Abstract
Mate choice for major histocompatibility complex (MHC) compatibility has been found in several taxa, although rarely in birds. MHC is a crucial component in adaptive immunity and by choosing an MHC-dissimilar partner, heterozygosity and potentially broad pathogen resistance is maximized in the offspring. The MHC genotype influences odour cues and preferences in mammals and fish and hence olfactory-based mate choice can occur. We tested whether blue petrels, Halobaena caerulea, choose partners based on MHC compatibility. This bird is long-lived, monogamous and can discriminate between individual odours using olfaction, which makes it exceptionally well suited for this analysis. We screened MHC class I and II B alleles in blue petrels using 454-pyrosequencing and quantified the phylogenetic, functional and allele-sharing similarity between individuals. Partners were functionally more dissimilar at the MHC class II B loci than expected from random mating (p = 0.033), whereas there was no such difference at the MHC class I loci. Phylogenetic and non-sequence-based MHC allele-sharing measures detected no MHC dissimilarity between partners for either MHC class I or II B. Our study provides evidence of mate choice for MHC compatibility in a bird with a high dependency on odour cues, suggesting that MHC odour-mediated mate choice occurs in birds.
Keywords: major histocompatibility complex, MHC class I, MHC class II B, 454-pyrosequencing, genetic compatibility
1. Introduction
The major histocompatibility complex (MHC) plays a crucial role in vertebrate adaptive immunity [1]. The extraordinary polymorphism found in this gene cluster is thought to be mainly driven by pathogen-mediated balancing selection although mate choice for compatibility, could be an additional mechanism for maintaining MHC diversity [2–5]. Mate choice for compatibility requires disassortative mating between genetically dissimilar/compatible individuals to gain heterozygote advantages beyond what can be obtained by choice of heterozygous mates (which will eventually lead the population to genetic equilibrium) [2]. MHC heterozygosity is associated with increased disease resistance and survival [6–8], and MHC compatibility in human couples is known to decrease the rate of spontaneous abortions [9,10]. MHC-based mate choice could therefore be beneficial. In several species of fish and mammals, the MHC genotype is detectable by olfactory cues (personal odours) and these cues may guide the choice of a partner (reviewed in [11,12]).
Mate choice for MHC compatibility has been found in several taxa, e.g. humans [13], other primates [14,15], mice [16,17], lizards [18,19] and fish [20,21], although not in all studies or examined species (reviewed in [3,12,22,23]). MHC compatibility and other male quality characters may interact in the female mate choice decision, i.e. an MHC-dependent mate choice may only be found when there are small differences between male quality characters [24]. In birds, direct benefits of a social mate, like territory quality or male feeding rate, are known to be very important [25] and it is likely that mate choice for MHC compatibility (an indirect benefit) may be undetectable or absent in many avian systems. Zelano & Edwards [26] suggested that an avian genetically based mate-choice system for MHC compatibility would be most probable in long-lived species that engage in lifelong monogamy and have good olfaction (to permit an MHC-based olfactory discrimination of potential partners). All these characteristics are found among procellariiform birds (petrels) [27–29]. The single mate choice event in petrels is likely to be subject to strong selection since making a good mate choice for life is crucial to maximize fitness. An MHC-dependent mate choice is therefore predicted in this avian family [26,30].
The blue petrel, Halobaena caerulea (Procellariiformes), is an Antarctic burrow nesting bird that breeds in dense colonies on small islands in the Southern Ocean [31]. Foraging is odour-guided [32], and the underground nest and partner is identified solely by olfactory cues [33–35]. In particular, the ability to recognize the partner and avoid self in favour of other individuals in odour choice tests [35] raises the question whether the blue petrel could have a genetically based mate-choice system for MHC compatibility. The blue petrel MHC genes are organized in, at least, eight class I and two class II B loci where most genes are transcribed [36].
We test the hypothesis that blue petrels choose MHC-compatible partners, with the assumptions that MHC heterozygosity increases fitness directly (or is an indicator of overall heterozygosity) and that blue petrels can evaluate the MHC genotype of potential partners. We predict that MHC genotypes between partners should be more dissimilar than MHC genotypes from random pairs.
We analyse both MHC class I and class II B loci to investigate potential differences between the two classes in relation to mate choice. The different gene expression patterns and functions of the two classes of MHC justify a parallel investigation; class I molecules are expressed on all nucleated cells and present peptides from intracellular pathogens, e.g. many viruses, whereas MHC class II molecules are expressed on immune cells and present peptides from extracellular pathogens, e.g. many bacteria [1]. The analyses are based on high-coverage 454-pyrosequencing of MHC class I and II B amplicons in a multi-loci setup in blue petrels with known partners. The DNA sequences were analysed with phylogenetical and functional distance-based methods and the percent difference (PD) in allele sharing was established. The genotyped partners were contrasted to random expectations.
