Summary
Viral nucleic acids often trigger an innate immune response in infected cells. Many viruses, including hepatitis C virus (HCV), have evolved mechanisms to evade intracellular recognition. Nevertheless, HCV-permissive cells can trigger a viral RNA-, TLR7- and cell contact-dependent compensatory interferon response in nonpermissive plasmacytoid dendritic cells (pDCs). Here we report that these events are mediated by transfer of HCV RNA-containing exosomes from infected cells to pDCs. The exosomal viral RNA transfer is dependent on the endosomal sorting complex (ESCRT) machinery and on Annexin A2, an RNA-binding protein involved in membrane vesicle trafficking, and it is suppressed by exosome release inhibitors. Further, purified concentrated HCV RNA-containing exosomes are sufficient to activate pDCs. Thus, vesicular sequestration and exosomal export of viral RNA may serve both as a viral strategy to evade pathogen-sensing within infected cells and as a host strategy to induce an unopposed innate response in replication-nonpermissive by-stander cells.
Introduction
Upon sensing invading viruses, host cells trigger signaling events that ultimately lead to the secretion of type I interferons (IFNs) and to the expression of an array of IFN-stimulated genes (ISGs) (Kawai et al., 2009). ISGs are important effectors that suppress viral spread by blocking the viral life cycle at multiple levels (Sadler et al., 2008). Specific motifs, Pathogen Associated Molecular Patterns (PAMPs), including viral nucleic acids, are recognized by pattern recognition receptors (PRRs) that mobilize the innate immune response (Kawai et al., 2009). These receptors can be cytoplasmic, e.g. retinoic inducible gene-I (RIG-I)-like receptors (RLRs) (Kawai et al., 2009; Loo et al., 2011) and NOD-like receptors (NLRs) (Kawai et al., 2009), or endosomal, e.g. Toll-like receptors (TLRs) (Blasius et al., 2010; Kawai et al., 2011). Thus, the current concept of virus-induced innate immune signaling is that recognition of viral nucleic acids typically occurs within cells that are either productively infected or have internalized viral particles (Blasius et al., 2010; Loo et al., 2011).
Many viruses deploy mechanisms designed to avert, subvert, or evade pathogen-sensing pathway (Versteeg et al., 2010). For example, the hepatitis C virus (HCV) has evolved a potent strategy that precludes type 1 interferon (IFN) induction by infected hepatocytes (Cheng et al., 2006; Liang et al., 2008). Specifically, the NS3-4A protease of HCV inactivates, by proteolytic cleavage, MAVS/IPS-1/cardif, the signaling adapter of the RLR sensing pathway (Foy et al., 2005; Horner et al., 2011; Li et al., 2005b; Loo et al., 2006; Meylan et al., 2005). Furthermore, cleavage of the TIR-domain-containing adapter-inducing IFNβ (TRIF) adapter by NS3-4A abrogates signaling transduction induced by endosomal TLR3 (Li et al., 2005a; Wang et al., 2009). However, despite these inhibitory mechanisms, HCV infection strongly triggers the expression of ISGs in the liver of experimentally HCV-infected chimpanzees (Bigger et al., 2001; Bigger et al., 2004; Su et al., 2002) and of naturally HCV infected humans (Alter et al., 2000). These results therefore suggest the existence of alternative pathogen-sensing mechanisms that aren’t opposed by viral evasion mechanisms.
We (Takahashi et al., 2010) and, subsequently, others (Dental et al., 2012; Zhang et al., 2012) recently reported that HCV-infected cells trigger plasmacytoid dendritic cells (pDCs) to produce type 1 IFN. We found that pDC-activation occurs in a cell-cell contact-, and Toll-like receptor (TLR)-7-dependent manner that is dependent on the intracellular HCV RNA level of cocultured infected cells (Takahashi et al., 2010). In addition, we showed that the induction of the IFN response by pDCs is independent of virus production since HCV subgenomic replicon (SGR) cells that replicate the viral RNA but cannot produce infectious virus particles can also trigger pDCs to produce type 1 IFN (Takahashi et al., 2010).
Many cell types release membrane-enclosed microvesicles into the extracellular milieu, the best-characterized of which are 40–100 nm endosome-derived secreted vesicles (exosomes) (Thery et al., 2009) that have been shown to transfer functional microRNA and mRNA to recipient cells (Kosaka et al., 2010; Meckes et al., 2010; Pegtel et al., 2010; Skog et al., 2008; Valadi et al., 2007). Based on previous reports showing that the supernatant of HCV SGR cells contain subgenomic viral RNA (Pietschmann et al., 2002), that exosome-like structures are present in the supernatant of HCV-infected cells (Gastaminza et al., 2010), and that HCV RNA-containing exosomes are detected in the plasma of HCV-infected patients (Masciopinto et al., 2004), we reasoned that HCV RNA might be secreted within exosomal vesicles from HCV infected cells and trigger IFNα production by pDCs.
Here we report that exosomes are a previously unrecognized PAMP-carrier that can mediate cell-to-cell transfer of immunostimulatory viral RNA from infected cells to cocultured pDCs and trigger the production of type 1 IFN. In addition, we define the cellular mechanisms responsible for biogenesis of the PAMP-containing exosomes. These results demonstrate a previously unsuspected aspect of the innate host response in which infected cells, whose pathogen-sensing machinery is inhibited by viral proteins, utilize exosomes to transfer viral RNA to neighboring non-infectible cells that are specifically designed to respond to viral RNA.
Results
Exosomal transfer of viral RNA from HCV infected and HCV SGR cells to pDCs triggers IFNα production
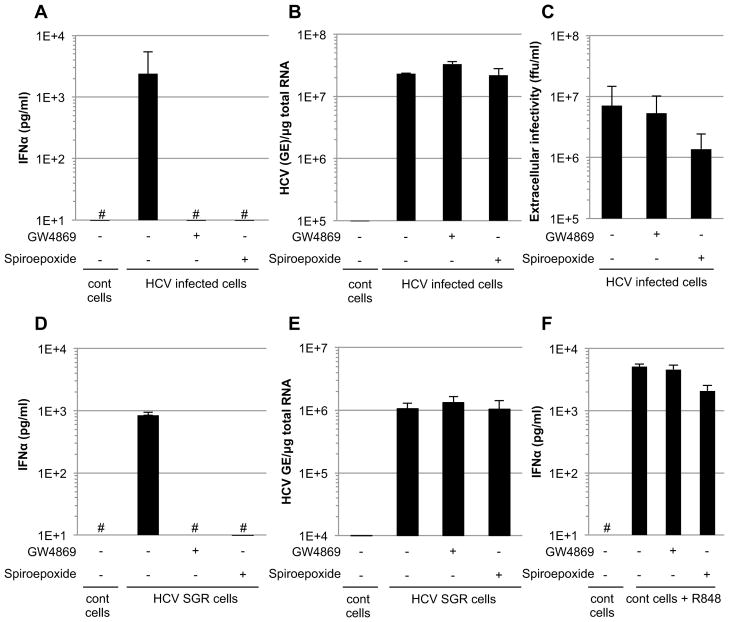
HCV infected Huh-7.5.1c2 (Pedersen et al., 2007) cells were cocultivated with primary, human, peripheral blood-derived pDCs in the presence or absence of two structurally unrelated neutral sphingomyelinase inhibitors (GW4869 and Spiroepoxide) that are well-known inhibitors of exosome release (Chairoungdua et al., 2010; Kogure et al., 2011; Kosaka et al., 2012; Kosaka et al., 2010; Trajkovic et al., 2008; Yuyama et al., 2012). Both GW4869 and Spiroepoxide completely abolished (> 2 log reduction) IFNα secretion by the pDCs (Figure 1A), without reducing the intracellular HCV RNA content of the infected cells (Figure 1B) and with little (<5-fold) or no reduction of infectious HCV particle production (Figure 1C). Similar results were also obtained using Huh-7.5.1 cells and parental Huh-7 cells (Figure S1A–C). Remarkably, both compounds also abrogated IFNα production by pDCs cocultured with HCV SGR cells (Figure 1D) in a dose dependent manner (Figure S1D) that correlated with the magnitude of inhibition of exosome production (Figure S1F) and exosomal HCV RNA (Figure S1G) release. Neither compound inhibited HCV RNA replication in the SGR cells (Figures 1E and S1E) nor did they inhibit pDC IFNα production that was triggered by R848 (a specific TLR7 agonist) (Kawai et al., 2009) (Figure 1F and S1D), thus ruling out potential non-specific effects of these compounds on pDCs.
