Abstract
This paper reports a method for fabricating multilayer microfluidic protein crystallization platforms using different materials to achieve X-ray transparency and compatibility with crystallization reagents. To validate this approach, three soluble proteins, lysozyme, thaumatin, and ribonuclease A were crystallized on-chip, followed by on-chip diffraction data collection. We also report a chip with an array of wells for screening different conditions that consume a minimal amount of protein solution as compared to traditional screening methods. A large number of high quality isomorphous protein crystals can be grown in the wells, after which slices of X-ray data can be collected from many crystals still residing within the wells. Complete protein structures can be obtained by merging these slices of data followed by further processing with crystallography software. This approach of using an x-ray transparent chip for screening, crystal growth, and X-ray data collection enables room temperature data collection from many crystals mounted in parallel, which thus eliminates crystal handling and minimizes radiation damage to the crystals.
Keywords: X-ray transparency, microfabrication, protein crystallization, structure determination, Crystallography
1. Introduction
Microfluidic platforms have gained widespread use in diverse fields like chemical synthesis, enzymatic and DNA analysis [1, 2], proteomics [3, 4], point-of-care, medical and clinical diagnostics [5–7], energy conversion [8, 9], and protein / pharmaceutical crystallization [10–14]. Microfluidics offers several advantages in terms of reduced sample size and easy preparation, fine control over transport phenomena on the microscale, ease of scalability, detection and sample analysis on a single, integrated platform [15, 16]. A large fraction of microfluidic devices reported in the literature to date have been fabricated using poly(dimethylsiloxane) (PDMS) and glass, which allow for rapid prototyping [13, 17, 18]. Issues still exist, however, with respect to compatibility with different chemistries, temperature and pressure variation, as well as amenability to integration of analysis techniques. Many alternative materials and combination of materials have been explored for the fabrication of microfluidic platforms with the goal to overcome these issues for specific applications [19, 20]. Typical challenges encountered in these efforts are bonding of different materials, interfacing with ancillary equipment, and the assembly of chips comprised of highly integrated, complex designs.
The focus of this work is on the application of microfluidic platforms in structural biology, specifically for protein crystallography. Protein structure determination involves handling of small sample volumes, fine control over transport properties during crystallization, and requires the collection and analysis of X-ray diffraction data from the crystals formed [21–23]. A microfluidic approach has the potential to meet these requirements. For example, we recently reported a microfluidic array chip for solid form screening of candidate pharmaceuticals [14], but this chip lacks X-ray transparency. The first step in protein crystallization involves screening a protein against a wide range of precipitants to identify suitable crystallization condition(s). Once one ore more conditions have been identified, traditional methods require the crystals to be manually harvested, cryo-cooled and mounted in an X-ray beam for structure determination, all one crystal at a time. These challenging tasks, especially when crystals are small and fragile, often damage the crystal, affecting the quality of data collected. Several of the microfluidic platforms reported in the literature have tried to address these challenges, however most of them still require the crystal to be manually harvested before data collection [12, 13, 24, 25]. X-ray transparent platforms have also been reported, but they have been limited to relatively simple, single-layer designs [11, 19, 26, 27] that do not take advantage of the integrated fluid handling capabilities of multilayer microfluidics or require cutting out the section containing the crystal from the whole chip for further analysis [28].
Here we report on the fabrication of a microfluidic platform that allows for (i) screening up to 100 crystallization conditions while consuming minimal amount of protein solution, (ii) on-chip X-ray data collection from the protein crystals grown, while (iii) retaining the fluid routing and manipulation capabilities of integrated, valve-based high throughput microfluidic systems. This is achieved by assembling a thin hybrid microfluidic chip comprised of layers of cyclic olefin copolymer (COC) and PDMS. We validate the utility of this chip and its use by crystallizing the soluble proteins lysozyme, thaumatin and ribonuclease A on-chip, followed by X-ray diffraction data collection from the crystals grown while they still reside within the microfluidic platform mounted as a whole in the X-ray beam.
2. Materials and methods
2.1 Protein solutions
Hen egg white lysozyme (Sigma) was dissolved in 50 mM sodium acetate (Sigma-Aldrich) at pH 4.6 with 20% (w/v) glycerol (Fisher Scientific) at a concentration of ~100 mg/mL. Lysozyme concentrations were determined by UV absorbance measurements (Lambda 650 UV-Vis spectrophotometer, Perkin Elmer) at 280 nm using an extinction coefficient of 2.64 mL/(mg-cm) [29]. For proof-of-concept crystallography experiments precipitant solutions of 1M and 2M NaCl (Aldrich) in 50 mM sodium acetate, pH 4.6 with 20% (w/v) glycerol were prepared. For screening experiments Hampton Crystal Screen chemicals were used directly (Hampton Research).
Thaumatin from Thaumatococcus daniellii (Sigma) was dissolved in 100 mM NaH2PO4 (EMD Chemicals) at pH 6.5 at a concentration of 82 mg/mL. The protein concentration was determined by UV absorbance measurements at 280 nm using an extinction coefficient of 1.25 mL/(mg-cm) [30]. A precipitant solution of 30% (w/v) Na/K tartrate (Malinckrodt) and 20% w/v glycerol in 100 mM NaH2PO4 pH 7.0 was used [31].
Ribonculease A (R-5500, Sigma) from bovine pancreas was dissolved in 100 mM sodium acetate at pH 4.5 at a concentration of 229 mg/mL. The protein concentration was determined by UV absorbance measurements at 280 nm using an extinction coefficient of 0.70 mL/(mg-cm).[32] A precipitant solution of saturated NaCl in 100 mM sodium acetate at pH 4.5 was used [33].
2.2 Fabrication and operation of microfluidic platforms
Silicon wafers were patterned using negative photoresist SU8-25 and SU8-2050 (MicroChem). The microfluidic platforms consisted of different layers of polymers. The control layer was fabricated with COC (TOPAS Advanced Polymers Inc, 4 mil). Patterning of the COC was performed via hot embossing at 175°C (Tg+50°C), using a master made out of a high temperature epoxy resin (Conapoxy FR 1080, 83:100 hardener: epoxy, by mass) using a hot press (Carver hot press, model 3851-0).
The fluid layer was fabricated out of PDMS (General Electric RTV 615, Part A/B). Inlets were drilled using a 750 µm drill bit (McMaster Carr). Bonding of the fluid and control layer (see SI for further details) was done using a 1% (v/v) solution of 3-glycidoxypropyltrimethoxysilane (GPTMS, Sigma Aldrich) and a 1% (v/v) solution of 3-aminopropyltrimethoxysilane (APTMS, Sigma Aldrich). The completed assembly was placed on a flat COC substrate prior to setting up of crystallization trials. The use of vacuum for the operation of the devices eliminated the need to bond the fluid layer irreversibly to the COC substrate.
All solutions were introduced on-chip by pipetting 2–4 µL of protein/precipitant solution on the inlet ports, then pulling the fluid into the chip by actuation of the appropriate valves by applying vacuum through a manifold (Cole Parmer) attached to a vacuum pump (GAST; Model DOA-P704-AA). The vacuum was applied onto the chip through 24 gauge AWG, thin walled PTFE tubing (Cole Parmer) connected to a block of PDMS, which was positioned over the appropriate control line inlet. Once the chip was filled, all the inlets were covered with Crystal Clear Tape (Hampton HR4-511) to prevent evaporation and the trials were incubated. Even though some water is lost by absorption into the thin layer of PDMS, crystallization trials could easily be performed for a week, in for most wells for two weeks, before solvent loss started to affect the crystallization outcomes, for example by precipitation of amorphous material.
