Abstract
The tumor suppressor gene FBXW7 is mutated in numerous types of human cancers leading to loss of its function and/or expression. However the clinic significance of FBXW7 alterations remains largely unknown. Here, we carried out a meta-analysis of 10 gene expression microarray studies for a total 1900 patients of breast cancer with clinic information to evaluate the prognostic impact of FBXW7 mRNA expression. The FBXW7 mRNA levels significantly reduced in breast cancer compared to normal tissues. In addition, significant difference in the FBXW7 mRNA levels was found among molecular subtypes (normal-like, luminal A, luminal B, ERBB2 and basal). ERBB2 and basal tumors had significantly lower average FBXW7 mRNA level than normal-like tumors, whereas luminal A and B tumors have the lowest average FBXW7 mRNA level. The patients with higher FBXW7 mRNA level significantly increased disease-free survival, particularly in the group of patients with ER negative and basal subtype tumors. Moreover, higher FBXW7 mRNA level also significantly increased overall survival in the patients with ER negative tumors. But we strikingly found opposite effect of FBXW7 expression on overall survival in different subtypes. The patients with higher FBXW7 mRNA level significantly decreased overall survival in normal-like subtype while the patients with higher FBXW7 mRNA level significantly increased overall survival in ERBB2 and Basal subtype. Taken together, our results suggest that FBXW7 mRNA levels were a prognostic factor for disease-free and overall survival according to ER status and molecular subtypes.
Keywords: Breast cancer, FBXW7, meta-analysis, transcriptional profile, prognosis
Introduction
Breast cancer is the most frequent malignancy in women worldwide (1, 2) and has had a major impact on both health and the economy of the United States (1). While some improvements have been made in diagnosis and treatment of breast cancer, the prognosis and survival for most patients has not dramatically changed. Many women who are at low risk of disease progression are subjected to unnecessary, aggressive therapy from which they are unlikely to benefit, because of inadequacy in reliable predictors of outcome. Many patients in the poor prognosis category either fail to respond to available treatments, or show transient responses followed by development of drug resistance. Furthermore, breast tumors display remarkable biological and clinical heterogeneity. Developing a better understanding of the molecular basis of such heterogeneity is urgently required for individualized treatment for patients based on molecular profiling.
Several endpoints including overall survival (OS) and disease-free survival (DFS) have been used to assess the clinic benefit of cancer treatments. OS has long been the gold standard primary endpoint for the demonstration of clinical benefit. OS denotes the proportion of patients remain alive at a specified time after treatment, which takes into account death due to any cause including both related and unrelated to the cancer. However, DFS is increasingly being used as the primary endpoint of most studies testing the benefits of potential cancer therapies since DFS is observed earlier than OS and is statistically sensitive to real treatment benefit. DFS denotes the proportion of patients remain free of disease at a specified time after treatment.
F-box and WD repeat domain containing 7 (FBXW7) protein encodes a substrate adaptor for an E3 SCF ubiquitin ligase complex and negatively regulates the abundance of different oncoproteins, including c-Myc (3, 4), c-Jun (5), cyclin E (6, 7), different members of the Notch family (8, 9), Aurora-A (10–12), mTor (13, 14), KLF5 (15, 16), and MCL-1 (17, 18). These observations indicate that FBXW7 lies at the nexus of many pathways which control cell growth, cell differentiation, and tumorigenesis. FBXW7 as a human tumor suppressor gene is further supported by the discovery of FBXW7 gene mutations in cancers from a wide spectrum of human tissues with overall 6% point mutation frequency (19, 20). A recent study showed that breast cancer patients with low FBXW7 mRNA levels had poorer prognoses than those with high expression when analyzing breast cancer-specific survival, whereas FBXW7 mRNA levels did not affect DSF (21). However, the clinical significance of FBXW7 alteration in human cancer still remains largely unknown. In this study, we conducted a meta-analysis of pooled transcriptional data from 10 studies to evaluate the prognostic impact of FBXW7 expression level in human breast cancer by assessing its association with DFS and OS in further depth by considering the heterogeneity of breast cancer.
Materials and methods
Transcriptional datasets used in study
We used 2 previously published transcriptional profiling datasets that contained both normal and breast cancer samples and 10 breast cancer datasets that included clinical and gene expression data (Table 1). The normal and primary human breast tumor samples in these datasets had been profiled with an Affymetrix microarray assay (either HG-U133A or HG U133 Plus 2.0) or Agilent oligo microarray (22–33) (Table 1). The process data from GEO website were downloaded for analysis. In total, data on DFS and OS were available for 1900 and 889 of 1935 patients, respectively, in 10 datasets (Table 1).