2. Material and methods
(a). Birds and sample preparation
A study colony of about 80 nest burrows of blue petrels, Halobaena caerulea, was studied during seven successive field seasons (December–January) between 2001 and 2009 (excluding season 2004–2005). The species is widespread around the Kerguelen archipelago (southern Indian Ocean), and many of its islands are common breeding sites for the blue petrels. The study colony is situated on the small island ‘Ile Verte’ (49°51′ S, 70°05′ E). Most birds from these nests are ringed and burrows have been fitted with a closable aperture above the incubating chamber to facilitate capture. Removing birds from the burrow for a brief time does not appear to affect incubation behaviour or the hatchability of the eggs. In this study, 55 pairs (55 males and 55 females) were included in the analyses (for full description of the screening, see the electronic supplementary material, S1). Pairs with the highest number of seasons spent together were chosen in the cases where the partner had been replaced (n = 7). Blood was collected from all individuals. The samples (0.1–0.5 ml of blood) were collected with syringe from the brachial vein and were kept in Queen's lysis buffer (0.5 ml) (10 mM Tris-HCl, 10 mM NaCl, 10 mM EDTA, 1% n-lauroylsarcosine, pH8.0), stored at +4°C until DNA extraction.
DNA was extracted from the blood samples, stored in Queen's buffer, according to the manufacturer's protocol using the DNeasy Blood & Tissue Kit (Qiagen) and DNA was kept at −20° until used.
(b). 454-pyrosequencing of major histocompatibility complex class I and II B
Class-specific primers for amplification of variable regions of the MHC class I and class II B genes in the blue petrel were designed in conserved flanking regions identified by sequencing of longer stretches of the genes [36]. PCR amplifications of class I and II B alleles were run separately. For each MHC class, a set of 56 individually tagged primer pairs were designed (the individual tags differed with at least three and on average five nucleotides). Both the forward and the reverse primer were tagged and had the following design: 5'CC-7 base ID-class-specific primer and were used for MHC amplification of each individual (see the electronic supplementary material, S1). Technical duplicates were performed for 11 individuals. The PCR products were purified, quantified and pooled. The amount of each target was adjusted for sequence coverage of 1500× per individual for MHC class I targets and 700× coverage for class II B targets. The 454-pyrosequencing was performed on a Roche 454 GS FLX platform following the manufacturer's instructions at Genoscope, Evry, France.
(c). Filtering of sequence data
The 454-pyrosequencing data were filtered to remove low-quality sequences (see the electronic supplementary material, S1), any reads representing artefactual MHC alleles or putatively non-functional sequences. Sequences with presence of the two primers (tolerating a maximum of two errors on each primer) with correct tags on both sides were kept and then all sequence reads with an occurrence lower than 10 in the whole dataset were removed (the low occurrence is a potential indication of artefactual sequences). Identical sequences were detected with the web-application seqeqseq (http://mbio-serv2.mbioekol.lu.se/apps/seqeqseq.html) and the number of reads for identical sequences were summarized for each individual with the web-application mergeMatrix (http://mbio-serv2.mbioekol.lu.se/apps/mergeMatrix.html). To remove more artefactual sequences derived from either PCR or sequencing errors, we used two of the customizable bioinformatic tools in the web-application popMatrix (http://mbio-serv2.mbioekol.lu.se/apps/popMatrix.html) designed for filtering of high-throughput sequencing data according to Galan et al. [37] and Babik et al. [38]. Data from individuals with low total sequence abundance potentially do not cover the full MHC genotype and was hence discarded. Erroneous, e.g. chimeric sequences, were considered to appear at a lower frequency than ‘true’ sequences and low relative abundance sequences within an individual was therefore discarded. The technical duplicates of 11 samples were used to determine the cut-off values for the filtering of the MHC I and II B datasets, respectively. This was done separately since the number of loci are different for the two MHC classes. Filter 1 (minimum total sequence abundance/individual) was set to 200 for MHC I and 175 for MHC II B. Filter 3 (minimum relative abundance of a sequence/individual) was set to 2 per cent for MHC I and 3 per cent for MHC II B. Also, only alleles that were verified in at least two independent PCR reactions and with an open reading frame (potentially expressed genes) were kept. Finally, before using the dataset in further analyses, additional potential chimeric sequences were identified per individual by eye in BioEdit v. 7.0.9 [39]. Potential PCR-chimeras were eliminated if they were a combination of two parental sequences and had lower than half the read number compared with any of the two parental sequences.
(d). Data analysis
The genetic distances between individuals were calculated separately for the two MHC classes by the following approach. A maximum-likelihood tree (one for each MHC class) was inferred for all verified and translated sequences using the RaxML software (v. 7.0.4) under the PROTMIX model and the JTT substitution matrix, with default settings [40]. These trees (MHC class I, electronic supplementary material, figure S1; MHC class II B, electronic supplementary material, figure S2) were used as references from which the phylogenetic distances between individual's translated MHC-sequence repertoires were calculated. For example, two individuals having exactly the same MHC alleles will share the same nodes and branches from root to terminal leafs in the reference tree, consequently they will have a phylogenetic distance of 0 per cent. In contrast, two individuals with sequences from different clades in the reference tree will only share nodes and branches from the basal parts of the phylogenetic topology, consequently they will have a phylogenetic distance more than 0 per cent. Accordingly, the pairwise distances between all individuals were estimated by using UniFrac, a phylogenetic comparison tool originally developed for measuring genetic distances between microbial communities [41,42].
The amino acid positions in each MHC molecule that bind to antigens, the peptide-binding regions (PBRs), were inferred from earlier studies [43,44] and were previously shown for blue petrel MHC sequences [36]. The chemical binding properties of the amino acids in the PBR of the verified sequences were described by five physico-chemical descriptor variables (z-descriptors) for each amino acid [45], and the resulting matrix was used to construct alternative maximum-likelihood trees with contml in the PHYLIP-package, v. 3.69. These trees represent clusters of functionally rather than evolutionary similar MHC sequences (MHC class I, electronic supplementary material, figure S3; MHC class II B, electronic supplementary material, figure S4). Functional MHC distances between individuals were calculated as described for the phylogenetic distances (UniFrac) but the RaxML-tree was replaced with the functional contml-tree.
