Abstract
AT-rich interactive domain 1A (ARID1A) has emerged as a new tumor suppressor in which frequent somatic mutations have been identified in several types of human cancers. Although most ARID1A somatic mutations are frame-shift or nonsense mutations that contribute to mRNA decay and loss of protein expression, 5% of ARID1A mutations are in-frame insertions or deletions (indels) that involve only a small stretch of peptides. Naturally occurring in-frame indel mutations provide unique and useful models to explore the biology and regulatory role of ARID1A. In this study, we analyzed indel mutations identified in gynecological cancers to determine how these mutations affect the tumor suppressor function of ARID1A. Our results demonstrate that all in-frame mutants analyzed lost their ability to inhibit cellular proliferation or activate transcription of CDKN1A, which encodes p21, a downstream effector of ARID1A. We also showed that ARID1A is a nucleocytoplasmic protein whose stability depends on its subcellular localization. Nuclear ARID1A is less stable than cytoplasmic ARID1A because ARID1A is rapidly degraded by the ubiquitin-proteasome system in the nucleus. In-frame deletions affecting the consensus nuclear export signal reduce steady-state protein levels of ARID1A. This defect in nuclear exportation leads to nuclear retention and subsequent degradation. Our findings delineate a mechanism underlying the regulation of ARID1A subcellular distribution and protein stability and suggest that targeting the nuclear ubiquitin-proteasome system can increase the amount of the ARID1A protein in the nucleus and restore its tumor suppressor functions.
Introduction
Mutations in AT-rich interactive domain 1A (ARID1A) have been detected in a wide variety of human cancers, most frequently in endometrium-related carcinomas (e.g., ovarian clear cell carcinoma, ovarian endometrioid carcinoma, and uterine endometrioid carcinoma) [1–5] and subtypes of gastric carcinoma associated with microsatellite instability or Epstein-Barr virus infection [6]. Most ARID1A mutations are nonsense or out-of-frame mutations, which lead to inactivation of the protein products; therefore, ARID1A is thought to be a tumor suppressor. To directly demonstrate the role of ARID1A in tumorigenesis, in a previous study, we restored wild-type ARID1A expression in ovarian cancer cells that harbor deleterious ARID1A mutations. Our results showed that ARID1A expression is sufficient to suppress cellular proliferation and tumor growth in mice [7]. Moreover, silencing ARID1A expression in nontransformed epithelial cells enhances cellular proliferation and tumorigenicity in a mouse tumor xenograft model [7]. At the molecular level, ARID1A forms a complex with BRG1, and this complex directly interacts with p53 and regulates transcription of downstream effectors including CDKN1A (p21) and SMAD3 through collaboration with p53 [7].
We analyzed sequence mutations of ARID1A reported in different types of neoplasms and found that 5% of the mutations were small in-frame insertions or deletions (indels). These indel mutations may play a role in tumor suppression but may not directly affect protein expression as occurred with nonsense or frameshift mutations that lead to mRNA or protein instability. Therefore, in-frame indels represent unique models for characterizing unknown mechanism(s) in the regulation of ARID1A activity. Our results demonstrate that ARID1A in-frame mutations alter cellular proliferation and CDKN1A (p21) transcription and reveal mechanisms underlying the regulation of ARID1A subcellular distribution and protein stability.
Materials and Methods
DNA Constructs
The vectors pLenti-puro and pcDNA6-V5/His.b containing wild-type full-length ARID1A have been described previously [7]. Mutant constructs DupPP, DupQ, DelL, and Del9 were generated by site-directed mutagenesis using the pCDNA6-V5/His.b-ARID1A plasmid as template [7]. To generate the Del5A construct, the ARID1A N-terminal fragment was amplified by polymerase chain reaction (PCR) with T7 promoter primer and Del5A reverse primer (5′-GCCGCCGCAGCCCCCCAACACTG-3′) using pcDNA6-V5/His.b-ARID1A as template. This fragment was then digested with NheI and cloned into the pcDNA6-V5/His.b-ARID1A plasmid between the NheI site and blunted NotI site. These five constructs were subcloned into the pLenti-puro vector for production of lentivirus, as previously described [7]. All ARID1A constructs used in this study contain V5/His tags at the C terminus. The human influenza hemagglutinin (HA)-tagged ubiquitin vector used in this study was previously described [8].
