Abstract
The enlargement of cetacean brain size represents an enigmatic event in mammalian evolution, yet its genetic basis remains poorly explored. One candidate gene associated with brain size evolution is the abnormal spindle-like microcephaly associated (ASPM), as mutations in this gene cause severe reductions in the cortical size of humans. Here, we investigated the ASPM gene in representative cetacean lineages and previously published sequences from other mammals to test whether the expansion of the cetacean brain matched adaptive ASPM evolution patterns. Our analyses yielded significant evidence of positive selection on the ASPM gene during cetacean evolution, especially for the Odontoceti and Delphinoidea lineages. These molecular patterns were associated with two major events of relative brain size enlargement in odontocetes and delphinoids. It is of particular interest to find that positive selection was restricted to cetaceans and primates, two distant lineages both characterized by a massive expansion of brain size. This result is suggestive of convergent molecular evolution, although no site-specific convergence at the amino acid level was found.
Keywords: cetaceans, ASPM, positive selection, brain size enlargement
1. Introduction
There has been a dramatic expansion in brain size during mammalian evolution [1,2]. Primates and cetaceans are the most remarkable examples of such massive brain size expansion, especially of the cerebral cortex—the brain area associated with higher cognitive functions [3]. Previous research indicates that the evolution of larger brain sizes in these groups has been driven by selection for life in increasingly complex social environments and the need for highly developed cognitive abilities [4,5]. Accordingly, the primate brain, and especially the human brain, has been recognized as one of the most striking evolutionary adaptations [6,7]. However, it is still unclear whether this is also the case for cetaceans.
Cetaceans diverged from terrestrial artiodactyls approximately 56–53 million years ago (Ma) [8], representing one of the most enigmatic events in evolution. Early cetaceans (called archaeocetes) diversified through amphibious stages to become fully aquatic by 40 Ma [9]. Extant cetaceans, consisting of highly diversified species subdivided into two suborders, Mysticeti (large rorqual and baleen whales) and Odontoceti (dolphins, porpoises and toothed whales), evolved from archaeocetes at about 34 Ma and dispersed into the world's oceans and estuaries and even some rivers [10,11]. It has been proposed that the transition from land to water selected for the development of distinct, and in some cases more complex, physiology, morphology, sensorial and cognitive traits, such as underwater vision [12] and hearing ability [13], and comprehension of artificial communication systems [14]. Therefore, relatively larger brain sizes were favoured during this transition. Indeed, paleontological and neuroanatomical data have confirmed that modern cetacean brains are among the largest across all mammals both in absolute and relative size (expressed relative to body size as encephalization level or quotient, EQ [1]). Almost all odontocetes (toothed whales) have above average levels of encephalization compared with relative terrestrial mammals. In particular, some odontocetes possess EQs in the range of 4–5, second only to modern humans (EQ ≈ 7) and significantly higher than any of the modern nonhuman anthropoid primates (highest EQ ≈ 3.3) [15,16]. Recent studies have contributed substantially towards the identification of the putative genetic basis underlying the enlarged brain of primates [6,7,17,18]. The most striking finding in this regard was that positive selection or accelerated evolution was found in a group of genes associated with primary microcephaly (MCPH), a developmental defect characterized by a severe reduction in brain size. Seven such loci (MCPH 1–MCPH 7) with recessive mutations that lead to MCPH have been identified to date (reviewed in [19]). Of these, MCPH 5 (or abnormal spindle-like microcephaly associated, ASPM) is essential for normal mitotic spindle function in embryonic neuroblasts, with ASPM mutations being the most common cause of MCPH in a clinical sample of humans [17,19,20]. Additionally, the functions of Drosophila and mouse orthologues of ASPM have also certified that mutations in ASPM reduce brain size (reviewed in [19]). Interestingly, there is strong evidence that some nucleotide changes in ASPM were subject to positive selection in the lineage leading to humans, consistant with a possible role of ASPM in the evolutionary enlargement of human brain size [6,17,20]. Still, the investigation of the genetic basis underlying brain size evolution in cetaceans has just begun very recently, although the adaptive significance and anatomical basis of their brain diversity has long been studied [5,15,16,21,22]. A recent study investigated the MCPH1 gene in cetaceans [23], but did not find compelling evidence of an association between its evolution and brain size enlargement in the group.
In this study, the ASPM gene was investigated in representative species of major cetacean lineages and compared with corresponding sequences from other mammals to determine its contribution to brain size enlargement during cetacean evolution. First, we tested whether positive selection on the ASPM gene across cetacean phylogeny corresponded to brain size expansion events in this group. Second, in comparison with homologous sequences of other mammals, we evaluated whether different cetacean clades or other mammals have experienced different selective regimes, particularly whether cetaceans and primates had similar adaptations (or convergent evolution) at the molecular level.
2. Material and methods
(a). Taxonomic coverage
Fourteen cetacean species from eight families, including nine species of the superfamily Delphinoidea, which includes the highest EQ, were sequenced (see the electronic supplementary material, table S1). In addition, ASPM sequences of nine primates and one artiodactyl (Hippopotamus Hippopotamus amphibius) were downloaded from GenBank (see the electronic supplementary material, table S1). We also retrieved Ensembl-predicted ASPM sequences from published genomes (http://genome.ucsc.edu/ and http://asia.ensembl.org/index.html) of an additional eight mammal species from six orders. In total, 32 ASPM sequences of high quality and integrity were used (see the electronic supplementary material, table S1 for information on sequence data and accession numbers).