To quantify any dissimilarity in allele composition on a pure shared/non-shared allele basis in pairs versus randomly set-up pairs, the PD was calculated: PD = Vab/(Fa + Fb), where Vab = total number of variable alleles present in individuals a and b (shared alleles count as 1), F = number of alleles within an individual.
To test if partners are more dissimilar at the MHC than expected from random pairs, differences in MHC amino acid distance, functional distance and allele sharing (PD) between true pairs and randomly set-up pairs were analysed. Bootstrap confidence tests were performed with 10 000 permutations. We let each female ‘randomly pair’ with one of the males without re-sampling in each round of the permutations and then a distance-average was calculated for each permutation (class I: females n = 50, males n = 50; class II B: females n = 48, males n = 48; in total 55 pairs, where 43 pairs are the same for both the class I and II analyses).
3. Results
(a). Major histocompatibility complex screening with 454-pyrosequencing
After removing potentially artefactual alleles and only keeping alleles that were verified in at least two independent PCR reactions with an open reading frame (potentially functional genes), we obtained 156 different MHC class I alleles (maximum number of alleles/individual = 16, mean/individual = 10.8 ± 2.4 s.d., n = 100) and 97 different MHC class II B alleles (maximum number of alleles/individual = 6, average 3.8 ± 0.7 s.d., n = 96) represented in the examined individuals (the sequences will be provided on request). After filtering, the reproducibility of alleles between technical duplicates (n = 11) was on average 80 per cent for MHC class I and 99 per cent for MHC class II B.
(b). Phylogenetic distances
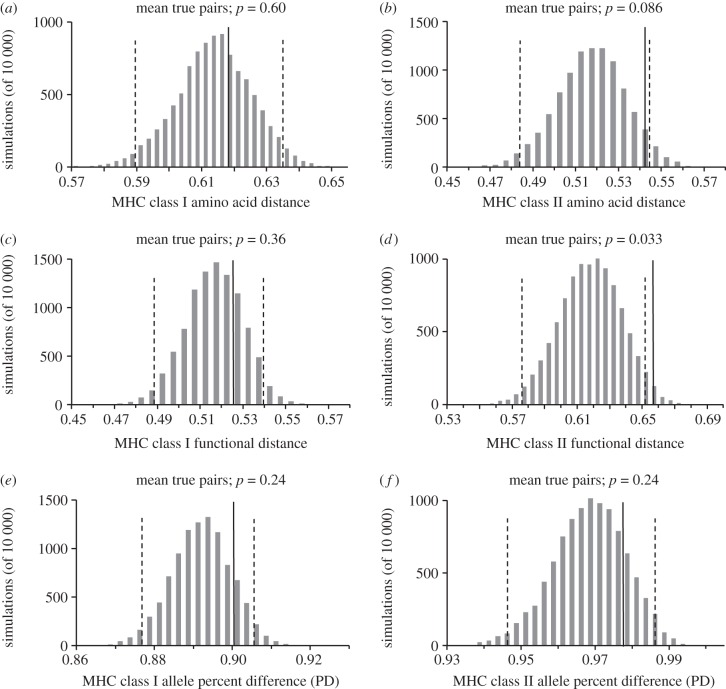
The difference in average total amino acid distances (based on the MHC-allele repertoire in each individual) was compared between females and males in true versus randomly picked pairs (with 10 000 permutations). For neither MHC class I nor class IIB, there was any difference in average amino acid distance between true and random pairs (bootstrap confidence two-tailed test; MHC class I, p = 0.60; MHC class II B, p = 0.086; figure 1a,b).
Figure 1.
Distribution of mean MHC phylogenetic distance, PBR functional distance and allele PD from 10 000 simulations of random pairings between blue petrel pair-females and all analysed males (grey bars) with two-tailed 95% CI (broken lines) compared with the mean distance/difference of true pairs (filled line). For MHC class I analyses, true pairs (n = 50), for MHC class II B analyses, true pairs (n = 48). (a) MHC class I phylogenetic amino acid distance (exon 3 sequences). (b) MHC class II B phylogenetic amino acid distance (exon 2 sequences). (c) MHC class I functional distance of exon 3 PBR [43]. (d) MHC class II B functional distance of exon 2 PBR [44]. (e) MHC class II PD. (f) MHC class B PD.
(c). Functional distances
The functional binding specificities of the amino acids in the PBR of an MHC molecule determine what pathogen antigens that can be bound and detected in that particular molecule. The average functional distance of each individuals' complete MHC repertoire was compared between true and random pairs, and again there was no difference for MHC class I between mean distances of true and random pairs (bootstrap confidence test, p = 0.36; figure 1c). However, the average true pair was significantly more distant/dissimilar functionally at the MHC class II B loci, than would be expected from random matings (bootstrap confidence test, p = 0.033; figure 1d).
(d). Allele sharing
The PD in shared MHC alleles between partners was not different between true and random pairs for either MHC class I (bootstrap confidence test, p = 0.24) or class II B (bootstrap confidence test, p = 0.24; figure 1e,f).
