Abstract
Plasminogen and plasmin tether to cell surfaces through ubiquitously expressed and structurally quite dissimilar family of proteins, as well as some nonproteins, that are collectively referred to as plasminogen receptors. Of the more than one dozen plasminogen receptors that have been identified, many have been shown to facilitate plasminogen activation to plasmin and to protect bound plasmin from inactivation by inhibitors. The generation of such localized and sustained protease activity is utilized to facilitate numerous cellular responses, including responses that depend on cellular migration. However, many cells express multiple plasminogen receptors and numerous plasminogen receptors are expressed on many different cell types. Furthermore, several different plasminogen receptors can be used to support the same cellular response, such as inflammatory cell migration. Here, we discuss the perplexing issue: why are there so many different Plg-Rs?
1. Introduction
Plasminogen receptors (Plg-Rs) are a broadly distributed and heterogeneous group of cell surface proteins that share a common feature, the ability to interact with plasminogen (Plg) and plasmin. The list in Table 1, not necessarily all inclusive, identifies 12 different Plg-Rs. Many of these Plg-Rs are expressed by many different cell types, and many are present on the same cell type. Indeed, the number of Plg binding sites on any particular cell type can be extraordinarily high (range from 105 to 107 Plg binding sites per cell). The similarities among these Plg-Rs are very limited and appear to rest only on their ability to be expressed at cell surfaces where they can display their Plg and Plm binding function. Nevertheless, this binding function allows many different Plg-Rs to orchestrate diverse biological responses including fibrinolysis, inflammation, wound healing, and angiogenesis. The question then arises as to why there are so many Plg-Rs and whether there is a plausible explanation for this extensive functional redundancy? This paper will consider these basic questions. As a forewarning, we do not purport to provide clear answers to these questions but hopefully our speculations will be challenging and stimulating.
Table 1.
Plg-Rs on various cell types.
| Plg-Rs | Cell types | C-terminal lysine | Major cellular localization | Secretory pathways |
|---|---|---|---|---|
| (1) Annexin A2 | Endothelial cells, monocytoid lineage | Absent∗ | Cytosol and or nucleus | Translocation depends on p11 and phosphorylation; activity of L-type like Ca2+ channels and intracellular Ca2+; associates with plasma membrane via phosphatidylserine. |
| (2) Actin | Endothelial cells, carcinoma, catecholaminergic cells, PC-3, HT1080 | Absent | Cytoskeleton | Not known |
| (3) Amphoterin | Neuronal cells | Absent | Cytoplasmic and extracellular | Not known |
| (4) αVβ3 | Endothelial cells | Absent | Integral membrane protein | Classical endoplasmic reticulum and Golgi pathway |
| (5) αMβ2 | Neutrophils, monocytes, macrophages | Absent | Integral membrane protein | Classical endoplasmic reticulum and Golgi pathway |
| (6) αIIbβ3 | Platelets, RA synovial fibroblasts | Absent | Integral membrane protein | Classical endoplasmic reticulum and Golgi pathway |
| (7) Cytokeratin 8 | Hepatocellular, breast carcinoma | Present | Cytoskeleton | Not known |
| (8) α-Enolase | Monocytes, neutrophils, carcinoma, lymphoid, myoblast neurons | Present | Cytosol | L-type-like Ca2+ channel and intracellular Ca2+ |
| (9) Histone 2B | Neutrophils, monocytoid cells, endothelial cells | Present | Nucleus | L-type-like Ca2+ channel and intracellular Ca2+. Associates with plasma membrane via phosphatidylserine and heparin sulfate |
| (10) P11 | Endothelial cells, HT1080 cells | Present | Cytosol and or nucleus | L-type-like Ca2+ channel and intracellular Ca2+. Associates with multiple plasma membrane binding partners, including annexin 2 |
| (11) Plg-RKT | Monocytes, macrophages, neuronal cells | Present | Integral membrane protein | Classical endoplasmic reticulum and Golgi pathway |
| (12) TATA-binding protein-interacting protein | Monocytoid cells | Present | Nucleus | Not known |
∗requires cleavage to bind Plg [8].
