Abstract
Differential 18O/16O stable isotope labeling of peptides that relies on enzyme-catalyzed oxygen exchange at their carboxyl termini in the presence of H218O has been widely used for relative quantitation of peptides/proteins. The role of tryptic proteolysis in bottom-up shotgun proteomics and low reagent costs, has made trypsin-catalyzed 18O post-digestion exchange a convenient and affordable stable isotope labeling approach. However, it is known that trypsin-catalyzed 18O exchange at the carboxyl terminus is in many instances inhomogeneous/incomplete. The extent of the 18O exchange/incorporation fluctuates from peptide to peptide mostly due to variable enzyme-substrate affinity. Thus, accurate calculation and interpretation of peptide ratios are analytically complicated and in some regard deficient. Therefore, a computational approach capable of improved measurement of actual 18O incorporation for each differentially labeled peptide pair is needed. In this regard, we have developed an algorithmic method that relies on the trapezoidal rule to integrate peak intensities of all detected isotopic species across a particular peptide ion over the retention time, which fits the isotopic manifold to Poisson distributions. Optimal values for manifold fitting were calculated and then 18O/16O ratios derived via evolutionary programming. The algorithm is tested using trypsin–catalyzed 18O post-digestion exchange to differentially label bovine serum albumin (BSA) at a priori determined ratios. Both, accuracy and precision are improved utilizing this rigorous mathematical approach. Utilizing this algorithmic technique, we demonstrate the effectiveness of this method to accurately calculate 18O/16O ratios for differentially labeled BSA peptides, by accounting for artifacts caused by a variable degree of post-digestion 18O exchange. We further demonstrate the effectiveness of this method to accurately calculate 18O/16O ratios in a large scale proteomic quantitation of detergent resistant membrane microdomains (DRMMs) isolated from cells expressing wild-type HIV-1 Gag and its non myristylated mutant.
Keywords: quantitation, 18O/16O stable isotope labeling, variable/incomplete 18O exchange
INTRODUCTION
Protease catalyzed 18O/16O stable isotope labeling is a simple method used to differentially label two biologically distinct samples that allows relative quantitation of proteins on a large-scale basis using mass spectrometry (MS)-based proteomics.1, 2 This approach relies on trypsin-catalyzed exchange of two 16O atoms for two 18O atoms at the C-terminal carboxyl group of tryptic peptides, resulting in a mass shift of 4 daltons between singly charged, differentially labeled peptides observed in MS1 spectra.3 It has been demonstrated that 18O exchange can be decoupled from the protein digestion step, allowing the labeling conditions to be optimized separately.4 This strategy is currently adopted as a standard 18O/16O labeling procedure, allowing minimal consumption of 18O water and simplifying sample handling.1
In comparison to other stable isotope labeling techniques, 18O/16O stable isotope labeling offers several advantages. First, this labeling technique does not target peptides containing particular amino acids and does not require an additional affinity-based step for labeled peptide enrichment (e.g. ICAT).5 Second, in contrast to metabolic labeling techniques (e.g. SILAC),6 18O/16O labeling is amenable to clinically relevant samples, including human plasma/serum or fresh-frozen tissue specimens.7 Third, during the 18O/16O labeling process, every proteolytic fragment is labeled according to enzyme-specific cleavage pattern (e.g. trypsin, Glu-C). For this reason 18O/16O labeling is well suited for amount-limited samples including laser capture microdissected (LCM) specimens8 obtained from fresh-frozen and formalin-fixed tissue slices or plasma membrane specimens.9 Finally, the low cost of reagents along with the ability to be combined with other stable isotope labeling methods, represents additional comparative advantages of 18O/16O labeling.10
However, it is known that the two C-terminal 16O atoms are not always completely exchanged for two 18O atoms during the enzyme catalyzed 18O exchange. This is primarily due to variable peptide-specific enzyme reaction rates Km11 and the fact that the majority of labeling experiments are carried out in the presence of 95% enriched H218O. Thus, the isotopic envelope depicting peptide doublets characterized by variable 18O exchange rate shows a complex pattern.12 Additionally, the accurate computation of 18O/16O ratios for peptides featuring variable/incomplete 18O exchange is not straightforward. It is for these reasons, along with experimental efforts focused on optimization of labeling conditions,12–14 that several computational methods have been developed for analysis of MS data, in attempt to rectify incomplete 18O incorporation.12,15–23
Therefore, we now report and demonstrate results of an algorithm for improved accuracy and reproducibility of 18O/16O ratios computation capable of recognizing and accounting for peptide clusters exhibiting variable/incomplete 18O exchange. Our method applies the trapezoidal rule to integrate peak intensities of all detected isotopic species across a particular peptide ion over the retention time. Then it fits the isotopic manifold to Poisson distributions and derives fitting parameters via evolutionary programming. This then allows calculation of more accurate 18O/16O peptide ratios especially in the presence of variable 18O incorporation. The utility of this algorithm for quantitative proteomics is demonstrated using samples with varied complexity, including: 18O labeled bovine serum albumin (BSA) at a priori determined ratios, and 18O/16O labeled lipid raft membrane proteins from HeLa cells expressing HIV-1 Gag protein in a study of Gag trafficking.
MATERIALS AND METHODS
Reagents
Bovine serum albumin (BSA) and ammonium bicarbonate (NH4HCO3) were purchased from Sigma (St. Louis, MO). Sequencing grade trypsin was obtained from Promega (Madison, WI). H218O (95 % pure) was purchased from Cambridge Isotope Laboratories, Inc (Andover, MA). Bicinchonic acid (BCA) protein assay reagent kit was purchased from Pierce (Rockford, IL). Trifluoroacetic acid (CF3CO2H) and formic acid (CHOOH) were purchased from Fluka (Milwaukee, WI). High-performance liquid chromatography (HPLC)-grade methanol (CH3OH) and acetonitrile (CH3CN) were obtained from EM Science (Darmstadt, Germany). All solutions were prepared using water purified by a Nanopure II system (Dubuque, IA).
Transfection of HeLa cells and isolation of detergent-resistant membrane microdomains (DRMMs)
To express full-length Gag proteins in HeLa cells in the non-biohazardous context, we transfected approximately 2.5 × 106 HeLa cells with either pCMVNLGagPolRRE/PR− or pCMVNLGagPolRRE/PR−/1GA, along with pCMV-Rev (a kind gift from Dr. S. Venkatesan) and pCMV-Vphu (a kind gift from Dr. K. Strebel), using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as described in the manufacturer’s instruction. When co-transfected with Rev, pCMVNLGagPolRRE/PR− expresses wild type Gag and GagPol precursor polyproteins, whereas pCMVNLGagPolRRE/PR−/1GA expresses nonmyristylated derivatives of these polyproteins. pCMV-Vphu encodes the codon-optimized Vpu protein that facilitates release of virus-like particles.24 Twenty hours later, homogenates of transfected cells were prepared, post-nuclear supernatants of the homogenates were treated with 0.25% Triton X-100, and the detergent-resistant membrane fractions were isolated by equilibrium flotation centrifugation, as described previously.25 These fractions were diluted 18 fold with 25 mM (NH4)2CO3 and subjected to ultracentrifugation at 100,000×g for 2 h. Pellets were resuspended in 1.1 mL of 25 mM (NH4)2CO3 and stored at − 80 °C for subsequent proteomic analysis.