Figure 1. Inhibition of pDC activation triggered by HCV infected and SGR cells by exosome release inhibitors.
(A) Quantification of IFNα in supernatants of pDCs cocultured with HCV infected Huh-7.5.1c2 cells (MOI of 1 for 48 hours) that were treated or not with exosome-release inhibitors GW4869 or Spiroepoxide (10 and 5 μM, respectively) throughout the course of the coculture. Uninfected Huh-7.5.1c2 cells are referred to as control (cont) cells. The hatch marks (#) indicate results below the limit of detection of the IFNα ELISA (i.e 12.5 pg/ml). In parallel, the intracellular HCV RNA levels (B) and extracellular infectious virus production (C) of HCV infected cells treated with exosome release inhibitors were analyzed. (A–C) Results are representative of 2 independent experiments each performed in triplicate. Error bars represent the mean ± SD. (D) Quantification of IFNα in supernatants of pDCs cocultured with HCV SGR Huh-7.5.1c2 cells after treatment with exosome-release inhibitors GW4869 or Spiroepoxide (treatment with 10 and 5 μM, respectively) throughout the course of the coculture. HCV-negative Huh-7.5.1c2 cells served as controls (cont cells). (E) Intracellular HCV RNA levels in HCV SGR cells treated for 24-hours with exosome release inhibitors. (F) Parallel pDC/control cell cocultures were incubated with the TLR-7 ligand agonist R848 and supernatants were analyzed for IFNα. Results are representative of 3 independent experiments each performed in triplicate. Bar graphs depict the mean ± SD. The treatment conditions (i.e. incubation time and concentration) were exactly the same for the analysis of IFNα production, HCV RNA replication and extracellular infectious virus production. See also Figure S1.
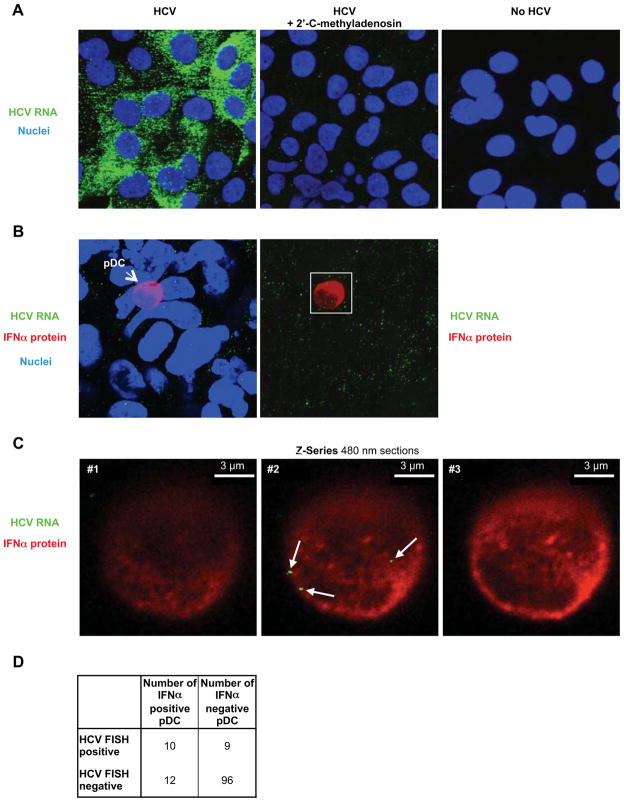
The SGR cell results indicated that the pDC immunostimulatory signal could be transmitted via a virus particle-independent pathway. To corroborate this assumption and to determine if HCV RNA is transmitted to cocultured pDCs even in the absence of virus production, we used a highly sensitive HCV RNA-specific fluorescence in situ hybridization (FISH) assay to monitor the presence of HCV RNA in infected cells, SGR cells, and cocultivated pDCs. As expected, the intensity of the HCV RNA signal was much stronger in infected cells (Figure 2A, left panel) than in SGR cells (Figure 2B, left panel) reflecting their ~10-fold greater content of HCV RNA (data not shown). Nonetheless, HCV RNA (green) was readily detected in pDCs after 20 hours of cocultivation with HCV SGR cells (Figure 2C, middle panel). Combined HCV RNA FISH and IFNα immunostaining revealed that HCV RNA was detected in 45.5% (10/22) of IFNα-positive pDCs and 8.6% (9/105) IFNα-negative pDCs after cocultivation with SGR cells (Figure 2D) but it was never detectable (0/61) in pDCs cocultured with HCV-negative cells (not shown). Collectively, these results suggest that both HCV infected cells and SGR cells release HCV RNA-containing exosomes that transfer HCV RNA to cocultured pDCs and trigger IFNα production. The exosomal nature of the transfer process is underscored by the transfer of HCV RNA to pDCs by SGR cells in which virus particle production cannot occur.
Figure 2. HCV RNA is transferred from HCV SGR cells to pDCs.
(A) Specificity of HCV RNA detection (green) by fluorescence in situ hybridization analysis (FISH) in HCV-infected Huh-7.5.1c2 cells that expressed high levels of HCV RNA (left panel), that were treated with the HCV polymerase inhibitor 2′-C-methyladenosine (5 μM for 24 hours) (middle panel) and in uninfected Huh-7.5.1c2 cells (no HCV, right panel). Nuclei are stained with Hoechst dye (blue). Projection (B) and consecutive Z-axis sections (C) of pDC cocultured with HCV SGR Huh-7.5.1c2 cells, 3.5-fold magnification of the white box in (B). HCV RNA (green), IFNα protein (red), nuclei (blue). HCV RNAs inside pDC are indicated (white arrows). Of note, FISH HCV RNA signals are lower in HCV SGR cells because they contain about 10-fold less HCV RNA than infected cells and in addition, FISH signals are reduced when combined with immunofluorescent staining (data not shown). Similar results were obtained in 5 independent experiments. (D) Summary table showing the fraction of total cocultured pDCs that contains HCV RNA.
HCV RNA is selectively packaged within exosomes that trigger pDC IFNα production
Exosomes are endosome-derived secreted vesicles (Thery et al., 2009) that are formed by membrane invagination within multivesicular bodies (MVB) and exocytosed by fusion of the MVB membrane with the cell surface membrane (Bobrie et al., 2011; Thery et al., 2009). Since HCV RNA-containing exosomes cannot be readily distinguished or physically separated from infectious HCV particles by conventional biophysical techniques because they share similar buoyant density and sedimentation velocity (Lindenbach et al., 2005; Meckes et al., 2011), we used HCV SGR cells in all subsequent studies in order to study the exosome-mediated transfer process without the potentially confounding impact of coexisting virus particles. We view those SGR cell-based results as representative of HCV infected cells since both infected and SGR cells trigger IFNα production by pDCs in a TLR-7-dependent and cell-cell contact-dependent manner (Takahashi et al., 2010) that, in both cases, is antagonized by exosome release inhibitors (Figure 1).
