Summary
The current genetic and recombination maps of the cat have less than 3,000 markers and a resolution limit greater than 1 Mb. To complement the first generation domestic cat maps, support higher resolution mapping studies, and aid genome assembly in specific areas as well as in the whole genome, a 15,000Rad radiation hybrid (RH) panel for the domestic cat was generated. Fibroblasts from the female Abyssinian cat that was used to generate the cat genomic sequence were fused to a Chinese hamster cell line (A23), producing 150 hybrid lines. The clones were initially characterized using 39 STR and 1536 SNP markers. The utility of whole genome amplification (WGA) in preserving and extending RH panel DNA was also tested using ten STR markers; no significant difference in retention was observed. The resolution of the 15,000Rad RH panel was established by constructing framework maps across ten different 1 Mb regions on different feline chromosomes. In these regions, two-point analysis was used to estimate RH distances, which compared favorably with the estimation of physical distances. The study demonstrates that the 15,000Rad RH panel constitutes a powerful tool for constructing high-resolution maps, having an average resolution of 40.1 kb per marker across the ten 1 Mb regions. In addition, the RH panel will complement existing genomic resources for the domestic cat, aid in the accurate reassemblies of the forthcoming cat genomic sequence, and support cross-species genomic comparisons.
Introduction
Every genome needs a good map [Lewin et al. 2009]. The accuracy and density of genetic map construction for any particular species has improved with the evolution of molecular biology, cell culture, and genetic and genomic techniques. Chromosomal abnormalities, somatic cell hybrid maps, and family-based linkage analyses are techniques that have increased map density and can decipher gene order. Radiation hybrid (RH) mapping provided an additional leap forward as polymorphic markers were not required and resolution could be manipulated by radiation dosage. Sanger-based and next-generation sequencing now suggest the finest levels of resolution, density, and order, although correct assembly is complicated and not error free. Overall, each independent technique has benefitted from the previously built framework maps and all mapping efforts have combined to improve accuracy of orders and distances of genes in the genome.
Successful RH mapping in humans [Cox et al. 1989] pioneered RH mapping efforts in many species such as, rat, dog, cow, sheep, horse, pig, and macaque [Chowdhary et al. 2002; Laurent et al. 2007; Murphy et al. 2001; Priat et al. 1998; Watanabe et al. 1999; Womack et al. 1997; Yerle et al. 1998] and non-mammalian vertebrate models including the zebrafish [Geisler et al. 1999; Hukriede et al. 1999]. RH panels were generated for several species initially as lower resolution RH panels, generally less than 5,000Rad for initial genome map construction; later, higher resolution panels, generally 10,000Rad or greater, were developed for several species and used in fine mapping and genome assembly. For example, RH maps for the human genome have increased from 3,000Rad to 50,000Rad, improving resolution from approximately 1.2 Mb to an average of 94 kb between ordered markers [Gyapay et al. 1996; Hudson et al. 1995; Olivier et al. 2001]. Comparisons to other mapping techniques, such as recombination maps and genome assemblies, have indicated that each method has value and that cross-method comparisons will help to generate the ultimate map, correct in orientation, order, and distance between loci.
The domestic cat has become a useful model organism for the analysis of many diseases and disorders that occur in the human population [Lyons et al. 2004; Menotti-Raymond et al. 2010; Meurs et al. 2007; Rah et al. 2005]. To facilitate the cat’s role as a model for human disease, genetic and genomic resources for the cat have been in production for nearly three decades, progressing from somatic cell hybrid panels [O’Brien and Nash 1982], interspecies and intra-species linkage maps [Menotti-Raymond et al. 1999; Grahn et al. 2005; Cooper et al. 2006], integration with radiation hybrid panels [Menotti-Raymond et al. 2003b; Sun et al. 2001], and currently to complete genome sequencing.
Next-generation sequencing techniques are more efficient in the capture of regions of the genome that had been missed by earlier technologies [Wheeler et al. 2008], however, the assembly of sequence data generated by non-Sanger based, massively parallel sequencing technologies faces unique challenges due to the short read lengths produced [Pop 2009] and higher error rates [Xu et al. 2009]. The existing Sanger-based genome assembly is necessary as a scaffold to map new sequence data [Wheeler et al. 2008]. Although an excellent initial resource, the coverage of the cat genome captured by the 1.9x sequencing effort of the domestic cat was approximately 60% [Pontius et al. 2007]; additional light sequencing of the cat genome for SNP discovery increased depth to 3x and expanded coverage to approximately 80% of the genome [Mullikin et al. 2010]. Despite the expanded coverage, low contig and short scaffold N50s of 4.6 kb and 162 kb, respectively, suggest that significant gaps in the initial cat sequence assemblies could prevent the formation of continuous contigs with significant length. Next-generation sequencing of the domestic cat to obtain 10 – 13x coverage is nearing completion (see the Felis catus entry in http://www.genome.gov/10002154). However, since the feline 3x sequence contains significant gaps in coverage and multiple unaligned contigs and the scaffold was based on the canine assembly, accurate assembly of the deeper non-Sanger based feline genome sequence will be highly challenging. RH maps have played an important role in facilitating the process of whole genome sequencing and assembly for human, mouse, rat, dog and cattle [Lander et al. 2001; Waterston et al. 2002; Gibbs et al. 2007; Lindblad-Toh et al. 2005; Bovine Genome Sequencing and Analysis Consortium et al. 2009]. To date, the available maps for the cat have less than 3,000 markers [Davis et al. 2009; Menotti-Raymond et al. 2009] with a resolution of ~1 Mb, thus the generation of a high-density map is of great importance to improve the power of studies that require fine-mapping and physical mapping approaches to identify regions of interest. A high-density and high-resolution genome map could strongly support the placement of contigs and resolve placement of the singleton contigs that have yet to be assigned to a chromosome.
To complement the previous physical maps and further resolve the structure of the domestic cat genome, reported is the construction and initial characterization of a 15,000Rad whole-genome RH panel suitable for high-resolution mapping.
