Abstract
Structural DNA Nanotechnology uses unusual DNA motifs to build target shapes and arrangements. These unusual motifs are generated by reciprocal exchange of DNA backbones, leading to branched systems with many strands and multiple helical domains. The motifs may be combined by sticky ended cohesion, involving hydrogen bonding or covalent interactions. Other forms of cohesion involve edge-sharing or paranemic interactions of double helices. A large number of individual species have been developed by this approach, including polyhedral catenanes, a variety of single-stranded knots, and Borromean rings. In addition to these static species, DNA-based nanomechanical devices have been produced that are targeted ultimately to lead to nanorobotics. Many of the key goals of structural DNA nanotechnology entail the use of periodic arrays. A variety of 2D DNA arrays have been produced with tunable features, such as patterns and cavities. DNA molecules have be used successfully in DNA-based computation as molecular representations of Wang tiles, whose self-assembly can be programmed to perform a calculation. Four years ago, on the fiftieth anniversary of the double helix, the area appeared to be at the cusp of a truly exciting explosion of applications; this was a correct assessment, and much progress has been made in the intervening period.
Keywords: Holliday junctions, branched DNA, unusual DNA motifs, sticky-ended cohesion, DNA-based computation, DNA objects, DNA polyhedra, DNA nanomechanical devices, 2D DNA arrays, DNA parallelograms, DNA double crossover molecules, DNA triple crossover molecules, paranemic cohesion, edge-sharing, reciprocal exchange, DNA complementarity, DNA supercoiling, DNA architecture, nanoelectronics, DNA crystals, DNA origami, self-replicable objects, autonomous machines, translation devices, nanoparticle organization, polymer scaffolding, algorithmic assembly
Introduction
This is an update of an article written in 2003,1 the fiftieth anniversary of the Watson-Crick2 model for double helical DNA. The impact of this model during the past half-century has been immense. Indeed, the double helix has become a cultural icon of our civilization in much the same way that the pyramids of Egypt, the temples of Greece, the cathedrals of medieval Europe and the great wall of China were icons of previous eras. The simplicity and elegance of the molecule nature evolved to perpetuate and express genetic information has revolutionized genetics, and has had a similar impact in other areas ranging from medicine to forensics. All of these applications are predicated on the complementarity of the two strands of DNA, rooted in the hydrogen bonded base pairing between adenine (A) and thymine (T) and between guanine (G) and cytosine (C). The DNA double helix is inherently a nanoscale object; its diameter is about 20 Å (2 nm) and the separation of the bases is 3.4 Å; the helical periodicity is 10–10.5 nucleotide pairs per turn, or ~3.5 nm per turn. Here, we will discuss making complex materials with nanoscale features from DNA; this pursuit is termed structural DNA nanotechnology. This area began in the early 1980s3, and had made significant progress by 2003; the intervening period has seen remarkable progress, which will be summarized here, along with the basic principles, which are largely the same.
What purposes would be served by producing DNA-based constructs? We expect these systems can be applied to several practical ends: The initial motivating goal for this research is that spatially periodic networks are crystals. If we can build stick-figure crystalline cages on the nanometer scale, they could be used to orient other biological macromolecules as guests inside those cages, thereby rendering their 3D structures amenable to diffraction analysis3. This notion is illustrated in Figure 1a, which shows a DNA 'box' hosting macromolecular guests. Similarly, the same types of crystalline arrays could be used to position and orient components of molecular electronic devices with nanometer-scale precision4. An example of this type of application is shown in Figure 1b. Two DNA branched junctions have pendent from them a nano-wire. When their sticky ends cohere, the nanowires are organized as well. DNA-based nanomechanical devices can lead to a nanometer-scale robotics and to very smart materials, materials that respond to specific stimuli by particular spatial transitions. Structural DNA nanotechnology creates motifs that can be useful for DNA-based computation and for the algorithmic assembly of materials.5 Algorithmic assembly holds great promise for the highly controlled construction of materials with designed features and target sizes in one, two or three dimensions.
Figure 1. Applications of DNA Periodic Arrays.
(a) Biological Macromolecules Organized into a Crystalline Array. A cube-like box motif is shown, with sticky ends protruding from each vertex. Attached to the vertical edges are biological macromolecules that have been aligned to form a crystalline arrangement. The idea is that the boxes are to be organized into a host lattice by sticky ends, thereby arranging the macromolecular guests into a crystalline array, amenable to diffraction analysis. (b) Nanoelectronic Circuit Components Organized by DNA. Two DNA branched junctions are shown, with complementary sticky ends. Pendent from the DNA are molecules that can act like molecular wires. The architectural properties of the DNA are seen to organize the wire-like molecules, with the help of a cation, which forms a molecular synapse.
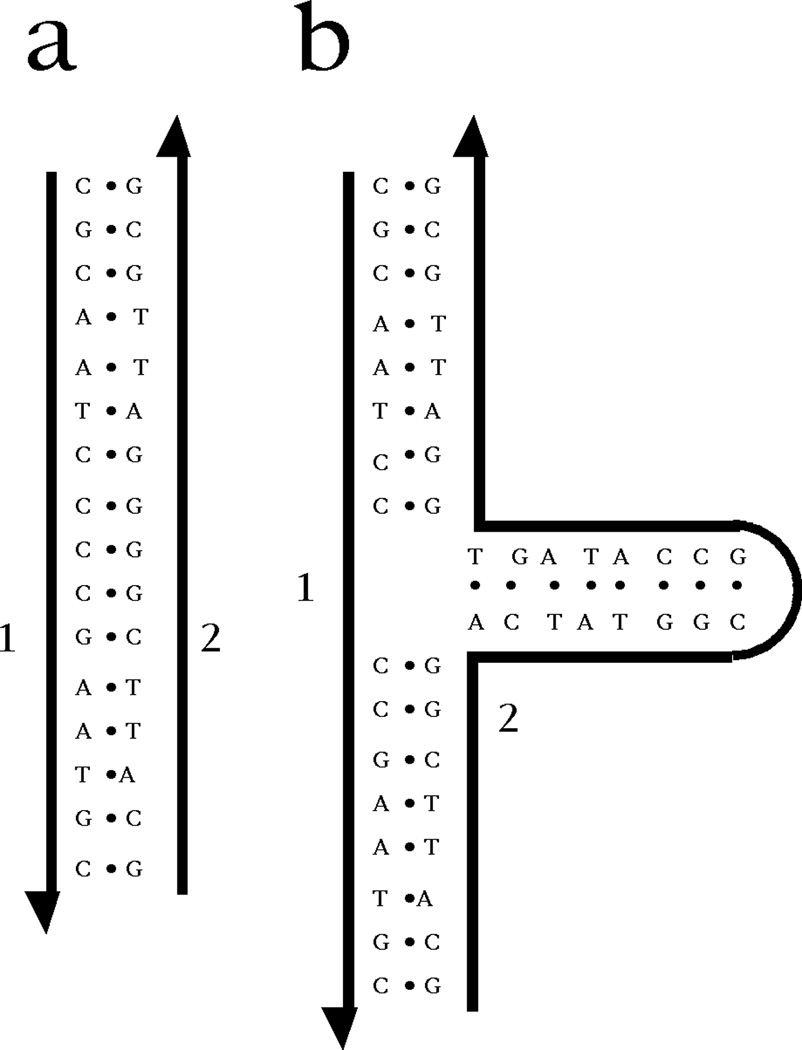
A key, if often unnoticed, feature of the double helix is that its axis is linear, not in the geometric sense of being a straight line, but in the topological sense that it is unbranched. The biological relevance of this fact is that only a linear complement to a DNA strand is well-defined.6 This point is illustrated in Figure 2. Panel (a) illustrates a DNA duplex molecule, where each base of strand 2 is paired to strand 1. By contrast, panel (b) shows a molecule in which strand 2 contains a hairpin that produces a 3-arm branched junction. Every base in strand 1 has a complement in strand 2, just as in (a). However, the complement is not well-defined. The third (horizontal) arm of the junction could have any length, without affecting the complementarity of strand 2 for strand 1. Thus, the replication protocol used by DNA polymerase would not be effective in replicating this structure exactly, even though it would produce complements to the individual strands. Nevertheless, biology does use branched DNA as ephemeral intermediates in the processes of replication, recombination and repair.
Figure 2. Inexact Complementarity in Branched DNA.
On the left is a DNA duplex, labeled 'a'; its strands are numbered 1 and 2. On the right is a 3-arm branched junction, labeled 'b'. Its strands are also labeled 1 and 2, and strand 1 is identical to strand 1 of a. Every nucleotide of strand 1 in b is complemented by a nucleotide in strand 2 of b, but there are many nucleotides in the branch whose sequences are irrelevant to the complementarity between strands 2 and 1. The only well-defined complement to strand 1 is strand 2 of a.