4. Discussion
Blue petrels choose mates with functionally dissimilar MHC class II B genes (figure 1d). This result supports mate compatibility theory where a genetically dissimilar mate should be chosen to maximize offspring heterozygosity [2,46,47]. A plausible explanatory driver for such disassortative mating is the benefit of high offspring MHC polymorphism that potentially allows recognition of a broader variety of pathogens compared with low MHC polymorphism [7,48,49]. Additionally, because MHC genes are highly polymorphic they can provide a system for kin recognition that generates further selection through the benefit of inbreeding avoidance [11,12]. Given the ecology of the blue petrel where individuals return to their nestling areas to breed with only one partner for life, such a mechanism would be beneficial [26]. Contrary to the choice for an MHC class II B-dissimilar partner, there was no evidence for disassortative mating on functional MHC class I characteristics (figure 1c). Neither was there any evidence for phylogenetic nor allele-sharing difference between partners for either MHC class I or II B (figure 1). Potentially, these measures are too blunt to detect any differences between partners and non-partners since the information used may not reflect biologically relevant differences.
The odour preferences for partners in blue petrels [35], suggests that an MHC-dissimilar partner could be detected using olfaction. Since a functionally different MHC genotype is chosen, the differences are likely to be perceivable between individuals. Hence, there are strong indications for an MHC-dependent odour-mediated mate choice in a bird, which to our knowledge has not been shown before. The link between personal odour, MHC and mate choice has been described in several taxa [12,50,51], whereas in birds such link has not been obvious, possibly because many examined species (e.g. passerines) are not known for extensive chemical communication or do not have well-developed olfactory systems [27,29]. Whether this could explain the lack of MHC-based mate choice among studied passerines [52] can only be speculated. On the other hand, alternative mating strategies are found in passerines, with an increased success in extra-pair matings when the social mate is MHC similar [53–55]. In the serial monogamous great frigatebird, Fregata minor (Pelicaniformes), a disassortative mating pattern for two MHC class II B loci was recently shown [56]. The addition of our findings, a remotely related species, may indicate that this mate choice strategy is a common feature among seabirds. However, unlike the blue petrel, odour-mediated mate discrimination has not yet been established for the great frigatebird [56].
The main source for individual olfactory discrimination in the blue petrel is the uropygial gland lipids (that are used for preening of the feathers) that differ chemically between individuals [33,57,58]. The interindividual differences of the odours could, given our results, be at least partly explained by individual MHC profiles as described in e.g. rodents [59,60]. Also the olfactory driven avoidance of self and preference for others, that were shown experimentally in the blue petrel [35], may well be mediated by MHC-dependent odour cues, especially since related individuals that de facto share more MHC alleles, smell more alike than unrelated individuals [61]. MHC-dependent odours have been suggested to indicate kinship also in e.g. mice and fish [62,63], which may also apply to these birds.
The contrasting mating pattern on MHC class I (not different from random expectations) and class II B (disassortative) in blue petrels could be an effect of the different targets of MHC class I (intracellular pathogens, mainly virus) and class II (extracellular pathogens, mainly bacteria) [1]. In the light of olfactory-driven mate choice, bacteria are known to influence individual odour [64], and therefore it is plausible that this could contribute to the proximate link between partner selection and MHC class II alleles as opposed to MHC class I alleles. MHC-dependent odour cues have been hypothesized to originate directly from the microbial communities that are harboured by different MHC types [65] or indirectly from microbial community metabolites [22]. In mice, the MHC profile contributes to the particular microbial community found in their scent marks [66]. The uropygial gland lipids, in the blue petrel, may also contain microbes that could modify these odour components to accentuate individual odours. Moreover, degradation of the gland components, which may be attributed to microbial breakdown, has been shown on the feathers of blue petrels [58]. Consequently, if the uropygial gland or feather microflora is determined by the MHC profile, it could contribute to the personal odour signature.
Our high-coverage screening data of two multi-loci MHC gene families derived from 454-pyrosequencing give the advantage of possibly representing all MHC class I and II B loci in each investigated individual. We are confident that the choice of method has helped to genotype the individuals more precisely than would have been possible with direct sequencing or cloning. Nevertheless, the reproducibility of the duplicated MHC class I samples was lower (80%) than for the MHC class II B samples (99%) which means that MHC class I alleles may have been missed in the genotyping. This in turn could have obscured any disassortative mating patterns on MHC class I. Still, if the differences were pronounced we should have detected it since there are on average 10.8 MHC class I alleles per individual.
It should be noted that our MHC class II result is based on one of the subunits of the MHC class II molecule (the β-unit) and not the MHC class II A (α-subunit), which may also influence choice on MHC class II. Future investigations could elucidate if the two subunits are equally important in partner choice. With the higher sequence coverage, we found evidence for at least three MHC class II B loci in the blue petrel genome which is one more than found with traditional methods [36]. However, we were not able to assign individual MHC alleles to specific loci, which is a well-known limitation when working with non-model birds [67]. To circumvent this problem, we adopted a novel approach for MHC comparison of all alleles simultaneously by using a tool originally developed for molecular distance assessment of microbial communities [41]. This approach might actually better mirror the reality in the choice situation as opposed to fine-scaled choice on particular loci, since all transcribed (functional) loci probably contribute to any perceivable MHC profile.
Whether the MHC class II dissimilarity between partners is a way to pursue genetic benefits in terms of pathogen resistance for the offspring or if MHC is used as an indicator for inbreeding avoidance deserves further investigation in this species. By developing protocols for genome-wide sequence comparisons, the MHC dissimilarity between partners could be contrasted to genome differences to address this question.