Cell Culture and Treatment
The ovarian surface epithelial cell line OSE4 [9] and ovarian clear cell carcinoma cell line OVISE [10] were cultured in RPMI-1640 medium supplemented with 10% FBS. The ovarian clear cell carcinoma cell line ES2 and human embryonic kidney cell line HEK293FT (Invitrogen, Grand Island, NY) were maintained in high-glucose Dulbecco's modified Eagle's medium with 10% FBS. Lentivirus expressing ARID1A was produced in HEK293FT cells using the second-generation packaging system pSPAX2 (Addgene plasmid 12260) and pMD2.G (Addgene plasmid 12259). Lentiviral titer was determined by using quantitative PCR (qPCR) to measure viral RNA content in the viral supernatant [11]. Lipofectamine 2000 and Lipofectamine LTX/plus reagents (Invitrogen) were used to transfect the DNA constructs into HEK293FT cells and OSE4 cells, respectively. The proteasome inhibitor MG132 (Boston Biochem, Cambridge, MA) was used at 5 µM, protein synthesis inhibitor cycloheximide (Sigma, St Louis, MO) at 10 µg/ml, and nuclear export inhibitor leptomycin B (Sigma) at 10 ng/ml.
Cell Proliferation and Flow Cytometry Analysis
Cell proliferation was evaluated in 96-well plates as described previously [7]. To determine the number of cells in different phases of cell cycle, we pulse labeled cells with 10 µM 5-ethynyl-2′-deoxyuridine (EdU) for 2 hours using the Click-iT EdU kit (Invitrogen) according to the manufacturer's instruction. The EdU was subsequently detected with Alexa Fluor 647 azide, and DNA was stained with CellCycle 405-blue. Cells were then analyzed by flow cytometry (BD LSR II flow cytometer; BD Biosciences, San Jose, CA).
In Vivo Ubiquitination Assay
To determine the ubiquitination of ARID1A, we followed a previously published method [12]. Briefly, HEK293FT cells were cotransfected with vectors expressing wild-type ARID1A or mutant Del9 or DelL along with the HA-tagged ubiquitin expression vector. Forty-eight hours after transfection, the cells were lysed in urea lysis buffer (8 M urea, 100 mM NaH2PO4, 20 mM imidazole, pH 7.5). The lysates were then incubated with 20 µl of nickel-nitrilotriacetic acid (Ni-NTA) agarose resin (Qiagen, Valencia, CA) for 30 minutes at room temperature. Proteins bound to the resin were washed with urea lysis buffer, boiled in Laemmli buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by immunoblot analysis.
Cell Fractionation, Immunoprecipitation, and Chromatin Immunoprecipitation
Nuclear and cytoplasmic lysates were prepared by using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Thermo Scientific, Rockford, IL) with slight modifications to the manufacturer's protocol. Briefly, instead of using the nuclear extraction reagent provided, the nuclear pellet was solubilized by sonication in a 1:1 mixture of 2x Laemmli buffer and NP-40 buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40). For coimmunoprecipitation of exportin 1 (XPO1) and ARID1A, ES2 cells were treated with dimethyl 3,3′-dithiobispropionimidate (Thermo Scientific) to cross-link proteins and then lysed in NP-40 buffer with 0.5% Triton. Antibody complexes were pulled down using Protein A/G (1:1) magnetic beads (Dynabeads; Invitrogen).
Chromatin immunoprecipitation was performed with anti-V5 antibodies (V8012, Sigma) using chromatin from OSE4 cells transfected with the ARID1A DNA constructs. After transfection, OSE4 cells were sequentially fixed with ethylene glycol bis(succinimidylsuccinate) (Thermo Scientific) and 1% formaldehyde and lysed with chromatin immunoprecipitation lysis buffer [1% sodium dodecyl sulfate, 10 mM EDTA, and 50 mM Tris (pH7.5)]. Binding of ARID1A to the CDKN1A promoter was assessed by qPCR as previously described [7].