2.3 Visualization of crystallization experiments and setup of traditional crystallization trials
Crystallization experiments were set up and visualized using either a stereomicroscope (Leica, MZ12.5) equipped with a digital camera (Leica, DFC295) or a computer controlled imaging system comprised of an optical microscope (Leica Z16 APO) equipped with an auto-zoom lens (Leica 10447176), a digital camera (Leica DFC280), and a motorized x-y stage (Semprex KL66) controlled by Image Pro Plus (Media Cybernetics). Periodically birefringent images of the wells were taken.
Traditional microbatch-under-oil crystallization trials were set up combining 2 µL each of protein and precipitant solutions in a Greiner well plate (Hampton Research) at room temperature. Crystals were harvested using crystal mounts (Mitegen). Crystallization trials of lysozyme were performed in traditional well plates, large well devices, and array chips at 4°C or at room temperature. Crystallization trials for thaumatin and ribonuclease A were performed at room temperature.
2.4 X-ray data collection
The microfluidic chips were mounted on a standard magnetic goniometer mount (Hampton Research) with an attached metal tube into which a slit was cut. A set-screw was used to secure samples. Diffraction data was collected either at room temperature or under cryogenic conditions. Cryo-cooling of samples was achieved by direct immersion into liquid nitrogen. Various sample-to-detector distances were used based on the quality of the crystal present. Typically data was collected using 1° steps with a 1s exposure at an X-ray energy of 12.7 keV (λ = 0.979Å) at Argonne National Lab (LS-CAT). Data from multiple crystals in the microfluidic devices was collected over a range of 10° (−5° to +5° from the normal) and an optimal subset of the frames was subsequently merged to obtain a complete dataset. Bench-top diffraction experiments were performed at the George L. Clark X-ray Facility at the University of Illinois using a General Area Diffraction Detector System (GADDS; Bruker) equipped with a four circle diffractometer and HiStar multiwire area detector. A rotating anode generator (Bruker M18XHF22) operating at 40 kV and 60 mA was used with a graphite monochromator supplying a Cu Kα radiation beam (λ = 1.54Å or 8.048 keV). The sample to detector distance is ~18 cm. Data was collected in a coupled mode where 2ω = 2θ such that multiple frames could be collected over a wider range of 2θ. Typically two such frames were collected, spanning the range of 2θ from 0° to 40°, up to a resolution of 2.3 Å.
Analysis of X-ray diffraction data collected at the synchrotron was performed using HKL2000 software for indexing, refinement, integration, and scaling (HKL Research) [34]. Diffraction data collected at the University of Illinois was analyzed using GADDS software (version 4.1.08, Bruker AXS) and To-pas 3 (Bruker AXS). Subsequent processing of crystallography datasets was done using the CCP4 suite of programs [35]. Electron density maps were displayed using COOT [36]. Molecular replacement [37] for lysozyme was done using PDB structure 193L as the model [38].
3. Results and discussion
3.1 X-ray compatibility and materials characterization
In designing an X-ray compatible microfluidic device for use in protein crystallography three main considerations with respect to the interaction between materials and X-rays must be taken into account: (i) attenuation and (ii) scattering of X-rays passing through device materials, and (iii) the strength of diffraction resulting from a crystal. Attenuation originates from the absorption of photons by the material, thereby decreasing the intensity of both the incident X-ray beam and the resulting diffracted X-rays. Scattering is an elastic redirection of photons based on the internal structure of the material and can affect the signal-to-noise ratio. The strength of the diffraction signal from a crystal is related to not only the degree of order within the crystal, but also the packing density and the size of the crystal [39–43].
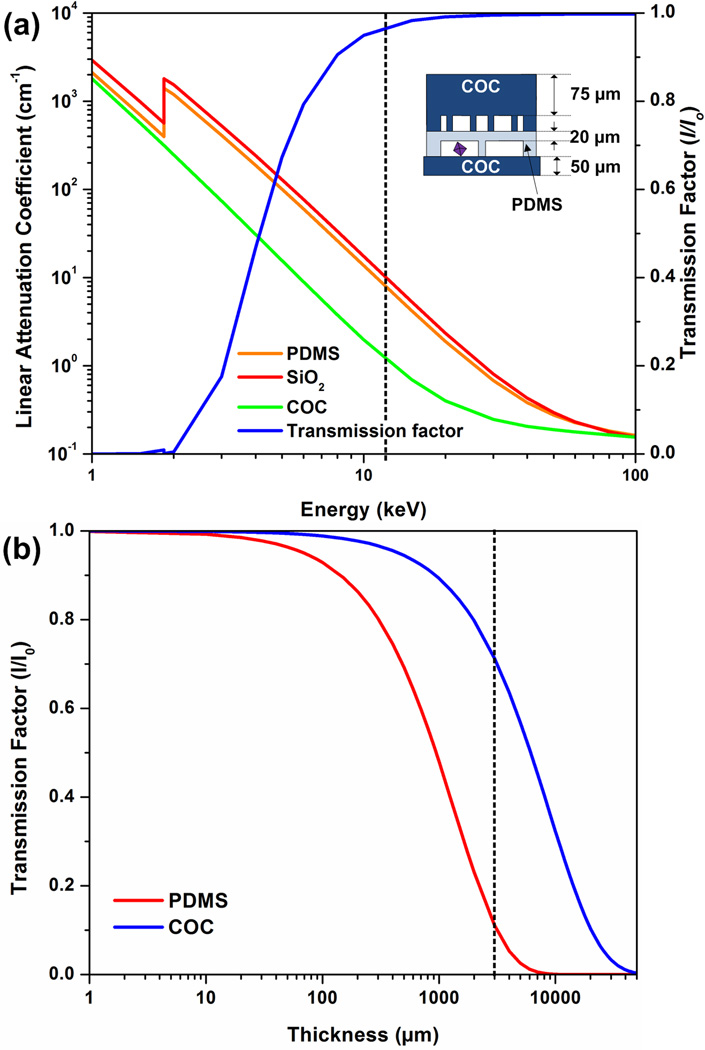
Figure 1a shows the variation of the linear attenuation coefficient with the X-ray energy. In general, the linear attenuation coefficient is lower at higher X-ray energy. At relevant energies for X-ray data collection (here 12.4 keV), COC attenuates X-rays seven times less than PDMS and SiO2. Figure 1b shows the transmission factor for COC and PDMS as a function of film thickness. A thickness exceeding 3 mm is typical for conventional microfluidic devices, at which point the transmission factor is less than 10% (Figure 1b). However for the hybrid COC-PDMS device as reported here, the transmission factor was more than 90% (Figure 1a, dotted line shows data collection energy of 12.4 keV). An analysis of both PDMS and COC shows that these materials produce a characteristic scattering pattern, seen in the form of rings in magnified diffraction data. However this scattering occurs at relatively low resolution and does not impact the collection of high resolution diffraction data (see the SI for more detail).
Figure 1.
(a) Linear attenuation coefficient for PDMS, COC, and SiO2 (quartz) as a function of photon energy. The transmission factor I/I0 as a function of photon energy for a typical device architecture as shown in the schematic. (b) Transmission factor I/I0 as a function of film thickness for PDMS and COC. The transmission factor was calculated at photon energy of 12.4 keV, or a wavelength of 1Å.
3.2 Design and operation of microfluidic devices
The microfluidic device presented here enables the screening of protein solution against different precipitants, and also allows for structure determination of the protein once a suitable crystallization condition has been found. The main challenge here is to create a microfluidic chip that is still capable of fluid routing and compartmentalization to allow screening of many conditions, while also being X-ray transparent.
