Abstract
Flexibility in biological systems is seen as an important driver of macro-ecosystem function and stability. Spatially constrained endosymbiotic settings, however, are less studied, although environmental thresholds of symbiotic corals are linked to the function of their endosymbiotic dinoflagellate communities. Symbiotic flexibility is a hypothesized mechanism that corals may exploit to adapt to climate change. This study explores the flexibility of the coral–Symbiodinium symbiosis through quantification of Symbiodinium ITS2 sequence assemblages in a range of coral species and genera. Sequence assemblages are expressed as an index of flexibility incorporating phylogenetic divergence and relative abundance of Symbiodinium sequences recovered from the host. This comparative analysis reveals profound differences in the flexibility of corals for Symbiodinium, thereby classifying corals as generalists or specifists. Generalists such as Acropora and Pocillopora exhibit high intra- and inter-species flexibility in their Symbiodinium assemblages and are some of the most environmentally sensitive corals. Conversely, specifists such as massive Porites colonies exhibit low flexibility, harbour taxonomically narrow Symbiodinium assemblages, and are environmentally resistant corals. Collectively, these findings challenge the paradigm that symbiotic flexibility enhances holobiont resilience. This underscores the need for a deeper examination of the extent and duration of the functional benefits associated with endosymbiotic diversity and flexibility under environmental stress.
Keywords: coral, Symbiodinium, flexibility, generalist, specifist
1. Introduction
Variety in endosymbiotic communities provides a setting for interactions between endosymbionts within and among cells that have the potential to influence processes among host cells in both positive and negative directions. The paradigm that flexibility in the form of functional diversity equates to resilience, stability, greater functional range and adaptive potential is heavily grounded in ecological studies [1–3], but has yet to be investigated in detail at both the functional and the genetic level in endosymbioses. Endosymbiotic communities represent extreme examples of such scenarios, with hosts (usually macro-eukaryotes) housing communities of micro-organisms (micro-eukaryotes or bacteria) within the boundaries of their own cell membranes and tissues. The implications of flexibility in spatially constrained, densely populated environments may be particularly profound in driving the structure and function of the community [3]. Cnidarian–dinoflagellate symbioses are provocative subjects for such an investigation because they are taxonomically complex associations, whose stability and functional integrity underpins the persistence of corals and coral reef ecosystems through time.
Coral reefs are often described as sentinel ecosystems for their sensitivity to climate change stressors such as increasing seawater temperatures and ocean acidification as well as local impacts of anthropogenic activities [4,5]. The sensitivity of reef-building corals to these stressors is driven, to a large degree, by physiological constraints imposed by their unions with endosymbiotic dinoflagellates in the genus Symbiodinium [6–8]. The genus Symbiodinium is classified into nine major taxonomic lineages (A–I; [9]) that each contain multiple types. Symbiodinium clades and within-clade types exhibit patterns of association with specific coral taxa, and patterns in the distribution of Symbiodinium types across space and environment indicate differences in their physiological thresholds within the genus [10,11]. Indeed, shifts in the taxonomic composition of the communities of endosymbiotic dinoflagellates harboured by reef-building corals have been implicated as one mechanism that might confer resistance through tolerance, or resilience through adaptation, to changes in the environment [12].
The identification of functional differences in Symbiodinium physiology prompted the introduction of the adaptive bleaching hypothesis (ABH, [12]), which states that ‘bleaching provides an opportunity for the host to be repopulated with a different type of partner; frequent stress tends to favour a stress-resistant combination’. It has been further posited that this could be achieved by switching the existing symbiotic community for a new and better-adapted type, or by shuffling the relative proportions of the existing types within the community, increasing the abundance of previously cryptic types better adapted to the stress event [13]. Recent research has emphasized the importance of Symbiodinium shuffling in withstanding changes in global climate, such as temperature-induced bleaching [14,15]. While it has been suggested that the flexibility in Symbiodinium–coral interactions is largely beneficial [13] through improved holobiont thermal resistance [14], there are indications that this may not be the case for all species or temporal scales [7,8,16].
Much of the work characterizing Symbiodinium has focused on dominant Symbiodinium types in hosts, or those that are readily cultured from a host. As a result, the general perception has been that only a few corals are capable of hosting multiple Symbiodinium types, and that corals generally exhibit low endosymbiotic flexibility [17]. However, in recent years and with the application of more sensitive and more inclusive molecular approaches, our understanding of the complexity of cnidarian–Symbiodinium symbiosis has grown significantly [9], and many corals have been identified as hosting multiple types of Symbiodinium [18]. Furthermore, in some coral species, the Symbiodinium communities are now known to vary widely over space and time [19,20]. To date, however, symbiotic flexibility has not been quantitatively compared in a broad range of coral taxa, and the ecological performance of corals has never been linked to the levels of endosymbiotic flexibility of the host.
The goals of this study were to characterize, quantify and compare the flexibility of a range of coral taxa for Symbiodinium and map this trait onto the ecological resilience and biological attributes of the coral host documented in the literature. Flexibility here is defined as the ability to have a varied Symbiodinium sequence assemblage, or sequence community. Importantly, our study uses a comparative approach among samples and taxa, by assessing differences in Symbiodinium sequence assemblages among coral species. These data provide the capacity to link patterns in the Symbiodinium sequence assemblages that represent the endosymbiotic flexibility in corals, with the documented physiological susceptibility of coral holobionts.
2. Material and methods
(a). Sample processing
Corals samples were collected from fringing reef, lagoon reef or forereef habitats around the island of Moorea (figure 1; 1–24 m; electronic supplementary material, tables S1 and S2). DNA was extracted from coral fragments using the Qiagen DNA Easy kit (Qiagen, CA). Following extraction, polymerase chain reaction (PCR) amplification of the nuclear ribosomal partial 5.8S, internal transcribed spacer 2 (ITS2) and partial 28S rDNA regions was carried out using the ITS-DINO forward primer [21] and ITS2rev2 reverse primer [22]. To minimize cross hybridization of the primers with the coral host, touchdown PCR thermocycling conditions (25 µl) were used as follows: (i) 95°C for 10 min; (ii) 25 cycles 94°C for 30 s, 65°C for 30 s (decreasing the annealing temperature 0.5°C for every cycle after cycle 1) and 72°C for 1 min; (iii) 14 cycles of 94°C for 30 s, 52°C for 30 s and 72°C for 1 min; and (iv) final extension of 72°C for 10 min. PCR products were purified using the QIAquick PCR Purification Kit, ligated into pGEM-T Easy vector and transformed using α-select gold efficiency competent cells. Clone libraries were grown overnight on selective LB agar plates containing ampicillin, IPTG (isopropyl-β-d-thiogalactopyranoside, Fermentas, MD) and XGAL. Positive inserts were identified via PCR screening with M13 primers, purified with exonuclease I in shrimp alkaline phosphatase and sequenced on an automated sequencer (BigDye Terminator chemistry) with a target of 10 sequences per sample.
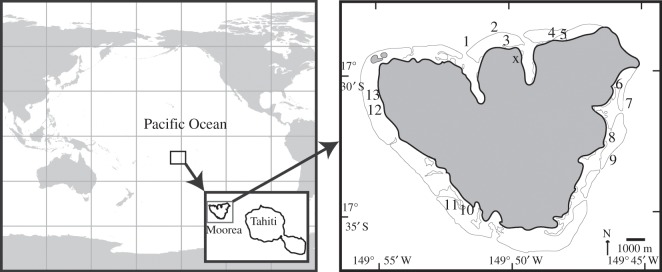
Figure 1.
Map displaying location of Moorea and 13 sampling sites (see electronic supplementary material, tables S1 and S2) around the island. Sampling sites include forereef, lagoon and fringing reef types.