Materials and methods
Cell line fusion and panel construction
A 15,000Rad RH panel was generated using a fibroblast donor primary culture derived from a female Abyssinian cat fused with an A23 thymidine kinase (TK-) deficient Chinese hamster fibroblast cell line. Fusion, isolation, selection, and initial cloning procedures were as previously described [Chowdhary et al. 2002] with the following modification: 30 sequential doses of 500 cGy (rad) were administered to the donor cells for a total absorbed dose of 150Gy (15,000 cGy) using a 6MV photon beam produced by a linear accelerator (Varian 2100C, Varian Corporation, Palo Alto CA). Dosages were calculated using a computer software program (IMSure, version 1.22, Prodigm, Inc. Chico, CA). Hybrid cell lines (N = 204) were expanded in T25 flasks (Thermo Fisher Scientific, Rochester NY USA) in HATO medium containing DMEM medium with 10% FBS, 1X antibiotic-antimycotic (Invitrogen, Carlsbad CA USA), 1X HAT (Invitrogen), and 1X Ouabain (Sigma-Aldrich, St. Louis MO USA). At the second passage (p2), approximately two thirds of the cells of each hybrid clone were cryopreserved in freezing medium containing 10% DMSO; the remaining cells were returned to T25 flasks (p3), allowed to reach confluency, then expanded in triplicate T75 flasks (p4) and harvested and cryopreserved at confluency. One hundred and fifty clones were successfully cultured and expanded to p4. DNA was isolated from approximately 10 μl of each cell pellet using the Qiagen DNeasy kit (Valencia, CA USA) according to manufacturer’s protocol. In addition, each DNA sample was whole-genome amplified using the Qiagen REPLI-g Mini Kit (Valencia, CA USA) following the amplification of genomic DNA from blood or cells protocol with 3 μl extracted DNA as the template.
Cat DNA retention studies
Isolated DNA from RH clones was tested for total DNA concentration using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE USA) (supplementary table 1). Thirty-nine STR markers [Menotti-Raymond et al. 2003a; Menotti-Raymond et al. 1999; Menotti-Raymond et al. 2003b] (supplementary table 2) were multiplexed and tested with the 150 RH clones as previously described [Lipinski et al. 2008]. For a validation test, ten markers were tested on both WGA and non-WGA DNA samples (supplementary table 3). In each reaction, 2 μl WGA DNA (diluted 1:10) or non-WGA DNA was amplified as previously described [Lipinski et al. 2008]. All STR genotypes were analyzed using STRand software ([Toonen and Hughes 2001], http://www.vgl.ucdavis.edu/STRand) for analysis using semi-automatic calling of alleles. An internal size standard (GeneScan 500 LIZ, Applied Biosystems) was used to determine allele size for each STR. Allele ranges for each marker were established from genotyping the DNA of the donor cat. Additionally, both WGA and non-WGA DNA from five clones were genotyped with 1,536 SNPs via the Illumina GoldenGate assay as described below (data not shown). Retention frequencies for non-WGA and WGA DNA samples were tested for significant difference between the two conditions using Pearson’s chi square test [Pearson 1900].
SNP typing for map construction
Whole-genome amplified DNA from the 150 cell lines and the host hamster and donor cat DNA samples were genotyped for 1,536 SNPs using the Illumina GoldenGate genotyping assay analyzed on an Illumina® BeadStation 500G ([Oliphant et al. 2002]; http://www.illumina.com) following the manufacturer’s protocol at the Genetics Core Laboratory at the Texas Biomedical Research Institute. SNPs were selected within random 1 Mb regions on 10 different cat chromosomes (A1, A2, B3, C2, D1, D2, D4, E2, F2 and X) using data from the Felis catus sequencing effort [Mullikin et al. 2010]. Each region contains ~ 153 SNPs, having a geometric distribution of the SNPs in the region, averaging one SNP per 6 kb. GenomeStudio 2010.2 version software was used to manually score the presence or absence of loci across the RH panel clones. Data was filtered by considering the quality of the SNPs and the quality of the clones, as previously described [McKay et al. 2007]. For the primary filtering, SNPs were not scored in the clones if the SNP successfully amplified in the hamster and could not be distinguished from the cat or if the SNP failed to amplify in the donor cat DNA. Calling thresholds for the SNPs that amplified within a clone were absent (0) for call scores below 0.10, unknown (−) for call scores between 0.1 and 0.15, and present (1) for call scores above 0.15. Finally, secondary filtering removed markers with an “unknown” rate of more than 5%.
RH map construction
The filtered data from each region was used to construct linkage groups and framework maps in CarthaGene-1.2 [de Givry et al. 2005]. Markers were tested to detect duplicate RH vectors using the “merge” command. For markers within a given 1 Mb region, linkage groups were formed by two-point analysis with a threshold LOD score > 3.0. Framework maps were constructed using the “buildfw” command with a saving threshold and an adding threshold of 3. The resulting maps were refined with the commands “flips” (7 1 1), “polish”, and “greedy” (3 1 1 15 25) and tested by “robustness” (−3). For each chromosomal region, ten maps with the highest likelihood were examined to confirm the reliability of major changes detected within each region.
Results
RH panel characterization
Two hundred and four initial cat-hamster fusion lines were identified and propagated, 150 clones were successfully cultured to at least passage 4 and were further characterized to develop the RH panel. To verify accurate representation of clone DNA in WGA samples, ten STRs were genotyped on WGA and non-WGA template DNA from the same clones (supplementary table 3). Average retention frequencies (RFs) between non-WGA and WGA DNA were 28.7% and 27.3%, respectively, and the percentage of clones with greater than 10% RF were 96.7% and 92.7%, respectively (table 1). Comparing presence or absence of each marker for each clone, 3% discordancy was detected between the WGA and non-WGA DNA (supplementary table 3). In addition, five non-WGA DNA clones were tested on 1536 SNPs and no significant differences were detected (data not shown).
Table 1.
Retention frequencies for the feline 15,000Rad RH panel calculated by STR and SNP analysis.
| Non-WGA Test STR | WGA Test STR | STR | SNP | |
|---|---|---|---|---|
| N markers | 10 | 10 | 39 | 794 |
| Average RF | 28.7% | 27.4% | 21% | 29.4% |
| N clones < 10% RF | 5 | 11 | 18 | 14 |
| N clones 10% – 25% RF | 37 | 63 | 53 | 62 |
| N clones > 25% RF | 108 | 76 | 79 | 74 |
|
| ||||
| Total clones | 150 | 150 | 150 | 150 |
|
| ||||
| % of clones > 10% RF | 96.7% | 92.7% | 88% | 90.7% |
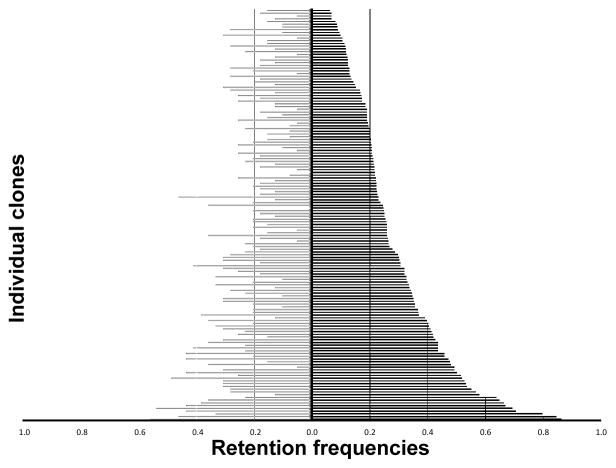
Each clone was tested with 39 STR markers distributed across all 18 autosomes and the X chromosome for initial characterization of the panel. Marker RFs are summarized in table 1 and the data matrix is presented in supplementary table 4. Marker RFs ranged from 0.00% to 54%, with an average of 21% for the panel. Clones C027 and C184 were not positive for any STRs, and clone C087 had the highest RF at 54%. Of the 150 clones tested, 88% had RFs greater than 10%. No markers were located near the selectable TK and HPRT loci. Figure 1 shows the graphical representation of RFs for each clone summarized in supplementary table 1, and supplementary fig. 1 illustrates the distribution of markers within clones.
