Abstract
Treatment with sigma1 receptor (Sigma1) ligands can inhibit cell proliferation in vitro and tumor growth in vivo. However, the cellular pathways engaged in response to Sigma1 ligand treatment that contribute to these outcomes remain largely undefined. Here, we show that treatment with putative antagonists of Sigma1 decreases cell mass. This effect corresponds with repressed cap-dependent translation initiation in multiple breast and prostate cancer cell lines. Sigma1 antagonist treatment suppresses phosphorylation of translational regulator proteins p70S6K, S6, and 4E-BP1. RNAi-mediated knockdown of Sigma1 also results in translational repression, consistent with the effects of antagonist treatment. Sigma1 antagonist mediated translational repression and decreased cell size are both reversible. Together, these data reveal a role for Sigma1 in tumor cell protein synthesis, and demonstrate that small molecule Sigma1 ligands can be used as modulators of protein translation.
Keywords: sigma1 receptor, Sigma1 ligand, protein translation, cell mass, tumor
Introduction
The sigma1 receptor (Sigma1) is a unique 26-kilodalton integral membrane protein [1,2,3,4,5] that has been detected in the endoplasmic reticulum (ER), mitochondrion, nuclear membrane, and plasma membrane of various cell types in a range of tissue [1,5,6,7]. Initially thought to be an opioid receptor [8], Sigma1 is now considered a distinct protein unrelated to any traditional receptor [9]. Indeed, emerging evidence suggests that Sigma1 may function as a molecular chaperone [7]. A range of structurally diverse small molecule compounds binds Sigma1 with high affinity [10]. Many of these Sigma1 ligands have been categorized as antagonists and agonists based primarily on behavioral pharmacology assays [10]. However, at the cellular level, the signaling actions of Sigma1 antagonists versus agonists have not been clearly defined. Sigma1 is highly expressed in various tumor cell types, including breast and prostate tumor cell lines [11,12,13]. However, the role of Sigma1 in tumor cell biology remains unclear. Interestingly, some Sigma1 putative antagonists, but not putative agonists, inhibit tumor cell proliferation in vitro and inhibit tumor growth in mouse tumor xenograft experiments. Protracted antagonist treatment can lead to apoptotic cell death [13,14,15]. However, the intracellular signaling pathways activated in response to Sigma1 antagonist treatment, prior to apoptosis, remain largely unmapped.
The increased demand for protein synthesis associated with proliferation, growth (i.e., cell biomass), and survival render tumor cells particularly dependent on translation initiation [16,17,18,19]. In many cancers, tumorigenesis and tumor progression are driven by mutations in the growth control apparatus that produce aberrant levels or activity of factors involved in the highly regulated process of translation initiation, making components of this process attractive targets for chemotherapy [16,17,18,19].
While evaluating the cell growth inhibiting properties of Sigma1 ligands, we observed that treatment of several tumor cell lines with some Sigma1 ligands visibly diminished cell size. Based on these observations, we investigated whether translational repression through the inhibition of translation initiation contributes to the modulation of cell size in response to treatment with Sigma1 ligands.
Materials and Methods
Chemicals and cell lines
Sigma1 ligands IPAG (1-(4-Iodophenyl)-3-(2-adamantyl)guanidine), haloperidol, PRE084 (2-(4-Morpholinethyl) 1-phenylcyclohexanecarboxylate hydrochloride), and (+)SKF-10047 ([2S-(2a,6a,11R*]-1,2,3,4,5,6-hexahydro-6,11-dimethyl-3- (2-propenyl)-2,6-methano-3-benzazocin-8-ol hydrochloride), were purchased from Tocris. All chemical compounds were dissolved in DMSO (dimethyl sulfoxide).
All cell lines were obtained from ATCC. LNCaP cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (CellGro). All other cell lines were maintained in a 1:1 mixture of DMEM:F-12 with 4.5g/liter glucose, 10% FBS and penicillin/streptomycin.
