SUMMARY
Innate immune responses are characterized by precise gene expression whereby gene subsets are temporally induced to limit infection, although the mechanisms involved are incompletely understood. We show that antiviral immunity in Drosophila requires the transcriptional pausing pathway, including Negative Elongation Factor (NELF) that pauses RNA Polymerase II (Pol II) and Positive Elongation Factor b (P-TEFb), which releases paused Pol II to produce full length transcripts. We identify a set of genes that is rapidly transcribed upon arbovirus infection, including components of antiviral pathways (RNA silencing, autophagy, JAK/STAT, Toll, and Imd) and various Toll receptors. Many of these genes require P-TEFb for expression and exhibit pausing-associated chromatin features. Furthermore, transcriptional pausing is critical for antiviral immunity in insects, as NELF and P-TEFb are required to restrict viral replication in adult flies and vector mosquito cells. Thus, transcriptional pausing primes virally-induced genes to facilitate rapid gene induction and robust antiviral responses.
INTRODUCTION
The innate immune system is an ancient, highly conserved mode of defense against pathogens and the sole method of protection for invertebrates and plants. A critical aspect of innate immunity is the rapid activation of gene expression programs to generate effectors that restrict pathogens (Medzhitov and Horng, 2009; Smale, 2010). The best-characterized example is the lipopolysaccharide (LPS)-induced macrophage response, which is classically described in two stages: the rapid protein synthesis-independent induction of immediate-early genes (termed the primary response), followed by the subsequent protein-synthesis-dependent induction of secondary response genes (Medzhitov and Horng, 2009; Smale, 2010). Studies have implicated the step of transcription initiation in regulating these waves of gene expression, in both mammals (Medzhitov and Horng, 2009) and insects (Boutros et al., 2002; Hoffmann, 2003). In this mode of regulation, pathogen recognition leads to activation of specific transcription factors that recruit RNA Polymerase II (Pol II) and general transcription factors to promoters, thereby inducing gene expression (Medzhitov and Horng, 2009; Roeder, 2005). While this is often considered canonical, how other transcriptional regulatory mechanisms play a role in orchestrating immune responses are less clear, particularly across a diverse range of pathogens and hosts. Furthermore, inducible host programs that control pathogens in disease-transmitting insect vectors, particularly viruses, are not well defined.
Transcriptional pausing is a mode of gene regulation that occurs as a step in the transcription cycle downstream of initiation (Sims et al., 2004). Recent studies suggest that a subset of inducible genes can also be tightly regulated at this step (Nechaev and Adelman, 2011; Saunders et al., 2006). Initially described to negatively regulate a handful of genes, including Drosophila heat shock loci, recent studies reveal that a larger set of genes are positively regulated by this step of the transcription cycle (Bernstein et al., 2006; Boyer et al., 2006; Gilchrist et al., 2010; Gilchrist et al., 2008; Lee et al., 2006; Muse et al., 2007; Saunders et al., 2006; Wang et al., 2007; Zeitlinger et al., 2007). At these loci, Pol II is recruited and engaged in transcription, but only synthesizes short, abortive precursor transcripts (Nechaev and Adelman, 2011). Pol II is paused and unable to transition into productive elongation by associating with Negative Elongation Factor (NELF) and DRB-sensitivity Factor (DSIF). This process competes with nucleosomes for occupancy in the promoter-proximal region, thereby keeping these loci more accessible for future activation (Gilchrist et al., 2010). Upon stimulation, Pol II is released from pausing by recruitment of Positive Elongation Factor (P-TEFb), leading to the phosphorylation of NELF, DSIF and Serine-2 of the Pol II CTD (Pol II Ser-2-P). This causes the rapid transition to the elongating form of Pol II (Pol II Ser-2-P/Ser-5-P) and functional mRNA production (Nechaev and Adelman, 2011). In some systems, open histone marks near the promoter region, including Histone 3 Lysine 4 trimethylation (H3K4me3), are associated with transcriptionally paused loci (Guenther et al., 2007; Hargreaves et al., 2009). Collectively, these studies indicate that transcriptional pausing promotes an open chromatin state near the transcription start site for some inducible genes, thereby potentiating their future activation. A subset of mammalian LPS-dependent primary response genes is regulated by pausing (Adelman et al., 2009; Hargreaves et al., 2009). Whether this is evolutionarily conserved or required for antiviral defense is unknown.
Drosophila melanogaster is a powerful genetic organism to study how transcriptional mechanisms govern innate responses (Ganesan et al., 2011; Hultmark, 2003; Kemp and Imler, 2009; Lemaitre and Hoffmann, 2007; Wasserman, 2004). Not only was transcriptional pausing initially discovered and extensively characterized in Drosophila (Gilmour and Lis, 1986; Nechaev and Adelman, 2011), its sole reliance on innate defenses provides a robust model system for study (Cherry and Silverman, 2006; Hultmark, 2003; Lemaitre and Hoffmann, 2007; Sabin et al., 2010). Indeed, Toll was discovered using Drosophila, leading to subsequent discovery of the mammalian Toll-Like Receptors (TLRs) (Fitzgerald and Chen, 2006; Lemaitre and Hoffmann, 2007; Lemaitre et al., 1996). This system has also been used to study antiviral immunity of insect vectors, as many human arthropod-borne viruses (arboviruses) can infect and replicate in flies, including the Alphavirus Sindbis virus (SINV), Rhabdovirus Vesicular Stomatitis virus (VSV), Bunyavirus Rift Valley Fever Virus (RFV), and Flavivirus Kunjin (KUN) (Chotkowski et al., 2008; Filone et al., 2010; Rose et al., 2011; Sabin et al., 2009; Shelly et al., 2009). While transcriptional programs against bacteria and fungi are well-established in insect models (Ferrandon et al., 2007), much less is known about the host programs that restrict viruses (Sabin et al., 2010). Some antiviral transcriptional pathways, including the classic JAK/STAT pathway, have found to be conserved in Drosophila (Avadhanula et al., 2009; Costa et al., 2009; Dostert et al., 2005; Zambon et al., 2005), but are insufficient to account for all antiviral defenses.
To discover additional antiviral transcriptional mechanisms, we used RNAi screeningagainst disparate arboviruses in Drosophila and identified multiple components of the transcriptional pausing pathway (including NELF, DSIF, and P-TEFb) as essential mediators of insect antiviral defense. Genome-wide transcriptional profiling led us to characterize a complex virally-induced genetic program, including components of known antiviral pathways (Toll, Imd, JAK/STAT, autophagy, and RNA silencing) and a number of Toll receptors, including one recently found to play a role in antiviral defense (Lemaitre and Hoffmann, 2007; Nakamoto et al., 2012; Sabin et al., 2010). This transcriptional response is rapid and consists of two classes: translation-independent and dependent genes. Furthermore, we find that over half of this response relies on the pausing-release factor P-TEFb and has biochemical features of inducible paused loci, including the promoter-proximal enrichment of pausing machinery (Pol II, NELF, and DSIF) and NELF-dependent basal synthesis of short, abortive transcripts. We also demonstrate that transcriptional pausing controls infection at the organismal level, as NELF and P-TEFb restrict viral replication in flies. We extend our findings to mosquitoes, the natural hosts of some arboviruses, and find that NELF and P-TEFb restrict infection in Aedes aegypti cells. Our data collectively suggest that transcriptional pausing enhances promoter accessibility of virally-responsive loci to allow for rapid activation upon infection. Once induced, this program produces a robust and multifaceted response to restrict viral infection.