Table 1.
Information of gene expression datasets used in this study
| Dataset* | GEO access number or web location | Number of patients | % DFS data available | % OS data available | Reference |
|---|---|---|---|---|---|
| 1 | GSE1456 | 159 | 100 | 100 | (22) |
| 2 | GSE2603 | 99 | 82.8 | 0 | (23) |
| 3 | GSE6532 | 327 | 96.0 | 0 | (24) |
| 4 | GSE3494 | 108& | 100 | 100 | (25) |
| 5 | GSE7390 | 198 | 100 | 100 | (26) |
| 6 | GSE11121 | 197 | 100 | 0 | (27) |
| 7 | GSE12093 | 136 | 100 | 0 | (28) |
| 8 | GSE2034 | 286 | 100 | 0 | (29) |
| 9 | ArrayExpress: E-TABM-158 | 130 | 99.2 | 99.2 | (30) |
| 10 | NKI (http://microarray-pubs.stanford.edu/wound_NKI/) | 295 | 100 | 100 | (31) |
| 11 | GSE17080 | 185 | 0 | 0 | (32) |
| 12 | GSE3744 | 47 | 0 | 0 | (33) |
Dataset 1 to 9, 11 and 12 are profiled using Affymetrix microarray, Dataset 11 using Agilent Oligo microarray. Dataset 1–10 are used to evaluate prognostic impact of FBXW7 expression. Dataset 1–10 was used for analysis of disease-free survival in Figure 2, 3, 5 and 6. Dataset 1, 4, 5, 9 and 10 was used for analysis of overall survival in Figure 2, 3, 5 and 6. Dataset 11 and 12 contains both normal and breast cancer samples.
Probe 218751_s_at and 19539 (Genebank NM_018315) were used to measure FBXW7 mRNA expression in Affymetrix and Agilent GeneChip, respectively. In each dataset, a sample si in the set S was defined as “FBXW7 Low,” “FBXW7 Intermediate”, or “FBXW7 High” using the rule:
If si ≤ (mean [S]−0.5 × standard deviation [S]), assign Low.
If si ≥ (mean [S]+0.5 × standard deviation [S]), assign High.
Otherwise, assign Intermediate. The same rule has been used in our previous study of PER3 (34). This method allowed us to compare relative FBXW7 mRNA expression levels across the different platforms of microarrays and all datasets fused as a single group of patients.
Statistical analysis
The difference in FBXW7 mRNA expression levels between normal and breast cancers was analyzed by Mann-Whitney U. Kruskal-Wallis test was used to assess the difference in FBXW7 mRNA expression levels among molecular subtypes. Kaplan-Meier plots were constructed and a long-rank test was used to determine differences among disease-free and overall survival according to FBXW7 mRNA levels or molecular subtypes. All analyses were performed by SPSS 11.5.0 for Windows. A two-tailed p-value of less than 0.05 was considered to indicate statistical significance.
Results
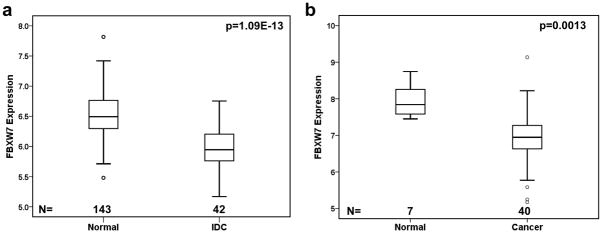
We first examined FBXW7 gene expression in two datasets that contained both normal and breast cancer samples (Dataset 11 and 12 in Table 1). The levels of FBXW7 mRNA in invasive ductal carcinomas (IDC) were statistically significantly lower than these in the normal breast ducts (p=1.09E-13) (Figure 1a). In other dataset it was also found that the level of FBXW7 mRNA was significantly lower in breast cancers than in normal breast tissue (p=0.0013) (Figure 1b). These findings were consistent with that FBXW7 plays a tumor suppressive role in breast cancer.
Figure 1.
FBXW7 expression level in human breast cancer and normal tissues. FBXW7 mRNA expression is assessed by Affymetrix microarray. FBXW7 expression is measured as log2 (probe intensities). In both dataset GSE10780 (dataset 11 in Table 1) (a) and GSE3844 (dataset 12 in Table 1) (b), FBXW7 mRNA expression level is significantly reduced in breast tumors in comparison to normal breast tissues.
