Abstract
SLC9A9 (solute carrier family 9, member 9, also known as Na+/H+ exchanger member 9 (NHE9)) has been implicated in human attention deficit hyperactivity disorder (ADHD), autism, and rat studies of hyperactivity and inattentiveness. SLC9A9 is a membrane protein that regulates the luminal pH of the recycling endosome. We recently reported the interactions of SLC9A9 with two molecules: calcineurin homologous protein (CHP) and receptor for activated C-kinase 1 (RACK1). We also reported two novel SLC9A9 mutations and abnormal gene expression profiles in the brains of an inattentive type rat model of ADHD (WKY/NCrl rat). In this study, we further examined the expression and relationship of SLC9A9 and nine additional genes (CHP, RACK1, CaM, PPP3R1, PPP1R10, PKCm, CaMKI, NR2B, PLCb1) that may directly or indirectly interact with SLC9A9 in the hippocampus of the WKY/NCrl rat and the spontaneous hypertensive rat (SHR) model of the combined type of ADHD. We found that the expression levels of these genes were significantly correlated, suggesting that they may be co-regulated. Principal component analysis identified two main factors that accounted for 94% of the expression variance of the ten genes. Significant differences were found for both factors across the three different rat strains. The two ADHD rat models (WKY/NCrl and SHR rats), although different from each other in adulthood, showed similar profiles in adolescence. Both models were significantly different from WKY/NHsd control rats at both ages. The expression abnormalities of each gene were evaluated and their roles in cell signaling processes such as calcium signaling and protein phosphorylation are discussed. Our results suggest that abnormalities in SLC9A9 -mediated signaling pathways could contribute to the ADHD phenotype of two rat models (WKY/NCrl and SHR/NCrl), and that the perturbation of the SLC9A9-network is age-dependent.
Keywords: SLC9A9, ADHD, CHP, RACK1, hippocampus, gene expression
INTRODUCTION
SLC9A9, a member of the Na+/H+ exchanger (NHE), is a transmembrane protein, residing mostly in the recycling endosome. It plays a key role in regulating the pH of endosomes [1, 2]. The endocytic pathway is crucial for the trafficking and membrane distribution of many synaptic proteins such as neurotransmitter receptors and transporters. For example, endosome-dependent trafficking of glutamate receptors is a critical component of long-term potentiation (LTP)[3, 4]. Endocytosis-dependent trafficking of the dopamine transporter (DAT, or SLC6A3) is the major determinant of presynaptic membrane-associated DAT, thus regulating dopamine synapses [5]. Therefore, we believe that SLC9A9 may have the potential to regulate neuronal activities.
Recently, SLC9A9 has been implicated in attention deficit hyperactivity disorder (ADHD), autism (with epilepsy) and rat studies of hyperactivity and inattentiveness. SLC9A9 was first implicated in ADHD by a clinical report of an extended family in which ADHD co-segregated with a pericentric inversion of Chromosome 3 that disrupted both DOCK3 and SLC9A9 [6]. Probands from that family presented with an ADHD-like phenotype with mental retardation. Subsequently, SNPs in SLC9A9 were identified with significant findings in several association studies of ADHD [7–10]. SLC9A9 has also been implicated in autism. For example, a heterozygous stop codon mutation (R423X) that removes the C-terminal of SLC9A9 was found in patients with autism and epilepsy [11].
Evidence from animal studies also supports the involvement of SLC9A9 in animal models of ADHD. Although several molecular and genetic manipulations may produce hyperactive animals, hyperactivity alone is insufficient for the animal to qualify as a model of ADHD. Based on a wider range of criteria – behavioral, genetic and neurobiological – the spontaneously hypertensive rat (SHR) obtained from Charles River, Germany (SHR/NCrl) at present constitutes the best validated animal model of ADHD combined subtype (ADHD-C), and the Wistar Kyoto substrain obtained from Harlan, UK (WKY/NHsd) is its most appropriate control [12, 13]. Two inbred substrains have been created from a cross between SHR and WKY/NHsd: the WKHA/N rat with selection for high spontaneous activity and low systolic blood pressure and the WKHT/N with selection for normal spontaneous activity and high systolic blood pressure [14–17]. QTL mapping of the WKHA/N identifying chromosomal regions responsible for the phenotype, revealed a single genome-wide significant QTL on chromosome 8, designated the Act QTL [18, 19]. SLC9A9 is located within this region and close to the highest predicted LOD score. This region of the rat genome is homologous to the region in human chromosome 3 where the pericentric inversion disrupting SLC9A9 was found in the report from Silva and colleagues [6]. Moreover, the homologous region in mouse chromosome 9 that also contains the gene SLC9A9 has also been reported to contain activity-related QTL by several independent studies [20–22].
We recently reported the behavioral and genetic characterization of a new rat model (WKY/NCrl) of the primarily inattentive subtype of ADHD [13, 23]. These rats demonstrated severe impairment in sustained attention, but normal activity level and impulsiveness. We also reported that the inattentive WKY/NCrl rats were genetically divergent from the isogenic WKY strain (WKY/NHsd) we had used as control strains[23]. These divergent regions included the region of chromosome 8 containing SLC9A9. Resequencing of this region discovered two co-transmitted non-synonymous mutations in exon 16 of the gene SLC9A9 in the WKY/NCrl rats [24].
There are currently 10 known proteins in the Na+/H+ exchanger family, all of which contain 10–14 transmembrane domains forming the Na+/H+ exchanging pore and a long intracellular C-terminal interacting with various signaling molecules, such as calmodulin (CaM)[25], calcineurin-homologous protein (CHP) [26], the receptor for activated kinase C (RACK1) [27], phosphatidylinositol 4,5-bisphosphate (PIP2)[28]and others [29]. The two mutations that we identified in the inattentive WKY/NCrl rats are located in the juxtamembrane C-terminal that also appeared to be putative binding regions for several molecules. We found that the mutations affected SLC9A9 interaction with CHP [24]. Notably, the ADHD-associated pericentric inversion of chromosome 3 and the autism-associated R423X mutation would completely remove all the potential interactions of the SLC9A9 C-terminal. It is not known what functions may be affected by these mutations that affect the protein’s C-terminal. At the molecular level, we believe that the protein-protein interaction network of the NHE proteins, particularly their C-terminals, represents a mechanism by which an abnormality in a single gene could affect multiple pathways and functions. For example, we have shown that SLC9A9 C-terminals interact with CHP and RACK1 [24]. It is not clear whether SLC9A9 also interacts with these other NHE binding partners such as PIP2 and CaM. It is noteworthy that many of these binding molecules participate in intracellular Ca2+ signaling cascades and protein phosphorylation/dephosphorylation, key cellular mechanisms underlying synaptic transmission and neuronal plasticity.