RESULTS
NELF Restricts Viral Infection In Drosophila Cells
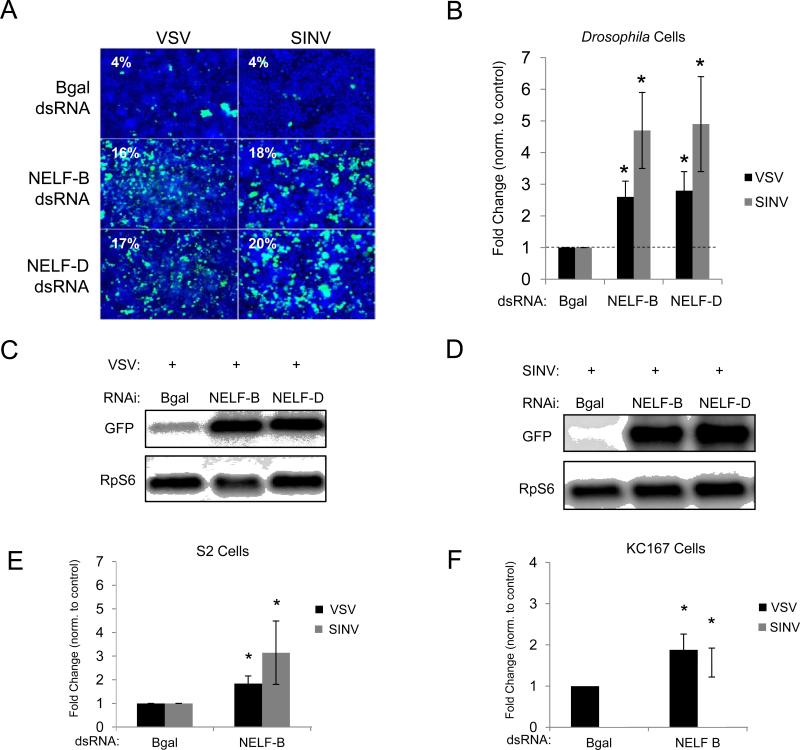
To identify host factors that broadly restrict viral infection, we performed a small-scale RNAi screen in Drosophila cells using disparate arthropod-borne RNA viruses (VSV and SINV) whose natural cycle involves transmission between insects and vertebrates (Rose et al., 2011; Sabin et al., 2009; Weaver and Barrett, 2004). Depletion of two components of the four subunit NELF complex, which is involved in transcriptional pausing (Sims et al., 2004), increased infection by these viruses (S.C., unpublished data). To further investigate NELF's role in antiviral defense, we generated independent dsRNAs targeting alternative regions of the identified subunits, NELF-B and NELF-D (TH1). We found that depletion of NELF-B or NELF-D with an independent dsRNA similarly increases susceptibility of cells to SINV and VSV infection compared to non-targeting controls, as measured by percent infection (Figure 1A). The infection percentage is determined by fluorescence microscopy of GFP, a reporter expressed from the genome of both viruses (Burnham et al., 2007; Ramsburg et al., 2005). Quantification reveals >2-fold increase in VSV and SINV infection (Figure 1B). Northern blot analysis shows efficient knockdown (Figures S1A and S1B). We also measured viral replication by Northern blot and found increased VSV mRNA levels in NELF-B or NELF-D-depleted cells (Figure 1C). Similarly, we found that SINV RNA levels are increased in NELF-deficient cells (Figure 1D). Furthermore, we found that NELF-B can also restrict VSV and SINV infection in Drosophila S2 and Kc167 cells (Figures 1E, 1F, S1C, and S1D). From these findings, we conclude that NELF restricts viral pathogens from disparate families in a variety of Drosophila cell lines.
Figure 1. NELF restricts viral infection in Drosophila cells.
(A) Drosophila cells were treated with dsRNA against a control (Bgal), NELF-B, or NELF-D. Infected cells expressing a VSV-encoded GFP (MOI=0.2) or SINV- encoded GFP (MOI=5) are shown in green, nuclei in blue. (B) Quantification of images in (A) as normalized to controls. Mean ± S.D. of three independent experiments shown; *p < 0.05. Northern blot analysis of cells pretreated with the indicated dsRNAs and infected with (C) VSV or (D) SINV. (E) S2 and (F) Kc167 cells were treated as in (A). Quantification of images as percentage of infection, normalized to controls. Mean ± S.D. of three independent experiments shown; *p < 0.05. See also Figure S1.
Transcriptional Pausing Is Required For Defense Against Disparate Arboviral Pathogens
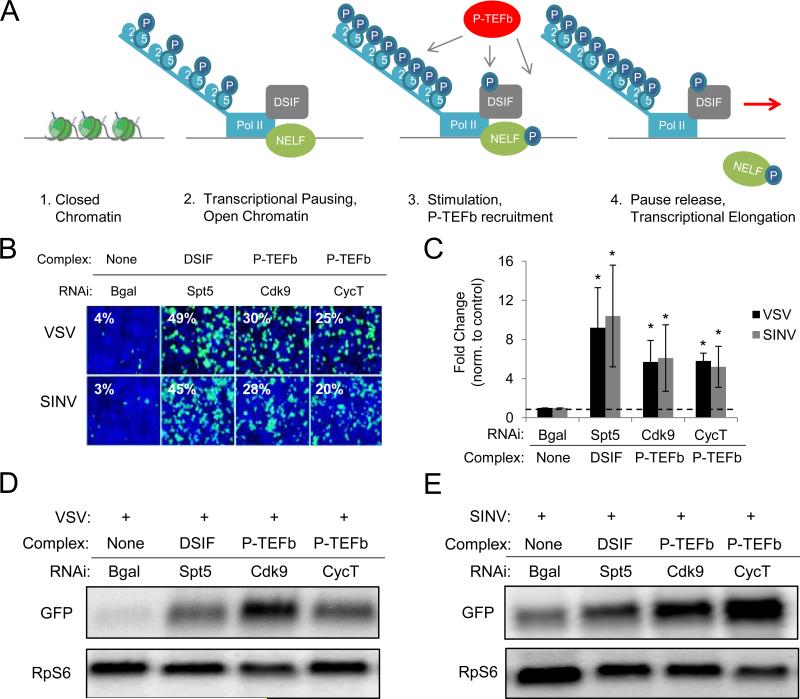
To determine whether NELF's antiviral activity is mediated through involvement in the transcriptional pausing pathway, we tested whether DSIF or P-TEFb, two complexes required for pausing and release, are antiviral against VSV and SINV (Figure 2A). DSIF, comprised of Spt4 and Spt5, binds NELF and helps facilitate polymerase pausing about 50 base pairs downstream of the transcription start site (Saunders et al., 2006). P-TEFb, comprised of Cdk9 and CyclinT (CycT), releases this pause to promote transcriptional elongation and functional mRNA production (Nechaev and Adelman, 2011; Saunders et al., 2006). We find that both of these complexes restrict VSV and SINV infection, as their depletion causes a significant increase in infection (Figures 2B and 2C). In addition, Northern blotting indicates that both VSV and SINV mRNA levels are increased in cells depleted of DSIF or P-TEFb subunits (Figures 2D and 2E). We also found that P-TEFb is antiviral in S2 and Kc167 cells (Figures S2A and S2B). Lastly, we determined whether this pathway is antiviral against other medically relevant arboviruses and found that P-TEFb restricts KUN and RFV infection in Drosophila cells, along with the natural Drosophila pathogen Drosophila C virus (DCV) (Figures S2C and S2D).
Figure 2. Transcriptional pausing and release restricts viral infections in Drosophila cells.
(A) Schematic of transcriptional pausing and release. (B) Drosophila cells treated with the indicated dsRNAs were challenged with VSV (MOI=0.2) or SINV (MOI=5) and monitored by fluorescence microscopy. (C) Quantification of images in (A). Mean ± S.D. of three independent experiments shown; *p < 0.05. Northern blot analysis of cells pretreated with the indicated dsRNAs and infected with (D) VSV or (E) SINV. See also Figure S2.