Immunoblots and antibodies
Cells were seeded approximately 24 hours prior to the start of drug treatment in most assays. Treated cells were lysed and proteins extracted in a modified RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate and 0.1% SDS) supplemented with 10% glycerol (volume/volume), Complete protease inhibitor cocktail (Roche), and Halt phosphatase inhibitor cocktail (Pierce). Approximately 10-20μg of detergent soluble protein were resolved on NOVEX 10% or 10-20% polyacrylamide Tris-glycine gels (Invitrogen) and subsequently immunoblotted onto PVDF membrane. Immunoblots were performed in a 20 mM Tris-buffered 137 mM saline solution (pH 7.6) containing 0.1% Tween-20 (polyoxyethylene (20) sorbitan monolaurate) and 5% (weight/volume) blotting grade non-fat dry milk (Bio Rad). All washes were performed in the same buffer without the milk. The anti-phospho-threonine 389-p70S6K, anti-phospho-serine 235/236-ribosomal S6, anti-phospho-serine 65-4E-BP1, anti-4E-BP1, and anti-eIF4E antibodies were all purchased from Cell Signaling Technology. The mouse anti-β-actin antibody was purchased from Santa Cruz Biotechnology. The rabbit polyclonal Sigma1 antibody against a peptide corresponding to murine Sigma1 residues 137-159 (KEGTTKSEVFYPGETVVHGPGEA) was generated in our laboratory in collaboration with the Memorial Sloan-Kettering Monoclonal Antibody Core Facility. The Lumigen PS-3 enhanced chemiluminescence kit (GE Healthcare) or Illumina enhanced chemiluminescence kit (Millipore) was used to reveal immunoblotted proteins.
Cap binding assay
To evaluate changes in cap-dependent translation initiation we performed a pull-down assay using m7GTP-sepharose beads (GE Healthcare) that mimic 5’ mRNA cap and thus can be used to isolate 5’ cap bound protein complexes [20]. Following 24 hour treatment with Sigma1 ligands, we collected and lysed treated cells in the modified RIPA buffer described above, and incubated the detergent soluble fraction for 3 hours with pre-washed m7GTP-sepharose beads (GE Healthcare) at 4°C with constant rotation, washed 3 times with the same modified RIPA buffer, and bound proteins were eluted from the beads by heating 5 minutes at 95°C.
Cell size determinations and flow cytometry based cell size assays
Cell size was quantified by flow cytometry using a Becton Dickinson FACS Calibur flow cytometer with Cell Quest software. Forward scatter height (FSC-H) was used as an indicator of cell size as described [21,22]. For cell size experiments, cells were seeded onto 10 cm tissue culture plates at 106 cells/plate and treated 24 hours later with 10μM Sigma1 ligand. Cells were harvested for flow cytometry at the indicated times. To harvest cells, plates were washed once with PBS without magnesium and calcium, and then incubated at room temperature for 5 min in 3 mL of PBS/EDTA (2.5 mM), and gently detached from the plates, transferred to 15 mL conical tubes. The cell pellets were washed once in PBS containing 1% FBS, and resuspended in 0.5 mL of PBS, and fixed by adding 5 mL of 88% ethanol (80% final) dropwise, while gently vortexing. Prior to flow cytometry, the fixed cells were washed once with PBS/1% FBS, and then incubated at 37°C for 30 min in propidium iodide/RNase A solution (10 μg/mL propidium iodide in 0.76 mM sodium citrate at pH 7.0; 250 μg/mL RNase A in 10 mM Tris-HCl, 15 mM NaCl at pH 7.5) diluted into PBS/1% FBS. The mean forward scatter height (FSC-H) of the G1-phase population was determined as a measure of relative cell size. Single cells were gated away from aggregated cells using an FL2-width versus FL-2 area dot plot. Mean FSC-H ± S.E. was calculated from at least 4 independent determinations. For each FSC-H determination, FACS analysis was performed on 10,000 single cells.
siRNA mediated knockdown of Sigma1
For most cell lines, 10 nmoles of human Sigma1 siRNA (Santa Cruz Biotechnology) per 100,000 cells was transfected with INTERFERin transfection reagent (PolyPlus), and 48 hours later, cells were reseeded, allowed to attach and recover for 16 hours and transfected again. Twenty-four hours following the second transfection of Sigma1 siRNA, cells were treated with Sigma1 ligand for 16 hours. Transfection of Control-A siRNA (Santa Cruz Biotechnology) was performed in parallel, using the conditions described above.
Statistical analysis
Statistical significance was determined by one-way ANOVA followed by Bonferroni's post-test using Prism software (GraphPad).