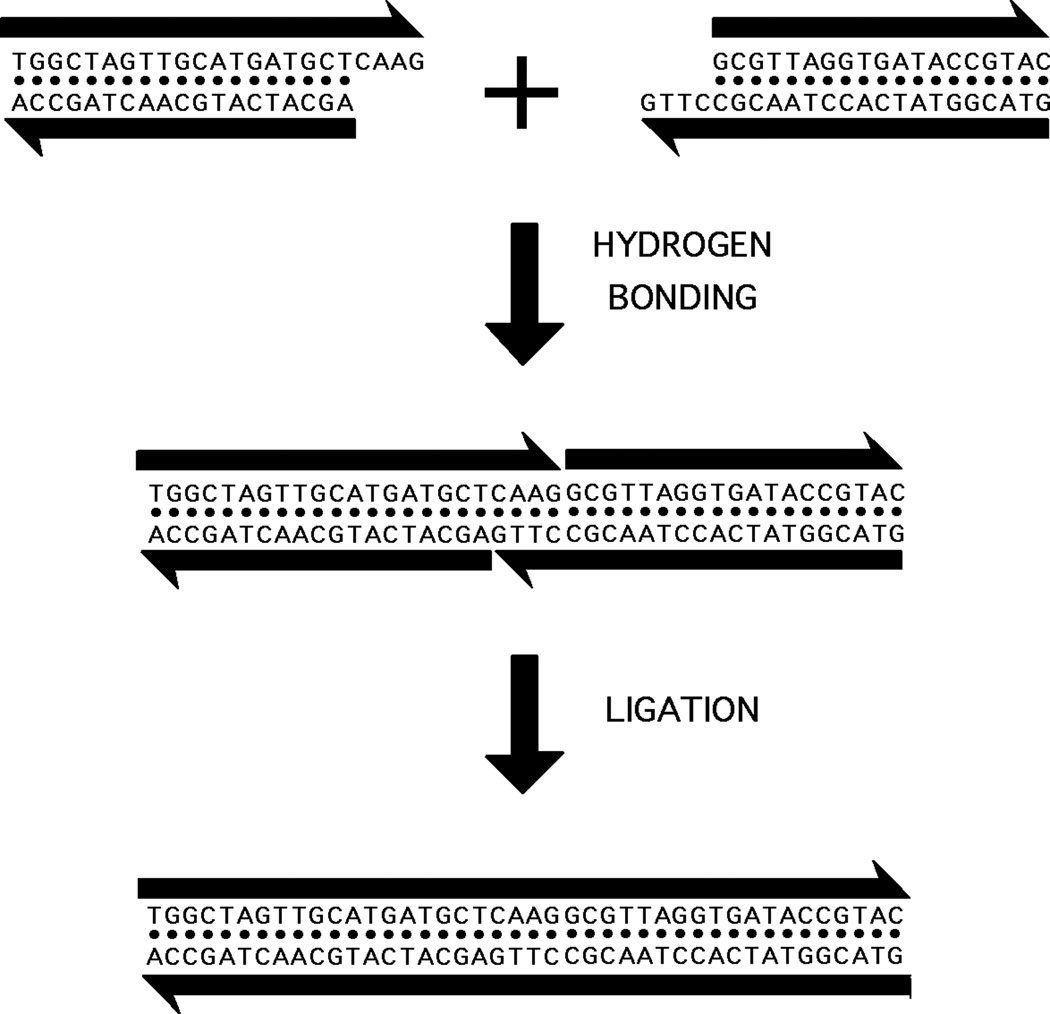
Regardless of the difficulties that branched DNA might present as genetic material, it is extremely valuable for structural DNA nanotechnology, because linear DNA molecules do not lead to inherently interesting arrangements of DNA in two or three dimensions. Genetic engineers have used sticky-ended cohesion and ligation for nearly 35 years to produce linear recombinant DNA molecules.7 These molecules are of value for their sequences and the gene products to which they can lead, but structurally speaking, they consist of long lines or circles, perhaps including some catenanes or knots. However, if one combines sticky-ended cohesion with branched DNA species, it is possible to produce many complex structures and topologies with relatively little effort. Figure 3 illustrates the nature of sticky ended cohesion: Two double helices with complementary single-stranded overhangs will cohere to form a complex under the appropriate solution conditions.
Figure 3. Sticky Ended Cohesion.
Two linear double helical molecules of DNA are shown at the top of the figure. The antiparallel backbones are indicated by the black lines terminating in half-arrows. The half-arrows indicate the 5'-->3' directions of the backbones. The right end of the left molecule and the left end of the right molecule have single-stranded extensions ('sticky ends') that are complementary to each other. The middle portion shows that, under the proper conditions, these bind to each other specifically by hydrogen bonding. The bottom panel shows that they can be ligated to covalency by the proper enzymes and cofactors.
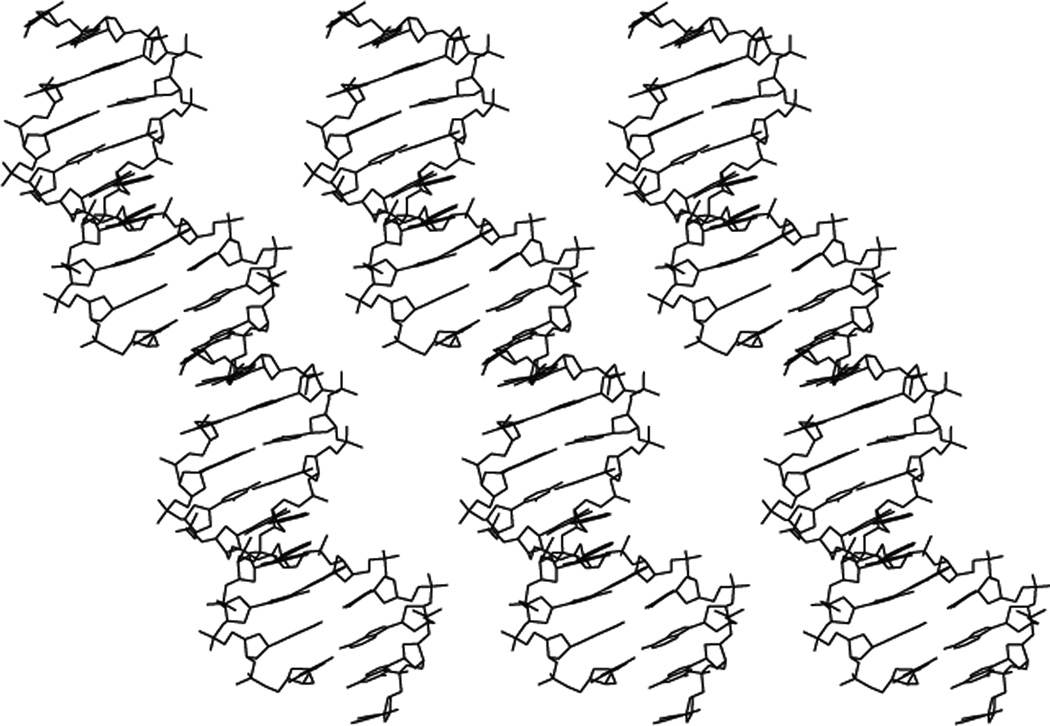
The recognition of one strand of DNA by another strand is common to many types of DNA nanotechnology and to molecular biology. The special feature of structural DNA nanotechnology is that it takes advantage of the well-defined structure at the point where the two helices cohere. Figure 4 shows the crystal structure of a DNA decamer that forms continuous helices within the periodic lattice.8 In contrast to many other crystals of DNA decamers, these decamers are held together by sticky ends at their 5' ends. The key point of the crystal structure is that the 3D conformation of the DNA in the vicinity of the sticky end is the same as the rest of it: It is all B-DNA, the classical structure. Thus, if two DNA molecules are held together by sticky ends, and we know the coordinates of one of them, we know the coordinates of the one to which it is binding by sticky-ended cohesion. This fact is exploited extensively in structural DNA nanotechnology. If one wished to use some other biologically-based recognition system, such as antibody-antigen interactions, it would be possible, but one would have to work out the spatial relationships of the two partners in the binding reaction for every pair, say by a crystal structure. By contrast, the many possible DNA sticky ends (4N for N-base sticky ends) have structures that are virtually identical. Other modes of cohesion are being introduced into structural DNA nanotechnology. These include PX cohesion,9–11 edge-sharing,12 and lateral cohesion.13 However, the detailed structural parameters of these forms of cohesion can only be estimated at this time.It is easy to generate branched DNA molecules by the logic used in biological systems, reciprocal exchange of strands.14 An example of reciprocal exchange is shown in Figure 5a, where a filled strand and an outlined strand trade portions of their strands. Figure 5b illustrates a number of motifs derived in this fashion that have proved to be important in structural DNA nanotechnology. At the left is the Holliday junction (HJ), the biological intermediate in genetic recombination,15 resulting from a single reciprocal exchange between double helices. To its right is the double crossover (DX) molecule,16 an intermediate in meiosis,17 derived by two exchanges between double helices. On the right of the DX molecule is the triple crossover molecule (TX), generated by two reciprocal exchanges between a helix of the DX molecule and another double helix (LaBean et al., 2000).18 The two strands of the double helix have opposite polarities. Exchange can occur between strands of the same polarity or strands of opposite polarity. The HJ, DX and TX molecules shown are all illustrated to have been the result of exchanges between strands of opposite polarity. The PX structure on the right of the TX molecule illustrates crossovers between strands of the same polarity.14 Exchange in the PX motif occurs at every possible point where the strands are juxtaposed. The JX2 variant to the right of the PX molecule lacks two crossovers in the middle; it has been used in a robust nanomechanical device.19 It is important to recognize that for the past decade or so it has been possible to design the shapes of DNA motifs,20 through the use of inherently rigid motifs, including the DX, TX, PX, JX2 6-helix bundle, 8-helix bundle;13,21 the tensegrity triangle,22–24 and deltahedra.10,25
Figure 4. The Crystal Structure of Infinite DNA Helices Held Together by Sticky Ends.
