Abstract
Gastric cancers arise through a multistep process characterized by the progressive accumulation of molecular alterations in which genetic and epigenetic mechanisms have been implicated. Gastric cancer is one of the human malignancies in which aberrant promoter CpG island hypermethylation is frequently found. Helicobacter pylori and Epstein-Barr virus, which are known carcinogens for gastric cancer, are closely associated with enhanced hypermethylation of CpG island loci in gastric non-neoplastic epithelial cells and cancer cells, respectively. Aberrant CpG island hypermethylation occurs early in the multistep cascade of gastric carcinogenesis and tends to increase with the step-wise progression of the lesion. Approximately 400 genes that are actively expressed in normal gastric epithelial cells are estimated to be inactivated in gastric cancers as a result of promoter CpG island hypermethylation. In this review, a variety of information is summarized regarding CpG island hypermethylation in gastric cancer.
Keywords: CpG island, DNA methylation, Gastric cancer, Intestinal metaplasia
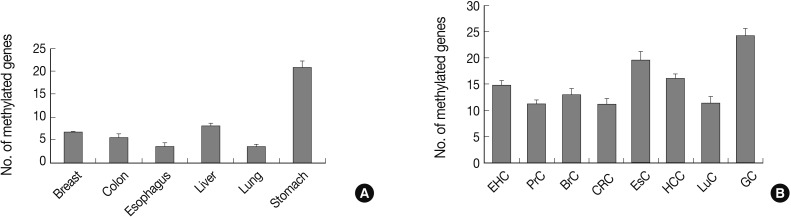
The field of epigenetics describes information transmission through cell division of heritable changes in a phenotype that does not involve DNA sequence changes. CpG island hypermethylation, histone modification, and transmitted chromatin structure are the underlying mechanisms for epigenetic transmission, and CpG island hypermethylation is a key component for altered gene expression associated with human cancers. CpG islands are DNA segments that are at least 0.5 kb in size, rich in G:C and CpG content, and found in approximately 70% of human gene promoters.1 Promoter CpG islands are usually unmethylated in normal cells, with the exception of those on an inactive X chromosome or associated with imprinted genes. Although the cause is unclear, promoter CpG island hypermethylation can occur in association with cancer development and aging. Promoter CpG island hypermethylation is found in virtually all human cancer tissue types and acts as an important mechanism for the inactivation of tumor suppressor genes and tumor-related genes.2,3 Gastric cancer is one of the human cancers in which promoter CpG island hypermethylation is frequently found.2,4 In our preliminary study, which analyzed 41 candidate genes in major types of human cancers and cancer-associated normal tissues, gastric cancer demonstrated a significantly higher number of methylated genes than those of other human cancer tissue types (Fig. 1). In addition, gastric cancer-associated normal mucosa also exhibited the highest number of methylated genes when compared to normal tissue from other organs, including the lung, breast, colon, and liver (Fig. 1). These facts suggest the possibility that aberrant CpG island hypermethylation is more involved in the carcinogenesis of gastric cancer than in that of other human cancer tissue types.
Fig. 1.
Bar graphs display the number of methylated genes in cancer-associated normal tissue (A) and cancer tissue (B). The error bars indicate the standard error of the mean. Forty-one genes are analyzed for their methylation status in tissue samples from eight human cancer tissue types and six types of cancer-associated normal tissue using the MethyLight assay. EHC, extrahepatic bile duct cancer; PrC, prostate adenocarcinoma; BrC, breast adenocarcinoma; CRC, colorectal adenocarcinoma; EsC, esophageal adenocarcinoma; HCC, hepatocellular carcinoma; LuC, lung adenocarcinoma; GC, gastric adenocarcinoma.
GASTRIC CANCER METHYLATION CHANGE
How many genes are known to be methylated in gastric cancer?
In 1996, Lee et al.5 reported for the first time that p16 was inactivated in gastric cancer by promoter CpG island hypermethylation rather than by genetic mutation. Since then, many researchers have demonstrated cancer-specific hypermethylation and inactivation of candidate genes in gastric cancer tissues and have correlated this hypermethylation with clinicopathologic features. However, the number of genes demonstrated to be inactivated by promoter CpG island hypermethylation was limited until the application of array-based genome-scale DNA methylation analysis for gastric cancers. With the application of 5-aza-2'-deoxycytidine (DAC) treatment and oligonucleotide microarrays, Yamashita et al.6 estimated that approximately 421 genes are silenced by promoter CpG island hypermethylation in a gastric cancer cell line (AGS). Considering the fact that promoter CpG island hypermethylation is exaggerated in a cancer cell line compared with primary cancer tissue and that the AGS cell line has an increased rate of de novo methylation because of over-expression of DNMT3b, the actual number of genes silenced by promoter CpG island hypermethylation can be estimated to be less than 421 in primary gastric cancers. Our team also performed bead array-based expression analysis of gastric cancer cell lines before and after DAC treatment with subsequent confirmation of CpG island hypermethylation of the candidate genes by methylation-specific PCR. We found 140 novel genes that are silenced by promoter CpG island hypermethylation in primary gastric cancer tissue (Jung et al. in preparation).