In order to understand the functional networks that SLC9A9 may play a role in and how the mutations of SLC9A9 may perturb the other signaling pathways in neurons, we examined the transcriptional relationship of SLC9A9 with the putative interacting molecules in the hippocampus of two rat strains that model different aspects of the ADHD phenotype. This study is a continuation from our previous gene expression analysis of SLC9A9 in different ADHD rat models, but it is further expanded to an analysis of a network of genes that may directly or indirectly interact with SLC9A9. We focused on the hippocampus for the real time quantitative PCR (qPCR) analysis of the expression of these genes, but also explored the relationship of genes in other brain regions relevant to ADHD (the prefrontal cortex and the substantia nigra and ventral tegmental area (SN_VTA)) using a set of existing microarray data. We chose hippocampus for three reasons. First, SLC9A9 expression in hippocampal neurons has been reported to change in response to modulation of neuronal activity [11]. Secondly, the endosomal protein trafficking pathway is crucial for hippocampal long term potentiation (LTP) [3, 4]. Finally, these potential SLC9A9 -interacting molecules participate in calcium signaling and protein phosphorylation, which are also essential signaling events for hippocampal LTP. We hypothesized that there might be synchronized expression abnormalities in the SLC9A9 gene network. If so, these may be responsible for the behavioral abnormalities seen in ADHD and autism and in rat models of ADHD.
METHODS
Animals
15 male rats at 28 days old (WKY/NCrl=6, SHR/NCrl=6 and WKY/NHsd=3) and 16 adult (~65 days old, WKY/NCrl=5, SHR/NCrl=3, and WKY/NHsd=8) male rats were used in the PCR-based analysis featured in this study. These rats were from two sources: WKY/NCrl and SHR/NCrl rats from Charles River (Sulzfeld, Germany), and WKY/NHsd control rats from Harlan Europe, (Blackthorn, UK). Day 28 reflects the transition point between late childhood and early adolescence in rats. For simplicity, we refer to this age group as “adolescent” in the rest of the article. In this study, we also analyze microarray data generated from a separate cohort of 40 male SHR/NCrl, WKY/NHsd, WKY/NCrl, Sprague Dawley (SD) and Wistar rats. In that study, 8 animals were used at 65 days of age, and the RNA from multiple brain regions compared using the Rat RAE230 2.0 GeneChip (Affymetrix). Those rats were also housed, sacrificed and dissected in the same laboratory in Norway, and shipped to the SUNY laboratory for RNA extraction and microarray analysis. All animal procedures were approved by the Norwegian Animal Research Authority (NARA), and the animals were housed and euthanized at the laboratory facilities at University of Oslo (Oslo, Norway). The experiments were conducted in accordance with the laws and regulations controlling experimental procedures in live animals in Norway and the European Union, including minimizing the number of animals and their suffering. The dissected hippocampus were preserved in RNAlater or AllProtect (Qiagen, CA), and shipped at room temperature to SUNY Upstate Medical University (Syracuse, NY) for the expression analysis.
Expression analysis with quantitative real time PCR (qPCR)
The RNA extraction of the brain tissue and reverse-transcription were done previously [24]. Briefly, total RNA was extracted using the RNeasy mini kit (Qiagen, Valencia, CA). Equal amounts of RNA from each rat were reverse-transcribed using Quantitect Reverse Transcription (RT) kit (Qiagen). Diluted RT reactions were used for qPCR in a Roche LightCycler® 480 Real-Time PCR system using Roche LightCycler 480 SYBR Green I Master reagents. Three genes (CycA, Hprt1, and Ywhaz), whose expression were found to be very stable across different strains and age groups in rat brain tissues including the hippocampus, were used as reference genes for normalization of gene expression [30] (unpublished data). Relative expression levels (in log 2 scale) were calculated based on differences in the number of cycles required to reach the threshold for target amplicon detection (Ct) versus the geometric mean of the three reference genes (the ΔCt method).
SLC9A9 gene expression and a synaptic marker, the synaptophysin (SYP) gene, were examined in our previous studies. The nine additional genes included in the current study are three potential binding partners of SLC9A9 (CHP, RACK1 and CaM) and several molecules that may directly interact with these binding partners and are involved in signaling pathways critical for synaptic plasticity and learning and memory: a type B calcineurin (protein phosphatase 3, regulatory subunit B, alpha isoform, PPP3R1), protein phosphatase 1 subunit 10 (PPP1R10), protein kinase C, mu (PKCm), calcium/calmodulin-dependent protein kinase 1 (CaMKI), glutamate NMDA receptor 2B subunit (NR2B), and phospholipase C, beta 1 (PLCb1). The primers for these genes are listed in the Supplementary Table 1.
Because developmental differences in neuronal numbers and synaptic connections between different ages and strains of animals could be a confounding factor for gross expression analysis, we calculated the gene/SYP ratio (a log 2 difference) to estimate the relative amount of gene expression per synapse. SYP was chosen because its expression is correlated with the basic differentiation process of neurons such as proliferation, fiber outgrowth and the formation of synapses [31–33]. The level of SYP expression was normalized against the GM of the 3 reference genes (ΔCt method). Then, the gene/SYP ratio was calculated as the difference between the specific ΔCt of each individual gene vs. the ΔCt of SYP (ΔΔCt method). All statistical analysis were performed on gene/SYP ratios.
Statistical Analyses
The statistical analyses were performed in STATA 12. Principal Factors Factor Analysis (PFFA) was used to assess the associations among transcripts. The main principal factors were subjected to linear regression analysis and Wald tests for the effects of strain, age and the strain by age interaction. We assessed all 10 individual genes together for the effects of strain, age and the strain by age interaction using seemingly unrelated regression (SUR) analyses with small sample corrections. This method provides joint estimates from several regression models when the dependent variables in these models are correlated. The strain by age interactions were included in our regression model only if there was a significant effect. If there was no significant strain by age interaction, this term was excluded from the model and the results from the main effects model were reported. Any significant main effects were followed up by computing pair-wise comparisons using Wald tests.