Fig. 1.
RH clone retention frequencies estimated by STR and SNP analysis. Each bar represents one clone. Estimates of RF calculated by STR analysis are indicated by gray bars to the left on the graph, and estimates of RF calculated by SNP analysis are indicated by black bars to the right on the graph.
Further characterization of the 150 clones was accomplished via high-throughput SNP genotyping. Of the 1536 SNPs included in the Illumina GoldenGate assay, 921 (59.96%) passed primary filtering and were successfully typed. Seventy-five SNPs (4.88 %) failed in the donor cat and 540 SNPs (35.15%) were positive in hamster, thus genotyping was not possible. Of the 921 SNPs that passed primary filtering, 127 SNPs failed secondary filtering, resulting in 794 SNPs available for analysis. For the SNP markers, the average RF was 29.4%, and the percentage of clones with greater than 10% RF was 90.7% (table 1). Clone RFs estimated by SNP genotyping are summarized in supplementary table 1 and illustrated in fig. 1. The clones that were negative for all STRs had RFs > 20% for the SNPs. A general trend of similar RFs was observed between STRs and SNPs (fig. 1).
RH map construction
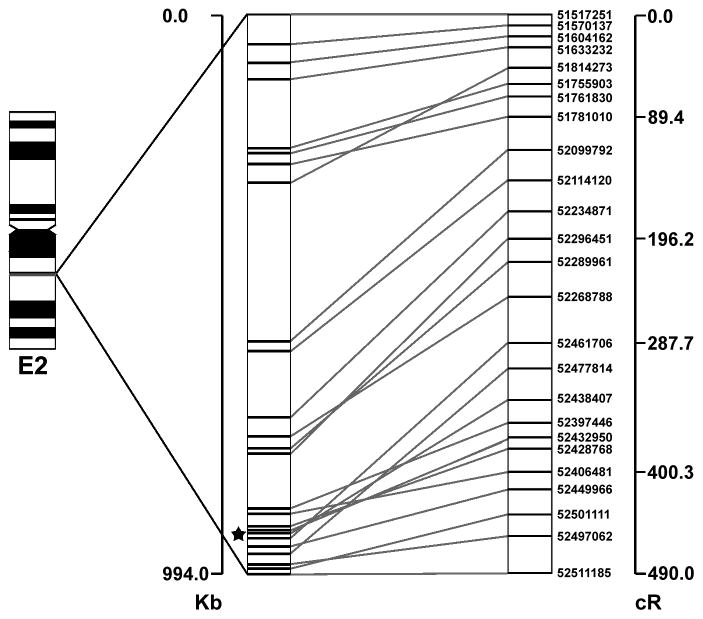
The 794 SNPs that passed quality control were used to construct framework RH maps (table 2). No SNPs had identical RH vectors as determined by the “merge” command. The 794 markers formed ten linkage groups, one per chromosome region. Framework maps included 181 of the 794 (22.76%). The RH maps with best likelihoods obtained from the ten initial 1 Mb regions are shown in supplementary fig. 2 and summarized in table 2. A representative map for Chromosome E2 is presented as fig. 1. LOD scores to the next nearest marker in a framework map were generally above 20, ranging from 12.7 – 37.7. Major inversions were suggested on chromosomes A2, B3, D1, and D2, which were consistent across all 10 highest likelihood maps for each region. The overall SNP order, beside the noted inversions, was conserved across all maps. However, in every map, some SNPs showed a different location in the RH map as compared to the positions identified by the sequence assembly. The length of the RH maps ranged from 178.9 cR15,000 to 489 cR15000 with an average kb to cR ratio of 2.4 kb/cR15,000 and an average inter-marker distance of 40.1 kb (table 2). Chromosome E2 (fig. 2) and D4 were the most densely mapped chromosomes, with 25 markers assigned to each 1 Mb region and lengths of 489 cR15,000 and 425.4 cR15,000. Conversely, B3 was the most sparsely mapped chromosome, with 11 markers on the framework map and a length of 178.9 cR15,000. The average map lengths in kb and in cR15,000 as well kb/cR15,000 are shown in table 2.
Table 2.
High-throughput SNP genotyping summary and framework map marker summary.
| SNPs | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Chr. | SNPs No. | Failed in donor cat | Positive in hamster | > 5% Unknown | No. for map | No. in FWM | Map Length (kb) | Map length cR15000 | kb/cR15000 | kb/marker | Number of genes |
|
|
|||||||||||
| A1 | 154 | 4 | 51 | 49 | 48 | 18 | 955 | 351.5 | 2.7 | 53.1 | ~3 |
| A2 | 155 | 9 | 69 | 6 | 71 | 19 | 491 | 267 | 1.8 | 25.8 | ~6 |
| B3 | 153 | 5 | 76 | 7 | 67 | 11 | 164 | 178.9 | 0.9 | 14.9 | ~7 |
| C2 | 154 | 12 | 52 | 21 | 69 | 14 | 805 | 206.5 | 3.9 | 57.5 | ~7 |
| D1 | 154 | 3 | 69 | 8 | 74 | 19 | 345 | 211.2 | 1.6 | 18.2 | ~3 |
| D2 | 152 | 4 | 67 | 4 | 77 | 17 | 751 | 250.6 | 3 | 44.2 | ~18 |
| D4 | 153 | 6 | 32 | 9 | 106 | 25 | 864 | 425.4 | 2 | 34.6 | ~18 |
| E2 | 154 | 7 | 40 | 7 | 100 | 25 | 994 | 489 | 2 | 39.8 | ~2 |
| F2 | 154 | 12 | 45 | 13 | 84 | 18 | 905 | 324.4 | 2.8 | 50.3 | ~4 |
| X | 153 | 13 | 39 | 3 | 98 | 15 | 944 | 264 | 3.6 | 62.9 | ~6 |
|
| |||||||||||
| Total | 1536 | 75 | 540 | 127 | 794 | 181 | 8947 | 2968 | ~74 | ||
| Average | 18.1 | 894.7 | 296.9 | 2.4 | 40.1 | ~7.4 | |||||
Fig. 2.