Tryptic digestion and 18O/16O labeling
The differential 18O/16O labeling of BSA and DRMMs were performed using slightly modified method, previously described.26 Briefly, proteins were first digested by trypsin at an enzyme/protein ratio of 1:50 overnight at 37 °C and lyophilized to dryness by Speed Vac (Thermo Savant, Holbrook, NY). Then the digest was aliquoted and resuspended in 20 % CH3OH/50 mM NH4HCO3 prepared in 95 % H218O or H216O respectively, and incubated with trypsin overnight at 37 °C using an enzyme/protein ratio of 1:20. After labeling, the enzyme activity was terminated by boiling in a water bath for 10 min. Digests were cooled on ice followed by acidification to pH 3 using TFA. The individually digested peptide pools H218O and H216O were mixed accordingly and lyophilized. The solely 18O labeled BSA digest was also lyophilized in the same fashion. The lyophilized digests were stored at − 80 °C and dissolved in 0.1 % TFA before LC-MS analysis.
Strong cation exchange (SCX) fractionation of differentially 16O/18O-Labeled DRMMs
The SCX of differentially labeled DRMMs digest was performed as previously described.27 Briefly, the 18O/16O-labeled digest from HeLa cell DRMM was reconstituted in 100 μL of 45% (v/v) CH3CN containing 0.1% (v/v) FA immediately prior to SCX chromatography. The digest was resolved into 14 fractions using a microcapillary HPLC system (Model 1100, Agilent Technologies Inc., Palo Alto, CA) on a 1 × 150 mm, 5 μm, 300 Å, polysulfoethyl A SCX column (PolyLC, Inc., Columbia, MD). Mobile phase A was 45% (v/v) CH3CN and mobile phase B was 45% (v/v) CH3CN containing 0.5 M ammonium formate (pH 3). Peptide fractions were eluted with an ammonium formate/multistep gradient at a flow rate of 200 μL/min as follows: 1% B/0–2 min, 10% B/62 min, 62% B/82 min, 100% B/85 min.
Nanoflow reversed-phase liquid chromatography (nanoRPLC) - tandem mass spectrometry (MS2)
NanoflowRPLC-MS2 analyses were performed in triplicates using an Agilent 1100 nanoflow LC system coupled with hybrid linear ion trap-Fourier transform ion cyclotron resonance (LIT-FTICR) mass spectrometer (LTQ FT Ultra from Thermo Electron, San Jose, CA). Microcapillary RPLC column (75 μm i.d. × 10 cm fused silica capillary with a flame pulled tip) was in-house slurry-packed with 5 μm, 300 Å pore size, Jupiter C-18 stationary phase (Phenomenex, Torrance, CA). After sample injection, the column was washed for 30 min with 98 % mobile phase A (0.1 % formic acid in water) at 0.5 μL/min. BSA peptides were eluted using a linear gradient of 2 % mobile phase B (0.1 % formic acid in ACN) to 42 % solvent B in 40 minutes at 0.25 μL/min, then to 98 % B for an additional 10 min. DRMM peptides from SCX fractions were eluted using a linear gradient of 2 % to 60 % solvent B in 100 minutes at 0.25 μL/min, then to 98 % B for an additional 10 min. The LIT-FTICR-MS was operated in a data dependent mode in which each full MS1 scan was followed by seven MS2 scans wherein the seven most abundant molecular ions were dynamically selected for collision-induced dissociation using normalized collision energy of 35 %.
MS data analysis and interpretation
Acquired mass spectra were searched against BSA protein sequence for BSA and human database with appended Gag protein sequence for HeLa cell DRMM using the SEQUEST algorithm implemented in the BioWorks 3.2 application (Thermo Electron, San Jose, CA), using 10 ppm precursor ion tolerance and 0.5 Da for fragment ions. Only fully tryptic peptides with up to two miscleavages were considered as positive identification. Charge state dependent cross correlation (Xcorr) ≥ 2.1 for [M+H]1+, ≥ 2.5 for [M+2H]2+ and ≥ 3.2 for [M+3H]3+ were used for BSA peptides and Xcorr ≥ 1.9 for [M+H]1+, ≥ 2.2 for [M+2H]2+ and ≥ 3.1 for [M+3H]3+ were used in DRMM samples.
Calculation of 18O/16O ratios using the XPRESS software
The ratios of heavy 18O labeled and light 16O labeled peptides were calculated using the XPRESS algorithm implemented in the BioWorks™ package (Version 3.2). XPRESS reports 18O/16O ratios for differentially labeled proteins as an average of ratios of all identified peptides unique to the protein under evaluation, except for C-terminal peptides.
RESULTS
Computation of 18O/16O ratios by fitting an isotope manifold using a Poisson distribution along with evolutionary programming
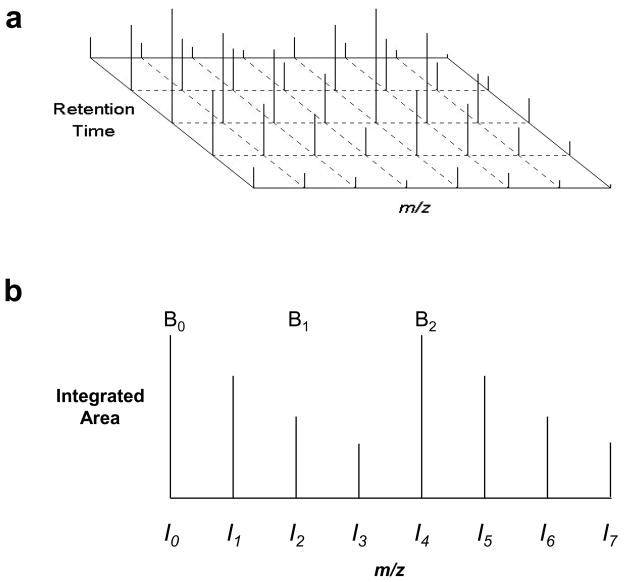
The LC-MS profile for each MS experiment was extracted from the respective raw spectral data using a conversion function in BioWorks™. Principal extracted variables included: identities of differentially labeled peptides, their respective precursor ion masses, charge states and retention times throughout the LC-MS run/analysis. These parameters provide the location of respective MS1 spectra for identified peptide ions. Once the MS1 spectra of these peptides were located, isotopic species for each respective peptide cluster were defined by sequential examination of MS1 spectra in both directions, obtaining the envelopes for each isotopic species, as shown in Figure 1a. For each direction, the MS1 spectra were examined and the envelope was stopped for all isotopic species when less than Ni isotopic intensities were observed. In this investigation, the Ni, minimal vector length, was set to two, meaning that two of the four isotopic peaks for a given precursor ion had to be present to incorporate these MS1 intensities in the envelope. The retention time at which the envelope was stopped was then assigned as the start and end point. The intensities at the start and end point were given an arbitrary value of zero to correct background noise.
Figure 1.
