Abstract
The extent of tight junction (TJ) formation is one of many factors that regulate motility, invasion, and metastasis. Claudins are required for the formation and maintenance of TJs. Claudin-3 (CLDN3) and claudin-4 (CLDN4) are highly expressed in the majority of ovarian cancers. We report here that CLDN3 and CLDN4 each serve to constrain the growth of human 2008 cancer xenografts and limit metastatic potential. Knockdown of CLDN3 increased in vivo growth rate by 2.3-fold and knockdown of CLDN4 by 3.7-fold in the absence of significant change in in vitro growth rate. Both types of tumors exhibited increase in birth rate as measured by Ki67 staining and decrease in death rate as reflected by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining. Knockdown of either claudin did not alter expression of other TJ protein but did reduce TJ formation as measured by transepithelial resistance and paracellular flux of dextran, enhance migration and invasion in in vitro assays, and increase lung colonization following intravenous injection. Knockdown of CLDN3 and CLDN4 increased total lung metastatic burden by 1.7-fold and 2.4-fold, respectively. Loss of either CLDN3 or CLDN4 resulted in down-regulation of E-cadherin mRNA and protein, increased inhibitory phosphorylation of glycogen synthase kinase-3β (GSK-3β), and activation of β-catenin pathway signaling as evidenced by increases in nuclear β-catenin, the dephosphorylated form of the protein, and transcriptional activity of β-catenin/T-cell factor (TCF). We conclude that both CLDN3 and CLDN4 mediate interactions with other cells in vivo that restrain growth and metastatic potential by sustaining expression of E-cadherin and limiting β-catenin signaling.
Introduction
The mechanisms that control intercellular adhesion are central to the process of invasion and metastasis. Normal epithelial cells are held together by tight junctions (TJs), adherens junctions (AJs), and gap junctions. These serve two roles: they mechanically link cells, and they generate signals that are sent to the interior of the cell to report on the extent of contact with neighbors and the extracellular matrix [1,2]. One of the hallmarks of malignant transformation of epithelia is that these connections, particularly TJ, are lost [3–5]. Disassembly or remodeling of TJs can cause loss of cell polarity and an increase in motility and invasiveness [6–8], and there is an association between the loss of cell-cell adhesion structures and metastasis in many epithelial cancers [9].
Normal ovarian surface cuboidal epithelial cells do not express either claudin-3 (CLDN3) or claudin-4 (CLDN4); however, CLDN3 and CLDN4 are expressed at high levels in as many as 92% of ovarian cancers [10,11]. Initial studies based on the idea that the ovarian cancers arose from the ovarian surface cuboidal epithelium concluded that CLDN3 and CLDN4 were upregulated in ovarian cancers [12–16]. However, recent studies favor the concept that these tumors arise from the distal Fallopian tube, which expresses both of these claudins [17–20]. Recent expression profiling studies have demonstrated a wide range of CLDN3 and CLDN4 expression among epithelial ovarian cancers with a substantial fraction having low-level expression [21]. Such low expression has been linked to a mesenchymal pattern and poor prognosis in breast [22], esophageal [23], colon [24,25], and pancreatic carcinoma [26]. What role these proteins serve in ovarian cancers is largely unknown and data from studies addressing this question have yielded conflicting results (reviewed in [27–29]). For example, in one study, low CLDN3 protein expression was found to be associated with a trend toward poor survival in 115 primary ovarian carcinomas [16]. However, in another study, high CLDN3 was correlated with shorter survival [10]. It is generally believed that claudin expression is deregulated in cancer in a complex and organ-dependent manner, and what role these proteins serve in ovarian cancers remains poorly defined.
The concept that the claudins of the TJ can regulate tumor cell behavior has precedence in the well-documented ability of E-cadherin, the major structural protein of the AJ, to do this. Loss of E-cadherin is a hallmark of the epithelial-to-mesenchymal transition (EMT) through which many investigators believe tumor cells must pass to become metastatic [8,30,31]. Passage through the EMT is widely reported to result in increased growth rate, enhanced metastatic potential, drug resistance, and the acquisition of stem cell characteristics [32]. The EMT generates cells with properties of stem cells [33–35]. In ovarian cancer cell lines, knockdown of E-cadherin has been reported to increase tumor cell growth and constitutive activation of phosphoinositide 3-kinase (PI3K)/Akt signaling. Silencing of E-cadherin releases membrane-bound β-catenin and enhances nuclear β-catenin signaling, an effect augmented by inactivation of GSK-3β by the PI3K/Akt pathway [36]. Loss of E-cadherin staining is common as ovarian cancer progresses and this is associated with more extensive peritoneal dissemination [37] as a result of up-regulation of the α5 integrin that results in enhanced invasiveness and adhesion to the peritoneal surface [38]. Thus, although the role of CLDN3 and CLDN4 found in TJ is not well defined, the role of a major structural protein of the AJ in controlling tumor cell behavior is well established and provides a precedent for analysis of the influence of claudins.
There are now several lines of indirect evidence indicating that loss of CLDN3 and CLDN4 expression is associated with poor prognosis. Delocalization of claudins from membranes is common among transformed cells, and in ovarian cancer, this is associated with increased migration and invasion [39,40]. Disruption of cell-cell adhesions correlates with progression, invasion, and metastasis in breast cancer [9,41,42], and low expression of CLDN4 is associated with poor prognosis in some subtypes of breast [22,43,44], pancreatic [26], and colon cancers [24,25]. We report here that both CLDN3 and CLDN4 individually control the growth rate and metastasis potential of cancer cells, processes that are fundamental to the biology and clinical management of this disease. Constitutive knockdown of the expression of either CLDN3 or CLDN4 in the putative ovarian cancer 2008 cells increases in vivo growth rate and enhances metastasis in a nu/nu mouse model. The magnitude of these effects is quite large and is accompanied by alterations in adhesion, migration, invasion, E-cadherin, and activity of the canonical Wnt signaling pathway through β-catenin.
Materials and Methods
Cells, Cell Culture, and Transfection
The human cell lines 2008 and HEY were grown in RPMI 1640 supplemented with 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin. Both of these cell lines were reported to be isolated from patients with ovarian cancer [45,46]. However, DNA fingerprinting has disclosed that the 2008 line may be related to the cervical carcinoma cell line ME-180 (ATCC HTB-33) and that the HEY cell line may be related to the neuronal cell line CRL-10442 (ATCC HCN-1A). Their CLDN3 or CLDN4 knockdown or overexpressing sublines were cultured in the presence of 10 µg/ml puromycin. Cultures were maintained at 37°C in a humidified atmosphere of 5% CO2 and 95% air. For measurement of β-catenin transcriptional activity, the cells were co-transfected with the firefly luciferase TOPflash TCF reporter and control plasmid pCMXβgal by using the FuGENE transfection reagent (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. Twenty-four hours after transfection, cells were lysed in potassium phosphate buffer containing 1% Triton X-100, and light emission was detected in the presence of luciferin by using a microtiter plate luminometer (MicroBeta TriLux, Gaithersburg, MD). The luciferase values were normalized for variations in transfection efficiency by using the β-galactosidase internal control and are expressed either as relative luciferase units or as fold stimulation of luciferase activity compared with the designated control cultures. All of the transfection results represent means of a minimum of three independent transfections assayed in duplicate (±SEM).
