Abstract
We recently demonstrated that lack of Furin-processing of the N-cadherin precursor (proNCAD) in highly invasive melanoma and brain tumor cells results in the cell-surface expression of a nonadhesive protein favoring cell migration and invasion in vitro. Quantitative polymerase chain reaction analysis of malignant human brain tumor cells revealed that of all proprotein convertases (PCs) only the levels of Furin and PC5A are modulated, being inversely (Furin) or directly (PC5A) correlated with brain tumor invasive capacity. Intriguingly, the N-terminal sequence following the Furin-activated NCAD site (RQKR↓DW161, mouse nomenclature) reveals a second putative PC-processing site (RIRSDR↓DK189) located in the first extracellular domain. Cleavage at this site would abolish the adhesive functions of NCAD because of the loss of the critical Trp161. This was confirmed upon analysis of the fate of the endogenous prosegment of proNCAD in human malignant glioma cells expressing high levels of Furin and low levels of PC5A (U343) or high levels of PC5A and negligible Furin levels (U251). Cellular analyses revealed that Furin is the best activating convertase releasing an ∼17-kDa prosegment, whereas PC5A is the major inactivating enzyme resulting in the secretion of an ∼20-kDa product. Like expression of proNCAD at the cell surface, cleavage of the NCAD molecule at RIRSDR↓DK189 renders the U251 cancer cells less adhesive to one another and more migratory. Our work modifies the present view on posttranslational processing and surface expression of classic cadherins and clarifies how NCAD possesses a range of adhesive potentials and plays a critical role in tumor progression.
Introduction
Of the many processes that occur during tumor progression, invasion and the formation of secondary metastases are the most clinically relevant but the least well understood at the molecular level. Yet, it is now well established that changes in surface expression of classes of cell adhesion molecules correlate with malignant transformation [1,2]. Reduction of intercellular adhesive interactions of tumor cells is necessary to promote detachment from the main tumor mass and to alter tumor-host cell interactions necessary for migration and invasion [3].
Classic cadherins are key cell adhesion molecules in epithelia that mediate Ca2+-dependent intercellular interactions [4,5] and are strongly implicated in tumor development [6–8]. It has been shown that a decrease in cadherin expression is linked to metastatic tumor behavior [9–12]. Loss of E-cadherin (ECAD) expression and/or function appears to be necessary for tumor metastasis [1] and correlates with high tumor grades and poor prognosis [13–15]. Contrary to ECAD, during the progression of several types of carcinomas, an up-regulation of neural cadherin (NCAD; also known as cadherin 2) expression has been shown to occur and this correlates with increased tumor cell motility and metastasis [2,3,16,17]. In many cancers, a switch from ECAD to NCAD accompanies a highly malignant phenotype [6,13,16,18,19]. However, other tumors, such as primary gliomas, do not express ECAD at any stage of development but upregulate NCAD compared to brain parenchyma [20].
An important determinant of cadherin adhesive activity is the proteolytic processing of its prosegment, which contains at its C terminus a long and flexible linker region connecting it to the first extracellular domain of NCAD [6]. The removal of the prosegment is needed to expose a critical Trp at the second residue following the cleavage site (P2′ position) in mature NCAD. This Trp is functionally implicated in the homophilic dimerization of cell-surface NCAD resulting in efficient cell-cell adhesion [21] and the formation of a complex between the cytosolic catenins and the cytoplasmic tail of type 1 transmembrane cadherins [22,23]. The prosegment is composed of 138 amino acids (aa) and lacks the essential structural features required for adhesion [21]. Following N-glycosylation in the ER and trafficking to the Golgi, the prodomain is thought to be cleaved in the trans-Golgi network (TGN) and/or at the cell surface by one or more members of the proprotein convertase (PC) family. At the C terminus of their prosegments, classic cadherins contain a consensus cleavage site for members of the PC family (RXKR↓) [24].
The basic aa-specific mammalian subtilisin-like PCs are a family of Ca2+-dependent endoproteases, responsible for the activation of precursor proteins by cleavage at a consensus recognition site (R/K-(2X)-R/K↓; X = 0–4 aa). The PCs are PC1 (also known as PC1/3), PC2, Furin, PC4, PC5 [also known as PC5/6A and found in two isoforms, namely, a soluble PC5A (913 aa) and a longer (1860 aa) membrane-bound mostly intestinal PC5B isoform], PACE4, and PC7 [24,25]. Whereas PC1 and PC2 are important in the neural and endocrine pathway, and PC4 only functions in germinal cells, Furin, PACE4, PC5A, and PC7 have a wide tissue distribution and proteolytically process precursors in the constitutive secretory pathway [25]. Furin has catalytic activity primarily in the TGN, at the cell surface and/or endosomes [26], whereas in many instances PC5A and PACE4 exert their functions mostly at the cell surface [bound to heparin sulfate proteoglycans (HSPGs)] or extracellularly [25,27], and PC7 seems to act exclusively close to the cell surface [28]. Studies have demonstrated that NCAD [29] and ECAD [30] can be cleaved by Furin to release their prodomains and expose their critical Trp for adhesion.
What role do PCs play in tumor development? PCs are involved in the processing and activation of key molecules in tumor progression, invasion, and metastasis, such as growth factors and membrane-type metalloproteinases [31–34]. Studies have shown that Furin and PACE4 are overexpressed in tumor cell lines as well as in primary human malignancies, and inhibition of these PCs correlates with decreased tumorigenesis [31,34–38]. Conversely, cadherins, such as NCAD, are rendered adhesively active by Furin processing, resulting in tight cell-cell adhesion and suppression of tumor progression [29]. However, some reports show an opposite effect of Furin and PACE4 in breast cancer cell invasion [39], Furin in the growth of hepatocellular carcinoma [40], and a protective effect of PC5 in intestinal tumorigenesis [41]. Thus, in some cancer models, Furin, PC5, and/or PACE4 may act in similar or opposite fashion.
NCAD is also implicated in physiological invasion processes during embryonic development, including neural growth cone extension [42–45], gastrulation [42,46], and migration of neural crest cells over large distances [47,48]. It is thus conceivable that differentially processed cell-surface NCAD plays an important functional role in these physiological processes, as also supported by the presence of unprocessed precursor of N-cadherin (proNCAD) on the surfaces of neurites in the developing brain [49].
We recently showed that during malignant melanoma transformation and in highly invasive glioma, e.g., U251 cells, in addition to mature NCAD, significant amounts of nonadhesive uncleaved proNCAD are present on the cell surface, promoting tumor cell migration and invasion [29]. In contrast, in the less invasive U343 cells, most of proNCAD was processed by endogenous Furin. The present study extends our original findings and presents a novel role of PC5A in controlling cell-cell adhesion and invasion through an atypical processing of proNCAD into a nonadhesive and pro-invasive form.
Materials and Methods
Cell Culture and Transfections
HeLa cells were purchased from American Type Culture Collection (Manassas, VA). The human U343 malignant glioma cell line was generously provided by A. Guha (University of Toronto, Toronto, Ontario), and the human U251 malignant glioma cell line was provided by R. Bjervig (University of Bergen, Bergen, Norway). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS (Invitrogen, Burlington, Ontario) and maintained at 37°C in a humidified atmosphere of 5% CO2. Lipofectamine Plus transfection reagent (Invitrogen) was used to transfect cells.
Constructs
Wild-type (WT) NCAD-myc or mutant proNCAD-myc were previously described [29]. V5-tagged Furin, PC5A, PC5A-ΔCRD, PACE4, PACE4-ΔCRD, and PC7 constructs were described elsewhere [27,50,51]. An NCAD myc-tagged mutated at S2 (NCAD-II) was generated using QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), according to the manufacturer's instructions. Mutagenesis primers are included in Table W1.
Antibodies and Reagents
The following primary antibodies were used for Western immunoblot analysis and immunofluorescence: anti-proN [21], mouse monoclonal anti-myc (clone 9E10; Sigma-Aldrich, Oakville, ON), rabbit polyclonal anti-Furin and anti-PC5A (generated in-house), and fluorescent-conjugated secondary antibodies (Millipore, Billerica, MA). Fluorescence mounting media (DAKO, Burlington, ON) was used to mount coverslips on glass slides.
Immunoblot analysis
Protein samples were resolved on a 4% to 15% linear gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Bio-Rad, Mississauga, Ontario), transferred to nitrocellulose, blocked with 5% milk protein, and incubated overnight with primary antibodies at 4°C. Blots were then incubated with HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA), and routine washes were carried out. Blots were developed with the chemiluminescence system (Pierce Biotechnology, Rockford, IL). Densitometric analysis was carried out using the ImageJ software (National Institutes of Health, Bethesda, MD). Bands were boxed and background signal was subtracted from their relative intensities. Intensity values were normalized to reference values (loading control).