Example of intensities for a given isotopic manifold across multiple MS1 spectra (a). Example of the integrated areas for peaks within a given isotopic manifold (b).
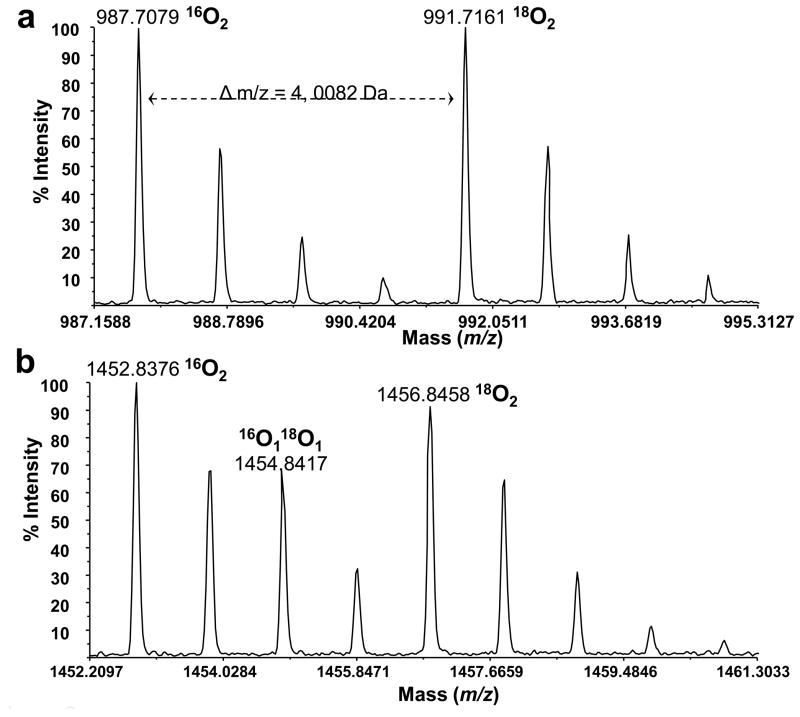
Following the determination of the start and end points, the peak area was calculated using the trapezoidal rule. For a given peptide, each isotopic species in the isotopic cluster was calculated, resulting in up to eight integrated areas (I0 through I7), one for each isotopic species, as shown in Figure 1b. Evidently, the last four integrated areas (I4 through I7), depicting isotopic envelope of heavy labeled isotopomer, can only originate from the 18O2 tagged sample, which is referred to as the B-sample. Figure 2a shows differentially labeled isotopic cluster exhibiting complete exchange of two 16O atoms for two 18O atoms in the presence of 18O water. Correspondingly, intensities of the first four peaks (I0 through I3) can originate either from the 16O isotopomer (i.e. incubated with trypsin in the presence of H216O, which is referred to as the A-sample), or in part from the singly 18O1 tagged isotopomer present in the B-sample due to incomplete C-terminal labeling (Figure 2b). While earlier investigations fit the series of integrated peak areas using the set of Gaussian functions, we propose that the manifold of isotopic intensities should follow a Poisson distribution as described below:
| (1) |
Figure 2.
MS1 of differentially labeled tryptic isotopomeric peptide pair indicating complete 18O exchange/incorporation (a). MS1 of differentially labeled convoluted peptide clusters indicating variable/incomplete 18O exchange/incorporation exemplified by the presence of 18O1 isotopomer (b).
In this expression λ is the expected number of heavy isotopes present in the peptide and k is the observed number. A given isotopic envelope is possibly perturbed to a small extent by oxygen or sulfur. This situation exists since 0.20% of all oxygen atoms are 18O and 4.29% of all sulfur atoms are 34S, then a corresponding Poisson distribution will cause the intensities to change for (m+2)/z.
In a typical 16O/18O labeling experiment, a fraction f of all peptides is 18O tagged. Defining our nomenclature, if the total amount of a given peptide species is B, the total unlabeled amount of this peptide is B0, the total amount of singly 18O1 tagged isotopomer is B1, and the total amount of doubly 18O2 tagged isotopomer is B2 (Figure 1b). In the same way, based on the reaction kinetics and the assumption that the exchange of the two oxygen atoms is independent of each other, their relations may be expressed as following:
| (2) |
| (3) |
| (4) |
| (5) |
Assuming that there can only be at most three substitutions that cause a mass shift of one and one substitution that causes a mass shift of two from the naturally occurring isotopes, the calculated intensity of the eight peaks (I0 to I7) within the isotopic envelope can be given by the following expressions:
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
Substituting equation 3, 4 and 5 into these equations reduces the parameters down to five: A, B, f, λ1 and λ2. Then all calculated areas are divided by I0 and the first ratio becomes the unity, thereby reducing the number of parameters to four: B/A, f, λ1 and λ2.
The optimal values of these adjustable parameters used to fit the manifold of integrated area ratios were determined using evolutionary programming. Each parameter was initially set to a random value, while its fitness was determined within a reasonable range. This was done Npop times to create an initial population of putative solutions. In each generation, each putative solution was used to generate a new putative solution by copying the parameter values and then randomly changing them by a small amount. This new putative solution is evaluated and its parameter values and fitness were stored in an offspring population. The parent and offspring populations are then merged and all putative solutions were ordered from lowest to highest fitness. After Ngen generations the process stopped and the final Npop solutions with the highest fitness were examined. All putative solutions with an average error within 10% of the best solution were used to find average values for each of the parameters, which then represented the optimal values for the parameters. In this investigation, the population size, Npop, was set to 2000, the process was run for 4000 generations (Ngen=4000), and the result that yielded the smallest average error was reported.
Assessment of completeness of 18O incorporation
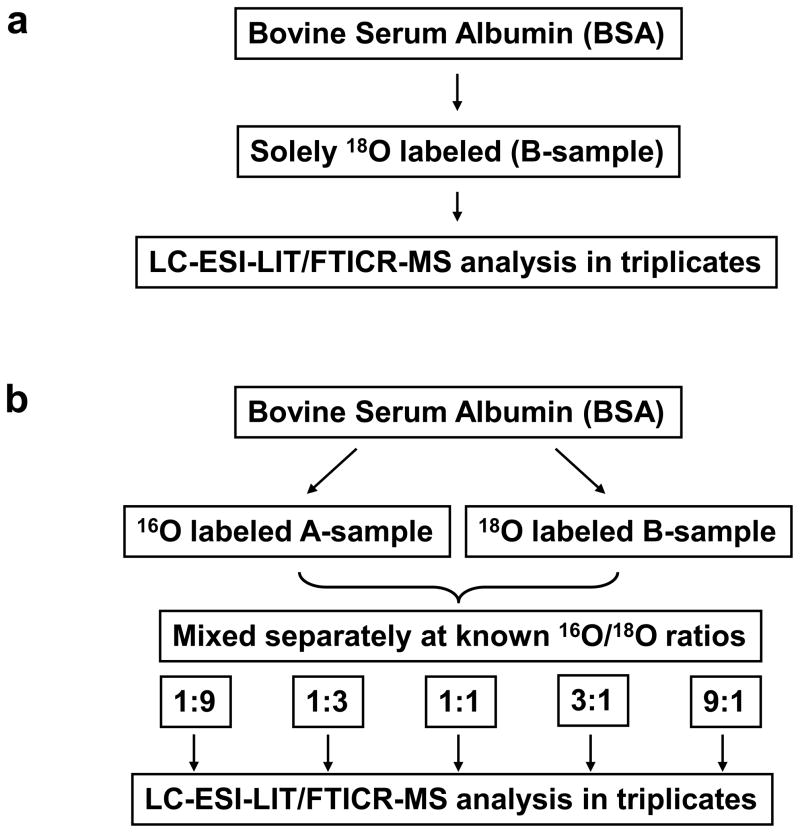
To study the completeness of 18O incorporation we used the BSA as a model protein and labeled it in the presence of H218O. To determine how thorough the 18O exchange is and to assess the extent of variable 18O incorporation, we analyzed solely 18O labeled BSA digest (B-sample) to obtain empirical values of f for each identified BSA peptide (Figure 3a). Table 1 summarizes results obtained by LC-LIT/FTICR analysis of 18O labeled BSA digest.