Framework RH map of feline chromosome E2. Ideogram of the chromosome is shown on the left, the estimated position of each marker is shown in the center, and the framework 15,000Rad map is shown on the right. The star indicates one large rearrangement in marker order between the sequence data and the RH map.
Discussion
The domestic cat is increasingly recognized as a robust animal model for human diseases. Expanding resources, particularly the genome sequencing projects and the resulting SNP discoveries, should facilitate candidate gene identification and analyses. The anticipated deeper coverage should accelerate investigations of simple phenotypes and enable analyses of more complex traits in the cat. However, mutation detection studies are hampered by incorrect gene orders caused by statistical fluctuations in genetic maps and incorrect genome assemblies. Even with very high coverage-derived assemblies of humans and mice, imperfections and artifacts in gene order and distances are a recognized hazard [Kong et al. 2002], which can be resolved with the generation of a high-resolution high-density RH map [Marques et al. 2007].
Besides the genome sequence, many excellent resources are currently available for the domestic cat supporting the identification of diseases and traits. The current cat 5,000Rad RH map contains over 2,000 markers with a resolution of ~1 Mb, which supported the 1.9x and 3x cat genome assemblies. The use of a more robust whole genome assembly from the most closely related species is a common practice for comparative assemblies, as has been demonstrated in the bovine genome [Prasad et al. 2007]. The dog scaffold was used where available to direct the cat assembly below the 1 Mb level, introducing bias towards the dog genome in the cat gene order and inherently incorporating errors into the cat sequence assembly [Pontius et al. 2007]. Thus, a high-resolution gene map for the cat, developed by an independent technology such as the radiation hybrid technique, should augment genome assembly for the cat, thereby facilitating fine mapping and candidate gene studies.
The overall goal was to produce a high-resolution RH map of the cat to facilitate genome assembly. The same Abyssinian cat was used to generate the genome sequence and construct the RH panel to alleviate concerns regarding individual sequence rearrangements and variations due to repeat regions. The cat RH 15,000Rad panel is composed of 150 RH clones, which supports mapping of closely spaced markers. Marker retention has been shown to vary across the genome in RH panels [Cox et al. 1990; James et al. 1994; Walter et al. 1994] and can be estimated with different technologies, such as STRs and high-throughput SNP genotyping. An initial retention frequency (RF) estimate of 21% was determined by testing 39 STRs, with 88% of clones having greater than 10% RF. As expected, this RF estimate is lower than the initial estimate of the 5,000Rad panel [Murphy et al. 1999] but similar to RFs obtained from other species RH panels with high radiation dose, such as the 12,000Rad ovine panel, which reported 87.5% of clones with RF greater than 10% [Laurent et al. 2007]. Retention frequencies were also estimated from the SNP analysis and illustrate the limitations of typing a high-resolution RH panel with few markers. The RF estimate from SNP analysis is higher than that estimated by STR analysis, with 29.4% RF and 90.7% of clones greater than 10% RF. By testing more loci, a more complete estimate of RF can be calculated, which reflects the higher retention expected from using a low-passage RH panel. Both STR and SNP analyses demonstrate the fluctuation in retention in different regions of the genome. Using these data, a subset of 94 clones could easily be selected that has an optimum average retention frequency across the cat genome.
The newer SNP genotyping technologies should support the construction of high-density genetic maps. For this study, Illumina technology was used to genotype 1536 SNPs on the cat RH panel. The initial selection of SNPs did not consider conservation to hamster, implying the amplification of some SNPs was expected in the hamster background. Approximately 35% of SNPs amplified in the hamster, precluding their analysis in the cat RH panel. In addition, this analysis of the SNPs is the first genotyping of these particular loci in the cat, thus a 5% failure rate, as determined by failure in the donor cat DNA, was expected due to design and assay failure. Approximately 8% of SNPs failed to amplify robustly, resulting in ambiguous genotyping. Overall, 49% (794) of SNPs were suitable for map construction, supporting the rapid development of a high-density map.
The current 5,000Rad RH panel has a resolution limit exceeding 1 Mb, thus, below this level, the cat genome assembly does not have a secondary mapping method to support assembly or resolve discrepancies. The ten framework maps constructed had an average inter-marker distance of 40.1 kb, suggesting a significant 25-fold increase in resolution as compared to the 5,000Rad feline RH map, and will support genome assembly below 1 Mb. Conservative framework maps for the 15,000Rad cat RH panel were constructed with 181 SNPs, averaging 18 markers covering an average of 900 kb. A majority of markers in the framework maps had consistent order with the suggested SNP positioning. However, several large inversions on five of 10 chromosomes were identified, which were supported by all 10 maps with the highest likelihoods. Each 1 Mb region also had smaller potential inversions. The SNPs were selected and positioned from unpublished versions of the cat genome assembly, therefore the more robust assembly may show more concordance with the RH maps. In addition, large-scale SNP calling methods may need refinement to improve accuracy and objectivity in assigning call scores for RH data, as amplification levels fluctuate unpredictably between markers and minor differences in call rates for even one marker can dramatically change the final marker order of a map.
A recognized challenge of RH panels is that as fused RH clones continue to grow and divide, donor DNA is progressively lost [Karere et al. 2010], continually changing the profile of markers present at each passage. Thus, to be comparable, gene maps must be constructed using DNA from the same passage for a particular clone, which often limits the amount of DNA available for mapping. In addition, large-scale culture steps are required to prepare DNA in sufficient quantities for distribution to multiple laboratories and for large-scale marker typing. The efficiency of RH mapping should be increased by using early passage WGA DNA samples, which have higher retention frequencies and sufficient amounts of DNA. Moreover, WGA may also amplify low-copy fragments that would have otherwise been lost to the analysis. To avoid a significant reduction in the retention frequency, and to make the resource available for the community, each clone was whole genome amplified. Promising results with WGA DNA were obtained on a 10,000Rad panel for the Rhesus macaque [Karere et al. 2010] and a 3,000Rad panel for the gilthead seabream [Senger et al. 2006]. In this cat RH panel, WGA was a reliable method for genome amplification as retention frequencies were comparable between WGA and non-WGA DNA, and only 3% discordancy was detected between STR amplification in WGA versus non-WGA DNA. The low requirements of DNA concentration and sample volume for SNP array technologies and the extensive amplification of RH clone DNA by WGA suggests that the cat 15,000Rad panel should be a sufficient resource for additional mapping on higher density arrays and fine mapping specific regions.