2. So Many Plg-Rs: Do Different Plg-Rs Bind Plg Differently?
Almost all of the Plg-Rs listed in Table 1 engage the lysine binding sites (LBS) of Plg and Plm by virtue of a C-terminal lysine or by presenting an internal amino acid residue in a context that mimics a C-terminal lysine. As a consequence of a common mechanism of engagement, Plg-Rs are projected to enhance Plg activation by either urokinase plasminogen activator (uPA) or tissue plasminogen activator (tPA), to enhance the catalytic activity of plasmin and to protect bound plasmin from inactivation by plasmin inhibitors [1–3]. Indeed, several Plg-Rs have been reported to have one or more of these functional attributes [4–6]. Also, with a similar mechanism of binding, the affinities of the various Plg-Rs for Plg should be similar. The context of the LBS binding residue within a Plg-R might be influenced by adjacent amino acids or local conformation and thereby influence the affinity of specific subset of Plg-Rs for Plg. However, even for Plg-Rs that utilize an internal residue rather than a C-terminal lysine to engage Plg, affinities for the ligand appear to be similar (~1 μM), [7]. One potential exception to this assertion could be the annexin A2/p11 heterotetramer, where the proximity of multiple Plg binding sites within a single molecular species could enhance affinity substantially. To support this possibility or other reports of higher-affinity Plg-Rs, variability in ligand preparations used (e.g., presence of Lys-Plg in Glu-Plg preparation) must be controlled. Furthermore, since ligand availability seems not to be limiting (Plg is present at high concentrations), differences in apparent affinity may have less impact than anticipated.
3. So Many Plg-Rs: Do Different Cell Types Use Different Plg-Rs?
Not all Plg-Rs are expressed on all cell types. As an example of a Plg-R with a restricted cellular distribution, integrin αMβ2 is a Plg-R [7] and its expression is confined to leukocytes. However, leukocytes express many other Plg-Rs, including annexin A2/p11, which has long been promulgated as the major Plg-R on endothelial cells (ECs). Indeed, inactivation of either the annexin A2 or p11 genes does affect EC-dependent responses, including angiogenesis, tumorogenesis, fibrinolysis, and inflammation [9–12]. However, ECs do express other Plg-Rs. As an illustrative example, histone H2B, a high-abundance Plg-R on monocytoid cells, is also readily detected on the surface of HUVEC (Figure 1). In Figure 1, H2B was detected on the surface of HUVEC by a cell-surface biotinylation approach [13] in which the cells were surface labeled with biotin, lysed, and the biotinylated proteins were isolated on streptavidin beads and then identified by western blotting with specific antibodies (see Figure 1 and its legend for details). H2B was labeled with biotin, whereas p65, a control intracellular protein, was not even though both H2B and p65 were readily detected in the whole cell lysates of HUVEC. H2B associates with the surface of monocytoid cells by binding to phosphatidylserine (PS) [14].Annexin V, another PS binding protein displaces the H2B from the surface of monocytoid cells [14] and also chases biotin labeled H2B from the surface of HUVEC (see Figure 1). Biotinylation also labeled H2B on the surface of microvascular endothelial cells as well as on large-vessel endothelial cells (not shown). As an independent approach, we confirmed the presence of H2B on the surface of HUVECs by flow cytometry. Also, α-enolase, the first identified Plg-R, has been implicated in the binding of Plg to microparticles released from ECs [15]. Thus, in addition to annexin A2/p11, other Plg-Rs have been detected on endothelial cells. Hence, the notion of the preeminence of a specific Plg-R on a particular cell type does not seem tenable.
Figure 1.
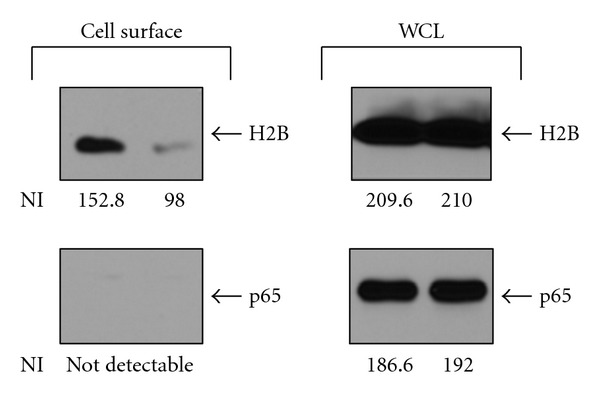
H2B exposure on the surface of endothelial cells. Human umbilical vein endothelial cells (HUVECs) were either untreated or treated with annexin V (250 nM) for 48 hr. Cells were surface labeled with biotin, and the biotinylated proteins were isolated using streptavidin-conjugated beads. H2B and p65 (a transcription factor with a cytosolic and nuclear localization) that were bound and eluted from the streptavidin beads were detected by western blotting with a rabbit anti-H2B or rabbit anti-p65. The absence of biotinylated p65 serves as a control for surface labeling of H2B. Band intensities of the western blots were analyzed using Kodak ID 3.6 software, and net intensity (NI) of each band is indicated below each lane. In each set of two lanes, the right-hand lane is in the presence of annexin V and the left-hand lane in its absence. (WCL: whole cell lysates).