3.2.1 Array chip design
As discussed above, we chose to replace the traditionally thick layers of PDMS with thin films of COC because of its lower X-ray attenuation profile and beneficial material properties. However, COC is not suitable for the integration of valves into a microfluidic device because it does not have the same degree of flexibility as PDMS, an essential property for typical pneumatic valves [44]. To retain the ability to route and compartmentalize fluids on-chip, we use a thin layer of PDMS sandwiched between COC layers. This hybrid approach eliminates the bulk of PDMS usually found in traditional devices. Using a 20 µm PDMS layer sandwiched between two COC layers has only a minimal effect on the transmission of X-rays at energies relevant for X-ray data collection (Figure 1a).
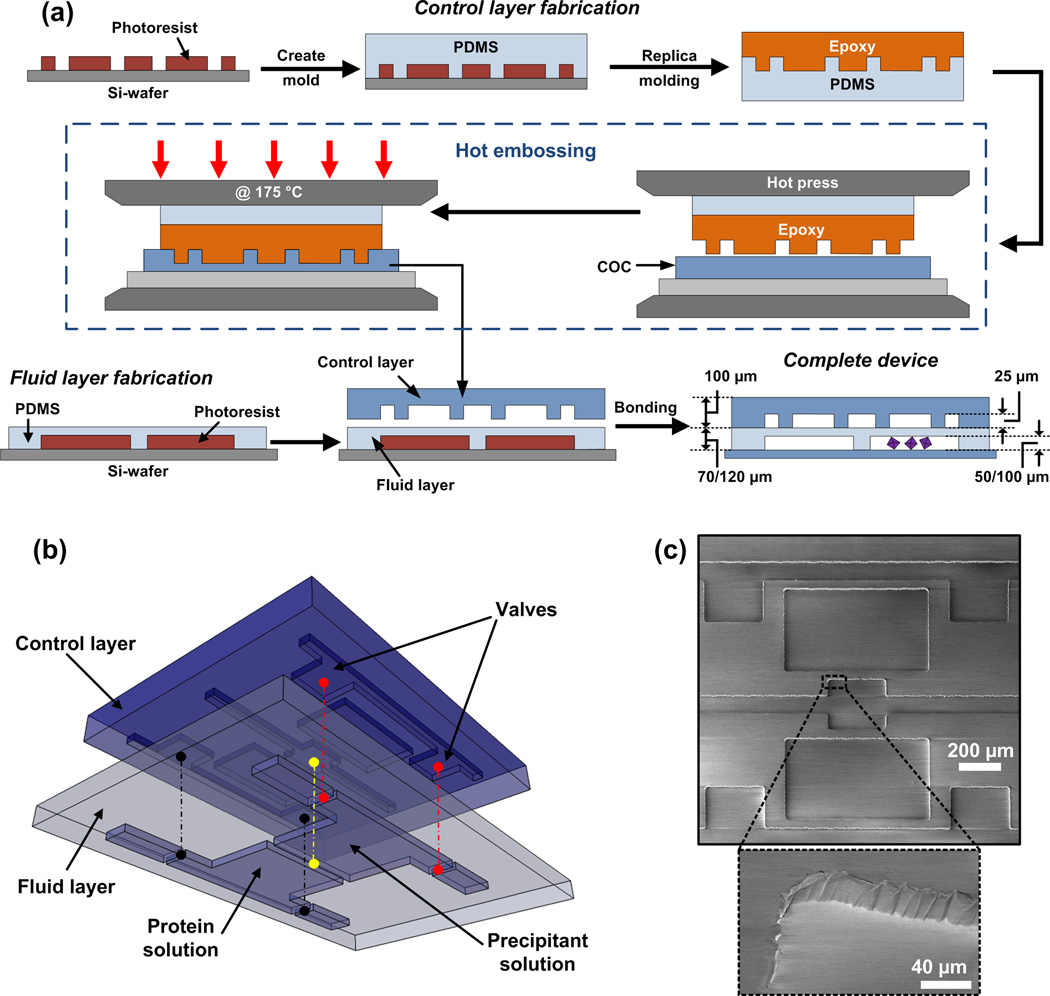
The hybrid microfluidic chips are comprised of a thin PDMS fluid layer sandwiched between and bonded to a COC control layer and flat COC substrate (Figure 2a). The control layer is fabricated via hot embossing against an epoxy master. This epoxy master is a fabricated by first creating a photoresist-on-silicon master using standard photolithography, followed by replication of this silicon master into a PDMS mold, which in turn is replicated in epoxy [20]. Sheets of COC (100 µm) were patterned with this epoxy master by hot embossing at 175°C (Figure 2a). The COC control layers have 25 µm deep and 50 µm wide negative relief patterns as the control lines. Figure 2c shows the fidelity of pattern transfer into COC. Inlet holes for actuation of valves were then drilled in the COC control layer.
Figure 2.
(a) Fabrication scheme for the hybrid COC-PDMS chip. The inverse pattern for the control layer is patterned onto a Si-wafer using negative photoresist to create a master. PDMS (10:1) was poured over the master mold for the control layer. High temperature epoxy (FR-1080) was poured on the PDMS mold and cured at 120 °C for 4 hours, after which it was gently lifted off the PDMS mold. The epoxy master was then placed on top of a COC sheet (100 µm) and sandwiched between a PDMS backing layer and a glass slide and the assembly is placed inside a hot press. The entire assembly was heated up to 175 °C in the hot press and maintained at that temperature for 5 minutes to pattern the control layer. A thin fluid layer was made out of PDMS (15:1) and patterned using standard soft lithographic techniques. Holes were drilled for the control layer inlets (not shown) following which, the control and fluid layer were aligned and bonded using a silane based chemical treatment (see Figure S3). Holes were drilled for the fluid layer inlets (not shown) and the assembled device was placed on a thin COC substrate. (b) A 3D perspective view of the control and fluid layer being aligned with the colored lines corresponding to the points in in the control layer and fluid layer that are aligned. (c) SEM image of the COC control layer fabricated via hot embossing of the epoxy master demonstrating the excellent pattern transfer.
The fluid layer is fabricated in PDMS using standard soft lithography [44], with the COC control layer being bonded to the 70 µm thick fluid layer before being lifted from its photoresist mater [45]. A perspective view of this assembly is shown in Figure 2b. Inlet holes for the fluid layer were then drilled through this composite assembly. Lastly, this COC-PDMS assembly was placed on a 50 µm thick COC sheet to close the fluid channels. Both the 24-well array chip (Figure 2) and the larger, single well chip for proof-of-principle testing (Figure 3) were fabricated in this way. More details on the fabrication and assembly procedures can be found in the Materials and Methods section and the Supplementary Information.