The ultimate map of a genome is not just the complete sequence of the organism, but also the correctly assembly of the sequence with chromosomal assignments and orientation. Thus, the production of a whole-genome sequence does not dismiss the need for accurate and efficient mapping techniques. The cat genome sequencing coverage is shallow at ~3x, and the highly fragmented sequence assembly further highlights the need for high-resolution RH maps in the species. This cat 15,000Rad RH panel should provide support for the continued refinement of the domestic cat genome assembly, particularly since the deeper cat sequence was developed with shorter read technologies that have higher error rates. With a comprehensive high-density, high-resolution RH map, marker order rearrangements can be identified and resolved and cross-species genome comparisons can be accomplished, providing a powerful tool for inferring genome function and evolutionary history.
Supplementary Material
Acknowledgments
Funding for this project was provided by NIH-NCRR RR016094, the Center for Companion Animal Health at UC Davis, the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health, and the George and Phyllis Miller Feline Health Fund of the San Francisco Foundation. This investigation was partially conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR013556 from NCRR, NIH. Cat genome sequence for SNP detection was provided by Dr. Wesley Warren of the Genome Institute at Washington University School of Medicine. The authors would like to thank Paul Lathrop and Hasan Alhaddad for technical support and advice.
References
- Elsik CG, Tellam RL, Worley KC, Gibbs RA, Muzny DM, Weinstock GM, Adelson DL, Eichler EE, Elnitski L, Guigo R, Hamernik DL, Kappes SM, Lewin HA, Lynn DJ, Nicholas FW, Reymond A, Rijnkels M, Skow LC, Zdobnov EM, Schook L, Womack J, Alioto T, Antonarakis SE, Astashyn A, Chapple CE, Chen HC, Chrast J, Camara F, Ermolaeva O, Henrichsen CN, Hlavina W, Kapustin Y, Kiryutin B, Kitts P, Kokocinski F, Landrum M, Maglott D, Pruitt K, Sapojnikov V, Searle SM, Solovyev V, Souvorov A, Ucla C, Wyss C, Anzola JM, Gerlach D, Elhaik E, Graur D, Reese JT, Edgar RC, McEwan JC, Payne GM, Raison JM, Junier T, Kriventseva EV, Eyras E, Plass M, Donthu R, Larkin DM, Reecy J, Yang MQ, Chen L, Cheng Z, Chitko-McKown CG, Liu GE, Matukumalli LK, Song J, Zhu B, Bradley DG, Brinkman FS, Lau LP, Whiteside MD, Walker A, Wheeler TT, Casey T, German JB, Lemay DG, Maqbool NJ, Molenaar AJ, Seo S, Stothard P, Baldwin CL, Baxter R, Brinkmeyer-Langford CL, Brown WC, Childers CP, Connelley T, Ellis SA, Fritz K, Glass EJ, Herzig CT, Iivanainen A, Lahmers KK, Bennett AK, Dickens CM, Gilbert JG, Hagen DE, Salih H, Aerts J, Caetano AR, Dalrymple B, Garcia JF, Gill CA, Hiendleder SG, Memili E, Spurlock D, Williams JL, Alexander L, Brownstein MJ, Guan L, Holt RA, Jones SJ, Marra MA, Moore R, Moore SS, Roberts A, Taniguchi M, Waterman RC, Chacko J, Chandrabose MM, Cree A, Dao MD, Dinh HH, Gabisi RA, Hines S, Hume J, Jhangiani SN, Joshi V, Kovar CL, Lewis LR, Liu YS, Lopez J, Morgan MB, Nguyen NB, Okwuonu GO, Ruiz SJ, Santibanez J, Wright RA, Buhay C, Ding Y, Dugan-Rocha S, Herdandez J, Holder M, Sabo A, Egan A, Goodell J, Wilczek-Boney K, Fowler GR, Hitchens ME, Lozado RJ, Moen C, Steffen D, Warren JT, Zhang J, Chiu R, Schein JE, Durbin KJ, Havlak P, Jiang H, Liu Y, Qin X, Ren Y, Shen Y, Song H, Bell SN, Davis C, Johnson AJ, Lee S, Nazareth LV, Patel BM, Pu LL, Vattathil S, Williams RL, Jr, Curry S, Hamilton C, Sodergren E, Wheeler DA, Barris W, Bennett GL, Eggen A, Green RD, Harhay GP, Hobbs M, Jann O, Keele JW, Kent MP, Lien S, McKay SD, McWilliam S, Ratnakumar A, Schnabel RD, Smith T, Snelling WM, Sonstegard TS, Stone RT, Sugimoto Y, Takasuga A, Taylor JF, Van Tassell CP, Macneil MD, Abatepaulo AR, Abbey CA, Ahola V, Almeida IG, Amadio AF, Anatriello E, Bahadue SM, Biase FH, Boldt CR, Carroll JA, Carvalho WA, Cervelatti EP, Chacko E, Chapin JE, Cheng Y, Choi J, Colley AJ, de Campos TA, De Donato M, Santos IK, de Oliveira CJ, Deobald H, Devinoy E, Donohue KE, Dovc P, Eberlein A, Fitzsimmons CJ, Franzin AM, Garcia GR, Genini S, Gladney CJ, Grant JR, Greaser ML, Green JA, Hadsell DL, Hakimov HA, Halgren R, Harrow JL, Hart EA, Hastings N, Hernandez M, Hu ZL, Ingham A, Iso-Touru T, Jamis C, Jensen K, Kapetis D, Kerr T, Khalil SS, Khatib H, Kolbehdari D, Kumar CG, Kumar D, Leach R, Lee JC, Li C, Logan KM, Malinverni R, Marques E, Martin WF, Martins NF, Maruyama SR, Mazza R, McLean KL, Medrano JF, Moreno BT, More DD, Muntean CT, Nandakumar HP, Nogueira MF, Olsaker I, Pant SD, Panzitta F, Pastor RC, Poli MA, Poslusny N, Rachagani S, Ranganathan S, Razpet A, Riggs PK, Rincon G, Rodriguez-Osorio N, Rodriguez-Zas SL, Romero NE, Rosenwald A, Sando L, Schmutz SM, Shen L, Sherman L, Southey BR, Lutzow YS, Sweedler JV, Tammen I, Telugu BP, Urbanski JM, Utsunomiya YT, Verschoor CP, Waardenberg AJ, Wang Z, Ward R, Weikard R, Welsh TH, Jr, White SN, Wilming LG, Wunderlich KR, Yang J, Zhao FQ Bovine Genome Sequencing and Analysis Consortium. The genome sequence of taurine cattle: A window to ruminant biology and evolution. Science. 2009;324:522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary BP, Raudsepp T, Honeycutt D, Owens EK, Piumi F, Guerin G, Matise TC, Kata SR, Womack JE, Skow LC. Construction of a 5000(rad) whole-genome radiation hybrid panel in the horse and generation of a comprehensive and comparative map for eca11. Mamm Genome. 2002;13:89–94. doi: 10.1007/s00335-001-2089-8. [DOI] [PubMed] [Google Scholar]
