Figure S2. Equilibrium dialysis binding curves of the SMK RNA to [3 H]SAM. Chamber A contained 100 nM [3H]SAM (50 μCi). Chamber B contained a variable amount of SMK RNA (seven concentrations from 0.01 to 100 μM). Equilibration between the two chambers was performed for at least 2 h at 20 °C with rotary agitation. Partitioning of the [3H]SAM in each chamber was then measured by scintillation counting. Each data point was measured in triplicates and the standard deviations are shown.
ACKNOWLEDGEMENT
Work in the Ke lab and Henkin lab was supported by Public Health Service grants from NIH (GM086766 and GM63615, respectively). We thank Dr. Kevin Weeks for the generous gift of SHAPE reagent 1M7. The authors declare no financial conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Current Opinion in Structural Biology. 2005;15:342–8. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Grundy FJ, Henkin TM. From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements. Critical Reviews in Biochemistry and Molecular Biology. 2006;41:329–38. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- 3.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends in genetics : TIG. 2004;20:44–50. doi: 10.1016/j.tig.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Winkler WC. Metabolic monitoring by bacterial mRNAs. Arch Microbiol. 2005;183:151–9. doi: 10.1007/s00203-005-0758-9. [DOI] [PubMed] [Google Scholar]
- 5.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annual Review of Microbiology. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 6.Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Molecular microbiology. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 7.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature structural biology. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 8.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5052–6. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, Neph S, Tompa M, Ruzzo WL, Breaker RR. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic acids research. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberg Z, Regulski EE, Hammond MC, Barrick JE, Yao Z, Ruzzo WL, Breaker RR. The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. RNA (New York, N.Y.) 2008;14:822–8. doi: 10.1261/rna.988608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs RT, Grundy FJ, Henkin TM. The SMK box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nature structural & molecular biology. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 13.Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR. Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. RNA (New York, N.Y.) 2008;14:685–95. doi: 10.1261/rna.937308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome biology. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poiata E, Meyer MM, Ames TD, Breaker RR. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA (New York, N.Y.) 2009;15:2046–56. doi: 10.1261/rna.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs RT, Grundy FJ, Henkin TM. S-adenosylmethionine directly inhibits binding of 30S ribosomal subunits to the SMK box translational riboswitch RNA. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4876–4880. doi: 10.1073/pnas.0609956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 18.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–33. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nature Structural & Molecular Biology. 2008;15:177–82. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 20.Lu C, Smith AM, Fuchs RT, Ding F, Rajashankar K, Henkin TM, Ke A. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nature Structural & Molecular Biology. 2008 doi: 10.1038/nsmb.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson RC, Smith AM, Fuchs RT, Kleckner IR, Henkin TM, Foster MP. Tuning Riboswitch Regulation through Conformational Selection. Journal of Molecular Biology. 2011;405:926–38. doi: 10.1016/j.jmb.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA (New York, N.Y.) 2009;15:1314–21. doi: 10.1261/rna.1536209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat Protoc. 2009;4:1413–21. doi: 10.1038/nprot.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gherghe CM, Mortimer SA, Krahn JM, Thompson NL, Weeks KM. Slow conformational dynamics at C2′-endo nucleotides in RNA. J Am Chem Soc. 2008;130:8884–5. doi: 10.1021/ja802691e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–19. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 27.Zuker C, Lodish HF. Repetitive DNA sequences cotranscribed with developmentally regulated Dictyostelium discoideum mRNAs. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:5386–90. doi: 10.1073/pnas.78.9.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuster P, Fontana W, Stadler PF, Hofacker IL. From sequences to shapes and back: a case study in RNA secondary structures. Proc Biol Sci. 1994;255:279–84. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- 29.Xayaphoummine A, Bucher T, Isambert H. Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots. Nucleic Acids Research. 2005;33:W605–10. doi: 10.1093/nar/gki447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xayaphoummine A, Bucher T, Thalmann F, Isambert H. Prediction and statistics of pseudoknots in RNA structures using exactly clustered stochastic simulations. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15310–5. doi: 10.1073/pnas.2536430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. Journal of Bacteriology. 2008;190:823–33. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wabiko H, Ochi K, Nguyen DM, Allen ER, Freese E. Genetic mapping and physiological consequences of metE mutations of Bacillus subtilis. Journal of Bacteriology. 1988;170:2705–10. doi: 10.1128/jb.170.6.2705-2710.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. Journal of the American Chemical Society. 2007;129:4144–5. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 34.Smith AM, Fuchs RT, Grundy FJ, Henkin TM. The SAM-responsive S(MK) box is a reversible riboswitch. Molecular Microbiology. 2010;78:1393–402. doi: 10.1111/j.1365-2958.2010.07410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu C, Ding F, Chowdhury A, Pradhan V, Tomsic J, Holmes WM, Henkin TM, Ke A. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. Journal of Molecular Biology. 2010;404:803–18. doi: 10.1016/j.jmb.2010.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–97. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ke A, Doudna JA. Crystallization of RNA and RNA-protein complexes. Methods (San Diego, Calif.) 2004;34:408–14. doi: 10.1016/j.ymeth.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–6. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 39.Wilkinson KA, Merino EJ, Weeks KM. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNA(Asp) transcripts. Journal of the American Chemical Society. 2005;127:4659–67. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- 40.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) Journal of the American Chemical Society. 2005;127:4223–31. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 41.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA (New York, N.Y.) 2005;11:344–54. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller J. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y. USA: 1972. [Google Scholar]
- 43.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 44.Henkin TM, Chambliss GH. Genetic mapping of a mutation causing an alteration in Bacillus subtilis ribosomal protein S4. Molecular and General Genetics. 1984;193:364–9. doi: 10.1007/BF00330694. [DOI] [PubMed] [Google Scholar]
- 45.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith AM, Fuchs RT, Grundy FJ, Henkin TM. The SAM-responsive S(MK) box is a reversible riboswitch. Mol Microbiol. 2010;78:1393–402. doi: 10.1111/j.1365-2958.2010.07410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Quantified SHAPE reactivity profile of the SMK RNA in the presence or absence of SAM. Blue bars, profile of the apo SMK RNA; yellow bars, profile of the SAM-bound form. Residue numbers and structure motifs are indicated. Standard deviations were generated from five sets of independent measurements.
Figure S2. Equilibrium dialysis binding curves of the SMK RNA to [3 H]SAM. Chamber A contained 100 nM [3H]SAM (50 μCi). Chamber B contained a variable amount of SMK RNA (seven concentrations from 0.01 to 100 μM). Equilibration between the two chambers was performed for at least 2 h at 20 °C with rotary agitation. Partitioning of the [3H]SAM in each chamber was then measured by scintillation counting. Each data point was measured in triplicates and the standard deviations are shown.