The timing of hypermethylation in multistep gastric carcinogenesis
Promoter CpG island hypermethylation is now recognized to be an important mechanism responsible for the inactivation of tumor suppressor genes or tumor-related genes. If promoter CpG island hypermethylation of some genes plays an important role in the malignant transformation of gastric epithelial cells, this pattern of hypermethylation should be found in premalignant lesions of the stomach, including gastric adenomas and intestinal metaplasia. In order to determine the frequency and timing of hypermethylation during multistep gastric carcinogenesis, Kang et al. analyzed multistep lesions of the stomach, in regards to their methylation status, in five genes7 or 12 genes8 using methylation-specific PCR or in 25 genes using MethyLight analysis;9 they demonstrated that promoter CpG island hypermethylation occurs early in multistep gastric carcinogenesis and accumulates during progression of the gastric lesion along the multistep carcinogenesis pathway. During multistep gastric carcinogenesis, there is a steep rise in the number of methylated genes when progressing from chronic gastritis to intestinal metaplasia, which was a consistent finding in a series of studies.7-9 Regardless of the status of Helicobacter pylori infection, the number of methylated genes in intestinal metaplasia was significantly higher than that found in chronic gastritis without intestinal metaplasia.9 This suggests that intestinal metaplasia is an epigenetically altered lesion. However, even in chronic gastritis without intestinal metaplasia, promoter CpG island hypermethylation occurs in association with H. pylori infection10 and aging.11,12
Helicobacter pylori infection-associated DNA hypermethylation
H. pylori has been designated as a human class I carcinogen for gastric malignancy by the International Agency for Research on Cancer. Although the exact mechanism of H. pylori-associated gastric carcinogenesis is unknown, long-standing bacterial infection, perpetuated chronic inflammation, and sustained mucosal epithelial cell proliferation are thought to produce a carcinogenic environment. Chan et al.10 were the first to demonstrate H. pylori-associated hypermethylation in the gastric epithelia, which was supported by subsequent studies demonstrating that the eradication of H. pylori infection results in a reversal of the methylation status of multiple CpG island loci.13-15 Thus, it is plausible that aberrant methylation induced by H. pylori infection may contribute to H. pylori infection-associated gastric carcinogenesis. It has been reported that interleukin 1 beta can modulate CpG island methylation through the activation of DNA methyltransferase.16 In an in vitro study, interleukin 1 beta siRNA blocked H. pylori-induced methylation of the CDH1 promoter CpG island locus in a gastric cancer cell line.17 In an animal model experiment by the Ushijima team, H. pylori infection resulted in the induction of CpG island hypermethylation of candidate genes, and the eradication led to marked decreases in methylation levels in the candidate genes. However, the suppression of inflammation by treatment with the immunosuppressive drug cyclosporine blocked the induction of DNA methylation in the candidate genes. These findings suggest that the infection-associated inflammatory response, rather than H. pylori itself, was responsible for the induction of altered DNA methylation.18 In a subsequent gerbil study, neutrophilic inflammation caused by treatment with ethanol or NaCl did not induce DNA methylation in candidate genes, whereas chronic inflammation caused by H. pylori or H. felis infection led to altered methylation in candidate genes. This finding suggests that it is not the inflammation itself, but rather specific types of inflammation, that are necessary for methylation induction.19
Aging-related hypermethylation vs inflammation-related hypermethylation
Challenging traditional thought regarding the lack of CpG island methylation in normal tissues, a recent study indicated that 4-8% of CpG island loci are methylated in the genomic DNA of human blood, brain, muscle, and spleen tissue.20,21 Additionally, normal cells have been shown to acquire hypermethylation in an aging-related manner: aging-related methylation was first demonstrated for the oestrogen receptor (ER) gene by Issa et al.22 and has subsequently been demonstrated in multiple genes by Ahuja et al.23 In the stomach, Waki et al.11 reported aging-related methylation of CDH1, MLH1, and p16 in non-neoplastic gastric epithelia. However, because CpG island hypermethylation can be induced by chronic inflammation in the stomach and because the prevalence of H. pylori infection increases with age,24 the interplay between aging and chronic inflammation is complicated by H. pylori infection. Chan et al.25 found that CDH1 methylation was associated with age in the stomach, the presence of chronic gastritis, and H. pylori infection using a univariate analysis, but H. pylori infection was the only independent factor associated with CDH1 methylation in the multivariate analysis. Furthermore, Maekita et al.26 reported that there was no aging-related hypermethylation in healthy gastric mucosa. However, a current study by our team supports the presence of aging-related hypermethylation in the gastric mucosa because the number of methylated genes was significantly higher in H. pylori-negative adult stomach samples than in H. pylori-negative pediatric samples.27 In WI-38 human embryonic lung fibroblasts, p16 was found to undergo spontaneous promoter CpG island hypermethylation during passage of these cells in culture,28 which supports the presence of age-related hypermethylation. Thus, we cannot exclude the possibility that aging-related methylation occurs in non-neoplastic gastric epithelia without H. pylori infection.