We found that machinery that helps to pause Pol II (NELF, DSIF) and P-TEFb that alleviates this pause both have antiviral phenotypes (Figures 1 and 2), suggesting that these complexes act concertedly to promote antiviral defense. This is consistent with recent studies reporting that NELF-mediated pausing positively regulates gene expression by promoting open chromatin structure in the promoter-proximal region of genes, facilitating their future activation by P-TEFb (Adelman et al., 2009; Hargreaves et al., 2009). This is in contrast to other genes, most classically the heat shock loci, where NELF and DSIF negatively regulate gene expression (Wu et al., 2003). Altogether, these data suggest that NELF's antiviral activity is mediated through the transcriptional pausing pathway to positively control gene expression, through regulation of antiviral gene induction.
Viral Infection Induces A Rapid Antiviral Host Response
To identify virally-induced genes that are regulated by transcriptional pausing, we first analyzed a published Drosophila microarray study that found two-thirds of approximately 250 NELF-dependent genes are positively controlled by NELF (Gilchrist et al., 2008). One immune-associated gene called Tep II was genetically dependent on NELF for its basal expression and had enrichment of pausing machinery near its promoter (Gilchrist et al., 2008). There was increased nucleosome occupancy in the promoter region of Tep II upon NELF depletion, explaining how the loss of NELF could lead to reduced expression (Gilchrist et al., 2008). This is provocative, because Tep II is a complement-related gene that is induced by SINV infection in Drosophila and mosquito cells (Mudiganti et al., 2010).
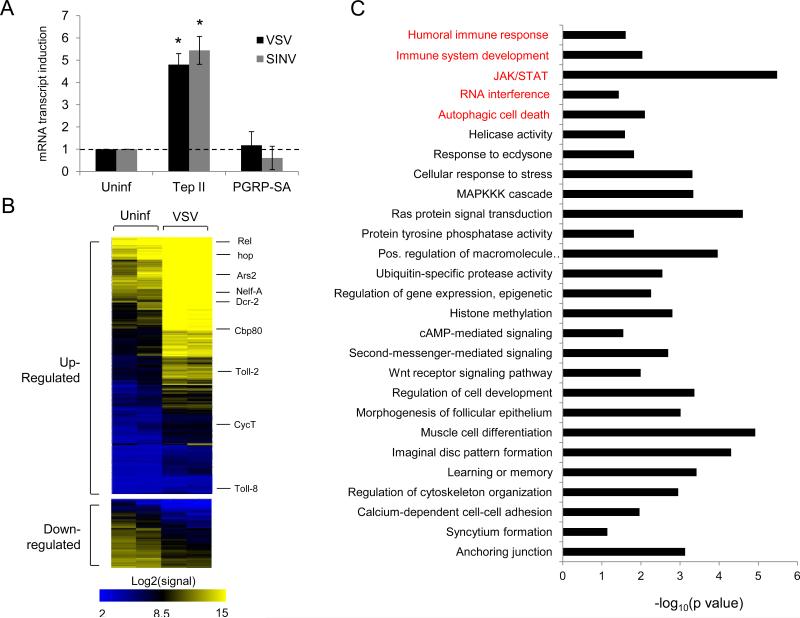
Based on these findings, we first tested whether virus infection leads to Tep II induction (Gilchrist et al., 2008). We also tested Peptidoglycan recognition protein SA (PGRP-SA), a bacterial-recognition protein of the Toll pathway, that was shown to be genetically dependent on NELF, but its biochemical status had been unexplored (Gilchrist et al., 2008; Gottar et al., 2002). Given that pausing-regulated genes are often rapidly induced (Rasmussen and Lis, 1995; Zeitlinger et al., 2007), we challenged Drosophila cells with virus and monitored gene induction at 4 hours post-infection. We found that both VSV and SINV can induce Tep II expression at this early time-point post-infection, as measured by RT-qPCR (Figure 3A). This is prior to the initiation of viral replication at approximately 6 hours post-infection (Figure 3A, not shown) (Dezélée et al., 1987; Gliedman et al., 1975). In contrast, we found that PGRP-SA is not induced (Figure 3A), suggesting that only a subset of NELF-regulated genes is responsive to viral infection.
Figure 3. An antiviral transcriptional program is rapidly induced by viral infection.
(A) Drosophila cells were infected with VSV (MOI=10) or SINV (MOI=25) for 4 hours. RT-qPCR was performed for Tep II and PGRP-SA, normalized to Rp49, and shown compared to uninfected controls. Mean ± S.D. of three independent experiments is shown; *p < 0.005. (B) Heat map of raw signal levels for genes differentially expressed at 4 hours post VSV-infection (MOI=10), performed in biological duplicates (q<0.005). Shown are 636 transcripts (540 upregulated, 96 downregulated) with at least 2.8-fold change in VSV-infected cells. Genes of interest are shown on the right. (C) Enriched GO terms for the 540 virally induced genes (p<0.05). See also Tables S1A-D.
Next, we set out to identify the full spectrum of virally induced genes that may be pausing-regulated. To this end, we performed global gene expression profiling of Drosophila cells that were either uninfected or infected with VSV for 4 hours. We profiled two independent experiments, identifying 540 upregulated and 96 downregulated genes with at least a 2.8-fold change in mRNA levels (q<0.005; Figure 3B, Tables S1A and S1B). Gene Ontology (GO) enrichment analysis revealed that several known immune and antiviral pathways are over-represented within this dataset (p<0.05, Figure 3C, Tables S1C and D), including humoral immune response and immune system development (Figure 3C and Table S1C). We also identified components of major antiviral pathways known to restrict viruses in Drosophila including RNA interference, JAK/STAT, Imd, and autophagy pathways (Figure 3C and Table S1D) (Avadhanula et al., 2009; Costa et al., 2009; Dostert et al., 2005; Sabin et al., 2010; Shelly et al., 2009; Zambon et al., 2005). Interestingly, we also found that transcriptional pausing pathway components NELF-A and CycT are induced (Figure 3B and Table S1D). Altogether, our findings suggest that this rapidly induced early transcriptional program has antiviral effector function.