Figure 3.
Scheme of experimental design – The LC-MS analysis of solely 18O labeled BSA (a). The LC-MS analysis of differentially 16O/18O labeled BSA digest mixed at known 18O/16O ratios.
Table 1.
The average f value of BSA peptides in a triplicate LC-MS2 analysis of solely 18O labeled BSA digest. The tagged C-termini are marked by “[“. *Average ± standard deviation was calculated for all K or R ending peptides.
| Peptide | MH+ | z | No. of f | Average |
|---|---|---|---|---|
| K.DLGEEHFK[.G | 978.4658 | 2 | 3 | 0.76 |
| K.DLGEEHFK[.G | 978.4658 | 1 | 3 | 0.78 |
| R.FKDLGEEHFK[.G | 1253.629 | 3 | 3 | 0.71 |
| R.FKDLGEEHFK[.G | 1253.629 | 2 | 3 | 0.75 |
| K.AEFVEVTK[.L | 926.496 | 2 | 3 | 0.71 |
| K.AEFVEVTK[.L | 926.496 | 1 | 3 | 0.74 |
| K.HLVDEPQNLIK[.Q | 1309.724 | 2 | 3 | 0.66 |
| K.HLVDEPQNLIK[.Q | 1309.724 | 1 | 3 | 0.95 |
| K.LVNELTEFAK[.T | 1167.639 | 1 | 3 | 0.97 |
| K.KQTALVELLK[.H | 1146.722 | 3 | 3 | 0.96 |
| K.YLYEIAR[.R | 931.5015 | 2 | 3 | 0.93 |
| K.YLYEIAR[.R | 931.5015 | 1 | 3 | 0.96 |
| R.KVPQVSTPTLVEVSR[.S | 1643.946 | 2 | 3 | 0.98 |
| R.RHPEYAVSVLLR[.L | 1443.82 | 3 | 2 | 0.9 |
| K.VPQVSTPTLVEVSR[.S | 1515.851 | 2 | 3 | 0.92 |
| R.RHPEYAVSVLLR[.L | 1443.82 | 2 | 3 | 0.91 |
| R.HPEYAVSVLLR[.L | 1287.719 | 3 | 3 | 0.97 |
| K.LGEYGFQNALIVR[.Y | 1483.803 | 1 | 2 | 0.99 |
| K.DAFLGSFLYEYSRR[.H | 1727.852 | 2 | 1 | 0.9 |
| K.DAFLGSFLYEYSR[.R | 1571.751 | 2 | 3 | 0.71 |
| R.MPCTEDYLSLILNR[.L | 1671.821 | 2 | 2 | 0.9 |
| K.LVTDLTK[.V | 793.4797 | 1 | 2 | 0.85 |
| R.KVPQVSTPTLVEVSR[.S | 1643.946 | 3 | 2 | 0.94 |
| K.LVNELTEFAK[.T | 1167.639 | 2 | 1 | 0.99 |
| K.AEFVEVTKLVTDLTK[.V | 1696.95 | 2 | 1 | 0.97 |
| K.AEFVEVTKLVTDLTK[.V | 1696.95 | 3 | 1 | 0.93 |
| K.VPQVSTPTLVEVSR[.S | 1515.851 | 3 | 1 | 0.93 |
| All K-ending peptides | 35 | 0.82 ± 0.12* | ||
| All R-ending peptides | 31 | 0.92 ± 0.08* |
The peptide sequence, mass and charge are listed in the first three columns. The fourth column contains the number of calculated f values for that peptide. If a given fragment was observed in all three LC-LIT/FTICR runs then there are a total of three calculated f values indicated in the fourth column. The fifth column contains the arithmetic average of the f values. The last two rows of Table 1 summarize the average and standard deviation of f values for all peptides that end in either lysine or arginine.
Although 50% of the identified peptides did not show detectable unlabeled B0 peak, the rest showed B0 peak to variable extent. The average percentage of detectable B0 peaks in 18O labeled BSA was around 3%, which is expected because 95% pure 18O water was used. In contrast, the average percentage of 18O1 mono-labeled peptide isotopomers (B1) was as high as 21%. In one extreme example the mono-labeled 18O1 isotopomer of DAFLGSFLYEYSR peptide reached 45%, which is almost as intense as the 18O2 dual-labeled isotopomer. We also observed that the labeling efficiency for lysine-ending peptides is different from arginine-ending peptides. On average, the f value is significantly higher (z-score = 5.778, P < 4.0 × 10−9) if the isotopic substitution occurs at an arginine residue.
Algorithm validation using LC-MS analysis of differentially 18O/16O labeled BSA digest
We used BSA as a model protein to assess the utility the utility of the developed algorithm for 18O/16O-based quantitative proteomics. Figure 3b depicts experimental approach designed to test the algorithm by analyzing the five differentially 18O/16O labeled BSA samples, mixed using a priori 18O/16O ratios: 9:1, 3:1, 1:1, 1:3 and 1:9. The 16O-labeled BSA sample was conveniently labeled as the A-sample while the 18O-labeled BSA was labeled as the B-sample. After combining A-sample and B-sample using predetermined ratios, these samples were analyzed in triplicate by LC-LIT/FTICR. The 18O/16O ratios at mixing rate of 3:1, 1:1 and 1:3 were determined using three different analytical approaches for each of the three runs. Table 2 contains summary information and statistics on the computed 18O/16O ratios for differentially labeled BSA digests.
Table 2.