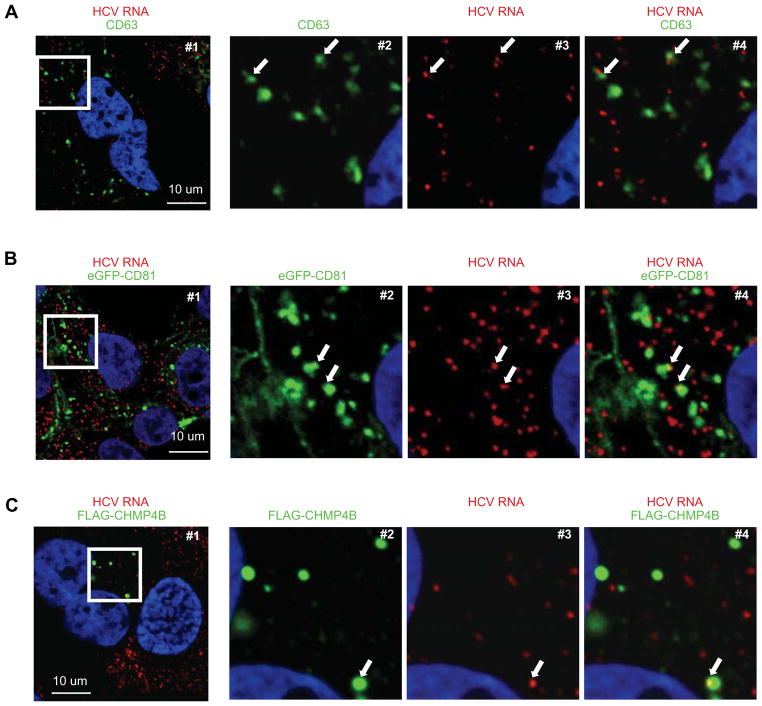
First, we asked if there was evidence that HCV RNA might colocalize in HCV SGR cells with proteins (e.g. CD63, CD81, and CHMP4B) that are known to be enriched in exosomes (Bobrie et al., 2011; Thery et al., 2009). As shown in Figure 3, HCV RNA colocalized with a subset of CD63-, CD81-, and CHMP4B-positive signals in HCV SGR cells, consistent with the notion that HCV RNA could be incorporated into intracellular vesicles destined to become exosomes.
Figure 3. Colocalization of HCV RNA, exosome markers in HCV SGR cells.
Detection of HCV RNA (red), and in green CD63 (A), eGFP-CD81 (B) and FLAG-CHMP4B proteins (C), and nuclei (blue) in HCV SGR Huh-7.5.1c2 cells. Panels 2-to-4 correspond to a 3.6-fold magnification of the white box in #1 in each row. The arrows indicate HCV RNA in CD63-, CD81-, or CHMP4B-positive compartments. The frequencies of HCV SGR cells in which HCV RNA co-localized with CHMP4B, CD81 and CD63 were 25/29, 38/56 and 17/24 respectively. Results are representative of 3 independent experiments.
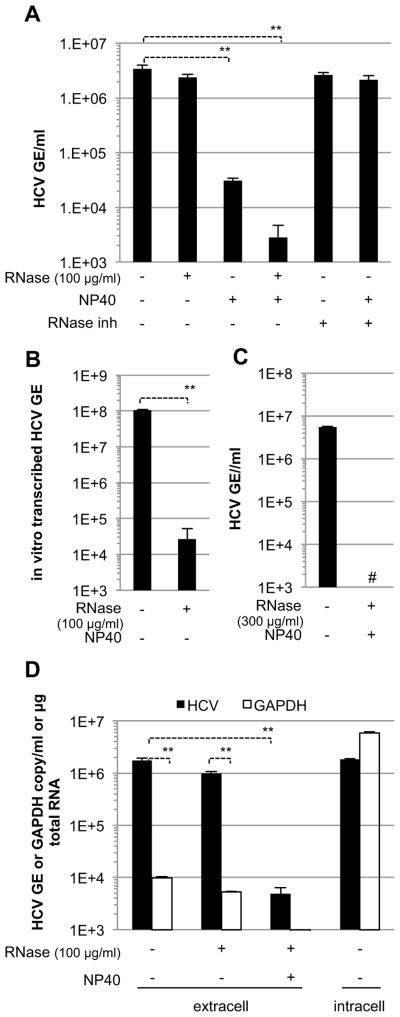
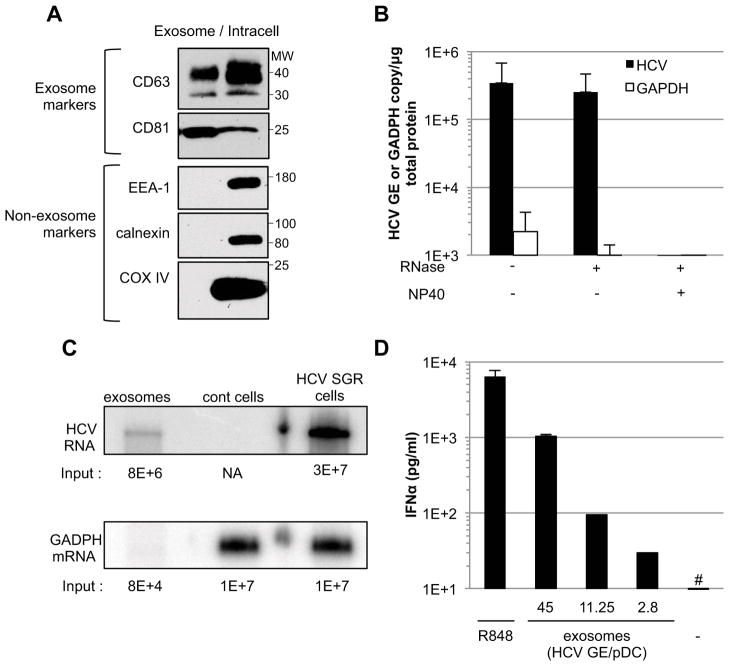
Next, we asked if membrane-bound HCV RNA is released into the supernatant of HCV SGR cells that do not produce HCV structural proteins and, thus, cannot produce virus particles. As shown in Figures 4A-C, the supernatant of HCV SGR cells contained >106 copies per ml of HCV RNA that was resistant to digestion by ribonuclease A (RNase) unless it was also exposed to NP40. These results suggested that the secreted HCV RNA was contained within lipid membrane-bound vesicles. Additionally, we showed that the extracellular HCV RNA was not merely present in cellular debris since it was >100-fold more abundant than the extracellular GAPDH mRNA content in the same supernatants (Figure 4D) while the intracellular HCV RNA content was less abundant than GAPDH mRNA in the corresponding SGR cells (Figure 4D).
Figure 4. RNase-resistant HCV RNA is secreted from HCV SGR cells.
HCV RNA levels in pre-cleared supernatants from HCV SGR Huh-7.5.1c2 cells (n=4, mean ± SD) (A), (n=2, mean ± SD) (C) and in vitro transcribed HCV RNA (n=3, mean ± SD) (B) were treated with RNase, NP40 and/or RNase inhibitor, as indicated. GE, genome equivalents. The hatch marks (#) indicate results below the detection limit of the assay (i.e. 500 HCV GE/ml). (D) HCV and GADPH RNA levels in pre-cleared supernatants from HCV SGR Huh-7.5.1c2 cells (extracell) either untreated or treated with RNase, NP40 and untreated SGR cell RNA (intracell), as indicated. Error bars represent the mean ± SD (n=3), Paired t-test student, ** p<0.005.