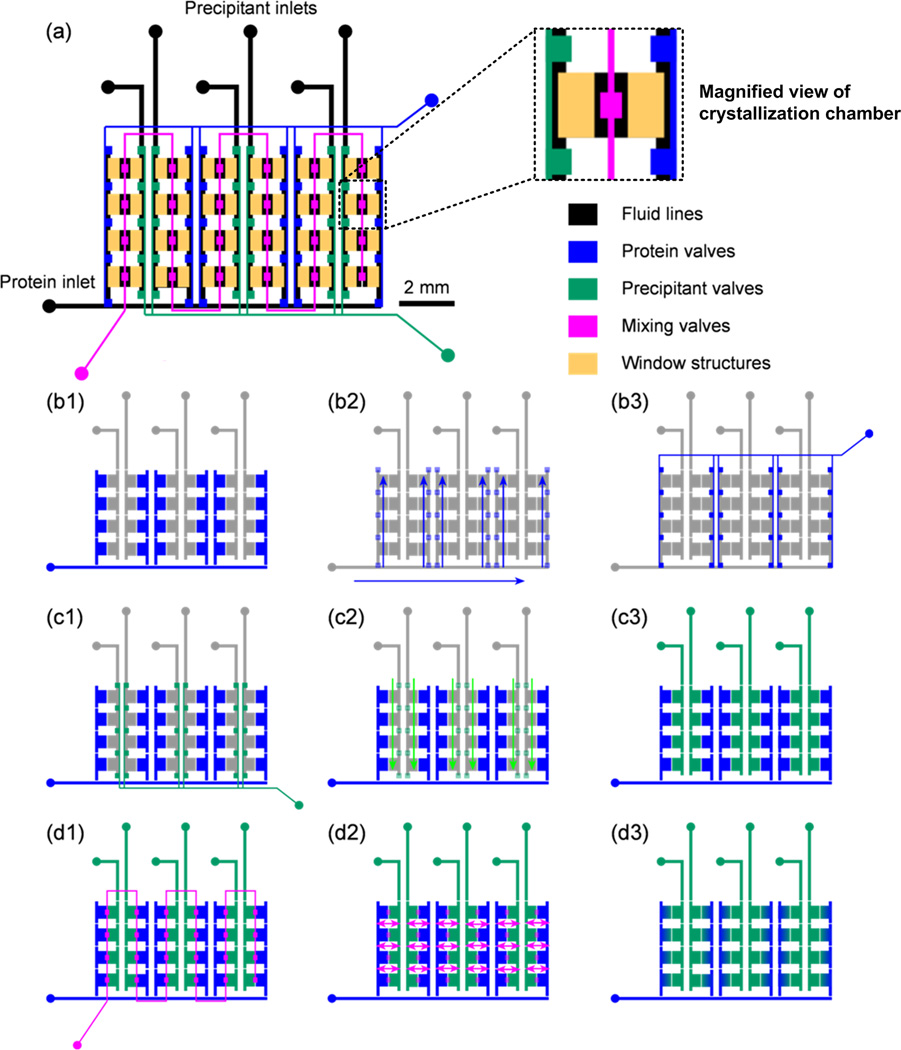
Figure 3.
Schematic of the design and operation of a 24-well hybrid array chip. (a) Complete chip with the fluid layer shown in black, and the various components of the control layer are shown in respective color according to function. The window structures are present to decrease the total material present in the path of the X-ray beam. (b) Blue valves are actuated to fill in protein. (c) Green valves are actuated to fill in precipitant. (d) Pink valves between the protein and precipitant chambers are actuated to allow diffusion between the chambers over time.
The array chips consist of a series of separate half-wells for protein and precipitant solutions arranged in columns. Each of these individual wells is a separate crystallization trial (Figure 3a) and is isolated from the rest of the wells using a series of normally-closed valves (Figure 3a, green and blue valves) [46]. Since these valves are closed at rest, the chip can be transported easily after filling without disturbing the crystallization trials. Each half-well contains 50 nL of solution and the entire chip uses just 1.4 µL of protein solution for a 24-well design. These adjacent half wells are separated by a normally closed valve that allows mixing between the protein and precipitant solution through free interface diffusion. The time required to mix two solutions in adjacent wells can be estimated using Fick’s law, t = x2/(4D), where×is the combined length along which the two solutions are mixing and D is the diffusivity of the diffusing species (protein in this case, approximately ~10−6 cm2/s). For a mixing length of around 1 mm, the time needed to mix two solutions is approximately 20 minutes. The mixing time can be adjusted for other protein solutions with different diffusivities.
3.2.2 Array chip operation
The two half-wells are filled independently of each other using dedicated valve lines (blue and green valves for the aqueous protein and precipitant solutions respectively; see Figure 3a) for each set of halfwells arranged in columns. These valves are closed at rest thus isolating the various chambers. The mixing valves (pink in Figure 3a) between adjacent half-wells enable the protein and precipitant solution to mix by diffusion. The entire filling process is illustrated in Figure 3. First 2 µL of protein solution is pipetted over the inlet of the protein line and vacuum is applied to the protein valve line (blue) which allows for dead-end filling of the protein solution into the series of protein half-wells (Figure 3b). Once the protein solution has been introduced, the protein valve set is closed, isolating the solutions in the protein chambers. Next, 1 µL droplets of precipitant solution are pipetted over the inlets of each of the six precipitant. Vacuum is then applied via the precipitant valve lines (green) and the precipitant solutions flow into the appropriate half-wells (Figure 3c). After closing the precipitant valve set, thus locking the precipitants in their respective chambers, the inlets for filling and actuation are sealed with Crystal clear tape, preventing evaporation of the filled solutions. Next, the mixing valves (pink) located between the protein and precipitant chambers are actuated, allowing sets of two adjacent solutions to mix by free interface diffusion (Figure 3d). We allowed the solutions to mix for 20 minutes for all experiments performed here. After mixing, the mixing valves are closed, the inlets of these control lines are sealed with Crystal Clear tape, and the chips are incubated at either 4°C or room temperature for 8–48 hours. Previously, we have used a similar microfluidic array chip for solid form screening of candidate pharmaceuticals [14], but this platform lacked X-ray transparency.
3.2.3 Single-well chip design, operation, and proof-of-concept experiments
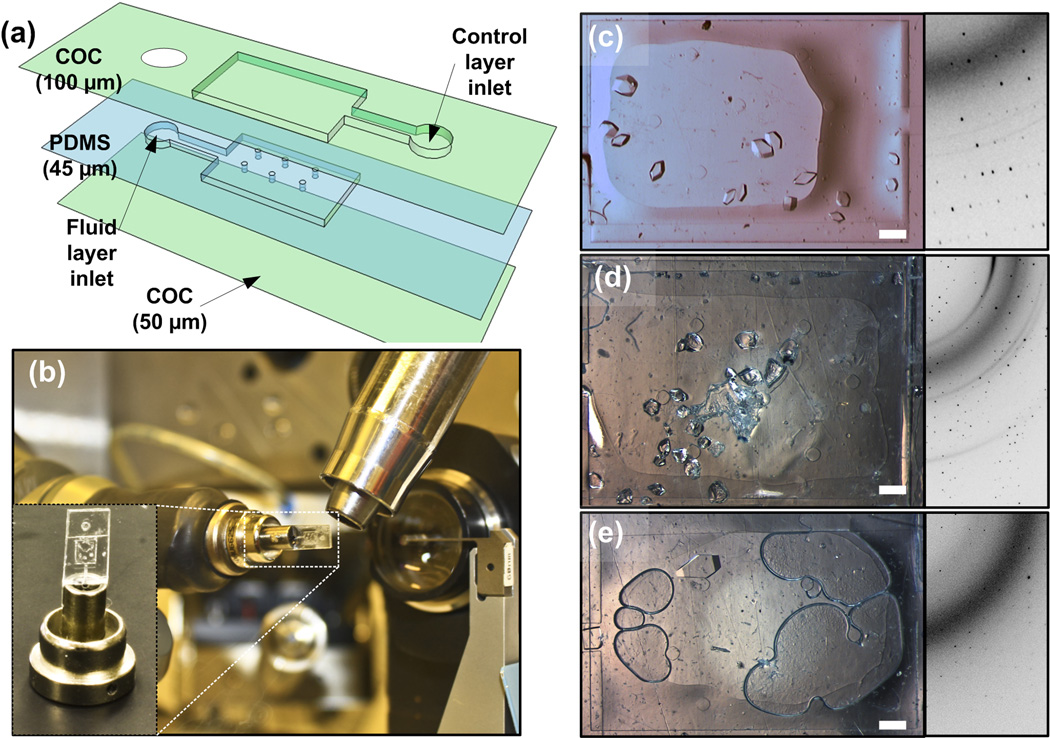
Before completing the full design and fabrication of the 24-well array chip, we wished to confirm that indeed high quality diffraction data can be collected from crystals residing inside a microfluidic well comprised of COC layers as well as a thin PDMS layer (Figure 4a). These chips for validating the X-ray compatibility of the materials are made using a similar fabrication scheme described earlier (Section 3.2.1). They consist of a large rectangular 2.36 mm by 3.36 mm by 25 µm wells with a volume of 0.2 µL. Six posts were fabricated within the fluidic well chamber to provide support for the thin PDMS membrane. The COC control layer consisted of an identical sized chamber aligned directly above the fluid well (Figure 4a). Pre-mixed solutions of protein and precipitant (see Materials and Methods) were introduced into the chip by pipetting a drop over the inlet of the fluid layer and pulling it into the fluid layer well by applying vacuum to the inlet of the control layer (Figure 4a). The chip was allowed to incubate for 48–72 hours.