The compartmentalization of specific Plg-Rs to select locations on the cell surface could provide a mechanism to distinguish the function of one Plg-R from another. Several integrins serve as Plg-Rs (Table 1) and integrins do localize to the leading edge of migrating cells [16], and uPAR also localizes to the leading edge of migrating cells [17]. Furthermore, annexin 2 has been localized to the leading edge of migrating retinal glial cells and malignant glioma cell [18, 19]. Thus, an advantageous microenvironment may be created in which one Plg-R is particularly proficient in Plg activation. However, recent data have suggested that cell-surface-bound Plg can be efficiently activated or even more efficiently activated by uPA bound to another cell than that on the same cell [20]. The boost in efficiency of Plg activation gained by localization on a single cell may be offset by the restricted diffusion or orientation of the Plg activator on the cell surface. Thus, localization of certain Plg-Rs to a specific microdomain on the cell surface and the functional advantage of such localization remain a possibility. We did note a uniform distribution of several Plg-Rs, as well as bound Plg, on monocytoid cells by confocal microscopy although changes in distribution under stimulated conditions were not tested [13]. A common mechanism dependent on L-type like calcium channels has been implicated in translocation of several Plg-Rs to the surface of monocytoid cells [21], but the mechanisms by which these Plg-R tether to the cell surface are distinct [14]. Hence, Plg-Rs could compartmentalize on the cell surface.
4. So Many Plg-Rs: Are Plg-Rs Differentially Regulated on Cells?
It is well established that Plg binding to cells can be markedly modulated; changes in Plg binding capacity of specific cell types can increase 3- to 20-fold in response to specific stimuli. Cellular events and responses that can induce such changes include oncogenic transformation (breast and adenocarcinoma cancer) [22, 23] differentiation (monocytes, adipocytes) [21, 24], agonist stimulation, (leukocytes, endothelial cells, platelets) [7, 25–27], adhesion (monocytoid cells) [28], and apoptosis (monocytoid cells) [14]. In addition, Plg binding can be enhanced by proteolysis of existing cell surface proteins to generate new C-terminal lysines [29, 30]. This latter mechanism for exposing new Plg-Rs can be triggered by plasmin itself and depends on the availability of uPA on the cell surface [31]. Thus, even though a cell type can express multiple Plg-Rs, a subset of Plg-Rs may be differentially upregulated and utilized to mediate a specific cellular response.
The data in Figure 2 provides an illustrative example of how different Plg-Rs maybe utilized by the same cell in responding to different stimuli. THP-1 monocytoid cells were either stimulated to undergo differentiation using vitamin D3 + IFNγ or apoptosis using camptothecin. Consistent with our prior report [13, 14], the cells respond to these stimuli by markedly upregulating their Plg binding capacity. In association with differentiation, Plg binding increased by 3.3-fold. Of the Plg-Rs analyzed by FACS, enolase, annexin2, p11, and H2B, surface expression increased most markedly for H2B (4.7-fold) in response to differentiation. In response to apoptosis induced by camptothecin, Plg binding increased by 10-fold. While surface localization of H2B did increase significantly (4.6-fold), much more striking was the 20-fold upregulation of p11 in the camptothecin-treated THP-1 cells. This pattern of enhanced p11 expression was also observed in U937 monocytoid cells treated with camptothecin, where 5.8-fold increase of Plg binding was associated with 6.3-fold increase in p11 expression. Of note, these increases in p11 expression on apoptotic cells were not paralleled by substantial increases of the annexinA2 subunit. In the camptothecin-treated THP-1 cells, surface expression of the annexinA2 subunit increased by 2.8-fold and for U397 cells, the increase was 2.3-fold. As explanations for this disproportional upregulation of p11, the subpopulation of annexinA2 molecules that escort p11 to the cell surface may not react with the antibody used in this analysis, or the anti-p11 may selectively penetrate apoptotic cells, which are known to be leaky [32]. A more interesting possibility is that a portion of the p11 that becomes surface expressed is in a free form or is associated with other binding partners. Besides annexin2, other plasma membrane proteins, NaV1.8 sodium channel, TASK1 potassium channel, TRPV5/TRPV6 channels, and cathepsin B [33] have been shown to interact with p11, could assist in its transport to the cell surface, and may still further extend the repertoire of Plg-Rs expressed by monocytoid cells.