Finally, exosomes were prepared from the supernatants of HCV SGR cells and analyzed for their protein and RNA content and their ability to trigger IFNα production by pDCs. As shown in Figures 5A, S1F and S2A the concentrated exosome preparations were specifically enriched in the exosome markers CD63 and CD81 relative to non-exosomal cellular proteins EEA1 (an endosome marker), Calnexin (an ER marker), COX IV (a mitochondrial marker) and ARF1 (a Golgi marker). Importantly, the concentrated exosome preparations contained >105 copies of HCV RNA (per microgram of total protein) that were >150-fold concentrated relative to the amount of HCV RNA in the corresponding unconcentrated SGR cell supernatants (i.e. about 1–2×108 versus 1–2×106 GE/ml respectively) (Figure S1G and 4A, respectively), >100-fold enriched relative to GAPDH mRNA (Figure 5B), and fully protected from RNase digestion unless they were also treated with NP40 (Figure 5B). Interestingly, Northern blot analysis of the exosome-associated RNA revealed that the HCV RNA co-migrated with the dominant HCV RNA species present in the corresponding HCV SGR cells (Figure 5C), suggesting that it represented a full length HCV SGR genome (i.e. 8024 nucleotides). Importantly, the viral NS4B and NS5A proteins and calnexin, all of which are known to be present in HCV replication complexes (Backes et al., 2010; El-Hage et al., 2003), were not detected in concentrated exosome preparations (Figure S2A). Collectively, the foregoing results suggest that the RNAse-resistant extracellular HCV RNA reflects the release of viral RNA-containing exosomes from the SGR cells rather than HCV replication complexes that, theoretically, could be present in cellular debris in the culture supernatants. Additionally, as shown in Figure S2B, buoyant density separation analysis revealed that the extracellular HCV RNA co-fractionated with exosome markers (CD81 and CD63) and it displayed the expected density profile of exosomes (1.10–1.19 g/ml) (Meckes et al., 2011) rather than HCV replicase complexes (>1.17 g/ml) (Ferraris et al., 2010; Villanueva et al., 2010) or apoptotic bodies (1.24–1.28 g/ml) (Meckes et al., 2011). Importantly, highly concentrated exosomes prepared from HCV SGR cells triggered IFNα production by pDCs in a dose-dependent manner (Figure 5D). Collectively, these results suggest that HCV RNA is selectively packaged in exosomes that transfer it to pDCs that respond by secreting IFNα.
Figure 5. Exosomes prepared from HCV SGR cell supernatants trigger IFNα production by pDCs.
(A) Comparison of exosome (CD63, CD81) and non-exosome (EEA-1, calnexin, COX IV) markers in exosomes prepared from HCV SGR Huh-7.5.1c2 cell supernatants and from the corresponding whole cell lysates (intracell) by immunoblotting. The molecular weight markers (MW, in kDa) are indicated to the right. (B) Exosome preparations were treated with NP40 and/or RNase and RT-qPCR results are expressed as HCV GE or GAPDH mRNA copies per μg of total protein. Error bars represent the mean ± SD (n=3). (C) Northern Blot analysis. The input copy number of HCV and GADPH RNA determined by RT-qPCR is indicated below each blot. NA, not applicable; Huh-7.5.1c2 cells served as negative control (cont cells) (D) pDCs (4×105 cells) were incubated with concentrated exosomes at varying HCV GE/pDC ratios or with R848, a TLR7 agonist. The hatch marks (#) indicate results below the limit of detection of the IFNα ELISA (12.5 pg/ml). Results are representative of 2 independent experiments. Error bars represent the mean ± SD. See also Figure S2.
ESCRT proteins are required for HCV infected and HCV SGR cells to trigger pDC IFNα production
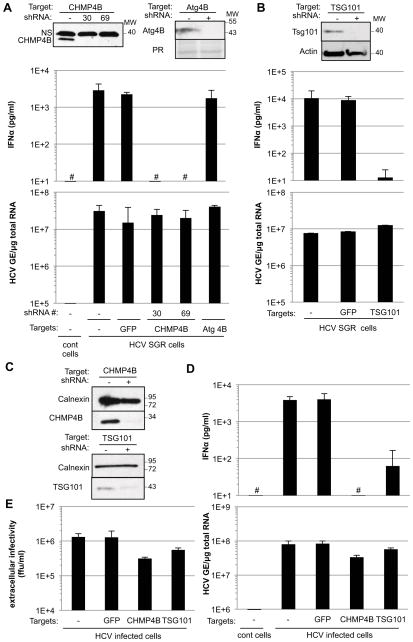
The endosomal sorting complex required for transport (ESCRT) machinery is known to be required for exosome biogenesis (Baietti et al., 2012; Fevrier et al., 2004; Tamai et al., 2010). To determine if it is also required for generation of the pDC-activating signal by HCV infected Huh-7.5.1c2 cells and SGR cells, we used lentivirus vector-based shRNA transduction to inhibit the expression of chromatin modifying protein 4B (CHMP4B) (Figure 6A, upper left panel), a component of complex III of the ESCRT machinery (Hurley et al., 2010). Importantly, the ability of CHMP4B down-regulated HCV SGR cells to stimulate IFNα production by cocultured pDCs was strongly inhibited (Figure 6A, middle panel, lanes 4 and 5) while their ability to support HCV replication was unchanged (Figure 6A, lower panel, lanes 4 and 5). In contrast, shRNAs that target either an irrelevant (GFP) (Figure 6A, middle panel, lane 3) or an unrelated non-ESCRT protein (Atg4B) (Figure 6A, middle panel, lane 6) did not impair IFNα production by cocultured pDCs. To confirm these results, we asked if tumor susceptibility gene 101 (TSG101) expression, a critical component of complex I of the ESCRT machinery (Hurley et al., 2010) was required for the HCV SGR cells to trigger pDC IFNα production. Again, downregulation of TSG101 expression in HCV SGR cells (Figure 6B, upper panel) abolished their ability to trigger pDC IFNα production (Figure 6B, middle panel) without inhibiting HCV RNA replication (Figure 6B, lower panel). These results strongly suggest that MVBs play a key role in the production of the pDC-activating signal by SGR cells.
Figure 6. ESCRT proteins are required for production of the pDC-activating signal by HCV SGR and HCV infected cells.
The impact of shRNA-mediated CHMP4B down-regulation, using two different shRNAs (#30 and #69, described in Table S1) (A) and shRNA-mediated TSG101 down-regulation (B) on expression of the corresponding proteins (upper panels), levels of IFNα in HCV SGR Huh-7.5.1c2 cell-pDC coculture supernatants (middle panels) and intracellular HCV RNA in Huh-7.5.1c2 SGR cells (lower panels) was compared with HCV SGR cells transduced with shRNA against GFP and an shRNA against an unrelated cellular protein (Atg4B) or non-transduced HCV SGR cells (-). HCV-negative Huh-7.5.1c2 cells served as controls (cont cells). The hatch marks (#) indicate results below the limit of detection of the IFNα ELISA (12.5 pg/ml). NS, non-specific band; PR, ponceau red staining; MW, molecular weight marker. Results are representative of 2 independent experiments each performed in triplicate. Error bars represent the mean ± SD. (C–E) CHMP4B and TSG101 down-regulated Huh7.5.1c2 cells, infected by HCV (MOI of 5 for 48 hours) were cocultured with pDCs for 20 hours. The levels of CHMP4B and TSG101 expression (C), IFNα in coculture supernatants (D, upper panel), intracellular HCV RNA (D, lower panel) and titration of HCV infectious particles in supernatants of the infected cells (E) were compared with control HCV infected cells transduced with shRNA against GFP. Uninfected Huh-7.5.1c2 cells served as controls (cont cells). Results are representative of 2 independent experiments each performed in triplicate. Error bars represent the mean ± SD. See also Figure S3.
Importantly, we confirmed these results by demonstrating that shRNA-mediated downregulation of CHMP4B and TSG101 expression in HCV infected cells (Figure 6C) either abrogated or markedly impaired (about 60 fold) their ability to trigger pDC IFNα production (Figure 6D, upper panel), and - consistent with previous reports (Ariumi et al., 2011; Corless et al., 2010; Tamai et al., 2012) - with little or no impact on HCV RNA replication (Figure 6D, lower panel), and inhibition, although modestly (i.e. <2–4 fold), of infectious virus production (Figure 6E). Similar results were obtained using Huh-7.5.1 cells and parental Huh-7 cells (Figure S3). Collectively, these results indicate that the ESCRT machinery is required for HCV infected and SGR cells to activate pDCs, suggesting that MVBs are required for this process and reinforcing the notion that the pDC activating signal is exosomal HCV RNA.