Figure 4.
(a) Design of simple large well microfluidic chip. Numbers in brackets represent the total height of each layer. The feature size in both the COC and PDMS layers is 25 µm. (b) Optical micrograph of an X-ray transparent large well device mounted on a beamline at LS-CAT, Argonne National Lab with the inset showing a close-up of the modified magnetic goniometer mount on which the device is mounted. Optical micrographs of microfluidic chips with crystals of (c) lysozyme (d) ribonuclease A and (e) thaumatin. On-chip diffraction data (collected at LS-CAT, Argonne National Lab) from each of the crystals at cryogenic conditions is displayed alongside as well. Scale bars correspond to 300 µm.
To test the effectiveness of the chip materials for on-chip X-ray data collection, we first grew crystals of model proteins lysozyme (Figure 4c), ribonuclease A (Figure 4d), and thaumatin (Figure 4e) in single well chips. The whole chips were then mounted onto a goniometer mount (Figure 4b, see Materials and Methods) and taken to Argonne National Lab (LS-CAT). Figure 4c-e shows on-chip diffraction data collected from these chips from the different model proteins. Even though a scatter ring from the materials is present, good quality, circular diffraction spots are still clearly visible, sufficient for structure determination.
3.3 Screening and on-chip protein structure determination
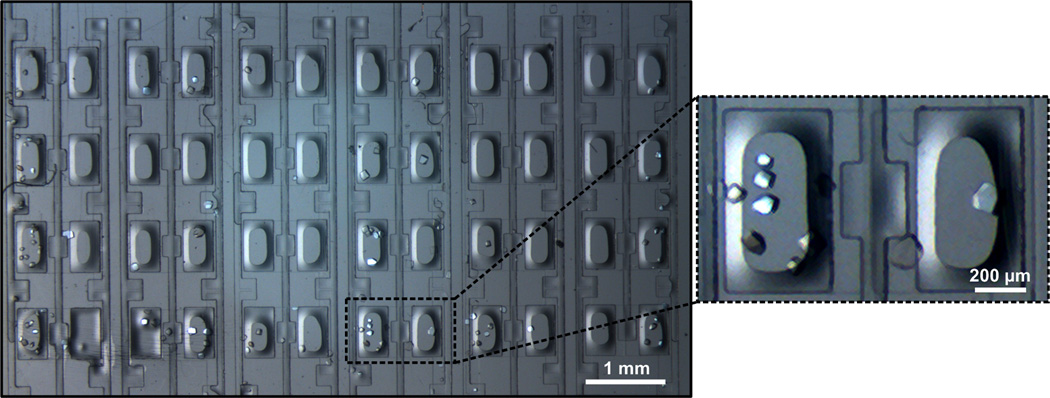
Identification of suitable crystallization conditions by screening protein solution against a wide variety of crystallization mixtures (cocktail of salts, buffers and precipitants) is the first step in the process of protein structure determination. Figure 5 shows a 24-well array chip that allows for the testing of six different precipitant solutions in quadruplicate. A series of array chips was used to test each of the individual conditions multiple times and the results were compared to those obtained using a traditional microbatch wellplate.
Figure 5.
Optical micrograph of crystals of lysozyme grown on a 24-well hybrid array chip, with a magnified view of one of the crystallization chambers.
The microfluidic array chips were validated for crystallization screening experiments by testing solutions of lysozyme against the 50 condition Crystal Screen kit (Hampton Research) at room temperature. After one week crystals were observed in 32 out of the 50 conditions in the array chips (Table 1) compared to only 26 hits in the microbatch wellplate. X-ray compatibility of the chip allowed us to easily identify whether the hits were indeed protein crystals or just salt crystals using a local X-ray source (Bruker APEX). A comparison of the results obtained in our well plates versus the traditional microbatch method shows that 21 of the conditions produced crystals on both platforms while 5 conditions yielded crystals uniquely in the microbatch wellplates and 11 hits were observed uniquely in the microfluidic chips. The variability between these results can be explained by (i) differences in the method of mixing of protein and precipitant solutions; (ii) slow concentration of solutions in the microfluidic chips over time; and (iii) the stochastic nature and variability of crystallization trials [47–49]. Note that typical microbatch wellplate experiments need 2 µL of protein solution per condition whereas a whole 24-well array chip can be filled using less than 2 µL of protein solution.
Table 1.
Crystallization results of 100 mg/mL lysozyme in 50mM sodium acetate with 20% glycerol against the 50 condition Hampton crystal screen at RT.
|
Condition number |
On−chipa/off−chipb |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| 1−10 | −c/− | −/+d | −/− | −/− | −/− | +/+ | +/+ | +/− | +/+ | +/+ |
| 11−20 | +/+ | +/− | −/− | −/+ | +/+ | +/− | +/+ | +/+ | +/− | +/+ |
| 21−30 | +/− | +/− | +/− | −/− | +/+ | −/− | +/+ | +/+ | +/+ | +/+ |
| 31−40 | +/+ | +/− | +/− | +/+ | +/+ | −/− | −/+ | −/+ | −/+ | +/+ |
| 41−50 | +/+ | −/− | +/− | −/− | +/− | +/− | −/− | +/+ | −/− | −/− |
Crystallization trials set up on a 24-well array chip.
Trials set up on a microbatch tray under oil.
“−” indicates absence and
“+” indicates presence of crystals.
Strategy for on-chip data collection
Once crystals have grown on-chip, we want to collect X-ray data with the crystals still residing in the microfluidic wells. In tradition crystallography, crystals are manually looped out, cryo-cooled, mounted in a cryo-stream, and rotated in the X-ray beam for data collection. The dimensions and design of the array chips makes rotation of the chip for the collection of a complete dataset very difficult. The chip is also too large to fit within typical cryo-streams on crystallography beamlines, necessitating data collection at room temperature.
The inability to utilize cryo-cooling to mitigate the effects of radiation damage [50] led us to adopt an alternative strategy where we collect small wedges of data (10 degrees) from a large number of crystals at room temperature and later merge these wedges to form a complete dataset. The use of small wedges of data limits the time of exposure, and thus the extent of radiation damage. These wedges can be merged to form a complete dataset, provided that non-isomorphism between crystals is minimal and the orientation of the grown crystals is mostly random. This method has been used previously to obtain structural information from tiny or fragile crystals or crystals which suffer from excessive radiation damage [51, 52].
The advantage of applying this data collection strategy to crystals grown in a microfluidic chip is the ease by which a large number of crystals can be generated. Furthermore, the fine control of transport on the microfluidic scale allows for improved reproducibility both in crystal quality and isomorphism between crystals grown in different wells or even in different chips. Analysis of lysozyme crystals grown on-chip demonstrated that 98% of the crystals (47 out of 48) showed a coefficient of variation (standard deviation divided by the average) in the unit cell dimensions of ~0.1%.
Analysis of on-chip collected X-ray data
We collected and analyzed room temperature X-ray data of lysozyme crystals grown in 24-well chips using the same crystallization conditions as used for the proof-of-principle experiments. For comparison, we also collected X-ray data at cryogenic temperatures on a lysozyme crystal grown in a wellplate and mounted using a loop with Table 2 showing a comparison of the important crystallographic statistics between the two methods.