(b). Amplification and sequencing of cetacean ASPM
We designed primers for the two largest ASPM exons (exons 3 and 18; approx. 60% of the transcribed ASPM protein) based on the alignment of the genomic data for the cow, horse and bottlenose dolphin (Tursiops truncatus). These exons were selected as they are characterized by the highest concentration of non-synonymous substitutions, possess most of the mutations that cause human primary microcephaly [17,20] and encompass the main functional sites of the protein [24].
Genomic DNA was extracted from myologic samples by using the DNeasy tissue kit (Qiagen), following the manufacturer's protocol. Polymerase chain reaction (PCR) was carried out in a total volume of 50 μl comprising 2.5 mM MgCl2, 10 mM Tris–HCl (pH 8.4), 50 mM KCl, 0.2 mM of each dNTP, 0.4 μM of each primer, 1.0 unit Ex-Taq DNA polymerase (Takara) and 10–100 ng DNA template. The amplification profile consisted of 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C and 30 s at 72°C, with a final extension of 8 min at 72°C. The amplified PCR products were purified and sequenced in both directions with an ABI 3730 automated genetic analyser.
(c). Cetacean ASPM gene polymorphism
Nucleotide and deduced amino acid sequences of the mammalian ASPM gene were aligned using the Clustal X v. 1.83 program [25]. Average pairwise nucleotide distances (Kimura 2-parameter model, K2P) and Poisson-corrected amino acid distances were computed using MEGA v. 5 [26]. Standard errors of estimates were obtained by running 1000 bootstrap replicates.
(d). Test for selection on the ASPM gene
The non-synonymous to synonymous rate ratio ω (dN/dS) indicates changes in selective pressures, where ω = 1, ω < 1 and ω > 1 correspond to neutral evolution, purifying and positive selection, respectively [27]. The ω ratio was estimated using a codon-based maximum-likelihood method implemented in CODEML program of PAML v. 4.4 package [28]. A well-accepted phylogeny of Laurasiatheria and primates was used as the input tree in all analyses. The topology of Laurasiatherian mammals was based on analyses of approximately 2.1 M base pairs (bp) of 1608 genes from 15 mammalian species [29] and approximately 1.4 M bp of 110 genes from 15 cetacean species [30], whereas the phylogeny of primates was based on approximately 8 M bp of 54 nuclear genes from 186 species [31]. We also used the concatenated two-exon ASPM dataset to estimate the phylogenetic relationships using maximum-likelihood (ML) and Bayesian inference (see the electronic supplementary material, appendix S1). The resulting tree was similar to the well-accepted phylogeny, with only some minor differences within Delphinidae (see the electronic supplementary material, appendix S1 and figure S1). The analyses of selection using the ASPM tree produced results nearly identical to those obtained using the well-accepted phylogeny; hence, only the latter analysis is reported.
A combination of branch, site and clade models was used to analyse the datasets including all mammals or only cetaceans. In the case of the branch model, the ‘free-ratios’ model (M1), which assumes an independent ω ratio for each branch, was compared with the ‘one-ratio’ model (M0), which assumes the same ω ratio for all branches [32]. Subsequently, to identify those sites under positive selection for the ASPM gene, site models in which ω can vary among sites were implemented. Specially, two pairs of site models were tested: M1a (nearly neutral: ω0 < 1, ω1 = 1) versus M2a (positive selection: ω0 < 1, ω1 = 1, ω2 > 1) and M8a (nearly neutral; beta distribution: 0 < ω0 < 1 and ω1 = 1) versus M8 (positive selection; beta distribution: 0 < ω0 < 1 and ω1 > 1) [33] (details on models M1a and M2a in [34]). Finally, to detect divergent selection acting on groups of related key taxa, a pair model (Clade model C versus M1a) was implemented in each dataset. Three site classes are assumed in the clade model C, which includes two clades, i.e. focal clade and the background clade. Site class 0 and 1 separately represent purifying selection (0 < ω0 < 1) and neutral evolution (ω1 = 1), whereas in site class 2, branches in the two clades are evolving with ω2 and ω3 (ω2 ≠ ω3), respectively [34]. Because clade model C does not perform well without an outgroup [28], hyrax as well as cow + hippopotamus were used as outgroups for the datasets, including ‘all mammals’ and ‘all cetacean’, respectively. In the latter case, clade models were separately undertaken for odontocetes (branch A), delphinoids (branch B), delphinids (branch C) and mysticetes (branch D), whereas, in the former case, the clade models were determined for all cetaceans (branch I), primates (branch II), Whippomorpha (whale + hippo, branch III), carnivores (branch IV) and eulipotyphlans (branch V). In addition, the selection pattern on the cetacean clade was estimated, considering all mammals with the exception of primates in order to exclude the effect of primates.
The significance of differences between the two nested models was evaluated using likelihood ratio tests (LRTs) by calculating twice the log-likelihood (2ΔL) of the difference following a chi-square distribution, and the degrees of freedom were the difference in the number of free parameters between models. For all PAML-based analyses, all models corrected transition and transversion rates and codon usage biases (F3 × 4). Different starting ω values were also used to avoid the local optima on the likelihood surface [35]. Considering that selection analyses including alignment gaps (setting clean data = 0) was basically the same as that of removing alignment gaps (setting clean data = 1, see the electronic supplementary material, table S2); thus, only the former analysis is presented here.