- Cooper MP, Fretwell N, Bailey SJ, Lyons LA. White spotting in the domestic cat (felis catus) maps near kit on feline chromosome b1. Anim Genet. 2006;37:163–165. doi: 10.1111/j.1365-2052.2005.01389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR, Burmeister M, Price ER, Kim S, Myers RM. Radiation hybrid mapping: A somatic cell genetic method for constructing high-resolution maps of mammalian chromosomes. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- Cox DR, Pritchard CA, Uglum E, Casher D, Kobori J, Myers RM. Segregation of the huntington disease region of human chromosome 4 in a somatic cell hybrid. Genomics. 1989;4:397–407. doi: 10.1016/0888-7543(89)90347-9. [DOI] [PubMed] [Google Scholar]
- Davis BW, Raudsepp T, Pearks Wilkerson AJ, Agarwala R, Schaffer AA, Houck M, Chowdhary BP, Murphy WJ. A high-resolution cat radiation hybrid and integrated fish mapping resource for phylogenomic studies across felidae. Genomics. 2009;93:299–304. doi: 10.1016/j.ygeno.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Givry S, Bouchez M, Chabrier P, Milan D, Schiex T. Carhta gene: Multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics. 2005;21:1703–1704. doi: 10.1093/bioinformatics/bti222. [DOI] [PubMed] [Google Scholar]
- Geisler R, Rauch GJ, Baier H, van Bebber F, Bross L, Dekens MP, Finger K, Fricke C, Gates MA, Geiger H, Geiger-Rudolph S, Gilmour D, Glaser S, Gnügge L, Habeck H, Hingst K, Holley S, Keenan J, Kirn A, Knaut H, Lashkari D, Maderspacher F, Martyn U, Neuhauss S, Neumann C, Nicolson T, Pelegri F, Ray R, Rick JM, Roehl H, Roeser T, Schauerte HE, Schier AF, Schönberger U, Schönthaler HB, Schulte-Merker S, Seydler C, Talbot WS, Weiler C, Nüsslein-Volhard C, Haffter P. A radiation hybrid map of the zebrafish genome. Nature genetics. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y, Dinh HH, Dugan-Rocha S, Fulton LA, Gabisi RA, Garner TT, Godfrey J, Hawes AC, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Kirkness EF, Cree A, Fowler RG, Lee S, Lewis LR, Li Z, Liu YS, Moore SM, Muzny D, Nazareth LV, Ngo DN, Okwuonu GO, Pai G, Parker D, Paul HA, Pfannkoch C, Pohl CS, Rogers YH, Ruiz SJ, Sabo A, Santibanez J, Schneider BW, Smith SM, Sodergren E, Svatek AF, Utterback TR, Vattathil S, Warren W, White CS, Chinwalla AT, Feng Y, Halpern AL, Hillier LW, Huang X, Minx P, Nelson JO, Pepin KH, Qin X, Sutton GG, Venter E, Walenz BP, Wallis JW, Worley KC, Yang SP, Jones SM, Marra MA, Rocchi M, Schein JE, Baertsch R, Clarke L, Csuros M, Glasscock J, Harris RA, Havlak P, Jackson AR, Jiang H, Liu Y, Messina DN, Shen Y, Song HX, Wylie T, Zhang L, Birney E, Han K, Konkel MK, Lee J, Smit AF, Ullmer B, Wang H, Xing J, Burhans R, Cheng Z, Karro JE, Ma J, Raney B, She X, Cox MJ, Demuth JP, Dumas LJ, Han SG, Hopkins J, Karimpour-Fard A, Kim YH, Pollack JR, Vinar T, Addo-Quaye C, Degenhardt J, Denby A, Hubisz MJ, Indap A, Kosiol C, Lahn BT, Lawson HA, Marklein A, Nielsen R, Vallender EJ, Clark AG, Ferguson B, Hernandez RD, Hirani K, Kehrer-Sawatzki H, Kolb J, Patil S, Pu LL, Ren Y, Smith DG, Wheeler DA, Schenck I, Ball EV, Chen R, Cooper DN, Giardine B, Hsu F, Kent WJ, Lesk A, Nelson DL, O’Brien WE, Prufer K, Stenson PD, Wallace JC, Ke H, Liu XM, Wang P, Xiang AP, Yang F, Barber GP, Haussler D, Karolchik D, Kern AD, Kuhn RM, Smith KE, Zwieg AS. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Grahn RA, Lemesch BM, Millon LV, Matise T, Rogers QR, Morris JG, Fretwell N, Bailey SJ, Batt RM, Lyons LA. Localizing the x-linked orange colour phenotype using feline resource families. Anim Genet. 2005;36:67–70. doi: 10.1111/j.1365-2052.2005.01239.x. [DOI] [PubMed] [Google Scholar]
- Gyapay G, Schmitt K, Fizames C, Jones H, Vega-Czarny N, Spillett D, Muselet D, Prud’Homme J-F, Dib C, Auffray C, Morissette J, Weissenbach J, Goodfellow PN. A radiation hybrid map of the human genome. Human Molecular Genetics. 1996;5:339–346. doi: 10.1093/hmg/5.3.339. [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Stein LD, Gerety SS, Ma J, Castle AB, Silva J, Slonim DK, Baptista R, Kruglyak L, Xu SH, Hu X, Colbert AME, Rosenberg C, Reeve-Daly MP, Rozen S, Hui L, Wu X, Vestergaard C, Wilson KM, Bae JS, Maitra S, Ganiatsas S, Evans CA, DeAngelis MM, Ingalls KA. An sts-based map of the human genome. Science. 1995;270:1945–1954. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- Hukriede NA, Joly L, Tsang M, Miles J, Tellis P, Epstein JA, Barbazuk WB, Li FN, Paw B, Postlethwait JH, Hudson TJ, Zon LI, McPherson JD, Chevrette M, Dawid IB, Johnson SL, Ekker M. Radiation hybrid mapping of the zebrafish genome. Proceedings of the National Academy of Sciences. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James MR, Richard CW, 3rd, Schott JJ, Yousry C, Clark K, Bell J, Terwilliger JD, Hazan J, Dubay C, Vignal A, et al. A radiation hybrid map of 506 sts markers spanning human chromosome 11. Nat Genet. 1994;8:70–76. doi: 10.1038/ng0994-70. [DOI] [PubMed] [Google Scholar]
- Karere GM, Lyons LA, Froenicke L. Enhancing radiation hybrid mapping through whole genome amplification. Hereditas. 2010;147:103–112. doi: 10.1111/j.1601-5223.2010.02166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Laurent P, Schibler L, Vaiman A, Laubier J, Delcros C, Cosseddu G, Vaiman D, Cribiu EP, Yerle M. A 12 000-rad whole-genome radiation hybrid panel in sheep: Application to the study of the ovine chromosome 18 region containing a qtl for scrapie susceptibility. Animal genetics. 2007;38:358–363. doi: 10.1111/j.1365-2052.2007.01607.x. [DOI] [PubMed] [Google Scholar]