Viral Infection Induces A Transcriptionally Complex Antiviral Host Response
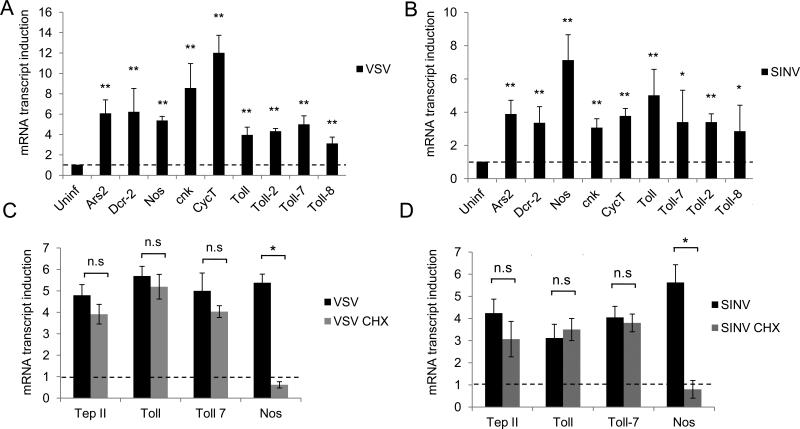
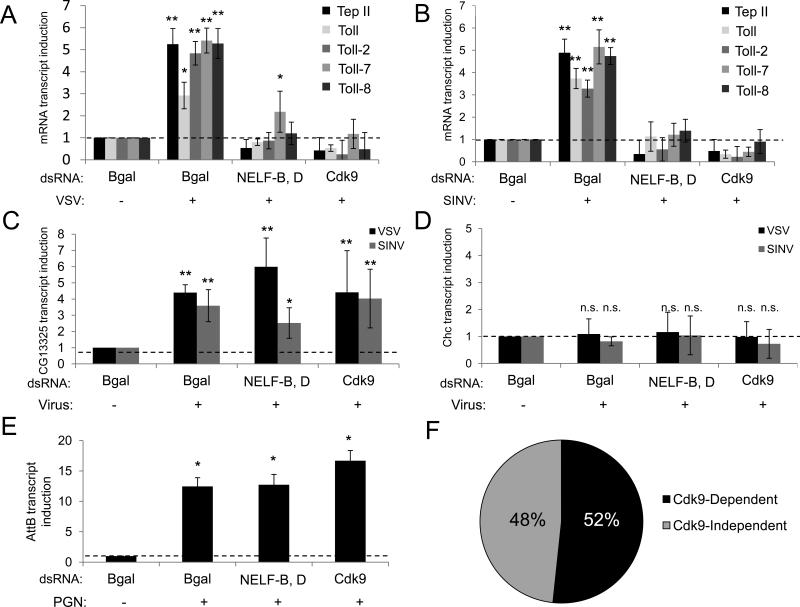
Next, we tested whether these virally responsive genes are inducible by both VSV and SINV. We selected candidates from our dataset with known antiviral function (Ars2, Dcr-2), genes not previously implicated in Drosophila antiviral immunity (Ago1, cnk, Nos), and the CycT subunit of the P-TEFb complex (Figure 2). We found that both viruses induce these genes at 4 hours post-infection (Figures 4A and 4B). Since two Toll receptors (Toll-2 and Toll-8) were VSV-induced in our profiling data, we also tested the panel of nine Drosophila Toll receptors. We found that Toll, Toll-2, Toll-7, and Toll-8 are inducible by both VSV and SINV, while the remaining Tolls are not inducible by both viruses (Figures 4A and 4B, not shown).
Figure 4. Viral infection triggers a rapid and transcriptionally complex antiviral expression program.
Drosophila cells were infected with (A) VSV (MOI=10) or (B) SINV (MOI=25). RT-qPCR was performed for the indicated genes at 4 h.p.i., normalized to Rp49, and shown relative to uninfected controls. Mean ± S.D. of three independent experiments is shown; **p < 0.005, *p<0.05. (C-D) Drosophila cells were untreated or pre-treated with 10 μg/ml Cycloheximide (CHX) and infected as in (A, B). RT-qPCR was performed for the indicated genes. Mean ± S.D. of three independent experiments is shown; *p<0.005.
Many rapidly inducible, pausing-regulated genes are activated in the absence of protein synthesis and are a subset of primary response genes (Herschman, 1991; Shaw and Kamen, 1986; Yamamoto and Alberts, 1976). To determine whether the induction of our candidate genes requires new protein synthesis, we compared their transcript levels upon viral infection in the presence and absence of Cycloheximide (CHX), a translation inhibitor. We found that Tep II, Toll, and Toll-7 are inducible by VSV and SINV in a translation-independent manner (Figure 4C and 4D). In contrast, Nos was virally induced in a Cycloheximide-dependent manner (Figure 4C and 4D). Hence, our findings suggest this early virally induced gene expression program is multifaceted, involving both primary responses and secondary responses.
Some Virally Induced Genes Are NELF And P-TEFb-dependent
To determine whether the induction of these translation-independent genes requires the transcriptional pausing machinery, we first depleted NELF (NELF-B, D) or P-TEFb (Cdk9) by RNAi, challenged Drosophila cells with VSV or SINV, and monitored their induction by RT-qPCR. We found that VSV- and SINV-induced expression of Tep II, Toll, Toll-2, Toll-7 and Toll-8 are attenuated upon NELF or P-TEFb knockdown (Figures 5A and 5B). In addition, we found that the basal levels of Tep II and Toll, but not Toll-7 or Toll-8, are reduced upon NELF and P-TEFb-depletion (Figure S3A). Expression levels of housekeeping genes Clathrin Heavy Chain (Chc) and Rp49 are unaffected by the loss of pausing factors, with or without viral infection (Figures 5D and S3B). This is consistent with reports suggesting that transcriptional pausing is a step in the transcription cycle of many constitutively active housekeeping genes, but not necessarily rate-limiting for their expression (Gilchrist et al., 2008). Hence, the requirement for NELF and P-TEFb are not necessarily to maintain basal levels per se.
Figure 5. A subset of virally induced genes is NELF and P-TEFb-dependent.
(A-D) Drosophila cells were treated with the indicated dsRNAs and infected with VSV (MOI=10) or SINV (MOI=25). RT-qPCR of the indicated genes at 4 h.p.i., normalized to Rp49, and shown as relative to uninfected controls. Mean ± S.D. of three independent experiments is shown; **p< 0.005, *p<0.05. (E) Cells were treated with PGN for 6 hours. AttB expression was measured and normalized as stated above. Mean ± S.D. of three independent experiments is shown; *p<0.005. (F) Analysis of Cdk9-dependency of the 540 VSV-induced genes (279 genes, ≥1.5 fold down-regulation). Black and gray indicate the percentage of Cdk9-dependent and independent genes, respectively. See also Figure S3.
We also found that pausing regulates only a subset of the response since not all virus-induced genes are NELF- and P-TEFb-dependent. One example is CG13325 (Figure 5C), a VSV-induced gene from our profiling dataset that is also found to be induced by DCV in flies (Dostert et al., 2005). In addition, we tested whether NELF or P-TEFb is required for induction of antimicrobial peptides (AMPs) Diptericin B (DptB) and Attacin B (AttB), which are classic peptidoglycan (PGN)-inducible genes that require NFkappaB (Boutros et al., 2002; Ganesan et al., 2011). We found their induction to be independent of NELF and P-TEFb (Figures 5E and S3C). These findings demonstrate that transcriptional pausing controls specific subsets of pathogen-induced genes.
To characterize the spectrum of virally induced genes that require pausing factors, we performed genome-wide profiling and found that P-TEFb regulates about 52% of the 540 virally induced genes (≥ 1.5 fold down-regulation, 279 genes, Figure 5F, and Table S1A). This is a highly significant enrichment compared to published studies showing that only 1-5% of the transcriptome is affected by the loss of NELF or P-TEFb in Drosophila and mammalian systems using similar approaches (Garriga et al., 2010; Gilchrist et al., 2008; Sun and Li, 2010; Yu et al., 2008). Altogether, our findings suggest that transcriptional pausing plays a major role in regulating the virally induced gene expression program.