(b). DNA sequence alignment and analysis
Alignment and inspection of raw sequences were completed using Sequencher v. 4.7 and Bioedit v. 5.0.9 [23]. Sequences with single polymorphisms were included only in the downstream analysis if recovered from three or more independent clone libraries (host samples). Symbiodinium sequences with single polymorphisms found in less than three clone libraries were likely due to PCR error, and were converted to the closest sequence in the dataset that occurred greater than three times [11]. Sequences with novel identities as assessed by BLAST results from GenBank were given a new alpha-numeric identifier (sensu [11]), based on the distance from the closest previously documented haplotype (e.g. C15 variant = C15.x). All sequences are available in GenBank under the accession numbers HE578975–HE579042 (table 1).
Table 1.
GenBank accession numbers for all Symbiodinium sequences recovered in the survey of coral hosts of Moorea, French Polynesia. First accession number is from the current study, whereas a second accession number denotes previous identification by the listed reference.
| sequence type | accession no. | novel | reference |
|---|---|---|---|
| A1 | HE578975, AF333505 | no | this study, [10] |
| A1.3 | HE578976 | yes | this study |
| A1.4 | HE578977 | yes | this study |
| A3 | HE578978, AF333507 | no | this study, [10] |
| C1 | HE578979, AF333515 | no | this study, [10] |
| C1.1 | HE578980, DQ480600 | no | this study, [24] |
| C1.6 | HE578981, FJ461493 | no | this study, [11] |
| C1.8 | HE578982, EU074955 | no | this study, [25] |
| C1.10 | HE578983 | yes | this study |
| C1.11 | HE578984 | yes | this study |
| C1.12 | HE578985 | yes | this study |
| C1.13 | HE578986 | yes | this study |
| C1.14 | HE578987 | yes | this study |
| C1.15 | HE578988 | yes | this study |
| C1.16 | HE578989, EU074883 | no | this study, [25] |
| C1.17 | HE578990 | yes | this study |
| C1.18 | HE578991 | yes | this study |
| C1.19 | HE578992 | yes | this study |
| C1.20 | HE578993 | yes | this study |
| C1.21 | HE578994, EU074889 | no | this study, [25] |
| C1.22 | HE578995 | yes | this study |
| C1b | HE578996, AY239363 | no | this study, [26] |
| C1b.1 | HE578997 | yes | this study |
| C1b.2 | HE578998 | yes | this study |
| C1b.3 | HE578999 | yes | this study |
| C1f | HE579000, AY258490 | no | this study, [27] |
| C3 | HE579001, FN298467 | no | this study, [28] |
| C3b | HE579002, AF499791 | no | this study, [29] |
| C3.15 | HE579003, FJ529649 | no | this study, [30] |
| C3.16 | HE579004, EU786015 | no | this study, [31] |
| C3.17 | HE579005 | yes | this study |
| C3.18 | HE579006 | yes | this study |
| C3.19 | HE579007 | yes | this study |
| C3.20 | HE579008 | yes | this study |
| C3.21 | HE579009 | yes | this study |
| C3.22 | HE579010 | yes | this study |
| C3.23 | HE579011 | yes | this study |
| C3.24 | HE579012 | yes | this study |
| C15 | HE579013, AM748552 | no | this study, [22] |
| C15.6 | HE579014, FN563472 | no | this study, [32] |
| C15.7 | HE579015 | yes | this study |
| C15.8 | HE579016 | yes | this study |
| C15.9 | HE579017, FN563475 | no | this study, [32] |
| C15.12 | HE579018 | yes | this study |
| C15.13 | HE579019 | yes | this study |
| C15.14 | HE579020 | yes | this study |
| C15.15 | HE579021 | yes | this study |
| C15.16 | HE579022 | yes | this study |
| C15.17 | HE579023 | yes | this study |
| C15.18 | HE579024 | yes | this study |
| C15.19 | HE579025 | yes | this study |
| C15.20 | HE579026 | yes | this study |
| C21 | HE579027, AY239372 | no | this study, [26] |
| C21.12 | HE579028, FJ461514 | no | this study, [11] |
| C21.17 | HE579029 | yes | this study |
| C21.18 | HE579030 | yes | this study |
| C21.19 | HE579031 | yes | this study |
| C21.20 | HE579032 | yes | this study |
| C21.21 | HE579033 | yes | this study |
| C42 | HE579034, AY765402 | no | this study, [27] |
| C45 | HE579035, AY239364 | no | this study, [26] |
| D1 | HE579036, AF334660 | no | this study, [10] |
| D1.8 | HE579037 | yes | this study |
| D1.9 | HE579038, AF174559 | no | this study, [33] |
| D1.10 | HE579039 | yes | this study |
| D1.11 | HE579040 | yes | this study |
| D1.12 | HE579041 | yes | this study |
| D1a | HE579042, AF499802 | no | this study, [29] |
(c). Statistical analysis
To determine coverage of the clone libraries, and to test for equal coverage across host taxa, coverage estimates were determined following Stat et al. [34], and coverage values were tested with one-way ANOVA among host genera. To compare Symbiodinium sequence assemblages among coral taxa, relative abundance data were square root-transformed, and a Bray–Curtis similarity coefficient [35] was calculated for all samples, ranging from 0 (indicating dissimilarity) to 100 (indicating identical assemblages). Analysis of similarity (ANOSIM) of the Bray–Curtis coefficients was carried out using primer v. 6.0 [36,37], and the two-dimensional ordination of the samples was represented with non-metric multi-dimensional scaling (MDS), or a spatial representation of the relative similarities between Symbiodinium sequence assemblages of coral samples. ANOSIM was used to test the hypotheses that there were no differences in the Symbiodinium sequence assemblages among species or genera.
To describe the Symbiodinium sequence assemblage for each sample, we included both genetic distance and frequency of symbiont occurrence in a single metric, or an index of flexibility (Fl). This index was calculated based on a modification of parasites' specificity for their hosts from parasitological research [38,39]. The index values range from 0 to ∞, where larger values indicate greater flexibility in the coral and the presence of a variety of Symbiodinium sequences that are genetically dissimilar, and lower values indicate lower flexibility in the coral and the presence of one or a few Symbiodinium sequences that are genetically similar. The index is described by the following equation:
In this equation, ωij is equal to the uncorrected pairwise genetic distance between Symbiodinium sequences; pi is equal to the relative abundance of the first Symbiodinium haplotype; pj is equal to the relative abundance of the second Symbiodinium haplotype. The product of ω(pipj) are summed across haplotypes i = 1 … n, and j = 1 … n (i.e. across all possible pairs of Symbiodinium haplotypes within a sample), normalized to the sum of the product of the relative abundance of each pair of haplotypes (pipj). For within-clade comparisons, uncorrected pairwise genetic distances of the ITS2 region were calculated using mega v. 4.0 [40]. Within-clade alignments were based on all non-redundant ITS2 A1, C1 and D1 sequence hits using the Symbiodinium database, SYM-BLAST (http://131.204.120.103/srsantos/symbiodinium/blast/blast_cs.html), in addition to the existing alignments from the current study. Alignments were created using ClustalW [41] as well as further manual alignment, and resulted in alignment of 101, 431 and 53 sequences for clades A, C and D, respectively. Therefore, genetic distances used for the index of flexibility calculations span the range of genetic distance documented within each Symbiodinium clade, and scale the genetic distance of our sequences to appropriate within-clade divergence. Similarly, average uncorrected pairwise genetic distance was calculated for the nine clades of Symbiodinium (A–I, [9]) based on the sequences of a portion of the relatively conserved nuclear ribosomal array (nr28S sequences, D1–D3 region, using the program mega v. 4.0 [40]. As the variation in ITS2 sequences between clades is too great for accurate alignment of sequences of all clades, and owing to the differences between markers (ITS2 versus nr28S), the specificity values are calculated separately within a clade (W) and among clades (A), and summed. Flexibility of the host for its Symbiodinium sequences was calculated for each coral sample. To statistically group coral genera in terms of their flexibility, the Gower % similarity distance measure [42] was used on the column-standardized values of flexibility. Flexibility groupings were calculated by UPGMA clustering [43] of the % similarity matrix among taxa in primer v. 6.0 [36,37].