5. Conclusions
Our results support mate choice for MHC class II B-compatibility between social mates of blue petrels, providing evidence for MHC-dependent mate choice in birds. In contrast, this pattern was not found for MHC class I. The reasons for choosing an MHC-dissimilar partner include heterozygote advantage in pathogen resistance and/or inbreeding avoidance. The monogamous lifestyle of blue petrels makes an accurate mate choice particularly important. An odour-mediated mate choice is highly plausible in this species due to the occurrence of olfactory discrimination of individual odours and also odour preference for the partner in choice experiments. A potential mechanism for detection of MHC-dissimilar mates is that MHC-dependent bacteria (the target of MHC class II B) determine individual odour signatures. Future investigations should focus on detecting links between the individual odour signature, the bird microflora and the individual MHC profile in this species.
Acknowledgements
This work was supported by the Institut Polaire Francais Paul-Emile Victor (IPEV program ETHOTAAF no. 354 to F.B.), the Agence Nationale de la Recherche Francaise (AMBO ANR-08-BLAN-0117-01 to F.B.) and the Swedish Research Council (VR, 621-2006-4551 to H.W.). All aspects of the study were performed according to guidelines established by the IPEV and the CNRS for the ethical treatment of animals and complied with current French regulations. We are grateful to Jerome Mardon, Aurore Malapert and all the fieldworkers who helped through the program ETHOTAAF over the years. We thank Jörgen Ripa for writing a bootstrap script and Charlie Cornwallis as well as two anonymous reviewers for helpful suggestions on an earlier draft of this manuscript.
References
- 1.Janeway C. A. J., Travers P., Walport M., Shlomchik M. J. 2001. Immunobiology, the immune system in health and disease, 5th edn New York, NY: Garland Science [Google Scholar]
- 2.Tregenza T., Wedell N. 2000. Genetic compatibility, mate choice and patterns of parentage: invited review. Mol. Ecol. 9, 1013–1027 10.1046/j.1365-294x.2000.00964.x (doi:10.1046/j.1365-294x.2000.00964.x) [DOI] [PubMed] [Google Scholar]
- 3.Jordan W. C., Bruford M. W. 1998. New perspectives on mate choice and the MHC. Heredity 81, 127–133 10.1046/j.1365-2540.1998.00428.x (doi:10.1046/j.1365-2540.1998.00428.x) [DOI] [PubMed] [Google Scholar]
- 4.Bernatchez L., Landry C. 2003. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J. Evol. Biol. 16, 363–377 10.1046/j.1420-9101.2003.00531.x (doi:10.1046/j.1420-9101.2003.00531.x) [DOI] [PubMed] [Google Scholar]
- 5.Jeffery K. J. M., Bangham C. R. M. 2000. Do infectious diseases drive MHC diversity? Microbes Infect. 2, 1335–1341 10.1016/S1286-4579(00)01287-9 (doi:10.1016/S1286-4579(00)01287-9) [DOI] [PubMed] [Google Scholar]
- 6.Worley K., Collet J., Spurgin L. G., Cornwallis C., Pizzari T., Richardson D. S. 2010. MHC heterozygosity and survival in red junglefowl. Mol. Ecol. 19, 3064–3075 10.1111/j.1365-294X.2010.04724.x (doi:10.1111/j.1365-294X.2010.04724.x). [DOI] [PubMed] [Google Scholar]
- 7.Carrington M., et al. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283, 1748–1752 10.1126/science.283.5408.1748 (doi:10.1126/science.283.5408.1748) [DOI] [PubMed] [Google Scholar]
- 8.Penn D. J., Damjanovich K., Potts W. K. 2002. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl Acad. Sci. USA 99, 11 260–11 264 10.1073/pnas.162006499 (doi:10.1073/pnas.162006499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedrick P. W. 1988. HLA-sharing recurrent spontaneous abortion and the genetic hypothesis. Genetics 119, 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ober C., Hyslop T., Elias S., Weitkamp L. R., Hauck W. W. 1998. Human leukocyte antigen matching and fetal loss: results of a 10 year prospective study. Hum. Reprod. 13, 33–38 10.1093/humrep/13.1.33 (doi:10.1093/humrep/13.1.33) [DOI] [PubMed] [Google Scholar]
- 11.Penn D. J., Potts W. K. 1999. The evolution of mating preferences and major histocompatibility complex genes. Am. Nat. 153, 145–164 10.1086/303166 (doi:10.1086/303166) [DOI] [PubMed] [Google Scholar]
- 12.Penn D. J. 2002. The scent of genetic compatibility: sexual selection and the major histocompatibility complex. Ethology 108, 1–21 10.1046/j.1439-0310.2002.00768.x (doi:10.1046/j.1439-0310.2002.00768.x) [DOI] [Google Scholar]