Field cancerization
Many studies have shown that methylation levels or frequencies of multiple genes are higher in gastric mucosa from gastric cancer patients than in mucosa from non-cancer subjects.11,26,29 In a stomach with enhanced CpG island hypermethylation, the affected cells may have a growth-selective advantage imparted by the expressional loss of the methylated genes, which may predispose the cells to acquiring further genetic or epigenetic defects that lead to neoplasia. Synchronous multiple gastric cancers, constituting 4-9% of gastric cancers, occur at an older age and are more commonly associated with an extensive distribution of intestinal metaplasia in the background mucosa compared with single gastric cancer.30,31 In a recent study, non-neoplastic gastric mucosa from synchronous gastric cancer patients was found to have more hypermethylated genes than non-neoplastic gastric mucosa from patients with a single gastric cancer, which suggests that enhanced hypermethylation in the background gastric mucosa might contribute to the development of multiple gastric cancers.32 Multiple gastric cancers are not only a genetic model but also an epigenetic model for field cancerization. Based on the findings that methylation levels in gastric mucosa are significantly increased in cases with a single gastric cancer and even more so in cases with multiple gastric cancers, it has been suggested that quantitative information regarding methylation levels of specific genes may serve as a biomarker for predicting an individual's risk for developing gastric cancer.32,33 However, it should be noted that increased methylation levels or frequencies in gastric mucosa from gastric cancer patients may reflect a higher prevalence of H. pylori infection and intestinal metaplasia (compared with gastric mucosa from non-cancer subjects).34,35 Because both H. pylori infection and intestinal metaplasia are closely linked with increased CpG island hypermethylation, further clarification is needed regarding whether non-neoplastic gastric mucosa without intestinal metaplasia from H. pylori-negative gastric cancer patients harbors more hypermethylated genes than non-neoplastic gastric mucosa from H. pylori-negative non-cancer subjects. In Park et al.'s study,9 no significant differences were observed in the number of methylated genes or methylation levels of 25 individual genes in chronic gastritis tissue from cancer and non-cancer patients after normalization of the confounding factors of H. pylori and intestinal metaplasia. Because synchronous multiple gastric cancers tend to be associated with widespread intestinal metaplasia when compared with single gastric cancers, cancer-associated gastric mucosa might show higher methylation frequencies or higher methylation levels of multiple CpG island loci in multiple synchronous gastric cancer patients than in single gastric cancer patients.
Prognostic implications of individual gene methylation
Promoter CpG island hypermethylation can be utilized as a tumor biomarker for the detection of tumor cells in gastric juice or serum or for the prediction of clinical outcomes. A dozen DNA methylation markers have been reported to be closely associated with worse or better clinical outcomes in gastric cancer patients; MAL or COX2 methylation was correlated with better clinical outcomes in gastric cancer patients,36,37 whereas DAPK, TMS1, IQGAP2, SOX2, CACNA2D3, DKK-3, TFPI2, and Cystatin methylation has been correlated with worse clinical outcomes in gastric cancer patients.38-44 However, most of the studies that have evaluated these DNA methylation markers for their prognostic implication used methylation-specific polymerase chain reaction (MSP). Although MSP is a highly sensitive method for detecting one methylated allele in 10,000 unmethylated alleles,45 it is unreliable for detecting low levels of methylation. Another issue related to these DNA methylation markers is that a validation study was not performed to prove their utility as a prognostic marker. The final issue concerns concordance among individual gene hypermethylation: hypermethylation of one specific gene tends to be concordant with that of another individual gene; thus, better or worse survival observed in gastric cancers with hypermethylation of an individual gene may not be attributed to hypermethylation of that particular gene. Rather, survival may be related to concordant hypermethylation of multiple CpG island loci, namely CpG island methylator phenotype (CIMP).
CIMP-positive gastric cancer
CIMP refers to a subset of malignancies that is characterized by widespread hypermethylation of multiple promoter CpG island loci. Since the CIMP concept was first introduced for the molecular pathways of colorectal cancers (CRCs) by Dr. Issa's group,46 many investigators have attempted to characterize the clinicopathological and molecular features of CIMP-positive CRCs and have found a close association with proximal colon location, older age at onset, poor differentiation, microsatellite instability (MSI), and BRAF mutations.47-49 Similarly, the presence of CIMP-positive gastric cancers has been reported by the same group,50 which did not find distinct clinicopathologic features of CIMP-positive gastric cancers, with the exception of a close association with MSI. Since then, several researchers have attempted to characterize the clinicopathological features of CIMP-positive gastric cancers using variable methodologies for DNA methylation analysis and their own CIMP marker panels; this has led to controversial results.51-56 Despite variable findings, relatively common findings include close associations of CIMP-positive gastric cancers with diffuse type-histology and better clinical outcomes. Recently, with the exclusion of MSI-positive gastric cancers and Epstein-Barr virus (EBV)-positive gastric cancers from the analysis, Park et al.57 found characteristic clinicopathologic features of CIMP-positive gastric cancers that were defined as tumors with methylation of 13 or more markers during the analysis of the 16 cancer-specific DNA methylation markers; these defined CIMP-positive gastric cancers tended to show distinct clinicopathologic features, including a proclivity toward diffuse or mixed-type histology, poor differentiation, infiltrative gross types, and higher cancer stages.