ANXA2 is required for HCV infected and HCV SGR cells to trigger pDC IFNα production
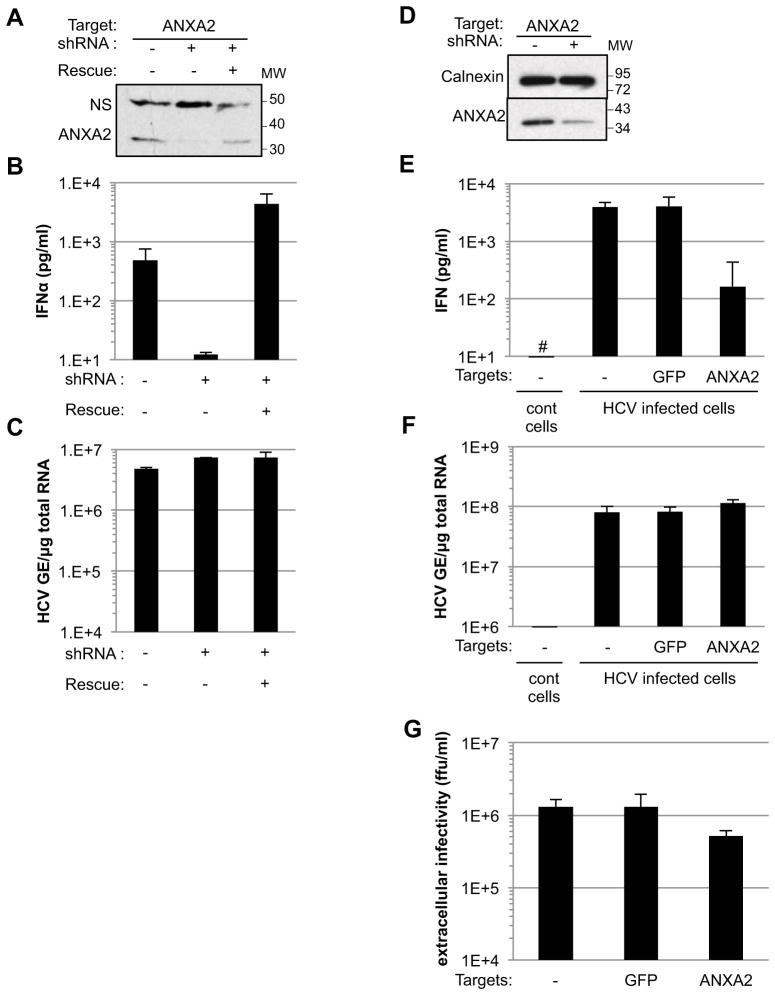
Annexin A2 (ANXA2) is an RNA-binding protein (Aukrust et al., 2007; Filipenko et al., 2004) that plays a key role in membrane vesicle trafficking and MVB biogenesis (Gerke et al., 2005; Mayran et al., 2003; Morel et al., 2009). Interestingly, recent reports indicate that ANXA2 is recruited to HCV replication sites where it regulates virus particle production (Backes et al., 2010; Saxena et al., 2012). Therefore, we used shRNA mediated knock-down to determine if ANXA2 is required for HCV infected Huh-7.5.1c2 cells and SGR cells to trigger IFNα production by co-cultured pDCs. As shown in Figure 7, ANXA2 downregulation in HCV SGR cells (Figure 7A) abolished their ability to trigger IFNα production by co-cultured pDCs (Figure 7B) without inhibiting HCV RNA replication (Figure 7C), the latter confirming the results of Backes et al (Backes et al., 2010). The ANXA2 requirement for pDC activation was confirmed by restoration of pDC-activation by ANXA2 shRNA-downregulated SGR cells by ectopic over-expression of an ANXA2 encoding shRNA-resistant RNA (Figure 7B). Consistent with these results, down-regulation of ANXA2 expression in HCV infected cells (Figure 7D) decreased their ability to trigger IFNα production by co-cultured pDCs by more than 20-fold (Figure 7E), without inhibiting HCV RNA replication (Figure 7F) or significantly (< 2.5-fold) suppressing infectious virus production (Figure 7G). Similar results were obtained using Huh-7.5.1 cells and parental Huh-7 cells (Figure S4). Altogether, these results demonstrated that ANXA2 is required for both HCV SGR cells and HCV infected cells to trigger IFNα production by pDCs, possibly by orchestrating the recruitment of appropriate cellular and/or viral components to generate the HCV RNA-containing exosomes that trigger the pDC IFN response.
Figure 7. ANXA2 is required for HCV infected and SGR cells to trigger IFNα production by pDCs.
shRNA-mediated downregulaton of ANXA2 protein in HCV SGR Huh-7.5.1c2 cells (A), strongly reduced IFNα production by cocultured pDCs (B) but had no impact on HCV replication (C). All results were compared with ANXA2 shRNA-transduced HCV SGR Huh-7.5.1c2 cells that ectopically express an shRNA-resistant variant of ANXA2 (Rescue). NS, non specific band; MW, molecular weight markers. Results are representative of 2 independent experiments each performed in triplicate. The levels of down-regulation of ANXA2 expression in HCV infected Huh-7.5.1c2 cells (MOI of 5 for 48 hours) (D), of IFNα in HCV infected cell-pDC coculture supernatants (E), of intracellular HCV RNA in HCV infected cells (F), and of HCV infectious particles in supernatants of the infected cells (G) were compared with HCV infected cells transduced with an shRNA against GFP. HCV-negative Huh-7.5.1c2 cells served as controls (cont cells). The hatch marks (#) indicate results below the limit of detection of the IFNα ELISA (12.5 pg/ml). Error bars represent the mean ± SD. See also Figure S4.
Discussion
Plasmacytoid dendritic cells function as sentinels of viral infection, predominantly via recognition of single-stranded RNA and unmethylated CpG-containing DNA (CpG) by endosomal TLR7 and TLR9 respectively (Reizis et al., 2011), and they are a major IFNα producing cell type in vivo (Asselin-Paturel et al., 2003; Cisse et al., 2008; Kumagai et al., 2007; Reizis et al., 2011). The pDC response to these stimuli typically occurs when they are infected by viruses such as respiratory syncytial virus, influenza virus, lymphocytic choriomeningitis infection (Blasius et al., 2010; Hornung et al., 2004; Lee et al., 2007; Loo et al., 2011; Macal et al., 2012), resulting in the rapid secretion of type 1 IFN in a response that is up to 1,000-fold more vigorous than other similarly infected cell types (Liu, 2005).
In contrast to these conventional mechanisms for pDC-activation, here we define a host strategy that assures recognition of infectious agents that, on one hand, defeat the pathogen-sensing machinery in the cells they infect and, on the other hand, do not productively infect pDCs (Decalf et al., 2007). Using HCV as a model, we demonstrate that HCV-permissive cells that do not produce type 1 IFN because the HCV NS3/4A protease interrupts the RLR and TLR signaling pathways in the cells (Cheng et al., 2006; Foy et al., 2005; Horner et al., 2011; Li et al., 2005a; Li et al., 2005b; Liang et al., 2008; Loo et al., 2006; Meylan et al., 2005; Wang et al., 2009) can selectively package immunostimulatory viral RNA within exosomes that deliver their RNA cargo to neighboring, HCV-nonpermissive (Decalf et al., 2007) pDCs in a concentration-dependent manner that triggers an antiviral IFN response. We suggest that this mechanism may have evolved to protect the host against the pathogens that, like HCV, can blunt the IFN response in productively infected cells (Versteeg et al., 2010).
To unambiguously dissect the exosomal transmission of immunostimulatory HCV RNA without the potentially confounding effect of HCV RNA-containing virus particles, in many experiments we used SGR cells that replicate a subgenomic fragment of viral RNA that encodes only the HCV nonstructural proteins and, thus, cannot assemble virus particles. Those results, and others performed in HCV infected cells using exosome release inhibitors and shRNA downregulation of the ESCRT machinery and ANXA2, reveal that immunostimulatory HCV RNA is produced by SGR cells and infected cells, packaged within exosomes, and transferred to cocultured pDCs which it triggers to produce type I IFN. Exosome formation is initiated in multivesicular bodies (MVBs) by invagination of endosomal membranes and generation of intraluminal vesicles (ILV) (Al-Nedawi et al., 2008; Bobrie et al., 2011; Couzin, 2005; Fevrier et al., 2004; Liegeois et al., 2006). Importantly, HCV RNA colocalized with CD63, CD81 and CHMP4B proteins - known to be enriched in MVB (Figure 3), suggesting that HCV RNA is directed to this cellular compartment by an as yet unknown mechanism. The ESCRT machinery regulates membrane budding and ILV formation at the late endosome level (Henne et al., 2011; Raiborg et al., 2009) and ESCRT components are thus involved in exosome biogenesis (Baietti et al., 2012; Fevrier et al., 2004; Tamai et al., 2010). Therefore, one might speculate that ESCRT proteins - such as Tsg101 and CHMP4B - might contribute to the processes of membrane bending and abscission that direct HCV RNA to ILVs and exosomes. Alternatively, Tsg101 and CHMP4B might be indirectly involved in endosomal compartments that require ESCRTs for their architectural and functional integrity (Doyotte et al., 2005; Razi et al., 2006).