Microarray Analyses
3 brain regions were included in this analysis, the hippocampus, the prefrontal cortex (PFC) and the substantia nigra and ventral tegmental area (SN_VTA). RNAs from 8 rats were pooled in equal quantities for each brain region and each strain, and processed in one Affymetrix Rat Genome 230.2 GeneChip according to standard protocols, for a total of 15 arrays. The arrays for SHR/NCrl, WKY/NHsd, and Sprague Dawley (SD) rats have been used in previous analysis and are available at Gene Expression Omnibus (GEO) repository (record # GSE12457 [34]). After scanning, the .cel files were imported into Partek Genomics Suite 6.6 (Partek Inc. St. Louis, MO, USA) using GCRMA normalization. Because there is only one pooled sample per brain region per strain, we could not consider the effects of both strain and region statistically. Also because these data had not been generated to demonstrate strain effects, these effects were confounded by processing date. Thus, we removed the effects of strain/processing date in Partek [35], in order to improve the sample size and data quality for analyzing brain region effects. By doing so, we have 5 arrays for each of the three rat brain regions represented by rats from 5 different strains. Normalized log 2 expression values of probes for the same 10 genes of interest, as well as SYP and the housekeeping genes (CycA, Hprt1, and Ywhaz), were extracted from the array data and imported into Stata 12 for additional analysis. We also included one of the two probes for SLC6A3 (dopamine transporter, DAT), that demonstrated 5 fold higher expression in SN_VTA region than the other two regions. We used linear regression to normalize the expression of genes of interest to the housekeeping genes and SYP (at the probe level). We have found previously that this method results in similar normalization effects as the delta CT approach in real time PCR data. Subsequently, PFFA was applied to extract the first two main factors for each individual brain region, as well as the three regions combined. The rotated factor loadings were plotted for each analysis.
RESULTS
Principal factor analysis of real time PCR data
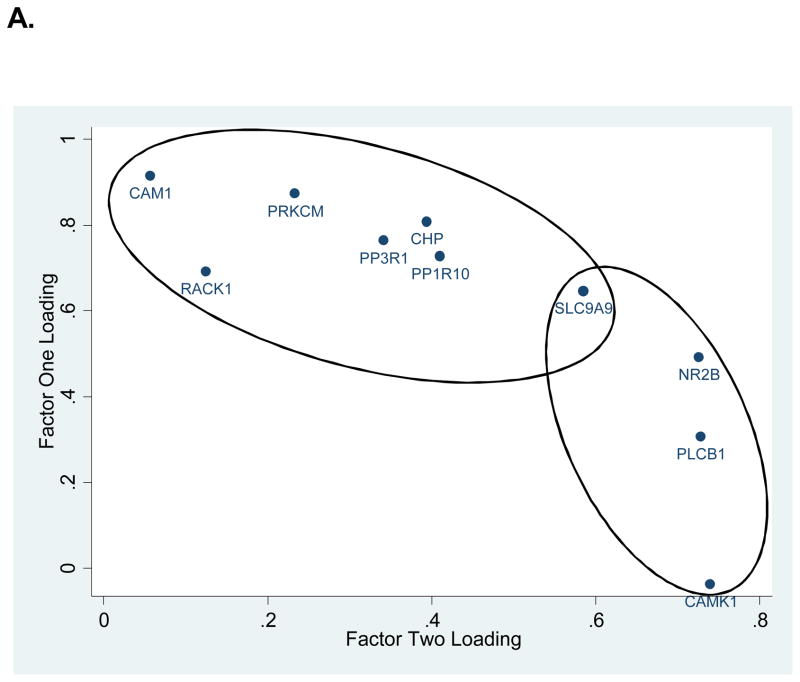
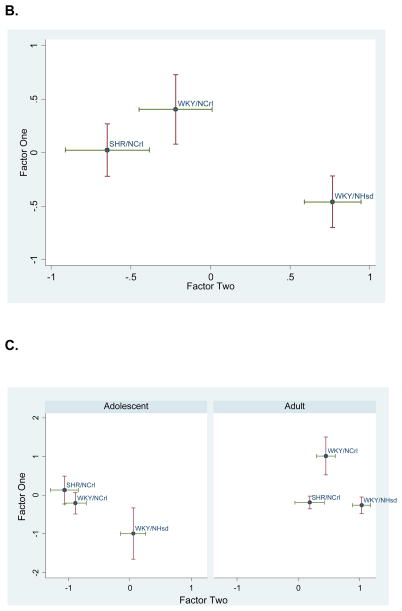
Two factors with eigenvalues greater than one were identified. These explained 79% and 15%, of the variance, respectively, and together accounted for 94% of the variance in gene expression. Figure 1A shows the factor loadings for each transcript. Notably, SLC9A9 loaded equally on both factors. The other 9 genes appeared to segregate into two groups (indicated by the shaded ovals and rectangles in Figure 1A), with each group representing mainly one factor. Linear regression analysis of these two principal factors revealed no strain by age interactions, significant strain effects for factors one (F(2, 28) = 3.73, p=0.0366) and two (F(2, 28)= 12.38, p=0.0001) and significant age effects for factor two (F(1, 28) =56.71, p<0.0001). The strain effects for both factors are plotted in Figure 1B. Figure 1C further plots this expression profile in two age groups. It shows that both ADHD animal models differed similarly from the WKY/NHsd control group at the adolescent age; at the adult age the two ADHD animal models did not differ significantly from one another and were both different from the control group. Regression analysis results for both factor scores are summarized in Tables 1 and 2. Table 3 shows further pairwise strain comparisons using both factor scores.
Figure 1. Principal Factors Factor Analysis (PFFA) results.
A. Factor loadings for all genes were plotted in 2D. Ovals group the genes with factor loading predominantly in one of the two factors. Notice that SLC9A9 loads equally on both factors. B. Factor scores for all animals are plotted as group means with one standard error in two dimensions. C. Factor scores separated by age and strain. Note that the two ADHD rat models (SHR/NCrl and WKY/NCrl) were significantly deviated from the control WKY/NHsd rats, and that the relative separation between the three strains appeared to vary by age.
Table 1.