Shown are three DNA double helices that consist of an infinite repeat of a decamer. The decamers are held together by complementary sticky ends, flanked by breaks in the DNA backbone. The key point is that the DNA around the sticky ends is the same as the rest of the DNA; it is all B-DNA, so the structure of any sticky-ended complex is known with high precision.
Figure 5. Motif Generation and Sample Motifs.
(a) Reciprocal Exchange of DNA Backbones. Two strands are shown on the left, one filled, and one unfilled. Following reciprocal exchange, one strand is filled-unfilled, and the other strand is unfilled-filled. (b) Key Motifs in Structural DNA Nanotechnology. On the left is a Holliday junction (HJ), a 4-arm junction that results from a single reciprocal exchange between double helices. To its right is a double crossover (DX) molecule, resulting from a double exchange. To the right of the DX is a triple crossover (TX) molecule, that results from two successive double reciprocal exchanges. The HJ, the DX and the TX molecules all contain exchanges between strands of opposite polarity. To the right of the TX molecule is a paranemic crossover (PX) molecule, where two double helices exchange strands at every possible point where the helices come into proximity. To the right of the PX molecule is a JX2 molecule that lacks two of the crossovers of the PX molecule. The exchanges in the PX and JX2 molecule are between strands of the same polarity.
Figure 6 illustrates the HJ single crossover junction structure in a convenient branched arrangement, much like the intersection two roads. Each of the four double helical arms has been tailed by a sticky end. The arrangement on the right shows four of these junctions assembled to form a quadrilateral by sticky-ended cohesion. In addition to the sticky ends that pair within each edge of the quadrilateral, there are also sticky ends visible on the outside of the structure, so that this arrangement could be extended to yield two-dimensional crystalline arrays. It is important to realize that the parallel-line representation of DNA used in this drawing is only a convenience. The double helical nature of DNA really makes this a 3D system, rather than a 2D system. Thus, in principle, branched DNA molecules tailed in sticky ends can be assembled into 3D objects and lattices.
Figure 6. The Combination of Branched Motifs and Sticky Ends.
At the left is a 4-arm branched junction with sticky ends, labeled X and its complement X', Y and its complement Y'. On the right four such molecules are combined to produce a quadrilateral. The sticky ends on the outside of the quadrilateral are available so that the structure can be extended to form a 2D lattice.
Why use DNA for this purpose? The key reason has already been discussed: Sticky-ended cohesion of DNA is the most predictable intermolecular interaction of any known molecular system, in the senses of affinity, diversity and structure. Furthermore, the advent of structurally robust motifs really makes the system very much like molecular versions of children's rigid construction systems, such as Tinker toys, Lego or Meccano. In addition, convenient automated chemistry exists for the synthesis of DNA molecules of particular sequences,26 both conventional nucleotides and specially derivatized variants, such as biotinylated nucleotides or branched residues. There are numerous enzymes that can be used with DNA: Ligases to join cohesive ends covalently;7 restriction enzymes that can be used both synthetically27 and analytically;28,29 exonucleases for the removal of failure products,28 and topoisomerases that can be used to diagnose topological problems.30 DNA is a relatively stiff molecule, with a persistence length of about 50 nm under standard conditions.31 There is an external code on DNA so that its sequence may be read from the grooves, even though the double helix is intact.32 The functional group density of DNA is very high; thus, it may be derivatized at a density limited only by the nucleotide repeat.33 In addition, the gene therapy and antisense enterprises have produced large numbers of variants on the theme of DNA (e.g., ref. 34). Although it is most convenient to prototype nanoconstructs with DNA, many applications are likely to benefit from the use of variants such as peptide nucleic acids;35 nevertheless, one must be aware that such variants are likely to entail modifications of the structural parameters.36
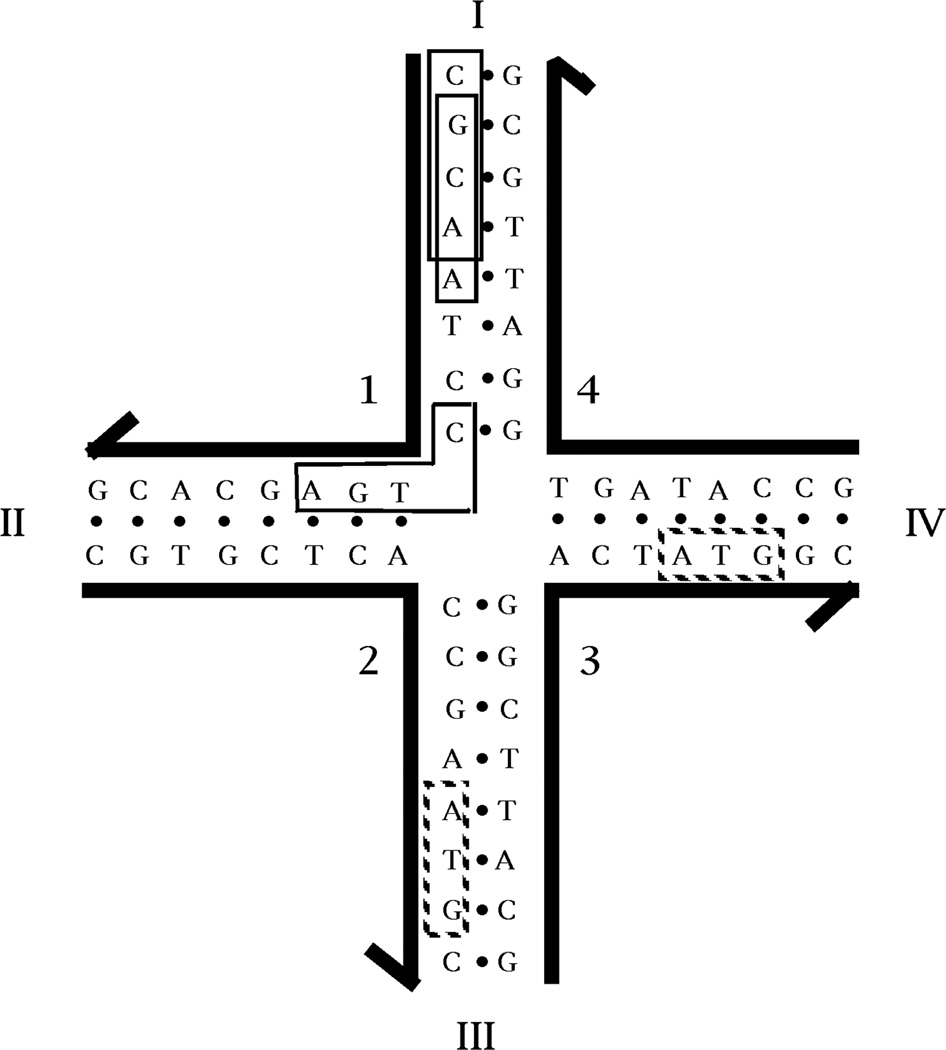
How do we choose sequences for DNA constructs? The key to any successful molecular engineering approach is to design the components of a construction not just so that they are capable of yielding the product, but also to ensure, insofar as possible, that no other product will be competitive with the target. Ultimately, one must estimate the thermodynamics of all possible sequences and select the one most likely to lead to the intended product.37. Empirically, we have found that adequate sequences can be generated by the approximation to this approach called Sequence Symmetry Minimization, which is shown in Figure 7.3,38 This drawing illustrates a 4-arm junction; its arms are each 8 nucleotides long, so each of its strands contains 16 nucleotides. We divide the 16 nucleotides into a series of overlapping elements (13 tetramers in this case). We insist that each of these elements, such as the boxed CGCA and GCAA, be unique. In addition, we also insist that any element that spans a bend (such as the boxed CTGA) not have its simple contiguous Watson-Crick complement (TCAG) anywhere in the sequence. With these constraints, competition with the target octamers can come only from trimers, such as the ATG sequences in the dotted boxes. It should be clear that this approach assumes that double helices are the most favorable structures that DNA can form, and that maximizing double helix formation will lead to the successful formation of branch points. The success of this method relies on the cooperativity of DNA double helix formation.