To evaluate the probabilities of positively selected sites on ASPM for the cetacean species examined, we first used a Bayes empirical Bayes (BEB) analysis to calculate posterior probabilities of positively selected sites implemented in the CODEML program of PAML v. 4.4 package [28]. Those sites with a posterior probability > 0.8 were considered as candidates for selection. We also used the fixed effects likelihood (FEL) method implemented in the DATAMONKEY web server [36] to infer positive selection sites with the default settings and a significance level of 0.2. The DATAMONKEY has the advantage that they can improve the estimation of the dN/dS ratio by incorporating variation in the rate of synonymous substitution [36]. Finally, only positively selected sites detected by both the M8 and FEL methods were used to estimate conservative or radical changes between ancestral and present ASPM sequences. Ancestral ASPM sequences were inferred based on empirical Bayesian methods implemented in the CODEML program of the PAML v. 4.4 package [28]. Conservative or radical non-synonymous substitutions were estimated according to charge, polarity and volume.
3. Results
(a). Characterization of the cetacean ASPM gene
All ASPM gene sequences from the cetacean species were intact, without premature stop codons or frame shift mutations, thus suggesting the presence of a functional ASPM protein. The sequences from the two exons of the ASPM genes (exon 3 with 1494 bp and a predicted amino acid sequence of 498 amino acid (aa), and exon 18, with 4710 bp and 1570 aa) were examined in all 14 cetacean species. In total, 6204 bp (2068 aa) were sequenced, representing 59.48 per cent of the coding region according to the human ASPM gene (3477 aa). An alignment of 2068 aa revealed a total of 394 variable sites (19.05%) and two separate amino acid deletions in the minke whale (Balaenoptera acutorostrata) and Omura's whale (B. omurai; see the electronic supplementary material, figure S2). In addition, the alignment of the dataset including the 14 newly sequenced cetaceans and the published sequences of 17 additional mammal species revealed that protein translations ranged from 1896 aa (in the horse) to 2081 aa (in the macaque).
(b). Signatures of positive selection
The site model analyses of all mammals (dataset I: 31 species) showed that models that incorporate selection (i.e. M2a and M8) fitted significantly better than neutral models (i.e. M1a and M8a; table 1). With model M8, the most stringent model implemented in PAML, a small proportion of codons (4.98% or 103 codons) was estimated to be under selection, with a ω value of 1.906. Of those sites under positive selection, 16 and 14 were identified by the BEB approach with posterior probabilities above 0.8 and 0.9, respectively. When only the cetaceans (dataset II: 14 species) were considered, the associated LRT reached statistical significance (M8a versus M8: p = 0.050) or is very close to statistical significance (M2a versus M1a: p = 0.052) in the site model analysis, with an estimated ω value of 5.780–5.983 at 0.87–2.47% sites at this loci (table 1). For M8, 10 codons were identified by the BEB approach with posterior probabilities above 0.8, whereas 17 codons were identified as under selection by FEL method. Of these putative positively selected sites, nine were picked simultaneously by both methods. These nine codons were therefore further investigated their radical or conservative nature (table 2). Seven of these (77.78%) were identified as having undergone radical changes: six in the suborder Odontoceti (including three in the superfamily Delphinoidea) and one in the suborder Mysticeti.
Table 1.
CODEML analyses of selective pattern on the ASPM gene in mammals (including alignment gaps: clean data = 0).
| models | −lnL | models compared | 2ΔLnL | d.f. | p-value | parameter estimates |
|---|---|---|---|---|---|---|
| dataset I: all mammals (31 sequences) | ||||||
| branch models | ||||||
| M0: one-ratio | 41 060.847 | ω = 0.440 | ||||
| M1: free-ratio | 41 006.339 | M0 versus M1 | 109.016 | 59 | < 0.001 | ω variation for each branch |
| site models | ||||||
| M1a (nearly neutral) | 40 619.593 | p0 = 0.628, p1 = 0.372, κ(kappa) = 3.69, ω0 = 0.189, ω1 = 1 | ||||