Table 2.
Crystallographic data statistics from the analysis of lysozyme crystals.
| Parameter | On-Chipa | Traditional (Cryogenic)b |
|---|---|---|
| Unit Cell Dimensions | a = b = 79.693Å c = 37.781Å |
a = b = 78.817Å c = 37.025Å |
| Space Group | P43212 | P43212 |
| Observations (Unique) | 783,994 (18,352) | 223,433 (17,510) |
| Resolution | 50 – 1.55Å | 50 – 1.55Å |
| Rsym | 0.064 (0.362) | 0.052 (0.102) |
| Mosaicity | 0.03° – 0.08° | 0.21° – 0.34° |
| Redundancy | 22.9 (5.7) | 7.7 (7.7) |
| Completeness | 98.1% (83.4%) | 99.7% (100%) |
| I/σ | 51.4 (3.9) | 42.3 (19.4) |
| # of Frames | 363 | 100 |
|
Refinement | ||
| R (Rfree) | 0.164 (0.227) | 0.173 (0.276) |
|
Ramachandran Statistics | ||
| Most Favored | 96.1% (122) | 96.1% (122) |
| Allowed | 3.9% (5) | 3.9% (5) |
| Disallowed | 0.0% (0) | 0.0% (0) |
Merging of small datasets from "multiple crystals" analyzed on-chip within a 24-well device at room temperature.
The "traditional" sample was grown using microbatch techniques and mounted using a standard crystal mount for cryogenic data collection. Reported values are for all hkls. Values in parenthesis represent the value for the highest resolution shell except where indicated for the number of observations as compared to unique reflections and R (Rfree) and for the Ramachandran statistics here the number in parenthesis indicates the number of residues in a given region. Data was analyzed over the range of 50 – 1.55Å to enable a direct comparison with the data collected from merging the diffraction data taken from multiple crystals at RT.
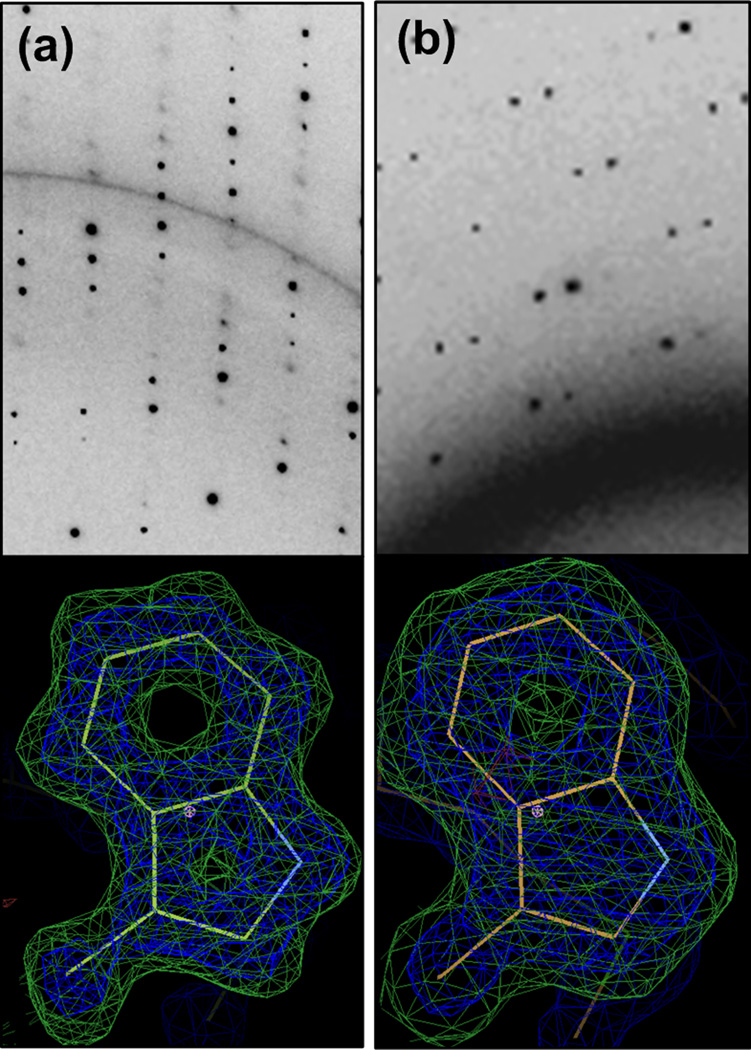
Analysis of the crystallographic parameters and statistical measures of quality for both data collection and structural refinement via molecular replacement show that the quality of the data was not significantly affected either by collection at room temperature or because of the merging of data from multiple crystals. The diffraction data was analyzed over the range of 50 – 1.55 Å (the diffraction limit for the on-chip data) to enable direct comparison between the other parameters. Other data collection parameters such as Rsym, completeness, redundancy, and I/σ and structural refinement parameters such as R/Rfree are comparable for the two cases, despite differences in crystal size, resolution, and signal-to-noise resulting from the presence of device materials. Interestingly, due the absence of both physical handling and cryo-cooling, the mosaicity for the room temperature merged data is nearly an order of magnitude lower (i.e., better) than the mosaicity of the single-crystal cryogenic datasets. Figure 6 compares the diffraction patterns and electron density maps obtained from the traditional, single-crystal cryogenic data and the on-chip, multi-crystal, room temperature data. The high resolution data obtained in each of these cases is evident from the electron density maps of, e.g., aromatic amino acid side chains with the rings clearly visible.
Figure 6.
X-ray diffraction data collected at LS-CAT, APS from (a) a cryo-cooled lysozyme crystal grown using the traditional microbatch method and mounted on a standard crystal mount, (b) a lysozyme crystal on a hybrid array chip at room temperature. Corresponding figures below show an omit map for the Trp 111 residue, the calculated map is displayed in blue (σ = 2.0) and the difference map is in green (σ = 3.0).
4. Conclusions
In summary, the microfluidic platform reported here allows for crystallization screening and subsequent on-chip X-ray data collection of crystallized proteins. The fabrication scheme uses a combination of COC and PDMS layers, and yields a platform that retains fluid routing capabilities via pneumatic valving while also achieving X-ray transparency. The dense network of integrated pneumatic valves allows for fast and easy setup of the crystallization screens, which in turn facilitates the reproducible growth of a large number of isomorphous crystals. The valves are only opened by vacuum actuation during filling and mixing. The valves are closed in rest, so the individual crystallization trials are isolated during crystallization and data collection in the absence of connections with ancillary equipment. The whole chip can be transported to and mounted in the X-ray beam for collection of diffraction data wedges from many crystals still residing on chip at room temperature. By merging these wedges of data a complete data set for full structure determination is obtained. This approach eliminates the need for crystal harvesting and cryo-cooling, while it simultaneously avoids the detrimental effects of the effects of radiation damage associated with room temperature data collection.
The screening capabilities of the platform were highlighted using a standard screen, followed by comparison of the results with the same screen performed via a traditional crystallization method. The platform was validated by collecting data from several proteins like lysozyme, thaumatin and ribonuclease A. By solving the structure of lysozyme using only diffraction data from many crystals residing on-chip we confirmed that the strategy of merging wedges of data from many crystals to obtain a complete dataset can be applied to these microfluidic platforms.