Results
Treatment with Sigma1 putative antagonists diminishes cell mass
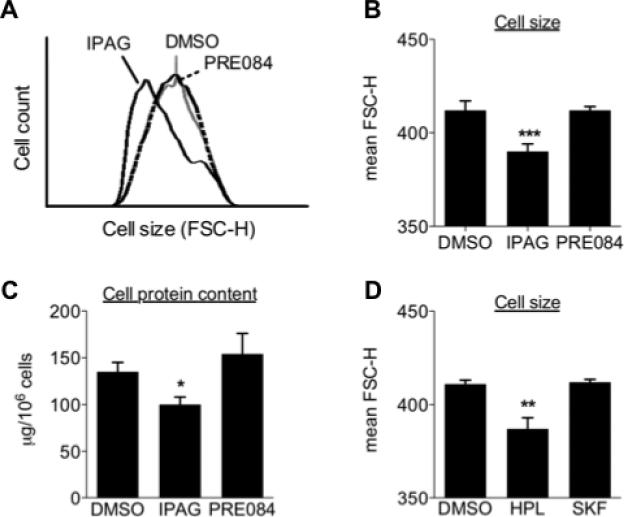
While evaluating the anti-proliferative and cytotoxic properties of Sigma1 ligands, we consistently observed that cells treated with Sigma1 putative antagonists appeared smaller than control or Sigma1 putative agonist-treated cells. We observed this effect with several cell lines that natively express Sigma1, including T47D, MCF7, and MDA-MB-468 breast adenocarcinoma cells (data shown for T47D in Fig. 1). To quantify apparent differences in cell size, we performed a widely used flow cytometry based assay that measures the forward scatter height (FSC-H) of cells as an indicator of size [21,22]. T47D cells were treated for 24 hours with 10 μM IPAG (Sigma1 putative antagonist) or 10 μM PRE084 (Sigma1 putative agonist), and subsequently analyzed by flow cytometry. The mean FSC-H of DMSO (control) and PRE084 treated T47D cells measured 412 ± 5 and 412 ± 2, respectively, whereas the mean FSC-H of IPAG treated cells was 390 ± 4 (Fig. 1B). Diminished cell size corresponded with diminished cell protein content. Protein content, measured as micrograms (μg) of protein per 106 cells (counted and isolated following treatment and immediately prior to protein extraction), of DMSO (control) and PRE084 treated T47D cells measured 135 ± 10 and 154 ± 22, respectively, whereas IPAG treatment produced a mean of 100 ± 8 μg per 106 cells (Fig. 1C). These data suggested that decreased cell size correlates with decreased cell mass.
Fig. 1. Sigma1 antagonist treatment is associated with diminished cell mass.
(A.) Histogram of representative experiment measuring FSC-H of G1 population of T47D cells treated for 24 hours with DMSO (vehicle control), 10μM IPAG (antagonist), or 10μM PRE084 (agonist). (B.) Cell size data generated from four independent determinations, and presented as mean FSC-H ± S.E. (standard error) of DMSO, 10μM IPAG and 10μM PRE084 treated T47D (**P < 0.01). (C.) Cellular protein content was determined by BCA assay and presented as micrograms (μg) of protein per 106 cells. Data are from four independent determinations (*P < 0.05). (D.) T47D cells treated for 24 hours with 10μM antagonist, haloperidol (HPL), or 10μM agonist, (+)SKF-10047 (SKF). Data are presented as mean FSC-H ± S.E., and are from four independent determinations (**P < 0.01).
Finally, we evaluated the cell size modulating activity of another Sigma1 antagonist and agonist. We observed a consistent decrease in cell size when T47D cells were treated with antagonist (haloperidol), but not agonist ((+)SKF-10047) (Fig. 1D).