| M2a (positive selection) | 40 605.499 | M2a versus M1a | 28.189 | 2 | <0.001 | p0 = 0.618, p1 = 0.365, p2 = 0.013, κ = 3.755, ω0 = 0.193, ω1 = 1, ω2 = 3.025 |
| M8a | 40 603.433 | p0 = 0.719, p1 = 0.281, p = 1.279, q = 3.750, K = 3.635, ω = 1 | ||||
| M8 (β and ω) | 40 587.596 | M8a versus M8 | 31.674 | 2 | <0.001 | p0 = 0.946, p = 0.737, q = 1.058, p1 = 0.054, K = 3.693 ω = 1.906 |
| clade model C (31 sequences + hyrax as outgroup) | ||||||
| M1a′ (with outgroup hyrax) | 43 291.790 | ω = 0.193 | ||||
| cetaceans only (branch I) | 43 277.239 | M1a′ versus branch I | 29.102 | 3 | <0.001 | ω0 = 0.197, p0 = 0.622, ω1 = 1, p1 = 0.363, ω2 = 3.076, ω3 = 1.470, p2 = 0.015 |
| primates only (branch II) | 43 278.271 | M1a′ versus branch II | 27.038 | 3 | <0.001 | ω0 = 0.197, p0 = 0.622, ω1 = 1, p1 = 0.364, ω2 = 2.527, ω3 = 3.782, p2 = 0.014 |
| whippomorpha only (branch III) | 43 264.914 | M1a′ versus branch III | 53.752 | 3 | <0.001 | ω0 = 0.047, p0 = 0.215, ω1 = 1, p1 = 0.299, ω2 = 0.297, ω3 = 0.436, p2 = 0.486 |
| carnivores only (branch IV) | 43 269.979 | M1a′ versus branch IV | 43.622 | 3 | <0.001 | ω0 = 0.038, p0 = 0.193, ω1 = 1, p1 = 0.304, ω2 = 0.309, ω3 = 0.276, p2 = 0.503 |
| eulipotyphlans only (branch V) | 43 270.040 | M1a′ versus branch V | 43.5 | 3 | <0.001 | ω0 = 0.036, p0 = 0.190, ω1 = 1, p1 = 0.305, ω2 = 0.310, ω3 = 0.287, p2 = 0.505 |
| clade model C (22 sequences, all mammals except primates) | ||||||
| M1a′ (with Hyrax as outgroup) | −35 922.456 | ω = 0.179 | ||||
| cetaceans only (branch I) | −35 901.087 | M1a″ versus branch I | 42.738 | 3 | <0.001 | ω0 = 0.148, p0 = 0.524, ω1 = 1, p1 = 0.340, ω2 = 0.384, ω3 = 1.778, p2 = 0.136 |
| dataset II: cetaceans only (14 sequences) | ||||||
| branch models | ||||||
| M0: one-ratio | 13 327.673 | ω = 0.622 | ||||
| M1: free-ratio | 13 313.734 | M1 versus M0 | 27.878 | 25 | 0.314 | ω variation for each branch |
| site models | ||||||
| M1a (nearly neutral) | 13 311.637 | ω0 = 0, ω1 = 1 | ||||
| M2a (positive selection) | 13 308.679 | M2a versus M1a | 5.916 | 2 | 0.052 | p0 = 0.431, p1 = 0.559, p2 = 0.011, K = 4.334, ω0 = 0.056, ω1 = 1, ω2 = 5.983 |
| M8a | 13 311.637 | p0 = 0.403, p1 = 0.597, p = 0.005, q = 2.304 K = 4.334, ω = 1 | ||||
| M8 (β and ω) | 13 308.661 | M8a versus M8 | 5.952 | 2 | 0.050 | p0 = 0.988, p = 0.099, q = 0.070, (p1 = 0.012) ω = 5.780 K = 4.332 |
| clade model C (14 sequences + cow and hippopotamus as outgroup) | ||||||
| M1a′ (with cow and hippopotamus as outgroup) | 18 082.652 | p0 = 0.576, p1 = 0.424, K = 3.828, ω0 = 0.157, ω1 = 1 | ||||
| toothed whales (branch A) | 18 075.535 | M1a′ versus branch A | 14.234 | 3 | 0.003 | ω0 = 0.257, p0 = 0.746, ω1 = 1, p1 = 0.076, ω2 = 1.308, ω3 = 2.018, p2 = 0.178 |
| delphinoids (branch B) | 18 067.867 | M1a′ versus branch B | 29.570 | 3 | <0.0001 | ω0 = 0.478, p0 = 0.586, ω1 = 1, p1 = 0.386, ω2 = 0.478, ω3 = 10.100, p2 = 0.029 |
| delphinids (branch C) | 18 053.851 | M1a′ versus branch C | 57.602 | 3 | <0.0001 | ω0 = 0.166, p0 = 0.590, ω1 = 1, p1 = 0.394, ω2 = 0.342, ω3 = 30.959, p2 = 0.016 |
| mysticetes (branch D) | 18 077.580 | M1a′ versus branch D | 10.144 | 3 | 0.017 | ω0 = 0.190, p0 = 0.608, ω1 = 1, p1 = 0.376, ω2 = 4.67, ω3 = 3.611, p2 = 0.016 |
Table 2.
Candidate amino acid sites under positive selection identified in cetaceans using different methods.
| AA position | maximum-likelihood method |
radical change | conservative change | cladec | |
|---|---|---|---|---|---|
| PAML M8a | FELb | ||||
| 21 | 0.925 | 0.108 | Lys–Gln Lys–Asn | — | O, D |
| 299 | 0.809 | 0.169 | Gly–Arg(2)d | — | O, D |
| 991 | 0.811 | 0.113 | Val–Ala | — | O, D |
| 1047 | 0.844 | 0.091 | — | Lys–Val | O, D |
| 1073 | 0.814 | 0.103 | Ala–Val | — | M |
| 1176 | 0.800 | 0.152 | Ala–Thr | — | O |
| 1253 | 0.800 | 0.126 | Val–Thr | — | O |
| 1901 | 0.819 | 0.143 | — | Thr–Ser | O, D |
| 2008 | 0.840 | 0.051 | Lys–Ser | O | |
aPosterior probabilities of the BEB analysis with p > 0.8 considered as candidates for selection.
bp-Values with significance levels of 0.2 considered as candidates for selection.
cClades in which amino acid substitution occurred, O, suborder Odontoceti. M, suborder Mysticeti; D, superfamily Delphinoidea.
dThe numbers within parentheses indicate the time of amino acid change.