Computed 18O/16O ratios for BSA mixed at predetermined ratios of 1:1, 3:1 and 1:3 using different adjustment.
| 18O:16O | Method | Average ± SD | |||
|---|---|---|---|---|---|
| Run 1 | Run 2 | Run 3 | All Runs | ||
| 1:1 | Allow f to vary | 1.50 ± 1.33 | 1.41 ± 0.57 | 1.24 ± 0.58 | 1.38 ± 0.88 |
| Filtered by f > 0.6 | 1.27 ± 0.48 | 1.41 ± 0.57 | 1.18 ± 0.51 | 1.29 ± 0.53 | |
| Predetermined f | 1.00 ± 0.52 | 1.18 ± 0.56 | 1.01 ± 0.47 | 1.06 ± 0.52 | |
| 3:1 | Allow f to vary | 4.15 ± 2.22 | 3.58 ± 1.56 | 3.70 ± 1.67 | 3.80 ± 1.82 |
| Filtered by f > 0.6 | 4.26 ± 2.31 | 3.53 ± 1.39 | 3.72 ± 1.69 | 3.83 ± 1.83 | |
| Predetermined f | 3.56 ± 5.84 | 2.68 ± 1.40 | 3.40 ± 1.96 | 3.23 ± 3.37 | |
| 1:3 | Allow f to vary | 0.52 ± 0.38 | 0.45 ± 0.25 | 0.42 ± 0.13 | 0.47 ± 0.29 |
| Filtered by f > 0.6 | 0.42 ± 0.12 | 0.44 ± 0.26 | 0.43 ± 0.13 | 0.43 ± 0.17 | |
| Predetermined f | 0.33 ± 0.12 | 0.34 ± 0.20 | 0.30 ± 0.15 | 0.33 ± 0.16 | |
The first method denoted as “Variable” allows all four parameters (B/A, f, λ1, and λ2) to vary in value in regard to their roles in determining the ratios of integrated areas in the manifold. The B/A ratio was calculated for each identified BSA peptide. Results in Table 2 list the mean B/A ratio and its standard deviation across these peptides. For the 1:1 mixture of BSA, the calculated B/A ratios were 1.50 ± 1.33, 1.41 ± 0.57, and 1.24 ± 0.58 for run 1, 2 and 3, respectively, and 1.38 ± 0.88 when averaged across all peptides in all runs. Though these values are reasonably good, the standard deviation is quite large. As seen in Table 2, for the 3:1 and 1:3 mixtures of BSA, this approach yields reasonable B/A ratios, although with relatively large standard deviations.
The second method denoted as “Filtered f > 0.6” is based on the observation that the experimentally determined f value for most peptides was ≥ 0.60. These results are also reported in Table 2. Therefore, this method uses the same results obtained by the first method, except it only accepts f values greater than 0.6. In general, the average B/A values showed improvement for the 1:1 and 3:1 mixtures exemplified in overall decrease in the standard deviation. The results for run 2 in the 1:1 mixture are the same as for the first method, which simply means that all of the calculated f values were above 0.60. Notably, the second method did not result in a significant improvement of average f values, but did improve dispersion measurements as reflected in most standard deviation scores.
The third method denoted as “predetermined f” relies on LC-MS/MS pre-run to analyze a portion of 18O labeled sample B before combining the 16O labeled and 18O labeled sample. This analysis permits the experimental determination of a f value for each identified peptide, along with an assessment of the extent of variable 16O incorporation. This approach fixes the values of f for every peptide identified in the pre-run as shown in Table 1. If a peptide is not contained in the pre-run list, the corresponding average f value from either lysine-ending or arginine-ending cohort shown in Table 1 is used This choice depends on terminal amino acid (i.e., lysine or arginine). Finally the manifold of the isotopic envelope are then reexamined by using experimentally obtained f values and varying only the parameters: B/A, λ1 and λ2. As shown in Table 2, the resulting B/A ratios were more accurate and precise than the ratios when f was allowed to vary as exemplified in SD values.
Table 3 shows the ratios obtained using our program, which allowed f values to vary versus those obtained by XPRESS for the following mix conditions: 9:1, 3:1, 1:1, 1:3 and 1:9. The ratios calculated using our algorithm is closer to true values as student t-test (P = 0.10) indicates that there is no significant difference. In contrast, the ratios calculated using XPRESS is significantly lower than true values (P = 0.01). In addition, the range of ratios from different peptides obtained using our algorithm is significantly decreased compared to the algorithm of XPRESS (P < 0.001).
Table 3.
Computed 18O/16O ratios in logarithmic scale for BSA mixed at predetermined ratios of 9:1, 3:1, 1:1, 1:3 and 1:9 using our algorithm by allowing f to vary and by XPRESS. The smallest and largest ratios are included in the parenthesis.
| Log2(18O:16O) | Ratio by allowing f to vary
|
Ratio by XPRESS
|
||
|---|---|---|---|---|
| Median | Range | Median | Range | |
| 2.19 | 2.45 | 2.48 (1.40~3.88) | 1.74 | 3.30 (−0.15~3.15) |
| 1.09 | 1.19 | 2.66 (−0.11~2.55) | 0.64 | 4.11 (−2.40~1.71) |
| 0 | 0.09 | 1.73 (−0.94~0.79) | −0.27 | 3.01 (−2.52~0.49) |
| −1.1 | −0.92 | 1.74 (−1.94~−0.20) | −1.26 | 3.17 (−3.17~0.00) |
| −2.2 | −2.26 | 1.76 (−3.21~−1.45) | −2.27 | 3.00 (−3.35~−0.35) |
Algorithm application to quantitative profiling of 16O/18O labeled HeLa DRMMs
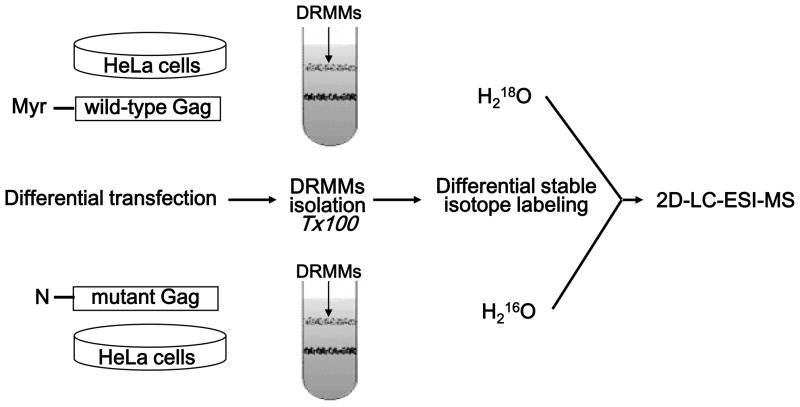
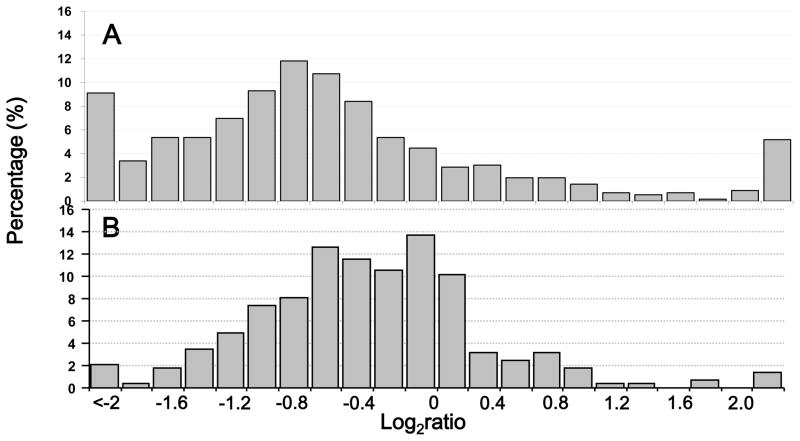