- 13.Chaix R., Cao C., Donnelly P. 2010. Is mate choice in humans MHC-dependent? PLoS Genet. 6, e1000184. 10.1371/journal.pgen.1000184 (doi:10.1371/journal.pgen.1000184). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Setchell J. M., Charpentier M. J. E., Abbott K. M., Wickings E. J., Knapp L. A. 2010. Opposites attract: MHC-associated mate choice in a polygynous primate. J. Evol. Biol. 23, 136–148 10.1111/j.1420-9101.2009.01880.x (doi:10.1111/j.1420-9101.2009.01880.x) [DOI] [PubMed] [Google Scholar]
- 15.Schwensow N., Eberle M., Sommer S. 2008. Compatibility counts: MHC-associated mate choice in a wild promiscuous primate. Proc. R. Soc. B 275, 555–564 10.1098/rspb.2007.1433 (doi:10.1098/rspb.2007.1433). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamazaki K., Boyse E. A., Mike V., Thaler H. T., Mathieson B. J., Abbott J., Boyse J., Zayas Z. A., Thomas L. 1976. Control of mating preferences in mice by genes in the major histo compatibility complex. J. Exp. Med. 144, 1324–1335 10.1084/jem.144.5.1324 (doi:10.1084/jem.144.5.1324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egid K., Brown J. L. 1989. The major histocompatibility complex and female mating preferences in mice. Anim. Behav. 38, 548–550 10.1016/S0003-3472(89)80051-X (doi:10.1016/S0003-3472(89)80051-X) [DOI] [Google Scholar]
- 18.Olsson M., Madsen T., Nordby J., Wapstra E., Ujvari B., Wittsell H. 2003. Major histocompatibility complex and mate choice in sand lizards. Proc. R. Soc. Lond. B 270(Suppl. 2), S254–S256 10.1098/rsbl.2003.0079 (doi:10.1098/rsbl.2003.0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller H. C., Moore J. A., Nelson N. J., Daugherty C. H. 2009. Influence of major histocompatibility complex genotype on mating success in a free-ranging reptile population. Proc. R. Soc. B 276, 1695–1704 10.1098/rspb.2008.1840 (doi:10.1098/rspb.2008.1840) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landry C., Garant D., Duchesne P., Bernatchez L. 2001. ‘Good genes as heterozygosity’: the major histocompatibility complex and mate choice in Atlantic salmon (Salmo salar). Proc. R. Soc. Lond. B 268, 1279–1285 10.1098/rspb.2001.1659 (doi:10.1098/rspb.2001.1659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Consuegra S., Garcia de Leaniz C. 2008. MHC-mediated mate choice increases parasite resistance in salmon. Proc. R. Soc. B 275, 1397–1403 10.1098/rspb.2008.0066 (doi:10.1098/rspb.2008.0066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penn D., Potts W. 1998. How do major histocompatibility genes influence odor and mating preferences? Adv. Immunol. 69, 411–435 10.1016/S0065-2776(08)60612-4 (doi:10.1016/S0065-2776(08)60612-4) [DOI] [PubMed] [Google Scholar]
- 23.Huchard E., Knapp L. A., Wang J., Raymond M., Cowlishaw G. 2010. MHC, mate choice and heterozygote advantage in a wild social primate. Mol. Ecol. 19, 2545–2561 10.1111/j.1365-294X.2010.04644.x (doi:10.1111/j.1365-294X.2010.04644.x) [DOI] [PubMed] [Google Scholar]
- 24.Roberts S. C., Gosling L. M. 2003. Genetic similarity and quality interact in mate choice decisions by female mice. Nat. Genet. 35, 103–106 10.1038/ng1231 (doi:10.1038/ng1231) [DOI] [PubMed] [Google Scholar]
- 25.Andersson M. 1994. Monographs in behavior and ecology: sexual selection, 599 p. Princeton, NJ: Princeton University Press. [Google Scholar]
- 26.Zelano B., Edwards S. V. 2002. An MHC component to kin recognition and mate choice in birds: predictions, progress, and prospects. Am. Nat. 160(Suppl. 6), S225–S237 10.1086/342897 (doi:10.1086/342897) [DOI] [PubMed] [Google Scholar]
- 27.Bang B. G., Cobb S. 1968. The size of the olfactory bulb in 108 species of birds. Auk 85, 55–61 [Google Scholar]
- 28.Warham J. 1996. The behaviour, population biology and physiology of the petrels. London, UK: Academic Press [Google Scholar]
- 29.Zelenitsky D. K., Therrien F. O., Ridgely R. C., McGee A. R., Witmer L. M. 2011. Evolution of olfaction in non-avian theropod dinosaurs and birds. Proc. R. Soc. B 278, 3625–3634 10.1098/rspb.2011.0238 (doi:10.1098/rspb.2011.0238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonadonna F., Nevitt G. A. 2004. Partner-specific odor recognition in an Antarctic seabird. Science 306, 835. 10.1126/science.1103001 (doi:10.1126/science.1103001) [DOI] [PubMed] [Google Scholar]
- 31.Brooke M. 2004. Albatrosses and petrels across the world. New York, NY: Oxford University Press Inc [Google Scholar]
- 32.Bonadonna F., Caro S., Jouventin P., Nevitt G. A. 2006. Evidence that blue petrel, Halobaena caerulea, fledglings can detect and orient to dimethyl sulfide. J. Exp. Biol. 209, 2165–2169 10.1242/jeb.02252 (doi:10.1242/jeb.02252) [DOI] [PubMed] [Google Scholar]
- 33.Mardon J., Saunders S. M., Anderson M. J., Couchoux C., Bonadonna F. 2010. Species, gender, and identity: cracking petrels’ sociochemical code. Chem. Senses 35, 309–321 10.1093/chemse/bjq021 (doi:10.1093/chemse/bjq021) [DOI] [PubMed] [Google Scholar]