Overall Analysis Results for Individual Genes and Factor Scores
| Overall Effect | Strain Effects | Age Effects | Interaction | |||
|---|---|---|---|---|---|---|
| Gene | χ2(2) | p -value | χ2(1) | p -value | χ2(2) | p -value |
| RACK1 | 13.77 | 0.001 | 5.03 | 0.02 | 0.46 | 0.79 |
| CaMK1 | 14.66 | 0.0007 | 8.28 | 0.004 | 7.73 | 0.02 |
| CHP | 0.71 | 0.70 | 9.38 | 0.0022 | 1.72 | 0.42 |
| PPP1R10 | 1.46 | 0.48 | 2.72 | 0.10 | 5.28 | 0.07 |
| PKCm | 8.7 | 0.01 | 12.02 | 0.0005 | 5.9 | 0.05 |
| PPP3R1 | 0.82 | 0.67 | 3.11 | 0.08 | 2.65 | 0.27 |
| PLCB1 | 14.97 | 0.0006 | 29.77 | <0.0001 | 1.61 | 0.45 |
| CaM | 6.48 | 0.04 | 4.35 | 0.04 | 2 | 0.37 |
| NR2B | 2.45 | 0.29 | 28.4 | <0.0001 | 2.14 | 0.34 |
| SLC9A9 | 3.72 | 0.16 | 19.14 | <0.0001 | 6.02 | 0.049 |
| Factor One | 3.73 | 0.04 | 3.82 | 0.06 | 1.99 | 0.16 |
| Factor Two | 12.38 | <0.0001 | 56.71 | <0.0001 | 0.45 | 0.64 |
Table 2.
Strain effects within age groups.
| Strain Effect | Adolescence | Adult | ||
|---|---|---|---|---|
| Gene | χ2(2) | p -value | χ2(2) | p -value |
| RACK1 | 2.83 | 0.24 | 14.19 | 0.0008 |
| CaMK1 | 15.46 | 0.0004 | 0.8 | 0.67 |
| CHP | 0.22 | 0.9 | 2.7 | 0.26 |
| PPP1R10 | 0.58 | 0.75 | 5.91 | 0.52 |
| PKCm | 3.81 | 0.15 | 13.03 | 0.0015 |
| PPP3R1 | 0.39 | 0.82 | 4.18 | 0.12 |
| PLCB1 | 3.54 | 0.17 | 11.89 | 0.0026 |
| CaM | 6.57 | 0.04 | 2.72 | 0.26 |
| NR2B | 0.06 | 0.97 | 8.22 | 0.02 |
| SLC9A9 | 0.57 | 0.75 | 7.64 | 0.02 |
| Factor One | 1.71 | 0.22 | 4.33 | 0.03 |
| Factor Two | 5.83 | 0.02 | 6.45 | 0.01 |
Table 3.
Strain Comparison Using Factor Scores.
| Strain Difference | Adolescence | Adult | Overall | |||
|---|---|---|---|---|---|---|
| Factor One | T | p -value | T | p -value | T | p -value |
| WKY/NCrl vs WKY/NHsd | 1.29 | 0.22 | 2.81 | 0.01 | 2.25 | 0.03 |
| SHR/NCrl vs WKY/NHsd | 1.85 | 0.09 | 0.12 | 0.9 | 1.17 | 0.25 |
| WKY/NCrl vs SHR/NHsd | 0.68 | 0.51 | 2.02 | 0.06 | 0.93 | 0.35 |
| Factor Two | T | p -value | T | p -value | T | p -value |
| WKY/NCrl vs WKY/NHsd | 2.80 | 0.02 | 2.71 | 0.01 | 3.23 | 0.003 |
| SHR/NCrl vs WKY/NHsd | 3.32 | 0.006 | 3.14 | 0.007 | 4.30 | <0.0001 |
| WKY/NCrl vs SHR/NHsd | 0.65 | 0.53 | 0.93 | 0.36 | 1.33 | 0.19 |
Expression changes for each individual gene
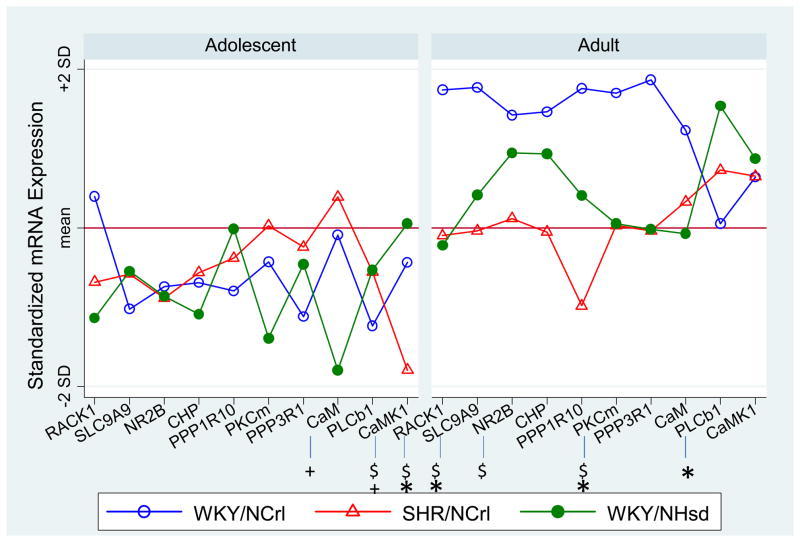
The SUR analyses (in Table 1) showed that only two transcripts showed significant strain by age interactions, CaMKI (χ2 (2) = 7.73, p=0.0209) and SLC9A9 (χ2 (2)= 6.02, p=0.0492). We found significant effects of strain for 5 of 10 transcripts and significant effects of age for 8 out of 10 transcripts. The 5 transcripts that showed significant strain effects are RACK1, CaMKI, PKCm, PLCB1, CaM. The results are summarized in Table 1. To clarify the nature of the interactions, we evaluated the strain effects within each age group (Table 2). Figure 2 plots the normalized mean expression level for these 10 genes separated by age group, with symbols highlighting the genes that demonstrated significant strain effects within each age group (p<0.05).
Figure 2.
Standardized expression level for each individual gene within age groups. Significant strain effects (p<0.05) were highlighted with by * (WKY/NCrl vs WKY/NHsd), +(SHR/NCrl vs WKY/NHsd), and $(WKY/NCrl vs SHR/NCrl). Note that the WKY/NCrl and SHR/NCrl rats represent models of the primarily inattentive and combined subtypes of ADHD, respectively.
Principal factor analysis of Microarray data
The first two main factors accounted for 74.5%, 69.1%, 70.6% or 56.5% of total variance for the Hippocampus, PFC, SN_VTA, or all three regions combined, respectively. The rotated loadings were plotted in Supplementary Figure 1. SLC9A9 loaded high on factor 2 for the PFC, factor 1 for the Hippocampus and SN_VTA regions, and equally on both factors for the three regions combined. Genes that were loaded high (positively and negatively) on the same factor as SLC9A9 consistently across three individual brain regions were RACK1, NR2B and CaMK1. In the SN_VTA, CHP, CaM, SLC6A3 and PPP3R1 also loaded high with SLC9A9 on factor 1. When the three brain regions were combined, we obtained a factor loading pattern that was very similar to the real time PCR data on hippocampus, in which SLC9A9 loaded equally on both factors and all other genes were loaded positively on one of the two main factors.