3. Results
In total, 1240 Symbiodinium ITS2 sequences were recovered from 132 samples representing 34 species, 14 genera and seven families of coral (see the electronic supplementary material, table S1). All 1240 ITS2 sequences were used to report Symbiodinium clade by host (figure 2). Owing to the very low number of sequences recovered from samples of Montastrea curta, this species was excluded from downstream analyses. The average number of sequences recovered from the remaining 128 host samples was 9.6, and a total of 1223 sequences were used in similarity and flexibility calculations and statistical analyses. Coverage of the clone libraries at the level of genus ranged from approximately 54 to 75 per cent and did not differ among genera (F12,115 = 1.01, p > 0.05).
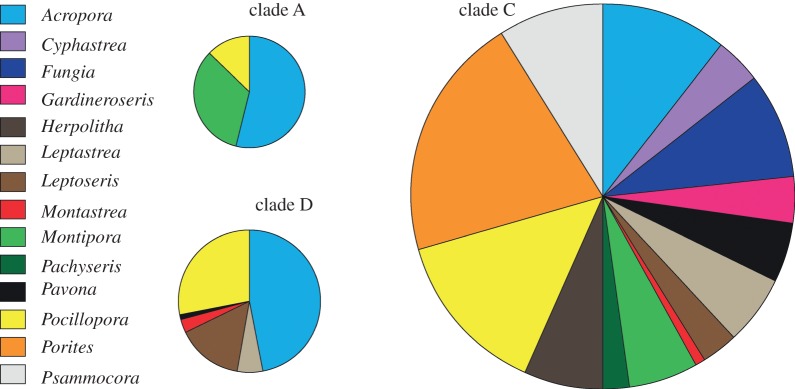
Figure 2.
Pie charts of the proportion of coral taxa hosting Symbiodinium from each of clades A, C and D. Area of the circles represents the abundance of sequences reported for each clade, from the total of 1240 sequences.
BLAST identity grouped the sequences into 68 different groups, representing 4, 7 and 57 Symbiodinium sequences from clades A, D and C, respectively (table 1 and electronic supplementary material, table S1). These included several commonly reported types (A1, A3, D1, D1a, C15, C45, C1, C1b, C3, C21), in addition to a number of novel ITS2 sequences (table 1). Ninety per cent of the corals sampled hosted one clade of Symbiodinium (n = 101 for C, 11 for D and 7 for A), 9 per cent hosted two (n = 9 for CD, 2 for AC and 1 for AD) and 1 per cent hosted three (n = 1 for ACD; see electronic supplementary material, table S1 for details). The similarities of Symbiodinium sequence assemblages among coral samples were assessed via calculation of Bray–Curtis similarity coefficients. MDS of the similarity coefficients showed separation in the Symbiodinium sequence assemblages among host taxa (figure 3 and electronic supplementary material, figure S1). ANOSIM of the Bray–Curtis coefficients provided support for the MDS ordination with significant differences in the Symbiodinium sequence assemblages among coral hosts at the level of species (R = 0.62, p = 0.001), and genus (R = 0.44, p = 0.001; electronic supplementary material, table S3). The average similarity of Symbiodinium assemblages within a genus ranged from approximately 11 per cent in Leptoseris to 50 per cent in Gardineroseris, and the average dissimilarity between genera ranged from approximately 55 to 100 per cent (see the electronic supplementary material, table S4).
Figure 3.
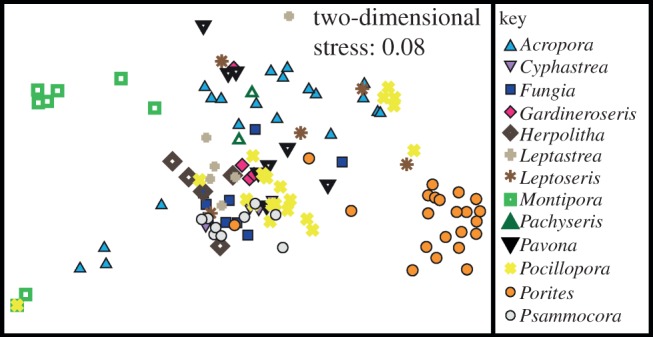
Multi-dimensional scaling (MDS) plot based on the Bray–Curtis similarity coefficients of square root-transformed, relative abundance of sequences in each coral sample (n = 128 corals). Not shown but included in the similarity and MDS analyses is one sample of Acropora cytherea, which hosts only Symbiodinium A1.3, and therefore is placed far to the right in a solitary position (see electronic supplementary material, figure S1). Visual exclusion of this sample for clarity of the positions of all other samples does not change the significance of the ANOSIM.
The diversity and genetic divergence of ITS2 sequence assemblages was collapsed into a single metric (index of flexibility) for the 128 coral samples. The distribution of values of flexibility, ranged from 0 to 0.2197, reflecting corals that were only host to a single type (i.e. 0), and those hosting multiple distantly related types of Symbiodinium from the most distantly related clades (i.e. 0.2197), respectively. Overall, seven coral samples hosted one sequence, and 121 corals hosted multiple sequences (see the electronic supplementary material, table S1). Acropora, Leptastrea, Leptoseris, Montipora, Pavona and Pocillopora had the highest values and variance, exhibiting the greatest flexibility with respect to Symbiodinium (figure 4). The high flexibility values for Acropora were driven by six corals each with large values, whereas the high flexibility in Montipora was driven by intersample variance in the presence of a single, but different, clade (A or C; figure 4 and electronic supplementary material, table S1). Cyphastrea, Fungia, Gardineroseris, Herpolitha, Porites and Psammocora had consistently low intersample variance and low flexibility values (figure 4). The patterns of low and high flexibility were supported with UPGMA clustering of percentage similarity in flexibility among genera. The corals clustered into two groups that were less than 26 per cent similar between groups and more than 78 per cent similar within a group. The high flexibility group (i.e. generalists) consisted of Acropora, Leptastrea, Leptoseris, Montipora, Pavona and Pocillopora, and the low flexibility group (i.e. specifists) consisted of Cyphastrea, Fungia, Gardineroseris, Herpolitha, Porites, Pachyseris and Psammocora (see the electronic supplementary material, figure S2).
Figure 4.
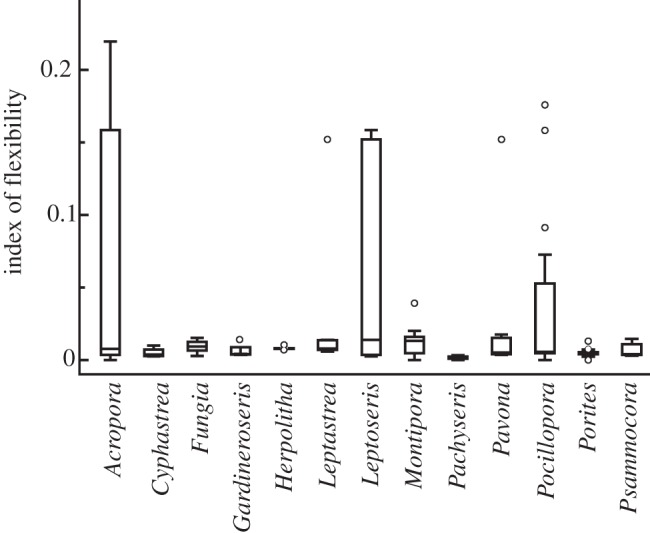
Index of flexibility for individual corals by genus (n = 128 corals and 1223 sequences). A value of 0 indicates a coral that forms a union with only one Symbiodinium sequence, values > 0 indicate a coral hosting multiple Symbiodinium sequences (see §2 for calculation details). Box plots display the median as the midline, and the upper and lower quartiles as the top and bottom lines of the boxes, respectively, with outliers displayed as circles.