- 34.Bonadonna F., Villafane M., Bajzak C., Jouventin P. 2004. Recognition of burrow's olfactory signature in blue petrels, Halobaena caerulea: an efficient discrimination mechanism in the dark. Anim. Behav. 67(Part 5), 893–898 10.1016/j.anbehav.2003.08.013 (doi:10.1016/j.anbehav.2003.08.013) [DOI] [Google Scholar]
- 35.Mardon J., Bonadonna F. 2009. Atypical homing or self-odour avoidance? Blue petrels (Halobaena caerulea) are attracted to their mate's odour but avoid their own. Behav. Ecol. Sociobiol. 63, 537–542 10.1007/s00265-008-0688-z (doi:10.1007/s00265-008-0688-z) [DOI] [Google Scholar]
- 36.Strandh M., Lannefors M., Bonadonna F., Westerdahl H. 2011. Characterization of MHC class I and II genes in a subantarctic seabird, the blue petrel, Halobaena caerulea (Procellariiformes). Immunogenetics 63, 653–666 10.1007/s00251-011-0534-8 (doi:10.1007/s00251-011-0534-8) [DOI] [PubMed] [Google Scholar]
- 37.Galan M., Guivier E., Caraux G., Charbonnel N., Cosson J.-F. 2010. A 454 multiplex sequencing method for rapid and reliable genotyping of highly polymorphic genes in large-scale studies. BMC Genomics 11, 296. 10.1186/1471-2164-11-296 (doi:10.1186/1471-2164-11-296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babik W., Taberlet P., Ejsmond M. J., Radwan J. 2009. New generation sequencers as a tool for genotyping of highly polymorphic multilocus MHC system. Mol. Ecol. Resour. 9, 713–719 10.1111/j.1755-0998.2009.02622.x (doi:10.1111/j.1755-0998.2009.02622.x) [DOI] [PubMed] [Google Scholar]
- 39.Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 [Google Scholar]
- 40.Stamatakis A., Hoover P., Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771 10.1080/10635150802429642 (doi:10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- 41.Lozupone C., Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 10.1128/aem.71.12.8228-8235.2005 (doi:10.1128/aem.71.12.8228-8235.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lozupone C., Lladser M. E., Knights D., Stombaugh J., Knight R. 2011. UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172 10.1038/ismej.2010.133 (doi:10.1038/ismej.2010.133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallny H.-J., et al. 2006. Peptide motifs of the single dominantly expressed class I molecule explain the striking MHC-determined response to Rous sarcoma virus in chickens. Proc. Natl Acad. Sci. USA 103, 1434–1439 10.1073/pnas.0507386103 (doi:10.1073/pnas.0507386103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown J. H., Jardetzky T. S., Gorga J. C., Stern L. J., Urban R. G., Strominger J. L., Wiley D. C. 1993. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364, 33–39 10.1038/364033a0 (doi:10.1038/364033a0) [DOI] [PubMed] [Google Scholar]
- 45.Sandberg M., Eriksson L., Jonsson J., Sjöström M., Wold S. 1998. New chemical descriptors relevant for the design of biologically active peptides: a multivariate characterization of 87 amino acids. J. Med. Chem. 41, 2481–2491 10.1021/jm9700575 (doi:10.1021/jm9700575) [DOI] [PubMed] [Google Scholar]
- 46.Brown J. L. 1997. A theory of mate choice based on heterozygosity. Behav. Ecol. 8, 60–65 10.1093/beheco/8.1.60 (doi:10.1093/beheco/8.1.60) [DOI] [Google Scholar]
- 47.Kempenaers B. 2007. Mate choice and genetic quality: a review of the heterozygosity theory. In Advances in the study of behavior (eds Brockmann H. J., Roper T. J., Naguib M., WynneEdwards K. E., Barnard C., Mitani J.), pp. 189–278 San Diego, CA: Elsevier Academic Press. [Google Scholar]
- 48.O'Connor S. L., et al. 2010. MHC heterozygote advantage in simian immunodeficiency virus-infected Mauritian cynomolgus macaques. Sci. Transl. Med. 2, 22ra18. 10.1126/scitranslmed.3000524 (doi:10.1126/scitranslmed.3000524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeshima S., Matsumoto Y., Chen J., Yoshida T., Mukoyama H., Aida Y. 2008. Evidence for cattle major histocompatibility complex (BoLA) class II DQA1 gene heterozygote advantage against clinical mastitis caused by Streptococci and Escherichia species. Tissue Antigens 72, 525–531 10.1111/j.1399-0039.2008.01140.x (doi:10.1111/j.1399-0039.2008.01140.x) [DOI] [PubMed] [Google Scholar]
- 50.Carroll L. S., Penn D. J., Potts W. K. 2002. Discrimination of MHC-derived odors by untrained mice is consistent with divergence in peptide-binding region residues. Proc. Natl Acad. Sci. USA 99, 2187–2192 10.1073/pnas.042244899 (doi:10.1073/pnas.042244899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wedekind C., Penn D. 2000. MHC genes, body odours, and odour preferences. Nephrol. Dial. Transplant. 15, 1269–1271 10.1093/ndt/15.9.1269 (doi:10.1093/ndt/15.9.1269) [DOI] [PubMed] [Google Scholar]