To evaluate algorithm’s utility for quantitative profiling of complex protein mixture and investigate HIV Gag trafficking to the plasma membrane, we analyzed differentially 16O/18O labeled DRMMs isolated from HeLa cells expressing wild-type Gag or a myristylation-defective Gag mutant, which does not bind DRMMs.25, 28 DRMMs proteins isolated from HeLa cells transfected with a plasmid encoding mutant Gag were digested and labeled in the presence of 16O water, while DRMMs proteins isolated from HeLa cells transfected with a wild-type Gag-expressing plasmid were labeled in the presence of 18O water (Figure 4). Following differential labeling, the digests were mixed at 1:1 ratio. Differentially labeled digest was then resolved via strong cation exchange (SCX) into fourteen fractions. Each fraction was then analyzed by LC-MS. Quantitative analysis using XPRESS algorithm resulted in identification/quantitation of a total of 1740 protein specific peptides (Supplementary Table S-1a) that allowed relative quantitation for a total of 558 unique proteins (Supplementary Table S-1b) with their ratios distribution shown in Figure 5a. In contrast, our algorithm reported identification/quantitation of a total of 726 protein specific peptides (Supplementary Table S-1b) that allowed relative quantitation for a total of 285 unique proteins (Supplementary Table S-1b) with their ratios distribution shown in Higher number of quantified peptides/proteins was obtained by XPPRES since this algorithm requires only monoisotopic peak to calculate area under the peak for a given isotopomer while our algorithm requires isotopic manifold. Consequently, considerable number of identified peptides was not quantified by our algorithm due to missing isotopic peaks in raw MS data. Figure 5b, exhibiting. Gaussian distribution around ratio 1, which is expected in large scale quantitative proteome studies. For proteins identified by two or more peptides, 29.1% were quantified by XPRESS and 28.8% were quantified by our algorithm. There was no significant difference in the coefficient of variation (CV) distribution between the two algorithms since the Pearson product-moment correlation coefficient was 0.999. Additionally, the median CV from XPRESS and our method were essentially the same (50% and 49% respectively). Importantly, more than 84% of peptides showed an increased ratio using our algorithm indicating the effectiveness of present algorithm to detect and account for incomplete labeling. Additionally, our approach is not influenced by cysteine or methionine residues, since there is no significant difference between peptides containing: cysteine, methionine, or neither (P > 0.99).
Figure 4.
Schematic outline depicting experimental design for quantitative 16O/18O-based proteomic analysis of DRMMs transfected with wild-type gag and a G1A mutant gag.
Figure 5.
The distribution of the percentage of unique proteins quantified by 18O labeling in the DRMMS of HeLa cells transfected with expression plasmids encoding the wild type Gag and 1GA mutant Gag within binned logarithms of protein abundance ratios obtained using XPRESS (A) and the developed algorithm (B).
In this study, eleven Gag peptides were quantified using our algorithm while twenty-two were quantified using XPRESS. Table 4 lists the peptides quantified by both algorithms. Of note, all of these peptides have higher ratios quantified by our algorithm versus XPRESS. For f values greater than 0.7, the increase is approximately 150%. In contrast, for f value less than 0.5, the average increase is around 300%, suggesting the necessity of detecting and accounting for incomplete 18O incorporation. Indeed, manual inspection of the MS scans of these peptides showed incomplete labeling at various levels. Among eleven Gag peptides only quantified by XPRESS five are repetitive identification and quantitation of the same peaks due to high peak intensity. The remaining peptides were not quantified using our algorithm due to missing isotopic peaks. The average 18O/16O ratio of Gag is 11.81 ± 7.37 using our algorithm while 8.30 ± 8.62 was reported by XPRESS. The quantitative results for Gag obtained by LC-MS analysis in this study in the context of differential 18O/16O stable isotope labeling are highly consistent with a well-accepted notion that N-terminal myristylation is essential for Gag-DRMMs association.29
Table 4.
Computed 18O/16O ratios for Gag protein in isolated DRMMs.
| Peptide | SCX Fraction | Ratio
|
f | ||
|---|---|---|---|---|---|
| Our program | XPRESS | Increase (%) | |||
| R.WIILGLNK[.I | 3 | 8.76 | 4.19 | 209 | 0.59 |
| R.FGEETTTPSQK[.Q | 3 | 18.58 | 12.48 | 148 | 0.76 |
| R.FGEETTTPSQK.Q | 3 | 7.50 | 5.77 | 130 | 0.73 |
| K.ELYPLASLR[.S | 3 | 5.43 | 1.20 | 452 | 0.42 |
| R.FGEETTTPSQK[.Q | 4 | 20.55 | 12.7 | 161 | 0.77 |
| R.FGEETTTPSQK[.Q | 4 | 8.78 | 5.59 | 157 | 0.74 |
| K.ELYPLASLR[.S | 5 | 6.96 | 2.00 | 347 | 0.47 |
| R.QILGQLQPSLQTGSEELR.S | 5 | 4.06 | 2.18 | 186 | 0.31 |
| K.ETINEEAAEWDR[.L | 5 | 13.46 | 3.48 | 386 | 0.34 |
| R.FGEETTTPSQK[.Q | 9 | 27.48 | 19.06 | 144 | 0.75 |
| R.FGEETTTPSQK[.Q | 9 | 8.38 | 4.92 | 170 | 0.73 |
DISCUSSION
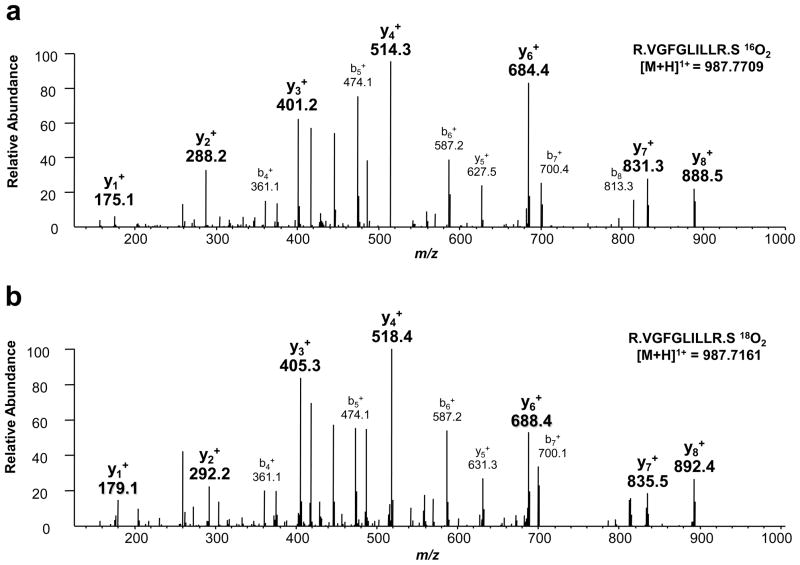
During a typical 18O/16O labeling experiment, trypsin binds covalently to C-terminal lysyl or arginyl-residues catalyzing a stepwise substitution of two C-terminal 16O atoms for two 18O atoms. The first hydrolytic reaction RC16ONHR′ + H218O → RC16O18O− + +H3NR′ is followed by a second hydrolytic reaction RC16O18O− + H218O → RC18O18O− + H216O. Two complete enzyme-catalyzed turnovers in the presence of the heavy isotopic form of water [H218O] result in a 4 Da mass increase in labeled peptides as shown in Figure 6a. This same difference is present in MS2 spectra of heavy 18O-labeled peptides. Figures 6a–b show a mass shift of 4 Da between singly charged y-fragments of light 16O2 isotopomer versus heavy 18O2 labeled isotopomer, thus confirming the presence of a C-terminal 18O2 tag.