Immunocytochemistry
Cells were plated onto poly-l-lysine-coated coverslips, fixed in 4% paraformaldehyde, permeabilized in 0.3% Triton X-100 and phosphate-buffered saline (PBS), and blocked in 5% BSA, 5% goat serum, and PBS. Cells were then incubated for 1 hour in primary antibody (Ab) diluted in 1% BSA, 0.02% Triton X-100, and PBS, followed by a 40-minute incubation in fluorescent-conjugated secondary antibodies. Three washes with PBS were performed before fixation, as well as following each step. Coverslips were mounted and examined by confocal laser microscopy using the Zeiss LSM 510 microscope with the Zen image acquisition software and 60x oil immersion objective. Images were acquired in the same plane of focus between comparisons.
For live cell staining, cells were plated on coverslips with primary Ab diluted in medium without serum at 4°C for 1 hour. The cells were washed with PBS and fixed in 3.7% paraformaldehyde. Following three washes with PBS, cells were incubated with fluorescent-conjugated secondary Ab diluted in 1% BSA, 0.02% Triton X-100, and PBS for 40 minutes at room temperature and washed, and coverslips were mounted and analyzed as above.
Cell Aggregation Assays
Aggregation assays were carried out as previously described [29]. Briefly, monolayer cultures were treated with 2 mM EDTA in PBS for 5 minutes at 37°C. Cells were then washed gently in Hepes-buffered Ca2+-Mg2+-free Hank's balanced salt solution supplemented with 1 mM CaCl2 and 1% BSA for 30 minutes at 37°C to dissociate the monolayer into single cells while leaving cadherins intact on the cell surface. Following cell dissociation, 5 x 105 cells per well were transferred to 24-well low-adherent dishes (VWR, Mississauga, Ontario) and brought up to a final volume of 0.5 ml of Hepes-buffered Ca2+-Mg2+-free Hank's balanced salt solution containing 1% BSA with or without 1 mM Ca2+. The plates were rotated at 80 rpm at 37°C for observation of aggregate formation during a 40-minute time course. At t = 0 minute, t = 20 minutes, and t = 40 minutes, 50 µl of the fixed aggregates was removed, placed on a slide, covered with a coverslip, and examined by light microscopy. The number of single cells was counted and the percentage of single cells during the aggregation assay was determined by the index Nt/N0, where Nt is the total number of single cells after a certain incubation time, and N0 is the total number of single cells at the initiation of incubation.
Detection of NCAD Proteolytic Products
N-terminal proNCAD segments were detected in the conditioned medium of cells using the proN Ab. Cells were transiently transfected with the appropriate construct(s); the conditioned medium was collected 48 hours later and concentrated (Millipore Amicon Filters), and total protein concentration was determined using a Lowry assay (Bio-Rad). Conditioned medium (15 µg of protein) was resolved on a 4% to 15% SDS-PAGE (Bio-Rad), and cleavage products were detected by Western immunoblot analysis as an ∼17-kDa band corresponding to cleavage at the S1 consensus site and an ∼20-kDa band representing cleavage at S2.
Wound-healing Migration Assays
To assess two-dimensional migration of tumor cell lines, we seeded 3 x 105 cells in six-well culture dishes. Before plating in these dishes, two parallel lines were drawn at the underside of the well with a marker, serving as fiducial marks for analysis of wound areas. The monolayer was ∼100% confluent on the day of analysis. The growth medium was first aspirated and replaced by calcium-free PBS to prevent death of cells at the edge of the wound by exposure to high calcium concentrations. Monolayers were disrupted with a parallel scratch wound made perpendicular to the marker lines with a fine pipette tip. Migration into the wound was observed using phase contrast microscopy on an inverted microscope with 5x objective. Images of random areas of the wound were sampled for cell counting at regular time intervals of areas flanking the intersections of the wound and the marker lines. The number of cells that migrated into the wound was determined by marking wound edges at t = 0 hour on the underside of the well and counting cells that migrated into the wound at specified time points appropriate for each cell line using Northern Eclipse software 6.0 (EMPIX Imaging Inc., Mississauga, Ontario).
Small Interfering RNA
Predesigned, small interfering RNA (siRNA) for Furin (#105594 and #112945), PC5A (#17520 and #144223), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and Cy3-labeled negative control #1 were purchased from Ambion (Austin, TX). U343 and U251 cells were transfected with Furin siRNA (80 nM) and PC5A siRNA (80 nM), respectively, using Lipofectamine plus reagent. Cells were used in experiments 3 days after transfection. Furin-targeted siRNA sequences have been described previously [29].
Reverse Transcriptase-Polymerase Chain Reaction
Reverse transcriptase-polymerase chain reaction (RT-PCR) was carried out to determine the expression of PCs, and GAPDH was used as a normalizing control. Real-time PCR was carried out to quantify PC5A and Furin expression relative to hS14 expression, as previously described [10]. Primers are listed in Table W1.
Results
The PCs Furin and PC5A Are Differentially Expressed in Glioma Cells
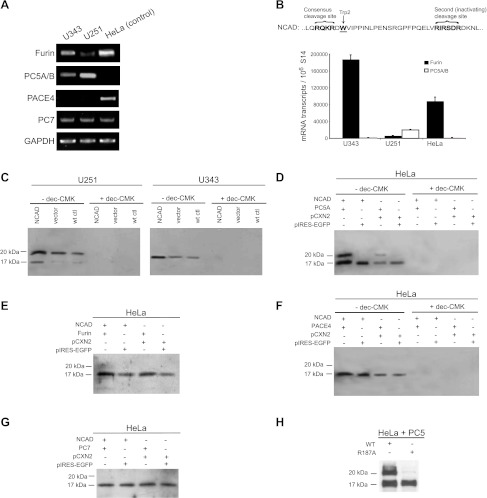
Classic cadherins are synthesized as inactive precursors, which become functionally mature proteins upon posttranslational modifications and cleavage. It has been shown that the PC Furin can cleave human proECAD at RQRR↓DW156 [30], rendering the molecule functionally adhesive by exposing Trp156. Similarly, Furin also cleaves mouse proNCAD at RQRR↓DW161 [21,30] at the C-terminal end of the prodomain, thereby exposing the critical Trp161 (Figure 1B).
Figure 1.
Furin and PC5A PCs mediate cleavage of NCAD at S1 and S2, respectively. (A) Semi-qRT-PCR was carried out to look at expression of PCs, and results show differential expression of Furin and PC5A (the major PC5A isoform is used here) in U343 and U251 cells. GAPDH expression served as reference. (B) Schematic diagram of precursor NCAD protein with the S1 cleavage site and the putative S2. Tryptophan at position 161, necessary for adhesion, is in bold and underlined. Real-time PCR was carried out to quantify Furin and PC5A expression. The results are plotted as number of mRNA messages/106 hS14 messages and show contrasting Furin and PC5A expression patterns in U343 and U251 cells. HeLa cells were used as a positive control for Furin and a negative control for PC5A. (C) U251 and U343 cells were transfected with WT NCAD vector or empty vector and incubated in the presence or absence of dec-RVKR-cmk. The conditioned medium was concentrated (20x) and run on a 15% gel, and NCAD cleavage peptides were detected with the proN Ab. Cleavage products with molecular masses of ∼17 kDa and ∼20 kDa corresponded to processing of NCAD at S1 and S2, respectively. NCAD was predominantly cleaved at S2 in U251 cells and exclusively at S1 in U343 cells. (D) HeLa cells were transiently transfected with NCAD +/- full-length PC5A or empty vector, and cells were incubated in the absence or presence of dec-RVKR-cmk. Cleavage products were detected in the conditioned medium as in C. (E–G) Transfections as in D were carried out. Furin was used in E, PACE4 was used in F, and PC7 was used in G. (H) HeLa cells were transiently transfected with PC5A and either WT NCAD or its R187A mutant (at the P1 position) position, and cleavage products in the conditioned medium were detected as above.