Figure 7. Sequence Symmetry Minimization Produces a Stable DNA Branched Junction.
The junction shown is composed of four strands of DNA, labeled with Arabic numerals. The 3' end of each strand is indicated by the half-arrows. Each strand is paired with two other strands to form double helical arms; the arms are numbered with Roman numerals. The hydrogen bonded base paring that forms the double helices is indicated by the dots between the bases. The sequence of this junction has been optimized to minimize symmetry and non-Watson-Crick base pairing. Because there is no homologous twofold sequence symmetry flanking the central branch point, this junction cannot undergo the branch migration isomerization reaction. At the upper part of arm I, two of the 52 unique tetrameric elements in this complex are boxed; these are CGCA and GCAA. At the corner of strand 1, the sequence CTGA is boxed. This is one of twelve sequences in the complex (3 on each strand) that span a junction. The complements to each of these 12 sequences are not present. Whereas tetrameric elements have been used to assign the sequence of this molecule, there is redundancy in the molecule amongst trimers, such as the ATG sequences shown in dotted boxes.
DNA Objects
Individual branched junctions have been constructed containing 3 arms,39 4 arms,40 5 arms and 6 arms,41 8 arms and 12 arms.42 A variety of studies have shown that the angles between the arms appear to be flexible.39.43–44 Consequently, the earliest constructions consisted of topological targets, rather than targets with specific 3D geometries. Both the level of control and the proofs of these structures are on the topological level. The connectivity of an object is the number of vertices to which every vertex is bonded via the edges. The first object with a non-trivial connectivity of 3 or greater was a DNA molecule whose helix axes were connected like the edges of a cube.28 The edges consisted of DNA double helices, and the vertices corresponded to the branch points of 3-arm junctions. There were two turns per edge, so each face consisted of a cyclic single strand linked twice to each of its four neighbors, forming a hexacatenane. Every edge of this molecule (shown in Figure 8a) contains a unique restriction site. It was constructed in solution, and proof of synthesis consisted of restricting the final hexacatenane to target subcatenanes.28,45
Figure 8. Ligated products from flexible DNA components.
(a) A Stick Cube and (b) a Stick Truncated Octahedron. The drawings show that each edge of the two figures contains two turns of double helical DNA. The twisting is confined to the central portion of each edge for clarity, but it actually extends from vertex to vertex. Both molecules are drawn as though they were constructed from 3-arm junctions, but the truncated octahedron has been constructed from 4-arm junctions, which has been omitted for clarity. (c-e) Deliberate Knots Constructed from DNA. The signs of the nodes are indicated. (c) A trefoil knot with negative nodes. (d) A figure-8 knot. (e) A trefoil knot with positive nodes. (f) Borromean Rings. Scission of any of the three rings shown results in the unlinking of the other two rings.
In the next development, a truncated octahedron was also constructed from DNA,29 this time using a solid-support based method.27 This molecule (shown in Figure 8b) is also a complex catenane. There are 14 faces to a truncated octahedron, so this molecule is a 14-catenane, with six strands corresponding to square faces and eight strands corresponding to hexagonal faces. Although the truncated octahedron is a 3-connected object, it was constructed with 4-arm junctions (not shown), in the hope that the extra helices could be used to connect the polyhedra into a macromolecular version of zeolite A. Enough material was produced to demonstrate the synthesis, but not to use it as starting material for lattice construction. In the last few years, has built an octahedron using PX cohesion10 and have built tetrahedra.25
In addition to these polyhedral catenanes, a series of strictly topological targets also have been produced. Knots and catenanes are characterized by the set of nodes seen when they are projected into a plane. The half-turn of DNA corresponds to a such node in any topological target. Consequently, it is easy to produce virtually any knot from DNA, with negative nodes being produced by conventional right-handed B-DNA and positive nodes being derived from left-handed Z-DNA.46 Figure 8c illustrates a trefoil knot with negative nodes, and Figure 8e illustrates a trefoil knot with positive nodes. Figure 8d shows a figure-8 knot with two negative nodes and two positive nodes. All of these knots (and an unknotted circle) have been produced from one strand of DNA by changing ligation conditions: The strand contained two regions that could turn into Z-DNA, but with different propensities; at non-Z promoting conditions the knot was negative nodes was produced, at conditions that weakly promote the B-Z transition, the figure-8 knot was produced and at strongly Z-promoting conditions the trefoil with positive nodes was produced.47 Figure 8f illustrates Borromean rings built from the combination of a right-handed B-DNA 3-arm junction connected to a left-handed Z-DNA 3-arm junction.48 Borromean rings have a special property: When any ring in a set of Borromean rings is cut, all the rings fall apart. This is an extremely difficult topology to produce from conventional organometallic systems, but it was relatively easy to make it from DNA. Borromean rings were recently built on the chemical scale by Stoddart and his colleagues.49
DNA Arrays
Being able to produce a series of topological targets, such as the knots, Borromean rings and polyhedral catenanes is of interest, but the greatest value will be derived when a variety of species can be attached to each other, particularly if there is functionality associated with them. The essence of structural DNA nanotechnological goals is to place specific functional species at particular loci, using the architectural properties of DNA. Functionality includes the use of periodic DNA arrays to scaffold molecular arrangements in other species, algorithmic assemblies that perform computations, and the development of DNA-based nanomechanical devices.
In addition to the intermolecular specificity described above, the key architectural property that is needed to build and demonstrate these arrays and devices is high structural integrity in the components; even if their associations are precise, the assembly from marshmallow-like components will not produce well-structured materials. As noted above, single-branched junctions, such as the HJ structure in Figure 6 are relatively flexible. Fortunately, the DX molecule is considerably more rigid, apparently stiffer than double helical DNA.20,50 As noted above, the TX and PX molecules appear to share this rigidity,18,19 along with the 6-helix and 8-helix bundle molecules.13,21 Consequently, it has been possible to use TX and PX molecules as the building blocks of both arrays and devices, and there is good reason to be optimistic about the helical bundles. As described below, arrays have proved to be useful as scaffolding for the hetero-molecules, as the basis of DNA computation by self-assembly and as frameworks to mount DNA nanomechanical devices.