qPCR and Western Blot Analysis
Total RNA was isolated using the RNeasy Plus Mini Kit (Qiagen). First-strand cDNA generated using the iScript cDNA synthesis kit (Bio-Rad) was served as a template for qPCR. qPCR was performed using the CFX96 Real-Time PCR Detection System (Bio-Rad) and SYBR Green I reagents (Invitrogen). Primers used to amplify ARID1A and CDKN1A (p21) were previously described [7]. The gene encoding human large ribosomal protein (RPLP0) was used for normalization (forward primer, 5′-GCAATGTTGCCAGTGTCTG-3′; reverse primer, 5′-GCCTTGACCTTTTCAGCAA-3′). The antibodies used for Western blot analysis were ARID1A (HPA005456, Sigma-Aldrich), p53 (FL-393, sc-6243; Santa Cruz Biotechnology, Santa Cruz, CA), p21 (DCS60, #2946; Cell Signaling Technology, Danvers, MA), glyceraldehyde-3-phosphate dehydrogenase (G9545, Sigma-Aldrich), V5 (R960-25, Invitrogen), histone H3 (ab1791; Abcam, Cambridge, MA), XPO1 (named CRM1 in yeast, sc-5595, Santa Cruz Biotechnology), and mouse anti-rabbit immunoglobulin G (IgG) (conformation specific; #3678, Cell Signaling Technology). Signal intensities on films were quantified using ImageQuant 5.2 software (Molecular Dynamics). Glyceraldehyde-3-phosphate dehydrogenase and p53 were used as loading controls for proteins in the cytoplasmic and nuclear fractions, respectively.
Results
In-frame Indel Mutations of ARID1A in Human Cancer
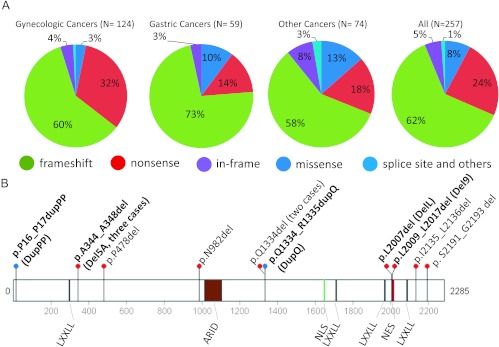
ARID1A mutations have been reported in a variety of human neoplasms [1–6,13,14]. In addition to those tumors in which ARID1A has already been sequenced, we sequenced an ovarian clear cell carcinoma specimen, 1013T, and found that it contained two heterozygous mutations: a frameshift mutation, p.1415Qfs, and an in-frame deletion, p.L2009_L2017del (Del9). Among the 257 somatic ARID1A mutations identified to date, we found that 62% were frameshift mutations, and 24% were nonsense mutations (Figure 1A). Thus, approximately 86% of ARID1A mutations would result in reduced ARID1A protein levels because of nonsense-mediated mRNA decay and/or degradation of incorrectly folded truncated proteins. A closer look at the ARID1A mutation spectrum revealed that 13 (5%) of 257 ARID1A mutations involved small indels (Figure 1B and Table 1). Ten of the 13 indels were unique, involving deletion or duplication at different locations in the ARID1A protein. Of note, three tumors harbored the p.A344_A348del mutation, two tumors harbored the p.Q1334del mutation, and two tumors harbored mutations affecting codons located at the leucine-rich (LxxxLxxLxL) nuclear export signal (NES) (Figure 1B).
Figure 1.
The spectrum of ARID1A mutations in human cancer. (A) Types of ARID1A mutations found in endometrium-related gynecologic cancer, gastric cancer, and other cancer types. Different mutation types are indicated by different colors. (B) Diagram of in-frame indel mutations. The five in-frame mutations identified in ovarian clear cell and endometrioid carcinomas are shown in bold font. The p.A344_A348del mutation was detected in three independent tumors, and the p.Q1334del mutation was detected in two tumors. LXXLL, leucine-rich steroid receptor binding motif; ARID, AT-rich interactive domain; NLS, nuclear localization signal; NES, nuclear export signal.
Table 1.
In-frame Indel Mutations of ARID1A in Human Carcinomas.
| Mutation | Tumor Type | Construct Name | Reference |
| p.A344_A348del | Ovarian endometrioid carcinoma | Del5A | [2] |
| p.P16_P17dupPP | Ovarian clear cell carcinoma | DupPP | [2] |
| p.Q1334_R1335dupQ | Ovarian clear cell carcinoma | DupQ | [1] |
| p.L2007del | Ovarian clear cell carcinoma | DelL | [2] |
| p.L2009_L2017del | Ovarian clear cell carcinoma | Del9 | This study |
| p.A344_A348del | Gastric carcinoma | Del5A | [6] |
| p.Q1334del | Gastric carcinoma | N/A | [6] |
| p.N982del | Colorectal carcinoma | N/A | [4] |
| p.Q1334del | Prostate carcinoma | N/A | [4] |
| p.I2135L2136del | Lung carcinoma | N/A | [4] |
| p.S2191_G2193del | Hepatocellular carcinoma | N/A | [13] |
| p.A344_A348del | Hepatocellular carcinoma | Del5A | [13] |
| p.P478del | Hepatocellular carcinoma | N/A | [14] |
Del, deletion; dup, duplication.