Although exosomal transfer of functional cellular mRNA and microRNA to recipient cells has been previously reported (Kosaka et al., 2010; Meckes et al., 2010; Pegtel et al., 2010; Skog et al., 2008; Valadi et al., 2007), to our knowledge, our demonstration that exosomes can mediate the transfer of immunostimulatory viral RNA to type I IFN-producing plasmacytoid dendritic cells in a cell contact-dependent manner reveals a previously unappreciated aspect of the innate immune response to viral infection. In addition, it is conceivable that exosome-mediated export of viral RNA may have a variety of unexpected and potentially alternative outcomes. For example, it could theoretically enhance viral clearance by purging viral RNA from the cell. Alternatively, it could have proviral effects by vesicular sequestration of otherwise immunostimulatory RNA from the cytosolic RNA sensing machinery of the cell. It is also conceivable that HCV RNA-containing exosomes might also transfer infectious viral genomes to cells (e.g. hepatocytes) that are permissive for viral replication, in which case it would facilitate cell-to-cell spread of an infection that is unconstrained by a neutralizing antibody response. Additional experiments will be required to test these interesting hypotheses.
We are intrigued by the fact that although pDC activation is exosome-mediated, it requires cell-cell contact because unconcentrated supernatants from either infected cells or SGR cells don’t trigger IFNα production by pDCs (Takahashi et al., 2010). This suggests that HCV infected cells and SGR cells secrete exosomes into the culture supernatant at concentrations that are below an excitatory threshold that could be reached in the intercellular space during cell-cell contact. This is corroborated by the fact that the maximal HCV/pDC ratio achieved by the coincubation of unconcentrated SGR cell supernatants with pDCs (i.e. ~0.2–0.5 HCV genomes/pDC) is not sufficient to trigger IFNα production (data not shown and (Takahashi et al., 2010)). In contrast, SGR cell supernatant derived exosome preparations containing at least 10-fold higher HCV RNA concentrations (i.e. HCV RNA/pDC ratio of at least 2) are required to trigger low level pDC IFNα production, and at least 45 HCV RNA copies per pDC are needed for robust pDC activation (Figure 5D). Interestingly, whereas HCV RNA was detectable by FISH in only ~10–15% of total pDC it was detectable in 3 times as many IFNα-positive pDCs (Figure 2D). The absence of HCV RNA in many IFNα-positive pDCs may imply that they were stimulated in a paracrine fashion by IFNα, as previously suggested (Takahashi et al., 2010). Importantly, the detection of HCV RNA in 8.6% of IFNα-negative pDCs may suggest either that independent subsets of pDCs may be differentially responsive to the HCV RNA stimulus, or that time is required for HCV RNA to trigger pDCs to produce enough IFNα to be detectable and they were observed before that threshold was reached.
The potential physiological relevance of the process described in this report is supported by the fact that exosome-mediated cell-cell transmission of siRNA occurs between hepatocytes in vivo (Pan et al., 2011), that HCV RNA-containing exosomes are detectable in the plasma of chronically HCV infected patients (Masciopinto et al., 2004), that pDCs are abundant in HCV infected liver (Lau et al., 2008), and that pDCs from HCV-infected patients respond normally to HCV infected cells (Takahashi et al., 2010). In principle, therefore, pDC IFN production triggered by exosomes released by neighboring HCV-infected hepatocytes could, theoretically at least, occur during natural HCV infection. Additional experiments will be necessary to formally test this hypothesis.
In summary, the current results illustrate a previously unsuspected aspect of the innate host response in which the vesicular transport machinery in a virus-infected cell produces immunostimulatory viral RNA-containing exosomes that are transferred at close range to professional IFN-producing cells whose ability to trigger an innate response is not opposed because they are not permissive for infection. These results may have broad implications for our understanding of the host-virus relationship because cell contact-dependent and TLR7-dependent pDC activation is now known to extend to cells that replicate alphaviruses (Takahashi et al., 2010), to HIV infection (Lepelley et al., 2011), and to a wide variety of human (e.g. Huh-7, HeLa) and animal (e.g. NIH3T3) cells that are infected by lymphocytic choriomeningitis virus (Wieland et al, manuscript in preparation). The current results provide a compelling rationale for examination of the role of RNA-containing exosomes in the delivery of these evolutionarily and structurally distinct viral RNAs to pDCs.
Experimental Procedures
Biological Materials
Unless otherwise indicated, a subclone of Huh-7.5.1 cells (Huh-7.5.1c2) (Dreux et al., 2009; Gastaminza et al., 2010; Pedersen et al., 2007) that were infected by the JFH-1 infectious molecular clone of HCV and Huh-7.5.1c2 cells that harbor the JFH-1 subgenomic replicon (SGR) (Kato et al., 2003) were used in this study. In selected experiments, results were confirmed using Huh-7.5.1 cells from which the Huh-7.5.1c2 cells were derived (Figures S1, S4 and S5), and using the parental Huh-7 cell line. All cells were maintained in DMEM (Cellgro, Mediatech, Herndon, VA) supplemented with 10% FBS (Cellgro), 10 mM HEPES, 100 units/ml penicillin, 100 mg/ml streptomycin and 2 mM L-glutamine (Invitrogen, Carlsbad, CA) in 5% CO2 at 37°C. Viral stocks of cell-culture adapted JFH-1 virus (D183) (Zhong et al., 2006) were prepared by infection of Huh-7.5.1c2 cells at a multiplicity of infection (MOI) of 0.01, as described previously (Dreux et al., 2009). Plasmacytoid dendritic cells (pDCs) were isolated from 450 ml of blood from healthy adult human volunteers after informed consent was obtained according to procedures approved by the Scripps Research Institute Human Research Committee. Briefly, PBMCs were isolated using Ficoll-Hypaque density centrifugation and pDCs were positively selected from PBMCs using BDCA-4-magnetic beads (MACS Miltenyi Biotec, Auburn, CA) according to the manufacturer’s instructions. The typical yields of PBMCs and pDCs were 500×106 and 2×106 cells, respectively, with a typical purity of >95% pDCs. Isolated pDCs were maintained in RPMI 1640 medium (Cellgro) supplemented with 10% FBS, 10 mM HEPES, 100 units/ml penicillin, 100 mg/ml streptomycin, 2 mM L-glutamine, non-essential amino acids and 1 mM sodium pyruvate at 37°C/5% CO2. Detailed descriptions of the reagents and constructs are provided as Supplemental Information.
Detection of HCV RNA in pDC-HCV SGR cell co-cultures by Fluorescent In Situ Hybridization (FISH) analysis
After isolation, pDCs were co-cultured with HCV SGR cells for 20 hours at 37°C. After 4% paraformaldehyde fixation at room temperature and PBS washing, HCV plus strand RNA was detected using a probe set that targets a region between nucleotide positions 2733 and 4870 in the JFH-1 genome (Panomics/Affymetrix, Santa Clara, CA) according to the manufacturer’s instructions. For immunostaining, mouse anti-IFNα (MACS Miltenyi Biotec, Auburn, CA) was diluted at 2 μg/ml, using blocking and binding buffers as previously described (Dreux et al., 2009). Nuclei were stained by Hoechst dye. Images were acquired with Zeiss LSM 710 laser scanning confocal microscope and analyzed with IMARIS (Bitplane Inc.) software. Details for the simultaneous detection of HCV RNA and CD63, CD81 and CHMP4B protein are provided as Supplemental Information.