- Lewin HA, Larkin DM, Pontius J, O’Brien SJ. Every genome sequence needs a good map. Genome Res. 2009;19:1925–1928. doi: 10.1101/gr.094557.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin C-W, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli K-P, Parker HG, Pollinger JP, Searle SMJ, Sutter NB, Thomas R, Webber C, Baldwin J, Abebe A, Abouelleil A, Aftuck L, Ait-Zahra M, Aldredge T, Allen N, An P, Anderson S, Antoine C, Arachchi H, Aslam A, Ayotte L, Bachantsang P, Barry A, Bayul T, Benamara M, Berlin A, Bessette D, Blitshteyn B, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Brown A, Cahill P, Calixte N, Camarata J, Cheshatsang Y, Chu J, Citroen M, Collymore A, Cooke P, Dawoe T, Daza R, Decktor K, DeGray S, Dhargay N, Dooley K, Dooley K, Dorje P, Dorjee K, Dorris L, Duffey N, Dupes A, Egbiremolen O, Elong R, Falk J, Farina A, Faro S, Ferguson D, Ferreira P, Fisher S, FitzGerald M, Foley K, Foley C, Franke A, Friedrich D, Gage D, Garber M, Gearin G, Giannoukos G, Goode T, Goyette A, Graham J, Grandbois E, Gyaltsen K, Hafez N, Hagopian D, Hagos B, Hall J, Healy C, Hegarty R, Honan T, Horn A, Houde N, Hughes L, Hunnicutt L, Husby M, Jester B, Jones C, Kamat A, Kanga B, Kells C, Khazanovich D, Kieu AC, Kisner P, Kumar M, Lance K, Landers T, Lara M, Lee W, Leger J-P, Lennon N, Leuper L, LeVine S, Liu J, Liu X, Lokyitsang Y, Lokyitsang T, Lui A, Macdonald J, Major J, Marabella R, Maru K, Matthews C, McDonough S, Mehta T, Meldrim J, Melnikov A, Meneus L, Mihalev A, Mihova T, Miller K, Mittelman R, Mlenga V, Mulrain L, Munson G, Navidi A, Naylor J, Nguyen T, Nguyen N, Nguyen C, Nguyen T, Nicol R, Norbu N, Norbu C, Novod N, Nyima T, Olandt P, O’Neill B, O’Neill K, Osman S, Oyono L, Patti C, Perrin D, Phunkhang P, Pierre F, Priest M, Rachupka A, Raghuraman S, Rameau R, Ray V, Raymond C, Rege F, Rise C, Rogers J, Rogov P, Sahalie J, Settipalli S, Sharpe T, Shea T, Sheehan M, Sherpa N, Shi J, Shih D, Sloan J, Smith C, Sparrow T, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Stone S, Sykes S, Tchuinga P, Tenzing P, Tesfaye S, Thoulutsang D, Thoulutsang Y, Topham K, Topping I, Tsamla T, Vassiliev H, Venkataraman V, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Yang S, Yang X, Young G, Yu Q, Zainoun J, Zembek L, Zimmer A, Lander ES. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lipinski MJ, Froenicke L, Baysac KC, Billings NC, Leutenegger CM, Levy AM, Longeri M, Niini T, Ozpinar H, Slater MR, Pedersen NC, Lyons LA. The ascent of cat breeds: Genetic evaluations of breeds and worldwide random-bred populations. Genomics. 2008;91:12–21. doi: 10.1016/j.ygeno.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons LA, Biller DS, Erdman CA, Lipinski MJ, Young AE, Roe BA, Qin B, Grahn RA. Feline polycystic kidney disease mutation identified in pkd1. J Am Soc Nephrol. 2004;15:2548–2555. doi: 10.1097/01.ASN.0000141776.38527.BB. [DOI] [PubMed] [Google Scholar]
- Marques E, de Givry S, Stothard P, Murdoch B, Wang Z, Womack J, Moore SS. A high resolution radiation hybrid map of bovine chromosome 14 identifies scaffold rearrangement in the latest bovine assembly. BMC genomics. 2007;8:254. doi: 10.1186/1471-2164-8-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay SD, Schnabel RD, Murdoch BM, Aerts J, Gill CA, Gao C, Li C, Matukumalli LK, Stothard P, Wang Z, Van Tassell CP, Williams JL, Taylor JF, Moore SS. Construction of bovine whole-genome radiation hybrid and linkage maps using high-throughput genotyping. Anim Genet. 2007;38:120–125. doi: 10.1111/j.1365-2052.2006.01564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Agarwala R, Schaffer AA, Stephens R, O’Brien SJ, Murphy WJ. Radiation hybrid mapping of 304 novel microsatellites in the domestic cat genome. Cytogenet Genome Res. 2003a;102:272–276. doi: 10.1159/000075762. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Lyons LA, Schaffer AA, Tomlin JF, Hutton MK, O’Brien SJ. A genetic linkage map of microsatellites in the domestic cat (felis catus) Genomics. 1999;57:9–23. doi: 10.1006/geno.1999.5743. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Roelke ME, Chen ZQ, Menotti KA, Sun S, Schaffer AA, Tomlin JF, Agarwala R, O’Brien SJ, Murphy WJ. Second-generation integrated genetic linkage/radiation hybrid maps of the domestic cat (felis catus) J Hered. 2003b;94:95–106. doi: 10.1093/jhered/esg008. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Schaffer AA, Tomlin JF, Eizirik E, Phillip C, Wells D, Pontius JU, Hannah SS, O’Brien SJ. An autosomal genetic linkage map of the domestic cat, felis silvestris catus. Genomics. 2009;93:305–313. doi: 10.1016/j.ygeno.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menotti-Raymond M, Deckman KH, David V, Myrkalo J, O’Brien SJ, Narfstrom K. Mutation discovered in a feline model of human congenital retinal blinding disease. Investigative ophthalmology & visual science. 2010;51:2852–2859. doi: 10.1167/iovs.09-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs KM, Norgard MM, Ederer MM, Hendrix KP, Kittleson MD. A substitution mutation in the myosin binding protein c gene in ragdoll hypertrophic cardiomyopathy. Genomics. 2007;90:261–264. doi: 10.1016/j.ygeno.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Mullikin JC, Hansen NF, Shen L, Ebling H, Donahue WF, Tao W, Saranga DJ, Brand A, Rubenfield MJ, Young AC, Cruz P, Sequencing NCP, Driscoll C, David V, Al-Murrani SW, Locniskar MF, Abrahamsen MS, O’Brien SJ, Smith DR, Brockman JA. Light whole genome sequence for snp discovery across domestic cat breeds. BMC Genomics. 2010;11:406. doi: 10.1186/1471-2164-11-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Menotti-Raymond M, Lyons LA, Thompson MA, O’Brien SJ. Development of a feline whole genome radiation hybrid panel and comparative mapping of human chromosome 12 and 22 loci. Genomics. 1999;57:1–8. doi: 10.1006/geno.1998.5695. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Page JE, Smith C, Desrosiers RC, O’Brien SJ. A radiation hybrid mapping panel for the rhesus macaque. The Journal of heredity. 2001;92:516–519. doi: 10.1093/jhered/92.6.516. [DOI] [PubMed] [Google Scholar]