Lentivirus-based Short Hairpin RNA Transduction
MISSION short hairpin RNA (shRNA) lentiviral transduction particles for the CLDN3 and CLDN4 knockdown experiments were obtained from Sigma (St Louis, MO). One of five CLDN3- or CLDN4-specific shRNA constructs (shRNA sequence targeting CLDN3: CCGGCGACCGCAAGGACTACGTCTACTC GAGTAGACGTAGTCCTTGCGGTCGTTTTTG; shRNA sequence targeting CLDN4: CCGGG CAACATTGTCACCTCGCAGACTCGAGTCTGCGAGGTGACAATGTTGCTTTTTG) and one “nontarget” construct were transduced separately into 2008 cells. The “nontarget” construct contained an shRNA sequence that did not target any known human gene and served as a scrambled negative control. Briefly, 2008 cells were transduced with the CLDN3- or CLDN4-specific shRNA lentiviral particles at a ratio of five particles to one cell for 24 hours in the presence of hexadimethrine bromide to improve transduction efficiency. Afterward, the medium containing viral particles was removed and replaced with fresh medium containing 10 µg/ml puromycin. Before lentiviral transduction, a puromycin titration was performed that identified 10 µg/ml as the minimum puromycin concentration that caused complete death of 2008 cells after a 5-day incubation. CLDN3 or CLDN4 knockdown was confirmed using quantitative reverse transcription-polymerase chain reaction (RT-PCR) and Western blot analysis by comparison to the parental 2008 cells and nontarget shRNA cells (designated here as 2008-scb).
Immunoblot Analysis, Immunofluorescence, Immunohistochemistry, and Confocal Microscopy
Immunoblot analysis was performed using the antibodies against CLDN3, CLDN4, occludin, JAM-1, ZO-1, E-cadherin, and β-catenin. For immunofluorescence analysis, cells were grown on polylysine-coated eight-well chamber slides, fixed in 3.7% formaldehyde, and permeabilized in phosphate-buffered saline containing 0.2% Triton X-100, and indirect immunofluorescence was performed as described previously [47]. The following antibodies were used: rabbit polyclonal anti-claudin-3 (Invitrogen, Carlsbad, CA), mouse monoclonal anti-claudin-4 (Abcam, Cambridge, MA), rabbit polyclonal anti-occludin (Abcam), mouse polyclonal anti-JAM-1 (Abcam), rabbit polyclonal anti-ZO-1 (Zymed, Grand Island, NY), rabbit polyclonal anti-E-cadherin (Santa Cruz Biotechnology, Santa Cruz, CA), and mouse monoclonal anti-β-catenin and anti-dephospho-β-catenin (Cell Signaling Technology, Danvers, MA). Immunohistochemistry was performed on xenografted tumors either imbedded in paraffin or frozen in Tissue-Tek OCT compound (Sakura Finetek USA, Torrance, CA) as described previously [48]. Sections were stained with anti-Ki67, anti-CD31, and anti-bromodeoxyuridine (BrdU) antibodies. Confocal microscopy was performed with a Zeiss LSM510 confocalmicroscope system (Carl Zeiss, Inc, Thornwood, NY). Cells were randomly selected for imaging taken in 0.5-µm sections. The number of Z-scan sections required to image a cell ranged between 15 and 25. Data are shown as superimposed (stacked) Z-scan images and as single confocal images.
Real-time PCR
RNA was extracted with TRIzol reagent (Invitrogen). First-strand cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen) and random primers. Real-time PCR was performed using the Bio-Rad iCycler iQ detection system in the presence of SYBR Green I dye (Bio-Rad Laboratories, Inc, Hercules, CA). β-Actin was used as reference gene and relative mRNA levels were determined using the 2(-ΔΔCt) method. A 1-unit difference of Ct value represents a two-fold difference in the level of mRNA.
Measurement of Transepithelial Electrical Resistance and Paracellular Permeability
One hundred thousand cells in 0.5 ml of complete medium were seeded into 12-mm transwell insert with a membrane pore size of 0.45 µm (Millipore, Carrigtwohill, Ireland) placed in 24-well plate containing 0.75 ml of the growth medium. Transepithelial electrical resistance (TER) was measured using an EVOM-Epithelial Voltohmmeter (World Precision Instruments, New Haven, CT). Readings were taken 3 days after seeding the cells. The TER values were calculated by subtracting the blank values from the sample values and normalized to the growth area of the monolayer.
The paracellular permeability assay measured the diffusion of fluorescein isothiocyanate (FITC)-labeled dextran (FITC-dextran, 4 kDa) across a cell layer grown on a membrane of transwell insert. The cells were seeded (1 x 105 cells per insert) and cultured to form a monolayer for 3 days. The culture medium was then replaced with prewarmed Ringer's solution a few minutes before the addition of FITC-dextran (1 mg/ml in Ringer's solution). Five hundred microliters of FITC-dextran was added to the upper chamber and 100 µl of the samples were removed from the lower chamber after 1 hour. Fluorescence was measured on a FLUOstar OPTIMA plate reader (Ex485 nm/Em544 nm). The diffusion of FITC-dextran across the insert without cells was also measured to serve as 100% permeability.
In Vivo Tumor Growth
Female nu/nu mice at age 5 to 6 weeks were purchased from Charles River Laboratories (Wilmington, MA). All work performed with animals was in accordance with and approved by the Institutional Animal Care and Use Committee at the University of California, San Diego. To determine the in vivo growth potential, the cells were inoculated subcutaneously (SC) on two sites of a nude mouse. Tumor volume was measured every 2 days and calculated using the formula: length x width2/2 and plotted as a function of time to generate the in vivo growth curves.
In Vitro Cell Migration and Invasion Assay
The invasive capabilities of the cells were determined using a modified Boyden chamber invasion assay [49]. Cells were cultured to about 80% confluence and serum starved overnight. Transwell inserts (BD Biosciences, San Jose, CA) of 8-µm pore size were coated with 150 µl of 1:60 diluted Matrigel (BD Biosciences). Three hundred thousand cells after overnight starvation were plated onto the top of each of the coated filters in 150 µl of serum-free medium. Three hundred microliters of the same medium containing 20% fetal calf serum was placed in the lower chamber to act as a chemoattractant. After 24 hours of culture, cells that did not migrate or invade through the pores of the transwell inserts will be manually removed with cotton swabs and the inserts were fixed in cold methanol for 10 minutes and then stained with 0.01% crystal violet in 20% ethanol. After washing thoroughly, colorimetric readings were taken at 595 nm. The experiment was performed twice with each sample in triplicate. To assess cell migration, assays were carried out as above except that cells were plated on top of uncoated (Matrigel-free) inserts.
Measurement of Migration Using Wound-healing/Scratch Assay
Cells were grown to confluence on 60-mm cell culture dishes and a scratch was made through the cell monolayer using a pipette tip. After washing with Hank's balanced salt solution, fresh culture medium was added and the cells were incubated at 37°C in a humid environment with 5% CO2. Wound closure was observed and photographed at 0, 12, and 24 hours after making the scratch to monitor the invasion of cells into the wounded area. The experiment was performed twice and assayed in triplicate.