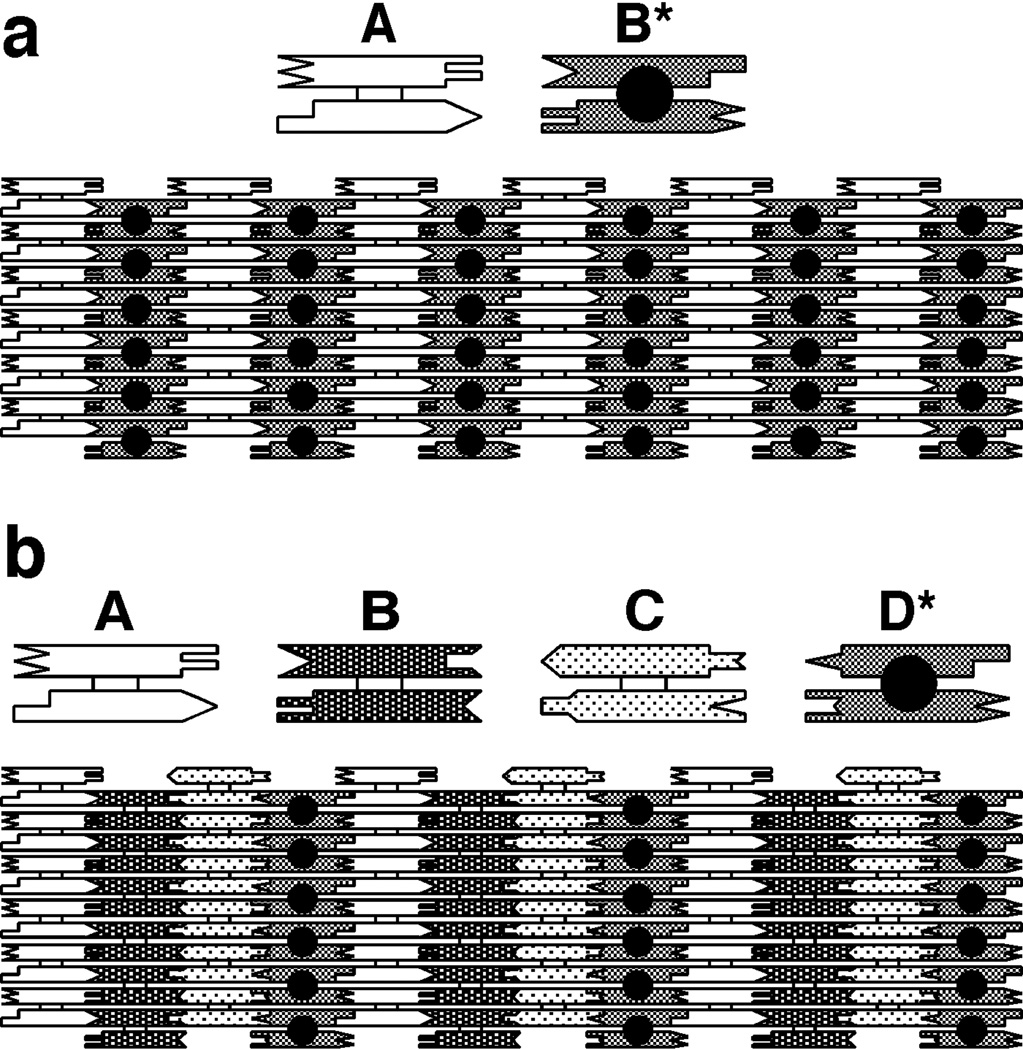
Figure 9 illustrates 2D arrangements that entail the use of DX molecules to produce periodic patterns. Figure 9a illustrates a two-component array that can tile the plane. One tile is a DX molecule labeled A and the second is a DX molecule labeled B*. B* contains another DNA domain that projects out of the plane of the helix axes. This other domain can serve as a topographic marker for the atomic force microscope when the AB* array is deposited onto the surface of mica. The dimensions of the two DX tiles in Figure 9a are about 4 nm tall × 16 nm wide × 2 nm thick. Thus, the B* markers in the 2D array shown should appear as stripe-like features separated by ~32 nm, which has been confirmed by experiment. Figure 9b shows a 4-tile arrangement that should produce stripes separated by ~64 nm, also confirmed by experiment.51 Thus, it is possible to design and produce patterns using DNA components; these patterns contain predictable features, based on the sticky-ended cohesion of individual motifs. The patterns can be modified by the enzymatic addition or removal of these features; non-enzymatic addition has also been demonstrated.52 In addition to forming arrays from DX molecules, it is also possible to produce periodic arrays from TX molecules.18 A variety of DNA parallelograms also have been used to produce 2D arrays.53–55 These motifs are produced by combining four HJ-like branched junctions. Unlike the DX, TX and PX molecules, the two domains of the HJ molecule are not parallel to each other. As a function of the sequence and backbone connections at the crossover point, they adopt angles of ~60°,53~−70°,54 or ~40°,55 thus producing a diversity of parallelogram angles. Substitution of a simple double helical motif with a DX motif has proved to be valuable in producing 2D motifs that have otherwise proved to be intractable. The principle of DX cohesion56 has been extended to a variety of motifs, including parallelograms, triangles, tensegrity triangles, and skewed triangles.24
Figure 9. Tiling the Plane with DX Molecules.
(a) A Two-Tile Pattern. The two helices of the DX molecule are represented schematically as rectangular shapes that terminate in a variety of shapes. The terminal shapes are a geometrical representation of sticky ends. The individual tiles are shown at the top of the drawing; the way tiles fit together using complementary sticky ends to tile the plane is shown at the bottom. The molecule labeled A is a conventional DX molecule, but the molecule labeled B* contains a short helical domain that protrudes from the plane of the helix axes; this protrusion is shown as a black dot. The black dots form a stripe-like feature in the array. The dimensions of the tiles are 4 nm × 16 nm in this projection. Thus, the stripe-like features should be about 32 nm apart. (b) A Four-Tile Pattern. The same conventions apply as in (a). The four tiles form an array in which the stripes should be separated by about 64 nm, as confirmed by AFM.
The promise of 2D DNA arrays to scaffold other species has been realized in a number of studies. DNA nanoparticles have been organized in linear arrays that sit on 2D assemblies by Kiehl and his colleagues.57,58 The work from that group includes the incorporation of differently sized particles into alternating linear arrays.59 2D arrays of multiple nanoparticles have also been scaffolded on 2D arrays.23 In addition to nanoparticles, 2D arrays have been used to organize DNAzymes,60 DNA tweezers61 and a robust nanomechanical DNA device (Ding & Seeman, 2006).62
DNA Nanomechanical Devices
Nanomechanical action is a central target of nanotechnology. The first DNA-based devices were predicated on structural transitions of DNA driven by small molecules. The initial DNA device entailed the extrusion of a DNA cruciform structure from a cyclic molecule.63 The system consisted of a DNA circle (~300 nucleotides) that contained a branch point capable of assuming five different positions, because there were four symmetric nucleotides at its base; the symmetry was eliminated beyond that point. The device is illustrated in Figure 10. The position of the branch point was controlled by the addition or removal of an intercalating dye that changes the supercoiling. This system was not very convenient to operate, and the large size of the DNA circle made it unwieldy to handle. Nevertheless, it demonstrated that DNA could form the basis of a two-state mechanical system.
Figure 10. A Torsionally Driven DNA Device.
On the left is a DNA circle that contains a fixed branch. There are four symmetric nucleotide pairs at the base of the branch, and these can undergo branch migration. With the normal twist of the DNA in the circle, the nucleotides are extruded from the circle. However, when the twist is decreased by the addition of ethidium, the nucleotides branch migrate to become part of the circle.
The second DNA-based device (Figure 11a) was a marked advance. It was relatively small, and included two rigid components, DX molecules like those used to make the arrays of Figure 9. The basis of the device was the transition between right-handed (conventional) B-DNA and left-handed Z-DNA. There are two requirements for the formation of Z-DNA, a 'proto-Z' sequence capable of forming Z-DNA readily (typically a (CG)n sequence), and conditions (typically high salt or molecules like Co(NH3)63+ that emulate the presence of high salt) to promote the transition.64 The sequence requirement provides control over the transition in space, and the requirement for special conditions provide control over the transition in time. As shown in Figure 11a, the device consists of two DX molecules connected by a (CG)10 double helical shaft that provides the proto-Z sequence. In B-promoting conditions, both of the DX helices not collinear with the shaft are on the same side of the shaft. In Z-promoting conditions, one of these helices ends up on the other side of the shaft. This difference is the result of converting a portion of the shaft to Z-DNA, which rotates one DX motif relative to the other by 3.5 turns. The movement was demonstrated by fluorescence resonance energy transfer (FRET) measurements that respond to the difference between dye separations on the DX motifs.65
Figure 11. DNA-Based Nanomechanical Devices.
(a) A Device Predicated on the B-Z Transition. The molecule consists of two DX molecules, connected by a segment containing proto-Z-DNA. The molecule consists of three cyclic strands, two on the ends drawn with a thin line, and one in the middle, drawn with a thick line. The molecule contains a pair of fluorescent dyes to report their separation by FRET. One is drawn as a filled circle, and the other as an empty circle. In the upper molecule, the proto-Z segment is in the B conformation, and the dyes are on the same side of the central double helix. In the lower molecule, the proto-Z segment is in the Z conformation, and the dyes are on opposite sides of the central double helix. The length of the proto-Z-DNA and its conformation are indicated at top and bottom by the two vertical lines flanking the conformation descriptor. (b) A Sequence-Dependent Device. This device uses two motifs, PX and JX2. The labels A, B, C and D on both show that there is a 180° difference between the wrappings of the two molecules. There are two strands drawn as thick lines at the center of the PX motif, and two strands drawn with thin lines at the center of the JX2 motif; in addition to the parts pairing to the larger motifs, each has an unpaired segment. These strands can be removed and inserted by the addition of their total complements (including the segments unpaired in the larger motifs) to the solution; these complements are shown in processes I and III as strands with black dots (representing biotins) on their ends. The biotins can be bound to magnetic streptavidin beads so that these species can be removed from solution. Starting with the PX, one can add the complement strands (process I), to produce an unstructured intermediate. Adding the set strands in process II leads to the JX2 structure. Removing them (III) and adding the PX set strands (IV) completes the machine cycle. Many different devices could be made by changing the sequences to which the set strands bind.