4. Discussion
Our results demonstrate a large difference in composition of endosymbiotic communities hosted by scleractinian corals from Moorea. Quantitative examination of the flexibility of scleractinian hosts for Symbiodinium sequence assemblages identified two groups of coral genera that can be classified as endosymbiotic ‘generalists’ (high flexibility) and ‘specifists’ (low flexibility; figure 4 and electronic supplementary material, figure S2). Notably, the differences in the patterns of ITS2 sequence data, we report here are consistent with results previously published in Stat et al. [11], who examined Symbiodinium sequence assemblages in a range of coral hosts from Johnston Atoll using the ITS2 and the low copy chloroplast 23S in parallel. Here, we use ITS2 sequence assemblages as tools to describe and compare the Symbiodinium communities in corals. Reflecting the known interpretational complexity of this multi-copy nature and intragenomically variable marker [34], we make no inference regarding the relationship between the number of sequence types present in the ITS2 assemblages and the number of independent biological entities in the Symbiodinium communities in these corals. Further, the differences detected using the sequence assemblage approach are also evident when only previously described dominant ITS2 types are examined, or identified to the clade level in our study, as well as within the broader literature ([44]; electronic supplementary material, figure S3). Specifically in our study, Acropora and Pocillopora are symbiotically flexible and host five previously described dominant Symbiodinium types from three different clades, while massive Porites colonies hosts a single type (C15) from a single clade.
To explore the potential functional implications of these very different symbiotic strategies (flexibility versus specificity), and to exploit the comparative opportunity afforded by these groupings, we examined the literature and evaluated other biological traits that associate with generalist and specifist coral hosts, with the goal of assessing the potential physiological and ecological implications of symbiotic generalists versus symbiotic specifists (table 2 and references therein). Generalist corals that are flexible with respect to Symbiodinium exhibit very different biological attributes when compared with corals that are specifists, or inflexible (table 2). Most surprising among these is the stark difference in environmental sensitivity. Many of the corals that exhibit high symbiotic flexibility (generalists), such as the acroporids and pocilloporids are stress sensitive and often described as ecological ‘losers’ [61]. In contrast, coral species with high symbiotic specificity (i.e. high fidelity with respect to Symbiodinium) such as massive environmentally resilient poritids are ecological ‘winners’ [62,63,69,70].
Table 2.
Biological traits associated with generalist and specifist Symbiodinium–coral associations.
| traits | generalist | specifist | references |
|---|---|---|---|
| scleractinian examples | Acropora and Pocillopora | massive Porites | — |
| symbiotic diversity | flexibility | fidelity | this study, [13,27] |
| colony-level morphological plasticity | high | low | [45–47] |
| symbiome biodiversity | high | low | [48–50] |
| variability in energy acquisition modes | high | low | [51–55] |
| symbiont transmission | variable | vertical | [56] |
| growth (linear extension) | high | low | [57–60] |
| sensitivity to environmental stress | high | low | [61–64] |
| lifespan | short | long | [65–68] |
The identification of the ecological pattern of endosymbiotic flexibility and stress susceptibility identified in tropical corals in our study conflicts with the broadly accepted idea that endosymbiotic flexibility enhances resilience in tropical reef coral–Symbiodinium symbiosis [12]. For example, ecological surveys of the bleaching severity of the corals of Moorea following the 1994 mass-bleaching event revealed that the per cent bleaching was highest in Acropora (approx. 90–100%) followed by Pocillopora (approx. 70–90%), Montipora (approx. 20–50%) and Porites (approx. 13–43%; [69]). Furthermore, the ‘winning’ and ‘losing’ taxa are relatively similar biogeographically [61,62] with the generalists Acropora and Pocillopora showing a higher propensity to bleach, and higher mortality than the specifists such as Porites [63]. This pattern also holds for the stress of ocean acidification [64], where there is a decrease in the diversity and complexity of the reef through the loss of Acropora and Pocillopora at low pH, and an increase in the relative abundance and dominance of massive Porites. Indeed, the higher symbiotic flexibility of the generalist species does not appear to be reflected in a holobiont function that is ecologically beneficial under challenging environmental conditions. Support for the benefits of fidelity in symbiotic assemblages is also paralleled in temperate symbioses, where low diversity of vertically transmitted Symbiodinium appears to promote stability in cnidarian–dinoflagellate symbiosis [71]. For example, anemones exposed to varying irradiances host genetically similar Symbiodinium and are ecologically stable, a trait likely maintained via maternal inheritance of genetically similar symbionts [72].
A number of other interesting features emerge from our data. For example, the composition of the endosymbiotic communities in the generalists Acropora and Pocillopora varies widely among individuals within and among species in these genera at the single sampling interval represented in our study (figure 4). Furthermore, this pattern is manifest in the broader literature where spatial and temporal variability in symbiotic flexibility in generalists is commonly reported. For example, the Symbiodinium type hosted by the pocilloporid Seriatopora hystrix varies among colonies within a single reef (2–27 m at Yonge Reef) and between two reef sites on the GBR (Yonge and Day Reefs; [20]). Likewise, temporal flexibility is common in Acropora, where an ontogenetic shift occurs from clade A at 10 days old, to clades A, C and D in 83-day-old juveniles, a distribution also found in adult Acropora longicyathus [19]. Generalists corals such as acroporids and pocilloporids are some of the most ubiquitous and speciose reef-building corals in the Pacific [73]. They are characterized by branching morphologies that are plastic with respect to the environment. These genera exhibit high recruitment and rapid growth following disturbances [74], attributes that frame their description as weedy species. Generalist corals also show intraspecific and intergeneric variability in their feeding strategies (autotrophy and heterotrophy), and they acquire Symbiodinium using both vertical and horizontal transmission strategies [56]. The highly flexible acroporids and pocilloporids are generally dominated by clade C Symbiodinium but also host clades A and D. The within-clade ITS2 types found in generalist corals are often found in symbiosis with a broad range of hosts and are, for the most part, basal members of their respective clades (A1, C1, C3 and D1). These are the Symbiodinium types often described as generalist [13,27] and opportunistic [16,75]. The broad biological flexibility and capacity of generalist corals to interact with a wide range of Symbiodinium types is clearly advantageous under stable conditions. However, under prolonged environmental stress, the promiscuity and symbiotic entrepreneurialism of flexible hosts may drive competitive interactions within the symbiosis that destabilize and impair the overall functional of the symbiotic interactions. Such a transition is likely to have dire fitness consequences for the coral host.
At the other end of the endosymbiotic flexibility spectrum are the specifist coral genera exemplified by Porites; corals that display high inter- and intra-specific fidelity in their symbiotic partnerships in our study (figure 4), as well as over space and time in other studies [32,76]. For example, massive Porites colonies associate with Symbiodinium C15 from depths of 1–17 m across the GBR, and in corals collected in Japan [77], Johnston Atoll [11] and American Samoa [32]. Massive Porites is exemplified by a greater stability and persistence over time and under stressful conditions [32,57,61,76], traits attributed to slow growth [57], thick tissues/high tissue biomass [61,63] and thermally tolerant Symbiodinium C15 [78,79]. Characteristics of specifist corals such as Porites include symbiosis with endosymbionts such as Symbiodinium C15, which is commonly transmitted vertically across generations in the host [56], a mechanism that promotes coevolution and integration with the host, and is one of the most highly derived members of the most derived clade C [9,80]. This high fidelity symbiosis results in a tight integration between a specifist coral and a symbiont, that leads to success under both stable and stressful environments, with fidelity promoting spatial and temporal stability of the holobiont and positive fitness benefits.