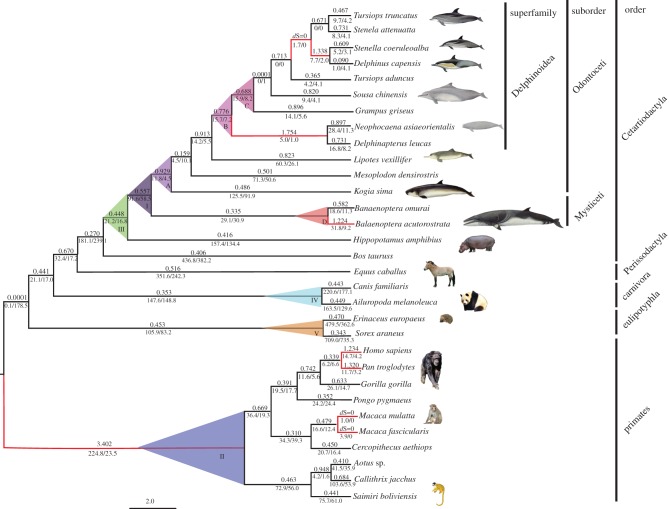
To test whether the evidence for positive selection was restricted to some specific lineages, several models were compared (table 1). In dataset I (all mammals), the free-ratio model was significantly better than the one-ratio model (p < 0.001, table 1), suggesting heterogeneous selective pressures on different lineages. Interestingly, infinite ω (dS = 0 and dN > 0) or ω values greater than one were restricted to those branches characterized by enlarged brain size, both for cetaceans and primates. In the former case, there were four branches: the ancestral branch of delphinids (T. truncatus, Stenella attenuata, Stenella coeruleoalba, Delphinus capensis), the ancestral branch of S. coeruleoalba + Delphinus capensis, the ancestral branch of Phocaenidae (Neophocaena asiaeorientalis) + Monodontidae (Delphinapterus leucas) and the branch leading to B. acutorostrata. In the case of primates, there were five branches: the ancestral branch of New World monkeys, Old World monkeys and Great Apes, and the branches leading to Homo sapiens, Pan troglodytes, Macaca mulatta and Macaca fascicularis (figure 1).
Figure 1.
ω Values for distinct evolutionary lineages of cetaceans and mammals, with a phylogenetic tree derived from Zhou et al. [29,30] and Perelman et al. [31]. The ω values for individual branch according to the free-ratio model are shown. Branches with ω > 1 are shown in red. The ω values of branches I–V and A–D (marked with different colours) were estimated according to the two-ratio models and clade models (with detailed results listed in table 1).
Clade models separately undertaken for cetaceans, primates, Whippomorpha, carnivores and eulipotyphlans revealed evidence of significant divergent selection. Of these clades, the ω value of the focal clade was greater than one only for cetaceans and primates, indicating the action of positive selection on both clades (table 1). When all mammal ASPM sequences (31 sequences and one outgroup) were considered, the ω value of the cetacean focal clade was lower than that of the background. However, when primates were excluded (21 sequences and one outgroup), the reverse was observed (table 1). When only the cetacean dataset (14 sequences and two outgroups) was considered, the ω values of the focal clade were higher than those of the background clade for odontocetes (2.018 versus 1.038), delphinoids (10.100 versus 0.478) and delphinids (30.959 versus 0.342; table 1).
4. Discussion
(a). Molecular evolution of the ASPM gene and brain size enlargement in cetaceans
Two events involving the enlargement of relative brain size in cetaceans were identified from anatomical research and computed tomography: one at the early origin of Odontoceti, and the second during the evolution of Delphinoidea [16]. Interestingly, the analyses of molecular evolution of the ASPM gene conducted in the present study showed evidence of positive selection matching these events. Clade models showed evidence of positive selection acting on odontocetes and delphinoids (table 1). Additionally, nine amino acid sites under positive selection were identified by the two ML approaches (M8 and FEL; table 2). Of these, seven were potentially radical changes associated with charge, polarity or volume of the amino acid. Most of such radically changed sites were found in suborder Odontoceti and/or Delphinoidea, whereas only one site occurred in suborder Mysticeti (table 2). Much higher rates of radical (as opposed to conservative) non-synonymous substitution identified in cetaceans (exact binomial test: p = 0.089) may have been taken as evidence of positive selection. More importantly, such radical amino acid changes at the ASPM gene occurred in toothed whales and some Delphinoidea species, which are further evidence for the association of adaptive ASPM evolution with cetacean brain size enlargement. Although only two exons were investigated, the selection detecting in the present study mainly relied on the PAML, which allows for detecting positive selection in specific codons as opposed to whole genes [37]. In such a case, the ascertainment bias may not be a problem because the selection signal is independent of the fragment length examined. Actually, seven other conservative exons (5–11) of the ASPM gene have also been obtained, and the additional exons did not have any effect on the pattern of selection (data not shown).