ARID1A In-frame Indel Mutants Lose Ability to Suppress Cellular Proliferation and Induce p21 Expression
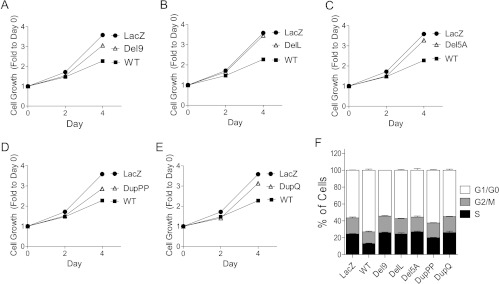
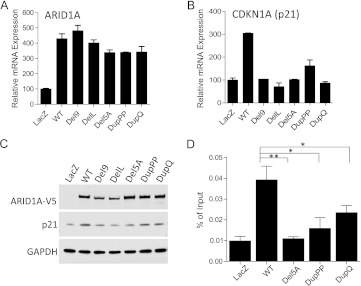
To determine whether in-frame indel mutations of ARID1A retain tumor suppressor function, we constructed expression vectors for all in-frame indel mutations identified in gynecological cancers (Del9, DelL, Del5A, DupPP, and DupQ; Table 1). For functional analysis of the mutants, we used the ovarian clear cell carcinoma cell line OVISE, because this cell line harbors biallelic ARID1A frameshift mutations, which results in undetectable ARID1A protein expression [1,7]. OVISE cells were transduced with lentivirus expressing wild-type ARID1A or one of the in-frame indel mutant constructs. Compared with control cells that expressed wild-type ARID1A, cells expressing in-frame indel mutants showed attenuated growth inhibition, as determined by growth curve analysis (Figure 2). Moreover, ectopic expression of the in-frame indel mutants failed to increase the proportion of cells at the G0/G1 phase, whereas G0/G1 phase arrest was observed in cells expressing wild-type ARID1A. In fact, cell cycle distribution in cells transfected with indel mutants was similar to that of cells transfected with the lacZ-expressing vector control (Figure 2F). Because CDKN1A (p21) is a direct target of the ARID1A chromatin remodeling complex, and p21 plays a role in the tumor suppressor function of ARID1A [7,15], we determined the effects of the in-frame indel mutations of ARID1A on p21 expression. ARID1A mRNA levels in cells transfected with wild-type ARID1A were similar to those of cells transfected with the indel mutants (Figure 3A); however, CDKN1A mRNA levels were markedly reduced in cells expressing the mutant constructs (Figure 3B). Western blot analysis was performed to determine whether the reduced CDKN1A transcription in cells expressing mutant ARID1A was because of reduced ARID1A protein levels. We found that that cells expressing the Del9 and DelL mutants showed reduced ARID1A protein expression, whereas cells expressing the DelA, DupPP, and DupQ mutants showed ARID1A protein levels similar to that of wild-type ARID1A (Figure 3C). Interestingly, p21 protein levels were decreased in cells transfected with each of the ARID1A in-frame indel mutants, including the mutations that did not alter ARID1A protein expression (i.e., DelA, DupPP, and DupQ; Figure 3C). We then evaluated whether chromatin remodeling complexes containing the DelA, DupPP, and DupQ mutant proteins were able to bind to CDKN1A promoter by performing ChIP-qPCR analysis. Compared with wild-type ARID1A, mutants DelA, DupPP, and DupQ exhibited significantly reduced binding activity to the CDKN1A promoter (Figure 3D).
Figure 2.
Loss of tumor suppressor function of ARID1A in-frame mutations. (A–E) OVISE cells, which do not express detectable levels of endogenous ARID1A protein, were infected with lentiviral vectors expressing wild-type or mutant ARID1A. Relative cell numbers were determined at different days after lentiviral transduction. (F) Cell cycle progression was analyzed by flow cytometry after EdU incorporation and DNA staining. The percentages of cells in different phases of cell cycle were determined.
Figure 3.