Virally Induced, P-TEFb-Dependent Genes Have Chromatin Features of Paused Loci
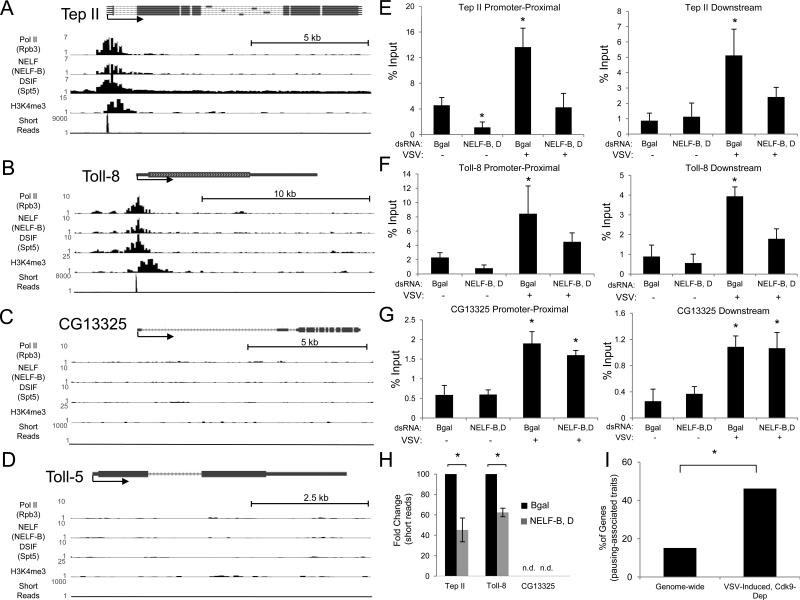
Several biochemical traits have been described for pausing-regulated inducible genes in the literature, including (1) promoter-proximal enrichment of NELF and DSIF, (2) production of short, abortive transcripts from the 5’ transcriptional start site, (3) promoter-proximal enrichment of the open chromatin mark H3K4me3, and (4) a peak of Pol II localization at the promoter, but largely absent from the body of the gene (Enderle et al., 2011; Gilchrist et al., 2010; Nechaev et al., 2010). Genome-wide studies have characterized these cellular features (Adelman et al., 2009; Enderle et al., 2011; Gilchrist et al., 2010) and we analyzed this data to determine whether virally responsive genes have these traits. Since Tep II has been shown to be regulated by transcriptional pausing both genetically and biochemically (Gilchrist et al., 2008), it serves as a proof-of-principle for our analysis (Figure 6A). We found that the Tep II promoter-proximal region is (1) enriched for the paused machinery (NELF, DSIF), (2) produces abundant short, abortive transcripts from the 5’ transcriptional start site, (3) exhibits promoter-proximal enrichment of H3K4me3, and (4) has a peak of Pol II restricted to the promoter-proximal region (Figure 6A). Next, we analyzed the Toll receptors and found that only the four pan-virally-inducible Toll receptors (Toll, Toll-7, Toll-2 and Toll-8) also have these features (Figures 6B, S4A, S4B, and S4C), while the remaining five Toll receptors do not (Figures 6D and S4D, data not shown). Furthermore, CG13325 also lacks the pausing-associated chromatin traits (Figure 6C), which is virally induced independent of NELF and P-TEFb (Figure 5C). PGN-inducible and NFkappaB-dependent AMP genes, DptB and AttA, also lacked these features (Figures S4E and S4F). As expected, housekeeping genes differ from the virally inducible pausing-regulated loci. Rp49 and Chc have Pol II occupancy spanning the body of the gene, indicative of robust functional mRNA production (Figures S4G and S4H). The virally induced genes that are NELF and P-TEFb-dependent lack this downstream occupancy, consistent with their low basal expression and dependence on transcriptional pausing for induction (Figures 4, 5, 6, and S4). Lastly, we analyzed available Pol II occupancy from Mod Encode for some of our pausing-regulated virally induced genes in other cell types (Celniker et al., 2009; Kharchenko et al., 2011). Tep II and Toll have the same basal promoter-proximal enrichment of Pol II in Kc167 and CME W1 cl.8+ cells (Figures S4I and S4J), suggesting that this pausing signature may be conserved.
Figure 6. Virally induced, P-TEFb-dependent genes have chromatin features of transcriptionally paused loci.
Mapping of Pol II (Rbp3), NELF (NELF-B), DSIF (Spt5), H3K4me3, and short RNA reads for (A) Tep II (B) Toll-8 (C) CG13325 and (D) Toll-5. ChIP of RNA Pol II for (E) Tep II (F) Toll-8 (G) CG13325 in cells treated with the indicated dsRNAs and either uninfected or infected with VSV (MOI=10). Primers span the promoter-proximal or downstream regions. The data is represented as a percentage of input. Mean ± SD for three independent experiments is shown; *p<0.05. (H) RT-qPCR of 5’ short reads for Tep II, Toll-8, and CG13325 with the indicated dsRNA treatment in Drosophila cells. Transcripts were normalized to Rp49 and shown as relative to controls. Mean ± S.D. of three independent experiments is shown; *p<0.05. (I) Comparison of VSV-induced, P-TEFb-dependent genes with pausing-associated chromatin features to the genome-wide distribution (*p<0.0001). See also Figure S4 and Table S2.
Next, we performed chromatin immunoprecipitation (ChIP) of RNA Pol II using an antibody that recognizes both initiating and elongating forms of Pol II (Ser-2-P/Ser-5-P) at virally induced loci in Drosophila cells. We found significant Pol II occupancy in the promoter-proximal regions of Tep II and Toll-8 in the absence of infection (Figures 6E and 6F), but little signal near the promoter of the NELF-independent gene CG13325 (Figure 6G). As expected, there is also little Pol II signal in the body of Tep II, Toll-8, and CG13325 prior to infection (Figures 6E, 6F, and 6G), consistent with our mRNA and ChIP analyses (Figures 5, 6A, 6B, and 6C). NELF-depletion leads to a reduction in Pol II occupancy near the Tep II promoter region (Figure 6E), consistent with our mRNA analysis and published findings (Gilchrist et al., 2008). We found that 5’ short reads are also detectable in uninfected Drosophila cells for Tep II and Toll-8, but not CG13325 (Figure 6H), as measured by RT-qPCR. Importantly, the synthesis of these short transcripts is NELF-dependent (Figure 6H). These findings, in addition to our genetic studies, suggest that NELF-dependent abortive transcription promotes Pol II occupancy at a basal state.
Moreover, we found that VSV infection triggers a significant increase in Pol II occupancy in both promoter-proximal and distal regions of Tep II, Toll-8, and CG13325 (Figures 6E, 6F, and 6G). This occupancy was NELF-dependent for Tep II and Toll-8, but not CG13325 (Figures 6E, 6F, and 6G). For Tep II and Toll-8, pausing helps facilitate Pol II release into the body of the genes upon viral infection and supports our finding of a genetic dependence on NELF for functional mRNA production upon viral infection (Figure 5). As a control, we tested the housekeeping gene RpL3 and found no significant difference in Pol II occupancy upon viral infection in the promoter-proximal or distal regions (not shown). Thus, we have biochemically and genetically identified a pausing-dependent, virally responsive gene expression program.
Lastly, we examined the pausing hallmarks at the chromatin level for the virally induced genes as a whole. Published genome-wide characterization of these features suggests that approximately 50% of the genome, including constitutively active housekeeping genes, have pausing traits to varying degrees (7466 genes). However, only 13% (1866 genes) are defined as strongly paused (Gilchrist et al., 2010). We similarly defined candidates as strongly paused if they had a statistically significant enrichment within +/- 500 kb of the transcription start site for at least three of these four traits: (1) NELF, (2) DSIF, (3) Pol II and (4) basal short transcript production. Our analysis reveals that 14% of the genome has these features (2225 genes, Figure 6I), consistent with published findings. Strikingly, we found that 47% of the P-TEFb-dependent genes are strongly paused at the chromatin level (130 genes, Figure 6I, Table S1A), a significant enrichment compared to the genome as a whole (p<0.0001, Figure 6I). GO enrichment analysis identified 16 categories within this set of genes (p<0.05, Figure S4K and Table S2). In addition to the immune response category that was also enriched within the dataset as a whole (Figure 3C and Table S2), other categories include MAPK signaling and genes involved in cytoskeleton organization (Table S2).
