Abstract
We present a mathematical and a computational framework for the modelling of cell motility. The cell membrane is represented by an evolving surface, with the movement of the cell determined by the interaction of various forces that act normal to the surface. We consider external forces such as those that may arise owing to inhomogeneities in the medium and a pressure that constrains the enclosed volume, as well as internal forces that arise from the reaction of the cells' surface to stretching and bending. We also consider a protrusive force associated with a reaction–diffusion system (RDS) posed on the cell membrane, with cell polarization modelled by this surface RDS. The computational method is based on an evolving surface finite-element method. The general method can account for the large deformations that arise in cell motility and allows the simulation of cell migration in three dimensions. We illustrate applications of the proposed modelling framework and numerical method by reporting on numerical simulations of a model for eukaryotic chemotaxis and a model for the persistent movement of keratocytes in two and three space dimensions. Movies of the simulated cells can be obtained from http://homepages.warwick.ac.uk/∼maskae/CV_Warwick/Chemotaxis.html.
Keywords: cell motility, chemotaxis, surface finite elements, reaction–diffusion systems
1. Introduction
Modelling the directional motility of cells is of great importance especially because of the central role-directed cell migration plays in several biological phenomena, such as embryonic development, cancer, tissue development and immune responses [1]. Broadly speaking, the motile cycle of a cell consists of the following processes: polarization where the cell develops a front and a back through the redistribution of proteins and lipids within the cell, protrusion at the leading edge of the cell pushing the front of the cell outwards, and retraction of the rear of the cell towards the leading edge [2]. Although the main aspects of the motile cycle appear deceptively simple, as further details are added to the modelling, various complexities arise. For example, in the case of chemotactic eukaryotic cells, the molecular mechanisms that govern gradient sensing and cell polarization are still not fully understood [3]. Furthermore, it is difficult to quantify the forces associated with motility and only recently has experimental progress been made in this direction [4,5]. Direct numerical simulation of cell motility necessitates the consideration of deformable surfaces [6,7] or multi-phase flow models [8,9], both of which are computationally challenging. Finally, the deformation of the cell surface is linked to the dynamics of actin and other cell-resident proteins, and these dynamics must be coupled in a consistent way with the evolution of the cell surface.
In this work, we present a mathematical framework for the modelling of cell motility and a numerical method for the simulation of such models. The approach we propose uses ideas of existing two-dimensional models but generalizes these and extends the modelling to the three-dimensional setting. It consists of partial differential equations, specifically those of reaction–diffusion type, posed on the cell boundary coupled with an evolution law for the cell membrane. Further, we discuss the inclusion of external forces and illustrate this with a phenomenological model for the interaction between a cell and a obstacle. We present a numerical method, based on evolving triangulated surfaces, that consists of an evolving surface finite-element method [10] for the approximation of the surface partial differential equation and a parametrized finite-element method [11] for the approximation of the surface evolution law.
It is our hope that the parametric approach we employ will be more efficient than other standard approaches such as phase field [6] or level set methods [12]. The reasoning behind this statement is that our methodology based on triangulated surfaces formulates the problem in one dimension less than other approaches, in which the equations are discretized in the embedding space. See, for example [13–16] and references therein for further discussion.
We consider two specific models for cell motility. First, a model for eukaryotic chemotaxis. Aspects of chemotaxis, such as changes in direction owing to splitting and biased generation of pseudopods as well as response to a changing chemotactic signal [17], are present in this surface-based model. We also present simulations of pseudopod-driven migration of a cell in three dimensions. We next consider a surface-based model for the persistent migration of fish keratocytes, presenting numerical simulations in two- and three dimensions. The surface-based model is able to capture the different shapes that characterize migrating keratocytes and the correlation between aspect ratio and cell speed [18].
A summary of the contributions of our study is as follows. We derive a rigorous mathematical framework for the modelling of cell motility and chemotaxis in two or three space dimensions, our modelling includes both surface tension and bending rigidity with volume conservation and allows the inclusion of external forces. We present a numerical method for the simulation of the model. Equations for and on the surface of the cell are discretized on a discrete surface. The efficacy of our methodology is illustrated by computer simulations of pseudopod-driven migration and persistent migration in three space dimensions and the simulation of cell migration in the presence of obstacles.
While a major part of this work is the investigation of modelling generalizations through numerical simulation, especially with respect to cell motility in three dimensions, our intention is to present a general modelling framework and numerical method that will be a potentially useful methodology for experimentalists and theoreticians alike in future studies of cell motility.
The remainder of our discussion proceeds as follows. In §2, we present our general modelling framework and our modelling assumptions. In §3, we present a numerical method for the approximation of surface evolution laws coupled with surface partial differential equations. In §4, we report the results of numerical simulations of a model for chemotaxis. In §5, we report the results of numerical simulations of a surface-based model for the persistent motion of cells such as fish keratocytes. In §6, we discuss the implications of our findings in the study of cell motility and possible applications of our methodologies in future studies. We provide the technical details of our modelling and the numerical methods we employ in the electronic supplementary material.
2. A surface-based model for cell polarization and movement
We consider models for cell motility and chemotaxis that consist of a geometric evolution law for a hypersurface representing the cell boundary coupled to a spatial pattern generator on the evolving surface describing polarization of the cell. The particular form of the spatial patterning mechanism we shall investigate is a Turing pattern generator, i.e. a semi-linear reaction–diffusion system (RDS). The use of Turing type systems to model biological pattern formation phenomena is widespread (see Murray [19] for a review), and recent numerical studies of Turing type systems on evolving surfaces show that while the key features of Turing mechanisms persist, such as spontaneous pattern formation and bifurcations owing to surface evolution, the geometry of the evolving surface strongly influences the patterns expressed [20,21]. We stress that our general modelling strategy and the numerical methods we employ can be generalized to other possible polarization mechanisms, such as gradient-based models or excitable network and wave-based models all of which effectively couple surface partial differential equations to a surface evolution law [3].
2.1. Geometric evolution model
The cell membrane is represented by an evolving hypersurface, with the movement of the cell determined by the interaction of various forces that act normal to the cell membrane. We consider external forces such as a protrusive force associated with the RDS species and a pressure that constrains the enclosed volume, as well as internal forces that arise from the reaction of the cells' surface to stretching and bending. We use the following force balance on the membrane, where we neglect the inertia of the membrane:
| 2.1 |
where  denotes the outward pointing unit normal to the surface Γ. We account for the following force contributions appearing in equation (2.1).
denotes the outward pointing unit normal to the surface Γ. We account for the following force contributions appearing in equation (2.1).
- — A protrusive force depending on the densities of chemical species resident on the membrane (cf. equation (2.9)) is denoted by
In the subsequent numerical simulations, we make the phenomenological modelling assumption that the force is proportional to the species densities and given by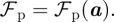
2.2 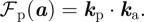 The sign of the component
The sign of the component 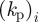 of the vector
of the vector 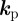 governs whether the ith species promotes protrusion (positive) or retraction (negative) of the cell membrane. For
governs whether the ith species promotes protrusion (positive) or retraction (negative) of the cell membrane. For 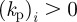 such a force may model the protrusive force generated by cross-linked filamentary actin in the vicinity of the cells' surface, while
such a force may model the protrusive force generated by cross-linked filamentary actin in the vicinity of the cells' surface, while 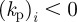 could correspond to the contraction force generated by actin bundles [22].
could correspond to the contraction force generated by actin bundles [22]. -
— Experimental studies suggest that while the cell surface area may exhibit variability during movement, the enclosed volume is relatively constant [18,23]. We take this fact into account as a hard constraint, which means that the cell is able to immediately counterbalance small volume changes on the time scale of the RDS and the boundary evolution. In the following, a corresponding Lagrange multiplier will be denoted by
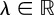 . It can be interpreted as a pressure difference between the interior and exterior of the cell. The corresponding force simply reads
. It can be interpreted as a pressure difference between the interior and exterior of the cell. The corresponding force simply reads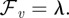
Note that the Lagrange multiplier λ is spatially constant and therefore models a spatially uniform force such that the enclosed volume is conserved.
- — We include a viscous force that opposes motion:
where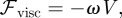
2.3 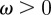 is a kinetic coefficient and V is such that
is a kinetic coefficient and V is such that 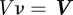 , where
, where 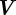 is the material velocity of the cell boundary (which we assume to be normal to the cell membrane). In the two-dimensional case, adhesion and de-adhesion may be modelled as an effective friction, i.e. a force, of the form (2.3), proportional to the local speed [22,24]. In the three-dimensional case, the situation is more complicated and we intend to address this subject in future work.
is the material velocity of the cell boundary (which we assume to be normal to the cell membrane). In the two-dimensional case, adhesion and de-adhesion may be modelled as an effective friction, i.e. a force, of the form (2.3), proportional to the local speed [22,24]. In the three-dimensional case, the situation is more complicated and we intend to address this subject in future work. — We write
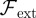 for any other external normal forces acting on the cell boundary, where we have interaction with the medium in mind. As a concrete example and in order to illustrate the versatility of the proposed approach for cell motility in the complicated environments encountered in vitro, in §4.1.3, we present a simple phenomenological model for the movement of cells in the presence of obstacle particles that the cell cannot invade but that it may push out of its way.
for any other external normal forces acting on the cell boundary, where we have interaction with the medium in mind. As a concrete example and in order to illustrate the versatility of the proposed approach for cell motility in the complicated environments encountered in vitro, in §4.1.3, we present a simple phenomenological model for the movement of cells in the presence of obstacle particles that the cell cannot invade but that it may push out of its way.- — Resistance of the cell boundary to stretching may be incorporated by means of a surface energy of the form:
where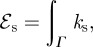
2.4 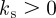 can be interpreted as a surface tension. The variation of the area functional is the mean curvature (we refer to Deckelnick et al. [25] for details of a derivation), hence the force arising from the surface energy is given by:
can be interpreted as a surface tension. The variation of the area functional is the mean curvature (we refer to Deckelnick et al. [25] for details of a derivation), hence the force arising from the surface energy is given by:
where H is the mean curvature of Γ.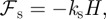
2.5 - — The lipid bilayer forming the basic component of the cell membranes also resists bending. We consider the established model of Helfrich [26] for the bending energy:
where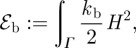
2.6 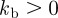 is the bending rigidity. The variation of the bending energy yields the force contribution
is the bending rigidity. The variation of the bending energy yields the force contribution
where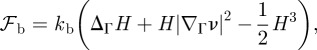
2.7 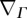 and ΔΓ denote the surface gradient and Laplace–Beltrami operator, respectively (electronic supplementary material). We refer to Willmore [27] for a derivation.
and ΔΓ denote the surface gradient and Laplace–Beltrami operator, respectively (electronic supplementary material). We refer to Willmore [27] for a derivation.