In Vivo Metastasis Study
To determine whether changes in migration and invasiveness detected in vitro translate into differences in metastatic potential in vivo, 6-week-old female nude mice were injected with 1 x 106 Ds-Red-expressing cells of 2008, CLDN3KD, or CLDN4KD in the tail vein. Mice were sacrificed and explored when they appeared premorbid. Postmortem external fluorescent images were obtained by a highly sensitive in vivo imaging system (IVIS 200; Caliper, Virginia Beach, VA). Each mouse underwent laparotomy and median sternotomy. Red fluorescent protein (RFP) expression was visualized in the whole-body image system, facilitating identification of metastatic tumors. After obtaining open images, lungs, liver, spleen, and kidneys were removed and were thoroughly examined for any evidence of metastasis using both fluorescence imaging and histopathologic analysis.
Statistical Analysis
All data are presented as the means ± SD. Statistical analysis was evaluated by the unpaired Student's t test, with reference to respective control and expressed as P values presented in the Results section.
Results
Suppression of CLDN3 or CLDN4 Expression Promotes Tumor Growth In Vivo
To investigate the role of CLND3 and CLDN4, we constitutively knocked down their expression in the human carcinoma cell line 2008 using shRNAi vectors. In the CLDN3KD cells, the CLDN3 mRNA level was reduced to 5% of control and the protein level to 26% of control. In the CLDN4KD cells, the CLDN4 mRNA level was reduced to 12% of control and the protein level to 16% of control. Transduction of 2008 cells with a vector expressing a scrambled shRNAi that did not target any human gene did not alter the mRNA or protein level of either CLDN3 or CLDN4 (Figure W1, A and B). Interestingly, we found that knocking down the expression of CLDN3 also decreased the expression of CLDN4 and vice versa. Knockdown of CLDN3 reduced CLDN4 mRNA to 14% of control and protein to 38%. Knockdown of CLDN4 reduced CLDN3 mRNA to 36% of control and protein to 49%. Rather than being due to an off-target effect of the two shRNAs, this can be explained by the fact that the promoters for both genes are sensitive to the same transcriptional repressors that are potentially activated when either one or the other mRNA is reduced [23,50,51].
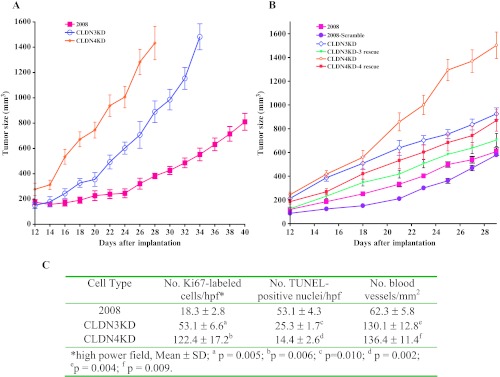
Knockdown of these claudins did not produce any change in the rate of proliferation of the cells in vitro (Figure W1C) and produced only minor perturbations of cell cycle phase distribution (Figure W1D). In contrast, as shown in Figure 1A, although tumors developed at all sites after SC inoculation, in vivo the CLDN3KD tumors grew 2.3-fold faster (P = 0.007), and the CLDN4KD tumor 3.7-fold (P = 0.001) faster, than the parental 2008 tumors. The effect of CLDN3 or CLDN4 knockdown on in vivo tumor growth was specific as reintroduction of an siRNA-resistant CLDN3 or CLDN4 construct into the knockdown cells significantly reduced the tumor growth (Figure 1B; P = 0.022, CLDN3KD vs CLDN3KD-3 rsc; P = 0.001, CLDN4KD vs CLDN4KD-4 rsc). As shown in Figure 1C, the birth rate in the CLDN3KD and CLDN4KD tumors, as reflected by Ki67 staining, was 2.9-fold and 6.7-fold higher in the CLDN3KD and CLDN4KD tumors, respectively. This was accompanied by a 2.0-fold and 3.7-fold reduction in death rate as evidenced by TUNEL staining and a doubling of the vascular density.
Figure 1.
CLDN3 and CLDN4 control the growth of ovarian tumor xenografts in vivo. (A) Relative growth rate of 2008 (■), CLDN3KD.(◊), and CLDN4KD (○) tumors after SC inoculation of 1 x 106 cells in nu/nu mice (n = 16 for each tumor type). (B) Re-expression of siRNA-resistant CLDN3 or CLDN4 in the claudin knockdown cells, designated here as CLDN3KD-3 rescue and CLDN4KD-4 rescue, slowed the tumor growth in vivo. Six synonymous point mutations were introduced in shRNA target regions in full-length CLDN3 or CLDN4 cDNA and the siRNA-resistant cDNAs were cloned into pLVX-mCherry vector. Following infection with lentivirus produced from this vector in HEK298T cells, 2008 cells expressing high levels of the protein were isolated by fluorescence-activated cell sorting (FACS) and used to inoculate SC in nu/nu mice (n = 16 for each tumor type). (C) Quantification of Ki67-positive and TUNEL-positive cells and density of CD31-positive vessels in 2008, CLDN3KD, and CLDN4KD tumors.
Knockdown of CLDN3 and CLDN4 Impairs TJ Formation and Cell Adhesion
The ability of the parental, CLDN3KD, and CLDN4KD ovarian cells to form TJ was examined by measuring transepithelial resistance. Knockdown of CLDN3 and CLDN4 produced only a marginal decrease in transepithelial resistance (Figure 2A), but this was associated with a significantly increased FITC-dextran flux (Figure 2B), reflecting a disruption of the integrity of the TJ complex. Thus, impairment of TJ formation was readily detectable in the CLDN knockdown cells. Failure of TJ formation in the 2008 knockdown cells led to investigation of the effect of CLDN3 and CLDN4 knockdown on attachment, migration, and invasion, all of which are modulated by the ability of cells to form adhesions with their substrates or their neighbors. Figure 2C shows that there was a 2.5-fold reduction in the fraction of CLDN3KD cells, and a 2.2-fold reduction in the fraction of CLND4KD cells, that had become attached to tissue culture dishes when measured at 6 hours.
Figure 2.
Effect of CLDN3 and CLDN4 expression on TJ formation and cell adhesion. (A) TER in the monolayers of 2008, CLDN3KD, and CLDN4KD cells 3 days after seeding onto the transwell inserts (n = 6). (B) Paracellular permeability of 4-kDa FITC-dextran measured on day 3 after seeding the cells onto the transwell inserts. Vertical bars, ±SEM, n = 6. *P = 0.008 and **P = 0.001 (vs 2008 control). (C) Relative adhesion quantified at 6 hours after seeding on uncoated tissue culture dishes. Vertical bars, ±SEM, n = 4. *P = 0.001 and **P = 0.003 (vs 2008 control).
Knockdown of CLDN3 and CLDN4 Increases Cell Migration and Invasion In Vitro
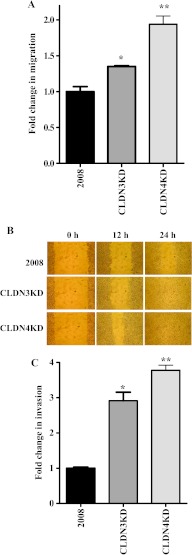
The effect on cell migration was compared using a Boyden two-chamber assay in which the cells were induced to migrate through a filter by the presence of FBS on the opposite side. As shown in Figure 3A, migration was enhanced 1.4-fold by the knockdown of CLDN3 and 1.9-fold by the knockdown of CLDN4. This motogenic phenotype was confirmed in a wound-healing/scratch assay, a widely accepted method for qualitative assessment of cell migration. The extent of wound closure can be taken as a direct measure of cell motility. As shown in Figure 3B, the CLDN3KD cells closed the wound more rapidly than the parental 2008 cells and the effect was even greater for the CLDN4KD cells. The effect on invasive potential was examined using a modified Boyden chamber invasion assay. The number of cells that invaded through a layer of Matrigel was analyzed at 24 hours after plating the cells on Matrigel-coated transwell inserts. As shown in Figure 3C, knockdown of CLDN3 or CLDN4 expression in 2008 cells significantly increased their invasive potential by a factor of 2.9-fold and 3.8-fold, respectively.