In the present work, we concentrated on the various forms of NCAD found in tumors. Accordingly, we evaluated the mRNA expression levels of the relevant constitutive basic aa-specific PCs, namely, Furin, PC5A, PACE4, and PC7 in a panel of various tumor cells to determine whether differences in levels of these enzymes might underlie the mechanism(s) leading to different forms of cell-surface NCAD. Because only 45% of the highly invasive glioma cells, compared to 70% of metastatic melanoma cells, expressed unprocessed proNCAD at the cell surface owing to the low expression of Furin [29], this suggested that there may be additional mechanisms associated with malignant glioma cell invasion. RT-PCR analyses revealed similar levels of PC7 in U343 and U251 cells, and expression of PACE4 was not detected in either cell line (Figure 1A). Interestingly, the mRNA levels of Furin were low in highly invasive U251 cells, relative to the less invasive U343 cells. In contrast, PC5 mRNA expression was high in U251 cells relative to U343 cells (Figure 1A). Quantitative RT-PCR (qRT-PCR) analyses showed that Furin is 36-fold more expressed than PC5A in U343 cells, whereas PC5A is 4-fold more expressed than Furin in U251 cells (Figure 1B). The relatively low Furin expression in U251 cells could be a plausible explanation for the previously observed presence of proNCAD at the surface of these cells [29]. The contrasting expression of PC5A was intriguing, especially because it was demonstrated that proECAD was processed in the Furin-deficient LoVo cells, indicating that another convertase could process cadherins [30], including proNCAD. Upon scrutiny of the mouse proNCAD sequence, we identified a second putative S2 site at RIRSDR↓DK189 (conserved in human proNCAD) following the primary Furin-cleavable S1 site at RQKR↓DW161 (Figure 1B). Should cleavage occur at S2, the ensuing NCAD-ΔN28 product lacking the N-terminal 28 aa of NCAD would have lost the critical Trp161 and likely be nonadhesive. Using an in-house-generated Ab against the prodomain of NCAD [29], we were surprised to find that the endogenous prodomain of NCAD in U343 and U251 cells exhibits a different molecular size. Thus, in U343 and U251 cells, it predominantly migrates on SDS-PAGE as an ∼17-kDa or an ∼20-kDa protein, respectively (Figure 1C), suggesting differential cleavage by endogenous proteases in these cells. To identify the cognate convertase at the S2 site (generating the ∼20-kDa prodomain), we first showed that incubation of cells with the general PC inhibitor decanoyl-RVKR-chloromethylketone (dec-RVKR-cmk) [25] completely abrogates both S1 and S2 cleavages, suggesting that both are accomplished by one or more PCs (Figure 1C).
To identify which PC is responsible for the generation of the ∼20-kDa prodomain following cleavage at S2, we expressed each PC and proNCAD in HeLa cells (that lack endogenous PC5A [52]). As control, we showed that PC5A and Furin can similarly cleave proPDGF-A [53] in these cells (not shown). The data showed that of all PCs, PC5A is the only convertase capable of generating the ∼20-kDa product from both endogenous and overexpressed proNCAD (Figure 1D), whereas Furin (Figure 1E) and PACE4 (Figure 1F) do not cleave proNCAD at the S2 site, and PC7 does not seem to cleave at either site, as the level of the ∼17-kDa product does not change upon expression of this enzyme (Figure 1G).
To unambiguously identify the important residues governing cleavage at the S2 site, we mutated the putative P1 residue Arg187 to Ala generating the sequence to RIRSDA↓DK189. Co-expression of this R187A mutant with PC5A in HeLa cells, lacking both endogenous NCAD [29] and PC5A expression [52], resulted in the generation of the ∼17-kDa product but not the ∼20-kDa product containing at its C terminus the first 28 aa of mature NCAD, confirming that the PC5A-mediated production of NCAD-ΔN28 by cleavage at position S2 is lost (Figure 1H).
Cell-surface PC5A Cleaves NCAD at S2 in Glioma and HeLa Cells
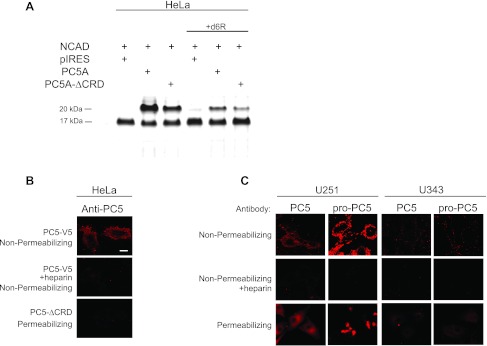
Proteolytic processing of proteins by PCs takes place in the TGN, cell surface, or endosomes [25,26]. To probe the subcellular location(s) where proNCAD processing by PC5A into NCAD-ΔN28 resulting in an ∼20-kDa prodomain takes place, we expressed in HeLa cells a truncated form of PC5A (PC5A-ΔCRD) lacking the C-terminal cysteine-rich domain (CRD) [27,50]. The results showed that co-expression of proNCAD and PC5A-ΔCRD substantially reduced PC5A-mediated proteolysis at S2 but did not abolish it (Figure 2A). This indicates that in large part cleavage at S2 takes place at the plasma membrane, as supported by previous studies showing that the CRD of PC5A mediates cell-surface anchoring of the enzyme through interactions with HSPGs [27,50]. To further support this conclusion, we demonstrated that incubations with the PC cell-surface inhibitor d6R [54,55] only partially abolished the proNCAD processing (Figure 2A), whereas dec-RVKR-cmk [54,55], a cell permeable PC inhibitor, completely blocked processing (Figure 1, C and D). Thus, proNCAD is processed by PC5A both at the cell surface and intracellularly, either at the TGN or following endocytosis.
Figure 2.
Processing of NCAD by PC5A and immunocytochemical localization of PC5A and its prosegment in U251 and U343 cells. (A) Transfections were done as in Figure 1 except that HeLa cells were transfected with either full-length PC5A (here, we only used the major PC5A isoform throughout [25]) or PC5A-ΔCRD. PC5A with a deleted CRD (PC5A-ΔCRD) cleaved NCAD to a much lesser extent compared to full-length PC5A. (B) Immunocytochemistry was carried out to look at PC5A localization in transfected HeLa cells both under nonpermeabilizing and permeabilizing conditions, as described [27]. PC5A C-terminally tagged with a V5 epitope was cloned in the pIRES2-EGFP vector, and hence, cells were probed with a monoclonal Ab-V5 [27]. Cells were transfected with either full-length PC5A, in the absence or presence of heparin and staining was carried out under nonpermeabilizing conditions to detect cell-surface localization. Cells were also transfected with PC5A-ΔCRD and stained under permeabilizing conditions to identify intracellular localization. (C) Immunocytochemistry of U251 and U343 cells demonstrate localization of endogenous PC5A and its inhibitory prosegment at the cell surface and intracellularly in a perinuclear site. An NT-PC5A Ab detected total PC5A protein, and an Ab against the PC5A prosegment (proPC5A) detected only the prosegment containing proteins [27,50]. Staining was carried out under nonpermeabilizing conditions with or without heparin or under permeabilizing conditions. Bar, 10 µm.
We then looked at the localization of PC5A in transfected HeLa cells (Figure 2B). Herein, the protein localized to the cell surface, as assayed by immunofluorescence under nonpermeabilizing conditions (Figure 2B, upper panel); however upon heparin treatment, PC5A was no longer located at the cell surface (Figure 2B, middle panel), befitting its cell-surface binding to HSPGs [27,50]. In addition, PC5A-ΔCRD did not localize to the cell surface of HeLa cells (Figure 2B, lower panel), as also reported for COS-1 cells [27]. Thus, HSPG-bound PC5A localizes to the cell surface [27,50] where it presumably cleaves most of proNCAD at the S2 site.
We next investigated the localization of endogenous PC5A by immunofluorescence and detected substantial levels of the enzyme at the cell surface of U251 cells but very low levels at the surface of U343 cells (Figure 2C, upper panel). As we observed upon PC5A overexpression in HeLa cells (Figure 2B), endogenous PC5A immunoreactivity was not detectable at the cell surface when heparin was added to the medium of glioma cells (Figure 2C, middle panel). Furthermore, as previously reported in COS-1 cells [27], immunoreactivity of the prosegment of PC5A can also be detected at the cell surface of U251 cells (but not U343 cells) with an Ab specific for the prosegment of PC5A [50]. This suggests that a fraction of PC5A is found in an inactive form noncovalently bound to its inhibitory prosegment at the surface of U251 cells, as reported for COS-1 cells [27]. Finally, under permeabilizing conditions, we note that in U251 cells endogenous immunoreactive PC5A and its prosegment mostly localize to a perinuclear compartment, likely the TGN (Figure 2C, bottom panel), similar to its intracellular localization in brain neurons [56]. We deduce that in this compartment a fraction of PC5A is still in an inactive form bound to its inhibitory prosegment. Thus, similar to COS-1 cells [27], we interpret the above data, combined with those in the literature describing the zymogen activation of PC5A [25,27], to mean that autocatalytic cleavage of proPC5A occurs in two steps, a first one in the ER where processing of its prosegment initially takes place at the first position, and then at the cell surface where the autocatalytic activation by cleavage at the second site in the prosegment finally releases the active form of PC5A [27]. On the basis of our present study, this general model seems to apply to two different glioma cell lines, as well as HeLa cells.