Although the large and well-developed cetacean brain has been hypothesized to be a direct product of adaptation to a fully aquatic lifestyle, the present analyses do not support this hypothesis. There was no evidence of positive selection acting on the ASPM gene in the latest common ancestor of whales + hippo (Whippomorpha, branch III, two-ratio branch model: ω = 0.292) and Cetacea (i.e. toothed whales + baleen whales, branch I, two-ratio branch models: ω = 0. 548), which separately represented the transition of the ancestral terrestrial cetaceans to the semi-aquatic habitat and the early stage of adaptation to a fully aquatic life. Actually, this is congruent with evidence from anatomy and computed tomography that the first enlargement of brain size did not occur immediately after the ancestral terrestrial artiodactyls occupied the aquatic environment and early adaptation of cetaceans to the full aquatic habitat [16]. Here, further evidence for this was provided by ω > 1 in the toothed whale clade (clade model C: ω = 2.018 in table 1), which suggested positive selection matching with the first brain size expansion event in the evolutionary history of cetaceans.
The hypothesis that the enlarged brain size of cetaceans was primarily a response to social forces and cognitive demands found, conversely, more support in the present analyses. Indeed, the events of relative brain size enlargement that occurred in the evolution of cetaceans coincide with the two rapid radiation periods associated with the origin of Odontoceti and the diversification of Delphinoidea [38]. Relatively larger brain sizes during these periods may have been favoured by enabling a higher degree of behavioural flexibility, hence the ability to cope with novel environmental conditions and larger and more complex social groups [21,39,40]. In addition, the larger brain sizes during the origin of Odontoceti might also be related to the emergence and development of echolocation to process high-frequency acoustic information, which is associated with a greatly enlarged high-frequency auditory system in toothed whales [41]. It is worth noting, however, that echolocation is likely not to be a primary or sole factor affecting brain size evolution, as bats also have the echolocation ability yet do not show exceptionally large brains.
Additionally, positive selection at the ASPM gene coinciding with brain size enlargements in cetaceans is contrasted with a recent study that analysed another primary microcephaly gene (i.e. MCPH1) in cetaceans but did not find significant association between high relative brain size and level of positive selection [23]. This suggested that the ASPM gene may have played a more important role than MCPH1 in the evolution of large brain in cetaceans. However, considering the lack of direct data on the biochemical function of ASPM in cetaceans, further research on the functional properties of this gene in cetaceans is necessary to determine its role in the brain size enlargement.
(b). Similar selective pressures on the ASPM gene in cetaceans and primates: convergent molecular evolution?
Although cetaceans and primates are highly diverged, different in habitat and neuroanatomical organization, they showed striking convergence in social behaviour and self-recognition ability [42]. Consequently, they both have significantly enlarged brain size [3]. Morino [42] proposed that cetaceans and primates have apparently undergone similar pressures for increased brain mass in their evolutionary history. However, this speculation has not yet been experimentally established at the molecular level. On the basis of molecular evolutionary analyses of the ASPM gene from cetaceans and related terrestrial mammals, the present study provided two lines of evidence to support the notion of similar selective pressures acting on the ASPM gene in primates and cetaceans. First, according to the results from the free-ratio model, the ω > 1 were restricted to some branches within cetaceans and primates (figure 1). Second, the clade models also suggested that positive selection was restricted to these two clades (table 1).
Thus, convergent evolution might underlie the observation of similar selective pressures acting on the ASPM gene in the cetaceans and primates, two mammalian groups highly diverged or distantly related with each other but both showed convergent brain size enlargement. However, in contrast to the strong evidence of molecular convergent evolution found in earlier studies that the motor protein Prestin of echolocating dolphins formed a well-supported group with that of echolocating bats at the amino acid level [43,44], the phylogenetic reconstruction of the ASPM gene conducted in the present study did not group cetaceans with primates at nucleotide or amino acid level (see the electronic supplementary material, figure S1). Moreover, site-specific convergent ASPM evolution was not detected in the two groups at the amino acid level. This is not unreasonable, considering that the evolution of brain size is most likely controlled by multiple genes; thus a relatively weak convergent effect might have occurred in individual ASPM genes. Further studies using more MCPH loci are necessary to determine whether and how brain size evolution in cetaceans and primates converged at the molecular level or whether different mutations could have similar functional effects.
Acknowledgements
We thank Mr Xinrong Xu for collecting the samples for several years; Tong Shen, Zhuo Chen and Wenhua Yu for helpful discussion; and Qi Wu for valuable comments on an early version of the manuscript. Financial support was provided by the National Natural Science Foundation of China (NSFC) to G.Y. (grant no. 30830016) and S.X.X. (grant no. 31000953), the Priority Academic Programme Development of Jiangsu Higher Education Institutions to G.Y. and S.X.X., Specialized Research Fund for the Doctoral Programme of Higher Education (grant no. 20103207120010), the Ministry of Education of China to S.X.X. and the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (grant no. 10KJB180002).