Figure 3.
CLDN3 and CLDN4 control tumor cell migration and invasion. (A) Relative migration through an uncoated filter toward serum-containing medium in a Boyden chamber assay. The migratory ability of cells was compared by measuring the number of cells migrating through the uncoated filters. Vertical bars, ±SEM, n = 6. *P = 0.035 and **P = 0.005 (vs 2008 control). (B) Relativemotility as determined by the ability of 2008, CLDN3KD, and CLDN4KD cells to close a wound made by creating a scratch through a lawn of confluent cells. (C) Relative invasion of cells through a layer of Matrigel coated on the filter of a Boyden chamber measured at 24 hours after seeding. Vertical bars, ±SEM, n = 6. *P = 0.004 and **P = 0.002 (vs 2008 control).
Conversely, we examined the effects of rescuing CLDN3 or CLDN4 expression by reintroduction of an siRNA-resistant CLDN3 or CLDN4 construct into the knockdown cells. In contrast to the knockdown cells, re-expression of CLDN3 or CLDN4 slowed down cell migration in wound-healing assay (Figure W2A) and decreased cell invasion through the Matrigel (Figure W2B).
CLDN3 and CLDN4 Control Invasive and Metastatic Ability
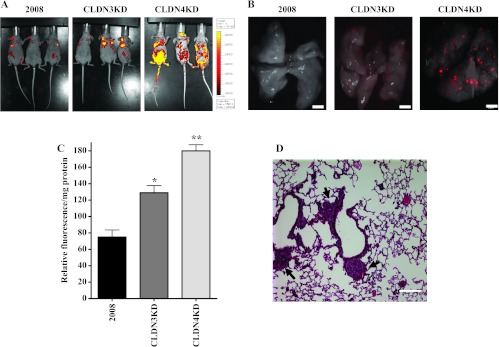
The effect of CLDN3 and CLDN4 knockdown in metastatic potential in vivo was examined by molecularly engineering the 2008, CLDN3KD, and CLDN4KD cells to express the Ds-Red fluorescent protein and then injecting 2 x 106 cells into the tail vein of nu/nu mice. This allowed the appearance of metastases to be serially monitored in living mice and their detection in organs freshly removed from the mouse by fluorescent imaging. Figure 4A shows fluorographs of animals obtained with an IVIS 200 imaging system on day 42 post-injection. Mice injected with either CLDN3KD or CLDN4KD cells had significantly more lung metastases compared to mice injected with 2008 cells. On necropsy, lungs of injected mice were removed and examined with an Olympus OV100 fluorescence microscope. As shown in Figure 4B, which presents representative images of fluorescent tumor cell in mouse lung 6 weeks after tail vein injection, knockdown of CLDN3 or CLDN4 remarkably increased the formation of metastases to the lung and other sites compared with 2008 cells. Quantification of total lung metastatic burden in the same sets of mice was performed by measurement of Ds-Red fluorescence in lung homogenates. Figure 4C shows that there was a 1.7-fold and a 2.4-fold increase of red fluorescence, respectively, in the lungs of CLDN3KD- and CLDN4KD-injected mice. Hematoxylin and eosin staining of fixed and paraffin-embedded tissue confirmed the presence of micrometastases in the lungs (Figure 4D). Thus, the anti-invasive effect of CLDN3 and CLDN4 detected in the Matrigel invasion assay was confirmed in vivo, and the results document a large enhancement of metastatic potential when either CLDN3 or CLDN3 is reduced.
Figure 4.
CLDN3 and CLDN4 control metastatic potential of ovarian cells injected IV in nu/nu mice. (A) Fluorographs of mice 42 days after tail vein injection of 2 x 106 2008, CLDN3KD, or CLDN4KD cells molecularly engineered to express Ds-Red showing more extensive metastases when CLDN3 and CLDN4 are knocked down. (B) Representative fluorescent images of mouse lung lobes showing that knockdown of CLDN3 and CLDN4 increases the extent of metastasis to lung when imaged at 42 days after tail vein injection. Scale bar, 2 mm. (C) Quantification of total lung metastasis burden in the five mice of each group. The lungs were dissected, homogenized, and diluted for quantification of metastasis by red fluorescence. Vertical bars, ±SEM, n = 10. *P = 0.012 and **P = 0.001 (vs 2008 control). (D) Hematoxylin and eosin staining of fixed and paraffin-embedded tissue confirmed the presence of micrometastases in the lungs of mice injected with the CLDN4KD cells. Scale bar, 100 µm.
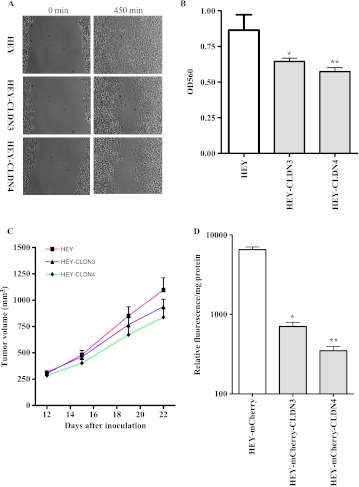
To determine whether increasing the expression of either CLDN3 or CLDN4 in a cell line that does not express either claudin would produce an opposite effect, we used the HEY human ovarian cell line in which CLDN3 and CLDN4 are undetectable. HEY cells were infected with lentiviral vectors expressing either CLDN3 or CLDN4, and constitutively expressing populations were tested for changes in cell migration using the wound-healing assay and for invasion in the Boyden chamber assay. Consistent with what was observed with 2008 and its CLDN3 or CLDN4 knockdown sublines, forced expression of either CLDN3 or CLDN4 in the HEY cells reduced cell migration and invasion (Figure 5, A and B). When inoculated subcutaneously (SC), HEY-CLDN3 and HEY-CLDN4 tumors grew 1.4-fold (P = 0.007) and 1.8-fold (P = 0.005) more slowly than the parental HEY tumors (Figure 5C). Expression of CLDN3 or CLDN4 in HEY cells significantly reduced their invasive potential as evidenced by the reduced ability to colonize the lungs of the immunocompromised mice (Figure 5D).
Figure 5.
Effect of claudin expression on cell migration and invasion in vitro as well as growth and metastasis in vivo. (A) Relative motility as determined by the ability of HEY, HEY-CLDN3, and HEY-CLDN4 cells to close a wound made by creating a scratch through a lawn of confluent cells. (B) Relative invasion of HEY, HEY-CLDN3, and HEY-CLDN4 cells through a Matrigel-coated layer on the filter of a Boyden chamber measured at 12 hours after seeding. Vertical bars, ±SEM, n = 6. *P = 0.031 and **P = 0.014 (vs HEY control). (C) Relative growth rate of HEY (■), HEY-CLDN3 (▴), and CLDN4 (◆) tumors after SC inoculation of 1 x 106 cells in nu/nu mice (n = 16 for each tumor type). (D) Quantification of total lung metastatic burden as determined by measurement of extractable mCherry fluorescence at 43 days after tail vein injection of mCherry-labeled HEY, HEY-CLDN3, and HEY-CLDN4 cells. Vertical bars, ±SEM, n = 10. *P = 0.009 and **P = 0.005 (vs HEY-mCherry control).