Figure 6.
MS2 spectra of differentially 16O/18O labeled peptide pair shown in Figure 2a. MS2 spectrum showing CID fragments of 16O labeled isotopomer (a). MS2 spectrum of 18O labeled isotopomer showing that all detected y ion fragments of this heavy labeled isotopomeric peptide exhibit a 4 Da difference when compared to corresponding y ion fragments of 16O labeled peptide (b). This finding confirms the presence of 18O2 tag at the
However, two complete enzyme turnovers are not always achieved. This results in an incomplete 18O exchange exemplified in co-occurrence of variable amount 18O1 isotopomeric species, as illustrated in Figure 2b. This was also confirmed by analyzing the solely 18O labeled BSA specimen sample that was used to assess the completeness of 18O exchange/labeling. The average percentage of 18O1 mono-labeled peptide isotopomers was found to be as high as 21%. In one extreme example the mono-labeled 18O1 isotopomer of DAFLGSFLYEYSR peptide reached 45%, which is almost as intense as the 18O2 dual-labeled isotopomer. Thus, labeling efficiency can vary and be dependent on the peptide sequence. As shown, the arginine-ending peptides appear to have a higher level of incomplete labeling than lysine-ending peptides. This is consistent with the previous observation, and is a consequence of the different substrate affinity of trypsin, for arginine and lysine ending peptides.30
To account and correct for the effect of inhomogeneous 18O incorporation, we developed an algorithmic method capable of determining the efficiency of 18O exchange. Specifically, we detect and account for partial 18O incorporation. Our approach involves curve fitting the integrated areas of the multifold peaks enclosed by the isotopic envelope and correcting for the incomplete labeling. The 18O/16O ratios are formed using a computational approach that employs a Poisson distribution along with a near optimization method that utilizes an evolutionary programming technique.
Compared to current computational methods for 18O/16O quantitative labeling, our approach offers advantages in several respects. First, the time window for peak integration is tailored for each identified peak instead of integrating it over a fixed region of the extracted ion chromatogram. This significantly improves the accuracy of isotopic peak intensities, and in particular for candidate peptide ions of low intensities. Second, we utilize an overlapping Poisson distribution model to fit isotopic manifolds, which better suits the natural physical properties of isotope distribution. Several earlier investigations have employed a set of Gaussian functions to fit the isotopic envelope of the identified peptides.16, 31 However, since the numbers of distinct isotope species for the manifold peaks are integers, we propose that the manifold of isotopic intensities should follow a Poisson distribution. This is also supported by our observation that the Gaussian distribution did not completely fit the integrated areas.
Third, evolutionary programming is employed to obtain optimized parameters for the fitting with several quality checkpoints to improve accuracy and reproducibility. The program requires that at least two of the four isotopic peaks for a given precursor ion must be present in the manifold envelope to be considered for quantitation. In addition, since λ1 is the expected number of 13C and λ2 is the expected number of 18O/34S present in a given peptide, these values are calculated for every identified peptide and serve as checkpoints for the quality of fitting.
It should be noted that when the BioWorks XPRESS algorithm is applied for 18O/16O ratio calculation, it assumes the complete 18O exchange for each differentially labeled peptide-ion pair. XPRESS calculates the 18O/16O ratio directly from the monoisotopic peaks located at a 4 Dalton difference from the monoisotopic peak of the identified peptide ion.32 Therefore, the XPRESS algorithm is not capable of accounting for variable/incomplete 18O incorporation.
As depicted in Table 1, a significant degree of variable/incomplete 18O incorporation is observed for certain tryptic peptides. If the presence of these 18O1 mono-labeled isotopomers is not taken into consideration for the 18O/16O ratios calculation, a variable degree of underestimation of the peptide/protein ratio is to be expected. Evidently, the average (18O/16O) ratio obtained by XPRESS is smaller than the real values (Table 3). These results are expected since XPRESS software simply divides the peak area exclusively from 18O2 tagged peptide isotopomers over that of 16O2 naturally tagged peptide species, overlooking the contribution of the mono-labeled peptide species. The relative standard deviation for calculated ratios using our developed algorithm was 13.0 ± 9.6 % compared to −23.5 ± 13.5 % using XPRESS. A significant increase in accuracy and reproducibility is obtained with our method. Of note, regarding proteins with heavy to light ratios less then, the difference between our program and XPRESS was not significant. This is understandable since the interference from relatively small amount of incomplete labeling to a relatively large third isotopic peak is minimal.
The best accuracy by our method was achieved when the f value was determined using a LC-MS/MS pre-run to analyze a portion of 18O labeled sample. However, this practice doubles instrument time and sample consumption, which makes it impractical in a large scale multiple dimensional separation or with very small samples. For this reason, the relationship between peptide sequence and the efficacy of 18O exchange reaction is currently under investigation. If f values can be predicted by the peptide sequences, then the accuracy and the speed of quantitative proteomics by 18O labeling will be much improved.
To further evaluate the robustness of our approach, it was applied to a more complex biological sample. This mixture contained 16O/18O labeled DRMMs isolated from HeLa cells differentially transfected with plasmids expressing mutant and wild-type HIV Gag protein, respectively. The process of retrovirus assembly requires that several viral constituents must localize to the plasma membrane of an infected cell and then assemble into a budding virus particle. Central to this process is the retroviral Gag polyprotein.33
To study the Gag trafficking mechanism, a mutant Gag protein with an amino-terminal myristylation site, which has been disrupted by amino acid substitution, was expressed in HeLa cells. The effects of this mutation on protein translocation to the plasma membrane were investigated. Indeed, the abundance of mutant Gag protein in DRMMs is significantly lower compared to that of wild type protein. This finding is in agreement with the role of previously proposed myristyl switch for the regulation of Gag membrane binding and subsequent western blotting experiment against Gag protein.28
Several proteins quantified by our algorithm are likely associated with Gag translocation, e.g. human CD59 protein is up-regulated by 2.0-fold, flotillin-1 is down-regulated by 0.4 times while flotillin-2 is up-regulated by 2.8 times. It has been reported that CD59 prevents assembly of the membrane attack complex in HIV transfection.34, 35 Flotillins are typical lipid raft associated membrane proteins and it has been found that the overall lipid composition of native HIV membranes resembles DRMMs.36 The changes in abundance of cellular proteins associated with DRMM upon the introduction of wild-type versus mutant Gag proteins are likely suggesting they are closely associated with Gag translocation and therefore the virus assembly, which is currently under investigation.