EBV-associated gastric cancer
In addition to H. pylori, EBV has also been recognized as a gastric cancer-causing infectious agent.58,59 EBV-associated gastric cancer, comprising nearly 10% of gastric cancers, are characterized by a younger age at onset, male predominance, proximal location, frequent association of lymphoid stroma, and a better prognosis.60 EBV-associated gastric cancer is a prototype of the CIMP-positive types of gastric cancer and exhibits consistently higher frequencies and levels of methylation in examined cancer-related methylation markers,53,57,61 which is independent of whether the EBV-associated gastric cancer is histologically lymphoepithelioma-like or ordinary.60 Because the number of methylated cancer-specific methylation markers is far higher in EBV-associated gastric cancers than in EBV-negative gastric cancers, EBV-associated aberrant hypermethylation is a global event. However, MLH1 methylation and resultant MSI are, if ever, rarely observed in EBV-associated gastric cancers. Although it can be speculated that the methylation machinery of EBV-associated gastric cancer is capable of recognizing its own methylation targets, it may be hypothesized that the presence of both CIMP and MSI could cause a growth disadvantage and subsequently lead to the negative selection of gastric cancer cells containing both CIMP and MSI. Tumor cells from EBV-associated gastric cancers display DNA methyltransferase I over-expression,62 which is closely associated with interleukin-1-beta (IL1B) over-expression60 or latent membrane protein 2A-associated phosphorylation of signal transducer and activator of transcription 3 (STAT3) in EBV-associated gastric cancers.63 IL1B is capable of increasing the expression of DNMT1 via the production of nitric oxide, and phosphorylated STAT3 binds to the DNMT1 promoter and induces transcription.
MSI-positive gastric cancer
The frequency of MSI-positive gastric cancer varies from 8% to 37%. In gastric cancer, MSI is mainly caused by promoter CpG island hypermethylation of the MLH1 gene. Somatic mutations of mismatch repair genes are very rare in sporadic gastric cancers. Thus, known clinicopathological features of MSI-positive gastric cancer, including female sex, older age of onset, antral location, ulcerofungating gross type by Borrmann's classification, intestinal type by Lauren classification, expanding type by Ming classification, and better survival64 represent features of sporadic MSI-positive gastric cancer. Since gastric cancer is an extracolonic lesion in Lynch syndrome, we have encountered MSI-positive gastric cancer without MLH1 methylation. Recently, our team has compared clinicopathological features between MSI-positive gastric cancers with and without MLH1 methylation (Kim et al. in preparation). Of the known clinicopathological features for MSI-positive gastric cancer, female preponderance, older age of onset and antral location do not correspond with Lynch syndrome-associated MSI-positive gastric cancer.65 Several studies have shown that MLH1 methylation occurs in premalignant stages, including intestinal metaplasia and gastric adenoma. Thus, MLH1 methylation has been considered an early event during multistep gastric carcinogenesis.7,9,66,67 However, a recent study of Ling et al.68 has shown that MSI can develop from MSI-low or the absence of MSI due to time-dependent accumulation of DNA methylation during progression of early stage-gastric cancer, which indicates that silencing of MLH1 due to promoter hypermethylation may appear as a later event during multistep gastric carcinogenesis. Unfortunately, the study did not investigate whether later acquisition of MLH1 methylation occurs as a phenomenon of CIMP. MLH1 methylation is unlikely to occur as an isolated sporadic event without concurrent hypermethylation of multiple gene promoter CpG island loci.
Epigenetic regulation of microRNA
A new class of small non-coding RNAs known as microRNAs (miRNA) have recently been discovered. Mature miRNAs, 21 to 30 nucleotide-sized, are cleaved from 70- to 100-nucleotide hairpin miRNA precursors in the cytoplasm by the RNase III enzyme Dicer. Base-pairing between the miRNA strand and the 3' untranslated regions of its target mRNAs (potentially hundreds of genes) directs RNA-induced silencing complex to either cause mRNA degradation or translational repression. Through these molecular mechanisms, miRNAs serve as key regulators of gene expression involved in crucial cellular processes, including development, proliferation, cellular differentiation, and apoptosis. Although the generation of miRNAs and their mode of action in regulating gene function have been intensively studied, the regulation of miRNA expression remains largely unclear. The means by which miRNA is regulated is somewhat complicated. Recently, Saito et al. demonstrated for the first time that miR-127 is regulated by DNA hypermethylation69 and since then, a dramatically increased number of studies have documented the epigenetic regulation of miRNAs.70 However, in gastric cancer, approximately 113 miRNAs have been demonstrated to be dysregulated compared with normal gastric mucosal tissues but a dozen miRNA genes have been shown to be downregulated by aberrant hypermethylation, including miR-512-5p,71 miR-375,72 miR-212,73 miR-181c,73 miR-196b,74 miR-137,75 miR-129-2,76 miR-124a-1, miR-124a-2, miR-124a-3,77 miR-34b, and miR-34c.78 In particular, Ando et al.77 demonstrated that miR-124a-1, -2, and -3 are frequently methylated in primary gastric cancer and in normal gastric mucosal tissues from healthy individuals with H. pylori infections. Among H. pylori-negative individuals, methylation levels are significantly higher in non-cancerous gastric mucosal tissues from gastric cancer patients than gastric mucosal tissues from healthy individuals, which suggest that methylation of miRNA genes comprises a field defect contributing to the pathogenesis of gastric cancer.77 Suzuki et al.78 also reported that miR-34b/c methylation is significantly associated with H. pylori infection among healthy individuals.