Figure 2.
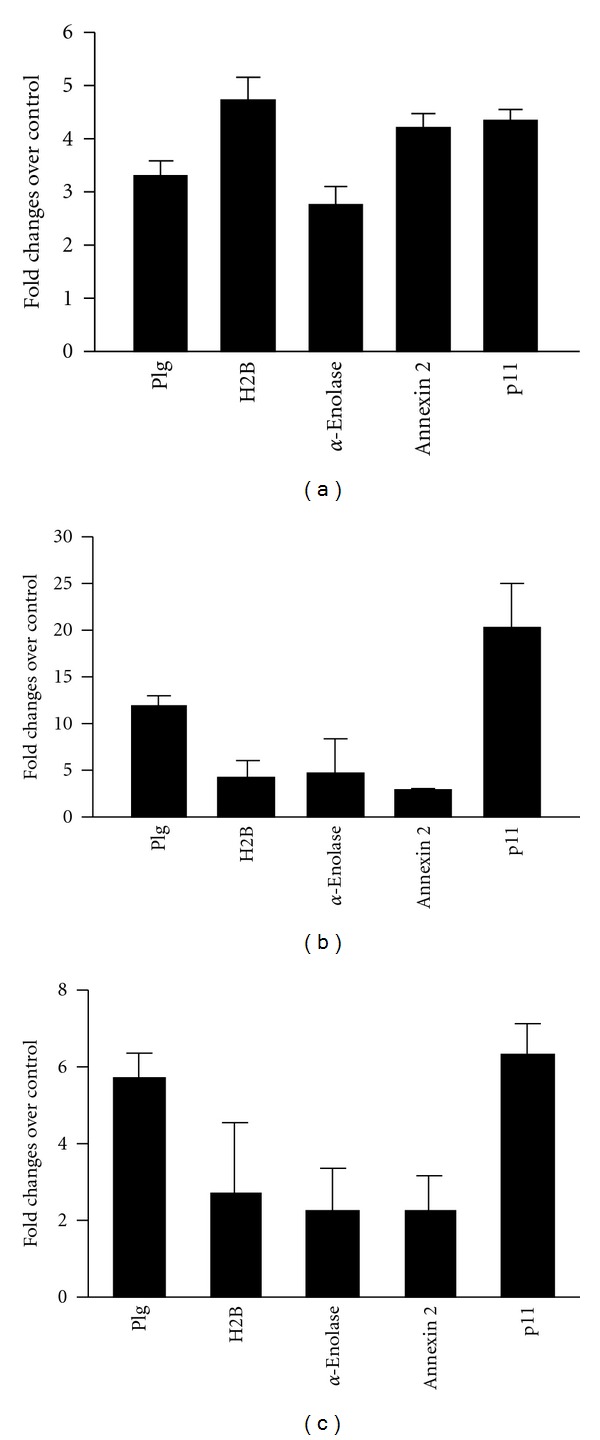
THP-1 (a), (b) and U937 (c) cells were either differentiated with IFNγ + VD3 for 48 h (a) or induced to undergo apoptosis with camptothecin for 24 h (b), (c). Cells are labeled with Alexa-488-Plg or anti-Plg-Rs antibodies against H2B, α-enolase, annexin A2, and p11 followed by Alexa-488-ant-rabbit IgG (c) and analyzed by FACS. Early apoptotic populations are used to analyze the data. Data are means ± SD of two to three independent experiments and presented as the fold change relative to untreated THP-1 or U937 cells.
In vivo data also support the proposition that different Plg-Rs mediate the response of the same cell type to different stimuli. In a thioglycollate-induced peritonitis model, an antibody to H2B that blocks Plg binding inhibited macrophage recruitment by ~50% while an antibody to α-enolase that also blocks Plg binding to its target produced less than 25% inhibition of macrophage recruitment [13]. In contrast, in an LPS-induced lung inflammation model, Plg binding to α-enolase overexpressing U937 cells produced a substantial enhancement of macrophage migration [34].