The profound differences in the symbiotic flexibility in corals on a reef, and the associations between symbiotic flexibility and coral environmental sensitivity raise fundamental questions that pertain to differences in interaction states in the coral–Symbiodinium endosymbiosis (i.e. mutualism versus parasitism). As reviewed by Thompson [81], high partner fidelity driven by few symbiotic options, and tight vertical transmission favours the evolution of reduced antagonism, or mutualism in symbioses, thereby increasing persistence of intimate and long-term mutualisms. This is clearly exemplified in the potentially obligate mutualism of massive Porites–Symbiodinium C15. Symbiodinium C15 is highly derived [80], unlikely to be free-living [28] and has not, to date, been successfully cultured [82], all evidence in support of an obligate mutualism. In stark contrast is the flexibility of generalist coral–Symbiodinium associations, and the opportunistic nature of these symbioses. Acropora, for example, is characterized by high symbiont flexibility [18,83] and horizontal symbiont transmission/acquisition [56,83], and therefore is exposed to a variety of free-living Symbiodinium options [28], many of which are currently in culture. Despite the short-term benefits to thermotolerance due to acquisition or shuffling [84–86], ecologically these holobionts are linked with reduced fitness and higher mortality under stressful conditions. Together, this suggests that the flexible symbioses lean towards relationships on the less beneficial end of the spectrum [16,75].
Here, we have highlighted two extremes, a specifist (massive Porites sp.) and a generalist (Acropora sp.). These represent some of the most important and abundant taxa on modern reefs and exhibit differences in their environmental sensitivity, making them major drivers of reef community composition. The ability to detect the range and flexibility of Symbiodinium sequences present within a host sample and quantify the flexibility of the host through studies such as ours, allows for the discovery of such links between endosymbiotic flexibility and environmental sensitivity. This results in the ability to generate new testable hypotheses for predictions of the stability of a range of Symbiodinium–holobiont combinations under various environmental stressors, particularly in a changing global climate.
The examination of coral–Symbiodinium interactions has expanded our understanding of diversity in Symbiodinium from one [87] to many [9,88,89], and from functional equality [87] to inequality [6,16,90]. Here, we link symbiotic stability to holobiont resistance, and symbiotic flexibility to holobiont sensitivity (table 2). The identification of symbiotic flexibility has promoted the hypothesis that there is potential for ‘adaptation’ of corals to environmental stress via the acquisition of new or shuffling of existing populations of Symbiodinium (sensu the ABH [12,13]). The rate at which natural selection can occur to produce a more tolerant holobiont, however, may be outpaced by the rapid rate of change in environmental conditions related to anthropogenic global warming and ocean acidification, and the increasing frequency of these stress events. Additionally, the outcome of this flexibility in acquisition, or ability to shuffle, may not always be beneficial to the holobiont (e.g. declines in growth and energy acquisition; [7,16,91]). Notably, the long-term benefits of this flexibility in symbiosis are unknown, as research to date has focused on short-term tolerance (less than or equal to one to two bleaching events), and implications across longer temporal scales have not been explored. It is likely that the reefs of tomorrow will be shaped by the resilient and resistant coral–Symbiodinium assemblages of today, which are dominated by those specifist coral genera associated with fidelity in their symbiotic unions (e.g. massive Porites sp.). A further examination of generalist and specifist Symbiodinium–coral unions is necessary to determine the range of benefits or costs to the holobiont associated with characteristics such as immune response, photosynthate production and release, symbiont population control and calcification. This information will improve our ability to determine the consequences of flexibility or specificity in the coral–Symbiodinium symbiosis in a future of environmental uncertainty and dire predictions for the maintenance of reef-building coral ecosystems.
Acknowledgements
R.D.G., M.S. and H.M.P. designed research; M.S. performed field research; H.M.P., X.P. and M.S. performed lab research; R.D.G. and H.M.P. contributed new reagents/analytic tools; H.M.P. analysed data; and H.M.P. and R.D.G. wrote the paper. We are thankful for assistance from the staff of the Richard B. Gump South Pacific Research Lab, as well as logistical support of Moorea Coral Reef Long-Term Ecological Research site (MCR-LTER) researchers and staff. We also thank Nicholas Fabina regarding discussions of quantifying flexibility, and two anonymous reviewers for their comments. This research was supported by funding from the National Science Foundation to the MCR-LTER (OCE 04-17412), the National Marine Sanctuary Program and Hawaii Institute of Marine Biology Reserve Partnership (memorandum of agreement 2005-008/66882), a postdoctoral fellowship to M.S. from the UWA-AIMS-CSIRO collaborative agreement, a grant to R.D.G. (OCE-0752604), as well as funds from the Gordon and Betty Moore Foundation. H.M.P. was supported by a fellowship from the US EPA (FP917199). The authors declare that they have no conflict of interest. This is a contribution of the MCR-LTER, and is contribution no. 1512 of the Hawaii Institute of Marine Biology and no. 8727 of SOEST.
References
- 1.Loreau M. 2000. Biodiversity and ecosystem functioning: recent theoretical advances. Oikos 91, 3–17 10.1034/j.1600-0706.2000.910101.x (doi:10.1034/j.1600-0706.2000.910101.x) [DOI] [Google Scholar]
- 2.Tilman D. 1994. Competition and biodiversity in spatially structured habitats. Ecology 75, 2–16 10.2307/1939377 (doi:10.2307/1939377) [DOI] [Google Scholar]
- 3.Nystrom M. 2006. Redundancy and response diversity of functional groups: implications for the resilience of coral reefs. Ambio 35, 30–35 [PubMed] [Google Scholar]
- 4.Hoegh-Guldberg O., et al. 2007. Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742 10.1126/science.1152509 (doi:10.1126/science.1152509) [DOI] [PubMed] [Google Scholar]
- 5.Pandolfi J. M., Connolly S. R., Marshall D. J., Cohen A. L. 2011. Projecting coral reef futures under global warming and ocean acidification. Science 333, 418–422 10.1126/science.1204794 (doi:10.1126/science.1204794) [DOI] [PubMed] [Google Scholar]
- 6.Rowan R. 2004. Thermal adaptation in reef coral symbionts. Nature 430, 742. 10.1038/430742a (doi:10.1038/430742a) [DOI] [PubMed] [Google Scholar]