Future perspective
Although it is well known that EBV-positive gastric cancer is featured with extensive hypermethylation of multiple genes, the mechanism leading to genome-wide hypermethylation is still unknown. Because EBV-positive gastric dysplasia or adenoma has never been reported, it seems likely that EBV-infected epithelial cells transform directly into malignant cells. DNMT1 elevation in association with EBV infection does not provide a satisfactory explanation for genome-wide extensive hypermethylation because in vitro transfection of DNMT1 in cell lines or DNMT1 elevation in association with H. pylori infection also does not lead to such extensive hypermethylation as seen in EBV-positive gastric cancer. If we can elucidate the mechanism of EBV-associated extensive hypermethylation, we will have a better understanding of how promoter CpG island hypermethylation occurs.
Although gastric cancer has shown a higher number of genes methylated compared with colorectal cancer, clinicopathological features of CIMP-positive gastric cancer are still obscure and marker panels diagnosing CIMP-positive gastric cancer are not established yet, which is in contrast to the situation in colorectal cancer. It is imperative to develop CIMP panel markers enabling the diagnosis of CIMP-positive gastric cancer and then to characterize clinicopathological features of CIMP-positive gastric cancer. In case these are accomplished, we expect to identify the precursor lesions of CIMP-positive gastric cancer and to delineate multistep morphological progression of CIMP-positive tumors.
CONCLUSION
Although molecular pathways and morphological pathways are not well established for gastric cancers when compared with colorectal cancers, rapid development of methylation analysis technology will enable us to take a glimpse at the landscape of epigenetic alterations occurring at each step of multistep gastric carcinogenesis, which will provide molecular insights on morphological progression pathways. In addition, epigenetic studies may offer great potential for the identification of tumor biomarkers that can be utilized to detect and diagnose gastric cancer at its earliest stages, to accurately assess an individual's risk for gastric cancer, or to predict the response to chemotherapy or clinical outcomes.
Acknowledgments
Supported by grant M10750030001-08N5003-00110 from the Korea Science and Engineering Foundation, funded by the Ministry of Education, Science and Technology (MEST); by grant 2010-0007579 from the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the MEST; and by grant 2009-0093820 from a Priority Research Centers Program through the NRF. Nam-Yun Cho contributed to this work through experiments.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 3.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 4.Park SY, Kim BH, Kim JH, et al. Methylation profiles of CpG island loci in major types of human cancers. J Korean Med Sci. 2007;22:311–317. doi: 10.3346/jkms.2007.22.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YY, Kang SH, Seo JY, et al. Alterations of p16INK4A and p15INK4B genes in gastric carcinomas. Cancer. 1997;80:1889–1896. doi: 10.1002/(sici)1097-0142(19971115)80:10<1889::aid-cncr3>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 6.Yamashita S, Tsujino Y, Moriguchi K, Tatematsu M, Ushijima T. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2'-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci. 2006;97:64–71. doi: 10.1111/j.1349-7006.2006.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang GH, Shim YH, Jung HY, Kim WH, Ro JY, Rhyu MG. CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res. 2001;61:2847–2851. [PubMed] [Google Scholar]
- 8.Kang GH, Lee S, Kim JS, Jung HY. Profile of aberrant CpG island methylation along multistep gastric carcinogenesis. Lab Invest. 2003;83:519–526. doi: 10.1097/01.lab.0000064704.53132.65. [DOI] [PubMed] [Google Scholar]
- 9.Park SY, Yoo EJ, Cho NY, Kim N, Kang GH. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol. 2009;219:410–416. doi: 10.1002/path.2596. [DOI] [PubMed] [Google Scholar]