Summing up the forces with their specific choices except for the external force, we obtain the following equation for the evolution of the cell boundary:
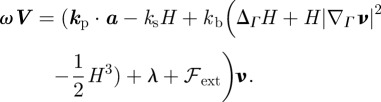 |
2.8 |
The variational formulation of the evolution law (2.8), which we use to construct a finite-element discretization, is given in the electronic supplementary material.
2.2. Cell polarization model
We consider an RDS posed on an evolving surface
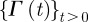 :
:
| 2.9 |
where 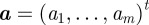 ,
, 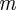 is the number of chemical species involved,
is the number of chemical species involved, 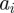 denotes the density of the ith chemical species,
denotes the density of the ith chemical species, 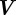 is the material velocity of the surface (cf. (2.3)),
is the material velocity of the surface (cf. (2.3)),
| 2.10 |
is the material derivative with respect to the velocity 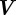 ,
, 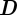 is a diagonal matrix of positive diffusion coefficients and
is a diagonal matrix of positive diffusion coefficients and 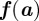 is the reaction. For details of the derivation we refer, for example, to Dziuk & Elliott [10] and Barreira et al. [21]. We assume in the following that the evolving hypersurface is closed so that no boundary condition is required. For the initial condition, we write
is the reaction. For details of the derivation we refer, for example, to Dziuk & Elliott [10] and Barreira et al. [21]. We assume in the following that the evolving hypersurface is closed so that no boundary condition is required. For the initial condition, we write
| 2.11 |
3. Discretization
Here, we describe the numerical methods we shall employ for the approximation of models for cell motility of the form described in §2. We keep the exposition relatively non-technical referring the interested reader to the electronic supplementary material for the technical details. We decouple the approximation of the surface evolution and the RDS by treating the RDS concentration explicitly in the surface update step. The numerical method is based on approximating the surface Γ(t) with a triangulated surface Γh(t), which stems from the method described in the seminal paper of Dziuk [28].
3.1. Definition (triangulated surface)
A triangulated surface Γh is a polygon or a polyhedron for d = 2 or 3, respectively, with linear edges for d = 2 and planar faces for d = 3, such that
| 3.1 |
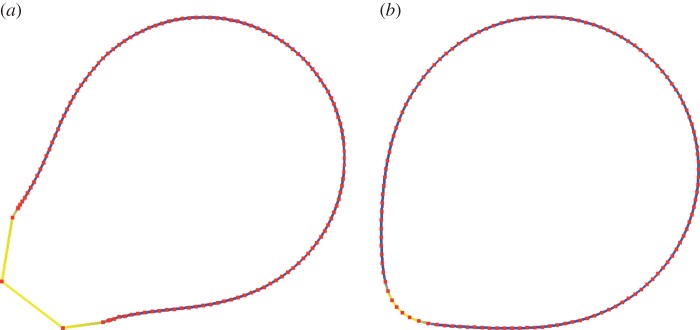
where  consists of a finite number of closed intervals (d = 2) or a finite number of closed non-degenerate triangles (d = 3). For the simulations on surfaces (d = 3), we make use of a quadratic triangulated surface. That is, a surface that consists of curvilinear triangles each of which is the image of a reference triangle under a quadratic map as illustrated in figure 1. We will use
consists of a finite number of closed intervals (d = 2) or a finite number of closed non-degenerate triangles (d = 3). For the simulations on surfaces (d = 3), we make use of a quadratic triangulated surface. That is, a surface that consists of curvilinear triangles each of which is the image of a reference triangle under a quadratic map as illustrated in figure 1. We will use  to denote both a triangulated surface and a quadratic triangulated surface interchangeably. So as no confusion arises, we stress that for the approximation of smooth curves, we consider triangulated (polygonal) discrete surfaces and for the approximation of smooth surfaces (d = 3) we consider quadratic triangulated surfaces. For details on triangulated surfaces, quadratic triangulated surfaces and approximation results, we refer to the earlier studies [10,29–32].
to denote both a triangulated surface and a quadratic triangulated surface interchangeably. So as no confusion arises, we stress that for the approximation of smooth curves, we consider triangulated (polygonal) discrete surfaces and for the approximation of smooth surfaces (d = 3) we consider quadratic triangulated surfaces. For details on triangulated surfaces, quadratic triangulated surfaces and approximation results, we refer to the earlier studies [10,29–32].
Figure 1.
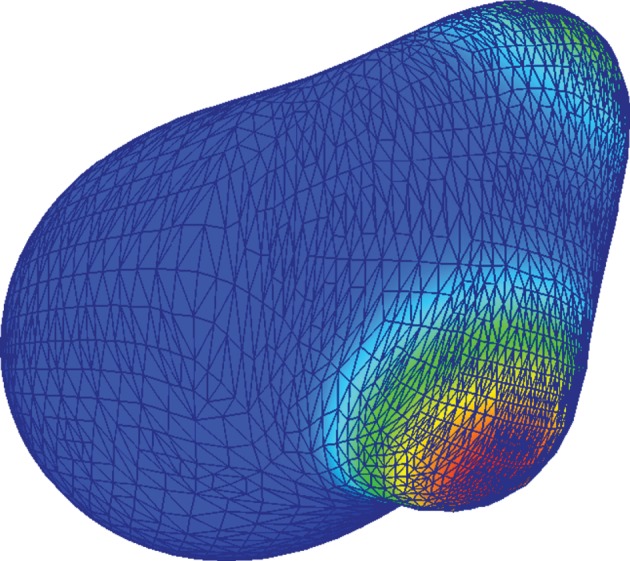
Snapshots of the triangulation during a simulation of cell motility using a quadratic triangulated approximation to a surface. (Online version in colour.)
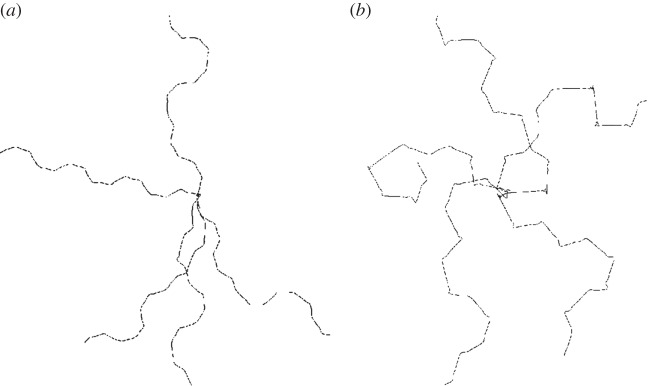
The evolution law (2.8) is discretized using a surface finite element described in detail in the electronic supplementary material. The method is based on the parametric finite-element methods for fourth-order geometric evolution equations derived in Dziuk [33] and Barrett et al. [11]. Under the proposed method, the movement of the nodes of the triangulation satisfies the evolution law in the normal direction and includes a tangential velocity (that leaves the evolution law unchanged), which gives highly desirable mesh-properties in practice. Figure 2 illustrates an example of the robustness of the proposed scheme in the approximation of large deformations over schemes where nodes are moved solely in the normal direction.
Figure 2.
Snapshots of the mesh during two simulations of cell motility using a scheme that moves the nodes only in the normal direction and the scheme that includes a tangential velocity. The large deformations result in meshes that are not suitable for computation with the former approach necessitating a remeshing step, while with the latter approach the mesh quality remains good throughout. (a) Movement in normal direction only; (b) tangential redistribution scheme. (Online version in colour.)
To approximate the RDS posed on the evolving surface, we employ a surface finite-element method based on the evolving surface finite-element method proposed by Dziuk & Elliot [10], where we account for the tangential velocity induced by the surface update scheme.
We also describe, in the electronic supplementary material, a framework for the inclusion of stochastic external signals into the model. In particular for the subsequent simulations of chemotaxis, we include a signal that is modelled by independent stochastic differential equations posed in each element (interval or triangle) which we approximate using the Euler–Maruyama method.
3.2. Software
The numerical methods were implemented using the adaptive finite-element toolbox Alberta [34] and the linear systems were solved using UMFPack [35], a direct solver for sparse linear systems.
4. Modelling pseudopod-driven chemotaxis
We investigate a model for eukaryotic chemotaxis originally proposed by Meinhardt [36]. The original model was posed at the discrete level and consisted of a three species RDS with a spatially varying local activator, a spatially varying local inhibitor, and a spatially constant global inhibitor. Meinhardt proposed that such a model could account for the polarization of chemotactic cells and the subsequent relocation and splitting of activator peaks in response to changing external signals. He did not, however, consider the mechanical aspects of the evolution of the cell membrane. Neilson et al. [7] investigated a continuous form of the model where the three species were all spatially dependent, approximating the model equations with a surface finite-element method for the RDS approximation and a level set method for the surface update. They have conducted detailed comparisons of their simulations using a level set method with experiments [37], as well as some preliminary investigations into robust computational methods, specifically short-time simulations using a surface finite-element method [16]. All their modelling and simulation was conducted in two dimensions, the model we consider extends the previous work by increasing the dimension, accounting for the bending energy and modelling obstacles.
In the original model posed by Meinhardt, the spatial independence of the global inhibitor is used in the derivation of the model. Since the global inhibitor is spatially constant, its concentration can be obtained by averaging, i.e. the use of a non-local term (the mean value of the local activator). We, therefore, consider the following transformation of Meinhardt's model from the spatially discrete fixed surface setting to a continuous evolving surface:
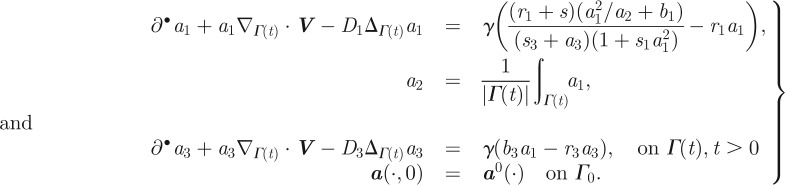 |
4.1 |
Here the ri, si and bi's are material parameters, γ is a scaling parameter that governs the overall timescale of the reaction rate and the term s(x,t) models both the underlying noise from the external media and a noisy chemotactic signal. The term involving the chemotactic signal feeds multiplicatively into the autocatalytic term in the activator equation. Weak signals are amplified owing to autocatalysis, thus the model provides a mechanism for gradient sensing of small chemotactic gradients. The RDS is coupled to the surface evolution law solely through the activator concentration a1, which promotes protrusion of the cell membrane. Adopting the notation of §2, the contribution of the RDS species to the evolution law (cf. (2.2)) reads
| 4.2 |
In fact, the activator and inhibitor are in phase and this is one potential drawback of the Meinhardt model as it presents no obvious mechanism for coupling surface concentrations to retraction of the cell membrane. One can show that the model (4.1) is equivalent to the model considered by Neilson et al. if the diffusivity of the global inhibitor in the Neilson model is sufficiently large relatively to the diffusivity of the other two species [38].