ARID1A mutants lose the ability to induce CDKN1A (p21) transcription. (A) Levels of ARID1A mRNA in cells expressing wild-type ARID1A or in-frame indel mutants. Vector expressing lacZ was used as a negative control. (B) Levels of CDKN1A (p21) mRNA in cells expressing wild-type ARID1A or in-frame mutants. Vector expressing lacZ was used as a negative control. (C) Western blot analysis shows protein levels of ARID1A and p21 in cells expressing wild-type ARID1A and in-frame indel mutants. GAPDH was used as a protein loading control. (D) Chromatin immunoprecipitation-qPCR analysis demonstrates the binding activity of wild-type ARID1A and in-frame indel mutants Del 5A, Dup2P, and DupQ to the CDKN1A promoter. One-way t test, *P < 0.05; **P < 0.01.
Degradation of ARID1A Protein by Ubiquitin-Proteasome System
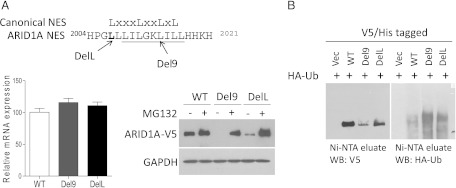
Because the DelL and Del9 mutants showed normal mRNA levels but decreased steady-state protein levels, we evaluated the posttranscriptional regulation of these two proteins. HEK293FT cells transfected with vectors expressing wild-type or mutant ARID1A protein tagged with the V5/His epitope were treated with the proteasome inhibitor MG132 or vehicle control [dimethyl sulfoxide (DMSO)]. We found that proteasome inhibition resulted in the accumulation of wild-type ARID1A protein, indicating that the proteasome pathway was involved in ARID1A degradation. In addition, in cells harboring the DelL and Del9 mutants, MG132 treatment fully restored ARID1A protein levels (Figure 4A). Taken together, our results suggest that the reduced steady-state protein levels of DelL and Del9 mutants were not related to translation efficiency or mRNA decay but to increased proteasomal degradation.
Figure 4.
Degradation of ARID1A by the ubiquitin-proteasome pathway. (A) Proteasome inhibition increases Del9 and DelL protein levels. The residue deleted in the DelL mutant is indicated in bold, and residues deleted in the Del9 mutant are underlined. ARID1A constructs with V5/His dual tags at the C terminus were introduced into HEK293FT cells. Twenty-four hours after transfection, cells were treated with MG132 (proteasome inhibitor) or DMSO (vehicle control) for an additional 16 hours. ARID1A protein levels were determined by Western blot analysis using an anti-V5 antibody (right), and ARID1A mRNA levels (left) were determined by qPCR analysis. (B) Enhanced ubiquitination of Del9 and DelL mutants as compared with the wild-type protein. HEK293FT cells were transfected with constructs expressing HA-tagged ubiquitin (HA-Ub) and ARID1A-V5/His. ARID1A proteins were enriched using Ni-NTA agarose beads. The eluates were subjected to Western blot analysis using anti-V5 and anti-HA antibodies.
Because ubiquitination typically precedes proteasomal degradation, we determined whether ARID1A degradation was mediated by the ubiquitin-mediated system. HA-tagged ubiquitin vector was transfected into HEK293FT cells expressing wild-type ARID1A, DelL, or Del9. The V5/His-tagged protein was enriched by purification with nickel affinity beads and analyzed by Western blot analysis using antiV5 and anti-HA antibodies. We found that cells expressing Del9 and DelL mutants showed higher levels of ARID1A ubiquitination than cells expressing wild-type ARID1A (Figure 4B, right panel). Conversely, lower levels of nonubiquitinated ARID1A were observed in cells expressing mutant proteins than in cells expressing wild-type ARID1A (Figure 4B, left panel). These findings suggest that the DelL and Del9 mutations lead to increased ubiquitination and subsequent proteasomal degradation, resulting in the observed reduction in steady-state ARID1A protein levels.