References
- 1.Jerison H. J. 1973. Evolution of the brain and intelligence. New York, NY: Academic Press [Google Scholar]
- 2.Rakic P. 1995. A small step for the cell, a giant leap for mankind, a hypothesis of neocortical expansion during evolution. Trends Neurosci. 18, 383–388 (doi:10.1016/0166–2236(95)93934-P) [DOI] [PubMed] [Google Scholar]
- 3.Clark D. A., Mitra P. P., Wang S. S. 2001. Scalable architecture in mammalian brains. Nature 411, 189–193 10.1038/35075564 (doi:10.1038/35075564) [DOI] [PubMed] [Google Scholar]
- 4.Dunbar R. I. M. 1993. Coevolution of neocortical size, group size and language in humans. Behav. Brain. Sci. 16, 681–735 10.1017/S0140525X00032325 (doi:10.1017/S0140525X00032325) [DOI] [Google Scholar]
- 5.Marino L., et al. 2007. Cetaceans have complex brains for complex cognition. PLoS Biol. 5, e139. 10.1371/journal.pbio.0050139 (doi:10.1371/journal.pbio.0050139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J. 2003. Evolution of the human ASPM gene, a major determinant of brain size. Genetics 165, 2063–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kouprina N., et al. 2004. Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLoS Biol. 2, 653–663 10.1371/journal.pbio.0020126 (doi:10.1371/journal.pbio.0020126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thewissen J. G. M., Cooper L. N., Clementz M. T., Bajpai S., Tiwari B. N. 2007. Whales originated from aquatic artiodactyls in the Eocene epoch of India. Nature 450, 1190–1194 10.1038/nature06343 (doi:10.1038/nature06343) [DOI] [PubMed] [Google Scholar]
- 9.Uhen M. 2010. The origin(s) of whales. Annu. Rev. Earth Planet. Sci. 38, 189–219 10.1146/annurev-earth-040809-152453 (doi:10.1146/annurev-earth-040809-152453) [DOI] [Google Scholar]
- 10.Thewissen J. G. M., Cooper L. N., George J. C., Bajpai S. 2009. From land to water, the origin of whales, dolphins, and porpoises. Evol. Educ. Outreach 2, 272–288 10.1007/s12052-009-0135-2 (doi:10.1007/s12052-009-0135-2) [DOI] [Google Scholar]
- 11.Fordyce R. E., de Muizon C. 2001. Evolutionary history of cetaceans, a review. In Secondary adaptations of tetrapods to life in water (eds Mazin J. M. & V. de Buffr′enil), pp. 169–233 München, Germany: Verlag Dr. Friedrich Pfeil [Google Scholar]
- 12.Mass A. M., Supin A. Y. 2007. Adaptive features of aquatic mammals’ eye. Anat. Rec. 290, 701–715 10.1002/ar.20529 (doi:10.1002/ar.20529) [DOI] [PubMed] [Google Scholar]
- 13.Nummela S., Thewissen J. G. M., Bajpai S., Hussain T., Kumar K. 2007. Sound transmission in archaic and modern whales, Anatomical adaptations for underwater hearing. Anat. Rec. 290, 716–733 10.1002/ar.20528 (doi:10.1002/ar.20528) [DOI] [PubMed] [Google Scholar]
- 14.Herman L. M. 2002. Exploring the cognitive world of the bottlenosed dolphin. In The cognitive animal, empirical and theoretical perspectives on animal cognition (eds Bekoff M., Allen C., Burghardt G. M.), pp. 275–283 Cambridge, MA: MIT Press [Google Scholar]
- 15.Marino L. 1998. A comparison of encephalization between odontocete cetaceans and anthropoid primates. Brain Behav. Evol. 51, 230–238 10.1159/000006540 (doi:10.1159/000006540) [DOI] [PubMed] [Google Scholar]
- 16.Marino L., McShea D., Uhen M. D. 2004. The origin and evolution of large brains in toothed whales. Anat. Rec. 281, 1247–1255 10.1002/ar.a.20128 (doi:10.1002/ar.a.20128) [DOI] [PubMed] [Google Scholar]
- 17.Dorus S., Vallender E. J., Evans P. D., Anderson J. R., Gilbert S. L., Mahowald M., Wyckoff G. J., Malcom C. M., Lahn B. T. 2004. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell 119, 1027–1040 10.1016/j.cell.2004.11.040 (doi:10.1016/j.cell.2004.11.040) [DOI] [PubMed] [Google Scholar]
- 18.Evans P. D., Anderson J. R., Vallender E. J., Gilbert S. L., Malcom C. M., Dorus S., Lahn B. T. 2004. Adaptive evolution of ASPM, a major determinant of cerebral cortical size in humans. Hum. Mol. Genet. 13, 489–494 10.1093/hmg/ddh055 (doi:10.1093/hmg/ddh055) [DOI] [PubMed] [Google Scholar]
- 19.Mahmood S., Ahmad W., Hassan M. J. 2011. Autosomal recessive primary microcephaly (MCPH): clinical manifestations, genetic heterogeneity and mutation continuum. Orphanet J. Rare Dis. 6, 39. 10.1186/1750-1172-6-39 (doi:10.1186/1750-1172-6-39) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bond J., et al. 2002. ASPM is a major determinant of cerebral cortical size. Nat. Genet. 32, 316–320 10.1038/ng995 (doi:10.1038/ng995) [DOI] [PubMed] [Google Scholar]
- 21.Marino L. 2007. Cetacean brains, how aquatic are they? Anat. Rec. 290, 694–700 10.1002/ar.20530 (doi:10.1002/ar.20530) [DOI] [PubMed] [Google Scholar]
- 22.Hof P., Chanis R., Marino L. 2005. Cortical complexity in cetacean brains. Anat. Rec. 287, 1142–1152 10.1002/ar.a.20258 (doi:10.1002/ar.a.20258) [DOI] [PubMed] [Google Scholar]