We model the stochastic term s(x,t) in equation (4.1) as the sum of an underlying noise term 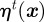 owing to, say, heterogeneity in the medium and a term
owing to, say, heterogeneity in the medium and a term 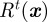 that models the cells' sensing of the chemotactic signal, i.e.
that models the cells' sensing of the chemotactic signal, i.e. 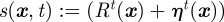 . The underlying noise term (present in all the simulations) satisfies the following Ornstein–Uhlenbeck (mean-reversion) stochastic process:
. The underlying noise term (present in all the simulations) satisfies the following Ornstein–Uhlenbeck (mean-reversion) stochastic process:
| 4.3 |
where Wt denotes the Wiener process. We discretize in space by assuming 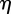 is constant over each finite element, and the Euler Maruyama method is used to approximate the solution numerically (for details see the electronic supplementary method).
is constant over each finite element, and the Euler Maruyama method is used to approximate the solution numerically (for details see the electronic supplementary method).
4.1. Results
For the results on curves, we took the unit circle as the initial steady state and used the reaction kinetic parameter values given in table 1. The parameter values are those Meinhardt used in his original study rescaled such that the diffusivity of the activator is 1.0, the only parameter we have tuned is the saturation of the activator concentration s1, which is smaller to account for dilution in the activator concentration owing to domain growth. For all the simulations on curves, we used the same equidistributed initial mesh with 1024 d.f. (further refinement did not change the solutions qualitatively) and a timestep of 10−5. The central processing unit (CPU) times of all the calculations on curves is in the order of minutes, for example, the two simulations reported in figure 3 had CPU times of just over 2000 s.
Table 1.
Parameter values for numerical experiments of the movement of two-dimensional cells with the Meinhardt kinetics, equation (4.1).
| reaction kinetic parameters | ||||||||
|---|---|---|---|---|---|---|---|---|
| D1 | D3 | γ | r1 | r3 | S1 | S3 | B1 | B3 |
| 1.0 | 7.0 | 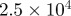 |
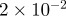 |
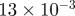 |
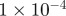 |
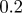 |
0.1 | 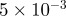 |
Figure 3.
Centroid trajectories of five cells migrating in the absence of a chemoattractant under different geometric evolution laws. In both cases, we see motion in a straight line for short times punctuated by sharp changes in direction corresponding to pseudopod splitting/decay. (a) Spider plots of cell centroid trajectories over the interval [0, 1.], under conserved surface tension evolution; for parameter values see table 1 and the text. (b) Spider plots of cell centroid trajectories over the interval [0, 1.], under a combination of surface tension and elastic evolution; for parameter values see table 1 and the text.
4.1.1. Random migration
Figure 3 shows the centroid trajectories of five cells migrating under two different geometric evolutions, surface tension evolution (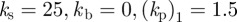 ) and combined surface tension-elastic evolution
) and combined surface tension-elastic evolution 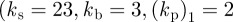 ), with no chemotactic signal and different realizations of the noise term. In both cases, the cells initially polarize and then migrate, with roughly two pseudopods at the front of the cell at any given time. The cells change direction via splitting and decay of pseudopods with one pseudopod splitting and persisting while the other decays. This leads to the characteristic linear motion over short times interspersed with sharp changes in direction corresponding to a pseudopod splitting/decay event, similar to that observed by Neilson et al. [7]. Under surface tension evolution, the cells maintain a characteristic shape (two pseudopods at the front of the cell with a valley between them), while the introduction of bending rigidity generates a greater variety of cell shapes and the cell no longer has a characteristic shape with varied banana-like shapes evident (the cell shapes resemble those in the simulations presented in figure 5). From the simulations, we observe that the diluting effect of protrusion plays an important role in destabilizing activator peaks and pseudopod splitting. By this, we refer to the fact that the formation of an activator peak results in protrusion of the cell membrane, which in turn leads locally to an decrease in activator density (as protrusion may be viewed as volumetric expansion). As the local maxima corresponding to the activator peak is reduced most at the tip of the peak, where protrusion is largest, this has the effect of increasing the propensity of activator peaks and hence pseudopods to split.
), with no chemotactic signal and different realizations of the noise term. In both cases, the cells initially polarize and then migrate, with roughly two pseudopods at the front of the cell at any given time. The cells change direction via splitting and decay of pseudopods with one pseudopod splitting and persisting while the other decays. This leads to the characteristic linear motion over short times interspersed with sharp changes in direction corresponding to a pseudopod splitting/decay event, similar to that observed by Neilson et al. [7]. Under surface tension evolution, the cells maintain a characteristic shape (two pseudopods at the front of the cell with a valley between them), while the introduction of bending rigidity generates a greater variety of cell shapes and the cell no longer has a characteristic shape with varied banana-like shapes evident (the cell shapes resemble those in the simulations presented in figure 5). From the simulations, we observe that the diluting effect of protrusion plays an important role in destabilizing activator peaks and pseudopod splitting. By this, we refer to the fact that the formation of an activator peak results in protrusion of the cell membrane, which in turn leads locally to an decrease in activator density (as protrusion may be viewed as volumetric expansion). As the local maxima corresponding to the activator peak is reduced most at the tip of the peak, where protrusion is largest, this has the effect of increasing the propensity of activator peaks and hence pseudopods to split.
Figure 5.
Response to a changing chemotactic signal. Initially, there is no signal with arrows indicating the time at which a signal is introduced and the signal direction. Note the two figures are not on the same scale and the cells have the same enclosed volume. (a) Chemotactic motion of a cell under conserved surface tension evolution, for parameter values, see table 1 and text. Cell outlines shown every 0.1 units of computational time over the interval [0, 1.8]. (b) Chemotactic motion of a cell under a combination of surface tension and elastic evolution with volume conservation, for parameter values, see table 1 and text. Cell outlines shown every 0.075 units of computational time over the interval [0, 1.725]. (Online version in colour.)
We may proceed to estimate some of the parameter values using available experimental data, which is readily available for dictyostelium cells [2]. The typical radius of a cell cross section is 4 µm, which sets the length scale for the computations. The maximum actin polymerization velocity (which is related to the non-dimensional parameter 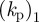 ) is approximately 0.1 m s−1, thus the value of
) is approximately 0.1 m s−1, thus the value of 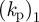 together with the maximum density of a1 in the simulations (approx. 30) sets the timescale for the simulations. Typical values of the surface tension are 10 pN µm−1, assuming a cell height of 0.1 µm, this sets a kinetic scale for the simulation. The remaining physically elevant parameters may thus be estimated and are given in table 2. Note that the timescale we refer to in table 2 corresponds to one unit of computational time. The length of the simulations in figure 3 is one computational time unit or 30 min in actual time and corresponds to roughly 20 pseudopod lifetimes (each change in direction in figure 3 represents a pseudopod splitting/decay event), thus the timescale of an individual pseudopod is around 90 s.
together with the maximum density of a1 in the simulations (approx. 30) sets the timescale for the simulations. Typical values of the surface tension are 10 pN µm−1, assuming a cell height of 0.1 µm, this sets a kinetic scale for the simulation. The remaining physically elevant parameters may thus be estimated and are given in table 2. Note that the timescale we refer to in table 2 corresponds to one unit of computational time. The length of the simulations in figure 3 is one computational time unit or 30 min in actual time and corresponds to roughly 20 pseudopod lifetimes (each change in direction in figure 3 represents a pseudopod splitting/decay event), thus the timescale of an individual pseudopod is around 90 s.
Table 2.
Physically relevant parameter values for simulations on curves.
| parameter | value | physical interpretation |
|---|---|---|
| r0 | 1 | initial radius of cell sets a length scale of 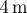
|
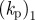 |
1.5–2 | coefficient of protrusive forcing term implies a timescale of 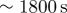
|
| D1 | 1.0 | implies a diffusivity of 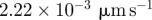
|
| D3 | 7.0 | implies a diffusivity of 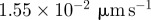
|
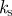 |
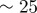 |
assuming a surface tension of 1 pN, this sets a kinetic scale for the simulations |
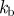 |
3 | implies a bending rigidity of approximately 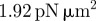
|
We note that other choices of the material parameters, specifically weaker surface tension, gives cells with more elongated shapes, larger protrusions, and cell bodies that appear less rounded.
4.1.2. Migration in the presence of a chemoattractant
We now include a chemoattractant in the model. We use the stochastic receptor model proposed by Neilson et al. [7] to model the noisy chemotactic signalling. For completeness, we state the essential details. At time 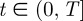 , they model the cells sensing of the chemotactic signal Rt with an Ornstein–Uhlenbeck stochastic process of the form:
, they model the cells sensing of the chemotactic signal Rt with an Ornstein–Uhlenbeck stochastic process of the form:
| 4.4 |
where Wt denotes the Wiener process, 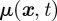 models the strength of the chemotactic signal,
models the strength of the chemotactic signal, 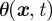 the rate of reversion to the mean μ, and
the rate of reversion to the mean μ, and 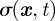 the variance. The mean μ is local and prescribed by the model, while the rate of reversion to the mean and variance is local too as they are chosen such that θ = 1/(1 − μ) and σ = cμ1/2 (for details, see Neilson et al. [7, §6.2.2]). To compare with Neilson et al. [7, §6.2.5], we model the signal such that if a chemoattractant is present, the mean μ varies from the base signal strength (at the back of the cell) of 0.5 to 0.5 + ρ at the front, where ρ > 0 represents the signal strength. For a given signal direction
the variance. The mean μ is local and prescribed by the model, while the rate of reversion to the mean and variance is local too as they are chosen such that θ = 1/(1 − μ) and σ = cμ1/2 (for details, see Neilson et al. [7, §6.2.2]). To compare with Neilson et al. [7, §6.2.5], we model the signal such that if a chemoattractant is present, the mean μ varies from the base signal strength (at the back of the cell) of 0.5 to 0.5 + ρ at the front, where ρ > 0 represents the signal strength. For a given signal direction 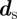 , the position of the rear of the cell
, the position of the rear of the cell 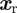 is such that
is such that 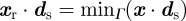 . We have also conducted experiments where
. We have also conducted experiments where 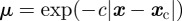 , where
, where 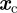 denotes the location of a static point source of chemoattractant and observe similar results. We discretize (in space) by assuming the mean μ is constant over each finite element, and hence θ and σ are also constant on each finite element.
denotes the location of a static point source of chemoattractant and observe similar results. We discretize (in space) by assuming the mean μ is constant over each finite element, and hence θ and σ are also constant on each finite element.
Figure 4 shows the trajectories of the centroids of five cells migrating leftwards in a linear chemotactic gradient of varying strength under conserved surface tension evolution (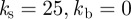 ,
, 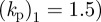 . The results are similar under the other geometric evolution considered (
. The results are similar under the other geometric evolution considered (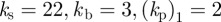 ) and are not illustrated. The migration of the cells with only the base signal and the signal strength set to zero is reported in figure 5a. We observe no clear directional preference similar to the migration observed in the absence of a chemoattractant. As the signal strength is increased (at a signal strength of 0.04 around 8% of the base signal), the cells start to exhibit a clear directional preference and successfully navigate up the chemotactic gradient. In table 3, we report on chemotaxis measures of 100 cells migrating under the six different signal strengths shown in figure 4. We state the average value over the 100 simulations (for each signal strength) of the following quantities, all evaluated at t = 0.5.