The reason that the B-Z device is not the ultimate DNA-based nanomechanical device is that it is activated by an unspecific molecule, Co(NH3)63+. Consequently, a number of such devices, embedded in an array, would all respond similarly, although some chemical nuance might be available to obtain differential responses.47 Thus, N two-state devices would result in essentially two structural states; clearly it would be of much greater utility to have N distinct 2-state devices capable of producing 2N structural states. It is evident that a sequence-dependent device would be an appropriate vehicle for the goal of achieving multiple states. The method for manipulating sequence-dependent devices was worked out by Yurke and his colleagues.66 It entails setting the state of a device by the addition of a 'set' strand that contains an unpaired tail. When the full complement to the set strand is added, the set strand is removed, and a different set strand may be added. This system has been adapted to the PX and JX2 motifs (Figure 5).19 Figure 11b shows how these two states can be interconverted by the removal of one pair of set strands (processes I and III) and the addition of the opposite pair (processes II and IV). The tops and bottoms of the two states differ by a half-rotation, as seen by comparing the A and B labels at the tops of the molecules, and the C and D labels at the bottoms of the molecules. A variety of devices can be produced by changing the sequences of the regions where the set strands bind.
A variety of walking devices have been produced.67,68 Mao and his colleagues have successfully produced the first autonomous DNA device.69 This device operates by having a DNAzyme with RNase activity relocate along a track containing RNA molecules. Dittmer & Simmel70 have extended control of the Yurke tweezers to a transcription system. A summary of DNA nanomechanical devices can be found in reviews by Seeman71 and by Bath and Turberfield.72 Recent notable systems include an array with variable dimensions,73 a nanomechanical device that translates DNA sequences into polymer assembly instructions74 and the incorporation of a sequence-dependent device into a 2D DNA array.62
DNA-Based Computation and Algorithmic Assembly
Experimental DNA-based computation was founded by Leonard Adleman in 1994.75 His approach is different from the use of DNA to scaffold nanoelectronic component assembly, suggested in Figure 1. Instead, Adleman combined the information in DNA molecules themselves, using standard biotechnological operations (ligation, PCR, gel electrophoresis and sequence-specific binding to derivatized beads) to solve a Hamiltonian path problem. This as a problem related to the traveling salesman problem (What is the optimal route for a salesman to visit N cities?). In the Hamiltonian path problem solved by Adleman, there is a defined start-point and end-point, and an incomplete set of routes between the cities; the problem is to establish whether there is a path between the start-point and end-point that visits all the cities. The idea behind DNA-based computation is that there exist certain classes of computational problems for which the parallelism of molecular assembly overcomes the slow speed of the required macroscopic manipulations. Many varieties of DNA-based computation have been proposed, and a number of them have been tested experimentally for relatively small cases. Space does not permit discussion of all of them.
Nevertheless, there is one approach to DNA-based computation that is relevant to our discussion of structural DNA nanotechnology. This was a method suggested by Winfree, who noticed that the system described above, branched junctions with sticky ends, could be a way to implement computation by 'Wang tiles' on the molecular scale.5 This is a system of tiles whose edges may contain one or more different markings; the tiles self-assemble into a mosaic according to the local rule that all edges in the mosaic are flanked by the same marking. Such a form of assembly can be shown to emulate the operation of a Turing machine, a general-purpose computer.76 A group of Wang tiles is shown on the left of Figure 12a, and an assembly of Wang tiles is shown on the right of Figure12a. The relationship between the sticky ends of a branched junction and the markings on a Wang tile is shown in Figure 12b.
Figure 12. DNA-Based Computation.
(a) Wang Tiles. On the left is a group of 16 Wang tiles. The edges of the tiles are flanked by a variety of patterns. These tiles assemble into the mosaic on the right according to the rule that each edge in the mosaic is flanked by the same pattern. The mosaic represents a calculation, adding 4 to 7 to obtain 11. The two addends are in the top row in the fourth and seventh column. The path through the calculation begins in the upper left corner, and continues on a diagonal until it encounters the vertical column in the fourth column. The path then switches to horizontal until the seventh column, and then again switches to the diagonal, terminating in the eleventh column. (After Grünbaum and Shephard, 1986). (b) The Relationship between Wang Tiles and Branched Junctions. The shadings are the same in both the tile and the sticky ends of the junction, indicating that the sticky ends on a branched junction can emulate a Wang tile. (c) The Components of a Cumulative XOR Calculation. TX tiles are shown as rectangles ending in sticky ends represented geometrically. The input x tiles are shown at the upper left; and the value of the tile is shown in the central domain. Initiator tiles C1 and C2 are shown in the upper right and the four possible y tiles are shown in the bottom row. The inputs of the y tiles is shown on their bottom domains. (d) The Self-Assembled Tiles. The strand structure of the TX tiles is illustrated on the upper left, with the reporter strand drawn with a thicker line. The assembly of tiles in a prototype calculation is shown, using the components illustrated in (c). The input 1, 1, 1, 0 produces an output of 1, 0, 1, 1 by successive binding of y tiles into the double sites created as the array assembles.
This form of DNA-based computation has been prototyped successfully in a 4-bit cumulative XOR calculation.77 The XOR calculation yields a 1 if the two inputs are different, and a 0 if they are the same. Figure 12c shows the components of this calculation. Each component is a TX molecule, schematized as three rectangles with geometrical shapes on their ends to represent complementarity. The input bits are 'x' tiles (upper left), and the output bits are 'y' tiles (bottom), and there are two initiator tiles, C1 and C2, as well (upper right). The upper left corner of Figure 5c shows the strand structure of the TX tiles; each strand contains a 'reporter strand' (drawn with a thicker line); the value of x and y tiles is set to 0 or 1 depending whether it contains a Pvu II or EcoR V restriction site, respectively. The yi tiles perform the gating function; there are four of them, corresponding to the four possible combinations of 0 and 1 inputs. The input involves the bottom domain (Figure 12c). The assembly of periodic arrays discussed above entails competition between correct and incorrect tiles for particular positions; by contrast, the competition here is between correct and partially correct tiles. For example, the yi−1 = 0 sticky end on the leftmost tile is the same as the yi−1 sticky end on the rightmost tile. In the cumulative XOR calculation, yi = XOR (xi,yi−1). The implementation of this formula is shown in Figure 12d. The xi tiles and the initiators are given longer sticky ends than the yi tiles, so they assemble a template first when the tiles are cooled. This creates a double site where the y1 tile can bind. This binding creates the double site where the y2 tile can bind, and so on. When the assembly is complete, the reporter strands are ligated together, creating a long strand that connects the input to the output through the initiator tiles. Partial restriction analysis of the resulting strand reveals that the correct answer is obtained almost exclusively.
Algorithmic assembly has been extended in recent years. Rothemund et al.78 have produced a 2D algorithmic assembly that yields a Sierpinski triangle. This is a 2D version of the XOR calculation described above. It is quite successful, having only a few errors in arrays that contain >102 tiles. In a similar fashion, Barrish et al.79 have implemented a 2D self-assembling system80 that is capable of counting; this one counts in binary to 8. Error correction in algorithmic assembly is an activity that is currently attracting a lot of effort.
Concluding Remarks
This article has discussed the current state of structural DNA nanotechnology, emphasizing the individual components, their assembly into objects and topological targets, into periodic and algorithmic arrays, and their manipulation as nanomechanical devices. Where is the field going? Four years ago, I pointed out that scaffolding of hetero-molecules would be key to the value of the field. This is a goal that has now been achieved in numerous examples noted above, particularly nanoparticles, DNAzymes, aptamers81 and polymers. This achievement suggests that nanorobotics, nanoelectronics and DNA-based control of chemical and polymer syntheses are within reach. I noted at the time that self-replicable molecules would be highly desirable, and this goal has been achieved by Shih and his colleagues,10 demonstrating a single-stranded self-replicable octahedron whose key edges are formed by PX cohesion.9 This is a major advance over previous oblique proposals.82,83
A development that has revolutionized the field is DNA origami, developed by Rothemund.84 He has expanded the usable scale of specific non-repeating structures from a few hundred nucleotides to around fifteen thousand. He has done by using a single-stranded virus for half the nucleotides in his construct, and complementing that sequence with several hundred short 'staple strands' to form repeating DX patterns. He has produced DNA structures that range from maps of the Western Hemisphere to smiley faces. This use of a long strand was anticipated by Yan and his colleagues,85 and to some extent by Shih's octahedron.10 Shih and his colleagues86 have recently used this approach to produce a pair of 6-helix bundle motifs that provide an organizing framework for NMR structure determination of membrane protein structure.
Among the unrealized goals is the extension of array-making capabilities from 2D to 3D, particularly with high order. Recent reports24,87 suggest that these goals are not unrealizable, but high order in 3D may require more groundwork in smaller structures before being successful in a routine fashion. Algorithmic assembly in 3D of the sort described in 2D above will lead ultimately to very smart materials, particularly if combined with nanodevices. Structural DNA nanotechnology remains a biokleptic pursuit, stealing genetic molecules from biological systems; ultimately, it must advance from biokleptic to biomimetic, not just using the central molecules of life, but improving on them, without losing their inherent power as central elements of self-assembled systems.