- 23.McGowen M. R., Montgomery S. H., Clark C., Gatesy J. 2011. Phylogeny and adaptive evolution of the brain-development gene microcephalin (MCPH1) in cetaceans. BMC Evol. Biol. 11, 98. (doi:10.1186/1471–2148-11-98) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponting C. P., Jackson A. P. 2005. Evolution of primary microcephaly genes and the enlargement of primate brains. Curr. Opin. Genet. Dev. 15, 241–248 10.1016/j.gde.2005.04.009 (doi:10.1016/j.gde.2005.04.009) [DOI] [PubMed] [Google Scholar]
- 25.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The Clustal X windows interface, flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 10.1093/nar/25.24.4876 (doi:10.1093/nar/25.24.4876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5, molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 10.1093/molbev/msr121 (doi:10.1093/molbev/msr121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nei M., Kumar S. 2000. Molecular evolution and phylogenetics. New York, NY: Oxford University Press [Google Scholar]
- 28.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 10.1093/molbev/msm088 (doi:10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 29.Zhou X., Xu S., Xu J., Chen B., Zhou K., Yang G. 2012. Phylogenomic analysis resolves the interordinal relationships and rapid diversification of the Laurasiatherian. Syst. Biol. 61, 150–164 10.1093/sysbio/syr089 (doi:10.1093/sysbio/syr089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X., Xu S., Yang Y., Zhou K., Yang G. 2011. Phylogenomic analyses and improved resolution of Cetartiodactyla. Mol. Phylogenet. Evol. 61, 255–264 10.1016/j.ympev.2011.02.009 (doi:10.1016/j.ympev.2011.02.009) [DOI] [PubMed] [Google Scholar]
- 31.Perelman P., et al. 2011. A molecular phylogeny of living primates. PLoS Genet. 7, e1001342. 10.1371/journal.pgen.1001342 (doi:10.1371/journal.pgen.1001342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15, 568–573 10.1002/evan.20046 (doi:10.1002/evan.20046) [DOI] [PubMed] [Google Scholar]
- 33.Yang Z., Nielsen R., Goldman N., Pedersen A. M. K. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155, 431–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z., Wong W. S. W., Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118 10.1093/molbev/msi097 (doi:10.1093/molbev/msi097) [DOI] [PubMed] [Google Scholar]
- 35.Suzuki Y., Nei M. 2001. Reliabilities of parsimony-based and likelihood-based methods for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 18, 2179–2185 10.1093/oxfordjournals.molbev.a003764 (doi:10.1093/oxfordjournals.molbev.a003764) [DOI] [PubMed] [Google Scholar]
- 36.Pond S. L. K., Frost S. D. W. 2005. Datamonkey, rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533 10.1093/bioinformatics/bti320 (doi:10.1093/bioinformatics/bti320) [DOI] [PubMed] [Google Scholar]
- 37.Yang Z., Nielsen R. 2002. Codon substitution models for detecting adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19, 908–917 10.1093/oxfordjournals.molbev.a004148 (doi:10.1093/oxfordjournals.molbev.a004148) [DOI] [PubMed] [Google Scholar]
- 38.Nikaido M., et al. 2001. Retroposon analysis of major cetacean lineages, the monophyly of toothed whales and the paraphyly of river dolphins. Proc. Natl Acad. Sci. USA 98, 7384–7389 10.1073/pnas.121139198 (doi:10.1073/pnas.121139198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allman J. M., McLaughlin T., Hakeem A. 1993. Brainweight and life-span in primate species. Proc. Natl Acad. Sci. USA 90, 118–122 10.1111/j.1365-2664.2010.01883.x (doi:10.1111/j.1365-2664.2010.01883.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reader S. M., Laland K. N. 2002. Social intelligence, innovation, and enhanced brain size in primates. Proc. Natl Acad. Sci. USA 99, 4436–4441 10.1073/pnas.062041299 (doi:10.1073/pnas.062041299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ridgway S. H. 2000. The auditory central nervous system of dolphins. In Hearing in whales and dolphins (eds Au W., Popper A., Fay R.), pp. 273–293 New York, NY: Springer [Google Scholar]
- 42.Marino L. 2002. Convergence of complex cognitive abilities in cetaceans and primates. Brain Behav. Evol. 59, 21–32 10.1159/000063731 (doi:10.1159/000063731) [DOI] [PubMed] [Google Scholar]
- 43.Liu Y., Cotton J. A., Shen B., Han X., Rossiter S. J., Zhang S. 2010. Convergent sequence evolution between echolocating bats and dolphins. Curr. Biol. 20, R53–R54 10.1016/j.cub.2009.11.058 (doi:10.1016/j.cub.2009.11.058) [DOI] [PubMed] [Google Scholar]
- 44.Li Y., Liu Z., Shi P., Zhang J. Z. 2010. The hearing gene prestin unites echolocating bats and whales. Curr. Biol. 20, R55–R56 10.1016/j.cub.2009.11.042 (doi:10.1016/j.cub.2009.11.042) [DOI] [PMC free article] [PubMed] [Google Scholar]