5. So, Why So Many Plg-Rs?
While the utilization of different Plg-Rs to orchestrate different cellular responses is supported by data cited above, blocking of several different Plg-Rs has been shown to markedly suppress what appears to be the same inflammatory response thioglycollate=induced peritonitis. The contribution of H2B (45% [13]), p11 (53% [12]), and Plg-RKT (58% [35]), either with antibodies or gene inactivation, exceeds 100%. Such extensive inhibition becomes even more incomprehensible since macrophage recruitment is decreased by only 65% in Plg−/− mice compared to wild-type littermates [36]. At least four explanations can be considered to explain such observations. First, these various Plg-Rs may exert an effect on macrophage recruitment unrelated to Plg. The effect of blockade of individual Plg-Rs in a Plg−/− background could be used to identify such functions. Second, a threshold of bound Plg must be attained in order for Plg to facilitate cell migration. No single Plg-R may harness sufficient Plg to reach this threshold, and, hence, cooperation among several Plg-Rs is required. Third, while many different Plg-Rs enhance Plg activation, the intracellular signaling responses that they elicit may be distinct. Cellular recruitment is a complex response requiring activation of many different intracellular signaling pathways. Different Plg-Rs may trigger distinct signaling events, and these pathways may need to cooperate to yield efficient migration. Blunting the signaling response elicited by occupancy of any one Plg-R may lead to suppressed signaling and diminished migration. Fourth, recruitment into the peritoneum is a temporally extended and multi-step response, and different Plg-Rs may come into play at different times and stages during the response. Hence, difference Plg-Rs may be utilized to achieve specific steps in the recruitment cascade.
6. Summary and Concluding Remarks
In this brief discussion, we have raised the question as to why there are so many Plg-Rs. With so many different receptors frequently, with many of them expressed on the same cell type, it is difficult to envision how the cell would prioritize its utilization among these multiple Plg-R. Affinity differences between Plg-Rs for their Plg and plasmin ligands could distinguish one receptor from another, but this can only be tested by direct comparisons among Plg-Rs. Utilization of specific Plg-Rs to mediate tissue specific or cell specific responses can also be envisioned, but such analyses again mandate comparative studies. In fact, in each of the explanations suggested above, to account for the profound role of many different Plg-Rs in what is globally visualized as a single cellular response, macrophage recruitment into the peritoneum, comparative studies are again needed. The goal of such comparative studies is not to prove that one particular Plg-R is better than another, but rather to help dissect the ways in which Plg orchestrates cell migration and other cellular responses in vivo.
Acknowledgments
The authors thank Sidney Jones from Plow Lab for assisting in endothelial experiments. This work was supported by NIH grant HL17964 (E. F. Plow) and an American Heart Association Scientist Development Grant 11SDG7390041(R. Das).
References
- 1.Plow EF, Herren T, Redlitz A, Miles LA, Hoover-Plow JL. The cell biology of the plasminogen system. FASEB Journal. 1995;9(10):939–945. doi: 10.1096/fasebj.9.10.7615163. [DOI] [PubMed] [Google Scholar]
- 2.Das R, Pluskota E, Plow EF. Plasminogen and its receptors as regulators of cardiovascular inflammatory responses. Trends in Cardiovascular Medicine. 2010;20(4):120–124. doi: 10.1016/j.tcm.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miles LA, Hawley SB, Baik N, Andronicos NM, Castellino FJ, Parmer RJ. Plasminogen receptors: the sine qua non of cell surface plasminogen activation. Frontiers in Bioscience. 2005;10(2):1754–1762. doi: 10.2741/1658. [DOI] [PubMed] [Google Scholar]