) and are not illustrated. The migration of the cells with only the base signal and the signal strength set to zero is reported in figure 5a. We observe no clear directional preference similar to the migration observed in the absence of a chemoattractant. As the signal strength is increased (at a signal strength of 0.04 around 8% of the base signal), the cells start to exhibit a clear directional preference and successfully navigate up the chemotactic gradient. In table 3, we report on chemotaxis measures of 100 cells migrating under the six different signal strengths shown in figure 4. We state the average value over the 100 simulations (for each signal strength) of the following quantities, all evaluated at t = 0.5.
— The chemotactic index (CI), defined as the cosine of the angle between a line connecting the present position of the cell centroid to the starting point and a line directly up the chemotactic gradient [39].
— The persistence length (PL) of the centroid trajectory in the x- and y-directions. The persistence length is taken to be the displacement in the chosen direction divided by the total length of the trajectory of the cell centroid [40].
— The squared displacement of the cell centroid from its initial position.
— The speed of the cell.
Figure 4.
Centroid trajectories of five cells migrating leftwards in the presence of a linear chemoattractant gradient under conserved surface tension evolution with varying signal strength (ρ).
Table 3.
Mean and s.d. (in parentheses) of chemotaxis measures at 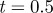 for 100 cells migrating as in figure 4.
for 100 cells migrating as in figure 4.
| signal strength | CI | PL (x) | PL (y) |
|---|---|---|---|
| 0 | n.a. | 0.4336 (0.2346) | 0.4601 (0.2442) |
| 0.02 | 0.7196 (0.2877) | 0.4938 (0.2188) | 0.3418 (0.1999) |
| 0.04 | 0.9423 (0.0742) | 0.6968 (0.1177) | 0.2005 (0.1163) |
| 0.06 | 0.9888 (0.0133) | 0.8510 (0.0511) | 0.1088 (0.0685) |
| 0.08 | 0.9860 (0.0120) | 0.8490 (0.0350) | 0.1288 (0.0676) |
| 0.10 | 0.9898 (0.0141) | 0.8489 (0.0272) | 0.0987 (0.0734) |
| signal strength |
squared displacement (arb. units) |
speed (arb. units) |
speed (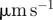 ) ) |
| 0 | 37.052 (14.5937) | 20.056 (1.9781) | 0.0445 |
| 0.02 | 34.430 (13.5045) | 19.799 (2.8624) | 0.0440 |
| 0.04 | 44.655 (11.9692) | 20.077 (2.8633) | 0.0446 |
| 0.06 | 66.011 (10.3248) | 21.156 (1.9327) | 0.0470 |
| 0.08 | 66.491 (6.8996) | 21.524 (1.9714) | 0.0478 |
| 0.10 | 67.387 (8.8005) | 21.435 (2.1560) | 0.0476 |
The data suggest that as the strength of the chemotactic signal is increased, the cells exhibit greater propensity for persistent migration up the chemical gradient with chemotactic indices similar to those observed experimentally in the case of Dictyostelium cells [39; 0.71–0.94]. The persistence length in the x-direction (up the chemotactic gradient) also increases with the signal strength, while the persistence length in the y-direction is reduced. We also note increasing the signal strength leads to larger displacements. The results suggest that for values of ρ ≥ 0.06, the cell is able to migrate successfully up the chemical gradient with all the reported statistics converging to similar means for further increases in the signal strength. The (physical) cell speeds are similar to those observed in migrating leukocytes [41, table 1] and Dictyostelium cells [39] (in both cases reported in µm min−1).
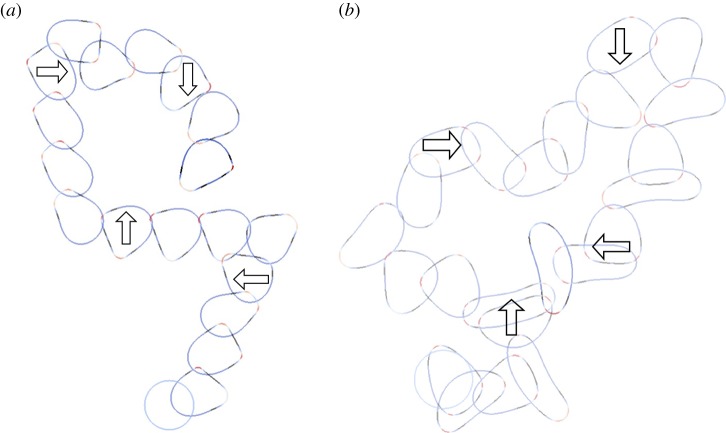
We now investigate the ability of this model to capture the ability of a cell to respond to a changing chemotactic signal. We use the same stochastic receptor model for the chemotactic signalling but now we change the direction of the signal at various stages in the evolution. Figure 5a,b shows snapshots of the cells shaded by activator concentration under the two different geometric evolutions (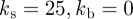 ,
,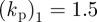 and
and 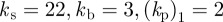 ) in response to a changing chemotactic signal. Initially, we include only the base signal with noise, i.e. the signal strength is set to zero. At the times in the evolution at which the arrows appear in the figure, we change the direction of the signal, with signal strength ρ = 0.1, to the direction indicated by the arrows. We see that under both geometric evolutions, the cell successfully responds to the changing signal, exhibiting a clear directional preference for movement in the direction of higher chemoattractant concentration. As a final example of response to a changing signal, we consider the case where the signal direction is changed by 180°. The results of such a simulation are shown in figure 6. We observe the cell successfully responds to the change in signal direction and does so via turning gradually through 180°. This corresponds to the so-called ‘hops’ (consecutive right/right or left/left splitting of pseudopods) that are an important mechanism for the reorientation of Dictyostelium cells moving in a direction more than 90° off the chemotactic gradient [42, fig. 4]. Under this model, we have however, thusfar, not observed the formation of de novo pseudopods towards the direction of increasing chemoattractant, which are another significant mechanism for major directional corrections [42].
) in response to a changing chemotactic signal. Initially, we include only the base signal with noise, i.e. the signal strength is set to zero. At the times in the evolution at which the arrows appear in the figure, we change the direction of the signal, with signal strength ρ = 0.1, to the direction indicated by the arrows. We see that under both geometric evolutions, the cell successfully responds to the changing signal, exhibiting a clear directional preference for movement in the direction of higher chemoattractant concentration. As a final example of response to a changing signal, we consider the case where the signal direction is changed by 180°. The results of such a simulation are shown in figure 6. We observe the cell successfully responds to the change in signal direction and does so via turning gradually through 180°. This corresponds to the so-called ‘hops’ (consecutive right/right or left/left splitting of pseudopods) that are an important mechanism for the reorientation of Dictyostelium cells moving in a direction more than 90° off the chemotactic gradient [42, fig. 4]. Under this model, we have however, thusfar, not observed the formation of de novo pseudopods towards the direction of increasing chemoattractant, which are another significant mechanism for major directional corrections [42].
Figure 6.
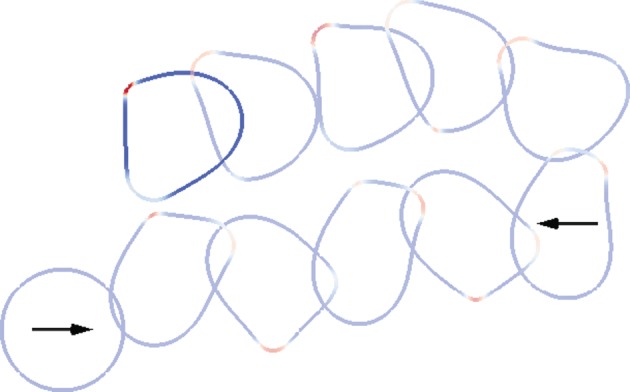
Response to a changing chemotactic signal. In this example, the direction of the chemotactic signal is changed by 180°. We observe that the cell turns through 180° and successfully responds to the changing signal, migrating up the new chemotactic gradient. Cell outlines shown every 0.1 units of computational time over the interval [0, 1]. (Online version in colour.)
4.1.3. Migration in the presence of obstacles
We now include an external force in the evolution law, which arises from a model for the migration of cells through a field of obstacles. We model the obstacle particles as rigid spherical bodies. The obstacle–cell interaction is described by a phenomenological repulsive force that points in the direction normal to the cell membrane with no tangential component. Unlike the models proposed by Hecht et al. [43] and Grima [44], the obstacles and the cell both move owing to mechanical interactions.