- 10.Chan AO, Lam SK, Wong BC, et al. Promoter methylation of E-cadherin gene in gastric mucosa associated with Helicobacter pylori infection and in gastric cancer. Gut. 2003;52:502–506. doi: 10.1136/gut.52.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waki T, Tamura G, Tsuchiya T, Sato K, Nishizuka S, Motoyama T. Promoter methylation status of E-cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am J Pathol. 2002;161:399–403. doi: 10.1016/S0002-9440(10)64195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang GH, Lee HJ, Hwang KS, Lee S, Kim JH, Kim JS. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol. 2003;163:1551–1556. doi: 10.1016/S0002-9440(10)63511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan AO, Peng JZ, Lam SK, et al. Eradication of Helicobacter pylori infection reverses E-cadherin promoter hypermethylation. Gut. 2006;55:463–468. doi: 10.1136/gut.2005.077776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung WK, Man EP, Yu J, et al. Effects of Helicobacter pylori eradication on methylation status of E-cadherin gene in noncancerous stomach. Clin Cancer Res. 2006;12:3216–3221. doi: 10.1158/1078-0432.CCR-05-2442. [DOI] [PubMed] [Google Scholar]
- 15.Ushijima T, Nakajima T, Maekita T. DNA methylation as a marker for the past and future. J Gastroenterol. 2006;41:401–407. doi: 10.1007/s00535-006-1846-6. [DOI] [PubMed] [Google Scholar]
- 16.Hmadcha A, Bedoya FJ, Sobrino F, Pintado E. Methylation-dependent gene silencing induced by interleukin 1beta via nitric oxide production. J Exp Med. 1999;190:1595–1604. doi: 10.1084/jem.190.11.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian X, Huang C, Cho CH, Hui WM, Rashid A, Chan AO. E-cadherin promoter hypermethylation induced by interleukin-1beta treatment or H. pylori infection in human gastric cancer cell lines. Cancer Lett. 2008;263:107–113. doi: 10.1016/j.canlet.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 18.Niwa T, Tsukamoto T, Toyoda T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430–1440. doi: 10.1158/0008-5472.CAN-09-2755. [DOI] [PubMed] [Google Scholar]
- 19.Hur K, Niwa T, Toyoda T, et al. Insufficient role of cell proliferation in aberrant DNA methylation induction and involvement of specific types of inflammation. Carcinogenesis. 2011;32:35–41. doi: 10.1093/carcin/bgq219. [DOI] [PubMed] [Google Scholar]
- 20.Illingworth R, Kerr A, Desousa D, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen L, Kondo Y, Guo Y, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 23.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- 24.Drumm B, Perez-Perez GI, Blaser MJ, Sherman PM. Intrafamilial clustering of Helicobacter pylori infection. N Engl J Med. 1990;322:359–363. doi: 10.1056/NEJM199002083220603. [DOI] [PubMed] [Google Scholar]
- 25.Chan AO, Lam SK, Wong BC, Kwong YL, Rashid A. Gene methylation in non-neoplastic mucosa of gastric cancer: age or Helicobacter pylori related? Am J Pathol. 2003;163:370–371. doi: 10.1016/s0002-9440(10)63663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maekita T, Nakazawa K, Mihara M, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12(3 Pt 1):989–995. doi: 10.1158/1078-0432.CCR-05-2096. [DOI] [PubMed] [Google Scholar]
- 27.Shin SH, Park SY, Ko JS, Kim N, Kang GH. Aberrant CpG island hypermethylation in pediatric gastric mucosa in association with Helicobacter pylori infection. Arch Pathol Lab Med. 2011;135:759–765. doi: 10.5858/2010-0140-OA.1. [DOI] [PubMed] [Google Scholar]
- 28.Barzily-Rokni M, Friedman N, Ron-Bigger S, Isaac S, Michlin D, Eden A. Synergism between DNA methylation and macroH2A1 occupancy in epigenetic silencing of the tumor suppressor gene p16(CDKN2A) Nucleic Acids Res. 2011;39:1326–1335. doi: 10.1093/nar/gkq994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung WK, Yu J, Ng EK, et al. Concurrent hypermethylation of multiple tumor-related genes in gastric carcinoma and adjacent normal tissues. Cancer. 2001;91:2294–2301. [PubMed] [Google Scholar]
- 30.Honmyo U, Misumi A, Murakami A, Haga Y, Akagi M. Clinicopathological analysis of synchronous multiple gastric carcinoma. Eur J Surg Oncol. 1989;15:316–321. [PubMed] [Google Scholar]
- 31.Marrano D, Viti G, Grigioni W, Marra A. Synchronous and metachronous cancer of the stomach. Eur J Surg Oncol. 1987;13:493–498. [PubMed] [Google Scholar]
- 32.Nakajima T, Maekita T, Oda I, et al. Higher methylation levels in gastric mucosae significantly correlate with higher risk of gastric cancers. Cancer Epidemiol Biomarkers Prev. 2006;15:2317–2321. doi: 10.1158/1055-9965.EPI-06-0436. [DOI] [PubMed] [Google Scholar]
- 33.Kaise M, Yamasaki T, Yonezawa J, Miwa J, Ohta Y, Tajiri H. CpG island hypermethylation of tumor-suppressor genes in H. pylori-infected non-neoplastic gastric mucosa is linked with gastric cancer risk. Helicobacter. 2008;13:35–41. doi: 10.1111/j.1523-5378.2008.00572.x. [DOI] [PubMed] [Google Scholar]