Residues Surrounding S2 Dictate the Efficiency of PC5A-mediated Processing of proNCAD
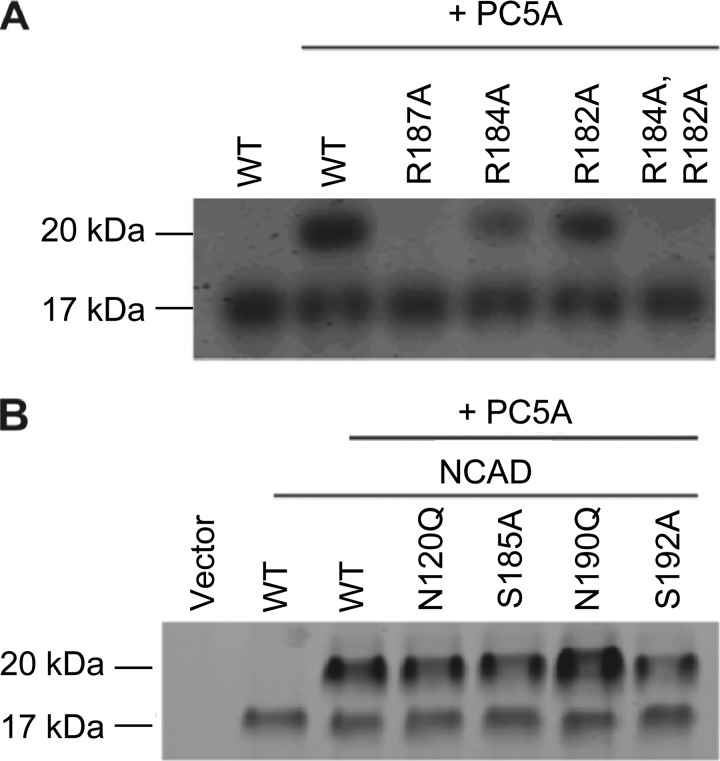
Residues adjacent to the consensus cleavage site (both at P2–P8 and P1′, PC2′) are known to influence the proteolytic efficiency of PCs through specific interactions with S and S′ subsites within the convertases, respectively [25,57–59]. Therefore, we carried out point mutations at the P1, P4, and P6 Arg of mouse proNCAD, corresponding to aa 187, 184, and 182, respectively. The P1 mutation R187A abolished PC5A-mediated proNCAD (Figure 3A), yielding an S2 cleavage-resistant cadherin (NCAD-II). The P4 substitution of R184A substantially diminished the proteolytic activity of PC5A at S2, without abolishing it, whereas the P6 mutant R182A only moderately reduced the proteolytic activity of PC5A at S2 (Figure 3A). However, the double P4, P6 substitution R184A and R182A completely abolished PC5A-mediated proteolysis at S2. These results demonstrate that the basic residues at the P1, P4, and P6 positions are important for PC5A-mediated proNCAD processing at S2, with the P1 Arg187 being the most critical, followed by the P4 and then P6 Arg. Thus, as seen with all other basic aa-specific convertases [25], at least two basic aa are necessary for proNCAD processing by PC5A at S2, namely, the critical P1 Arg and the presence of either a P4 or P6 Arg.
Figure 3.
Critical residues in the PC5A processing of NCAD. (A) HeLa cells were co-transfected with cDNAs coding for PC5A and either WT NCAD or its P1, P4, and P6 mutants. A control with no PC5A expression was included. Twenty-four hours after transfection, the media were analyzed by Western blot using the proN Ab. The migration positions of the NCAD prodomain with apparent molecular masses of ∼17 and 20 kDa are emphasized. (B) In a similar fashion, the PC5A processing of NCAD mutants at specific Asn or Ser residues is shown.
Proteins targeted to the plasma membrane get N- and/or O-glycosylated, and such posttranslational modifications are known to be important for cadherin function [60]. We tested the impact of glycosylation of residues in the vicinity of the S2 site. Our results demonstrate that N-glycosylation at the P3′ Asn190 at the consensus Asn190-X-Ser sequence partially inhibits PC5A-mediated cleavage at S2, because the N190Q mutant is better processed (Figure 3C). However, N120Q, S185A, and S192A have no effect on processing (Figure 3C). We conclude that N-glycosylation at the P3′ site reduces the extent of the PC5A-mediated processing of proNCAD at the S2 position. We also surmise that no O-glycosylation occurs in proximity to the S2 site, which would otherwise have reduced the extent of processing, as observed in multiple precursor proteins [61].
Cleavage of NCAD by Furin or PC5A Determines the Extent of Cellular Migration
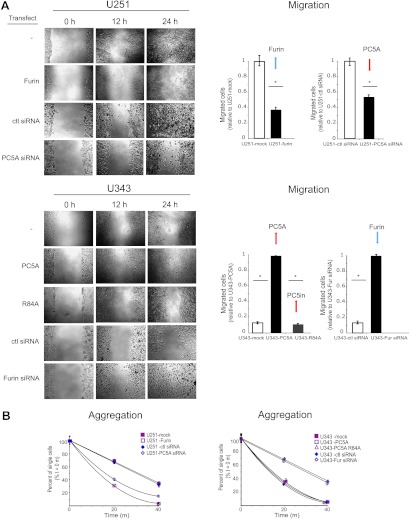
To test the functional significance of proNCAD processing, we carried out gain- and loss-of-function experiments, where U251 cells were stably transfected with Furin (Figures 4 and 5), and U343 cells (Figure 4) were stably transfected with PC5A or a catalytically inactive PC5A mutant lacking the second autocatalytic site (PC5A-R84A; [50]), resulting in its inability to process its inhibitory prosegment that remains noncovalently bound to the mature enzyme keeping it in an inactive state [27]. Our results demonstrate that compared to control transfections, overexpression of Furin in U251 cells resulted in an ∼65% decrease in the number of migrated cells at 12 hours in a wound-healing assay (Figure 4A) and these cells aggregated to a much greater extent compared to control ones (Figure 4B). In contrast, overexpression of PC5A in U343 cells resulted in a significant increase in the number of migrated cells compared to cells transfected with the control plasmid, and these cells formed very few small aggregates compared to control cells (Figure 4B). As expected, these effects were not seen with cells transfected with the inactive PC5A-R84A (Figure 4, A and B). Thus, the changes observed in migratory behavior are consistent with those that modulate cell aggregation.
Figure 4.
Migration and aggregation of U251 and U343 cells depends on the expression of Furin and PC5A. (A) A wound-healing assay was carried out with mock-transfected cells, U251-Furin cells, U343-PC5A cells, or U343 cells transfected with the PC5A-R84A catalytic mutant. In addition, this assay was carried out with U251 cells transfected with PC5A siRNA and with U343 cells transfected with Furin siRNA. Migration was monitored over a 24-hour period, and results were quantified as number of migrated cells at 12 hours. (B) An adhesion assay was carried out with the transfected glioma cells and siRNA-transfected cells. Cell aggregation was monitored over a 40-minute time period, and results were quantified as % single cells over time. Values are means ± SEM.
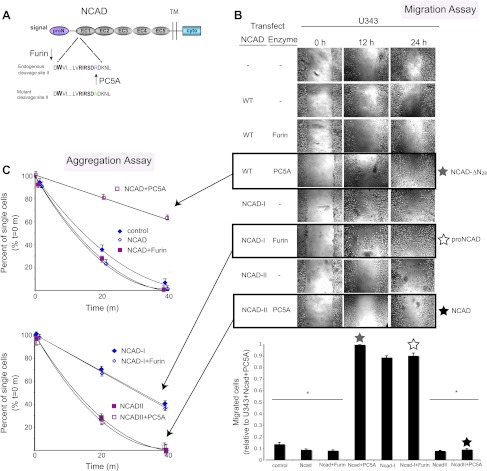
Figure 5.
Proprotein processing of NCAD by Furin or PC5A determines the extent of cellular migration. (A) Schematic diagram of precursor NCAD protein with the endogenous second cleavage site and the engineered mutant nonfunctional site (NCAD-II). Tryptophan at position 2, necessary for adhesion, is in bold. U343 cells were transiently transfected with WT NCAD, proN(Ncad-I), or NCAD mutated at the second cleavage site (NCAD-II), with or without Furin or PC5A, and wound-healing (B) and adhesion assays (C) were carried out to determine the functional effects of NCAD processing by Furin or PC5A at the consensus or the second cleavage site.
We also carried out knockdown experiments using siRNAs specific for PC5A or Furin. Cells were successfully transfected with siRNAs (Table W1), and RT-PCR demonstrated an ∼80% reduction of Furin mRNA levels in U343 cells and PC5A mRNA levels in U251 cells (Figure W1, B and C). Furin or PC5A siRNAs did not affect PC7 or NCAD mRNA levels (Figure W1B). In addition, GAPDH levels were not affected by Furin or PC5A siRNA but were reduced by a GAPDH-specific siRNA (Figure W1B). By immunocytochemistry, we also noted a decrease in Furin and PC5A levels in U343 and U251 cells, respectively, but there was no reduction in tubulin, nestin, or β-catenin expression (Figure W1D). Our siRNA results show that compared to control siRNA, knockdown of PC5A in U251 cells resulted in a significant decrease in cell migration (Figure 4A) and a corresponding increase in cell aggregation (Figure 4B). The effect on migration was somewhat less pronounced than that observed with Furin-transfected cells (Figure 4A, left panel). This can be explained by the fact that U251 cells transfected with PC5A-specific siRNA would still express proNCAD on their surface because of low endogenous Furin expression [29]. Knockdown of Furin in U343 cells resulted in a substantial increase in cell migration (Figure 4A) and decrease in cell aggregation (Figure 4B). Altogether, these results demonstrate that Furin expression appears to inhibit glioma cell migration, whereas PC5A expression promotes it.