Treatment with Sigma1 putative antagonists inhibits initiation of cap-dependent translation
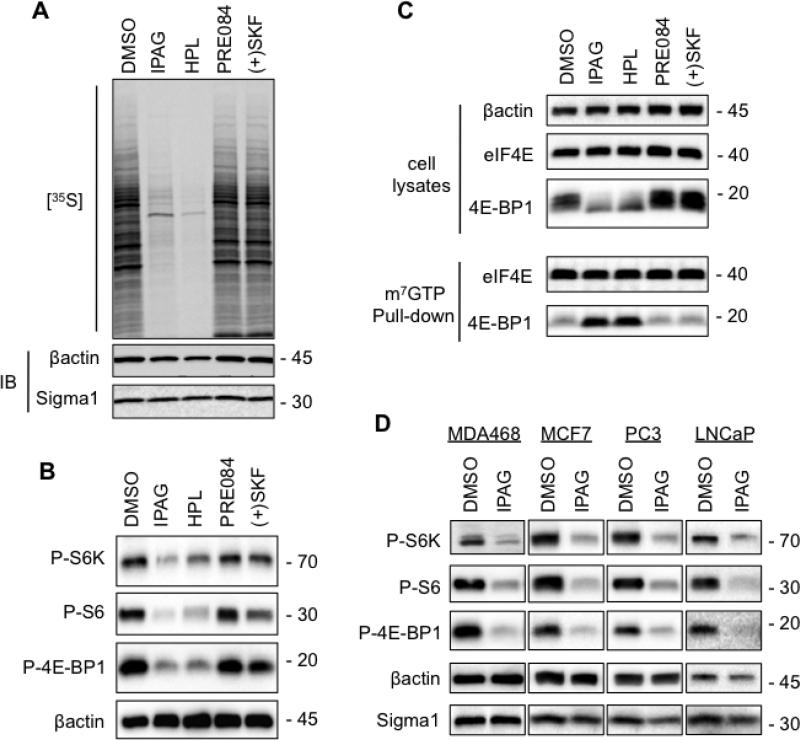
To determine whether translational repression contributes to Sigma1 antagonist treatment-associated decrease in cell mass, we first evaluated protein synthesis by [35S] metabolic labeling (Fig. 2A). Subsequent to 24 hour treatment with putative antagonists (IPAG, haloperidol) we observed a salient decrease in protein synthesis indicated by the amount of [35S] methionine and cysteine incorporated during the 1 hour [35S] pulse-label period, whereas treatment with putative agonists (PRE084, (+)SKF-10047) produced no detectable change compared to control (DMSO) (Fig. 2A). We confirmed Sigma1 antagonist-mediated translational repression by immunoblots demonstrating decreased levels of phospho-threonine 389-p70S6Kinase (P-S6K), phospho-serine 235/236-ribosomal S6 (P-S6), and phospho-serine 65-4E-BP1 (P-4E-BP1) (Fig. 2B). Inhibited phosphorylation of these downstream targets of AKT signaling is consistent with suppression of cap-dependent translation [16,17]. To confirm that cap-dependent translation is suppressed by antagonists, we performed a 5’-cap-binding assay. Dephosphorylation of 4E-BP1 allows it to bind to the eIF4E-mRNA cap complex, which prevents cap-dependent translation [20]. Following treatment with IPAG and haloperidol, the steady-state levels of eIF4E did not change, however, we observed a salient increase in 4E-BP1 bound to eIF4E by m7GTP-sepharose pull-down assay (Fig. 2C). Together, these data demonstrate that the same treatment conditions that diminish cell size correspond with inhibition of translation initiation.
Fig. 2. Inhibition of cap-dependent translation initiation mediated by Sigma1 antagonist.
(A.) T47D cells were treated for 24 hours with 10μM antagonists (IPAG, haloperidol) or 10μM agonists (PRE084, (+)SKF-10047). Subsequently, cells were pulse-labeled for 1 hour with 100μCi/ml [35S] methionine and cysteine. Protein extracts were resolved by SDS-PAGE, transferred onto nitrocellulose membrane and exposed to autoradiography film. (B.) Immunoblots of protein extracts from T47D cells treated for 24 hours with 10μM putative antagonists (IPAG, haloperidol) or putative agonists (PRE084, (+)SKF-10047). Phospho-threonine 389-p70S6K (P-S6K), phospho-serine 235/236 S6 (P-S6), and phospho-serine 65-4E-BP1 (P-4E-BP1). (C.) Cell lysates were precipitated with m7GTP-sepharose beads (pull-down) and subsequently immunoblotted with antibodies against 4E-BP1 and eIF4E to evaluate 4E-BP1 binding to the eIF4E-mRNA cap complex. The upper panel (cell lysates) demonstrates equivalent input into m7GTP-sepharose binding, as well as Sigma1 ligand mediated changes in 4E-BP1 phosphorylation profile. (D.) Immunoblots of protein extracts from breast (MDA468, MCF7) and prostate adenocarcinoma (PC3, LNCaP) cells treated for 24 hours with 10μM IPAG.
We observed IPAG mediated translational repression in several cancer cell lines (Fig. 2D). Interestingly, IPAG mediated translational repression did not appear to be influenced by PTEN status, as PTEN null and mutant (MDA468, LNCaP, PC3) cell lines were as responsive to Sigma1 antagonist mediated decrease in phospho-S6K, phospho-S6, and phospho-4E-BP1 levels as wild-type PTEN expressing PIK3CA mutant (T47D, MCF7) cell lines (Fig. 2).