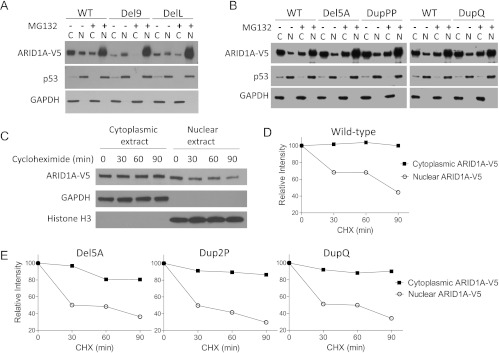
Protein Expression of Wild-type ARID1A and In-frame Indel Mutants after Proteasome Inhibition
The DelL and Del9 mutations occurred at the conserved NES motif, suggesting that these mutations inhibit nuclear export and facilitate proteasomal degradation in the nucleus. To test this possibility, we prepared cytosolic and nuclear fractions from OSE4 cells expressing wild-type ARID1A, DelL, or Del9. We found that cytoplasmic ARID1A protein levels were considerably lower in cells expressing DelL or Del9 than in cells expressing wild-type ARID1A, indicating that the nuclear export of these two mutants was inhibited (Figure 5A). In contrast, the cytoplasmic and nuclear distribution of ARID1A in cells expressing Del5A, DupPP, and DupQ was similar to that of cells expressing wild-type ARID1A (Figure 5B). After blocking proteasome activity with MG132, the DelL and Del9 mutant proteins accumulated in the nucleus to levels similar to that of wild-type ARID1A (Figure 5A). We also noted that cytoplasmic ARID1A levels were slightly reduced after MG132 treatment in all cells (expressing wild-type or mutant protein); this result may be because of nonspecific changes in the equilibrium distribution of proteins between the cytoplasm and nucleus on MG132 treatment.
Figure 5.
Localization of ARID1A protein in the nucleus and cytoplasm. (A) Steady-state levels of wild-type ARDI1A protein and the Del9 and DelL mutants in nuclear (N) and cytoplasmic (C) fractions. OSE4 cells were transfected with ARID1A constructs and treated with MG132 (proteasome inhibitor) or DMSO (vehicle control). Ectopic ARID1A levels were determined by Western blot analysis using an anti-V5 antibody. GAPDH and p53 served as loading controls for cytoplasmic and nuclear protein fractions, respectively. (B) Subcellular distribution of the three ARID1A in-frame indel mutants in which the NES was not affected. (C, D) The half-life of cytoplasmic and nuclear ARID1A was determined after inhibiting protein synthesis with cycloheximide (CHX). Histone H3 served as a nuclear protein loading control. (E) Assessment of the half-life of ARID1A mutant proteins in cytosolic and nuclear compartments.
Cytoplasmic Pool of ARID1A Is More Stable than the Nuclear Pool
To determine whether ARID1A degradation occurs primarily in the nucleus, we evaluated the stability of wild-type ARID1A protein in the cytosolic and nuclear compartments of OSE4 cells after inhibiting protein synthesis with cycloheximide. Western blot analysis of the cellular fractions showed no detectable decline in cytoplasmic ARID1A protein for up to 90 minutes, whereas the level of nuclear ARID1A protein decreased to 45% with a half-life of approximately 75 minutes (Figure 5, C and D). Using the same strategy, we further assessed the stabilities of cytosolic and nuclear ARID1A proteins from different mutants, Del5A, Dup2P, and DupQ. Del9 and DelL mutants were not included because of their low steady-state protein levels. As shown in Figure 5E, nuclear protein levels of all three ARID1A mutants decreased rapidly after protein synthesis inhibition, while their levels in cytosolic fraction remained relatively constant.
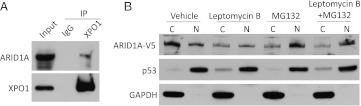
Next, we determined whether the cytoplasmic fraction of ARID1A was nascent protein that had yet to be imported into the nucleus or represented ARID1A exported from the nucleus. Because the leucine-rich NES interacts with XPO1 (also named CRM1 in yeast) to form an export complex [16], we determined whether ARID1A interacts with XPO1 by coimmunoprecipitation of these proteins. For this assay, we used ES2 cells, which express high levels of ARID1A [7]. ARID1A was readily detected in immunoprecipitates pulled down with the XPO1 antibody but not in immunoprecipitates pulled down with normal IgG (Figure 6A). Treatment with leptomycin B, a nuclear export inhibitor that targets XPO1, markedly reduced cytoplasmic levels of ARID1A protein (Figure 6B, lane 3). However, leptomycin B did not increase nuclear ARID1A levels, in part because of rapid protein degradation in the nucleus. In fact, in leptomycin B-treated cells, nuclear ARID1A protein level was restored by treatment with proteasome inhibitor MG132 (Figure 6B, lane 8).
Figure 6.