- 7.Little A. F., van Oppen M. J. H., Willis B. L. 2004. Flexibility in algal endosymbioses shapes growth in reef corals. Science 304, 1492–1494 10.1126/science.1095733 (doi:10.1126/science.1095733) [DOI] [PubMed] [Google Scholar]
- 8.Mieog J. C., Olsen J. L., Berkelmans R., Bleuler-Martinez S. A., Willis B. L., van Oppen M. J. H., Bruno J. F. 2009. The roles and interactions of symbiont, host and environment in defining coral fitness. PLoS ONE 4, e6364. 10.1371/journal.pone.0006364 (doi:10.1371/journal.pone.0006364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pochon X., Gates R. D. 2010. A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai'i. Mol. Phylogenet. Evol. 56, 492–497 10.1016/j.ympev.2010.03.040 (doi:10.1016/j.ympev.2010.03.040) [DOI] [PubMed] [Google Scholar]
- 10.LaJeunesse T. C. 2001. Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the ITS region: in search of a ‘species’ level marker. J. Phycol. 37, 866–880 10.1046/j.1529-8817.2001.01031.x (doi:10.1046/j.1529-8817.2001.01031.x) [DOI] [Google Scholar]
- 11.Stat M., Pochon X., Cowie R., Gates R. D. 2009. Specificity in communities of Symbiodinium in corals from Johnston Atoll. Mar. Ecol. Prog. Ser. 386, 83–96 10.3354/meps08080 (doi:10.3354/meps08080) [DOI] [Google Scholar]
- 12.Buddemeier R., Fautin D. 1993. Coral bleaching as an adaptive mechanism: a testable hypothesis. BioScience 43, 320–326 10.2307/1312064 (doi:10.2307/1312064) [DOI] [Google Scholar]
- 13.Baker A. C. 2003. Flexibility and specificity in coral-algal symbiosis: diversity, ecology and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34, 661–689 10.1146/annurev.ecolsys.34.011802.132417 (doi:10.1146/annurev.ecolsys.34.011802.132417) [DOI] [Google Scholar]
- 14.Berkelmans R., van Oppen M. J. H. 2006. The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc. R. Soc. B 273, 2305–2312 10.1098/rspb.2006.3567 (doi:10.1098/rspb.2006.3567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaJeunesse T. C., Smith R., Finney J., Oxenford H. 2009. Outbreak and persistence of opportunistic symbiotic dinoflagellates during the 2005 Caribbean mass coral ‘bleaching’ event. Proc. R. Soc. B 276, 4139–4148 10.1098/rspb.2009.1405 (doi:10.1098/rspb.2009.1405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stat M., Morris E., Gates R. D. 2008. Functional diversity in coral–dinoflagellate symbiosis. Proc. Natl Acad. Sci. USA 105, 9256–9261 10.1073/pnas.0801328105 (doi:10.1073/pnas.0801328105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goulet T. 2006. Most corals may not change their symbionts. Mar. Ecol. Prog. Ser. 321, 1–7 10.3354/meps321001 (doi:10.3354/meps321001) [DOI] [Google Scholar]
- 18.Baker A. C., Romanski A. M. 2007. Multiple symbiotic partnerships are common in scleractinian corals, but not in octocorals: comment on Goulet (2006). Mar. Ecol. Prog. Ser. 335, 237–242 10.3354/meps335237 (doi:10.3354/meps335237) [DOI] [Google Scholar]
- 19.Gómez-Cabrera M., Ortiz J. C., Loh W. K. W., Ward S., Hoegh-Guldberg O. 2008. Acquisition of symbiotic dinoflagellates (Symbiodinium) by juveniles of the coral Acropora longicyathus. Coral Reefs 27, 219–226 10.1007/s00338-007-0315-x (doi:10.1007/s00338-007-0315-x) [DOI] [Google Scholar]
- 20.Bongaerts P., Riginos C., Ridgway T., Sampayo E. M., van Oppen M. J. H., Englebert N., Vermeulen F., Hoegh-Guldberg O., Vollmer S. 2010. Genetic divergence across habitats in the widespread coral Seriatopora hystrix and its associated Symbiodinium. PLoS ONE 5, e10871. 10.1371/journal.pone.0010871 (doi:10.1371/journal.pone.0010871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pochon X., Pawlowski J., Zaninetti L., Rowan R. 2001. High genetic diversity and relative specificity among Symbiodinium-like endosymbiotic dinoflagellates in soritid foraminiferans. Mar. Biol. 139, 1069–1078 10.1007/s002270100674 (doi:10.1007/s002270100674) [DOI] [Google Scholar]
- 22.Pochon X., Garcia-Cuetos L., Baker A. C., Castella E., Pawlowski J. 2007. One-year survey of a single Micronesian reef reveals extraordinarily rich diversity of Symbiodinium types in soritid foraminifera. Coral Reefs 26, 867–882 10.1007/s00338-007-0279-x (doi:10.1007/s00338-007-0279-x) [DOI] [Google Scholar]
- 23.Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 [Google Scholar]
- 24.Reimer J. D., Takishita K., Maruyama T. 2006. Molecular identification of symbiotic dinoflagellates (Symbiodinium spp.) from Palythoa spp. (Anthozoa: Hexacorallia) in Japan. Coral Reefs 25, 521–527 10.1007/s00338-006-0151-4 (doi:10.1007/s00338-006-0151-4) [DOI] [Google Scholar]
- 25.Thornhill D. J., Lajeunesse T. C., Santos S. R. 2007. Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Mol. Ecol. 16, 5326–5340 10.1111/j.1365-294X.2007.03576.x (doi:10.1111/j.1365-294X.2007.03576.x) [DOI] [PubMed] [Google Scholar]
- 26.LaJeunesse T. C., Loh W. K. W., van Woesik R., Hoegh-Guldberg O., Schmidt G. W., Fitt W. K. 2003. Low symbiont diversity in southern Great Barrier Reef corals relative to those of the Caribbean. Limnol. Oceanogr. 48, 2046–2054 10.4319/lo.2003.48.5.2046 (doi:10.4319/lo.2003.48.5.2046) [DOI] [Google Scholar]
- 27.LaJeunesse T., Thornhill D. J., Cox E. F., Stanton F. G., Fitt W. K., Schmidt G. W. 2004. High diversity and host specificity observed among symbiotic dinoflagellates in reef coral communities from Hawaii. Coral Reefs 23, 596–603 [Google Scholar]
- 28.Pochon X., Stat M., Takabayashi M., Chasqui L., Chauka L. J., Logan D. D. K., Gates R. D. 2010. Comparison of endosymbiotic and free-living Symbiodinium (Dinophyceae) diversity in a Hawaiian reef environment. J. Phycol. 46, 53–65 10.1111/j.1529-8817.2009.00797.x (doi:10.1111/j.1529-8817.2009.00797.x) [DOI] [Google Scholar]
- 29.LaJeunesse T. C. 2002. Diversity and community structure of symbiotic dinoflagellates from Caribbean coral reefs. Mar. Biol. 141, 387–400 10.1007/s00227-002-0829-2 (doi:10.1007/s00227-002-0829-2) [DOI] [Google Scholar]
- 30.Sampayo E. M., Dove S., LaJeunesse T. C. 2009. Cohesive molecular genetic data delineate species diversity in the dinoflagellate genus Symbiodinium. Mol. Ecol. 18, 500–519 10.1111/j.1365-294X.2008.04037.x (doi:10.1111/j.1365-294X.2008.04037.x) [DOI] [PubMed] [Google Scholar]
- 31.Fay S., Weber M. X., Lipps J. H. 2009. The distribution of Symbiodinium diversity within individual host foraminifera. Coral Reefs 28, 717–726 10.1007/s00338-009-0511-y (doi:10.1007/s00338-009-0511-y) [DOI] [Google Scholar]
- 32.Barshis D. J., Stillman J. H., Gates R. D., Toonen R. J., Smith L. W., Birkeland C. 2010. Protein expression and genetic structure of the coral Porites lobata in an environmentally extreme Samoan back reef: does host genotype limit phenotypic plasticity? Mol. Ecol. 19, 1705–1720 10.1111/j.1365-294X.2010.04574.x (doi:10.1111/j.1365-294X.2010.04574.x) [DOI] [PubMed] [Google Scholar]