Transcriptional Pausing Restricts Viral Replication In Adult Flies And Mosquito Cells
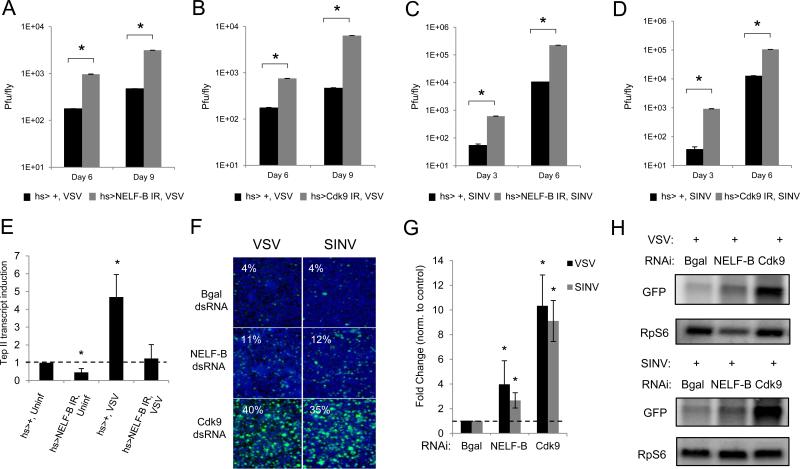
To determine whether transcriptional pausing plays an essential antiviral role at the organismal level, we depleted two major pathway components, NELF-B and Cdk9, in adult fliesusing inducible RNAi, as null mutants are lethal (Wang et al., 2010). We used the Gal4/UAS system to drive expression of UAS-inverted repeat transgenes (UAS-NELF-B IR or UAS-Cdk9 IR), which bear long hairpin dsRNA constructs to target the endogenous transcripts in vivo. To bypass developmental requirements, we drove the expression of hairpins using a heat shock (hs) promoter and found that we can deplete NELF-B or Cdk9 mRNAs (Figure S5). We challenged NELF-B-depleted, Cdk9-depleted, or control flies with carrier (PBS), VSV, or SINV, and monitored for viral replication using plaque assays. We found that NELF-B-deficient flies and Cdk9-deficient flies have significantly higher VSV and SINV titers compared to wildtype flies at two time points post-infection (Figures 7A, 7B, 7C, and 7D). To determine whether transcriptional pausing regulates a similar gene expression program in vivo, we challenged files with VSV and found that Tep II was induced 24 hours post-infection in a NELF-B-dependent manner (Figure 7E). These findings suggest that transcriptional pausing plays a critical role in Drosophila antiviral immunity at the organismal level.
Figure 7. NELF and P-TEFb restrict viral infection in flies and mosquito cells.
Adult flies of the indicated genotypes were challenged with (A and B) VSV or (C and D) SINV. Viral titers were measured with mean ± S.D. of three independent experiments shown; *p < 0.05. (E) Adult flies of the indicated genotypes were challenged with VSV. RT-qPCR of Tep II is shown, normalized to Rp49, and represented as relative to the uninfected mock-depleted controls. Mean ± S.D. of three independent experiments is shown; p<0.05. (F) Aedes aegypti Aag2 cells treated with the indicated dsRNAs were challenged with VSV (MOI=0.01) or SINV (MOI=0.5) and monitored by fluorescence microscopy (virus in green, nuclei in blue). (G) Quantification of images in (F) as normalized to controls. Mean ± S.D. of three independent experiments is shown; *p < 0.05. (H) Northern blot analysis of Aag2 cells pretreated with the indicated dsRNAs and infected with either VSV or SINV. See also Figure S5.
Lastly, to determine whether this antiviral mechanism is conserved in other insects, including mosquito vectors, we examined whether NELF and P-TEFb can restrict viral replication in mosquito cells. We used RNAi to deplete Aedes aegypti Aag2 cells of NELF-B (AAEL014752) or Cdk9 (AAEL013002) and compared the percentage of infected cells to non-targeting controls using microscopy. We found that NELF-B and Cdk9 restricted these two viruses in mosquito cells (Figures 7F and 7G). In addition, we used Northern blotting to measure viral RNA levels. We found VSV and SINV mRNA levels are increased in NELF-B or Cdk9-depleted cells as compared to control cells (Figure 7H). Our findings using both Drosophila and Aedes systems suggest that transcriptional pausing plays an important and conserved role in insect antiviral defense.
DISCUSSION
Through RNAi screening against disparate arboviral pathogens in Drosophila, we discovered that the transcriptional pausing pathway is broadly antiviral in insects (Figures 1, 2, and 7). This led us to characterize a rapidly-inducible host response that has components from all major known antiviral pathways, including RNA silencing genes Ars2, Cbp80, and Dcr-2 that control a range of RNA viruses in insects (Figure 3 and Table S1D) (Chotkowski et al., 2008; Galiana-Arnoux et al., 2006; Keene et al., 2004; Li et al., 2002; Mueller et al., 2010; Sabin et al., 2009; Wang et al., 2006; Zambon et al., 2006). 52% of this virus-induced response requires P-TEFb for activation (Figure 5F), which is highly overrepresented compared to the genome as a whole (Garriga et al., 2010; Gilchrist et al., 2010; Gilchrist et al., 2008; Sun and Li, 2010; Yu et al., 2008). We find that 47% of the P-TEFb-dependent response has multiple chromatin features of transcriptional pausing, which is also enriched compared to 14% of the genome as a whole (p<0.0001, Figure 6I). These strict biochemical criteria include the presence of the pausing machinery (Pol II, NELF, and DSIF) and the basal synthesis of 5’ short transcripts. Mechanistically, we find that RNA Pol II is enriched in the promoter-proximal region for the virally induced pausing-regulated genes and this occupancy is NELF-dependent (Figures 6E and 6F). At these loci, short abortive transcripts are synthesized in a NELF-dependent manner (Figure 6H). Upon viral infection, RNA Pol II is rapidly recruited downstream, leading to the production of full-length mRNAs (Figures 5 and 6). Moreover, we find that pausing is antiviral in various Drosophila cell lines, mosquito cells, and adult flies (Figures 1 and 7), suggesting that a transcriptional pausing-regulated gene expression program plays a broad and conserved role in insect antiviral defense.
Compared to bacterial infections, less is known about the transcriptional programs that restrict viral pathogens in insects and no rapid host responses have been characterized (Carpenter et al., 2009; Dostert et al., 2005; Ferrandon et al., 2007; Lemaitre and Hoffmann, 2007; Mudiganti et al., 2010; Roxström-Lindquist et al., 2004; Souza-Neto et al., 2009; Tsai et al., 2008). We discovered an early transcriptional response to viral infection that includes components of major antiviral pathways (Figures 3, 4, and Table S1) (Lemaitre and Hoffmann, 2007; Sabin et al., 2010). The highly conserved JAK/STAT signaling pathway is the best-studied antiviral transcriptional response. It is activated by and restricts DCV in adult flies and Dengue virus in adult mosquitoes, but has not shown to be active in cell culture (Dostert et al., 2005; Hedges and Johnson, 2008; Souza-Neto et al., 2009). We found that the Drosophila homolog of JAK (hop) is transcriptionally induced by viral infection along with 8 pathway-associated genes (Table S1D) (Dostert et al., 2005). However, induction of JAK/STAT-dependent target genes was not observed and STAT binding sites were not enriched within our dataset (data not shown) (Dostert et al., 2005). There are two NFkappaB-dependent signaling pathways in Drosophila, the Toll and IMD pathway, which are implicated in antiviral defense (Avadhanula et al., 2009; Costa et al., 2009; Ramirez and Dimopoulos, 2010; Zambon et al., 2005). We found that Toll itself and the IMD pathway component Relish were are induced by infection, but not other pathway components nor their canonical downstream AMP targets (Figures 3, 4, Table S1D) (Boutros et al., 2002; Busse et al., 2007; De Gregorio et al., 2002; Hoffmann, 2003; Irving et al., 2001; Pal et al., 2008). Moreover, our dataset lacks enrichment for NFkappaB binding sites (data not shown) (Ganesan et al., 2011). These findings suggest that our rapidly induced antiviral program is not likely dependent on JAK/STAT, Toll, or IMD pathways. Future work characterizing the upstream signals leading to P-TEFb recruitment may further shed light into this response.