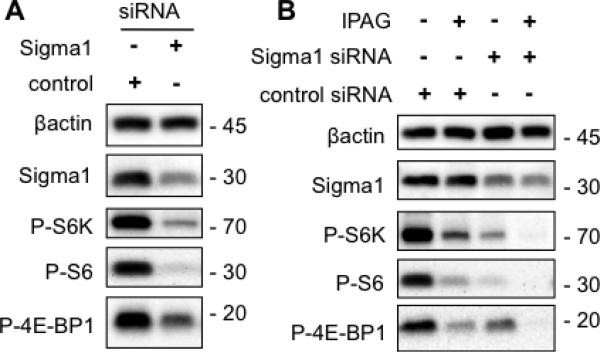
Sigma1 siRNA knockdown appears to mimic antagonist treatment (Fig. 3A), as siRNA mediated knockdown of Sigma1 (~70% knockdown) resulted in decreased levels of phospho-p70S6 kinase and phospho-S6, phospho-4E-BP1 (Fig. 3A). We were unable to recover viable cells in which Sigma1 knockdown was greater than 80%, suggesting that a certain minimal amount of Sigma1 may be necessary for tumor cell survival.
Fig. 3. Translational repression associated with siRNA mediated knockdown of Sigma1.
(A.) Approximately 4.5 days post-transfection, translational control markers phospho-threonine 389-p70S6K (P-S6K), phospho-serine 235/236 S6 (P-S6), and phospho-serine 65-4E-BP1 (P-4E-BP1) were evaluated in immunoblotted T47D protein extracts. (B.) Following transfection of control or Sigma1 siRNA, T47D cells were treated for 24 hours with 10μM IPAG and the translational control markers, above, were detected by immunoblot.
Antagonist treatment of cells in which Sigma1 had been knocked down resulted in nearly complete suppression of both phospho-S6 and phospho-4E-BP1 levels (Fig. 3B). This outcome was consistent in three independent determinations. It is possible that IPAG may act on a subpopulation of Sigma1 in the cell, and when Sigma1 is knocked down to minimal levels the remaining available Sigma1 are suppressed by antagonist. Consistent with this notion, treatment with antagonist does not alter Sigma1 steady-state protein levels in control or siRNA-transfected cells (Fig. 3B), suggesting that IPAG mediates its translational repressor actions by altering the activity of available Sigma1 and not by altering Sigma1 protein levels. However, the number of available Sigma1 may influence cellular protein translation capacity.
Sigma1 putative antagonist-mediated decrease in cell size and translational repression is reversible
One question that may provide important information regarding the potential therapeutic use of these compounds is whether or not the effects of the Sigma1 antagonist are reversible. That is, if the Sigma1 antagonist is withdrawn, would translation return to control conditions or would the treated cells continue to progress irrevocably toward cell death. We examined this by treating cells for 24 hours and examining the status of cell size and translation arrest markers following removal of the antagonist (Fig. 4). We refer to the post-drug removal period as recovery time.
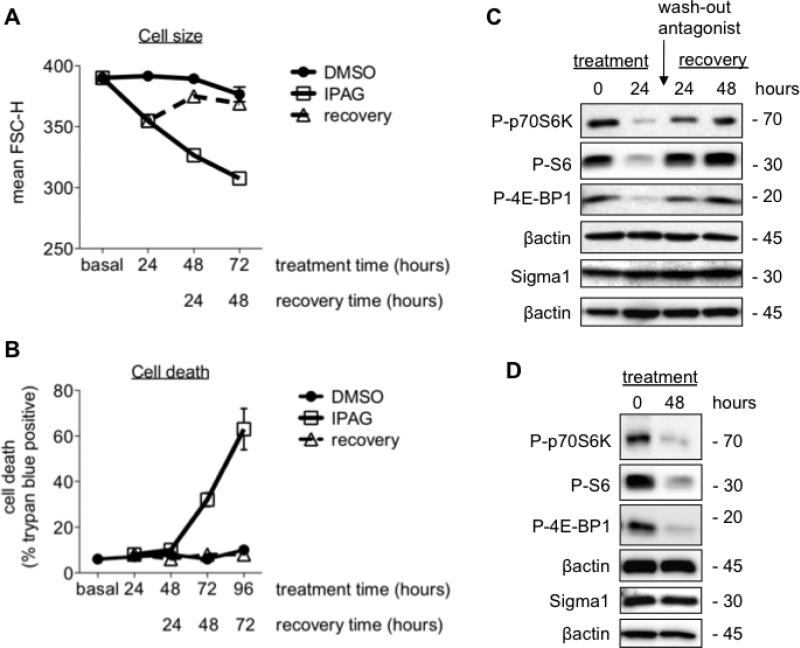
Fig 4. Reversibility of Sigma1 antagonist mediated decrease in cell size and translational repression.