The loss of transepithelial resistance that accompanied knockdown of CLDN3 or CLDN4 raised the question of their effect on the other major proteins found in TJ including occludin, JAM, and ZO-1. As shown in Figure W3A, Western blot analysis disclosed that there was a marginal decrease in the levels of occludin and ZO-1 but a moderate increase in the expression levels of JAM in the CLDN3KD or CLDN4KD cells. The effect on occludin was examined in more detail by immunocytochemical staining and laser scanning confocal microscopic analysis. This revealed that the intensity of staining for occludin was somewhat reduced in the CLDN3KD or CLDN4KD cells but that the relative membrane versus cytoplasmic intensity and subcellular location was not altered (Figure W3, B and C). These results indicate that knockdown of either CLDN3 or CLDN4 did not produce major changes in the level of these other TJ proteins.
CLDN3 and CLDN4 Regulate E-cadherin Expression and Affect β-catenin Signaling
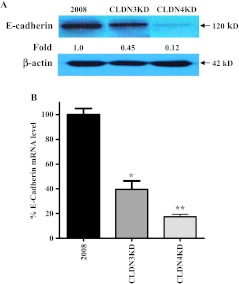
Suppression of E-cadherin expression is regarded as one of the main molecular events responsible for dysfunction in cell-cell adhesion. Most tumors have abnormal cellular architecture, and the loss of E-cadherin is a well-known prerequisite for tumor cell invasion. However, a direct protein-protein or genetic interaction between CLDN3 or CLDN4 and E-cadherin has not been reported in any organism and little is known about the regulation of E-cadherin expression by claudins in ovarian carcinomas. Interestingly, knockdown of CLDN3 expression in the ovarian carcinoma 2008 cells was accompanied by a 55% decrease in E-cadherin expression at the protein level and almost complete loss of expression in the CLDN4KD cells (Figure 6A). Measurement of E-cadherin mRNA levels by quantitative RT-PCR showed a 60 ± 11.8% reduction in the CLDN3KD cells and an 83 ± 3.5% decrease in the CLDN4KD cells (Figure 6B), indicating that E-cadherin is downregulated at the transcriptional level similar to the claudins themselves. It is noteworthy that the transcription factor Snail has been reported to bind to E-boxes in the promoters of both E-cadherin and claudins and suppress their expression [50,52,53].
Figure 6.
CLDN3 and CLDN4 regulate the expression of E-cadherin. (A) Western blot analysis showing reduced expression of E-cadherin in CLDN3KD and CLDN4KD cells. (B) The relative levels of E-cadherin mRNA measured by real-time RT-PCR in CLDN3KD and CLDN4KD cells compared with 2008 cells. Vertical bars, ±SEM, n = 5. *P = 0.006 and **P = 0.002 (vs 2008 control).
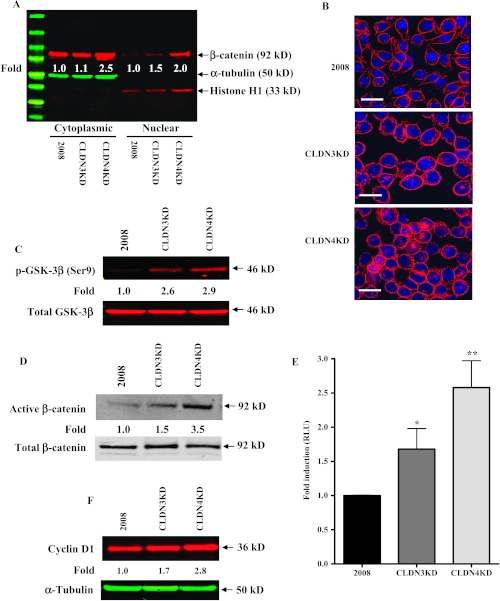
Since the intracellular domains of E-cadherin directly bind to and sequester cytoplasmic β-catenin, E-cadherin serves as a negative regulator that functionally blocks the β-catenin signaling pathway that guides cell fate decisions through the regulation of cell growth, motility, and survival [54]. This led us to investigate whether the cytoplasmic and/or nuclear accumulation of β-catenin was regulated by CLDN3 and CLDN4. In the CLDN3KD cells, there was a 1.1-fold and a 1.5-fold increase of β-catenin in the cytoplasm and nuclei, respectively, whereas increases of 2.5-fold and 2.0-fold were found in the CLDN4 knockdown cells (Figure 7A). Furthermore, immunocytochemical analysis with the same antibody confirmed increased levels of nuclear β-catenin in the knockdown sublines with the effect being greater in the CLDN4KD than the CLDN3KD cells (Figure 7B). Cytosolic β-catenin is targeted for degradation when phosphorylated by GSK-3β, the kinase primarily responsible for phosphorylating β-catenin. As phosphorylation of GSK-3β itself on Ser9 inhibits its activity and prevents targeting of β-catenin for degradation, we next determined whether the observed increase of β-catenin was correlated with the phosphorylation state of GSK-3β. Indeed, a significant increase in the level of Ser9-phosphorylated and therefore inactive GSK-3β was found in the CLDN3KD and CLDN4KD cells compared with the 2008 control (Figure 7C). In line with this, a significant increase in the active (dephosphorylated) pool of nuclear β-catenin was observed in CLDN3KD and CLDN4KD cells (Figure 7D). To further confirm the increase in nuclear β-catenin, we transfected cells with the TOPflash luciferase reporter construct to evaluate changes in β-catenin-regulated promoter activation. Transfection with this TCF- and β-catenin-dependent reporter plasmid demonstrated a 1.7-fold increase of reporter gene activity in the CLDN3KD cells and a 2.6-fold increase in the CLDN4KD cells (Figure 7E). An increase in the expression of the β-catenin/TCF-regulated target gene cyclin D1 was also seen in the CLDN3KD and CLDN4KD cells grown in vitro (Figure 7F) and in vivo (Figure W4). These results indicate that CLDN3 and CLDN4 regulate signaling through the E-cadherin/β-catenin pathway.
Figure 7.
Activation of β-catenin signaling on knockdown of CLDN3 and CLDN4. (A) Western blot analysis showing increased nuclear dephospho (active)-β-catenin levels in CLDN3KD and CLDN4KD cells. Relative levels of cytoplasmic and nuclear β-catenin were normalized against α-tubulin and histone H1, respectively, and expressed relative to the control from the mean of three separate analyses. (B) Immunocytochemical analysis of 2008, CLDN3KD, and CLDN4KD cells stained with antibody to β-catenin showing increased nuclear localization of β-catenin in the knockdown cells. (C) Western blot analysis showing increased GSK-3β phosphorylation in CLDN3KD and CLDN4KD cells. Relative levels of phospho-GSK-3β were normalized against total GSK-3β. The ratio in control 2008 cells is designated as 1. (D) Western blot analysis showing increased nuclear dephospho-β-catenin levels in CLDN3KD and CLDN4KD cells. Relative levels of active β-catenin (dephosphorylated form) were normalized against total β-catenin. The ratio in control 2008 cells is designated as 1. (E) Activation of β-catenin/TCF signaling in CLDN3KD and CLDN4KD cells. Cells were transiently co-transfected with the firefly luciferase TOPflash TCF reporter plasmid and the control plasmid pCMXβgal to normalize for transfection efficiency. The luciferase values were normalized for variations in transfection efficiency and are expressed as fold stimulation of luciferase activity compared with the 2008 control cultures. Vertical bars, ±SEM, n = 5. *P = 0.086 and **P = 0.016 (vs 2008 control). (F) Cyclin D1 protein levels in CLDN3KD and CLDN4KD cells relative to that in the 2008 cells were calculated densitometrically after normalization to the level of α-tubulin.