Interestingly, we found that three other Toll receptors (Toll-2, Toll-7, and Toll-8) are transcriptionally induced by VSV and SINV infection (Figure 4). We recently found that Toll-7 is antiviral against VSV via the autophagy pathway and 9 other autophagy-associated genes are also rapidly induced by viral infection (Table S1D) (Nakamoto et al., 2012; Shelly et al., 2009). Toll-2 may play a minor role in antibacterial immunity (Ligoxygakis et al., 2002; Williams et al., 1997). Toll-8 negatively regulates local immune responses to bacterial challenge (Akhouayri et al., 2011; Michel et al., 2001). Whether Toll-2, Toll-8, or additional Drosophila Toll receptors play roles in antiviral defense remains an open question. Furthermore, whether pausing regulates mammalian TLR expression or other pattern recognition receptors downstream of viral infection is unknown. Since our genome-wide profiling experiments reveal that known antiviral pathways are virally induced (Figures 3, 4, and Table S1D), additional genes within our gene-set may also play direct roles in antiviral immunity. Further exploration of these rapidly induced genes, both paused and non-paused, may reveal additional aspects of the innate immune arsenal.
One of the best-characterized innate immune transcriptional response is the LPS-induced primary and secondary response program in macrophages (Medzhitov and Horng, 2009; Smale, 2010). The primary response is independent of de novo protein synthesis and has differences in chromatin structure and CpG content compared to the secondary response (Hargreaves et al., 2009; Ramirez-Carrozzi et al., 2009). Recent studies reveal that a subset of these LPS-induced primary response genes, including TNF-alpha, is controlled by transcriptional pausing in vitro (Hargreaves et al., 2009). This rapid antiviral transcriptional program parallels many aspects of the LPS-induced macrophage response, as both translation-independent and translation-dependent targets were identified (Figure 4). Interestingly, some LPS-induced primary response genes, like the transcription factor IkappaBzeta, control downstream secondary responses (Yamamoto et al., 2004). Whether any of our virally induced primary response genes drive subsequent gene expression is an open question. We find that 32 genes are both pausing-regulated and involved in transcription as defined by annotated GO terms, including transcription factors and chromatin modifiers. Characterization of their potential roles in driving secondary responses may reveal insights into the sequential nature of these early immunologic events.
Transcriptional pausing restricts disparate arboviruses in cell culture and at the organismal level. Whether other insect immune responses are controlled by similar mechanismsis largely unknown (Gilchrist et al., 2012). Furthermore, whether pausing similarly controls antiviral defense in mammals is also unknown. Nevertheless, our findings in conjunction with published literature suggest that transcriptional pausing is associated with rapid gene induction and may serve as a robust mechanism for immediate defense against a variety of pathogens across diverse hosts (Adelman and Rogatsky, 2010; Hargreaves et al., 2009; Zeitlinger et al., 2007). Future work examining the role of transcriptional pausing in these various immunologic contexts may help define how paused genes are differentially activated and also lead to the development of therapeutic strategies for infectious diseases.
EXPERIMENTAL PROCEDURES
Detailed descriptions are provided in the Extended Experimental Procedures.
Cells, Viruses, and Reagents
Cells and viruses were grown and maintained as previously described (Rose et al., 2011; Shelly et al., 2009). Additional chemicals were obtained from Sigma.
RNAi and Viral Infections
RNAi was performed and cells were processed for microscopy or RNA at the indicated time points post infection, as described (Shelly et al., 2009).
RNA, Northern Blotting and RT-qPCR
Total RNA was processed for Northern blotting as described (Cherry et al., 2005). For RT-qPCR, cDNA was subject to PCR and analyzed by relative quantification, by normalizing to Rp49. Data is represented as relative mRNA expression compared to the untreated samples and displayed as the mean ± SD for three independent experiments.
Fly Infections and Titering
The indicated genotypes were infected and processed as described (Shelly et al., 2009).
ChIP
ChIPs were performed based on (Gilchrist et al., 2008). The data is represented as a percentage of each input and displayed as the mean ± SD for three independent experiments.
DNA Microarray Analysis
Affymetrix Drosophila GeneChip microarrays (Affymetrix) were analyzed. We calculated fold changes relative to uninfected controls and considered significant genes with at least a 2.8 fold change. For Cdk9-dependence, we calculated fold changes relative to VSV-infected samples and considered significant genes with ≥1.5 fold down-regulation.
Bioinformatics and Statistics
We mapped the Drosophila ChIP-on-chip data for total Pol II (Rpb3), NELF-B, and Spt5 (Gilchrist et al., 2010), H3K4me3 (Beisel et al. 2011), and the short RNA sequence reads (Nechaev et al., 2010). For H3K4me3 ChIP-seq data, the processed data files were directly utilized (Beisel et al. 2011). For 5’ short RNA sequence reads, raw sequences were aligned using the RNA-sequence alignment pipeline at http://www.cbil.upenn.edu/RUM/ (G.G, unpublished).
Supplementary Material
Highlights.
Insects mount a rapid antiviral transcriptional response
Genes from many antiviral pathways, including Toll receptors, are rapidly induced
Transcriptional pausing regulates this inducible gene expression program
The transcriptional pausing pathway restricts disparate arboviruses in insects
ACKNOWLEDGEMENTS
We thank R. Doms and S. Ross for critical reading of the manuscript; members of the Cherry lab for helpful discussions; G. Blobel and S. Master for helpful discussions; R. Zhou, G. Hannon and N. Perrimon for the screening library; K. Adelman, D. Gilchrist, D. Gilmour, R. Paro, and the Mod ENCODE project for publically available datasets. This work was supported by grants from the National Institutes of Health. (R01AI074951,U54AI057168) to SC. S.C. is a recipient of the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award. J. Xu is an HHMI International Student Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, Rogatsky I. RNA polymerase II stalling mediates cytokine gene expression. Cell Cycle. 2010;9:630–631. doi: 10.4161/cc.9.4.10841. [DOI] [PubMed] [Google Scholar]
- Akhouayri I, Turc C, Royet J, Charroux B. Toll-8/tollo negatively regulates antimicrobial response in the Drosophila respiratory epithelium. PLoS Pathog. 2011;7:e1002319. doi: 10.1371/journal.ppat.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 2009;5:e1000582. doi: 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Developmental Cell. 2002;3:711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N, Consortium HFA. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Burnham AJ, Gong L, Hardy RW. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology. 2007;367:212–221. doi: 10.1016/j.virol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Busse MS, Arnold CP, Towb P, Katrivesis J, Wasserman SA. A kappaB sequence code for pathway-specific innate immune responses. EMBO J. 2007;26:3826–3835. doi: 10.1038/sj.emboj.7601798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter J, Hutter S, Baines JF, Roller J, Saminadin-Peter SS, Parsch J, Jiggins FM. The transcriptional response of Drosophila melanogaster to infection with the sigma virus (Rhabdoviridae). PLoS One. 2009;4:e6838. doi: 10.1371/journal.pone.0006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S, Silverman N. Host-pathogen interactions in drosophila: new tricks from an old friend. Nat Immunol. 2006;7:911–917. doi: 10.1038/ni1388. [DOI] [PubMed] [Google Scholar]