DISCUSSION
Like other Na+/H+ exchangers, SLC9A9 appears to interact with a number of molecules to facilitate a local network of signaling pathways. In this study we examined the SLC9A9 gene network by assaying gene expression in the hippocampus of two strains of ADHD rat models in comparison with a control WKY rat strain using real time quantitative PCR(qPCR). The expression relationship of genes within the network was further explored in three different brain regions using a separate cohort of rat microarray data. Both results consistently showed that these genes are co-regulated and the qPCR results further showed that this network is differentially regulated in different strains of rat models for ADHD.
We chose three potential direct binding partners of the SLC9A9 : CHP, RACK1 and CaM. We reported previously that CHP and RACK1 bind to the SLC9A9 C-terminal. CaM is known to bind to NHE family members such as SLC9A1 [25]. Although it is not clear if CaM would interact with SLC9A9, the calmodulin target database (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/home.html) predicted a putative calmodulin binding site (145HAGYSLKKRHFFQNLG SIL163) for SLC9A9. This site is highly homologous to a putative binding site (animo acid 195–213) for SLC9A7, which has been shown to mediate the interaction of SLC9A7 with CaM [36].
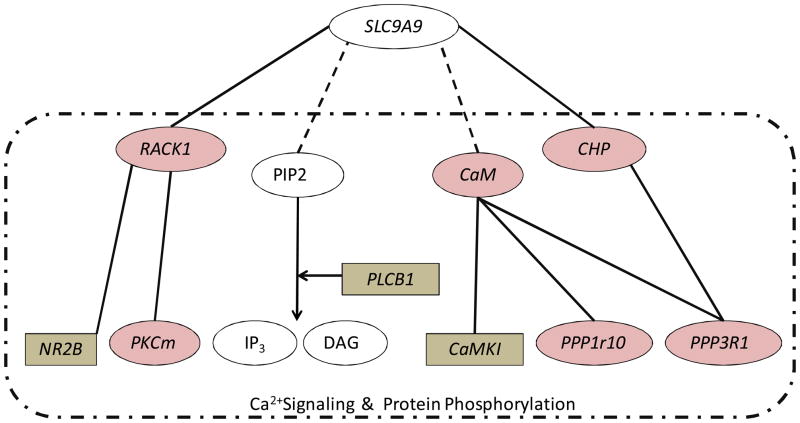
Six other molecules that may indirectly interact with SLC9A9 were also included in this expression analysis. The known or predicted interactions between these molecules are summarized in Figure 3. It is important to note that many of these are involved in intracellular Ca2+signaling and protein phosphorylation, essential signaling pathways for many cellular functions, particularly synaptic transmission and plasticity. For example, CHP and CaM, two Ca2+-binding proteins, can either inhibit or activate calcineurin (CaN), the main phosphotase that dephosphorylates membrane receptors and ion channels to counter-balance the phosphorylation of these proteins by CaM-dependent kinases. The phosphorylation of the N-methyl d-aspartate (NMDA) receptor is a crucial signaling component underlying long term changes in glutamate synaptic efficacy including both LTP and long-term depression (LTD) [37, 38]. Other molecules that we examined such as phospholipase C (PLC) can cleave a potential SLC9A9 -binding membrane phospholipid, PIP2, giving rise to the second messengers inositol 1,4,5-triphosphate (IP3) and diacylglycerol (DAG). IP3 binds to IP3 receptors and stimulates the release of Ca2+ ions from the endoplasmic reticulum (ER), whereas DAG is an activator of the Ca2+-sensitive protein kinase PKC.
Figure 3. SLC9A9 network examined in the current expression study.
Solid lines indicate known interactions, and dashed lines are predicted interactions. Shaded ovals are proteins whose expression loaded mainly on factor one. Shaded rectangles are the ones that loaded mainly on factor two. All molecules within the dash-lined box are known to play roles in intracellular Ca2+ signaling and/or protein phosphorylation
SLC9A9’s binding partner, RACK1, can bind to IP3 receptors (IP3R) and regulates Ca2+ release from intracellular stores by enhancing the affinity of IP3R binding for IP3 [39]. RACK1, in addition to being an adaptor for PKC, also binds to the dopamine transporter (DAT) [40]. DAT is upregulated in the brains of ADHD patients [41–43] and is the primary target of the stimulant drugs that treat ADHD [44, 45]. RACK1 mediated PKC activation regulates DAT phosphorylation and trafficking, thus maintaining appropriate levels of presynaptic membrane DAT and dopamine uptake[40]. In addition to its role in regulating the PKC pathway and ER-release of Ca2+, RACK1 also directly binds to the NMDA receptor subunit NR2B, and negatively regulates the phosphorylation of NR2B through Fyn kinase and decreases NMDA receptor-mediated currents in CA1 hippocampal slices [46]. Collectively, the SLC9A9 gene network may participate in intracellular signaling pathways that regulate synaptic transmission and plasticity, and lead to alterations in behavior such as attention, addiction, learning and memory. Supporting this idea, our microarray results showed that both the expression of NR2B and DAT were highly co-regulated with SLC9A9 in the specific rat brain regions examined, as suggested by their high loading on the same factors as SLC9A9 (Supplementary Figure 1). Consistent with this idea, abnormalities in hippocampal LTP and elevated DAT binding have been reported for SHR/NCrl and WKY/NCrl rats [47, 48].
It is noteworthy that SLC9A9 binding partners such as RACK1 and CaM are key signaling molecules with the capability of regulating or interacting with a wide variety of downstream signaling molecules. Although in this study we have only examined the expression of a few selected secondary interacting genes, our factor analysis results confirmed the relationship of these genes with SLC9A9 and suggested possible co-regulation of their expression. In addition, as a group (represented by the two main factors), the SLC9A9 signaling network showed a significantly different expression profile for the ADHD rat models in comparison with the WKY control strain (Figure 1B, 1C), indicating that dysregulation of the SLC9A9 signaling network may play a role in the pathophysiology of both ADHD animal models. This network may play a particular role in inattentiveness, which is a shared feature of the two rat models.