Expression of CLDN3 and CLDN4 in the Fallopian Tubes of Ovarian Cancer Patients
Many ovarian cancers express CLDN3 and CLDN4, but normal ovarian surface epithelium does not; this seems to suggest that low expression of these claudins is associated with the benign state and high expression with transformation. In light of the body of evidence indicating that most ovarian cancers arise in the distal Fallopian tube [17–20], we used histochemical analysis to examine the expression of CLDN3 and CLDN4 in both the tumor and distal Fallopian tube from the same patient in six cases of serous ovarian cancer. In all six cases, both CLDN3 and CLDN4 were abundantly expressed in both sites (Figure W5). This result is supportive of the concept that ovarian cancer arises in an epithelium that normally expresses both of these claudins. Our data suggest that those tumors that subsequently lose CLDN3 and CLDN4 expression can be expected to behave more aggressively.
Discussion
CLDN3 and CLDN4 are well known for their ability to affect TJ formation and control paracellular permeability in epithelial monolayers. The results reported here indicate that, in a fully malignant gynecologic cancer cell line, these two claudins individually have quite large effects on tumor growth rate and metastatic potential. The increased in vivo growth rate can be largely explained by an increased birth rate and decreased death rate of tumor cells and by an increase in vascular density, but the fact that knockdown of these claudins does not change the in vitro growth rate or significantly perturb cell cycle phase distribution indicates that these are the result of alterations in the interaction of the tumor cell with its in vivo environment rather than cell autonomous effects. Knockdown of CLDN3 or CLDN4 was accompanied by enhanced migration, increased invasion, and greater in vivo metastatic colonization of the lungs. That these effects were specific to the reduction in CLDN3 and CLDN4 was shown by the fact that re-expression of an siRNAi-resistant CLDN3 or CLDN4 mRNA in the knockdown cells, or forced expression of CLDN3 or CLDN4 in the HEY cells that do not express endogenous CLDN3 or CLDN4, slowed tumor growth and reduced lung metastases. It is noteworthy that the effect of knocking down CLDN4 was consistently greater than that of knocking down CLDN3 in the panel of assays reported here.
Most cells express multiple claudin isoforms that interact in a homotypic and heterotypic manner to regulate junctional permeability and confer selectivity and strength to the TJ. At least five pairs of claudin genes (CLDN3 and CLDN4, CLDN6 and CLDN9, CLDN8 and CLDN17, CLDN10 and CLDN15, and CLDN22 and CLDN24) have been found to be closely linked with respect to their close proximity in the human genome [55]. Whether this genomic arrangement leads to coordinate regulation is currently unknown, but, at least in the case of CLDN3 and CLDN4, coordinate expression has been reported in several normal and neoplastic tissues [27], and expression of these genes is frequently simultaneously elevated in a variety of cancers [39]. The similarity of the phenotype produced by knockdown of either CLDN3 or CLDN4 raised the question of whether these proteins function as a single unit whose effectiveness or stability is impaired when either is missing. The knockdown of CLDN3 was found to produce a significant reduction in the level of CLDN4 as determined by Western blot analysis (Figure W1B). Similarly, knockdown of CLDN4 substantially reduced the CLDN3 protein level. However, quantitative RT-PCR analysis of mRNA levels indicated that this was not just due to reduced stability of one protein in the absence of its putative partner since reduction in CLDN3 mRNA was accompanied by reduction in CLDN4 mRNA and vice versa (Figure W1A). Rather than being an off-target effect of the shRNAi, this is likely due to the fact that the promoters of both genes are sensitive to the same transcriptional repressors that are activated when either one or the other mRNA is reduced [23,50,51].
Knockdown of CLDN3 and CLDN4 was accompanied by reduced expression of E-cadherin at both the mRNA and protein levels. Knockdown of CLDN3 reduced E-cadherin mRNA by 60% and protein by 55%; the effect on CLDN4KD was greater with an 83% decrease in mRNA and a >95% reduction in protein. This suggests that a major component of the effect of CLDN3 and CLDN4 on E-cadherin expression is at the transcriptional level. Knockdown of CLDN3 and CLDN4 did not alter the expression of the proteins with which it is most intimately associated in TJs, suggesting that the reduction in E-cadherin was not part of a general failure of trafficking of proteins to sites of cell-cell contact. The mechanism by which loss of CLDN3 and CLDN4 trigger E-cadherin down-regulation remains to be defined, but it is noteworthy that loss of E-cadherin expression is a characteristic feature of cells that have gone through an EMT, and some of the transcription factors that control this process can act as transcriptional repressors of the E-cadherin promoter [52,56].
Loss of E-cadherin expression in polarized epithelial cells activates the β-catenin signaling pathway [54], and the down-regulation of E-cadherin that accompanied loss of CLDN3 or CLDN4 in the non-polarized malignant ovarian cancer cells produced the same effect. Knockdown of either CLDN3 or CLDN4 produced an increase in Ser9 phosphorylation of GSK-3β known to reduce the ability of GSK-3β to mediate the degradation of β-catenin. This was accompanied by an increase in total nuclear β-catenin as documented both by Western blot and immunocytochemical analyses, an increase in the active dephosphorylated form of β-catenin, enhanced β-catenin transcriptional activity as detected by the TOPflash reporter, and an increase in cyclin D1. In other cell systems, loss of E-cadherin is associated with increased signaling in other pathways as well [57]. Whether the increase in β-catenin by itself accounts for the enhanced 2008 growth rate and metastatic seeding or whether additional pathways are also involved remains to be determined. However, the results of this study indicate that both CLDN3 and CLDN4 function upstream of E-cadherin to limit growth rate, migration, invasion, and metastasis.
The majority of ovarian cancers express CLDN3 and CLDN4 at high levels [12,13,58]. The results reported here lead to the prediction that those ovarian cancers that do not express these claudins should behave more aggressively. As there is an increasing body of evidence suggesting that serous ovarian cancers arise from the distal Fallopian tubes rather than the ovarian surface epithelium, we have performed an immunohistochemical analysis of the expression of CLDN3 and CLDN4 in the Fallopian tubes of six patients with serous ovarian cancer. Consistent with the fact that most serous ovarian cancers express high levels of CLDN3 and CLDN4, we found that the distal Fallopian tubes of patients with serous ovarian cancer express these proteins in abundance. This new information provides a more logical explanation of the relationship between CLDN3 and CLDN4 expression and tumor behavior that parallels what is known about “high” versus “low” claudin breast cancers. As for ovarian carcinomas, serous ovarian cancer appears to arise in an epithelium that normally expresses CLDN3 and CLDN4 as such the distal Fallopian tube. While limited and contradictory evidence is available from studies of ovarian cancer [59,60], disruption of cell-cell adhesions correlates with progression, invasion, and metastasis in breast cancer [9,41,42], and low expression of CLDN4 is associated with poor prognosis in some subtypes of breast [22,43,44], pancreatic [26], and colon cancers [24,25]. Our data support the concept that those cancers that continue to express these proteins are less aggressive than those in which their expression is lost.