- 4.Cesarman GM, Guevara CA, Hajjar KA. An endothelial cell receptor for plasminogen/tissue plasminogen activator (t-PA). II. Annexin II-mediated enhancement of t-PA-dependent plasminogen activation. Journal of Biological Chemistry. 1994;269(33):21198–21203. [PubMed] [Google Scholar]
- 5.Redlitz A, Fowler BJ, Plow EF, Miles LA. The role of an enolase-related molecule in plasminogen binding to cells. European Journal of Biochemistry. 1995;227(1-2):407–415. doi: 10.1111/j.1432-1033.1995.tb20403.x. [DOI] [PubMed] [Google Scholar]
- 6.Kwon M, MacLeod TJ, Zhang Y, Waisman DM. S100A10, annexin A2, and annexin A2 heterotetramer as candidate plasminogen receptors. Frontiers in Bioscience. 2005;10(1):300–325. doi: 10.2741/1529. [DOI] [PubMed] [Google Scholar]
- 7.Pluskota E, Soloviev DA, Bdeir K, Cines DB, Plow EF. Integrin αMβ2 orchestrates and accelerates plasminogen activation and fibrinolysis by neutrophils. Journal of Biological Chemistry. 2004;279(17):18063–18072. doi: 10.1074/jbc.M310462200. [DOI] [PubMed] [Google Scholar]
- 8.Hajjar KA, Jacovina AT, Chacko J. An endothelial cell receptor for plasminogen/tissue plasminogen activator. I. Identity with annexin II. Journal of Biological Chemistry. 1994;269(33):21191–21197. [PubMed] [Google Scholar]
- 9.Phipps KD, Surette AP, O'Connell PA, Waisman DM. Plasminogen receptor S100A10 is essential for the migration of tumor-promoting macrophages into tumor sites. Cancer Research. 2011;71(21):6676–6683. doi: 10.1158/0008-5472.CAN-11-1748. [DOI] [PubMed] [Google Scholar]
- 10.Ling Q, Jacovina AT, Deora A, et al. Annexin II regulates fibrin homeostasis and neoangiogenesis in vivo. Journal of Clinical Investigation. 2004;113(1):38–48. doi: 10.1172/JCI200419684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Surette AP, Madureira PA, Phipps KD, Miller VA, Svenningsson P, Waisman DM. Regulation of fibrinolysis by S100A10 in vivo. Blood. 2011;118(11):3172–3181. doi: 10.1182/blood-2011-05-353482. [DOI] [PubMed] [Google Scholar]
- 12.O’Connell PA, Surette AP, Liwski RS, Svenningsson P, Waisman DM. S100A10 regulates plasminogen-dependent macrophage invasion. Blood. 2010;116(7):1136–1146. doi: 10.1182/blood-2010-01-264754. [DOI] [PubMed] [Google Scholar]
- 13.Das R, Burke T, Plow EF. Histone H2B as a functionally important plasminogen receptor on macrophages. Blood. 2007;110(10):3763–3772. doi: 10.1182/blood-2007-03-079392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das R, Plow EF. Phosphatidylserine as an anchor for plasminogen and its plasminogen receptor, Histone H2B, to the macrophage surface. Journal of Thrombosis and Haemostasis. 2011;9(2):339–349. doi: 10.1111/j.1538-7836.2010.04132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacroix R, Sabatier F, Mialhe A, et al. Activation of plasminogen into plasmin at the surface of endothelial microparticles: a mechanism that modulates angiogenic properties of endothelial progenitor cells in vitro. Blood. 2007;110(7):2432–2439. doi: 10.1182/blood-2007-02-069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nature Reviews Cancer. 2002;2(2):91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 17.Smith HW, Marshall CJ. Regulation of cell signalling by uPAR. Nature Reviews Molecular Cell Biology. 2010;11(1):23–36. doi: 10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- 18.Hayes MJ, Shao D, Bailly M, Moss SE. Regulation of actin dynamics by annexin 2. EMBO Journal. 2006;25(9):1816–1826. doi: 10.1038/sj.emboj.7601078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beckner ME, Chen X, An J, Day BW, Pollack IF. Proteomic characterization of harvested pseudopodia with differential gel electrophoresis and specific antibodies. Laboratory Investigation. 2005;85(3):316–327. doi: 10.1038/labinvest.3700239. [DOI] [PubMed] [Google Scholar]