Examining the strain differences for each individual gene by age, we found that CaM and CaMKI were the only two genes showed significant strain effect in adolescence (Figure 2 left). Although the strain effect was mostly due to the significantly deviated expression in the SHR rats, it is noteworthy that the changes seen for CaM and CaMK1 in adolescence (increases for CaM and decreases for CaMK1) were in the same direction for both ADHD models compared with the control WKY/NHsd rats. This suggests a similar abnormality of Ca2+/CaM/CaMK dependent signaling pathway in both models. The similarity between the two ADHD models was more clearly demonstrated by the highly similar profiles represented by the two principal factors at adolescence (Figure 1C left). More robust changes in gene expression and more genes with significant strain differences were observed in the adult age. At this later time point, the WKY/NCrl rats significantly differed from the control rats (Figure 2 right). However, a different set of genes showed significant effects of strain at the adult age. For example the significant findings for CaM and CaMK1 during adolescence were no longer observed in adulthood. Instead, we found significant strain differences for SLC9A9, RACK1, PLCb1, PKCm and NR2B genes in adulthood. Moreover, some genes with significant strain differences in the adult age were changed in the opposite directions for the SHR and WKY/NCrl rats in comparison with the control rats (Figure 2 Right). These strain differences were also revealed by factor scores (Figure 1C. Right. Table 3). This suggests that the SHR and WKY/NCrl rats may demonstrate different functional and behavioral abnormalities in adulthood due to different abnormal hippocampal gene expression. For example, the differential abnormality in expression of the RACK1/PKC pathway may affect DAT phoshorylation and trafficking differently in the animal models, supported by observation of different responses to methylphenidate treatment-induced normalization of DAT density in the two animal models [48]. However, we could not exclude the possibility that the different expression between the two models in adulthood was due to hypertension in adult SHR rats. Additional studies are needed to clarify this possibility.
A major limitation of the current study is the small sample size for the PCR data. This may have impaired our power to detect significant effects. For example, some of the genes that showed no significant strain or age effects, may have become statistically significant if we had had more animals. This limitation, however, was partly overcome by our network based analysis approach, which examined the expression of multiple genes and their relationship at once. Factor analysis not only extracts the common patterns shared by many different genes, it also helps to improve statistical power by limiting the number of statistical tests and creating summary factor scores that are more reliable than individual gene expression values. For example, multiple genes with small but non-significant changes can together lead to bigger and statistically significant effects, which may be captured by factor analysis. In addition, our microarray data set has more samples per brain region. Each of the five arrays comprised pooled RNA samples from 8 rats. With this larger sample size, we found similar patterns of factor loadings for the microarray data as we had for the PCR data, validating our PCR results, despite the small sample size.
In summary, SLC9A9 may interact with a variety of signaling molecules and participate in the intracellular signaling networks that are important for synaptic function. Taken together with our prior work, it appears that age-dependent dysregulation of gene expression of SLC9A9 and its signaling networks may contribute to ADHD phenotypes in the animal models we studied.
Supplementary Material
Supplementary Table 1. Primer sequences.
Supplementary Figure 1. Principal Factors Factor Analysis of microarray data from three brain regions: Prefrontal cortex, substantia nigra and vental tegmental area (SN_VTA) and hippocampus. or three brain region combined. Ovals group the genes with factor loading predominantly in one of the two factors in the combined brain region analysis. Notice that SLC9A9 loads equally on both factors, and appears most closely associated with NR2B, CAMK1, PPP1R10, and PRKCd across the three brain regions combined, although there were differences in the loadings within each region. Some of the genes were represented by multiple probes in the array, which were denoted by ‘_’ followed by a number.
Acknowledgments
We thank Karen L Gentile and Lu Liu for technical assistance. Cheryl Roe and Dr. Stephen Glatt for statistical advice. This study was supported by National Institutes of Health grant MH668877.
Footnotes
Conflict of interest
In previous years, Dr. Sagvolden (deceased) had received consulting fees or research support or has been on Advisory Boards or has been a speaker for: Shire, Janssen, and Eli Lilly. In the past year, Dr. Faraone received consulting income and/or research support from Shire, Otsuka and Alcobra and research support from the National Institutes of Health (NIH). In previous years, he received consulting fees or was on Advisory Boards or participated in continuing medical education programs sponsored by: Shire, McNeil, Janssen, Novartis, Pfizer and Eli Lilly. Dr. Faraone receives royalties from books published by Guilford Press: Straight Talk about Your Child’s Mental Health and Oxford University Press: Schizophrenia: The Facts. Drs. Middleton and Zhang-James, reported no biomedical financial interests or potential conflicts of interest.
Contributor Information
Yanli Zhang-James, Email: zhangy@upstate.edu.
Frank A. Middleton, Email: middletf@upstate.edu.
Stephen V Faraone, Email: sfaraone@childpsychresearch.org.