Let  denote the number of obstacles with centres
denote the number of obstacles with centres  and radii
and radii  . For the force acting on a point
. For the force acting on a point 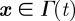 on the cell boundary owing to the interaction with obstacle i we postulate
on the cell boundary owing to the interaction with obstacle i we postulate
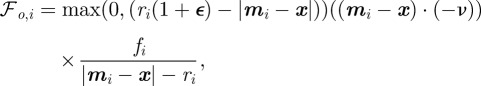 |
4.5 |
where 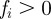 is a material coefficient and
is a material coefficient and 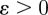 is a thickness parameter: the force is zero if the distance between the cell membrane and the obstacle boundary is bigger than
is a thickness parameter: the force is zero if the distance between the cell membrane and the obstacle boundary is bigger than 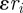 . The force becomes infinite as this distance approaches zero and then dominates any other forces on the cell membrane, thus preventing intersection of the cell and the obstacle. The external force acting on the cell boundary is given by:
. The force becomes infinite as this distance approaches zero and then dominates any other forces on the cell membrane, thus preventing intersection of the cell and the obstacle. The external force acting on the cell boundary is given by:
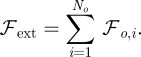 |
4.6 |
For the obstacle particles, we postulate a viscous motion law, too, where the reaction forces from the cell boundary 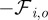 and obstacle–obstacle interactions are taken into account. We postulate
and obstacle–obstacle interactions are taken into account. We postulate
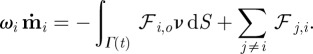 |
4.7 |
Here, the 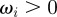 are positive kinetic coefficients related to the mass of the particle, the first term on the right-hand side modelling the cell–obstacle interaction is
are positive kinetic coefficients related to the mass of the particle, the first term on the right-hand side modelling the cell–obstacle interaction is
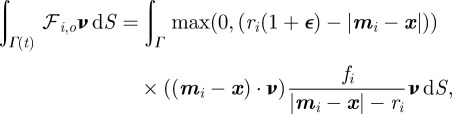 |
4.8 |
and 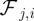 is the force from particle j exerted on particle i for which we postulate
is the force from particle j exerted on particle i for which we postulate
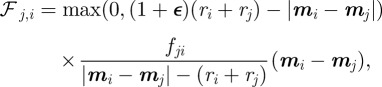 |
4.9 |
where the 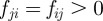 are material coefficients. Note that in the absence of the cell, the initial location of the obstacles is such that the sum of the forces
are material coefficients. Note that in the absence of the cell, the initial location of the obstacles is such that the sum of the forces 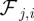 yields zero so that the particles do not move. Moreover, we have the following balance of forces exerted by the cell on the obstacles and forces on the cell membrane owing to the obstacles
yields zero so that the particles do not move. Moreover, we have the following balance of forces exerted by the cell on the obstacles and forces on the cell membrane owing to the obstacles
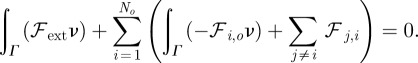 |
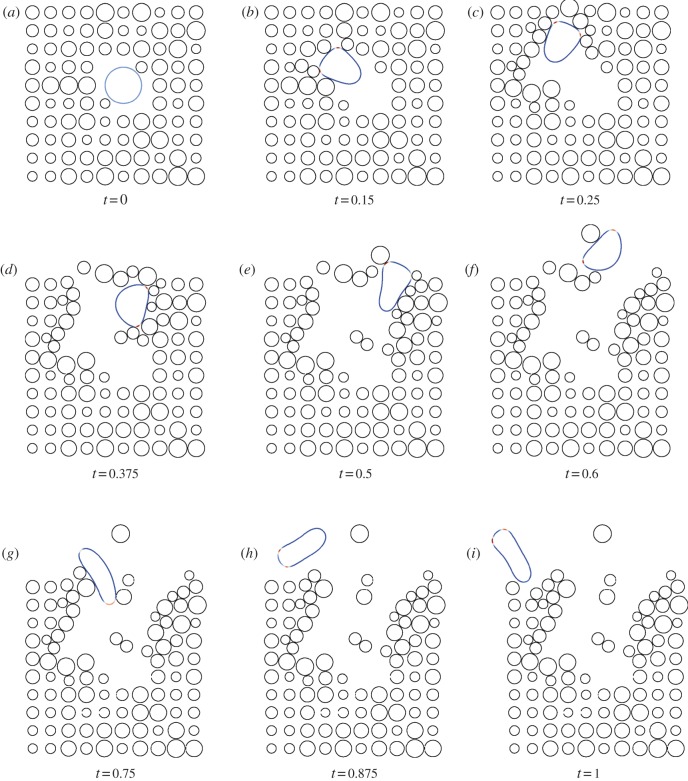
Figures 7 and 8 show a series of snapshots of cell migration through a field of obstacles, with parameter values as in table 1 and the two previously considered geometric evolutions. Our numerical experience suggests that under the simple model of cell obstacle interactions we have employed, the increase in computational time, even with a large number of obstacles, from the case of no obstacles is negligible. We include the forcing terms in the evolution law for the cell membrane and the obstacle centres given by equations (4.6)–(4.9), with parameter values 
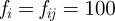 for all i, j and
for all i, j and 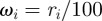 . We observe that the cell successfully migrates through the field of obstacles maintaining the characteristic shape as it deflects the obstacles. Our numerical experiments suggest that this behaviour is sensitive to the parameter values chosen in the repulsive potential (4.5). In particular, if we set the kinetic coefficient related to the mass of the obstacles
. We observe that the cell successfully migrates through the field of obstacles maintaining the characteristic shape as it deflects the obstacles. Our numerical experiments suggest that this behaviour is sensitive to the parameter values chosen in the repulsive potential (4.5). In particular, if we set the kinetic coefficient related to the mass of the obstacles 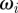 cf. equation (4.8) to be comparable in magnitude to the kinetic coefficient related to the mass of the cell (1 by assumption), which means the obstacles inhibit more strongly the protrusion of pseudopods, then pseudopod splitting no longer occurs and the cell exhibits persistent motion in the direction of an obstacle (not reported).
cf. equation (4.8) to be comparable in magnitude to the kinetic coefficient related to the mass of the cell (1 by assumption), which means the obstacles inhibit more strongly the protrusion of pseudopods, then pseudopod splitting no longer occurs and the cell exhibits persistent motion in the direction of an obstacle (not reported).
Figure 7.
Undirected migration (i.e. migration in the absence of a chemoattractant) in the presence of obstacles of a cell under conserved surface tension evolution; for parameter values see table 1 and text. (Online version in colour.)
Figure 8.
Undirected migration in the presence of obstacles of a cell under a combination of conserved surface tension and elastic evolution; for parameter values see table 1 and text. (Online version in colour.)
4.1.4. Migration of cells in three space dimensions
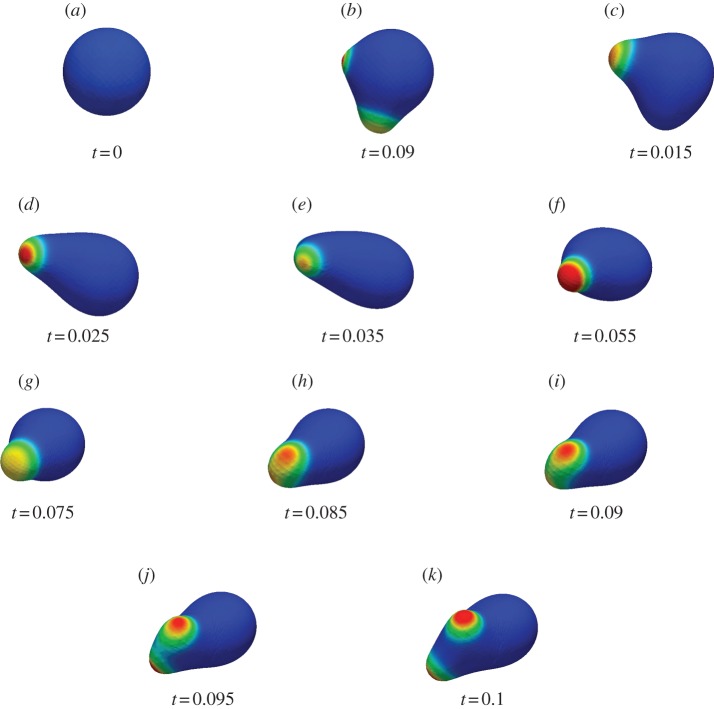
We now present results for the motion of three-dimensional cells in the absence of a chemoattractant. We took the unit sphere as the initial steady state and used the reaction kinetic parameter values given in table 4. We selected a timestep of 10−5 and used the adaptive strategy described in the electronic supplementary material with parameters NH = 0.5, Nh = 0.75, MH = 0.25 and Mh = 0.5. We considered an evolution law of the form (2.8) with parameters 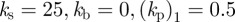 . As in table 2, we give a physical interpretation of the parameter values in table 5, assuming the radius of the spherical cell at rest is 1.17 µm. The model for the random signalling was the same as the model used in the two-dimensional case. Snapshots of the cell surface shaded by activator concentration are reported in figure 9. We see qualitatively similar behaviour to the case of curves with protrusion of activator peaks leading to pseudopod formation and the rest of the cell retracting behind the pseudopods. We also observe pseudopod splitting as the cell changes direction via biased generation and retraction of existing pseudopods. The simulation is considerably more challenging than the curve case considered previously and the total CPU time of the simulation was just over 27 h.
. As in table 2, we give a physical interpretation of the parameter values in table 5, assuming the radius of the spherical cell at rest is 1.17 µm. The model for the random signalling was the same as the model used in the two-dimensional case. Snapshots of the cell surface shaded by activator concentration are reported in figure 9. We see qualitatively similar behaviour to the case of curves with protrusion of activator peaks leading to pseudopod formation and the rest of the cell retracting behind the pseudopods. We also observe pseudopod splitting as the cell changes direction via biased generation and retraction of existing pseudopods. The simulation is considerably more challenging than the curve case considered previously and the total CPU time of the simulation was just over 27 h.
Table 4.
Parameter values for numerical experiments of the movement of three-dimensional cells with the Meinhardt kinetics (4.1).
| reaction kinetic parameters | ||||||||
|---|---|---|---|---|---|---|---|---|
| D1 | D3 | γ | r1 | r3 | s1 | s3 | b1 | b3 |
| 10 | 70 | 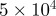 |
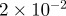 |
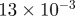 |
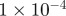 |
0.2 | 0.1 | 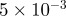 |
Table 5.
Physically relevant parameter values for simulation of migration of a cell in three dimensions.
| parameter | value | physical interpretation |
|---|---|---|
| r0 | 1 | initial radius of cell sets a length scale of 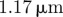
|
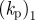 |
0.5 | coefficient of protrusive forcing term implies a timescale of approximately 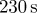
|
| D1 | 10 | implies a diffusivity of 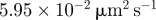
|
| D2 | 70 | implies a diffusivity of 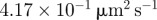
|
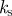 |
 |
assuming a surface tension of 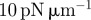 , sets kinetic scale , sets kinetic scale |
Figure 9.
Migration in the absence of a chemoattractant of a three-dimensional cell under conserved surface tension evolution; for parameter values see table 4 and text. (Online version in colour.)
5. Modelling the persistent motion of keratocytes
We present a model based on the general modelling framework described in §2 that seeks to capture the persistent motion of fish keratocytes. The cells deform rapidly into a temporally persistent shape and once in this shape move at a constant speed without changes in direction. Keren et al. [18] conducted an analysis of the shapes taken by moving keratocyte cells and propose a simple phenomenological model to account for the observed movement and cell shapes. Their results suggest that the steady-state shapes of the cell are broadly described by two modes and that cell shape, specifically the aspect ratio (length/width), is strongly correlated with the speed of motion. They also examined the actin distribution within the cell. Branched actin filaments promoting protrusion are concentrated at the fast-moving front and retraction promoting actin bundles are concentrated at the rear. The steady state appears stable to perturbations and if movement of the cell is disrupted, the cell rapidly regains its previous shape and speed of movement, usually moving in a new direction.