- 20.Dejouvencel T, Doeuvre L, Lacroix R, et al. Fibrinolytic cross-talk: a new mechanism for plasmin formation. Blood. 2010;115(10):2048–2056. doi: 10.1182/blood-2009-06-228817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das R, Burke T, Van Wagoner DR, Plow EF. L-type calcium channel blockers exert an antiinflammatory effect by suppressing expression of plasminogen receptors on macrophages. Circulation Research. 2009;105(2):167–175. doi: 10.1161/CIRCRESAHA.109.200311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranson M, Andronicos NM, O’Mullane MJ, Baker MS. Increased plasminogen binding is associated with metastatic breast cancer cells: differential expression of plasminogen binding proteins. British Journal of Cancer. 1998;77(10):1586–1597. doi: 10.1038/bjc.1998.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burtin P, Chavanel G, Andre-Bougaran J, Gentile A. The plasmin system in human adenocarcinomas and their metastases. A comparative immunofluorescence study. International Journal of Cancer. 1987;39(2):170–178. doi: 10.1002/ijc.2910390208. [DOI] [PubMed] [Google Scholar]
- 24.Hoover-Plow J, Yuen L. Plasminogen binding is increased with adipocyte differentiation. Biochemical and Biophysical Research Communications. 2001;284(2):389–394. doi: 10.1006/bbrc.2001.4984. [DOI] [PubMed] [Google Scholar]
- 25.Peterson EA, Sutherland MR, Nesheim ME, Pryzdial ELG. Thrombin induces endothelial cell-surface exposure of the plasminogen receptor annexin 2. Journal of Cell Science. 2003;116(12):2399–2408. doi: 10.1242/jcs.00434. [DOI] [PubMed] [Google Scholar]
- 26.Ratel D, Mihoubi S, Beaulieu E, et al. VEGF increases the fibrinolytic activity of endothelial cells within fibrin matrices: involvement of VEGFR-2, tissue type plasminogen activator and matrix metalloproteinases. Thrombosis Research. 2007;121(2):203–212. doi: 10.1016/j.thromres.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 27.Miles LA, Plow EF. Binding and activation of plasminogen on the platelet surface. Journal of Biological Chemistry. 1985;260(7):4303–4311. [PubMed] [Google Scholar]
- 28.Kim H-S, Plow EF, Miles LA. Regulation of plasminogen receptor expression on monocytoid THP- 1 cells by adherence to extracellular matrix proteins. Circulation. 1992;86(I-148) doi: 10.1074/jbc.271.39.23761. [DOI] [PubMed] [Google Scholar]
- 29.Camacho M, Fondaneche MC, Burtin P. Limited proteolysis of tumor cells increases their plasmin-binding ability. FEBS Letters. 1989;245(1-2):21–24. doi: 10.1016/0014-5793(89)80183-8. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez-Gronow M, Stack S, Pizzo SV. Plasmin binding to the plasminogen receptor enhances catalytic efficiency and activates the receptor for subsequent ligand binding. Archives of Biochemistry and Biophysics. 1991;286(2):625–628. doi: 10.1016/0003-9861(91)90090-6. [DOI] [PubMed] [Google Scholar]
- 31.Stillfried GE, Saunders DN, Ranson M. Plasminogen binding and activation at the breast cancer cell surface: the integral role of urokinase activity. Breast Cancer Research. 2007;9(1, article no. R14) doi: 10.1186/bcr1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Mullane MJ, Baker MS. Elevated plasminogen receptor expression occurs as a degradative phase event in cellular apoptosis. Immunology and Cell Biology. 1999;77(3):249–255. doi: 10.1046/j.1440-1711.1999.00823.x. [DOI] [PubMed] [Google Scholar]
- 33.Svenningsson P, Greengard P. p11 (S100A10)—an inducible adaptor protein that modulates neuronal functions. Current Opinion in Pharmacology. 2007;7(1):27–32. doi: 10.1016/j.coph.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Wygrecka M, Marsh LM, Morty RE, et al. Enolase-1 promotes plasminogen-mediated recruitment of monocytes to the acutely inflamed lung. Blood. 2009;113(22):5588–5598. doi: 10.1182/blood-2008-08-170837. [DOI] [PubMed] [Google Scholar]
- 35.Lighvani S, Baik N, Diggs JE, Khaldoyanidi S, Parmer RJ, Miles LA. Regulation of macrophage migration by a novel plasminogen receptor Plg-R KT. Blood. 2011;118(20):5622–5630. doi: 10.1182/blood-2011-03-344242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ploplis VA, French EL, Carmeliet P, Collen D, Plow EF. Plasminogen deficiency differentially affects recruitment of inflammatory cell populations in mice. Blood. 1998;91(6):2005–2009. [PubMed] [Google Scholar]