Going forward, the methodology reported here can be used to solve structure of novel proteins, especially those that (i) yield small, fragile crystals, (ii) have no known cryogenic crystallization conditions, and/or (iii) have been known to yield low resolution data using traditional cryogenic data collection. In addition to allowing for the routine collection of static structural information of proteins at biologically relevant temperatures as reported above, the fine control over transport phenomena coupled with on-chip analysis capabilities offered by these microfluidic platforms could enable a range of other biological studies. For example, precise microfluidic fluid handling could be utilized to enable dynamic structural studies based on the addition of various stimuli such as ligands, electrochemical agents, acid/base to affect the pH, or even changes temperature. Such studies have the potential to shed light on structural changes associated with protein function in unprecedented ways.
Supplementary Material
Acknowledgements
This work was funded by NIH (R01 GM086727) and a NIH Kirschstein Predoctoral Fellowship from the National Institute of Biomedical Imaging and Bioengineering (F31 EB008330). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817).
We would also like to thank Dr. Joseph Brunzelle, Dr. Keith Brister, and Dr. Spencer Anderson from LS-CAT at the Advanced Photon Source, Argonne National Laboratory for help in data collection at the 21-ID beamlines, Dr. Danielle Gray for assistance with X-ray experiments at the University of Illinois, as well as and Howard Robinson, Vivian Stojanoff, and Jean Jakoncic from the National Synchrotron Light Source at Brookhaven National Lab for their assistance with initial testing and development of these chips.
Biographies
Sudipto Guha received his B. Tech. degree in Chemical Engineering from the Indian Institute of Technology (IIT), Bombay. He is currently a PhD candidate at the University of Illinois at Urbana Champaign where his research focuses on the design and development of X-ray transparent microfluidic platforms for protein crystallography.
Sarah L. Perry received BS and MS degrees in Chemical Engineering as well as a BS in Chemistry from the University of Arizona, and her PhD degree from the University of Illinois at Urbana-Champaign. She is currently a post-doctoral researcher at the University of Chicago. Her research interests include the use of microfluidics for time resolved investigations of protein function and crystallogenesis as well as other self-assembling macromolecular systems.
Ashtamurthy S. Pawate received his B.Sc. in Physics, Chemistry and Math from Karnatak University, India, M.Sc. in Biotechnology from the Indian Institute of Technology (IIT), Bombay, and his Ph.D. in Biophysics and Computational Biology from the University of Illinois at Urbana-Champaign. He held a postdoctoral position at Sandia National Lab where he worked on metabolic pathway engineering of thermophilic ethanol-tolerant bacteria and diatoms. He is currently a postdoctoral researcher at the University of Illinois at Urbana-Champaign where he works on development of microfluidic platforms for biological applications.
Paul J.A. Kenis received his PhD in Chemical Engineering from Twente University, The Netherlands. After performing postdoctoral research at Harvard University he joined the faculty at the University of Illinois at Urbana-Champaign where he is currently a Professor and Head of the department of Chemical & Biomolecular Engineering, with affiliate appointments in the Beckman Institute, the Institute for Genomic Biology, the Frederick Seitz Material Research Laboratory, the Micro- & Nanotechnology Laboratory, and the departments of Bioengineering and Mechanical Science & Engineering. His research efforts include the development of microchemical systems: membraneless microfuel cells, microreactors (e.g. radiolabeling of biomolecules), microfluidic chips for pharmaceutical and membrane protein crystallization, platforms for biological cell studies (regenerative biology), sensors and valves for integrated microfluidic networks, and micro/nanofluidic tools for nanomanufacturing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schudel BR, Tanyeri M, Mukherjee A, Schroeder CM, Kenis PJA. Multiplexed detection of nucleic acids in a combinatorial screening chip. Lab on a Chip. 2011;11:1916–1923. doi: 10.1039/c0lc00342e. [DOI] [PubMed] [Google Scholar]
- 2.Zhang C, Xing D. Single-molecule DNA amplification and analysis using microfluidics. Chemical Reviews. 2010;110:4910–4947. doi: 10.1021/cr900081z. [DOI] [PubMed] [Google Scholar]
- 3.Lion N, Rohner TC, Dayon L, Arnaud IL, Damoc E, Youhnovski N, et al. Microfluidic systems in proteomics. ELECTROPHORESIS. 2003;24:3533–3562. doi: 10.1002/elps.200305629. [DOI] [PubMed] [Google Scholar]
- 4.Jebrail MJ, Wheeler AR. Let's get digital: Digitizing chemical biology with microfluidics. Current Opinion in Chemical Biology. 2010;14:574–581. doi: 10.1016/j.cbpa.2010.06.187. [DOI] [PubMed] [Google Scholar]
- 5.Wheeler TD, Zeng D, Desai AV, Onal B, Reichert DE, Kenis PJA. Microfluidic labeling of biomolecules with radiometals for use in nuclear medicine. Lab on a Chip. 2010;10:3387–3396. doi: 10.1039/c0lc00162g. [DOI] [PubMed] [Google Scholar]
- 6.Myers FB, Lee LP. Innovations in optical microfluidic technologies for point-of-care diagnostics. Lab on a Chip - Miniaturisation for Chemistry and Biology. 2008;8:2015–2031. doi: 10.1039/b812343h. [DOI] [PubMed] [Google Scholar]
- 7.Ohno KI, Tachikawa K, Manz A. Microfluidics: Applications for analytical purposes in chemistry and biochemistry. ELECTROPHORESIS. 2008;29:4443–4453. doi: 10.1002/elps.200800121. [DOI] [PubMed] [Google Scholar]
- 8.Whipple DT, Finke EC, Kenis PJA. Microfluidic reactor for the electrochemical reduction of carbon dioxide: The effect of pH. Electrochemical and Solid-State Letters. 2010;13:B109–B111. [Google Scholar]
- 9.Brushett FR, Hollinger AS, Markoski LJ, Kenis PJA. Microfluidic fuel cells as microscale power sources and analytical platforms. 2010:247–252. [Google Scholar]
- 10.Thorson MR, Goyal S, Schudel BR, Zukoski CF, Zhang GGZ, Gong Y, et al. A microfluidic platform for pharmaceutical salt screening. Lab on a Chip. 2011;11:3829–3837. doi: 10.1039/c1lc20645a. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Ismagilov RF. Protein Crystallization Using Microfluidic Technologies Based on Valves, Droplets, and SlipChip. Annual Review of Biophysics. 2010;39:139–158. doi: 10.1146/annurev.biophys.050708.133630. [DOI] [PubMed] [Google Scholar]
- 12.Perry SL, Roberts GW, Tice JD, Gennis RB, Kenis PJA. Microfluidic Generation of Lipidic Mesophases for Membrane Protein Crystallization. Crystal Growth & Design. 2009;9:2566–2569. doi: 10.1021/cg900289d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen CL, Skordalakes E, Berger JM, Quake SR. A robust and scalable microfluidic metering method that allows protein crystal growth by free interface diffusion. P Natl Acad Sci USA. 2002;99:16531–16536. doi: 10.1073/pnas.262485199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorson MR, Goyal S, Gong Y, Zhang GGZ, Zukoski CF, Kenis PJA. Microfluidic approach to polymorph screening through antisolvent crystallization. CrystEngComm. 2012;14:2404–2412. [Google Scholar]
- 15.Stone HA, Stroock AD, Ajdari A. Engineering flows in small devices: Microfluidics toward a lab-on-a-chip. Annual Review of Fluid Mechanics. 2004;36:381–411. [Google Scholar]