The observed behaviour of spontaneous polarization and subsequent development of a steady state stable to perturbations suggests a Turing type mechanism coupled to a surface evolution law could accurately capture the observed dynamics. Shao et al. [22] considered a membrane subject to surface tension, bending rigidity and forcing with volume conservation. The forcing strength was dependent on the concentrations of a two component RDS posed in the bulk of the cell. They present computational results, for two-dimensional cells with weak volume conservation (enforced via penalization), based on a phase-field method. Ziebert et al. [6] present a model for keratocyte movement, again based on a phase-field method, where they couple the surface evolution to a vector field that seeks to describe the polarization of the actin network. Studies suggest that branched filamentary actin and actin bundles are concentrated primarily near the cell membrane near areas of protrusion and retraction, respectively, while away from the cell surface the actin is in a remodelling phase between that of branched and bundled actin [18,45,46]. This suggests a surface model where the pattern formation process occurring on the cell membrane itself may be appropriate. We propose the activator-depleted substrate model [47]:
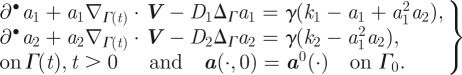 |
5.1 |
We first present results for curves, with material and RDS parameters given in table 6. We considered an initially circular cell with radius 1 centred at the origin. The initial condition for the RDS was taken as the linearly stable steady state 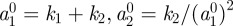 with a symmetry breaking perturbation of the form
with a symmetry breaking perturbation of the form 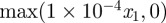 added to the initial condition of the a2 species. The specific form of the initial condition leads to cells that migrate only along the x-axis (we verified that the choice of other initial conditions only changed the direction of the movement). The hypothesis of Keren et al. [18] is that variability in the actin dynamics is the major factor governing the observed variations in shape and speed. To investigate this hypothesis, we propose that the a1 species in the RDS (5.1) corresponds to the density of retraction promoting actin bundles, while the a2 species corresponds to the density of protrusion promoting actin filaments, which is similar to the model considered in Shao et al. [22]. We can model variable actin dynamics by changing the constant k2 which can be interpreted as the growth rate of actin filaments. Increasing k2 leads to higher concentrations of a2 relative to a1, and thus should lead to faster moving cells with stronger forcing at the front.
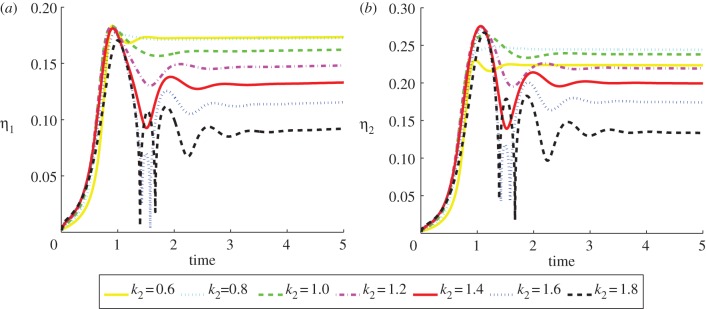
added to the initial condition of the a2 species. The specific form of the initial condition leads to cells that migrate only along the x-axis (we verified that the choice of other initial conditions only changed the direction of the movement). The hypothesis of Keren et al. [18] is that variability in the actin dynamics is the major factor governing the observed variations in shape and speed. To investigate this hypothesis, we propose that the a1 species in the RDS (5.1) corresponds to the density of retraction promoting actin bundles, while the a2 species corresponds to the density of protrusion promoting actin filaments, which is similar to the model considered in Shao et al. [22]. We can model variable actin dynamics by changing the constant k2 which can be interpreted as the growth rate of actin filaments. Increasing k2 leads to higher concentrations of a2 relative to a1, and thus should lead to faster moving cells with stronger forcing at the front.
Table 6.
Parameter values for numerical experiments of keratocyte movement with the RDS (5.1).
| reaction kinetic parameters |
surface evolution parameters |
||||||
|---|---|---|---|---|---|---|---|
| D1 | D2 | γ | k1 | 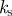 |
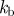 |
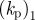 |
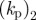 |
| 0.5 | 50 | 10 | 0.1 | 2 | 2 | −2 | 1 |
In all the simulations on curves, we used an initially equidistributed mesh with 1024 d.f. and a fixed timestep of 10−3. The CPU times were on the order of seconds with a typical simulation taking approximately 200 s. Figure 10a,b shows, for different values of k2, the initial position of the cells at time 0 and the cell positions and surface RDS concentrations at time 5 (by which time all the cells have reached a steady state with constant speed and time independent RDS concentrations). We see faster cell speeds and larger aspect ratios for increased values of k2, similar to the models where the RDS is posed in the bulk [22]. The shapes of the cells at steady state also resemble those observed experimentally [18]. In particular, we see the rounded ‘D’ shape in the right most cell corresponding to k2 = 0.6, and the much more elongated ‘canoe’ shape in the left most cell corresponding to k2 = 1.8. We report on the aspect ratio 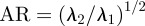 , as considered in Ziebert et al. [6], where for i = 1,2,
, as considered in Ziebert et al. [6], where for i = 1,2, 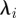 is as follows (the
is as follows (the 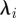 's are the eigenvalues of the diagonal 2 × 2 variance matrix of the cells centroid):
's are the eigenvalues of the diagonal 2 × 2 variance matrix of the cells centroid):
where 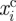 is the ith coordinate of the cells centroid. We also report on the deviation from reflection symmetry of the migrating cells as considered in Ziebert et al. [6]. This is measured by the following quantities (the non-zero components of the skewness tensor of the cells centroid scaled by a constant factor):
is the ith coordinate of the cells centroid. We also report on the deviation from reflection symmetry of the migrating cells as considered in Ziebert et al. [6]. This is measured by the following quantities (the non-zero components of the skewness tensor of the cells centroid scaled by a constant factor):
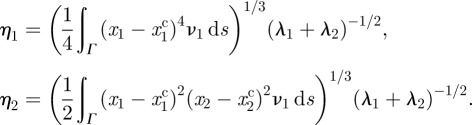 |
Figure 10.
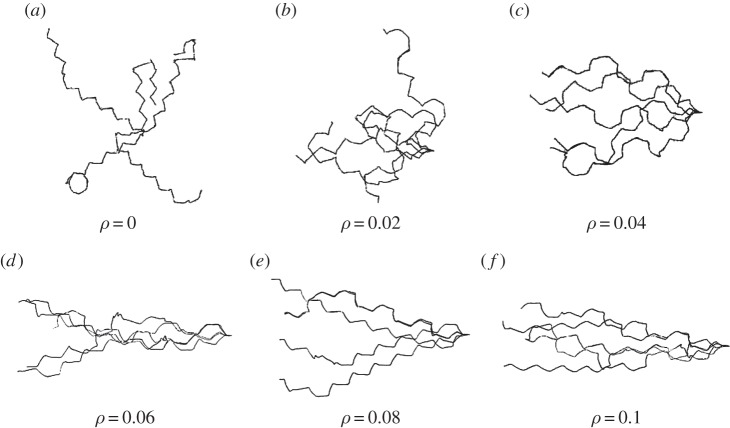
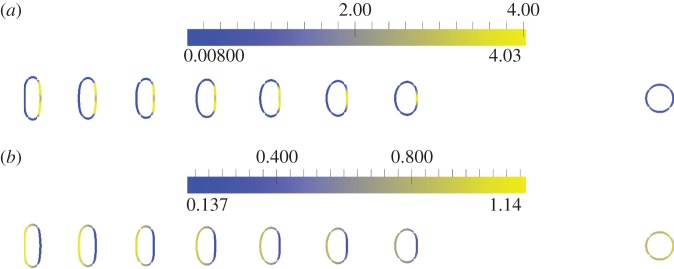
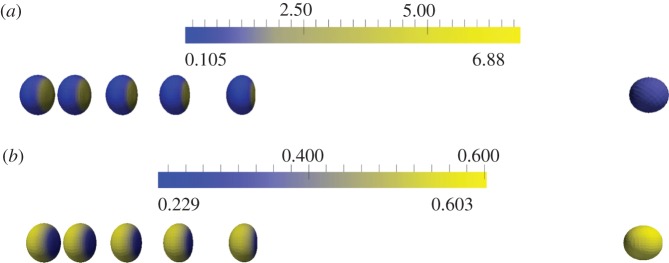
Initial position (at t = 0 right-hand cell) and persistent keratocyte-like migration of cells (at t = 5). The parameter k2 = 0.6, 0.8, 1.0, 1.2, 1.4, 1.6, 1.8 reading from right to left for the seven polarized (left hand) cells (cf. equation (5.1)) with the remaining parameters given in table 6. (a) Activator (a1) concentrations; (b) substrate (a2) concentrations. (Online version in colour.)
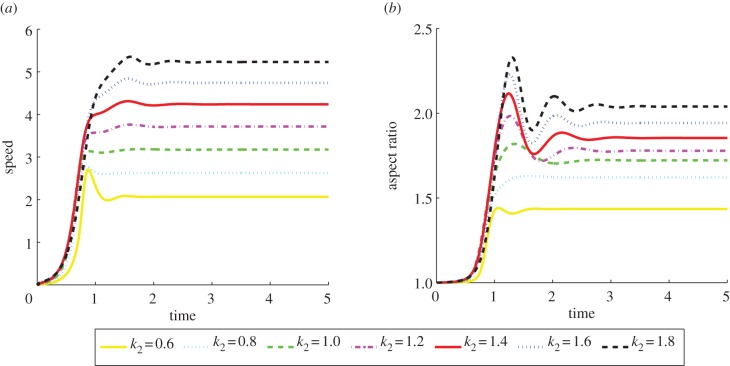
Figure 11 shows plots of the speed of the cell centroids and the aspect ratio of the cells both against time. We clearly see the positive relationship between aspect ratio and cell speed evident in the experimental studies. In physical units, the range of the speed at steady state of the cells shown in figure 10 is 0.178–0.445 µm s−1}. Both the speed and aspect ratios are similar to those observed in the experimental results reported in Keren et al. [18, fig. 4b]. Figure 12 shows values of the asymmetry measures against time. We see that the cells travelling at a slower speed exhibit larger deviations from reflection symmetry. This is in contrast to the results obtained under the model considered in Zibert et al. [6, fig. 4], where 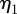 does increase as the speed of the cells decreases, but
does increase as the speed of the cells decreases, but 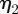 is positively correlated with speed. We note that for the first two simulations
is positively correlated with speed. We note that for the first two simulations 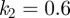 and
and 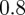 ,
, 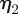 is negatively correlated with speed. As an ellipse satisfies
is negatively correlated with speed. As an ellipse satisfies 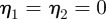 , it is not clear that the faster moving cells with larger aspect ratios, but more elliptical profiles, should exhibit larger deviations from reflection symmetry. We also observe that after a brief initial stage in which all the reported values oscillate, the values converge to a steady state as the cells travel in a persistent fashion with a fixed shape and a constant speed.
, it is not clear that the faster moving cells with larger aspect ratios, but more elliptical profiles, should exhibit larger deviations from reflection symmetry. We also observe that after a brief initial stage in which all the reported values oscillate, the values converge to a steady state as the cells travel in a persistent fashion with a fixed shape and a constant speed.
Figure 11.
The speed of the cell centroid and aspect ratio both versus time of the cells shown in figure 10. We observe a positive relationship between aspect ratio and speed. (Online version in colour.)
Figure 12.
Asymmetry measures versus time of the cells shown in figure 10. We observe larger deviations from reflection symmetry in the cells travelling at slower speeds. As the cells attain persistent shapes, the values converge to a steady state. (Online version in colour.)