To demonstrate the functional importance of proNCAD processing by PC5A or Furin, we used our previously reported NCAD-I mutant lacking the S1 site [29] and engineered an R187A mutant where the second S2 cleavage site was abolished (NCAD-II) but leaving the consensus S1 site RQKR↓DW161 intact (Figure 5A). We then carried out a series of transient transfections in U343 cells, which express low levels of PC5A, with either WT NCAD, NCAD-I, or NCAD-II, alone or in combination with Furin or PC5A. Compared to nontransfected cells, there is a small decrease in migration (Figure 5B) and a small increase in aggregation (Figure 5C) in cells transfected with WT NCAD. This was expected because U343 cells express high levels of Furin needed to process NCAD at the consensus S1 site. A 15% further decrease in migration and increase in aggregation was observed in cells transfected with WT NCAD and Furin (Figure 5, B and C). Importantly, when cells were transfected with NCAD-II and PC5A, the increase in migration detected with WT NCAD and PC5A was not observed (Figure 5B).
Discussion
Morphogenesis and tissue homeostasis depend on the assembly and regulation of cohesive intercellular junctions. Such key processes are finely regulated by calcium-dependent type 1 transmembrane proteins localized to intercellular junctions. Among these, the classic cadherins are the major cell adhesive molecules that modulate the architecture of intercellular junctions. The mature cell-surface localized forms of cadherins exert their cell-cell cohesion by homophilic interactions between adjacent cells. Such binding requires the homodimerization of their extracellular domains (ECs), arranged as five tandem structural segments named EC1 to EC5 (Figure 5A). The primary adhesive interface involves the mutual insertion of a conserved Trp at the second position from the N terminus of mature cadherins (Trp161 of mouse NCAD) into the hydrophobic pocket on EC1 domain (aa 160–267) of the apposing protein from neighboring cells [21]. Indeed, mutating Trp161 into Ala abolishes NCAD-mediated adhesion [62]. Another important structural element regulating the homophilic interaction of NCAD involves a salt bridge between Glu248 in EC1 and the free amine of the N-terminal Asp160 of mature NCAD [63]. Thus, the loss of the NH2-Asp-Trp161 dipeptide at the N terminus of NCAD would be expected to prevent NCAD homodimerization at the intercellular surface.
Unstable intercellular adhesion contributes to cellular detachment and metastatic dissemination of tumor cells. Among the various membrane-bound cadherins, NCAD is a key cell adhesion protein both in normal physiology and during tumorigenesis. It is thought that the strength of the intercellular adhesion is governed by the level of cell-surface NCAD and by the prevalence of its dimeric forms [64]. Up until recently, it was also believed that the N-terminal prodomain in classic cadherins is completely removed by an endoprotease within the late Golgi following association of the catenins, resulting in a mature adhesively competent molecule at the cell surface [4,22,23,65]. However, we recently showed that proNCAD can escape proper cleavage and be expressed as a precursor protein at the cell surface of aggressive brain tumor cells, as well as malignant melanoma cell lines and other human carcinoma cells, likely because of the low expression levels of its processing enzyme Furin [29]. Owing to the development of a specific NCAD prodomain Ab, this was the first reported case of persistence of the prodomain at intercellular surfaces.
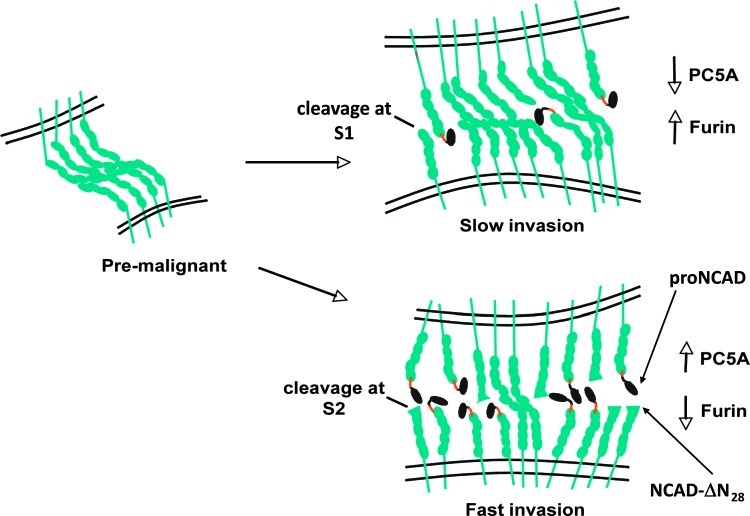
Herein, we report that during malignant transformation, proNCAD undergoes an alternative proteolytic processing by another convertase PC5A. Thus, we demonstrate that in highly invasive brain tumor cells NCAD can be processed by PC5A at an inactivating S2 cleavage site RIRSDR↓DK189. This PC-modulated mechanism results in a mixture of NCAD molecular forms at the cell surface with both proNCAD and NCAD-ΔN28 (Figure 6), functionally enhancing cellular migration and invasion (Figures 4 and 5). The enhanced levels of proNCAD at the cell surface [29] and cleavage at the second S2 site (this work) appear to be because of down-regulation of Furin and up-regulation of PC5A, respectively (Figures 1 and 6). The resulting NCAD-ΔN28, lacking the first 28 aa at the N terminus, would lose the critical N-terminal NH2-Asp-Trp161 sequence and is likely to be nonadhesive (Figures 4 and 5).
Figure 6.
Proposed schematic diagram depicting surface cadherin expression during tumor progression. When progressing from the premalignant to malignant stage, cells with a high proportion of proN and inactivated NCAD have a higher migratory and invasive potential compared to cells with a lower proportion of proNCAD and inactivated NCAD-ΔN28. This corresponds to higher levels of PC5A and lower levels of Furin and lower levels of PC5A and higher levels of Furin, respectively.
Classic cadherins can exist in a weakly adhesive monomeric form or as dimers, which are strongly adhesive [66,67]. We speculate that proNCAD and inactive NCAD-ΔN28 may disrupt NCAD arrays emanating from juxtaposed cells by intercalating between lateral dimers (Figure 6). It has been shown that the prodomain of ECAD prevents dimer formation and that dominant negative forms of cadherins with extracellular domain deletions prevent cadherin self-association. It is conceivable that by altering cadherin composition at the cell surface, the adhesive strength of nascent cell-cell contacts may be regulated, allowing for fine-tuning of malignant intercellular connections.
Would it be possible for proNCAD to be cleaved by Furin intracellularly and then inactivated by PC5A at the cell surface in cells expressing both enzymes? Sequential cleavage of a precursor protein by PCs has been previously demonstrated. The precursor of bone morphogenic protein 4 undergoes serial cleavage at two sites in its prodomain, and differential use of the upstream site determines the activity of the mature protein partially through regulating protein stability [68]. Perhaps in a proportion of highly aggressive tumor cells, NCAD gets cleaved sequentially by Furin and PC5A or proNCAD could be cleaved directly by PC5A at the cell surface. Preliminary results revealed that the PC5A-inactivated NCAD-ΔN28 is much less stable at the cell surface than mature active NCAD (not shown). Thus, the inactivation of NCAD results in the loss of the critical Trp161 for adhesion and decreased levels of cell-surface NCAD.
Mechanistically, we demonstrated that by determining the site of NCAD processing the convertases Furin and PC5A have opposing effects on the extent of intercellular adhesion and cellular motility (Figures 4 and 5). Perhaps a decrease in PC5A expression is an early event necessary for cells to associate with their neighbors and stop invading. It is conceivable that although PC5A cleavage of NCAD seems to be critical for invasion, the cleavage of other substrates by this convertase is also likely to be important for invasion or other traits acquired by highly malignant cells. Indeed, it has been shown that PCs are important for the activation of other cancer-related substrates. For example, in human head and neck squamous cell carcinoma and astrocytoma cells, Furin has been shown to be important for activation of substrates important in tumorigenesis, such as membrane type 1 matrix metalloproteinase, transforming growth factor-β, insulin-like growth factor-1 receptor, and vascular endothelial growth factor C [35,69,70]. Processing of the insulin-like growth factor-1 receptor by PCs including Furin and PC5A, as well as processing of proPDGF-A mainly by Furin, has also been shown to be important in human colon carcinoma cells [31,53].