T47D cells were treated with 10μM IPAG, and cell culture medium was changed with drug-free medium after treatment for 24 hours. Cells in “recovery” were treated as follows: cells were immediately washed with pre-warmed culture medium to remove residual drug (washed out). Recovery indicates T47D cell cultures in which drug-containing medium was removed and replaced with drug-free medium after 24 hour treatment with 10μM of IPAG (antagonist). Cells were collected and analyzed 24, 48, and 72 hours after removal of drug from cell culture medium FSC-H was measured at the indicated treatment time. (A.) T47D cells were either continuously treated with 10μM of IPAG (antagonist) for 96 hours (open square, solid line) and compared to DMSO treated cells (closed circle, solid line), or compared to cells that were treated with 10μM of IPAG (antagonist) for 24 hours and subsequently allowed to recover following removal of IPAG from cell culture medium (open triangle, broken line) (n=3, **P<0.01). The FSC-H of cells at the 48 hour recovery time point are not significantly different compared to control cells that have been treated for 72 hours with DMSO. Error bars are not shown since they are smaller than the size of the symbols. (B.) Cell death was monitored by Trypan blue exclusion assay following treatment with 10μM IPAG for the indicated times (n=3, **P<0.01). (C.) Restored protein synthesis 24 and 48 hours following IPAG wash-out. Protein extracted IPAG (10μM) treated T47D cells and evaluated for cell death in Figure 4B were resolved by SDS-PAGE and immunoblotted with antibodies against phospho-threonine 389-p70S6K (P-S6K), phospho-serine 235/236-S6 (P-S6), and phospho-serine 65-4E-BP1 (P-4EBP1). (D.) Suppressed protein synthesis following 48 hours of continuous treatment with 10μM IPAG.
Following 24 hours of treatment with 10μM IPAG, we removed the drug-containing cell culture medium and washed the treated cells twice with drug-free cell culture medium. Cells were subsequently cultured in drug-free medium and collected at the indicated times following medium replacement. After 24 hours, the mean FSC-H of IPAG treated T47D G1 cell cycle population decreased from 390 ± 3 to 355 ± 5 (P < 0.01) (Fig. 4B). Continued treatment with 10μM IPAG decreased the mean FSC-H to 327 ± 1 by 48 hours, and to 308 ± 3 by 72 hours of treatment (Fig. 4B). In contrast, the size of DMSO (vehicle control) treated T47D cells did not change significantly up to 72 hours of treatment with mean G1 population FCS-H of 392 ± 2, 389 ± 2, and 377 ± 10 after 24, 48, and 72 hours of treatment, respectively (Fig. 4B). This slight decrease in size by 72 hours also occurred with non-treated cells, suggesting an intrinsic decrease in T47D cell mass following extended periods of attachment or as a result of increased monolayer confluency and contact mediated growth inhibition. However, by removing IPAG from the cell culture medium after 24 hours of treatment, we observed a return toward control cell size, with mean FSC-H of 375 ± 3, 369 ± 2, and 366 ± 3 at the 24, 48, and 72 hour recovery time points, respectively (Fig. 4B).
During this time-course, no significant cell death was detected by up to 48 hours of continuous treatment with IPAG (Fig. 4B). Cell death was clearly detectable with 32 ± 3 % death by 72 hours of continuous treatment and 63 ± 9 % death by 96 hours of continuous treatment (Fig. 4B). This suggested that despite the change in cell mass, cell death pathways had not been engaged by up to 48 hours of treatment. In contrast, cell cultures in which IPAG was removed after 24 hours resembled DMSO controls, and no significant cell death was observed throughout the time-course (Fig. 4B). Here, our question was whether Sigma1 antagonist treatment activates irreversible signaling cascades toward cell death. On the contrary, we find that continuous, protracted antagonist treatment is required to produce cell death.
This led us to examine whether Sigma1 antagonist-induced translational repression would cause protein synthesis to return to control levels with a similar time-course (Fig. 4C). We found that protein synthesis markers progressively returned to control levels by 24-to-48 hours of recovery (Fig. 4C). In contrast, translation markers remained suppressed when IPAG was not washed-out (Fig. 4D).