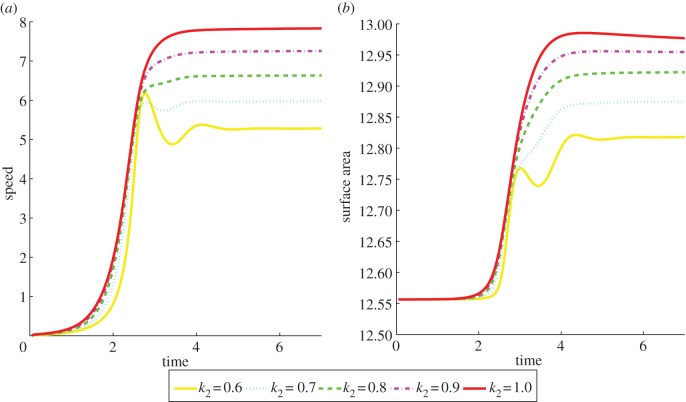
We also report on simulations of three-dimensional keratocyte motion. We took the unit sphere as the initial cell shape, the same initial conditions for the RDS concentrations as in the curve case, a fixed timestep of 10−4 and remaining parameter values for both the surface evolution and adaptive strategy as given in table 8. The CPU times were on the order of minutes with a typical simulation taking approximately 2000 s. Proceeding as in §4, we give physical interpretations of the parameter values for curves and surfaces in tables 7 and 9, respectively. Figure 13a,b shows a similar experiment to the one carried out for curves now on surfaces, specifically we report for different values of k2, the initial position of the cells at time 0 and the cell positions and surface RDS concentrations at time 5 (by which time all the cells have reached a steady state with constant speed and time-independent RDS concentrations). The gross behaviour is the same as the curve case, in that as the parameter k2 is increased from 0.6 to 1, the cells move faster at steady state and appear more elongated. Figure 14 shows plots of the speed of the cell centroids and the surface area of the cells both against time. We plot the surface area as it is proportional to the two, roughly equal, aspect ratios in the (x, y)-and (x, z)-directions. We observe the same positive relationship as in the curve case, with both surface area and speed converging to steady states. We have also verified that the aspect ratios converge to steady states with the aspect ratio in the (y, z)-direction approaching 1. Note for larger values of k2, the cells developed a self-intersection which is inadmissible under our modelling as it would correspond to a change in topology in the physical setting. Scenarios where one wishes to consider a topological change, or respectively, methods that avoid topological change, are a subject of our current research.
Table 8.
Parameter values for numerical experiments of three-dimensional keratocyte movement with the RDS (5.1) and surface evolution law (2.8).
| reaction kinetic parameters |
surface evolution parameters |
adaptive strategy parameters |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| D1 | D2 | γ | k1 | 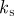 |
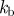 |
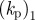 |
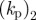 |
NH | Nh | MH | Mh |
| 1 | 100 | 20 | 0.1 | 1 | 1 | −0.7 | 0.35 | 0.5 | 1 | 0.25 | 0.5 |
Table 7.
Physically relevant parameter values for simulation of keratocyte movement in two-dimensions.
| parameter | value | physical interpretation |
|---|---|---|
| r0 | 1 | initial radius of cell sets a length scale of 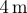
|
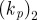 |
1 | coefficient of protrusive forcing term implies a timescale of approximately 45 s |
| D12 | 0.5 | implies the diffusivity of actin filaments of 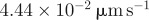
|
| D2 | 50 | implies the diffusivity of actin filaments of 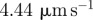
|
| k2 | 0.6–1.8 | implies an actin filament growth rate of 
|
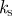 |
2 | assuming a surface tension of 1pN, sets the kinetic scale |
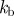 |
2 | implies a bending rigidity of approximately 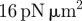
|
Table 9.
Physically relevant parameter values for simulation of keratocyte movement in three dimensions.
| parameter | value | physical interpretation |
|---|---|---|
| r0 | 1 | initial radius of cell sets a length scale of 1.17 µm |
| (kp)2 | 0.35 | coefficient of protrusive forcing term implies a timescale of approximately 4 s |
| D1 | 1.0 | implies the diffusivity of actin bundles of 0.342 µm2 s−1 |
| D2 | 100 | implies the diffusivity of actin filaments of 34.2 µm2 s−1 |
| k2 | 0.6–1.0 | implies an actin filament growth rate of 0.15 × 10−1–0.25 s−1 |
| ks | 1 | assuming a surface tension of 10 pN µm−1, sets the kinetic scale |
| kb | 1 | implies a bending rigidity of approximately 13.69 pN µm |
Figure 13.
Initial position (at t = 0 right-hand cell) and persistent keratocyte like migration of cells (at t = 5). The parameter k2 = 0.6, 0.7, 0.8, 0.9, 1.0 reading from right to left for the five polarized (left hand) cells (cf. equation (5.1)), for the remaining parameter values, see table 8. (a) Activator (a1) concentrations; (b) substrate (a2) concentrations. (Online version in colour.)
Figure 14.
The speed of the cell centroid and cell surface area both versus time of the cells shown in figure 13. We observe a positive relationship between surface area and speed. (Online version in colour.)
6. Conclusion
In this work, we have presented a computational framework for the modelling of cell motility. We propose a simple and consistent means of coupling cell movement with gradient sensing, polarization using surface partial differential equations (PDEs), and external forces. Our methods can be generalized to the modelling of more complex phenomena such as adhesion and crawling on a substrate or cell–cell interactions and we illustrate one such generalization with a concrete example of migration in the presence of obstacles.
A contribution of our study is the description of a numerical method for the simulation of cell movement that can account for the large deformations that arise in simulations of cell motility and that can be applied to the study of three-dimensional cell migration. The model equations consist of PDEs for and on surfaces and the numerical method seeks to approximate these equations on a discrete surface. Thus both the continuous and discrete problems are posed in one dimension less than the underlying spatial dimension, which in the case of the discrete problem typically means fewer degrees of freedom are needed than would be the case for embedded methods [10,13–16]. On curves, our experience is that the method maintains a mesh suitable for computation without the use of remeshing or adaptive mesh refinement. For the simulations of cell migration in three dimensions, we occasionally observed deterioration in the mesh quality, even with the redistribution of the vertices implicit in our numerical scheme, necessitating spatial adaptivity. An area of our ongoing research is the investigation of numerical methods robust to large deformations in the cell surface.
We consider a pseudopod-centred model for chemotaxis similar in form to that considered in Neilson et al. [37]. Unlike compass models [39], which are reasonable for cells with flexible polarity where a large gradient may induce pseudopods at any position on the cell membrane, pseudopod-centred models [17] are suitable for strongly polarized cells, where pseudopods are generated preferentially at the front with directional bias, owing to a chemotactic gradient, restricted primarily to small changes in direction [42]. The major contributions and imports of our study are the inclusion of bending rigidity, the inclusion of external forces, the observation that the gross behaviour, of pseudopod splitting, observed in Neilson et al. [37] for two-dimensional cells persists in three-dimensional simulations, and that the model remains qualitatively unchanged when one considers a two-component RDS, with a spatially constant global inhibitor, rather than a three component RDS with a biologically implausible non-local term. Our computational method based on surface finite elements extends the method in Neilson et al. [16] and is an alternative to the level set method considered in Neilson et al. [7,37]. The simulations illustrate that the model is capable of reproducing aspects of pseudopod-driven cell migration, described in Insall [17], in both two and three space dimensions. We report on many widely used chemotaxis measures and observe values similar to experimental observations. We also note that the simulations exhibit a dilution effect at the tip of a pseudopod, where the local maxima corresponding to an activator peak is reduced. This suggests experimental investigation of the relative importance of mechanical effects of membrane protrusion on the distribution of cell-resident proteins is warranted.
We also investigated a model for the motion of fish keratocytes. The model appears to reproduce some experimental observations of the shapes of motile keratocyte cells and the experimental observation of the correlation between cell shape and speed [18]. The computational model in Keren et al. [18] reproduces the velocity–aspect ratio relationship. However, unlike our model, both polarization and cell shapes are not explicitly modelled, with a parabolic actin profile at the leading edge assumed and the shape of the cell rear neglected. Studies [6,22] propose models where polarization is modelled by equations in the bulk of the cell which are coupled to an evolution law for the cell surface. The import of our study is to show that a surface RDS coupled to a surface evolution law gives qualitatively similar results. A further contribution is the use of surface finite elements rather than the phase-field method considered in Ziebert et al. [6] and Shao et al. [22]. This allows simulation of three-dimensional keratocyte migration, in which studies [6,22] both note is computationally expensive with the phase-field methodology. We do observe minor differences from Ziebert et al. [6], for example, in the measures of deviation from reflection symmetry.
Our numerical experience suggests that some aspects of cell migration and chemotaxis can be captured by the Schnakenberg RDS, equation (5.1). This RDS is considerably simpler from a mathematical analysis viewpoint than say the Meinhardt model, equation (4.1). One can show that the model is well posed on evolving (planar) domains [48], which is an open question even on fixed domains for the Meinhardt model. As the two components are out of phase, the model lends itself naturally to the case of a species that promotes protrusion (e.g. actin) and another that promotes retraction (e.g. myosin).
In this work, it is our intention to present a framework for future modelling rather than suggest any definitive models for cell migration. We hope that future studies will employ the framework we have set out to refine existing models for cell motility and make predictions based on numerical simulations that can be used to direct and inform experimental studies.
Acknowledgements
This research has been supported by the UK Engineering and Physical Sciences Research Council (EPSRC), grant no. EP/G010404.