Supplementary Material
Acknowledgments
We are grateful to Dennis Young of the University of California at San Diego Moores Cancer Center FACS facility for his expert assistance with FACS analysis and sorting. We also thank Dr Kersi Pestonjamasp for sharing his expertise on confocal microscopy and Dr Nissi Varki for pathology support.
Abbreviations
- CLDN3
claudin-3
- CLDN4
claudin-4
- TJ
tight junction
- EMT
epithelial-to-mesenchymal transition
Footnotes
This research was supported by the Clayton Medical Research Foundation. NIH grants P30 CA23100 and P30 NS047101 provided support for imaging resources. The authors declare no conflict of interest.
This article refers to supplementary materials, which are designated by Figures W1 to W5 and are available online at www.neoplasia.com.
References
- 1.Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol. 2003;4:225–236. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- 2.Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am J Physiol Cell Physiol. 2004;286:C1213–C1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- 3.Langbein L, Pape UF, Grund C, Kuhn C, Praetzel S, Moll I, Moll R, Franke WW. Tight junction-related structures in the absence of a lumen: occludin, claudins and tight junction plaque proteins in densely packed cell formations of stratified epithelia and squamous cell carcinomas. Eur J Cell Biol. 2003;82:385–400. doi: 10.1078/0171-9335-00330. [DOI] [PubMed] [Google Scholar]
- 4.Itoh M, Bissell MJ. The organization of tight junctions in epithelia: implications for mammary gland biology and breast tumorigenesis. J Mammary Gland Biol Neoplasia. 2003;8:449–462. doi: 10.1023/B:JOMG.0000017431.45314.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullin JM. Epithelial barriers, compartmentation, and cancer. Sci STKE. 2004;2004:pe2. doi: 10.1126/stke.2162004pe2. [DOI] [PubMed] [Google Scholar]
- 6.Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol. 2003;5:137–142. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
- 7.Kohler K, Zahraoui A. Tight junction: a co-ordinator of cell signalling and membrane trafficking. Biol Cell. 2005;97:659–665. doi: 10.1042/BC20040147. [DOI] [PubMed] [Google Scholar]
- 8.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Martin TA, Watkins G, Mansel RE, Jiang WG. Loss of tight junction plaque molecules in breast cancer tissues is associated with a poor prognosis in patients with breast cancer. Eur J Cancer. 2004;40:2717–2725. doi: 10.1016/j.ejca.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Choi YL, Kim J, Kwon MJ, Choi JS, Kim TJ, Bae DS, Koh SS, In YH, Park YW, Kim SH, et al. Expression profile of tight junction protein claudin 3 and claudin 4 in ovarian serous adenocarcinoma with prognostic correlation. Histol Histopathol. 2007;22:1185–1195. doi: 10.14670/HH-22.1185. [DOI] [PubMed] [Google Scholar]
- 11.Hudson LG, Zeineldin R, Stack MS. Phenotypic plasticity of neoplastic ovarian epithelium: unique cadherin profiles in tumor progression. Clin Exp Metastasis. 2008;25:643–655. doi: 10.1007/s10585-008-9171-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I, et al. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10:3291–3300. doi: 10.1158/1078-0432.CCR-03-0409. [DOI] [PubMed] [Google Scholar]
- 13.Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, Cho KR, Riggins GJ, Morin PJ. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000;60:6281–6287. [PubMed] [Google Scholar]
- 14.Rangel LB, Agarwal R, D'Souza T, Pizer ES, Alo PL, Lancaster WD, Gregoire L, Schwartz DR, Cho KR, Morin PJ. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res. 2003;9:2567–2575. [PubMed] [Google Scholar]
- 15.Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E, Anfossi S, Gokden M, Dunn D, Roman JJ, et al. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer. 2004;112:14–25. doi: 10.1002/ijc.20408. [DOI] [PubMed] [Google Scholar]
- 16.Heinzelmann-Schwarz VA, Gardiner-Garden M, Henshall SM, Scurry J, Scolyer RA, Davies MJ, Heinzelmann M, Kalish LH, Bali A, Kench JG, et al. Overexpression of the cell adhesion molecules DDR1, claudin 3, and Ep-CAM in metaplastic ovarian epithelium and ovarian cancer. Clin Cancer Res. 2004;10:4427–4436. doi: 10.1158/1078-0432.CCR-04-0073. [DOI] [PubMed] [Google Scholar]
- 17.Finch A, Shaw P, Rosen B, Murphy J, Narod SA, Colgan TJ. Clinical and pathologic findings of prophylactic salpingo-oophorectomies in 159 BRCA1 and BRCA2 carriers. Gynecol Oncol. 2006;100:58–64. doi: 10.1016/j.ygyno.2005.06.065. [DOI] [PubMed] [Google Scholar]
- 18.Karst AM, Levanon K, Drapkin R. Modeling high-grade serous ovarian carcinogenesis from the fallopian tube. Proc Natl Acad Sci USA. 2011;108:7547–7552. doi: 10.1073/pnas.1017300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y, Miron A, Drapkin R, Nucci MR, Medeiros F, Saleemuddin A, Garber J, Birch C, Mou H, Gordon RW, et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J Pathol. 2007;211:26–35. doi: 10.1002/path.2091. [DOI] [PubMed] [Google Scholar]
- 20.Levanon K, Ng V, Piao HY, Zhang Y, Chang MC, Roh MH, Kindelberger DW, Hirsch MS, Crum CP, Marto JA, et al. Primary ex vivo cultures of human fallopian tube epithelium as a model for serous ovarian carcinogenesis. Oncogene. 2010;29:1103–1113. doi: 10.1038/onc.2009.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshihara K, Tajima A, Yahata T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, Kudo Y, et al. Gene expression profile for predicting survival in advanced-stage serous ovarian cancer across two independent datasets. PLoS One. 2010;5:e9615. doi: 10.1371/journal.pone.0009615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szasz AM, Nemeth Z, Gyorffy B, Micsinai M, Krenacs T, Baranyai Z, Harsanyi L, Kiss A, Schaff Z, Tokes AM, et al. Identification of a claudin-4 and E-cadherin score to predict prognosis in breast cancer. Cancer Sci. 2011;102:2248–2254. doi: 10.1111/j.1349-7006.2011.02085.x. [DOI] [PubMed] [Google Scholar]
- 23.Lee KW, Lee NK, Kim JH, Kang MS, Yoo HY, Kim HH, Um SH, Kim SH. Twist1 causes the transcriptional repression of claudin-4 with prognostic significance in esophageal cancer. Biochem Biophys Res Commun. 2012;423:454–460. doi: 10.1016/j.bbrc.2012.05.140. [DOI] [PubMed] [Google Scholar]
- 24.Ersoz S, Mungan S, Cobanoglu U, Turgutalp H, Ozoran Y. Prognostic importance of claudin-1 and claudin-4 expression in colon carcinomas. Pathol Res Pract. 2011;207:285–289. doi: 10.1016/j.prp.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Matsuoka T, Mitomi H, Fukui N, Kanazawa H, Saito T, Hayashi T, Yao T. Cluster analysis of claudin-1 and -4, E-cadherin, and β-catenin expression in colorectal cancers. J Surg Oncol. 2011;103:674–686. doi: 10.1002/jso.21854. [DOI] [PubMed] [Google Scholar]