Discussion
The cellular protein synthesis apparatus is a compelling target for therapeutic intervention in cancer, and considerable effort has been made toward developing agents to inhibit its components [18,19,23,24,25]. However, redundancies and complex feedback mechanisms render therapeutic approaches to target growth regulatory pathways susceptible to resistance [23,24,25]. For example, cross-talk between PI3K-AKT-mTOR and Raf-MEK-ERK signaling enables mutually compensatory responses to the inhibition of either pathway individually [23]. The loss of functional PTEN, a key regulator of the PI3K-AKT-mTOR pathway, results in increased expression and activity of components of the cellular protein synthesis apparatus required to support increased growth (i.e., increase in biomass) and proliferation [18,19,23]. Selective inhibition of this signaling pathway can result in compensatory Raf-MEK-ERK mediated translation initiation by modulating the activity 4E-BP1 [23]. Recent work demonstrates that these pathways converge on key regulators of protein translation, including the translational repressor 4E-BP1 [23]. We find that neither loss of functional PTEN (MDA-MB-468, PC3, LNCaP) nor PIK3CA mutation (T47D, MCF7) alters IPAG mediated suppression of phospho-4E-BP1 levels, and thus translation initiation (Fig. 2).
The pharmacological actions of Sigma1 ligands have been defined primarily in behavioral assays to evaluate their actions in the central nervous system [10]. Therefore, the cellular mechanisms that define Sigma1 antagonists and agonists have remained unclear. Our work reveals that some small molecules with affinity for Sigma1 have translational repressor actions in tumor cell lines. Of the 4 ligands tested, the 2 putative antagonists inhibited translation and decreased cell size whereas the 2 putative agonists did not alter either process, under the conditions tested herein. It is unclear how a behaviorally defined putative antagonist mediates these cellular effects. Structure-activity relationship studies focused on protein synthesis and processing should provide insight into how small molecules with affinity for Sigma1 may be more appropriately categorized in terms of their mechanisms of action in tumor cells. It is also noteworthy that the high affinity Sigma1 putative agonists, PRE084 and (+)SKF10047, did not alter antagonist (IPAG) mediated translational repression (supplementary data). All of the compounds tested here bind to Sigma1 with similarly affinity, however, the precise binding sites for these compounds remain unclear. It is possible that these putative agonist and antagonists bind to distinct sites on Sigma1. Another potential explanation is that these compounds bind to different Sigma1 populations within the tumor cells, corresponding to Sigma1 in agonist or antagonist conformation. These essential questions will require further biochemical investigation. Furthermore, whether translational repression is induced directly by antagonist binding to Sigma1 or whether it is activated downstream of a broader signaling response to Sigma1 antagonist binding remains to be determined. However, our siRNA results demonstrate that these effects are indeed Sigma1 mediated (Fig. 3).
Our data suggest that Sigma1 may be a novel component of the protein synthesis and processing apparatus of tumor cells. We demonstrate translational repressor actions of Sigma1 antagonists and elucidate Sigma1 antagonist mediated signaling activity prior to cell death. A growing number of reports suggest that small molecule compounds targeting Sigma1 may be effective chemotherapeutic agents, however, most have focused on the cytotoxic properties of these compounds [13,15,26,27]. We demonstrate that Sigma1 ligands may be used as reversible modulators of tumor cell protein synthesis. Thus, our results demonstrate that Sigma1 ligands are not necessarily cytotoxic agents, and may be considerably more versatile.
Supplementary Material
HIGHLIGHTS.
Sigma1 ligand treatment mediates decrease in tumor cell mass.
Identification of a Sigma1 ligand with reversible translational repressor actions.
Demonstration of a role for Sigma1 in cellular protein synthesis.