- 34.Stemmermann GN. Intestinal metaplasia of the stomach: a status report. Cancer. 1994;74:556–564. doi: 10.1002/1097-0142(19940715)74:2<556::aid-cncr2820740205>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 35.Shimoyama T, Fukuda S, Tanaka M, Nakaji S, Munakata A. Evaluation of the applicability of the gastric carcinoma risk index for intestinal type cancer in Japanese patients infected with Helicobacter pylori. Virchows Arch. 2000;436:585–587. doi: 10.1007/s004289900179. [DOI] [PubMed] [Google Scholar]
- 36.Buffart TE, Overmeer RM, Steenbergen RD, et al. MAL promoter hypermethylation as a novel prognostic marker in gastric cancer. Br J Cancer. 2008;99:1802–1807. doi: 10.1038/sj.bjc.6604777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Maat MF, van de Velde CJ, Umetani N, et al. Epigenetic silencing of cyclooxygenase-2 affects clinical outcome in gastric cancer. J Clin Oncol. 2007;25:4887–4894. doi: 10.1200/JCO.2006.09.8921. [DOI] [PubMed] [Google Scholar]
- 38.Kato K, Iida S, Uetake H, et al. Methylated TMS1 and DAPK genes predict prognosis and response to chemotherapy in gastric cancer. Int J Cancer. 2008;122:603–608. doi: 10.1002/ijc.23143. [DOI] [PubMed] [Google Scholar]
- 39.Jin SH, Akiyama Y, Fukamachi H, Yanagihara K, Akashi T, Yuasa Y. IQGAP2 inactivation through aberrant promoter methylation and promotion of invasion in gastric cancer cells. Int J Cancer. 2008;122:1040–1046. doi: 10.1002/ijc.23181. [DOI] [PubMed] [Google Scholar]
- 40.Otsubo T, Akiyama Y, Yanagihara K, Yuasa Y. SOX2 is frequently downregulated in gastric cancers and inhibits cell growth through cell-cycle arrest and apoptosis. Br J Cancer. 2008;98:824–831. doi: 10.1038/sj.bjc.6604193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wanajo A, Sasaki A, Nagasaki H, et al. Methylation of the calcium channel-related gene, CACNA2D3, is frequent and a poor prognostic factor in gastric cancer. Gastroenterology. 2008;135:580–590. doi: 10.1053/j.gastro.2008.05.041. [DOI] [PubMed] [Google Scholar]
- 42.Yu J, Tao Q, Cheng YY, et al. Promoter methylation of the Wnt/beta-catenin signaling antagonist Dkk-3 is associated with poor survival in gastric cancer. Cancer. 2009;115:49–60. doi: 10.1002/cncr.23989. [DOI] [PubMed] [Google Scholar]
- 43.Jee CD, Kim MA, Jung EJ, Kim J, Kim WH. Identification of genes epigenetically silenced by CpG methylation in human gastric carcinoma. Eur J Cancer. 2009;45:1282–1293. doi: 10.1016/j.ejca.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Cao X, Dong W, et al. Cystatin M expression is reduced in gastric carcinoma and is associated with promoter hypermethylation. Biochem Biophys Res Commun. 2010;391:1070–1074. doi: 10.1016/j.bbrc.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 45.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 48.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10:13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim JH, Shin SH, Kwon HJ, Cho NY, Kang GH. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch. 2009;455:485–494. doi: 10.1007/s00428-009-0857-0. [DOI] [PubMed] [Google Scholar]
- 50.Toyota M, Ahuja N, Suzuki H, et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438–5442. [PubMed] [Google Scholar]
- 51.Enomoto S, Maekita T, Tsukamoto T, et al. Lack of association between CpG island methylator phenotype in human gastric cancers and methylation in their background non-cancerous gastric mucosae. Cancer Sci. 2007;98:1853–1861. doi: 10.1111/j.1349-7006.2007.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang MS, Uozaki H, Chong JM, et al. CpG island methylation status in gastric carcinoma with and without infection of Epstein-Barr virus. Clin Cancer Res. 2006;12:2995–3002. doi: 10.1158/1078-0432.CCR-05-1601. [DOI] [PubMed] [Google Scholar]
- 53.Kang GH, Lee S, Kim WH, et al. Epstein-barr virus-positive gastric carcinoma demonstrates frequent aberrant methylation of multiple genes and constitutes CpG island methylator phenotype-positive gastric carcinoma. Am J Pathol. 2002;160:787–794. doi: 10.1016/S0002-9440(10)64901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kusano M, Toyota M, Suzuki H, et al. Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein-Barr virus. Cancer. 2006;106:1467–1479. doi: 10.1002/cncr.21789. [DOI] [PubMed] [Google Scholar]
- 55.Oue N, Oshimo Y, Nakayama H, et al. DNA methylation of multiple genes in gastric carcinoma: association with histological type and CpG island methylator phenotype. Cancer Sci. 2003;94:901–905. doi: 10.1111/j.1349-7006.2003.tb01373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.An C, Choi IS, Yao JC, et al. Prognostic significance of CpG island methylator phenotype and microsatellite instability in gastric carcinoma. Clin Cancer Res. 2005;11(2 Pt 1):656–663. [PubMed] [Google Scholar]