Acknowledgments
I am grateful to all of my students, postdocs and collaborators for their contributions to the founding of structural DNA nanotechnology. This research has been supported by grants GM-29554 from NIGMS, grants DMI-0210844, EIA-0086015, CCF-0432009, CCF-0523290 and CTS-0548774, CTS-0608889 from the NSF, 48681-EL from ARO, DE-FG02-06ER64281 from DOE (Subcontract from the Research Foundation of SUNY), and a grant from the W.M. Keck Foundation.
References
- 1.Seeman NC. Structural DNA Nanotechnology: An Overview. In: Rosenthal Sandra J, Wright David W., editors. Methods in Molecular Biology 303: Bionanotechnology Protocols. Totowa, NJ: Humana Press; 2005. pp. 143–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watson JD, Crick FHC. A structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 3.Seeman NC. Nucleic acid junctions and lattices. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 4.Robinson BH, Seeman NC. The design of a biochip. Prot. Eng. 1987;1:295–300. doi: 10.1093/protein/1.4.295. [DOI] [PubMed] [Google Scholar]
- 5.Winfree E. On the computational power of DNA annealing and ligation. In: Lipton EJ, Baum EB, editors. DNA Based Computing. 1996. pp. 199–219. Am. Math. Soc., Providence. [Google Scholar]
- 6.Seeman NC. In the nick of space: Generalized nucleic acid complementarity and the development of DNA nanotechnology. Synlett. 2000;2000:1536–1548. [Google Scholar]
- 7.Cohen SN, Chang ACY, Boyer HW, Helling RB. Construction of biologically functional bacterial plasmids in vitro. Proc. Nat. Acad. Sci. (USA) 1973;70:3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu H, Dewan JC, Seeman NC. A DNA decamer with a sticky end: The crystal structure of d-CGACGATCGT. J. Mol. Biol. 1997;267:881–898. doi: 10.1006/jmbi.1997.0918. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Yan H, Shen Z, Seeman NC. Paranemic cohesion of topologically-closed DNA molecules. J Am. Chem. Soc. 2002;124:12940–12941. doi: 10.1021/ja026973b. [DOI] [PubMed] [Google Scholar]
- 10.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 11.Shen Z, Yan H, Wang T, Seeman NC. Paranemic crossover DNA: a generalized Holliday structure with applications in nanotechnology. J. Am. Chem. Soc. 2004;126:1666–1674. doi: 10.1021/ja038381e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan H, Seeman NC. Edge-sharing motifs in DNA nanotechnology. J. Supramol. Chem. 2003;1:229–237. (2001). [Google Scholar]
- 13.Kuzuya A, Wang R, Sha R, Seeman NC. six-helix and eight-helix DNA nanotubes assembled from half-tubes. NanoLetters. 2007 doi: 10.1021/nl070828k. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeman NC. DNA nicks and nodes and nanotechnology. NanoLetts. 2001;1:22–26. [Google Scholar]
- 15.Holliday R. A mechanism for gene conversion in fungi. Genet. Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 16.Fu T-J, Seeman NC. DNA double crossover structures. Biochem. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 17.Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 18.LaBean T, Yan H, Kopatsch J, Liu F, Winfree E, Reif JH, Seeman NC. The construction, analysis, ligation and self-assembly of DNA triple crossover complexes. J. Am. Chem. Soc. 2000;122:1848–1860. [Google Scholar]
- 19.Yan H, Zhang X, Shen Z, Seeman NC. A robust DNA mechanical device controlled by hybridization topology. Nature. 2002;415:62–65. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Yang X, Qi J, Seeman NC. Antiparallel DNA double crossover molecules as components for nanoconstruction. J. Am. Chem. Soc. 1996;118:6131–6140. [Google Scholar]
- 21.Mathieu F, Liao S, Mao C, Kopatsch J, Wang T, Seeman NC. Six-Helix Bundles Designed from DNA. NanoLetters. 2005;5:661–665. doi: 10.1021/nl050084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu D, Wang M, Deng Z, Walulu R, Mao C. Tensegrity: construction of rigid DNA triangles with flexible four-arm DNA junctions. J. Am Chem. Soc. 2004;126:2324–2325. doi: 10.1021/ja031754r. [DOI] [PubMed] [Google Scholar]
- 23.Zheng J, Constantinou PE, Micheel C, Alivisatos AP, Kiehl RA, Seeman NC. 2D nanoparticle arrays show the organizational power of robust DNA motifs. NanoLett. 2006;6:1502–1504. doi: 10.1021/nl060994c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Constantinou PE, Wang T, Kopatsch J, Israel LB, Zhang X, Ding B, Sherman WB, Wang X, Zheng J, Sha R, Seeman NC. Double cohesion in structural DNA nanotechnology. Org. Biomol. Chem. 2006;4:3414–3419. doi: 10.1039/b605212f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Rapid chiral assembly of rigid DNA building blocks form molecular fabrication. Science. 2005;310:1661–1664. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 26.Caruthers MH. Gene synthesis machines: DNA chemistry and its uses. Science. 1985;230:281–285. doi: 10.1126/science.3863253. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Seeman NC. A solid-support methodology for the construction of geometrical objects from DNA. J. Am. Chem. Soc. 1992;114:2656–2663. [Google Scholar]
- 28.Chen J, Seeman NC. The synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Seeman NC. The construction of a DNA truncated octahedron. J. Am. Chem. Soc. 1994;116:1661–1669. [Google Scholar]
- 30.Qi J, Li X, Yang X, Seeman NC. The ligation of triangles built from bulged three-arm DNA branched junctions. J. Am. Chem. Soc. 1996;118:6121–6130. [Google Scholar]
- 31.Hagerman PJ. Flexibility of DNA. Ann. Rev. Bioiphys. Biophys. Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 32.Seeman NC, Rosenberg JM, Rich A. Sequence specific recognition of double helical nucleic acids by proteins. Proc. Nat. Acad. Sci. (USA) 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu L, Lukeman PS, Canary JW, Seeman NC. Nylon/DNA: single-stranded DNA with a covalently stitched nylon lining. J. Am. Chem. Soc. 2003;125:10178–10179. doi: 10.1021/ja035186r. [DOI] [PubMed] [Google Scholar]
- 34.Freier SM, Altmann K-H. The ups and down of nucleic acid duplex stability. Nucl. Acids Res. 1997;25:4229–4243. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 36.Lukeman PS, Mittal A, Seeman NC. Two dimensional PNA/DNA arrays: estimating the helicity of unusual nucleic acid polymers. Chem. Comm. 2004;2004:1694–1695. doi: 10.1039/b401103a. [DOI] [PubMed] [Google Scholar]
- 37.Seeman N, Kallenbach NR. Design of immobile nucleic acid junctions. Biophys. J. 1983;44:201–209. doi: 10.1016/S0006-3495(83)84292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seeman NC. De novo design of sequences for nucleic acid structure engineering. J. Biomol. Struct. & Dyns. 1990;8:573–581. doi: 10.1080/07391102.1990.10507829. [DOI] [PubMed] [Google Scholar]
- 39.Ma R-I, Kallenbach NR, Sheardy RD, Petrillo ML, Seeman NC. Three arm nucleic acid junctions are flexible. Nucl. Acids. Res. 1986;14:9745–9753. doi: 10.1093/nar/14.24.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kallenbach NR, Ma R-I, Seeman NC. An immobile nucleic acid junction constructed from oligonucleotides. Nature. 1983;305:829–831. [Google Scholar]
- 41.Wang Y, Mueller JE, Kemper B, Seeman NC. The assembly and characterization of 5-Arm and 6-Arm DNA junctions. Biochem. 1991;30:5667–5674. doi: 10.1021/bi00237a005. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Seeman NC. The assembly and characterization of 8-arm and 12-arm DNA branched junctions. J. Am. Chem. Soc. 2007 doi: 10.1021/ja0693441. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrillo ML, Newton CJ, Cunningham RP, Ma R-I, Kallenbach NR, Seeman NC. Ligation and flexibility of four-arm DNA junctions. Biopolymers. 1988;27:1337–1352. doi: 10.1002/bip.360270902. [DOI] [PubMed] [Google Scholar]
- 44.Eis PS, Millar DP. Conformational distributions of a four-way DNA junction revealed by time-resolved fluorescence resonance energy transfer. Biochem. 1993;32:13852–13860. doi: 10.1021/bi00213a014. [DOI] [PubMed] [Google Scholar]