- 33.Brown B. E., Dunne R. P., Goodson M. S., Douglas A. E. 2002. Experience shapes the susceptibility of a reef coral to bleaching. Coral Reefs 21, 119–126 [Google Scholar]
- 34.Stat M., et al. 2011. Variation in Symbiodinium ITS2 sequence assemblages among coral colonies. PLoS ONE 6, e15854. 10.1371/journal.pone.0015854 (doi:10.1371/journal.pone.0015854) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bray R., Curtis J. 1957. An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 27, 325–349 10.2307/1942268 (doi:10.2307/1942268) [DOI] [Google Scholar]
- 36.Clarke K. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143 10.1111/j.1442-9993.1993.tb00438.x (doi:10.1111/j.1442-9993.1993.tb00438.x) [DOI] [Google Scholar]
- 37.Clarke K., Warwick R. 2001. Change in marine communities: an approach to statistical analysis and interpretation, 2nd edn Plymouth, UK: PRIMER-E. [Google Scholar]
- 38.Poulin R., Mouillot D. 2003. Parasite specialization from a phylogenetic perspective: a new index of host specificity. Parasitology 126, 473–480 10.1017/S0031182003002993 (doi:10.1017/S0031182003002993) [DOI] [PubMed] [Google Scholar]
- 39.Poulin R., Mouillot D. 2005. Combining phylogenetic and ecological information into a new index of host specificity. J. Parasitol. 91, 511–514 10.1645/GE-398R (doi:10.1645/GE-398R) [DOI] [PubMed] [Google Scholar]
- 40.Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (mega) software v. 4.0. Mol. Biol. Evol. 24, 1596–1599 10.1093/molbev/msm092 (doi:10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- 41.Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 10.1093/nar/22.22.4673 (doi:10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Legendre P., Legendre L. 1998. Numerical ecology. Amsterdam, The Netherlands: Elsevier Science [Google Scholar]
- 43.Sneath P., Sokal R. 1973. Numerical taxonomy. The principles and practice of numerical classification. San Francisco, CA: H. Freeman and Co [Google Scholar]
- 44.Franklin E. C., Stat M., Pochon X., Putnam H. M., Gates R. D. 2011. GeoSymbio: a hybrid, cloud-based web application of global geospatial bioinformatics and ecoinformatics for Symbiodinium–host symbioses. Mol. Ecol. Res. 12, 369–373 10.1111/j.1755-0998.2011.03081.x (doi:10.1111/j.1755-0998.2011.03081.x) [DOI] [PubMed] [Google Scholar]
- 45.Permata W. D., Hidaka M. 2005. Ontogenetic changes in the capacity of the coral Pocillopora damicornis to originate branches. Zool. Sci. 22, 1197–1203 10.2108/zsj.22.1197 (doi:10.2108/zsj.22.1197) [DOI] [PubMed] [Google Scholar]
- 46.Todd P. A. 2008. Morphological plasticity in scleractinian corals. Biol. Rev. 83, 315–337 10.1111/j.1469-185X.2008.00045.x (doi:10.1111/j.1469-185X.2008.00045.x) [DOI] [PubMed] [Google Scholar]
- 47.Kaniewska P., Anthony K. R. N., Hoegh-Guldberg O. 2008. Variation in colony geometry modulates internal light levels in branching corals, Acropora humilis and Stylophora pistillata. Mar. Biol. 155, 649–660 10.1007/s00227-008-1061-5 (doi:10.1007/s00227-008-1061-5) [DOI] [Google Scholar]
- 48.Vytopil E., Willis B. L. 2001. Epifaunal community structure in Acropora spp. (Scleractinia) on the Great Barrier reef: implications of coral morphology and habitat complexity. Coral Reefs 20, 281–288 10.1007/S003380100172 (doi:10.1007/S003380100172) [DOI] [Google Scholar]
- 49.Stella J. S., Jones G. P., Pratchett M. S. 2010. Variation in the structure of epifaunal invertebrate assemblages among coral hosts. Coral Reefs 29, 957–973 10.1007/s00338-010-0648-8 (doi:10.1007/s00338-010-0648-8) [DOI] [Google Scholar]
- 50.Gates R. D., Ainsworth T. D. 2011. The nature and taxonomic composition of coral symbiomes as drivers of performance limits in scleractinian corals. J. Exp. Mar. Biol. Ecol. 408, 94–101 10.1016/j.jembe.2011.07.029 (doi:10.1016/j.jembe.2011.07.029) [DOI] [Google Scholar]
- 51.Porter J. W. 1976. Autotrophy, heterotrophy, and resource partitioning in Caribbean reef-building corals. Am. Nat. 110, 731–742 10.1086/283100 (doi:10.1086/283100) [DOI] [Google Scholar]
- 52.Muscatine L., Porter J. W. 1977. Reef corals: mutualistic symbioses adapted to nutrient-poor environments. BioScience 27, 454–460 10.2307/1297526 (doi:10.2307/1297526) [DOI] [Google Scholar]
- 53.Edmunds P. J., Davies P. S. 1986. An energy budget for Porites porites (Scleractinia). Mar. Biol. 92, 339–347 10.1007/BF00392674 (doi:10.1007/BF00392674) [DOI] [Google Scholar]
- 54.Anthony K. R. N., Fabricius K. E. 2000. Shifting roles of heterotrophy and autotrophy in coral energetics under varying turbidity. J. Exp. Mar. Biol. Ecol. 252, 221–253 10.1016/S0022-0981(00)00237-9 (doi:10.1016/S0022-0981(00)00237-9) [DOI] [PubMed] [Google Scholar]
- 55.Houlbreque F., Ferrier-Pages C. 2009. Heterotrophy in tropical scleractinian corals. Biol. Rev. 84, 1–17 10.1111/j.1469-185X.2008.00058.x (doi:10.1111/j.1469-185X.2008.00058.x) [DOI] [PubMed] [Google Scholar]
- 56.Baird A. H., Guest J. R., Willis B. L. 2009. Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Ann. Rev. Ecol. Evol. Syst. 40, 551–571 10.1146/annurev.ecolsys.110308.120220 (doi:10.1146/annurev.ecolsys.110308.120220) [DOI] [Google Scholar]
- 57.Glynn P. W. 1993. Coral reef bleaching: ecological perspectives. Coral Reefs 12, 1–17 10.1007/BF00303779 (doi:10.1007/BF00303779) [DOI] [Google Scholar]
- 58.Gladfelter E. H., Monahan R. K., Gladfelter W. B. 1978. Growth rates of five reef-building corals in the northeastern Caribbean. Bull. Mar. Sci. 28, 728–734 [Google Scholar]
- 59.Huston M. 1985. Variation in coral growth rates with depth at Discovery Bay, Jamaica. Coral Reefs 4, 19–25 10.1007/BF00302200 (doi:10.1007/BF00302200) [DOI] [Google Scholar]
- 60.Tanzil J. T. I., Brown B. E., Tudhope A. W., Dunne R. P. 2009. Decline in skeletal growth of the coral Porites lutea from the Andaman Sea, South Thailand between 1984 and 2005. Coral Reefs 28, 519–528 10.1007/s00338-008-0457-5 (doi:10.1007/s00338-008-0457-5) [DOI] [Google Scholar]
- 61.Loya Y., Sakai K., Nakano Y., Sambali H., van Woesik R. 2001. Coral bleaching: the winners and the losers. Ecol. Lett. 4, 122–131 10.1046/j.1461-0248.2001.00203.x (doi:10.1046/j.1461-0248.2001.00203.x) [DOI] [Google Scholar]
- 62.Marshall P. A., Baird A. H. 2000. Bleaching of corals on the Great Barrier Reef: differential susceptibilities among taxa. Coral Reefs 19, 155–163 10.1007/s003380000086 (doi:10.1007/s003380000086) [DOI] [Google Scholar]
- 63.van Woesik R., Sakai K., Ganase A., Loya Y. 2011. Revisiting the winners and the losers a decade after coral bleaching. Mar. Ecol. Prog. Ser. 434, 67–76 10.3354/meps09203 (doi:10.3354/meps09203) [DOI] [Google Scholar]
- 64.Fabricius K. E., et al. 2011. Losers and winners in coral reefs acclimatized to elevated carbon dioxide concentrations. Nat. Clim. Change 1, 1–5 10.1038/nclimate1083 (doi:10.1038/nclimate1083) [DOI] [Google Scholar]
- 65.Soong K., Chen C. A., Chang J. C. 1999. A very large poritid colony at Green Island, Taiwan. Coral Reefs 18, 42. 10.1007/s003380050151 (doi:10.1007/s003380050151) [DOI] [Google Scholar]