- 57.Park SY, Kook MC, Kim YW, et al. CpG island hypermethylator phenotype in gastric carcinoma and its clinicopathological features. Virchows Arch. 2010;457:415–422. doi: 10.1007/s00428-010-0962-0. [DOI] [PubMed] [Google Scholar]
- 58.Imai S, Koizumi S, Sugiura M, et al. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc Natl Acad Sci U S A. 1994;91:9131–9135. doi: 10.1073/pnas.91.19.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fukayama M, Hayashi Y, Iwasaki Y, et al. Epstein-Barr virus-associated gastric carcinoma and Epstein-Barr virus infection of the stomach. Lab Invest. 1994;71:73–81. [PubMed] [Google Scholar]
- 60.Fukayama M, Hino R, Uozaki H. Epstein-Barr virus and gastric carcinoma: virus-host interactions leading to carcinoma. Cancer Sci. 2008;99:1726–1733. doi: 10.1111/j.1349-7006.2008.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang GH, Lee S, Cho NY, et al. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab Invest. 2008;88:161–170. doi: 10.1038/labinvest.3700707. [DOI] [PubMed] [Google Scholar]
- 62.Etoh T, Kanai Y, Ushijima S, et al. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am J Pathol. 2004;164:689–699. doi: 10.1016/S0002-9440(10)63156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hino R, Uozaki H, Murakami N, et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69:2766–2774. doi: 10.1158/0008-5472.CAN-08-3070. [DOI] [PubMed] [Google Scholar]
- 64.Kim H, An JY, Noh SH, Shin SK, Lee YC, Kim H. High microsatellite instability predicts good prognosis in intestinal-type gastric cancers. J Gastroenterol Hepatol. 2011;26:585–592. doi: 10.1111/j.1440-1746.2010.06487.x. [DOI] [PubMed] [Google Scholar]
- 65.Gylling A, Abdel-Rahman WM, Juhola M, et al. Is gastric cancer part of the tumour spectrum of hereditary non-polyposis colorectal cancer? A molecular genetic study. Gut. 2007;56:926–933. doi: 10.1136/gut.2006.114876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kang GH, Lee S, Kim JS, Jung HY. Profile of aberrant CpG island methylation along the multistep pathway of gastric carcinogenesis. Lab Invest. 2003;83:635–641. doi: 10.1097/01.lab.0000067481.08984.3f. [DOI] [PubMed] [Google Scholar]
- 67.Lee JH, Park SJ, Abraham SC, et al. Frequent CpG island methylation in precursor lesions and early gastric adenocarcinomas. Oncogene. 2004;23:4646–4654. doi: 10.1038/sj.onc.1207588. [DOI] [PubMed] [Google Scholar]
- 68.Ling ZQ, Tanaka A, Li P, et al. Microsatellite instability with promoter methylation and silencing of hMLH1 can regionally occur during progression of gastric carcinoma. Cancer Lett. 2010;297:244–251. doi: 10.1016/j.canlet.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 69.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 70.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011;278:1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 71.Saito Y, Suzuki H, Tsugawa H, et al. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene. 2009;28:2738–2744. doi: 10.1038/onc.2009.140. [DOI] [PubMed] [Google Scholar]
- 72.Tsukamoto Y, Nakada C, Noguchi T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 73.Wada R, Akiyama Y, Hashimoto Y, Fukamachi H, Yuasa Y. miR-212 is downregulated and suppresses methyl-CpG-binding protein MeCP2 in human gastric cancer. Int J Cancer. 2010;127:1106–1114. doi: 10.1002/ijc.25126. [DOI] [PubMed] [Google Scholar]
- 74.Tsai KW, Hu LY, Wu CW, et al. Epigenetic regulation of miR-196b expression in gastric cancer. Genes Chromosomes Cancer. 2010;49:969–980. doi: 10.1002/gcc.20804. [DOI] [PubMed] [Google Scholar]
- 75.Chen Q, Chen X, Zhang M, Fan Q, Luo S, Cao X. miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of Cdc42. Dig Dis Sci. 2011;56:2009–2016. doi: 10.1007/s10620-010-1536-3. [DOI] [PubMed] [Google Scholar]
- 76.Shen R, Pan S, Qi S, Lin X, Cheng S. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem Biophys Res Commun. 2010;394:1047–1052. doi: 10.1016/j.bbrc.2010.03.121. [DOI] [PubMed] [Google Scholar]
- 77.Ando T, Yoshida T, Enomoto S, et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124:2367–2374. doi: 10.1002/ijc.24219. [DOI] [PubMed] [Google Scholar]
- 78.Suzuki H, Yamamoto E, Nojima M, et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis. 2010;31:2066–2073. doi: 10.1093/carcin/bgq203. [DOI] [PubMed] [Google Scholar]