In summary, this work describes an algorithm amenable to automated interpretation and computation of 18O/16O ratios from spectra obtained by LC-MS/MS analysis of differentially labeled isotopomeric clusters. The algorithm is capable to accurately calculate 18O/16O ratios and eliminate artifacts caused by variable 18O exchange. It uses centroid peak data and is suitable for any high resolution MS platform. The algorithm has been successfully tested with the quantitation of a BSA protein standard and then applied to a comparative proteomic profiling of a more complex specimen.
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contracts HHSN261200800001E and NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the United States Government.
References
- 1.Reynolds KJ, Fenselau C. Curr Protoc Protein Sci. 2004;Chapter 23(Unit 23):24. doi: 10.1002/0471140864.ps2304s34. [DOI] [PubMed] [Google Scholar]
- 2.Ye X, Luke B, Andresson T, Blonder J. Brief Funct Genomic Proteomic. 2009;8:136–144. doi: 10.1093/bfgp/eln055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnolzer M, Jedrzejewski P, Lehmann WD. Electrophoresis. 1996;17:945–953. doi: 10.1002/elps.1150170517. [DOI] [PubMed] [Google Scholar]
- 4.Reynolds KJ, Yao X, Fenselau C. J Proteome Res. 2002;1:27–33. doi: 10.1021/pr0100016. [DOI] [PubMed] [Google Scholar]
- 5.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 6.Back JW, Notenboom V, de Koning LJ, Muijsers AO, Sixma TK, de Koster CG, de Jong L. Anal Chem. 2002;74:4417–4422. doi: 10.1021/ac0257492. [DOI] [PubMed] [Google Scholar]
- 7.Qian WJ, Monroe ME, Liu T, Jacobs JM, Anderson GA, Shen Y, Moore RJ, Anderson DJ, Zhang R, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Camp DG, 2nd, Smith RD. Mol Cell Proteomics. 2005;4:700–709. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zang L, Palmer Toy D, Hancock WS, Sgroi DC, Karger BL. J Proteome Res. 2004;3:604–612. doi: 10.1021/pr034131l. [DOI] [PubMed] [Google Scholar]
- 9.Stockwin LH, Blonder J, Bumke MA, Lucas DA, Chan KC, Conrads TP, Issaq HJ, Veenstra TD, Newton DL, Rybak SM. J Proteome Res. 2006;5:2996–3007. doi: 10.1021/pr0601739. [DOI] [PubMed] [Google Scholar]
- 10.Blonder J, Yu LR, Radeva G, Chan KC, Lucas DA, Waybright TJ, Issaq HJ, Sharom FJ, Veenstra TD. J Proteome Res. 2006;5:349–360. doi: 10.1021/pr050355n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedstrom L. Chem Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 12.Heller M, Mattou H, Menzel C, Yao X. J Am Soc Mass Spectrom. 2003;14:704–718. doi: 10.1016/S1044-0305(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 13.Wang YK, Ma Z, Quinn DF, Fu EW. Anal Chem. 2001;73:3742–3750. doi: 10.1021/ac010043d. [DOI] [PubMed] [Google Scholar]
- 14.Hajkova D, Rao KC, Miyagi M. J Proteome Res. 2006;5:1667–1673. doi: 10.1021/pr060033z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckel-Passow JE, Oberg AL, Therneau TM, Mason CJ, Mahoney DW, Johnson KL, Olson JE, Bergen HR., 3rd Bioinformatics. 2006;22:2739–2745. doi: 10.1093/bioinformatics/btl464. [DOI] [PubMed] [Google Scholar]
- 16.Halligan BD, Slyper RY, Twigger SN, Hicks W, Olivier M, Greene AS. J Am Soc Mass Spectrom. 2005;16:302–306. doi: 10.1016/j.jasms.2004.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horn DM, Zubarev RA, McLafferty FW. J Am Soc Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 18.Mason CJ, Therneau TM, Eckel-Passow JE, Johnson KL, Oberg AL, Olson JE, Nair KS, Muddiman DC, Bergen HR., 3rd Mol Cell Proteomics. 2007;6:305–318. doi: 10.1074/mcp.M600148-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Ramos-Fernandez A, Lopez-Ferrer D, Vazquez J. Mol Cell Proteomics. 2007;6:1274–1286. doi: 10.1074/mcp.T600029-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Shinkawa T, Taoka M, Yamauchi Y, Ichimura T, Kaji H, Takahashi N, Isobe T. J Proteome Res. 2005;4:1826–1831. doi: 10.1021/pr050167x. [DOI] [PubMed] [Google Scholar]
- 21.Stewart II, Thomson T, Figeys D. Rapid Commun Mass Spectrom. 2001;15:2456–2465. doi: 10.1002/rcm.525. [DOI] [PubMed] [Google Scholar]
- 22.Park SM, Hwang IK, Kim SY, Lee SJ, Park KS, Lee ST. Proteomics. 2006;6:1192–1199. doi: 10.1002/pmic.200500402. [DOI] [PubMed] [Google Scholar]
- 23.Mueller LN, Brusniak MY, Mani DR, Aebersold R. J Proteome Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen KL, llano M, Akari H, Miyagi E, Poeschla EM, Strebel K, Bour S. Virology. 2004;319:163–175. doi: 10.1016/j.virol.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Ono A, Waheed AA, Joshi A, Freed EO. J Virol. 2005;79:14131–14140. doi: 10.1128/JVI.79.22.14131-14140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blonder J, Hale ML, Chan KC, Yu LR, Lucas DA, Conrads TP, Zhou M, Popoff MR, Issaq HJ, Stiles BG, Veenstra TD. J Proteome Res. 2005;4:523–531. doi: 10.1021/pr049790s. [DOI] [PubMed] [Google Scholar]
- 27.Blonder J, Chan KC, Issaq HJ, Veenstra TD. Nat Protoc. 2006;1:2784–2790. doi: 10.1038/nprot.2006.359. [DOI] [PubMed] [Google Scholar]
- 28.Ono A, Freed EO. J Virol. 1999;73:4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindwasser OW, Resh MD. J Virol. 2001;75:7913–7924. doi: 10.1128/JVI.75.17.7913-7924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao X, Afonso C, Fenselau C. J Proteome Res. 2003;2:147–152. doi: 10.1021/pr025572s. [DOI] [PubMed] [Google Scholar]
- 31.Jorge I, Navarro P, Martinez-Acedo P, Nunez E, Serrano H, Alfranca A, Redondo JM, Vazquez J. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M800260-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moulder R, Filen JJ, Salmi J, Katajamaa M, Nevalainen OS, Oresic M, Aittokallio T, Lahesmaa R, Nyman TA. Proteomics. 2005;5:2748–2760. doi: 10.1002/pmic.200401187. [DOI] [PubMed] [Google Scholar]
- 33.Freed EO. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 34.Takefman DM, Spear GT, Saifuddin M, Wilson CA. J Virol. 2002;76:1999–2002. doi: 10.1128/JVI.76.4.1999-2002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu W, Yu Q, Hu N, Byrd D, Amet T, Shikuma C, Shiramizu B, Halperin JA, Qin X. J Immunol. 2009;184:359–368. doi: 10.4049/jimmunol.0902278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG. Proc Natl Acad Sci U S A. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.