- 45.Chen J, Seeman NC. The electrophoretic properties of a DNA cube and its sub-structure catenanes. Electrophor. 1991;12:607–611. doi: 10.1002/elps.1150120902. [DOI] [PubMed] [Google Scholar]
- 46.Seeman NC. The design of single-stranded nucleic acid knots. Molec. Eng. 1992;2:297–307. [Google Scholar]
- 47.Du SM, Stollar BD, Seeman NC. A synthetic DNA molecule in three knotted topologies. J. Am. Chem. Soc. 1995;117:1194–1200. [Google Scholar]
- 48.Mao C, Sun W, Seeman NC. Assembly of Borromean rings from DNA. Nature. 1997;386:137–138. doi: 10.1038/386137b0. [DOI] [PubMed] [Google Scholar]
- 49.Chichak KS, Cantrill SJ, Pease AR, Chiu SH, Cave GWV, Atwood JL, Stoddart JF. Molecular Borromean rings. Science. 2004;304:1308–1312. doi: 10.1126/science.1096914. [DOI] [PubMed] [Google Scholar]
- 50.Sa-Ardyen P, Vologodskii AV, Seeman NC. The flexibility of DNA double crossover molecules. Biophys. J. 2003;84:3829–3837. doi: 10.1016/S0006-3495(03)75110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 52.Liu F, Sha R, Seeman NC. Modifying the surface features of two-dimensional DNA crystals. J. Am. Chem. Soc. 1999;121:917–922. [Google Scholar]
- 53.Mao C, Sun W, Seeman NC. Designed two-dimensional DNA Holliday junction arrays visualized by atomic force microscopy. J. Am. Chem. Soc. 1999;121:5437–5443. [Google Scholar]
- 54.Sha R, Liu F, Millar DP, Seeman NC. Atomic force microscopy of parallel DNA branched junction arrays. Chem. & Biol. 2000;7:743–751. doi: 10.1016/s1074-5521(00)00024-7. [DOI] [PubMed] [Google Scholar]
- 55.Sha R, Liu F, Seeman NC. Atomic force measurement of the interdomain angle in symmetric Holliday junctions. Biochem. 2002;41:5950–5955. doi: 10.1021/bi020001z. [DOI] [PubMed] [Google Scholar]
- 56.Ding B, Seeman NC. Pseudohexagonal 2D DNA crystals from double crossover cohesion. J. Am. Chem. Soc. 2004;126:10230–10231. doi: 10.1021/ja047486u. [DOI] [PubMed] [Google Scholar]
- 57.Xiao S, Liu F, Rosen A, Hainfeld JF, Seeman NC, Musier-Forsyth KM, Kiehl RA. Self-assembly of nanoparticle arrays by DNA scaffolding. J. Nanopart. Res. 2002;4:313–317. [Google Scholar]
- 58.Le JD, Pinto Y, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA. Self-assembly of nanoelectronic component arrays by in situ hybridization to 2D DNA scaffolding. NanoLett. 2004;4:2343–2347. [Google Scholar]
- 59.Pinto YY, Le JD, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA. Sequence-encoded self-assembly of multiple-nanocomponent arrays by 2D DNA scaffolding. NanoLett. 2005;5:2399–2402. doi: 10.1021/nl0515495. [DOI] [PubMed] [Google Scholar]
- 60.Garibotti AV, Knudsen SM, Ellington AD, Seeman NC. Functional DNAzymes organized into 2D arrays. NanoLett. 2006;6:1505–1507. doi: 10.1021/nl0609955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chhabra R, Sharma J, Liu Y, Yan H. Addressable molecular tweezers for DNA-templated coupling reactions. NanoLett. 2006;6:978–983. doi: 10.1021/nl060212f. [DOI] [PubMed] [Google Scholar]
- 62.Ding B, Seeman NC. Operation of a DNA robot arm inserted into a 2D DNA crystalline substrate. Science. 2006;314:1583–1585. doi: 10.1126/science.1131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang X, Vologodskii AV, Liu B, Kemper B, Seeman NC. Torsional control of double stranded DNA branch migration. Biopolymers. 1998;45:69–83. doi: 10.1002/(SICI)1097-0282(199801)45:1<69::AID-BIP6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 64.Rich A, Nordheim A, Wang AH-J. The chemistry and biology of left-handed Z-DNA. Ann. Rev. Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- 65.Mao C, Sun W, Shen Z, Seeman NC. A DNA nanomechanical device based on the B-Z transition. Nature. 1999;397:144–146. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- 66.Yurke B, Turberfield AJ, Mills AP, Jr, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 67.Sherman WB, Seeman NC. A precisely controlled DNA bipedal walking device. NanoLett. 2004;4:1203–1207. [Google Scholar]
- 68.Shin J-S, Pierce NA. A synthetic DNA walker for molecular transport. J. Am. Chem. Soc. 2004;126:10834–10835. doi: 10.1021/ja047543j. [DOI] [PubMed] [Google Scholar]
- 69.Tian Y, He Y, Chen Y, Yin P, Mao C. Molecular devices - A DNAzyme that walks processively and autonomously along a one-dimensional track. Angew. Chemie, Int. Ed. 2005;44:4355–4358. doi: 10.1002/anie.200500703. [DOI] [PubMed] [Google Scholar]
- 70.Dittmer WU, Simmel FC. Transcriptional control of DNA-based nanomachines. NanoLett. 2004;4:689–691. [Google Scholar]
- 71.Seeman NC. From genes to machines: DNA nanomechanical devices. Trends Biochem. Sci. 2005;30:119–125. doi: 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bath J, Turberfield AJ. DNA nanomachines. Nature Nanotech. 2007;2:276–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 73.Feng L, Park SH, Reif JH, Yan H. A two-state DNA lattice switched by DNA nanoactuator. Angew. Chem. Int. Ed. 2003;42:4342–4346. doi: 10.1002/anie.200351818. [DOI] [PubMed] [Google Scholar]
- 74.Liao S, Seeman NC. Translation of DNA signals into polymer assembly instructions. Science. 2004;306:2072–2074. doi: 10.1126/science.1104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adleman L. Molecular computation of solutions to combinatorial problems. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 76.Grünbaum B, Shephard GC. Tilings & Patterns. New York: Freeman; 1986. [Google Scholar]
- 77.Mao C, LaBean T, Reif JH, Seeman NC. Logical computation using algorithmic self-assembly of DNA triple crossover molecules. Nature. 2000;407:493–496. doi: 10.1038/35035038. [DOI] [PubMed] [Google Scholar]
- 78.Rothemund PWK, Papadakis N, E Winfree E. Algorithmic self-assembly of DNA Sierpinski triangles. PLOS Biology. 2004;2:2041–2053. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barish RD, Rothemund PWK, Winfree E. Two computational primitives for algorithmic assembly: copying and counting. NanoLett. 2005;5:2586–2592. doi: 10.1021/nl052038l. [DOI] [PubMed] [Google Scholar]
- 80.Winfree E. Algorithmic self-assembly of DNA: Theoretical motivations and 2D assembly experiments. J. Biomol. Str. & Dyns. Conversat. 11. 2000;2:263–270. doi: 10.1080/07391102.2000.10506630. [DOI] [PubMed] [Google Scholar]
- 81.Lin CX, Katilius E, Liu Y, Zhang JP, Yan H. Self-assembled signaling aptamer DNA arrays for protein detection. Angew. Chemie Int. Ed. 2006;45:5296–5301. doi: 10.1002/anie.200600438. [DOI] [PubMed] [Google Scholar]
- 82.Seeman NC. The construction of 3-D stick figures from branched DNA. DNA and Cell Biology. 1991;10:475–486. doi: 10.1089/dna.1991.10.475. [DOI] [PubMed] [Google Scholar]
- 83.Eckardt HE, Naumann K, Pankau WM, Rein M, Schweitzer M, Windhab N, von Kiedrowski G. Chemical copying of connectivity. Nature. 2002;420 doi: 10.1038/420286a. 286-286. [DOI] [PubMed] [Google Scholar]
- 84.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 85.Yan H, LaBean TH, Feng L, Reif JH. Directed nucleation assembly of DNA tile complexes for barcode-patterned lattices. Proc. Nat. Acad. Sci. 2003;100:8103–8108. doi: 10.1073/pnas.1032954100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Douglas SM, Chou JJ, Shih WM. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Nat. Acad. Sci (USA) 2007;104:6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mao C, Constantinou PE, Liu F, Pinto Y, Kopatsch J, Lukeman PS, Wang T, Ding B, Yan H, Birktoft JJ, Sha R, Zhong H, Foley L, Wenzler LA, Sweet R, Becker M, Seeman NC. The design of self-assembled 3D DNA networks. In: Cahay M, Urquidi-Macdonald M, Bandyopadhyay S, Guo P, Hasegawa H, Koshida N, Leburton JP, Lockwood DJ, Seal S, Stella A, editors. Proc. Intl. Symp. on Nanoscale Devices, Materials, and Biological Systems; 206th Meeting of the Electrochemical Society; Honolulu, PV. 2005. pp. 509–520. 2004-XX. [Google Scholar]