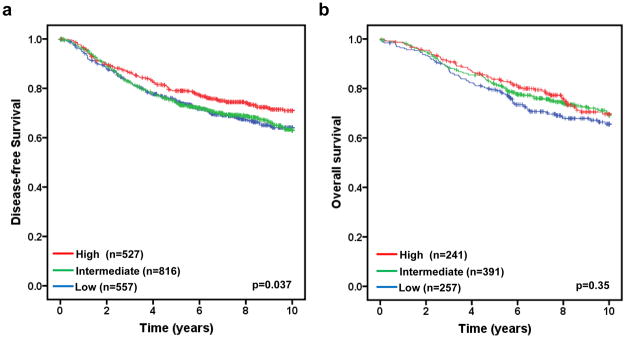
We next examined the association of FBXW7 mRNA levels with disease-free survival in 1900 breast tumor patients taken from 10 publicly available datasets (Table 1). To do so, we first divided the patients into different subgroups (“FBXW7 High”, “FBXW7 Intermediate”, and “FBXW7 Low”) according to FBXW7 mRNA levels in each dataset (details see Materials and Methods) and then pooled all datasets as a single group of patients. The DFS curves for all patients in each subgroup were shown in Figures 2a. Patients with high FBXW7 mRNA levels (“FBXW7 High”) significantly increased DFS than those with Intermediate (“FBXW7 Intermediate”) (p=0.019) or low (“FBXW7 Low”) (p=0.025) FBXW7 mRNA levels (p=0.037 obtained by long rank test among three groups) (Figure 2a). There is no difference in DFS curves between FBXW7 Intermediate and Low groups, suggesting there is no dose-dependent effect of FBXW7 expression on DFS. Surprisingly, there is no significant effect of FBXW7 mRNA levels on OS (p=0.35) (Figure 2b).
Figure 2.
Evaluating the prognostic impact of FBXW7 mRNA expression on disease-free and overall survival. Kaplan-Meier survival curves for breast cancer patients according to tumor expression of FBXW7 are presented. The patients from each cohort were divided into a group with High, Intermediate, or Low level of FBXW7 expression. The p values shown were obtained from a long-rank test among three groups. (a) Association of FBXW7 expression with disease-free survival. Dataset 1 to 10 in Table 1 were included in the analysis. (b) Association of FBXW7 expression with overall survival. Dataset 1, 4, 5, 9 and 10 in Table 1 were included. Other datasets were excluded due to that the information of overall survival was missing.
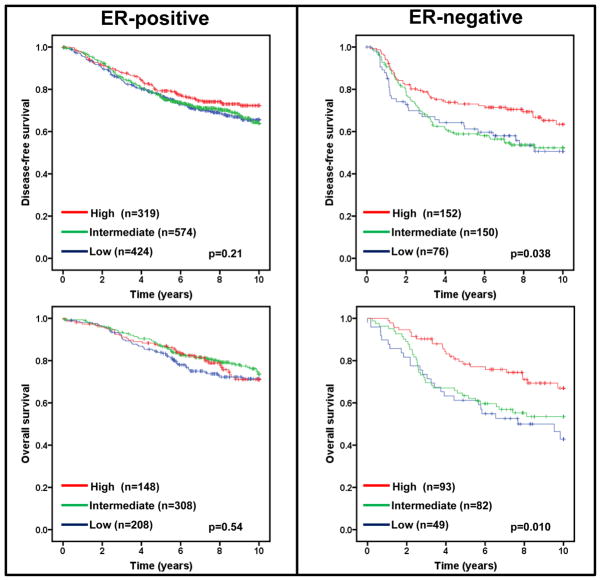
Estrogen receptor (ER) status in breast cancer is an important predictor of recurrence and greatly influences treatment regimens. If low expression of FBXW7 mRNA segregates with ER status, any effect of low FBXW7 expression could be confounded with the effect of ER status. We therefore performed a subset analysis of FBXW7 in ER-positive and ER-negative tumors. Lower levels of FBXW7 expression were significantly associated with shorter DFS and OS (p=0.038 and 0.010 respectively) in patients with ER-negative, but not ER-positive tumors (Figure 3).
Figure 3.
Effect of FBXW7 expression levels on DFS and OS according to ER status. Kaplan-Meier estimates of DFS and OS according to the FBXW7 expression are presented. The p values shown were obtained from a long-rank test among three groups.