- 66.Guzner B., Novoplansky A., Chadwick N. E. 2007. Population dynamics of the reef-building coral Acropora hemprichii as an indicator of reef condition. Mar. Ecol. Prog. Ser. 333, 143–150 10.3354/meps333143 (doi:10.3354/meps333143) [DOI] [Google Scholar]
- 67.Brown D. P., Basch L., Barshis D., Forsman Z., Fenner D., Goldberg J. 2009. American Samoa's island of giants: massive Porites colonies at Ta'u island. Coral Reefs 28, 735. 10.1007/s00338-009-0494-8 (doi:10.1007/s00338-009-0494-8) [DOI] [Google Scholar]
- 68.De'ath G., Lough J. M., Fabricius K. E. 2009. Declining coral calcification on the Great Barrier Reef. Science 323, 116–119 10.1126/science.1165283 (doi:10.1126/science.1165283) [DOI] [PubMed] [Google Scholar]
- 69.Hoegh-Guldberg O., Salvat B. 1995. Periodic mass-bleaching and elevated sea temperatures: bleaching of outer reef slope communities in Moorea, French Polynesia. Mar. Ecol. Prog. Ser. 121, 181–190 10.3354/meps121181 (doi:10.3354/meps121181) [DOI] [Google Scholar]
- 70.Gates R. D., Edmunds P. J. 1999. The physiological mechanisms of acclimatization in tropical reef corals. Am. Zool. 39, 30–43 [Google Scholar]
- 71.Muller-Parker G., Davy S. K. 2001. Temperate and tropical algal–sea anemone symbioses. Invert. Biol. 120, 104–123 10.1111/j.1744-7410.2001.tb00115.x (doi:10.1111/j.1744-7410.2001.tb00115.x) [DOI] [Google Scholar]
- 72.Bythell J. C., Douglas A. E., Sharp V. A., Searle J. B., Brown B. E. 1997. Algal genotype and photoacclimatory responses of the symbiotic alga Symbiodinium in natural populations of the sea anemone Anemonia viridis. Proc. R. Soc. Lond. B 264, 1277–1282 10.1098/rspb.1997.0176 (doi:10.1098/rspb.1997.0176) [DOI] [Google Scholar]
- 73.Veron J. E. N. 2000. Corals of the world. Townsville: Australian Institute of Marine Sciences [Google Scholar]
- 74.Grigg R. W., Maragos J. E. 1974. Recolonization of hermatypic corals on submerged lava flows in Hawaii. Ecology 55, 387–395 10.2307/1935226 (doi:10.2307/1935226) [DOI] [Google Scholar]
- 75.Stat M., Gates R. D. 2011. Clade D Symbiodinium in scleractinian corals, a ‘nugget’ of hope, a selfish opportunist, an ominous sign, or all of the above? J. Mar. Biol. 2011, 730715 [Google Scholar]
- 76.Stat M., Loh W. K. W., LaJeunesse T., Hoegh-Guldberg O., Carter D. 2009. Stability of coral–endosymbiont associations during and after a thermal stress event in the southern Great Barrier Reef. Coral Reefs 28, 709–713 10.1007/s00338-009-0509-5 (doi:10.1007/s00338-009-0509-5) [DOI] [Google Scholar]
- 77.Lajeunesse T. C., Bhagooli R., Hidaka M., deVantier L., Done T., Schmidt G. W., Fitt W. K., Hoegh-Guldberg O. 2004. Closely related Symbiodinium spp. differ in relative dominance in coral reef host communities across environmental, latitudinal and biogeographic gradients. Mar. Ecol. Prog. Ser. 284, 147–161 10.3354/meps284147 (doi:10.3354/meps284147) [DOI] [Google Scholar]
- 78.Fitt W. K., et al. 2009. Response of two species of Indo-Pacific corals, Porites cylindrica and Stylophora pistillata, to short-term thermal stress: the host does matter in determining the tolerance of corals to bleaching. J. Exp. Mar. Biol. Ecol. 373, 102–110 10.1016/j.jembe.2009.03.011 (doi:10.1016/j.jembe.2009.03.011) [DOI] [Google Scholar]
- 79.LaJeunesse T. C., Pettay D. T., Sampayo E. M., Phongsuwan N., Brown B., Obura D. O., Hoegh-Guldberg O., Fitt W. K. 2010. Long-standing environmental conditions, geographic isolation and host–symbiont specificity influence the relative ecological dominance and genetic diversification of coral endosymbionts in the genus Symbiodinium. J. Biogeogr. 37, 785–800 10.1111/j.1365-2699.2010.02273.x (doi:10.1111/j.1365-2699.2010.02273.x) [DOI] [Google Scholar]
- 80.Lajeunesse T. C. 2005. ‘Species’ radiations of symbiotic dinoflagellates in the Atlantic and Indo-Pacific since the Miocene–Pliocene transition. Mol. Biol. Evol. 22, 570–581 10.1093/molbev/msi042 (doi:10.1093/molbev/msi042) [DOI] [PubMed] [Google Scholar]
- 81.Thompson J. 1994. The coevolutionary process. Chicago, IL: University of Chicago Press [Google Scholar]
- 82.Krueger T., Gates R. D. 2012. Cultivating endosymbionts: host environmental mimics support the survival of Symbiodinium C15 ex hospite. J. Exp. Mar. Biol. Ecol. 413, 169–176 10.1016/j.jembe.2011.12.002 (doi:10.1016/j.jembe.2011.12.002) [DOI] [Google Scholar]
- 83.van Oppen M. J. H., Palstra F. P., Piquet A. M.-T., Miller D. J. 2001. Patterns of coral–dinoflagellate associations in Acropora: significance of local availability and physiology of Symbiodinium strains and host–symbiont selectivity. Proc. R. Soc. Lond. B 268, 1759–1767 10.1098/rspb.2001.1733 (doi:10.1098/rspb.2001.1733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baker A., Starger C., McClanahan T., Glynn P. W. 2004. Corals’ adaptive response to climate change. Nature 430, 741. 10.1038/430741a (doi:10.1038/430741a) [DOI] [PubMed] [Google Scholar]
- 85.Thornhill D., LaJeunesse T. C., Kemp D., Fitt W., Schmidt G. 2006. Multi-year, seasonal genotypic surveys of coral–algal symbioses reveal prevalent stability or post-bleaching reversion. Mar. Biol. 148, 711–722 10.1007/s00227-005-0114-2 (doi:10.1007/s00227-005-0114-2) [DOI] [Google Scholar]
- 86.Warner M., LaJeunesse T. C., Robison J., Thur R. 2006. The ecological distribution and comparative photobiology of symbiotic dinoflagellates from reef corals in Belize: potential implications for coral bleaching. Limnol. Oceanogr. 51, 1887–1897 10.4319/lo.2006.51.4.1887 (doi:10.4319/lo.2006.51.4.1887) [DOI] [Google Scholar]
- 87.Freudenthal H. D. 1962. Symbiodinium gen. nov. and Symbiodinium microadriaticum sp. nov., a Zooxanthella: taxonomy, life cycles and morphology. J. Eukaryot. Microbiol. 9, 45–52 10.1111/j.1550-7408.1962.tb02579.x (doi:10.1111/j.1550-7408.1962.tb02579.x) [DOI] [Google Scholar]
- 88.Rowan R., Powers D. 1991. A molecular genetic classification of zooxanthellae and the evolution of animal–algal symbioses. Science 251, 1348–1351 10.1126/science.251.4999.1348 (doi:10.1126/science.251.4999.1348) [DOI] [PubMed] [Google Scholar]
- 89.Stat M., Carter D., Hoegh-Guldberg O. 2006. The evolutionary history of Symbiodinium and scleractinian hosts: symbiosis, diversity, and the effect of climate change. Perspect. Plant Ecol. Evol. Syst. 8, 23–43 10.1016/j.ppees.2006.04.001 (doi:10.1016/j.ppees.2006.04.001) [DOI] [Google Scholar]
- 90.Sampayo E. M., Ridgway T., Bongaerts P., Hoegh-Guldberg O. 2008. Bleaching susceptibility and mortality of corals are determined by fine-scale differences in symbiont type. Proc. Natl Acad. Sci. USA 105, 10 444–10 449 10.1073/pnas.0708049105 (doi:10.1073/pnas.0708049105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cooper T. F., et al. 2011. Symbiodinium genotypic and environmental controls on lipids in reef building corals. PLoS ONE 6, e20434. [DOI] [PMC free article] [PubMed] [Google Scholar]