XPO1-dependent ARID1A nuclear export. (A) ES2 cell lysate was immunoprecipitated using a rabbit anti-XPO1 antibody or normal rabbit IgG. Total cell extracts (input) and immunoprecipitates were then probed using antibodies against XPO1 or ARID1A followed by a mouse anti-rabbit conformation-specific antibody. (B) Nuclear and cytoplasmic levels of ARID1A were determined in OSE4 cells transfected with wild-type ARID1A. Sixteen hours after transfection, cells were treated with leptomycin B (LMB) for an additional 24 hours; during the last 6 hours of incubation, MG132 was added to some of the cells as indicated. Protein levels of ARID1A-V5 were detected by Western blot analysis with an anti-V5 antibody. GAPDH and p53 served as loading controls for cytoplasmic and nuclear protein fractions, respectively.
Discussion
This study was undertaken to characterize all ARID1A in-frame indel mutations that have been identified in gynecological cancers. Because somatically acquired mutations are the result of natural selection for more advantageous tumor growth, structural changes in the mutant proteins may likely attenuate the tumor suppression function of ARID1A. In fact, we demonstrated that these indel mutants lack the ability to inhibit cellular proliferation or activate p21 transcription, the two main functions of wild-type ARID1A known so far [7]. We further showed that in-frame indel deletions involving the NES lead to the nuclear retention, increased ubiquitination, and subsequent degradation by the ubiquitin-proteasome pathway. The findings reported here demonstrate the regulatory mechanism involving ARID1A protein stability and set an example that studying naturally occurring mutations may elucidate the underlying biology of that protein.
In-frame indel mutations of ARID1A can be divided into two groups according to mechanisms underlying suppression of its tumor suppressor function. The first group of mutations is exemplified by Del9 and DelL, in which the NES sequence is disrupted by in-frame deletions, resulting in reduced nuclear export. The mutant proteins are more highly ubiquitinated, which may lead to more rapid degradation than wild-type ARID1A. The second group of mutant proteins includes DelA, DupPP, and DupQ, in which mutations do not occur at the NES or any known protein domain of ARID1A (Figure 1B). The amino acid changes in DelA, DupPP, and DupQ do not alter protein stability significantly; these mutant proteins bind with lower affinity to the CDKN1A promoter compared with wild-type ARID1A, suggesting that the residues deleted or duplicated in these three mutants are important for promoter binding. Alternatively, the residues may be important for interactions between ARID1A and the other switch/sucrose nonfermentable subunits in the ARID1A chromatin remodeling complex and mutations at these residues may disturb the stability of the whole protein complex.
In addition to the leucine-rich NES, which mediates the transport of large molecules through the nuclear pore complex, ARID1A also contains a classical nuclear localization signal [17], PVLKQRR (amino acids 1651–1657), which targets ARID1A protein to the nucleus. Therefore, it is likely that a highly dynamic balance between nuclear trafficking and nuclear proteasomal degradation is required to maintain the appropriate level of ARID1A in the nucleus, where it functions as a chromatin remodeler and tumor suppressor. Our data in Figure 5C indicate that ARID1A degradation happens mainly in the nucleus. It has been established that the cell nucleus contains the components of the ubiquitin-proteasome system, which include the ubiquitin-conjugating enzymes (E1, E2s, and E3s) and the 26S proteasome (including 19S and 20S subunits) [18]. The ubiquitin-proteasome system in the nucleus was shown to control the localization, quantity, and transcriptional activity of the transcriptional complexes [19]. For example, the proteolytic activity of the 20S proteasome was found to facilitate transcriptional elongation by mediating turnover of promoter-bound activators and allowing the initiating RNA polymerase to escape from the promoter [20,21]. We speculate that rapid ARID1A turnover in the nucleus might be integral to its dynamic function of chromatin remodeling and transcription regulation. So far, the ubiquitin ligase responsible for ARID1A ubiquitination has not been identified. However, a close homolog of ARID1A (ARID1B) is part of an E3 ligase complex that targets histone H2B [22], suggesting that ARID1A may ubiquitinate itself.
Although this study provides new insights into the effect of in-frame indels on the biological functions of ARID1A, several limitations of this study should be pointed out. First, we did not characterize all 10 unique in-frame indels identified to date but focused only on the five in-frame indels identified in gynecological cancer. It would be interesting to know whether the mutations that were not characterized in this report also show similar loss-of-function phenotypes. Second, the switch/sucrose nonfermentable complex containing ARID1A is composed of many subunits. Therefore, future studies are needed to understand how in-frame indels that do not involve the NES contribute to loss of function by determining if those mutations affect protein-protein interaction between ARID1A and other subunits in the complex or affect interaction between ARID1A complex and DNA. Finally, because ARID1A protein stability is largely regulated by the nuclear ubiquitin-mediated proteasome system, it would be interesting to test whether stabilizing nuclear proteins including ARID1A with specific ubiquitin-proteasome inhibitors exerts antitumor effects.