In general, inhibition of the convertases has been shown to decrease growth and tumorigenicity, as this group of proteases is known to process substrates important for tumor malignancy and cellular invasion [31,32,35,69]. However, recent studies demonstrated that PCs may, in some instances, be protective against tumorigenesis, cell migration, and metastases [40,41,71]. Our studies demonstrate that the effect of PC processing on tumor malignancy appears to be more complex than previously thought. Moreover, this complexity is certainly much greater in vivo compared to cells grown in vitro. The inactivation of a PC substrate at the plasma membrane is not unprecedented. Indeed, pro-endothelial lipase is retained at the cell-surface through HSPGs and was demonstrated to be inactivated by cleavage with full-length PACE4 and/or PC5A [72] but not by either of these PCs lacking their HSPG-binding CRDs, [27,50]. In our study, we demonstrate a second instance where the CRD of PC5A is necessary for cell-surface cleavage leading to inactivation of the NCAD likely in association with cell-surface HSPGs.
Down-regulation of the level of cell adhesion molecules and/or their dimerization represents one way by which cells reduce intercellular adhesions. In many carcinomas, epithelial junctions are no longer detectable because of the loss of close cell-cell contacts. This is an important step during the epithelial-to-mesenchymal transition (EMT) that is believed to occur during carcinogenesis [73]. Loss of cell-cell adhesion is followed by other important phenotypic changes during EMT such as loss of cell polarity and the acquisition of migratory and invasive properties. The loss of ECAD is considered to be a critical event in EMT, as ECAD repressors are capable of functioning as full EMT inducers in different cell types [74]. This is accompanied by an upregulation of NCAD in many types of carcinomas and the switch from ECAD to NCAD correlates with induced cellular motility [2,3,16]. It is believed that because of disruption of adhesion junctions, loss of ECAD allows malignant cells to detach from the epithelium and invade host tissue, whereas up-regulation of NCAD expression facilitates invasion of tumor cells by mediating adhesion of malignant cells to stromal or endothelial cells rather than epithelial cells [75].
Alternatively, as we have demonstrated, intercellular adhesion could be modulated by surface expression of a nonadhesive proNCAD [29], or NCAD-ΔN28 (this work). Thus, our results should lead to the revision of the current model of cadherin posttranslational processing and surface expression [3,16,18,76,77]. We propose that in later stages of tumor progression nonadhesive surface proNCAD and functionally inactivated NCAD-ΔN28 determine the degree of cell invasiveness and metastasis and shed light on the stages of malignancy during tumor progression. Consequently, the switch from ECAD to NCAD during EMT is in fact a switch to a mixture of NCAD molecules where only a certain proportion is functionally adhesive. In brain tumor cells, which do not undergo an ECAD-to-NCAD shift, the switch from mature NCAD to nonadhesive NCAD-ΔN28 molecules mediates detachment from the main tumor mass and migration over extensive distances in our in vitro assay (Figures 4 and 5). Preliminary data in an intracranial xenograft mouse model show that prevention of the formation of NCAD-ΔN28 results in a less aggressive phenotype compared to WT (in preparation).
We speculate that differential expression of PCs may be a common mechanism in many types of tumors to regulate cellular motility and perhaps other malignant traits and is perhaps central in developmental programs. In addition, precursor forms of other types of cadherins at the cell surface may play a similar role in tumor cell motility. Accordingly, alignment of the processing sites of various cell adhesion molecules (Figure W2) revealed that the seminal observation reported here may be more general than previously appreciated. Indeed, the Furin dibasic cleavage site and the alternative PC5A-mediated monobasic processing site characterized herein for proNCAD are found to be conserved in a number of cadherin family members including ECAD, P-cadherin, and cadherin-11 that play pivotal roles in cancer development and progression [76,78,79]. It remains to be defined if PC5A also mediates these inactivation processes and whether such a mechanism regulates the functions of these adhesion molecules both physiologically and/or pathologically. In that context, mining the ONCOMINE cancer gene expression database (http://www.oncomine.org) revealed that PC5A expression was significantly reduced in most tumors [41] but was actually upregulated in 3 of 10 tumor types including pancreatic ductal adenocarcinoma, glioblastoma multiforme, and anaplastic oligoastrocytoma/oligodendroglioma (Figure W3). On the basis of our present work, it is possible that in these latter types of tumors PC5A would also promote cancer metastasis.
Malignant primary brain tumors are extremely complex and heterogeneous microenvironments, characterized by subpopulations of highly invasive cells that infiltrate the normal brain parenchyma, often significant distances from the primary tumor mass [80]. Unlike most carcinomas, these tumors do not enter blood vessels and form metastases in distant organs. Instead, fatality is because of “micro-metastasis” within the brain, resulting in destruction of regions essential for survival of the patient [81]. Uncontrollable invasion leads to failure of treatment modalities such as radiotherapy and chemotherapy and recurrence following radical resection.
The mechanism proposed herein sheds light on putative novel treatment strategies with the potential to attenuate extensive infiltration of the brain parenchyma by malignant tumor cells and reduce the frequency of fatal recurrences following tumor resection. Furthermore, in cancers such as malignant melanoma and malignant glial tumors [29], characterization of the cell-surface forms of NCAD may serve as a prognostic tool for staging and progression of the disease. Our work may potentially lead to treatment strategies to control the metastatic spread of malignant brain carcinomas, through the development of specific PC5 inhibitors.
Supplementary Material
Abbreviations
- CRD
cysteine-rich domain
- ECAD
E-cadherin
- NCAD
N-cadherin
- PC
proprotein convertase
- proNCAD
precursor of N-cadherin
- WT
wild type
Footnotes
This work was supported in part by Canadian Institutes of Health Research (CIHR) grant MOP 44363, Canada Chair 216684, and a Strauss Foundation grant (to N.G.S.) and also by the Goals for Lily, the Alex Pavanel Family, the Raymonde and Tony Boeckh and the Maggie De Fontes Funds for Brain Tumour Research, and the Montréal English School Board. Funding was also obtained from the Franco Di Giovanni, B-Strong, and the Tony Colannino Foundations and the Brain Tumour Foundation of Canada. R.F.D.M. holds the William Feindel Chair of Neurooncology at the Montreal Neurological Institute. D.M. is supported by CIHR and RF Tomlinson Studentships. M.S.S. is supported by the Christian Geada Studentship, the McGill FOM Fellowship, and the Jeanne Timmons Costello Studentship.