- 26.Tsutsumi K, Sato N, Tanabe R, Mizumoto K, Morimatsu K, Kayashima T, Fujita H, Ohuchida K, Ohtsuka T, Takahata S, et al. Claudin-4 expression predicts survival in pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2012;19(suppl 3):491–499. doi: 10.1245/s10434-011-1970-2. [DOI] [PubMed] [Google Scholar]
- 27.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kominsky SL. Claudins: emerging targets for cancer therapy. Expert Rev Mol Med. 2006;8:1–11. doi: 10.1017/S1462399406000056. [DOI] [PubMed] [Google Scholar]
- 29.Ouban A, Ahmed AA. Claudins in human cancer: a review. Histol Histopathol. 2010;25:83–90. doi: 10.14670/HH-25.83. [DOI] [PubMed] [Google Scholar]
- 30.Klymkowsky MW, Savagner P. Epithelial-mesenchymal transition: a cancer researcher's conceptual friend and foe. Am J Pathol. 2009;174:1588–1593. doi: 10.2353/ajpath.2009.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 32.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu M, Casimiro MC, Wang C, Shirley LA, Jiao X, Katiyar S, Ju X, Li Z, Yu Z, Zhou J, et al. p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc Natl Acad Sci USA. 2009;106:19035–19039. doi: 10.1073/pnas.0910009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiman JM, Knutson KL, Radisky DC. Immune promotion of epithelial-mesenchymal transition and generation of breast cancer stem cells. Cancer Res. 2010;70:3005–3008. doi: 10.1158/0008-5472.CAN-09-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Mezencev R, Bowen NJ, Matyunina LV, McDonald JF. Isolation and characterization of stem-like cells from a human ovarian cancer cell line. Mol Cell Biochem. 2011;363:257–268. doi: 10.1007/s11010-011-1178-6. [DOI] [PubMed] [Google Scholar]
- 36.Lau MT, Klausen C, Leung PC. E-cadherin inhibits tumor cell growth by suppressing PI3K/Akt signaling via β-catenin-Egr1-mediated PTEN expression. Oncogene. 2011;30:2753–2766. doi: 10.1038/onc.2011.6. [DOI] [PubMed] [Google Scholar]
- 37.Yuecheng Y, Hongmei L, Xiaoyan X. Clinical evaluation of E-cadherin expression and its regulation mechanism in epithelial ovarian cancer. Clin Exp Metastasis. 2006;23:65–74. doi: 10.1007/s10585-006-9020-3. [DOI] [PubMed] [Google Scholar]
- 38.Sawada K, Mitra AK, Radjabi AR, Bhaskar V, Kistner EO, Tretiakova M, Jagadeeswaran S, Montag A, Becker A, Kenny HA, et al. Loss of E-cadherin promotes ovarian cancer metastasis via alpha 5-integrin, which is a therapeutic target. Cancer Res. 2008;68:2329–2339. doi: 10.1158/0008-5472.CAN-07-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–9606. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 40.Singh AB, Sharma A, Dhawan P. Claudin family of proteins and cancer: an overview. J Oncol. 2010;2010:541957. doi: 10.1155/2010/541957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osanai M, Murata M, Nishikiori N, Chiba H, Kojima T, Sawada N. Epigenetic silencing of occludin promotes tumorigenic and metastatic properties of cancer cells via modulations of unique sets of apoptosis-associated genes. Cancer Res. 2006;66:9125–9133. doi: 10.1158/0008-5472.CAN-06-1864. [DOI] [PubMed] [Google Scholar]
- 42.Naik MU, Naik TU, Suckow AT, Duncan MK, Naik UP. Attenuation of junctional adhesion molecule-A is a contributing factor for breast cancer cell invasion. Cancer Res. 2008;68:2194–2203. doi: 10.1158/0008-5472.CAN-07-3057. [DOI] [PubMed] [Google Scholar]
- 43.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J, Sahin A, Agarwal R, Joy C, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DiSaia PJ, Sinkovics JG, Rutledge FN, Smith JP. Cell-mediated immunity to human malignant cells. A brief review and further studies with two gynecologic tumors. Am J Obstet Gynecol. 1972;114:979–989. doi: 10.1016/0002-9378(72)90109-3. [DOI] [PubMed] [Google Scholar]
- 46.Selby PJ, Thomas JM, Monaghan P, Sloane J, Peckham MJ. Human tumour xenografts established and serially transplanted in mice immunologically deprived by thymectomy, cytosine arabinoside and whole-body irradiation. Br J Cancer. 1980;41:52–61. doi: 10.1038/bjc.1980.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan X, Lin X, Manorek G, Kanatani I, Cheung LH, Rosenblum MG, Howell SB. Recombinant CPE fused to tumor necrosis factor targets human ovarian cancer cells expressing the claudin-3 and claudin-4 receptors. Mol Cancer Ther. 2009;8:1906–1915. doi: 10.1158/1535-7163.MCT-09-0106. [DOI] [PubMed] [Google Scholar]
- 48.Holzer AK, Howell SB. The internalization and degradation of human copper transporter 1 following cisplatin exposure. Cancer Res. 2006;66:10944–10952. doi: 10.1158/0008-5472.CAN-06-1710. [DOI] [PubMed] [Google Scholar]
- 49.Albini A. Tumor and endothelial cell invasion of basement membranes. The matrigel chemoinvasion assay as a tool for dissecting molecular mechanisms. Pathol Oncol Res. 1998;4:230–241. doi: 10.1007/BF02905254. [DOI] [PubMed] [Google Scholar]
- 50.Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 51.Mironchik Y, Winnard PT, Jr, Vesuna F, Kato Y, Wildes F, Pathak AP, Kominsky S, Artemov D, Bhujwalla Z, Van Diest P, et al. Twist overexpression induces in vivo angiogenesis and correlates with chromosomal instability in breast cancer. Cancer Res. 2005;65:10801–10809. doi: 10.1158/0008-5472.CAN-05-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 53.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelialmesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 54.Foty RA, Steinberg MS. Cadherin-mediated cell-cell adhesion and tissue segregation in relation to malignancy. Int J Dev Biol. 2004;48:397–409. doi: 10.1387/ijdb.041810rf. [DOI] [PubMed] [Google Scholar]
- 55.Lal-Nag M, Morin PJ. The claudins. Genome Biol. 2009;10:235. doi: 10.1186/gb-2009-10-8-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 57.Qian X, Karpova T, Sheppard AM, McNally J, Lowy DR. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J. 2004;23:1739–1748. doi: 10.1038/sj.emboj.7600136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng TC, Manorek G, Samimi G, Lin X, Berry CC, Howell SB. Identification of genes whose expression is associated with cisplatin resistance in human ovarian carcinoma cells. Cancer Chemother Pharmacol. 2006;58:384–395. doi: 10.1007/s00280-005-0171-8. [DOI] [PubMed] [Google Scholar]
- 59.Agarwal R, D'Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res. 2005;65:7378–7385. doi: 10.1158/0008-5472.CAN-05-1036. [DOI] [PubMed] [Google Scholar]
- 60.Boylan KL, Misemer B, Derycke MS, Andersen JD, Harrington KM, Kalloger SE, Gilks CB, Pambuccian SE, Skubitz AP. Claudin 4 is differentially expressed between ovarian cancer subtypes and plays a role in spheroid formation. Int J Mol Sci. 2011;12:1334–1358. doi: 10.3390/ijms12021334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.