In summary, we demonstrated that in-frame indel mutations of ARID1A, like frameshift and nonsense mutations, are associated with the loss of ability to suppress cellular proliferation and activate CDKN1A (p21) transcription. These effects are mediated by increased protein degradation in the nucleus or reduced CDKN1A promoter binding. Our findings provide new evidence that ARID1A is a nucleocytoplasmic protein, and structural disruption of the NES results in retention of ARID1A in the nucleus, where it is subject to ubiquitin-proteasomal degradation. These results may have important implications for understanding the biology of ARID1A and its role as a tumor suppressor.
Abbreviations
- ARID
AT-rich interactive domain
- ChIP
chromatin immunoprecipitation
- XPO1
exportin 1
- DMSO
dimethyl sulfoxide
- EdU
5-ethynyl-2′-deoxyuridine
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HA
hemagglutinin
- indels
insertions/deletions
- NES
nuclear export signal
- qPCR
quantitative polymerase chain reaction
Footnotes
This study was supported by National Institutes of Health/National Cancer Institute grants R21CA165807, R01CA129080, and R01CA103937. B.G. is partly supported by a Program of Excellence Grant from the Ovarian Cancer Research Fund.
References
- 1.Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr, Vogelstein B, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan B, Mao TL, Panuganti PK, Kuhn E, Kurman RJ, Maeda D, Chen E, Jeng YM, Wang TL, Shih Ie M. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35:625–632. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones S, Li M, Parsons DW, Zhang X, Wesseling J, Kristel P, Schmidt MK, Markowitz S, Yan H, Bigner D, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100–103. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan AS, Tsui WY, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 7.Guan B, Wang TL, Shih Ie M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71:6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 9.Nitta M, Katabuchi H, Ohtake H, Tashiro H, Yamaizumi M, Okamura H. Characterization and tumorigenicity of human ovarian surface epithelial cells immortalized by SV40 large T antigen. Gynecol Oncol. 2001;81:10–17. doi: 10.1006/gyno.2000.6084. [DOI] [PubMed] [Google Scholar]
- 10.Gorai I, Nakazawa T, Miyagi E, Hirahara F, Nagashima Y, Minaguchi H. Establishment and characterization of two human ovarian clear cell adenocarcinoma lines from metastatic lesions with different properties. Gynecol Oncol. 1995;57:33–46. doi: 10.1006/gyno.1995.1097. [DOI] [PubMed] [Google Scholar]
- 11.Sastry L, Johnson T, Hobson MJ, Smucker B, Cornetta K. Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Ther. 2002;9:1155–1162. doi: 10.1038/sj.gt.3301731. [DOI] [PubMed] [Google Scholar]
- 12.Guan B, Pungaliya P, Li X, Uquillas C, Mutton LN, Rubin EH, Bieberich CJ. Ubiquitination by TOPORS regulates the prostate tumor suppressor NKX3.1. J Biol Chem. 2008;283:4834–4840. doi: 10.1074/jbc.M708630200. [DOI] [PubMed] [Google Scholar]
- 13.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 15.Nagl NG, Jr, Patsialou A, Haines DS, Dallas PB, Beck GR, Jr, Moran E. The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest. Cancer Res. 2005;65:9236–9244. doi: 10.1158/0008-5472.CAN-05-1225. [DOI] [PubMed] [Google Scholar]
- 16.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 17.Dingwall C, Sharnick SV, Laskey RA. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- 18.von Mikecz A. The nuclear ubiquitin-proteasome system. J Cell Sci. 2006;119:1977–1984. doi: 10.1242/jcs.03008. [DOI] [PubMed] [Google Scholar]
- 19.Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 20.Kinyamu HK, Chen J, Archer TK. Linking the ubiquitin-proteasome pathway to chromatin remodeling/modification by nuclear receptors. J Mol Endocrinol. 2005;34:281–297. doi: 10.1677/jme.1.01680. [DOI] [PubMed] [Google Scholar]
- 21.Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- 22.Li XS, Trojer P, Matsumura T, Treisman JE, Tanese N. Mammalian SWI/SNF-a subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol Cell Biol. 2010;30:1673–1688. doi: 10.1128/MCB.00540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]