Analysis of extracellular and intracellular RNA levels
Supernatants of 80% confluent HCV SGR cells were cleared by 2,000 x g centrifugation for 20 min at 4 °C followed by filtration through a 0.22 μm filter (Corning). Pre-cleared supernatants were treated with 0.5% NP40, 100 μg/ml RNAse and/or 0.4 U/μl RNAse inhibitors for one hour at room temperature with agitation. The efficiency of RNA extraction and RT-qPCR was controlled by adding in vitro transcribed Xef1α (x enopus transcription factor 1a) to supernatants diluted in guanidinium thiocyanate citrate buffer (GTC). Extracellular HCV RNA, Xef1α and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels were determined by reverse transcription-real-time quantitative PCR (RT-qPCR), as previously described (Dreux et al., 2009). Extracellular and intracellular HCV RNA levels were normalized for Xef1α and GAPDH RNA levels, respectively. The sequences of the primers are described in the Supplemental Table 1. A detailed description of the procedure used for Northern Blot Analysis is provided as Supplemental Information.
Exosome isolation
Supernatants were pre-cleared by sequential low and high-speed centrifugation as follows. Briefly, supernatants of 80% confluent HCV SGR cells were harvested twice 2 hours apart and clarified by centrifugation at 2,000 x g for 20 min (4°C), yielding supernatants that were further clarified by centrifugation at 10,000 x g for 30 min (4°C). Exosomes in the clarified supernatants were then pelleted at 110,000 x g for 2 hours at 4°C using a SW28 rotor, as previously described (Thery et al., 2006). The pelleted exosome preparations were resuspended in exosome-depleted medium. Approximately 0.25–0.5 μg of exosomal proteins were usually obtained from 1–2 × 106 cells. For each exosome isolation, the purity of the preparation was determined by Western blot analysis and described in Supplementary Information.
Downregulation by lentivirus vector-based shRNA transduction and expression rescue assay
The lentivirus vector-based shRNA were produced as described previously (Dreux et al., 2009) and details are provided in Supplementary Information. Sequences of the shRNA are provided in Table S1. ANXA2 expression was rescued by transduction of ANAX2 down-regulated cells with an MLV-based vector that permits expression of an ANXA2-shRNA resistant cDNA. Details are provided in Supplementary Information.
Coculture experiments
Unless otherwise indicated, 2 × 105 HCV SGR cells, infected cells, or parental cells were cocultured with 2 × 104 pDCs in 96-well round-bottom plates in a 37 °C/5% CO2 incubator. Twenty hours later, cell-culture supernatants were collected and IFNα levels were measured using a commercially available ELISA kit (PBL Interferon Source, Piscataway, NJ), following the manufacturer’s instructions.
Supplementary Material
Highlights.
HCV RNA is selectively packaged within exosomes and transferred to neighboring pDCs
Viral RNA-containing exosomes trigger interferon α production by pDCs
pDC activation by HCV infected cells is suppressed by exosome release inhibitors
Exosomal HCV RNA delivery is dependent on Annexin A2 and the ESCRT machinery
Acknowledgments
We thank Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan) for kindly providing the infectious JFH-1 molecular clone and replicon constructs, Charles M. Rice (Rockefeller University, New York) for Huh-7.5 cells from which the Huh-7.5.1 and Huh-7.5.1c2 cells were derived, and for anti-NS5A antibodies (9E10), Inder Verma (Salk Institute, La Jolla, CA) for lentiviral plasmids, François-Loïc Cosset (Ecole Normale Supérieure de Lyon, INSERM U758, France) for retroviral-based vectors, Weidong Zhong (Gilead Sciences) for the inhibitor 2′-C-methyladenosine, Michinori Kohara (Tokyo Metropolitan Institute of Medical Science, Tokyo) for providing the anti-NS4A/NS4B antibody. We are grateful to Wesley I. Sundquist (University of Utah, Salt Lake City) and Sandra Schmid (Scripps Research Institute, La Jolla, CA) for constructive comments and suggestions, to François-Loïc Cosset and Yvon Jaillais for critical reading of the manuscript and to our Scripps laboratory colleagues for encouragement and help. This work was supported by NIH grants AI079043 and AI088778, by a grant from the Institut Mérieux (Lyon, France), and by a grant from the “Agence Nationale pour la Recherche contre le SIDA et les Hépatites Virales” (ANRS). This is manuscript number 21522 from the Scripps Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- Alter H, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin Liver Dis. 2000;20:17–35. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- Ariumi Y, Kuroki M, Maki M, Ikeda M, Dansako H, Wakita T, Kato N. The ESCRT system is required for hepatitis C virus production. PLoS ONE. 2011;6:e14517. doi: 10.1371/journal.pone.0014517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Paturel C, Brizard G, Pin JJ, Briere F, Trinchieri G. Mouse strain differences in plasmacytoid dendritic cell frequency and function revealed by a novel monoclonal antibody. J Immunol. 2003;171:6466–6477. doi: 10.4049/jimmunol.171.12.6466. [DOI] [PubMed] [Google Scholar]
- Aukrust I, Hollas H, Strand E, Evensen L, Trave G, Flatmark T, Vedeler A. The mRNA-binding site of annexin A2 resides in helices C-D of its domain IV. J Mol Biol. 2007;368:1367–1378. doi: 10.1016/j.jmb.2007.02.094. [DOI] [PubMed] [Google Scholar]
- Backes P, Quinkert D, Reiss S, Binder M, Zayas M, Rescher U, Gerke V, Bartenschlager R, Lohmann V. Role of annexin A2 in the production of infectious hepatitis C virus particles. J Virol. 2010;84:5775–5789. doi: 10.1128/JVI.02343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Bigger CB, Brasky KM, Lanford RE. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J Virol. 2001;75:7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger CB, Guerra B, Brasky KM, Hubbard G, Beard MR, Luxon BA, Lemon SM, Lanford RE. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J Virol. 2004;78:13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Bobrie A, Colombo M, Raposo G, Thery C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. Exosome release of beta-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol. 2010;190:1079–1091. doi: 10.1083/jcb.201002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Zhong J, Chisari FV. Inhibition of dsRNA-induced signaling in hepatitis C virus-infected cells by NS3 protease-dependent and -independent mechanisms. Proc Natl Acad Sci U S A. 2006;103:8499–8504. doi: 10.1073/pnas.0602957103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, Holmberg D, Zweier C, den Hollander NS, Kant SG, Holter W, Rauch A, Zhuang Y, Reizis B. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless L, Crump CM, Griffin SD, Harris M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol. 2010;91:362–372. doi: 10.1099/vir.0.017285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzin J. Cell biology: The ins and outs of exosomes. Science. 2005;308:1862–1863. doi: 10.1126/science.308.5730.1862. [DOI] [PubMed] [Google Scholar]
- Decalf J, Fernandes S, Longman R, Ahloulay M, Audat F, Lefrerre F, Rice C, Pol S, Albert ML. Plasmacytoid dendritic cells initiate a complex chemokine and cytokine network and are a viable drug target in chronic HCV patients. J Exp Med. 2007;204:2423–2437. doi: 10.1084/jem.20070814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dental C, Florentin J, Aouar B, Gondois-Rey F, Durantel D, Baumert TF, Nunes JA, Olive D, Hirsch I, Stranska R. Hepatitis C virus fails to activate NF-kappaB signaling in plasmacytoid dendritic cells. J Virol. 2012;86:1090–1096. doi: 10.1128/JVI.05444-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyotte A, Russell MR, Hopkins CR, Woodman PG. Depletion of TSG101 forms a mammalian “Class E” compartment: a multicisternal early endosome with multiple sorting defects. J Cell Sci. 2005;118:3003–3017. doi: 10.1242/jcs.02421. [DOI] [PubMed] [Google Scholar]
- Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hage N, Luo G. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J Gen Virol. 2003;84:2761–2769. doi: 10.1099/vir.0.19305-0. [DOI] [PubMed] [Google Scholar]
- Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol. 2010;91:2230–2237. doi: 10.1099/vir.0.022186-0. [DOI] [PubMed] [Google Scholar]
- Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Filipenko NR, MacLeod TJ, Yoon CS, Waisman DM. Annexin A2 is a novel RNA-binding protein. J Biol Chem. 2004;279:8723–8731. doi: 10.1074/jbc.M311951200. [DOI] [PubMed] [Google Scholar]
- Foy E, Li K, Sumpter R, Jr, Loo YM, Johnson CL, Wang C, Fish PM, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaminza P, Dryden KA, Boyd B, Wood MR, Law M, Yeager M, Chisari FV. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J Virol. 2010;84:10999–11009. doi: 10.1128/JVI.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Horner SM, Liu HM, Park HS, Briley J, Gale M., Jr Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Schlender J, Guenthner-Biller M, Rothenfusser S, Endres S, Conzelmann KK, Hartmann G. Replication-dependent potent IFN-alpha induction in human plasmacytoid dendritic cells by a single-stranded RNA virus. J Immunol. 2004;173:5935–5943. doi: 10.4049/jimmunol.173.10.5935. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Yoshioka Y, Hagiwara K, Takeshita F, Ochiya T. Competitive interactions of cancer cells and normal cells via secretory microRNAs. J Biol Chem. 2012;287:1397–1405. doi: 10.1074/jbc.M111.288662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y, Takeuchi O, Kato H, Kumar H, Matsui K, Morii E, Aozasa K, Kawai T, Akira S. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity. 2007;27:240–252. doi: 10.1016/j.immuni.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Lau DT, Fish PM, Sinha M, Owen DM, Lemon SM, Gale M., Jr Interferon regulatory factor-3 activation, hepatic interferon-stimulated gene expression, and immune cell infiltration in hepatitis C virus patients. Hepatology. 2008;47:799–809. doi: 10.1002/hep.22076. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Lepelley A, Louis S, Sourisseau M, Law HK, Pothlichet J, Schilte C, Chaperot L, Plumas J, Randall RE, Si-Tahar M, Mammano F, Albert ML, Schwartz O. Innate sensing of HIV-infected cells. PLoS Pathog. 2011;7:e1001284. doi: 10.1371/journal.ppat.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005a;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Sun L, Seth RB, Pineda G, Chen Z. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005b;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ishida H, Lenz O, Lin TI, Nyanguile O, Simmen K, Pyles RB, Bourne N, Yi M, Li K, Lemon SM. Antiviral suppression vs restoration of RIG-I signaling by hepatitis C protease and polymerase inhibitors. Gastroenterology. 2008;135:1710–1718. e1712. doi: 10.1053/j.gastro.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Liegeois S, Benedetto A, Garnier JM, Schwab Y, Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J Cell Biol. 2006;173:949–961. doi: 10.1083/jcb.200511072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DTY, Weinman SA, Lemon SM, Michael Gale J. Viral and therapeutic control of IFN-β promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103:6001. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macal M, Lewis GM, Kunz S, Flavell R, Harker JA, Zuniga EI. Plasmacytoid Dendritic Cells Are Productively Infected and Activated through TLR-7 Early after Arenavirus Infection. Cell Host Microbe. 2012;11:617–630. doi: 10.1016/j.chom.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masciopinto F, Giovani C, Campagnoli S, Galli-Stampino L, Colombatto P, Brunetto M, Yen TS, Houghton M, Pileri P, Abrignani S. Association of hepatitis C virus envelope proteins with exosomes. Eur J Immunol. 2004;34:2834–2842. doi: 10.1002/eji.200424887. [DOI] [PubMed] [Google Scholar]
- Mayran N, Parton RG, Gruenberg J. Annexin II regulates multivesicular endosome biogenesis in the degradation pathway of animal cells. Embo J. 2003;22:3242–3253. doi: 10.1093/emboj/cdg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG, Jr, Raab-Traub N. Microvesicles and viral infection. J Virol. 2011;85:12844–12854. doi: 10.1128/JVI.05853-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG, Jr, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A. 2010;107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Morel E, Parton RG, Gruenberg J. Annexin A2-dependent polymerization of actin mediates endosome biogenesis. Dev Cell. 2009;16:445–457. doi: 10.1016/j.devcel.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Pan Q, Ramakrishnaiah V, Henry S, Fouraschen S, de Ruiter PE, Kwekkeboom J, Tilanus HW, Janssen HL, van der Laan LJ. Hepatic cell-to-cell transmission of small silencing RNA can extend the therapeutic reach of RNA interference (RNAi) Gut. 2011 Dec 23; doi: 10.1136/gutjnl-2011-300449. [Epub ahead of print] PMID 22198713. [DOI] [PubMed] [Google Scholar]
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietschmann T, Lohmann V, Kaul A, Krieger N, Rinck G, Rutter G, Strand D, Bartenschlager R. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J Virol. 2002;76:4008–4021. doi: 10.1128/JVI.76.8.4008-4021.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Razi M, Futter CE. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol Biol Cell. 2006;17:3469–3483. doi: 10.1091/mbc.E05-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis B, Bunin A, Ghosh HS, Lewis KL, Sisirak V. Plasmacytoid dendritic cells: recent progress and open questions. Annu Rev Immunol. 2011;29:163–183. doi: 10.1146/annurev-immunol-031210-101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena V, Lai CK, Chao TC, Jeng KS, Lai MM. Annexin A2 is involved in the formation of hepatitis C virus replication complex on the lipid raft. J Virol. 2012;86:4139–4150. doi: 10.1128/JVI.06327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, Chisari FV. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Asabe S, Wieland S, Garaigorta U, Gastaminza P, Isogawa M, Chisari FV. Plasmacytoid dendritic cells sense hepatitis C virus-infected cells, produce interferon, and inhibit infection. Proc Natl Acad Sci U S A. 2010;107:7431–7436. doi: 10.1073/pnas.1002301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai K, Shiina M, Tanaka N, Nakano T, Yamamoto A, Kondo Y, Kakazu E, Inoue J, Fukushima K, Sano K, Ueno Y, Shimosegawa T, Sugamura K. Regulation of hepatitis C virus secretion by the Hrs-dependent exosomal pathway. Virology. 2012;422:377–385. doi: 10.1016/j.virol.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Tamai K, Tanaka N, Nakano T, Kakazu E, Kondo Y, Inoue J, Shiina M, Fukushima K, Hoshino T, Sano K, Ueno Y, Shimosegawa T, Sugamura K. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem Biophys Res Commun. 2010;399:384–390. doi: 10.1016/j.bbrc.2010.07.083. [DOI] [PubMed] [Google Scholar]
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Versteeg GA, Garcia-Sastre A. Viral tricks to grid-lock the type I interferon system. Curr Opin Microbiol. 2010;13:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva RA, Jangra RK, Yi M, Pyles R, Bourne N, Lemon SM. miR-122 does not modulate the elongation phase of hepatitis C virus RNA synthesis in isolated replicase complexes. Antiviral Res. 2010;88:119–123. doi: 10.1016/j.antiviral.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Liang Y, Devaraj S, Wang J, Lemon SM, Li K. Toll-like receptor 3 mediates establishment of an antiviral state against hepatitis C virus in hepatoma cells. J Virol. 2009;83:9824–9834. doi: 10.1128/JVI.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuyama K, Sun H, Mitsutake S, Igarashi Y. Sphingolipid-modulated Exosome Secretion Promotes Clearance of Amyloid-beta by Microglia. J Biol Chem. 2012;287:10977–10989. doi: 10.1074/jbc.M111.324616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Kodys K, Babcock GJ, Szabo G. CD81/CD9 tetraspanins aid plasmacytoid dendritic cells in recognition of HCV-infected cells and induction of IFNalpha. Hepatology. 2012 May 10; doi: 10.1002/hep.25827. [Epub ahead of print] PMID: 22577054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Chung J, Stamataki Z, Isogawa M, Cheng G, McKeating JA, Chisari FV. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J Virol. 2006;80:11082–11093. doi: 10.1128/JVI.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.