This article refers to supplementary materials, which are designated by Table W1 and Figures W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Birchmeier W. Molecular aspects of the loss of cell adhesion and gain of invasiveness in carcinomas. Princess Takamatsu Symp. 1994;24:214–232. [PubMed] [Google Scholar]
- 2.Suyama K, Shapiro I, Guttman M, Hazan RB. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell. 2002;2:301–314. doi: 10.1016/s1535-6108(02)00150-2. [DOI] [PubMed] [Google Scholar]
- 3.Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147:631–644. doi: 10.1083/jcb.147.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozawa M, Kemler R. Correct proteolytic cleavage is required for the cell adhesive function of uvomorulin. J Cell Biol. 1990;111:1645–1650. doi: 10.1083/jcb.111.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeichi M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development. 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- 6.Christofori G. New signals from the invasive front. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- 7.Hsu MY, Meier FE, Nesbit M, Hsu JY, Van Belle BP, Elder DE, Herlyn M. E-cadherin expression in melanoma cells restores keratinocyte-mediated growth control and down-regulates expression of invasion-related adhesion receptors. Am J Pathol. 2000;156:1515–1525. doi: 10.1016/S0002-9440(10)65023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155–163. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- 9.Frixen UH, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Lochner D, Birchmeier W. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113:173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vleminckx K, Vakaet L, Jr, Mareel M, Fiers W, van Roy RF. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66:107–119. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 11.Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806–811. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- 12.Birchmeier W, Behrens J, Weidner KM, Frixen UH, Schipper J. Dominant and recessive genes involved in tumor cell invasion. Curr Opin Cell Biol. 1991;3:832–840. doi: 10.1016/0955-0674(91)90057-6. [DOI] [PubMed] [Google Scholar]
- 13.Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 14.Hirohashi S. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol. 1998;153:333–339. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christofori G, Semb H. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem Sci. 1999;24:73–76. doi: 10.1016/s0968-0004(98)01343-7. [DOI] [PubMed] [Google Scholar]
- 16.Islam S, Carey TE, Wolf GT, Wheelock MJ, Johnson KR. Expression of N-cadherin by human squamous carcinoma cells induces a scattered fibroblastic phenotype with disrupted cell-cell adhesion. J Cell Biol. 1996;135:1643–1654. doi: 10.1083/jcb.135.6.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazan RB, Kang L, Whooley BP, Borgen PI. N-cadherin promotes adhesion between invasive breast cancer cells and the stroma. Cell Adhes Commun. 1997;4:399–411. doi: 10.3109/15419069709004457. [DOI] [PubMed] [Google Scholar]
- 18.Hazan RB, Phillips GR, Qiao RF, Norton L, Aaronson SA. Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J Cell Biol. 2000;148:779–790. doi: 10.1083/jcb.148.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, Herlyn M. Dynamics of intercellular communication during melanoma development. Mol Med Today. 2000;6:163–169. doi: 10.1016/s1357-4310(00)01692-0. [DOI] [PubMed] [Google Scholar]
- 20.Asano K, Duntsch CD, Zhou Q, Weimar JD, Bordelon D, Robertson JH, Pourmotabbed T. Correlation of N-cadherin expression in high grade gliomas with tissue invasion. J Neurooncol. 2004;70:3–15. doi: 10.1023/b:neon.0000040811.14908.f2. [DOI] [PubMed] [Google Scholar]
- 21.Koch AW, Farooq A, Shan W, Zeng L, Colman DR, Zhou MM. Structure of the neural (N-) cadherin prodomain reveals a cadherin extracellular domain-like fold without adhesive characteristics. Structure. 2004;12:793–805. doi: 10.1016/j.str.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 22.Wahl JK, III, Kim YJ, Cullen JM, Johnson KR, Wheelock MJ. N-cadherin-catenin complexes form prior to cleavage of the proregion and transport to the plasma membrane. J Biol Chem. 2003;278:17269–17276. doi: 10.1074/jbc.M211452200. [DOI] [PubMed] [Google Scholar]
- 23.Shore EM, Nelson WJ. Biosynthesis of the cell adhesion molecule uvomorulin (E-cadherin) in Madin-Darby canine kidney epithelial cells. J Biol Chem. 1991;266:19672–19680. [PubMed] [Google Scholar]
- 24.Seidah NG, Chretien M. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 1999;848:45–62. doi: 10.1016/s0006-8993(99)01909-5. [DOI] [PubMed] [Google Scholar]
- 25.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11:367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 26.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer G, Hamelin J, Asselin MC, Pasquato A, Marcinkiewicz E, Tang M, Tabibzadeh S, Seidah NG. The regulated cell surface zymogen activation of the proprotein convertase PC5A directs the processing of its secretory substrates. J Biol Chem. 2008;283:2373–2384. doi: 10.1074/jbc.M708763200. [DOI] [PubMed] [Google Scholar]
- 28.Rousselet E, Benjannet S, Marcinkiewicz E, Asselin MC, Lazure C, Seidah NG. Proprotein convertase PC7 enhances the activation of the EGF receptor pathway through processing of the EGF precursor. J Biol Chem. 2011;286:9185–9195. doi: 10.1074/jbc.M110.189936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maret D, Gruzglin E, Sadr MS, Siu V, Shan W, Koch AW, Seidah NG, Del Maestro RF, Colman DR. Surface expression of precursor N-cadherin promotes tumor cell invasion. Neoplasia. 2010;12:1066–1080. doi: 10.1593/neo.10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posthaus H, Dubois CM, Laprise MH, Grondin F, Suter MM, Muller E. Proprotein cleavage of E-cadherin by furin in baculovirus over-expression system: potential role of other convertases in mammalian cells. FEBS Lett. 1998;438:306–310. doi: 10.1016/s0014-5793(98)01330-1. [DOI] [PubMed] [Google Scholar]
- 31.Khatib AM, Siegfried G, Prat A, Luis J, Chretien M, Metrakos P, Seidah NG. Inhibition of proprotein convertases is associated with loss of growth and tumorigenicity of HT-29 human colon carcinoma cells: importance of insulin-like growth factor-1 (IGF-1) receptor processing in IGF-1-mediated functions. J Biol Chem. 2001;276:30686–30693. doi: 10.1074/jbc.M101725200. [DOI] [PubMed] [Google Scholar]
- 32.Khatib AM, Siegfried G, Chretien M, Metrakos P, Seidah NG. Proprotein convertases in tumor progression and malignancy: novel targets in cancer therapy. Am J Pathol. 2002;160:1921–1935. doi: 10.1016/S0002-9440(10)61140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassi DE, Fu J, Lopez DC, Klein-Szanto AJ. Proprotein convertases: “master switches” in the regulation of tumor growth and progression. Mol Carcinog. 2005;44:151–161. doi: 10.1002/mc.20134. [DOI] [PubMed] [Google Scholar]
- 34.Coppola JM, Bhojani MS, Ross BD, Rehemtulla A. A small-molecule furin inhibitor inhibits cancer cell motility and invasiveness. Neoplasia. 2008;10:363–370. doi: 10.1593/neo.08166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassi DE, Lopez DC, Mahloogi H, Zucker S, Thomas G, Klein-Szanto AJ. Furin inhibition results in absent or decreased invasiveness and tumorigenicity of human cancer cells. Proc Natl Acad Sci USA. 2001;98:10326–10331. doi: 10.1073/pnas.191199198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D'Anjou F, Routhier S, Perreault JP, Latil A, Bonnel D, Fournier I, Salzet M, Day R. Molecular validation of PACE4 as a target in prostate cancer. Transl Oncol. 2011;4:157–172. doi: 10.1593/tlo.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scamuffa N, Siegfried G, Bontemps Y, Ma L, Basak A, Cherel G, Calvo F, Seidah NG, Khatib AM. Selective inhibition of proprotein convertases represses the metastatic potential of human colorectal tumor cells. J Clin Invest. 2008;118:352–363. doi: 10.1172/JCI32040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bassi DE, Zhang J, Cenna J, Litwin S, Cukierman E, Klein-Szanto AJ. Proprotein convertase inhibition results in decreased skin cell proliferation, tumorigenesis, and metastasis. Neoplasia. 2010;12:516–526. doi: 10.1593/neo.92030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lapierre M, Siegfried G, Scamuffa N, Bontemps Y, Calvo F, Seidah NG, Khatib AM. Opposing function of the proprotein convertases furin and PACE4 on breast cancer cells' malignant phenotypes: role of tissue inhibitors of metalloproteinase-1. Cancer Res. 2007;67:9030–9034. doi: 10.1158/0008-5472.CAN-07-0807. [DOI] [PubMed] [Google Scholar]
- 40.Huang Y-H, Lin K-H, Liao CH, Lai M-W, Tseng Y-H, Yeh C. Furin overexpression suppresses tumor growth and predicts a better postoperative disease-free survival in hepatocellular carcinoma. PLoS One. 2012;7:e40738. doi: 10.1371/journal.pone.0040738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun X, Essalmani R, Seidah NG, Prat A. The proprotein convertase PC5/6 is protective against intestinal tumorigenesis: in vivo mouse model. Mol Cancer. 2009;8:73. doi: 10.1186/1476-4598-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 43.Matsunaga M, Hatta K, Nagafuchi A, Takeichi M. Guidance of optic nerve fibres by N-cadherin adhesion molecules. Nature. 1988;334:62–64. doi: 10.1038/334062a0. [DOI] [PubMed] [Google Scholar]