- O’Brien SJ, Nash WG. Genetic mapping in mammals: Chromosome map of domestic cat. Science. 1982;216:257–265. doi: 10.1126/science.7063884. [DOI] [PubMed] [Google Scholar]
- Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. Beadarray technology: Enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques Suppl. 2002:56–58. 60–51. [PubMed] [Google Scholar]
- Olivier M, Aggarwal a, Allen J, Almendras aa, Bajorek ES, Beasley EM, Brady SD, Bushard JM, Bustos VI, Chu a, Chung TR, De Witte a, Denys ME, Dominguez R, Fang NY, Foster BD, Freudenberg RW, Hadley D, Hamilton LR, Jeffrey TJ, Kelly L, Lazzeroni L, Levy MR, Lewis SC, Liu X, Lopez FJ, Louie B, Marquis JP, Martinez Ra, Matsuura MK, Misherghi NS, Norton Ja, Olshen a, Perkins SM, Perou aJ, Piercy C, Piercy M, Qin F, Reif T, Sheppard K, Shokoohi V, Smick Ga, Sun WL, Stewart Ea, Fernando J, Tejeda, Tran NM, Trejo T, Vo NT, Yan SC, Zierten DL, Zhao S, Sachidanandam R, Trask BJ, Myers RM, Cox DR. A high-resolution radiation hybrid map of the human genome draft sequence. Science (New York, NY) 2001;291:1298–1302. doi: 10.1126/science.1057437. [DOI] [PubMed] [Google Scholar]
- Pearson K. London: 1900. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. [Google Scholar]
- Pontius JU, Mullikin JC, Smith DR, Lindblad-Toh K, Gnerre S, Clamp M, Chang J, Stephens R, Neelam B, Volfovsky N, Schaffer AA, Agarwala R, Narfstrom K, Murphy WJ, Giger U, Roca AL, Antunes A, Menotti-Raymond M, Yuhki N, Pecon-Slattery J, Johnson WE, Bourque G, Tesler G, O’Brien SJ. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17:1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop M. Genome assembly reborn: Recent computational challenges. Briefings in bioinformatics. 2009;10:354–366. doi: 10.1093/bib/bbp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Schiex T, McKay S, Murdoch B, Wang Z, Womack JE, Stothard P, Moore SS. High resolution radiation hybrid maps of bovine chromosomes 19 and 29: Comparison with the bovine genome sequence assembly. BMC genomics. 2007;8:310. doi: 10.1186/1471-2164-8-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priat C, Hitte C, Vignaux F, Renier C, Jiang Z, Jouquand S, Chéron A, André C, Galibert F. A whole-genome radiation hybrid map of the dog genome. Genomics. 1998;54:361–378. doi: 10.1006/geno.1998.5602. [DOI] [PubMed] [Google Scholar]
- Rah H, Maggs DJ, Blankenship TN, Narfstrom K, Lyons LA. Early-onset, autosomal recessive, progressive retinal atrophy in persian cats. Investigative ophthalmology & visual science. 2005;46:1742–1747. doi: 10.1167/iovs.04-1019. [DOI] [PubMed] [Google Scholar]
- Senger F, Priat C, Hitte C, Sarropoulou E, Franch R, Geisler R, Bargelloni L, Power D, Galibert F. The first radiation hybrid map of a perch-like fish: The gilthead seabream (sparus aurata l) Genomics. 2006;87:793–800. doi: 10.1016/j.ygeno.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Sun S, Murphy WJ, Menotti-Raymond M, O’Brien SJ. Integration of the feline radiation hybrid and linkage maps. Mamm Genome. 2001;12:436–441. doi: 10.1007/s003350010289. [DOI] [PubMed] [Google Scholar]
- Toonen RJ, Hughes S. Increased throughput for fragment analysis on an abi prism 377 automated sequencer using a membrane comb and strand software. Biotechniques. 2001;31:1320–1324. [PubMed] [Google Scholar]
- Walter MA, Spillett DJ, Thomas P, Weissenbach J, Goodfellow PN. A method for constructing radiation hybrid maps of whole genomes. Nat Genet. 1994;7:22–28. doi: 10.1038/ng0594-22. [DOI] [PubMed] [Google Scholar]
- Watanabe TK, Bihoreau MT, McCarthy LC, Kiguwa SL, Hishigaki H, Tsuji A, Browne J, Yamasaki Y, Mizoguchi-Miyakita A, Oga K, Ono T, Okuno S, Kanemoto N, Takahashi E, Tomita K, Hayashi H, Adachi M, Webber C, Davis M, Kiel S, Knights C, Smith A, Critcher R, Miller J, Thangarajah T, Day PJ, Hudson JR, Irie Y, Takagi T, Nakamura Y, Goodfellow PN, Lathrop GM, Tanigami A, James MR. A radiation hybrid map of the rat genome containing 5,255 markers. Nature genetics. 1999;22:27–36. doi: 10.1038/8737. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigó R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang S-P, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Womack JE, Johnson JS, Owens EK, Rexroad CE, Schläpfer J, Yang Y-P. A whole-genome radiation hybrid panel for bovine gene mapping. Mammalian Genome. 1997;8:854–856. doi: 10.1007/s003359900593. [DOI] [PubMed] [Google Scholar]
- Xu M, Fujita D, Hanagata N. Perspectives and challenges of emerging single-molecule DNA sequencing technologies. Small. 2009;5:2638–2649. doi: 10.1002/smll.200900976. [DOI] [PubMed] [Google Scholar]
- Yerle M, Pinton P, Robic a, Alfonso a, Palvadeau Y, Delcros C, Hawken R, Alexander L, Beattie C, Schook L, Milan D, Gellin J. Construction of a whole-genome radiation hybrid panel for high-resolution gene mapping in pigs. Cytogenetics and cell genetics. 1998;82:182–188. doi: 10.1159/000015095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.