References
- 1.Nakamura N, Tanaka S, Teko Y, Mitsui K, Kanazawa H. Four Na+/H+ exchanger isoforms are distributed to Golgi and post-Golgi compartments and are involved in organelle pH regulation. J Biol Chem. 2005;280(2):1561–72. doi: 10.1074/jbc.M410041200. [DOI] [PubMed] [Google Scholar]
- 2.Roxrud I, Raiborg C, Gilfillan GD, Stromme P, Stenmark H. Dual degradation mechanisms ensure disposal of NHE6 mutant protein associated with neurological disease. Exp Cell Res. 2009;315(17):3014–27. doi: 10.1016/j.yexcr.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD. Recycling endosomes supply AMPA receptors for LTP. Science. 2004;305(5692):1972–5. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- 4.Park M, Salgado JM, Ostroff L, Helton TD, Robinson CG, Harris KM, Ehlers MD. Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron. 2006;52(5):817–30. doi: 10.1016/j.neuron.2006.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melikian HE, Buckley KM. Membrane trafficking regulates the activity of the human dopamine transporter. J Neurosci. 1999;19(18):7699–710. doi: 10.1523/JNEUROSCI.19-18-07699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Silva MG, Elliott K, Dahl HH, Fitzpatrick E, Wilcox S, Delatycki M, Williamson R, Efron D, Lynch M, Forrest S. Disruption of a novel member of a sodium/hydrogen exchanger family and DOCK3 is associated with an attention deficit hyperactivity disorder-like phenotype. J Med Genet. 2003;40(10):733–40. doi: 10.1136/jmg.40.10.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lasky-Su J, Anney RJ, Neale BM, Franke B, Zhou K, Maller JB, Vasquez AA, Chen W, Asherson P, Buitelaar J, et al. Genome-wide association scan of the time to onset of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(8):1355–1358. doi: 10.1002/ajmg.b.30869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lasky-Su J, Neale BM, Franke B, Anney RJ, Zhou K, Maller JB, Vasquez AA, Chen W, Asherson P, Buitelaar J, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(8):1345–54. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- 9.Markunas CA, Quinn KS, Collins AL, Garrett ME, Lachiewicz AM, Sommer JL, Morrissey-Kane E, Kollins SH, Anastopoulos AD, Ashley-Koch AE. Genetic variants in SLC9A9 are associated with measures of attention-deficit/hyperactivity disorder symptoms in families. Psychiatr Genet. 2010;20(2):73–81. doi: 10.1097/YPG.0b013e3283351209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch KP, Faraone SV, Nguyen TT, Schafer H, Holmans P, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010;49(9):884–97. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321(5886):218–23. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sagvolden T. Behavioral validation of the spontaneously hypertensive rat (SHR) as an animal model of attention-deficit/hyperactivity disorder (AD/HD) Neurosci Biobehav Rev. 2000;24(1):31–9. doi: 10.1016/s0149-7634(99)00058-5. [DOI] [PubMed] [Google Scholar]
- 13.Sagvolden T, Johansen EB, Woien G, Walaas SI, Storm-Mathisen J, Bergersen LH, Hvalby O, Jensen V, Aase H, Russell VA, et al. The spontaneously hypertensive rat model of ADHD--the importance of selecting the appropriate reference strain. Neuropharmacology. 2009;57(7–8):619–26. doi: 10.1016/j.neuropharm.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendley ED, Atwater DG, Myers MM, Whitehorn D. Dissociation of genetic hyperactivity and hypertension in SHR. Hypertension. 1983;5(2):211–7. doi: 10.1161/01.hyp.5.2.211. [DOI] [PubMed] [Google Scholar]
- 15.Hendley ED, Wessel DJ, Van Houten J. Inbreeding of Wistar-Kyoto rat strain with hyperactivity but without hypertension. Behav Neural Biol. 1986;45(1):1–16. doi: 10.1016/s0163-1047(86)80001-2. [DOI] [PubMed] [Google Scholar]
- 16.Sagvolden T, Hendley ED, Knardahl S. Behavior of hypertensive and hyperactive rat strains. FASEB Journal. 1990;4:A1065–A1065. doi: 10.1016/0031-9384(92)90432-2. [DOI] [PubMed] [Google Scholar]
- 17.Sagvolden T, Hendley ED, Knardahl S. Behavior of hypertensive and hyperactive rat strains: Hyperactivity is not unitarily determined. Physiology and Behavior. 1992;52:49–57. doi: 10.1016/0031-9384(92)90432-2. [DOI] [PubMed] [Google Scholar]
- 18.Moisan MP, Courvoisier H, Bihoreau MT, Gauguier D, Hendley ED, Lathrop M, James MR, Mormede P. A major quantitative trait locus influences hyperactivity in the WKHA rat. Nat Genet. 1996;14(4):471–3. doi: 10.1038/ng1296-471. [DOI] [PubMed] [Google Scholar]
- 19.Moisan MP, Llamas B, Cook MN, Mormede P. Further dissection of a genomic locus associated with behavioral activity in the Wistar-Kyoto hyperactive rat, an animal model of hyperkinesis. Mol Psychiatry. 2003;8(3):348–52. doi: 10.1038/sj.mp.4001234. [DOI] [PubMed] [Google Scholar]
- 20.Miner LL, Marley RJ. Chromosomal mapping of the psychomotor stimulant effects of cocaine in BXD recombinant inbred mice. Psychopharmacology (Berl) 1995;122(3):209–14. doi: 10.1007/BF02246541. [DOI] [PubMed] [Google Scholar]
- 21.Mathis C, Neumann PE, Gershenfeld H, Paul SM, Crawley JN. Genetic analysis of anxiety-related behaviors and responses to benzodiazepine-related drugs in AXB and BXA recombinant inbred mouse strains. Behav Genet. 1995;25(6):557–68. doi: 10.1007/BF02327579. [DOI] [PubMed] [Google Scholar]
- 22.Grisel JE, Belknap JK, O’Toole LA, Helms ML, Wenger CD, Crabbe JC. Quantitative trait loci affecting methamphetamine responses in BXD recombinant inbred mouse strains. J Neurosci. 1997;17(2):745–54. doi: 10.1523/JNEUROSCI.17-02-00745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sagvolden T, Dasbanerjee T, Zhang-James Y, Middleton F, Faraone S. Behavioral and genetic evidence for a novel animal model of Attention-Deficit/Hyperactivity Disorder Predominantly Inattentive Subtype. Behav Brain Funct. 2008;4:56. doi: 10.1186/1744-9081-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang-James Y, Dasbanerjee T, Sagvolden T, Middleton FA, Faraone SV. SLC9A9 mutations, gene expression, and protein-protein interactions in rat models of attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(7):835–43. doi: 10.1002/ajmg.b.31229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertrand B, Wakabayashi S, Ikeda T, Pouyssegur J, Shigekawa M. The Na+/H+ exchanger isoform 1 (NHE1) is a novel member of the calmodulin-binding proteins. Identification and characterization of calmodulin-binding sites. J Biol Chem. 1994;269(18):13703–9. [PubMed] [Google Scholar]
- 26.Lin X, Barber DL. A calcineurin homologous protein inhibits GTPase-stimulated Na-H exchange. Proc Natl Acad Sci U S A. 1996;93(22):12631–6. doi: 10.1073/pnas.93.22.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohgaki R, Fukura N, Matsushita M, Mitsui K, Kanazawa H. Cell surface levels of organellar Na+/H+ exchanger isoform 6 are regulated by interaction with RACK1. J Biol Chem. 2008;283(7):4417–29. doi: 10.1074/jbc.M705146200. [DOI] [PubMed] [Google Scholar]