Acknowledgements
This work was supported, in part, by grants (DA06241 and DA02615) and a research scientist award (DA00220) to GWP and a postdoctoral fellowship (DA07274) to FJK from the National Institute on Drug Abuse, an award from the Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and The Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center (MSKCC), a grant from the Translational and Integrative Medicine Fund of MSKCC, and a grant from the National Genetics Foundation, and a core grant from the National Cancer Institute (CA008748) to MSKCC. We thank Dr. Francis Weiss-Garcia (MSKCC Monoclonal Antibody Core Facility) for assistance in generating the Sigma1 polyclonal antibody. We thank Dr. Paul Campbell for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanner M, Moebius FF, Flandorfer A, et al. Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proc Natl Acad Sci U S A. 1996;93:8072–8077. doi: 10.1073/pnas.93.15.8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kekuda R, Prasad PD, Fei YJ, et al. Cloning and functional expression of the human type 1 sigma receptor (hSigmaR1) Biochem Biophys Res Commun. 1996;229:553–558. doi: 10.1006/bbrc.1996.1842. [DOI] [PubMed] [Google Scholar]
- 3.Seth P, Fei YJ, Li HW, et al. Cloning and functional characterization of a sigma receptor from rat brain. J Neurochem. 1998;70:922–931. doi: 10.1046/j.1471-4159.1998.70030922.x. [DOI] [PubMed] [Google Scholar]
- 4.Seth P, Leibach FH, Ganapathy V. Cloning and structural analysis of the cDNA and the gene encoding the murine type 1 sigma receptor. Biochem Biophys Res Commun. 1997;241:535–540. doi: 10.1006/bbrc.1997.7840. [DOI] [PubMed] [Google Scholar]
- 5.Aydar E, Palmer CP, Klyachko VA, et al. The sigma receptor as a ligand-regulated auxiliary potassium channel subunit. Neuron. 2002;34:399–410. doi: 10.1016/s0896-6273(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 6.Jbilo O, Vidal H, Paul R, et al. Purification and characterization of the human SR 31747A-binding protein. A nuclear membrane protein related to yeast sterol isomerase. J Biol Chem. 1997;272:27107–27115. doi: 10.1074/jbc.272.43.27107. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi T, Su TP. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell. 2007;131:596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 8.Martin WR, Eades CG, Thompson JA, et al. The effects of morphine- and nalorphine-like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther. 1976;197:517–532. [PubMed] [Google Scholar]
- 9.Quirion R, Bowen WD, Itzhak Y, et al. A proposal for the classification of sigma binding sites. Trends Pharmacol Sci. 1992;13:85–86. doi: 10.1016/0165-6147(92)90030-a. [DOI] [PubMed] [Google Scholar]
- 10.Su TP, Hayashi T, Maurice T, et al. The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends Pharmacol Sci. 2010;31:557–566. doi: 10.1016/j.tips.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilner BJ, John CS, Bowen WD. Sigma-1 and sigma-2 receptors are expressed in a wide variety of human and rodent tumor cell lines. Cancer Res. 1995;55:408–413. [PubMed] [Google Scholar]
- 12.Aydar E, Palmer CP, Djamgoz MB. Sigma receptors and cancer: possible involvement of ion channels. Cancer Res. 2004;64:5029–5035. doi: 10.1158/0008-5472.CAN-03-2329. [DOI] [PubMed] [Google Scholar]
- 13.Spruce BA, Campbell LA, McTavish N, et al. Small molecule antagonists of the sigma-1 receptor cause selective release of the death program in tumor and self-reliant cells and inhibit tumor growth in vitro and in vivo. Cancer Res. 2004;64:4875–4886. doi: 10.1158/0008-5472.CAN-03-3180. [DOI] [PubMed] [Google Scholar]
- 14.Brent PJ, Pang G, Little G, et al. The sigma receptor ligand, reduced haloperidol, induces apoptosis and increases intracellular-free calcium levels [Ca2+]i in colon and mammary adenocarcinoma cells. Biochem Biophys Res Commun. 1996;219:219–226. doi: 10.1006/bbrc.1996.0208. [DOI] [PubMed] [Google Scholar]
- 15.Crawford KW, Bowen WD. Sigma-2 receptor agonists activate a novel apoptotic pathway and potentiate antineoplastic drugs in breast tumor cell lines. Cancer Res. 2002;62:313–322. [PubMed] [Google Scholar]
- 16.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 17.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 20.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 21.Fingar DC, Salama S, Tsou C, et al. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 23.She QB, Halilovic E, Ye Q, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010;18:39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carver BS, Chapinski C, Wongvipat J, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Yu D. PI(3)king apart PTEN's role in cancer. Clin Cancer Res. 2010;16:4325–4330. doi: 10.1158/1078-0432.CCR-09-2990. [DOI] [PubMed] [Google Scholar]
- 26.Ostenfeld MS, Fehrenbacher N, Hoyer-Hansen M, et al. Effective tumor cell death by sigma-2 receptor ligand siramesine involves lysosomal leakage and oxidative stress. Cancer Res. 2005;65:8975–8983. doi: 10.1158/0008-5472.CAN-05-0269. [DOI] [PubMed] [Google Scholar]
- 27.Piergentili A, Amantini C, Del Bello F, et al. Novel highly potent and selective sigma 1 receptor antagonists related to spipethiane. J Med Chem. 53:1261–1269. doi: 10.1021/jm901542q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.