- 44.Kiryushko D, Berezin V, Bock E. Regulators of neurite outgrowth: role of cell adhesion molecules. Ann N Y Acad Sci. 2004;1014:140–154. doi: 10.1196/annals.1294.015. [DOI] [PubMed] [Google Scholar]
- 45.Skaper SD. Neuronal growth-promoting and inhibitory cues in neuroprotection and neuroregeneration. Ann N Y Acad Sci. 2005;1053:376–385. doi: 10.1196/annals.1344.032. [DOI] [PubMed] [Google Scholar]
- 46.Hatta K, Takeichi M. Expression of N-cadherin adhesion molecules associated with early morphogenetic events in chick development. Nature. 1986;320:447–449. doi: 10.1038/320447a0. [DOI] [PubMed] [Google Scholar]
- 47.Derycke LD, Bracke ME. N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int J Dev Biol. 2004;48:463–476. doi: 10.1387/ijdb.041793ld. [DOI] [PubMed] [Google Scholar]
- 48.Tucker RP. Neural crest cells: a model for invasive behavior. Int J Biochem Cell Biol. 2004;36:173–177. doi: 10.1016/s1357-2725(03)00243-7. [DOI] [PubMed] [Google Scholar]
- 49.Reines A, Bernier LP, McAdam R, Belkaid W, Shan W, Koch AW, Seguela P, Colman DR, Dhaunchak AS. N-cadherin prodomain processing regulates synaptogenesis. J Neurosci. 2012;32:6323–6334. doi: 10.1523/JNEUROSCI.0916-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nour N, Mayer G, Mort JS, Salvas A, Mbikay M, Morrison CJ, Overall CM, Seidah NG. The cysteine-rich domain of the secreted proprotein convertases PC5A and PACE4 functions as a cell surface anchor and interacts with tissue inhibitors of metalloproteinases. Mol Biol Cell. 2005;16:5215–5226. doi: 10.1091/mbc.E05-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rousselet E, Benjannet S, Hamelin J, Canuel M, Seidah NG. The proprotein convertase PC7: unique zymogen activation and trafficking pathways. J Biol Chem. 2011;286:2728–2738. doi: 10.1074/jbc.M110.192344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Essalmani R, Hamelin J, Marcinkiewicz J, Chamberland A, Mbikay M, Chretien M, Seidah NG, Prat A. Deletion of the gene encoding proprotein convertase 5/6 causes early embryonic lethality in the mouse. Mol Cell Biol. 2006;26:354–361. doi: 10.1128/MCB.26.1.354-361.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siegfried G, Khatib AM, Benjannet S, Chretien M, Seidah NG. The proteolytic processing of pro-platelet-derived growth factor-A at RRKR86 by members of the proprotein convertase family is functionally correlated to plateletderived growth factor-A-induced functions and tumorigenicity. Cancer Res. 2003;63:1458–1463. [PubMed] [Google Scholar]
- 54.Kacprzak MM, Peinado JR, Than ME, Appel J, Henrich S, Lipkind G, Houghten RA, Bode W, Lindberg I. Inhibition of furin by polyarginine-containing peptides: nanomolar inhibition by nona-d-arginine. J Biol Chem. 2004;279:36788–36794. doi: 10.1074/jbc.M400484200. [DOI] [PubMed] [Google Scholar]
- 55.Susan-Resiga D, Essalmani R, Hamelin J, Asselin MC, Benjannet S, Chamberland A, Day R, Szumska D, Constam D, Bhattacharya S, et al. Furin is the major processing enzyme of the cardiac-specific growth factor bone morphogenetic protein 10. J Biol Chem. 2011;286:22785–22794. doi: 10.1074/jbc.M111.233577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villeneuve P, Seidah NG, Beaudet A. Immunohistochemical distribution of the prohormone convertase PC5-A in rat brain. Neuroscience. 1999;92:641–654. doi: 10.1016/s0306-4522(98)00763-5. [DOI] [PubMed] [Google Scholar]
- 57.Henrich S, Lindberg I, Bode W, Than ME. Proprotein convertase models based on the crystal structures of furin and kexin: explanation of their specificity. J Mol Biol. 2005;345:211–227. doi: 10.1016/j.jmb.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 58.Henrich S, Cameron A, Bourenkov GP, Kiefersauer R, Huber R, Lindberg I, Bode W, Than ME. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat Struct Biol. 2003;10:520–526. doi: 10.1038/nsb941. [DOI] [PubMed] [Google Scholar]
- 59.Komiyama T, VanderLugt B, Fugere M, Day R, Kaufman RJ, Fuller RS. Optimization of protease-inhibitor interactions by randomizing adventitious contacts. Proc Natl Acad Sci USA. 2003;100:8205–8210. doi: 10.1073/pnas.1032865100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo HB, Johnson H, Randolph M, Pierce M. Regulation of homotypic cell-cell adhesion by branched N-glycosylation of N-cadherin extracellular EC2 and EC3 domains. J Biol Chem. 2009;284:34986–34997. doi: 10.1074/jbc.M109.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gram Schjoldager KT, Vester-Christensen MB, Goth CK, Petersen TN, Brunak S, Bennett EP, Levery SB, Clausen H. A systematic study of site-specific GalNAc-type O-glycosylation modulating proprotein convertase processing. J Biol Chem. 2011;286:40122–40132. doi: 10.1074/jbc.M111.287912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leckband D, Prakasam A. Mechanism and dynamics of cadherin adhesion. Annu Rev Biomed Eng. 2006;8:259–287. doi: 10.1146/annurev.bioeng.8.061505.095753. [DOI] [PubMed] [Google Scholar]
- 63.Harrison OJ, Bahna F, Katsamba PS, Jin X, Brasch J, Vendome J, Ahlsen G, Carroll KJ, Price SR, Honig B, et al. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17:348–357. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka H, Shan W, Phillips GR, Arndt K, Bozdagi O, Shapiro L, Huntley GW, Benson DL, Colman DR. Molecular modification of N-cadherin in response to synaptic activity. Neuron. 2000;25:93–107. doi: 10.1016/s0896-6273(00)80874-0. [DOI] [PubMed] [Google Scholar]
- 65.Ozawa M. Lateral dimerization of the E-cadherin extracellular domain is necessary but not sufficient for adhesive activity. J Biol Chem. 2002;277:19600–19608. doi: 10.1074/jbc.M202029200. [DOI] [PubMed] [Google Scholar]
- 66.Shan W, Yagita Y, Wang Z, Koch A, Fex SA, Gruzglin E, Pedraza L, Colman DR. The minimal essential unit for cadherin-mediated intercellular adhesion comprises extracellular domains 1 and 2. J Biol Chem. 2004;279:55914–55923. doi: 10.1074/jbc.M407827200. [DOI] [PubMed] [Google Scholar]
- 67.Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 68.Cui Y, Hackenmiller R, Berg L, Jean F, Nakayama T, Thomas G, Christian JL. The activity and signaling range of mature BMP-4 is regulated by sequential cleavage at two sites within the prodomain of the precursor. Genes Dev. 2001;15:2797–2802. doi: 10.1101/gad.940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez DC, Bassi DE, Zucker S, Seidah NG, Klein-Szanto AJ. Human carcinoma cell growth and invasiveness is impaired by the propeptide of the ubiquitous proprotein convertase furin. Cancer Res. 2005;65:4162–4171. doi: 10.1158/0008-5472.CAN-04-2820. [DOI] [PubMed] [Google Scholar]
- 70.Mercapide J, Lopez DC, Bassi DE, Castresana JS, Thomas G, Klein-Szanto AJ. Inhibition of furin-mediated processing results in suppression of astrocytoma cell growth and invasiveness. Clin Cancer Res. 2002;8:1740–1746. [PubMed] [Google Scholar]
- 71.Nejjari M, Berthet V, Rigot V, Laforest S, Jacquier MF, Seidah NG, Remy L, Bruyneel E, Scoazec JY, Marvaldi J, et al. Inhibition of proprotein convertases enhances cell migration and metastases development of human colon carcinoma cells in a rat model. Am J Pathol. 2004;164:1925–1933. doi: 10.1016/S0002-9440(10)63753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin W, Fuki IV, Seidah NG, Benjannet S, Glick JM, Rader DJ. Proprotein convertases are responsible for proteolysis and inactivation of endothelial lipase. J Biol Chem. 2005;280:36551–36559. doi: 10.1074/jbc.M502264200. [DOI] [PubMed] [Google Scholar]
- 73.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 74.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 75.Qi J, Chen N, Wang J, Siu CH. Transendothelial migration of melanoma cells involves N-cadherin-mediated adhesion and activation of the beta-catenin signaling pathway. Mol Biol Cell. 2005;16:4386–4397. doi: 10.1091/mbc.E05-03-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomita K, van Leenders BA, van Leenders GJ, Ruijter ET, Jansen CF, Bussemakers MJ, Schalken JA. Cadherin switching in human prostate cancer progression. Cancer Res. 2000;60:3650–3654. [PubMed] [Google Scholar]
- 77.Li G, Satyamoorthy K, Herlyn M. N-cadherin-mediated intercellular interactions promote survival and migration of melanoma cells. Cancer Res. 2001;61:3819–3825. [PubMed] [Google Scholar]
- 78.Bussemakers MJ, Van Bokhoven BA, Tomita K, Jansen CF, Schalken JA. Complex cadherin expression in human prostate cancer cells. Int J Cancer. 2000;85:446–450. [PubMed] [Google Scholar]
- 79.Nakajima G, Patino-Garcia A, Bruheim S, Xi Y, San JM, Lecanda F, Sierrasesumaga L, Muller C, Fodstad O, Ju J. CDH11 expression is associated with survival in patients with osteosarcoma. Cancer Genomics Proteomics. 2008;5:37–42. [PubMed] [Google Scholar]
- 80.Bigner DD, Brown MT, Friedman AH, Coleman RE, Akabani G, Friedman HS, Thorstad WL, McLendon RE, Bigner SH, Zhao XG, et al. Iodine-131-labeled antitenascin monoclonal antibody 81C6 treatment of patients with recurrent malignant gliomas: phase I trial results. J Clin Oncol. 1998;16:2202–2212. doi: 10.1200/JCO.1998.16.6.2202. [DOI] [PubMed] [Google Scholar]
- 81.Burger PC. Morphologic correlates in gliomas: where do we stand? Monogr Pathol. 1990;32:16–29. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.