References
- 1.Bray D. 2001. Cell movements: from molecules to motility. London, UK: Routledge [Google Scholar]
- 2.Mogilner A. 2009. Mathematics of cell motility: have we got its number? J. Math. Biol. 58, 105–134 10.1007/s00285-008-0182-2 (doi:10.1007/s00285-008-0182-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jilkine A., Edelstein-Keshet L. 2011. A comparison of mathematical models for polarization of single eukaryotic cells in response to guided cues. PLoS Comput. Biol. 7, e10011210. 10.1371/journal.pcbi.1001121 (doi:10.1371/journal.pcbi.1001121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Del Alamo J. C., Meili R., Alonso-Latorre B., Rodríguez-Rodríguez J., Aliseda A., Firtel R. A., Lasheras J. C. 2007. Spatio-temporal analysis of eukaryotic cell motility by improved force cytometry. Proc. Natl Acad. Sci. USA 104, 13 343 – 13 348 10.1073/pnas.0705815104 (doi:10.1073/pnas.0705815104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lombardi M. L., Knecht D. A., Dembo M., Lee J. 2007. Traction force microscopy in Dictyostelium reveals distinct roles for myosin II motor and actin-crosslinking activity in polarized cell movement. J. Cell Sci. 120, 1624–1634 10.1242/jcs.002527 (doi:10.1242/jcs.002527) [DOI] [PubMed] [Google Scholar]
- 6.Ziebert F., Swaminathan S., Aranson I. S. 2012. Model for self-polarization and motility of keratocyte fragments. J. R. Soc. Interface 9, 1084–1092 10.1098/rsif.2011.0433 (doi:10.1098/rsif.2011.0433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neilson M. P., Mackenzie J., Webb S., Insall R. H. 2011. Modelling cell movement and chemotaxis pseudopod based feedback. SIAM J. Sci. Comput. 33, 1035–1057 10.1137/100788938 (doi:10.1137/100788938) [DOI] [Google Scholar]
- 8.Oliver J. M., King J. R., McKinlay K. J., Brown P. D., Grant D. M., Scotchford C. A., Wood J. V. 2005. Thin-film theories for two-phase reactive flow models of active cell motion. Math. Med. Biol. 22, 53–98 10.1093/imammb/dqh022 (doi:10.1093/imammb/dqh022) [DOI] [PubMed] [Google Scholar]
- 9.Alt W., Dembo M. 1999. Cytoplasm dynamics and cell motion: two-phase flow models. Math. Biosci. 156, 207–228 10.1016/S0025-5564(98)10067-6 (doi:10.1016/S0025-5564(98)10067-6) [DOI] [PubMed] [Google Scholar]
- 10.Dziuk G., Elliott C. M. 2007. Finite elements on evolving surfaces. IMA J. Numer. Anal. 27, 262–292 10.1093/imanum/drl023 (doi:10.1093/imanum/drl023) [DOI] [Google Scholar]
- 11.Barrett J. W., Garcke H., Nürnberg R. 2008. Parametric approximation of Willmore flow and related geometric evolution equations. SIAM J. Sci. Comput. 31, 225–253 10.1137/070700231 (doi:10.1137/070700231) [DOI] [Google Scholar]
- 12.Yang L., Effler J., Kutscher B., Sullivan S., Robinson D., Iglesias P. 2008. Modeling cellular deformations using the level set formalism. BMC Syst. Biol. 2, 68. 10.1186/1752-0509-2-68 (doi:10.1186/1752-0509-2-68) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landsberg C., Voigt A. 2010. A multigrid finite element method for reaction–diffusion systems on surfaces. Comput. Vis. Sci. 13, 177–185 10.1007/s00791-010-0136-2 (doi:10.1007/s00791-010-0136-2) [DOI] [Google Scholar]
- 14.Dziuk G., Elliott C. M. 2008. Eulerian finite element method for parabolic PDEs on implicit surfaces. Interfaces Free Boundaries 10, 464 [Google Scholar]
- 15.Saye R. I., Sethian J. A. 2011. The Voronoi implicit interface method for computing multiphase physics. Proc. Natl Acad. Sci. USA 108, 19 498–19 503 10.1073/pnas.1111557108 (doi:10.1073/pnas.1111557108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neilson M. P., Mackenzie J. A., Webb S. D., Insall R. H. 2010. Use of the parameterised finite element method to robustly and efficiently evolve the edge of a moving cell. Integr. Biol. 2, 687–695 10.1039/c0ib00047g (doi:10.1039/c0ib00047g) [DOI] [PubMed] [Google Scholar]
- 17.Insall R. H. 2010. Understanding eukaryotic chemotaxis: a pseudopod-centred view. Nat. Rev. Mol. Cell Biol. 11, 453–458 10.1038/nrm2905 (doi:10.1038/nrm2905) [DOI] [PubMed] [Google Scholar]
- 18.Keren K., Pincus Z., Allen G. M., Barnhart E. L., Marriott G., Mogilner A., Theriot J. A. 2008. Mechanism of shape determination in motile cells. Nature 453, 475–480 10.1038/nature06952 (doi:10.1038/nature06952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray J. D. 2003. Mathematical biology. Berlin, Germany: Springer [Google Scholar]
- 20.Venkataraman C., Sekimura T., Gaffney E. A., Maini P. K., Madzvamuse A.2011. Modeling Parr-Mark pattern formation during the early development of amago trout. Phys. Rev. E 84, 041 923. 10.1103/PhysRevE.84.041923 (doi:10.1103/PhysRevE.84.041923). [DOI] [PubMed] [Google Scholar]
- 21.Barreira R., Elliott C. M., Madzvamuse A. 2011. The surface finite element method for pattern formation on evolving biological surfaces. J. Math. Biol. 63, 1095–1119 10.1007/s00285-011-0401-0 (doi:10.1007/s00285-011-0401-0) [DOI] [PubMed] [Google Scholar]
- 22.Shao D., Rappel W. J., Levine H. 2010. Computational model for cell morphodynamics. Phys. Rev. Lett. 105, 108 104. 10.1103/PhysRevLett.105.108104 (doi:10.1103/PhysRevLett.105.108104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Traynor D., Robert Kay R. 2007. Possible roles of the endocytic cycle in cell motility. J. Cell Sci. 120, 2318–2327 10.1242/jcs.007732 (doi:10.1242/jcs.007732). [DOI] [PubMed] [Google Scholar]
- 24.Larripa K., Mogilner A. 2006. Transport of a 1d viscoelastic actin-myosin strip of gel as a model of a crawling cell. Phys. A Stat. Mech. Appl. 372, 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deckelnick K., Dziuk G., Elliott C. M. 2005. Computation of geometric partial differential equations and mean curvature flow. Acta Numer. 14, 139–232 10.1017/S0962492904000224 (doi:10.1017/S0962492904000224) [DOI] [Google Scholar]
- 26.Helfrich W. 1973. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. 28, 693–703 [DOI] [PubMed] [Google Scholar]
- 27.Willmore T. J. 1997. Riemannian geometry. Oxford, UK: Oxford University Press [Google Scholar]
- 28.Dziuk G. 1990. An algorithm for evolutionary surfaces. Numer. Math. 58, 603–611 10.1007/BF01385643 (doi:10.1007/BF01385643) [DOI] [Google Scholar]
- 29.Dziuk G., Elliott C. M. In press L2-estimates for the evolving surface finite element method. Math. Comp. [Google Scholar]
- 30.Elliott C. M., Stinner B. 2010. Modeling and computation of two phase geometric biomembranes using surface finite elements. J. Comput. Phys. 229, 6585–6612 10.1016/j.jcp.2010.05.014 (doi:10.1016/j.jcp.2010.05.014) [DOI] [Google Scholar]
- 31.Elliott C. M., Stinner B. In press Computation of two-phase biomembranes with phase dependent material parameters using surface finite elements. Commun. Comput. Phys. [Google Scholar]
- 32.Heine C. J. Isoparametric finite element approximation of curvature on hypersurfaces. Technical report. [Google Scholar]
- 33.Dziuk G. 2008. Computational parametric Willmore flow. Numer. Math. 111, 55–80 10.1007/s00211-008-0179-1 (doi:10.1007/s00211-008-0179-1) [DOI] [Google Scholar]
- 34.Schmidt A., Siebert K. G. 2005. Design of adaptive finite element software: the finite element toolbox Alberta. Berlin, Germany: Springer [Google Scholar]
- 35.Davis T. A. 2004. Algorithm 832: UMFPack v. 4.3—an unsymmetric-pattern multifrontal method. ACM Trans. Math. Softw. 30, 196–199 (doi:10.1145/992200.992206) [Google Scholar]
- 36.Meinhardt H. 1999. Orientation of chemotactic cells and growth cones: models and mechanisms. J. Cell Sci. 112, 2867–2874 [DOI] [PubMed] [Google Scholar]
- 37.Neilson M. P., Veltman D. M., van Haastert P. J. M., Webb S. D., Mackenzie J. A., Insall R. H. 2011. Chemotaxis: a feedback-based computational model robustly predicts multiple aspects of real cell behaviour. PLoS Biol. 9, e1000618. 10.1371/journal.pbio.1000618 (doi:10.1371/journal.pbio.1000618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amarasinghe D. P., Aylwin A., Madhavan P., Pettitt C. 2011. Biomembranes report. [Google Scholar]
- 39.Hecht I., Skoge M. L., Charest P. G., Ben-Jacob E., Firtel R. A., Loomis W. F., Levine H., Rappel W. J. 2011. Activated membrane patches guide chemotactic cell motility. PLoS Comput. Biol. 7, e1002044 10.1371/journal.pcbi.1002044 (doi:10.1371/journal.pcbi.1002044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng S. Y., Heilman S., Wasserman M., Archer S., Shuler M. L., Wu M. 2007. A hydrogel-based microfluidic device for the studies of directed cell migration. Lab. Chip 7, 763–769 10.1039/b618463d (doi:10.1039/b618463d) [DOI] [PubMed] [Google Scholar]
- 41.Ramsey W. S. 1972. Analysis of individual leucocyte behavior during chemotaxis. Exp. Cell Res. 70, 129–139 10.1016/0014-4827(72)90190-5 (doi:10.1016/0014-4827(72)90190-5) [DOI] [PubMed] [Google Scholar]
- 42.Bosgraaf L., Van Haastert P. J. M. 2009. Navigation of chemotactic cells by parallel signaling to pseudopod persistence and orientation. PLoS ONE 4, e6842. 10.1371/journal.pone.0006842 (doi:10.1371/journal.pone.0006842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hecht I., Levine H., Rappel W. J., Ben-Jacob E. 2011. ‘self-assisted’ amoeboid navigation in complex environments. PLoS ONE 6, e21955. 10.1371/journal.pone.0021955 (doi:10.1371/journal.pone.0021955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grima R. 2007. Directed cell migration in the presence of obstacles. Theor. Biol. Med. Model. 4, 2. 10.1186/1742-4682-4-2 (doi:10.1186/1742-4682-4-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bosgraaf L., Van Haastert P. J. M., Bretschneider T. 2009. Analysis of cell movement by simultaneous quantification of local membrane displacement and fluorescent intensities using quimp2. Cell Motil. Cytoskeleton 66, 156–165 10.1002/cm.20338 (doi:10.1002/cm.20338) [DOI] [PubMed] [Google Scholar]
- 46.Pollard T. D., Borisy G. G. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 10.1016/S0092-8674(03)00120-X (doi:10.1016/S0092-8674(03)00120-X) [DOI] [PubMed] [Google Scholar]
- 47.Lefever R., Prigogine I. 1968. Symmetry-breaking instabilities in dissipative systems II. J. Chem. Phys. 48, 1695–1700 10.1063/1.1668896 (doi:10.1063/1.1668896) [DOI] [Google Scholar]
- 48.Venkataraman C., Lakkis O., Madzvamuse A. 2012. Global existence for semilinear reaction–diffusion systems on evolving domains. J. Math. Biol. 64, 41–67 10.1007/s00285-011-0404-x (doi:10.1007/s00285-011-0404-x). [DOI] [PubMed] [Google Scholar]