- 16.Hansen C, Quake SR. Microfluidics in structural biology: smaller, faster… better. Current Opinion in Structural Biology. 2003;13:538–544. doi: 10.1016/j.sbi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 17.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu HK, Schueller OJA, et al. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 18.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane) Anal Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 19.Dhouib K, Malek CK, Pfleging W, Gauthier-Manuel B, Duffait R, Thuillier G, et al. Microfluidic chips for the crystallization of biomacromolecules by counter-diffusion and on-chip crystal X-ray analysis. Lab Chip. 2009;9:1412–1421. doi: 10.1039/b819362b. [DOI] [PubMed] [Google Scholar]
- 20.Mehta G, Lee J, Cha W, Tung YC, Linderman JJ, Takayama S. Hard Top Soft Bottom Microfluidic Devices for Cell Culture and Chemical Analysis. Anal Chem. 2009;81:3714–3722. doi: 10.1021/ac802178u. [DOI] [PubMed] [Google Scholar]
- 21.Stevens RC. High-throughput protein crystallization. Current Opinion in Structural Biology. 2000;10:558–563. doi: 10.1016/s0959-440x(00)00131-7. [DOI] [PubMed] [Google Scholar]
- 22.Ostermeier C, Michel H. Crystallization of membrane proteins. Current Opinion in Structural Biology. 1997;7:697–701. doi: 10.1016/s0959-440x(97)80080-2. [DOI] [PubMed] [Google Scholar]
- 23.Durbin SD, Feher G. Protein crystallization. Annual Review of Physical Chemistry. 1996;47:171–204. doi: 10.1146/annurev.physchem.47.1.171. [DOI] [PubMed] [Google Scholar]
- 24.Juarez-Martinez G, Steinmann P, Roszak AW, Isaacs NW, Cooper JM. High-throughput screens for postgenomics: Studies of protein crystallization using microsystems technology. Analytical Chemistry. 2002;74:3505–3510. doi: 10.1021/ac0112519. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Fu Q, Kors C, Stewart L, Nollert P, Laible P, et al. A plug-based microfluidic system for dispensing lipidic cubic phase (LCP) material validated by crystallizing membrane proteins in lipidic mesophases. Microfluidics and Nanofluidics. 2009;8:789–798. doi: 10.1007/s10404-009-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kisselman G, Qiu W, Romanov V, Thompson CM, Lam R, Battaile KP, et al. X-CHIP: an integrated platform for high-throughput protein crystallization and on-the-chip X-ray diffraction data collection. Acta Crystallographica Section D. 2011;67:533–539. doi: 10.1107/S0907444911011589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng JD, Clark PJ, Stevens RC, Kuhn P. In situ X-ray analysis of protein crystals in low-birefringent and X-ray transmissive plastic microchannels. Acta Crystallographica Section D. 2008;64:189–197. doi: 10.1107/S0907444907060064. [DOI] [PubMed] [Google Scholar]
- 28.Hansen CL, Classen S, Berger JM, Quake SR. A microfluidic device for kinetic optimization of protein crystallization and in situ structure determination. J Am Chem Soc. 2006;128:3142–3143. doi: 10.1021/ja0576637. [DOI] [PubMed] [Google Scholar]
- 29.Sophianopoulos AJ, Holcomb DN, Vanholde KE, Rhodes CK. Physical Studies of Lysozyme .1. Characterization. J. Biol Chem. 1962;237:1107–1112. [PubMed] [Google Scholar]
- 30.Charron C, Giege R, Lorber B. Structure of thaumatin in a hexagonal space group: comparison of packing contacts in four crystal lattices. Acta Crystallogr D. 2004;60:83–89. doi: 10.1107/s0907444903022613. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Jaramillo FJ, Garcia-Ruiz JM, Gavira JA, Otalora F. Crystallization and cryocrystallography inside X-ray capillaries. J Appl Crystallogr. 2001;34:365–370. [Google Scholar]
- 32.Tessier PM, Johnson HR, Pazhianur R, Berger BW, Prentice JL, Bahnson BJ, et al. Predictive crystallization of ribonuclease A via rapid screening of osmotic second virial coefficients. Proteins. 2003;50:303–311. doi: 10.1002/prot.10249. [DOI] [PubMed] [Google Scholar]
- 33.Talreja S, Perry SL, Guha S, Bhamidi V, Zukoski CF, Kenis PJA. Determination of the Phase Diagram for Soluble and Membrane Proteins. The Journal of Physical Chemistry B. 2010;114:4432–4441. doi: 10.1021/jp911780z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Chung H, Caffrey M. The Curvature Elastic-Energy Function of the Lipid-Water Cubic Mesophase. Nature. 1994;368:224–226. doi: 10.1038/368224a0. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Evans P, McCoy A. An introduction to molecular replacement. Acta Crystallographica Section D. 2008;64:1–10. doi: 10.1107/S0907444907051554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaney MC, Maignan S, Ries-Kautt M, Ducruix A. High-Resolution Structure (1.33 A) of a HEW Lysozyme Tetragonal Crystal Grown in the APCF Apparatus. Data and Structural Comparison with a Crystal Grown under Microgravity from SpaceHab-01 Mission. Acta Crystallographica Section D. 1996;52:505–517. doi: 10.1107/S090744499501674X. [DOI] [PubMed] [Google Scholar]
- 39.Caffrey M. Crystallizing Membrane Proteins for Structure Determination: Use of Lipidic Mesophases. Annual Review of Biophysics. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. [DOI] [PubMed] [Google Scholar]
- 40.Caffrey M. On the Mechanism of Membrane Protein Crystallization in Lipidic Mesophases. Crystal Growth & Design. 2008;8:4244–4254. doi: 10.1021/cg101384p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cherezov V, Liu J, Griffith M, Hanson MA, Stevens RC. LCP-FRAP Assay for Pre-Screening Membrane Proteins for In Meso Crystallization. Crystal Growth & Design. 2008;8:4307–4315. doi: 10.1021/cg800778j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W, Hanson MA, Stevens RC, Cherezov V. LCP-Tm: An Assay to Measure and Understand Stability of Membrane Proteins in a Membrane Environment. Biophysical Journal. 2010;98:1539–1548. doi: 10.1016/j.bpj.2009.12.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sennoga C, Heron A, Seddon JM, Templer RH, Hankamer B. Membrane-protein crystallization in cubo: temperature-dependent phase behaviour of monoolein-detergent mixtures. Acta Crystallogr D. 2003;59:239–246. doi: 10.1107/s0907444902020772. [DOI] [PubMed] [Google Scholar]
- 44.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 45.Tang L, Lee NY. A facile route for irreversible bonding of plastic-PDMS hybrid microdevices at room temperature. 2010;10:1274–1280. doi: 10.1039/b924753j. [DOI] [PubMed] [Google Scholar]
- 46.Mohan R, Schudel BR, Desai AV, Yearsley JD, Apblett CA, Kenis PJA. Design considerations for elastomeric normally closed microfluidic valves. Sensors and Actuators B: Chemical. 2011 [Google Scholar]
- 47.Newman J, Xu J, Willis MC. Initial evaluations of the reproducibility of vapor-diffusion crystallization. Acta Crystallographica Section D. 2007;63:826–832. doi: 10.1107/S0907444907025784. [DOI] [PubMed] [Google Scholar]
- 48.Goh L, Chen K, Bhamidi V, He G, Kee NCS, Kenis PJA, et al. A Stochastic Model for Nucleation Kinetics Determination in Droplet-Based Microfluidic Systems. Crystal Growth & Design. 2010;10:2515–2521. doi: 10.1021/cg900830y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talreja S, Kenis PJA, Zukoski CF. A kinetic model to simulate protein crystal growth in an evaporation-based crystallization platform. Langmuir. 2007;23:4516–4522. doi: 10.1021/la063734j. [DOI] [PubMed] [Google Scholar]
- 50.Garman EF, Schneider TR. Macromolecular cryocrystallography. Journal of Applied Crystallography. 1997;30:211–237. [Google Scholar]
- 51.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, et al. The 2.6 Angstrom Crystal Structure of a Human A(2A) Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, et al. High-resolution crystal structure of an engineered human beta(2)-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.