- Chotkowski HL, Ciota AT, Jia Y, Puig-Basagoiti F, Kramer LD, Shi PY, Glaser RL. West Nile virus infection of Drosophila melanogaster induces a protective RNAi response. Virology. 2008;377:197–206. doi: 10.1016/j.virol.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Jan E, Sarnow P, Schneider D. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS One. 2009;4:e7436. doi: 10.1371/journal.pone.0007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezélée S, Blondel D, Wyers F, Petitjean AM. Vesicular stomatitis virus in Drosophila melanogaster cells: lack of leader RNA transport into the nuclei and frequent abortion of the replication step. J Virol. 1987;61:1391–1397. doi: 10.1128/jvi.61.5.1391-1397.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler JL. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nature Immunology. 2005;6:946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- Enderle D, Beisel C, Stadler MB, Gerstung M, Athri P, Paro R. Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res. 2011;21:216–226. doi: 10.1101/gr.114348.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nature Reviews Immunology. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- Filone CM, Hanna SL, Caino MC, Bambina S, Doms RW, Cherry S. Rift valley fever virus infection of human cells and insect hosts is promoted by protein kinase C epsilon. PLoS One. 2010;5:e15483. doi: 10.1371/journal.pone.0015483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, Chen ZJ. Sorting out Toll signals. Cell. 2006;125:834–836. doi: 10.1016/j.cell.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Ganesan S, Aggarwal K, Paquette N, Silverman N. NF-κB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr Top Microbiol Immunol. 2011;349:25–60. doi: 10.1007/82_2010_107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga J, Xie H, Obradovic Z, Graña X. Selective control of gene expression by CDK9 in human cells. J Cell Physiol. 2010;222:200–208. doi: 10.1002/jcp.21938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Fromm G, dos Santos G, Pham LN, McDaniel IE, Burkholder A, Fargo DC, Adelman K. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev. 2012;26:933–944. doi: 10.1101/gad.187781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee CH, Ghosh SKB, Collins JB, Li LP, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes & Development. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gliedman JB, Smith JF, Brown DT. Morphogenesis of Sindbis virus in cultured Aedes albopictus cells. J Virol. 1975;16:913–926. doi: 10.1128/jvi.16.4.913-926.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of Inducible Gene Expression by Signal-Dependent Transcriptional Elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges LM, Johnson KN. Induction of host defence responses by Drosophila C virus. J Gen Virol. 2008;89:1497–1501. doi: 10.1099/vir.0.83684-0. [DOI] [PubMed] [Google Scholar]
- Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- Hultmark D. Drosophila immunity: paths and patterns. Curr Opin Immunol. 2003;15:12–19. doi: 10.1016/s0952-7915(02)00005-5. [DOI] [PubMed] [Google Scholar]
- Irving P, Troxler L, Heuer TS, Belvin M, Kopczynski C, Reichhart JM, Hoffmann JA, Hetru C. A genome-wide analysis of immune responses in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15119–15124. doi: 10.1073/pnas.261573998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene KM, Foy BD, Sanchez-Vargas I, Beaty BJ, Blair CD, Olson KE. RNA interference acts as a natural antiviral response to O'nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc Natl Acad Sci U S A. 2004;101:17240–17245. doi: 10.1073/pnas.0406983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp C, Imler JL. Antiviral immunity in drosophila. Curr Opin Immunol. 2009;21:3–9. doi: 10.1016/j.coi.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P, Bulet P, Reichhart JM. Critical evaluation of the role of the Toll-like receptor 18-Wheeler in the host defense of Drosophila. EMBO Rep. 2002;3:666–673. doi: 10.1093/embo-reports/kvf130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- Mudiganti U, Hernandez R, Brown DT. Insect response to alphavirus infection--establishment of alphavirus persistence in insect cells involves inhibition of viral polyprotein cleavage. Virus Res. 2010;150:73–84. doi: 10.1016/j.virusres.2010.02.016. [DOI] [PubMed] [Google Scholar]
- Mueller S, Gausson V, Vodovar N, Deddouche S, Troxler L, Perot J, Pfeffer S, Hoffmann JA, Saleh MC, Imler JL. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc Natl Acad Sci U S A. 2010;107:19390–19395. doi: 10.1073/pnas.1014378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, Gold B, Cherry S. Virus Recognition by Toll-7 Activates Antiviral Autophagy in Drosophila. Immunity. 2012 doi: 10.1016/j.immuni.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Wu J, Wu LP. Microarray analyses reveal distinct roles for Rel proteins in the Drosophila immune response. Dev Comp Immunol. 2008;32:50–60. doi: 10.1016/j.dci.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez JL, Dimopoulos G. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev Comp Immunol. 2010;34:625–629. doi: 10.1016/j.dci.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsburg E, Publicover J, Buonocore L, Poholek A, Robek M, Palin A, Rose JK. A vesicular stomatitis virus recombinant expressing granulocyte-macrophage colony-stimulating factor induces enhanced T-cell responses and is highly attenuated for replication in animals. J Virol. 2005;79:15043–15053. doi: 10.1128/JVI.79.24.15043-15053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. Short transcripts of the ternary complex provide insight into RNA polymerase II elongational pausing. J Mol Biol. 1995;252:522–535. doi: 10.1006/jmbi.1995.0517. [DOI] [PubMed] [Google Scholar]
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Rose PP, Hanna SL, Spiridigliozzi A, Wannissorn N, Beiting DP, Ross SR, Hardy RW, Bambina SA, Heise MT, Cherry S. Natural resistance-associated macrophage protein is a cellular receptor for sindbis virus in both insect and Mammalian hosts. Cell Host Microbe. 2011;10:97–104. doi: 10.1016/j.chom.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxström-Lindquist K, Terenius O, Faye I. Parasite-specific immune response in adult Drosophila melanogaster: a genomic study. EMBO Rep. 2004;5:207–212. doi: 10.1038/sj.embor.7400073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin LR, Hanna SL, Cherry S. Innate antiviral immunity in Drosophila. Current Opinion in Immunology. 2010;22:4–9. doi: 10.1016/j.coi.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nature Reviews Molecular Cell Biology. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Shaw G, Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Smale ST. Selective Transcription in Response to an Inflammatory Stimulus. Cell. 2010;140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc Natl Acad Sci U S A. 2009;106:17841–17846. doi: 10.1073/pnas.0905006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Li R. Human negative elongation factor activates transcription and regulates alternative transcription initiation. J Biol Chem. 2010;285:6443–6452. doi: 10.1074/jbc.M109.084285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CW, McGraw EA, Ammar ED, Dietzgen RG, Hogenhout SA. Drosophila melanogaster mounts a unique immune response to the Rhabdovirus sigma virus. Appl Environ Microbiol. 2008;74:3251–3256. doi: 10.1128/AEM.02248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hang S, Prazak L, Gergen JP. NELF potentiates gene transcription in the Drosophila embryo. PLoS One. 2010;5:e11498. doi: 10.1371/journal.pone.0011498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lee C, Gilmour DS, Gergen JP. Transcription elongation controls cell fate specification in the Drosophila embryo. Genes Dev. 2007;21:1031–1036. doi: 10.1101/gad.1521207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman SA. Nature's fortress against infection. Nat Immunol. 2004;5:474–475. doi: 10.1038/ni0504-474. [DOI] [PubMed] [Google Scholar]
- Weaver SC, Barrett AD. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol. 2004;2:789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MJ, Rodriguez A, Kimbrell DA, Eldon ED. The 18-wheeler mutation reveals complex antibacterial gene regulation in Drosophila host defense. EMBO J. 1997;16:6120–6130. doi: 10.1093/emboj/16.20.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes & Development. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto KR, Alberts BM. Steroid receptors: elements for modulation of eukaryotic transcription. Annu Rev Biochem. 1976;45:721–746. doi: 10.1146/annurev.bi.45.070176.003445. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Yamazaki S, Uematsu S, Sato S, Hemmi H, Hoshino K, Kaisho T, Kuwata H, Takeuchi O, Takeshige K, et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. 2004;430:218–222. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- Yu W, Ramakrishnan R, Wang Y, Chiang K, Sung TL, Rice AP. Cyclin T1-dependent genes in activated CD4 T and macrophage cell lines appear enriched in HIV-1 cofactors. PLoS One. 2008;3:e3146. doi: 10.1371/journal.pone.0003146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon RA, Nandakumar M, Vakharia VN, Wu LP. The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci U S A. 2005;102:7257–7262. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon RA, Vakharia VN, Wu LP. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.