- 52.Westerdahl H. 2004. No evidence of an MHC-based female mating preference in great reed warblers. Mol. Ecol. 13, 2465–2470 10.1111/j.1365-294X.2004.02238.x (doi:10.1111/j.1365-294X.2004.02238.x) [DOI] [PubMed] [Google Scholar]
- 53.Promerova M., Vinkler M., Bryja J., Polakova R., Schnitzer J., Munclinger P., Albrecht T. 2011. Occurrence of extra-pair paternity is connected to social male's MHC-variability in the scarlet rosefinch Carpodacus erythrinus. J. Avian Biol. 42, 5–10 10.1111/j.1600-048X.2010.05221.x (doi:10.1111/j.1600-048X.2010.05221.x) [DOI] [Google Scholar]
- 54.Richardson D. S., Komdeur J., Burke T., von Schantz T. 2005. MHC-based patterns of social and extra-pair mate choice in the Seychelles warbler. Proc. R. Soc. B 272, 759–767 10.1098/rspb.2004.3028 (doi:10.1098/rspb.2004.3028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freeman-Gallant C. R., Meguerdichian M., Wheelwright N. T., Sollecito S. V. 2003. Social pairing and female mating fidelity predicted by restriction fragment length polymorphism similarity at the major histocompatibility complex in a songbird. Mol. Ecol. 12, 3077–3083 10.1046/j.1365-294X.2003.01968.x (doi:10.1046/j.1365-294X.2003.01968.x) [DOI] [PubMed] [Google Scholar]
- 56.Juola F. A., Dearborn D. C. 2011. Sequence-based evidence for major histocompatibility complex-disassortative mating in a colonial seabird. Proc. R. Soc. B 279, 153–162 10.1098/rspb.2011.0562 (doi:10.1098/rspb.2011.0562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonadonna F., Miguel E., Grosbois V., Jouventin P., Bessiere J.-M. 2007. Individual odor recognition in birds: an endogenous olfactory signature on petrels’ feathers? J. Chem. Ecol. 33, 1819–1829 10.1007/s10886-007-9345-7 (doi:10.1007/s10886-007-9345-7) [DOI] [PubMed] [Google Scholar]
- 58.Mardon J., Saunders S. M., Bonadonna F. 2011. From preen secretions to plumage: the chemical trajectory of blue petrels’ Halobaena caerulea social scent. J. Avian Biol. 42, 29–38 10.1111/j.1600-048X.2010.05113.x (doi:10.1111/j.1600-048X.2010.05113.x) [DOI] [Google Scholar]
- 59.Brown R. E., Roser B., Singh P. B. 1989. Class I and class II regions of the major histocompatibility complex both contribute to individual odors in congenic inbred strains of rats. Behav. Genet. 19, 659–674 10.1007/BF01066029 (doi:10.1007/BF01066029) [DOI] [PubMed] [Google Scholar]
- 60.Novotny M. V., Soini H. A., Koyama S., Wiesler D., Bruce K. E., Penn D. J. 2007. Chemical identification of MHC-influenced volatile compounds in mouse urine. I. Quantitative proportions of major chemosignals. J. Chem. Ecol. 33, 417–434 10.1007/s10886-006-9230-9 (doi:10.1007/s10886-006-9230-9) [DOI] [PubMed] [Google Scholar]
- 61.Celerier A., Bon C., Malapert A., Palmas P., Bonadonna F. 2011. Chemical kin label in seabirds. Biol. Lett. 7, 807–810 10.1098/rsbl.2011.0340 (doi:10.1098/rsbl.2011.0340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olsén K. H., Grahn M., Lohm J., Langefors A. 1998. MHC and kin discrimination in juvenile Arctic charr, Salvelinus alpinus (L.). Anim. Behav. 56, 319–327 10.1006/anbe.1998.0837 (doi:10.1006/anbe.1998.0837) [DOI] [PubMed] [Google Scholar]
- 63.Yamazaki K., Beauchamp G. K., Kupniewski D., Bard J., Thomas L., Boyse E. A. 1988. Familial imprinting determines H-2 selective mating preferences. Science 240, 1331–1332 10.1126/science.3375818 (doi:10.1126/science.3375818) [DOI] [PubMed] [Google Scholar]
- 64.Leyden J. J., McGinley K. J., Holzle E., Labows J. N., Kligman A. M. 1981. Microbiology of the human axilla and its relationship to axillary odor. J. Invest. Dermatol. 77, 413–416 10.1111/1523-1747.ep12494624 (doi:10.1111/1523-1747.ep12494624). [DOI] [PubMed] [Google Scholar]
- 65.Howard J. C. 1977. H-2 and mating preferences. Nature 266, 406–408 10.1038/266406a0 (doi:10.1038/266406a0) [DOI] [Google Scholar]
- 66.Lanyon C. V., Rushton S. P., O'Donnell A. G., Goodfellow M., Ward A. C., Petrie M., Jensen S. P., Gosling L. M., Penn D. J. 2007. Murine scent mark microbial communities are genetically determined. FEMS Microbiol. Ecol. 59, 576–583 10.1111/j.1574-6941.2006.00252.x (doi:10.1111/j.1574-6941.2006.00252.x) [DOI] [PubMed] [Google Scholar]
- 67.Westerdahl H., Wittzell H., von Schantz T. 2000. Mhc diversity in two passerine birds: no evidence for a minimal essential MHC. Immunogenetics 52, 92–100 10.1007/s002510000256 (doi:10.1007/s002510000256) [DOI] [PubMed] [Google Scholar]