- 28.Aharonovitz O, Zaun HC, Balla T, York JD, Orlowski J, Grinstein S. Intracellular pH regulation by Na(+)/H(+) exchange requires phosphatidylinositol 4,5-bisphosphate. J Cell Biol. 2000;150(1):213–24. doi: 10.1083/jcb.150.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malo ME, Fliegel L. Physiological role and regulation of the Na+/H+ exchanger. Can J Physiol Pharmacol. 2006;84(11):1081–95. doi: 10.1139/y06-065. [DOI] [PubMed] [Google Scholar]
- 30.Bonefeld BE, Elfving B, Wegener G. Reference genes for normalization: a study of rat brain tissue. Synapse. 2008;62(4):302–9. doi: 10.1002/syn.20496. [DOI] [PubMed] [Google Scholar]
- 31.Grabs D, Bergmann M, Schuster T, Fox PA, Brich M, Gratz M. Differential expression of synaptophysin and synaptoporin during pre- and postnatal development of the rat hippocampal network. Eur J Neurosci. 1994;6(11):1765–71. doi: 10.1111/j.1460-9568.1994.tb00569.x. [DOI] [PubMed] [Google Scholar]
- 32.Bergmann M, Schuster T, Grabs D, Marqueze-Pouey B, Betz H, Traurig H, Mayerhofer A, Gratzl M. Synaptophysin and synaptoporin expression in the developing rat olfactory system. Brain Res Dev Brain Res. 1993;74(2):235–44. doi: 10.1016/0165-3806(93)90009-y. [DOI] [PubMed] [Google Scholar]
- 33.Bergmann M, Lahr G, Mayerhofer A, Gratzl M. Expression of synaptophysin during the prenatal development of the rat spinal cord: correlation with basic differentiation processes of neurons. Neuroscience. 1991;42(2):569–82. doi: 10.1016/0306-4522(91)90399-9. [DOI] [PubMed] [Google Scholar]
- 34.DasBanerjee T, Middleton FA, Berger DF, Lombardo JP, Sagvolden T, Faraone SV. A comparison of molecular alterations in environmental and genetic rat models of ADHD: a pilot study. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(8):1554–63. doi: 10.1002/ajmg.b.30877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inc P. Partek® software On-line Help, version 6.3, build 6.08.0110; Copyright ©2008. St. Louis, MO, USA: Removing Batch Effects in Partek. [Google Scholar]
- 36.Kagami T, Chen S, Memar P, Choi M, Foster LJ, Numata M. Identification and biochemical characterization of the SLC9A7 interactome. Mol Membr Biol. 2008;25(5):436–47. doi: 10.1080/09687680802263046. [DOI] [PubMed] [Google Scholar]
- 37.Belmeguenai A, Hansel C. A role for protein phosphatases 1, 2A, and 2B in cerebellar long-term potentiation. J Neurosci. 2005;25(46):10768–72. doi: 10.1523/JNEUROSCI.2876-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mansuy IM, Mayford M, Jacob B, Kandel ER, Bach ME. Restricted and regulated overexpression reveals calcineurin as a key component in the transition from short-term to long-term memory. Cell. 1998;92(1):39–49. doi: 10.1016/s0092-8674(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 39.Patterson RL, van Rossum DB, Barrow RK, Snyder SH. RACK1 binds to inositol 1,4,5-trisphosphate receptors and mediates Ca2+ release. Proc Natl Acad Sci U S A. 2004;101(8):2328–32. doi: 10.1073/pnas.0308567100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee KH, Kim MY, Kim DH, Lee YS. Syntaxin 1A and receptor for activated C kinase interact with the N-terminal region of human dopamine transporter. Neurochem Res. 2004;29(7):1405–9. doi: 10.1023/b:nere.0000026404.08779.43. [DOI] [PubMed] [Google Scholar]
- 41.Cheon KA, Ryu YH, Kim YK, Namkoong K, Kim CH, Lee JD. Dopamine transporter density in the basal ganglia assessed with [123I]IPT SPET in children with attention deficit hyperactivity disorder. Eur J Nucl Med Mol Imaging. 2003;30(2):306–11. doi: 10.1007/s00259-002-1047-3. [DOI] [PubMed] [Google Scholar]
- 42.Dougherty DD, Bonab AA, Spencer TJ, Rauch SL, Madras BK, Fischman AJ. Dopamine transporter density is elevated in patients with ADHD. Lancet. 1999;354(9196):2132–2133. doi: 10.1016/S0140-6736(99)04030-1. [DOI] [PubMed] [Google Scholar]
- 43.Dresel S, Krause J, Krause KH, LaFougere C, Brinkbaumer K, Kung HF, Hahn K, Tatsch K. Attention deficit hyperactivity disorder: binding of [99mTc]TRODAT-1 to the dopamine transporter before and after methylphenidate treatment. Eur J Nucl Med. 2000;27(10):1518–24. doi: 10.1007/s002590000330. [DOI] [PubMed] [Google Scholar]
- 44.Federici M, Geracitano R, Bernardi G, Mercuri NB. Actions of methylphenidate on dopaminergic neurons of the ventral midbrain. Biol Psychiatry. 2005;57(4):361–5. doi: 10.1016/j.biopsych.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 45.Capp PK, Pearl PL, Conlon C. Methylphenidate HCl: therapy for attention deficit hyperactivity disorder. Expert Review of Neurotherapeutics. 2005;5(3):325–331. doi: 10.1586/14737175.5.3.325. [DOI] [PubMed] [Google Scholar]
- 46.Yaka R, Thornton C, Vagts AJ, Phamluong K, Bonci A, Ron D. NMDA receptor function is regulated by the inhibitory scaffolding protein, RACK1. Proc Natl Acad Sci U S A. 2002;99(8):5710–5. doi: 10.1073/pnas.062046299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jensen V, Rinholm JE, Johansen TJ, Medin T, Storm-Mathisen J, Sagvolden T, Hvalby O, Bergersen LH. N-methyl-d-aspartate receptor subunit dysfunction at hippocampal glutamatergic synapses in an animal model of attention-deficit/hyperactivity disorder. Neuroscience. 2008 doi: 10.1016/j.neuroscience.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 48.Roessner V, Sagvolden T, Dasbanerjee T, Middleton FA, Faraone SV, Walaas SI, Becker A, Rothenberger A, Bock N. Methylphenidate normalizes elevated dopamine transporter densities in an animal model of the attention-deficit/hyperactivity disorder combined type, but not to the same extent in one of the attention-deficit/hyperactivity disorder inattentive type. Neuroscience. 2010;167(4):1183–91. doi: 10.1016/j.neuroscience.2010.02.073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Primer sequences.
Supplementary Figure 1. Principal Factors Factor Analysis of microarray data from three brain regions: Prefrontal cortex, substantia nigra and vental tegmental area (SN_VTA) and hippocampus. or three brain region combined. Ovals group the genes with factor loading predominantly in one of the two factors in the combined brain region analysis. Notice that SLC9A9 loads equally on both factors, and appears most closely associated with NR2B, CAMK1, PPP1R10, and PRKCd across the three brain regions combined, although there were differences in the loadings within each region. Some of the genes were represented by multiple probes in the array, which were denoted by ‘_’ followed by a number.