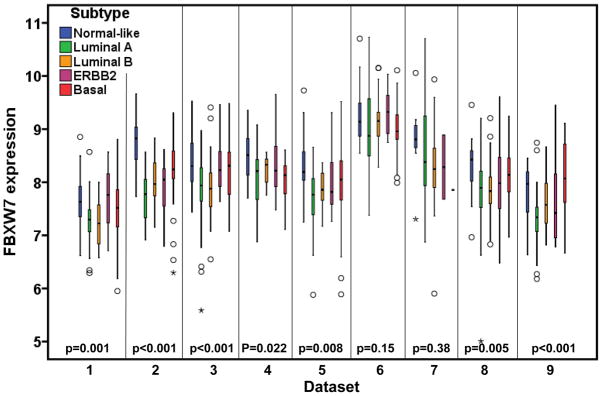
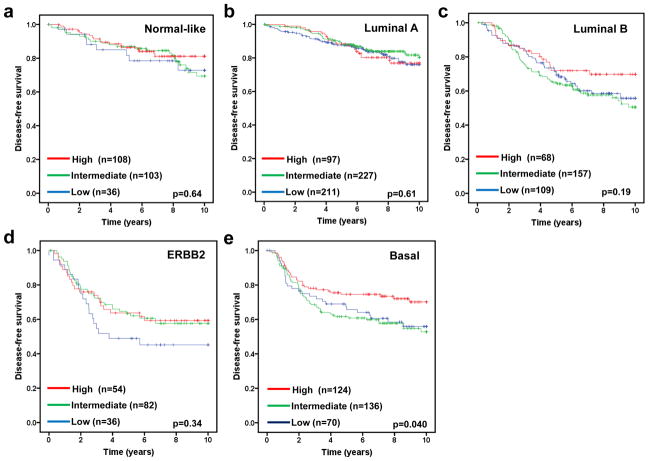
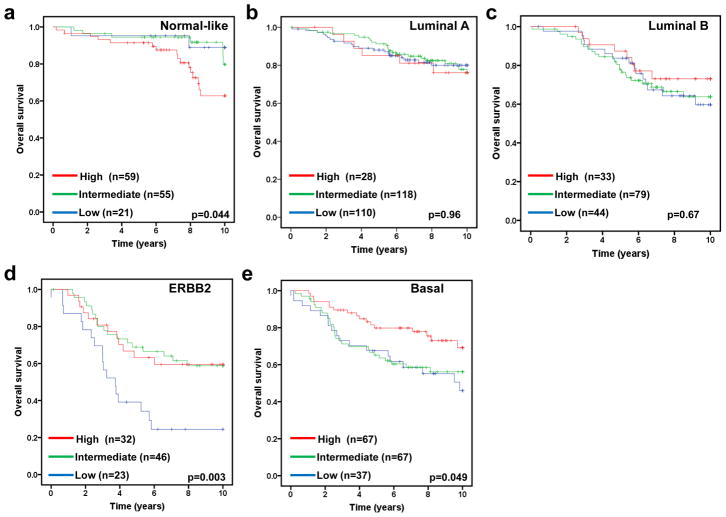
The molecular subtype of human breast cancer is another important prognostic factor. We next asked whether stratifying tumors according to their molecular subtype could reveal additional information of FBXW7 expression associated with breast cancer. Therefore the tumors were assigned using a nearest centroid classifier (35, 36). A subtype was only assigned if correlation with a target class was above 0.1. This resulted in samples assigned as normal-like (n=265), luminal A (n=550), luminal B (n=342), ERBB2 (n=193), basal (n=382), or Unclassified (n=282). The levels of FBXW7 expression were significantly different among different molecular subtypes (Figure 4). It was found surprisingly that luminal A tumors have the lowest average level of FBXW7 whereas the normal-like tumors have the highest average level of FBXW7 in most of datasets (Figure 4). We then performed a subset analysis of FBXW7 in each molecular subtype tumors. Of these groups, only in basal subtype, the patients with FBXW7 High significantly increased DFS (p=0.040) (Figure 5). There was a striking effect of FBXW7 expression on OS among different molecular subtypes (Figure 6). In normal-like subtype, the patients with FBXW7 High significantly decreased OS (p=0.044 obtained by long rank test among three groups) (Figure 6a), while in ERBB2 and basal subtype, the patients with FBXW7 High significantly increased OS (p=0.003 and 0.049, respectively) (Figure 6d and e).
Figure 4.
There are significant differences in FBXW7 expression among molecular subtypes. The p-values obtained from Kruskal-Wallis test by comparing the difference in FBXW7 expression among subtypes.
Figure 5.
Effect of FBXW7 expression levels on DFS according to molecular subtypes. Kaplan-Meier estimates of DFS according to the FBXW7 expression are presented. (a) Nomal-like; (b) Luminal A; (c) Luminal B; (d) ERBB2; and (e) Basal subtype. The p values shown were obtained from a long-rank test among three groups.
Figure 6.
Effect of FBXW7 expression levels on OS according to molecular subtypes. Kaplan-Meier estimates of OS according to the FBXW7 expression are presented. (a) Nomal-like; (b) Luminal A; (c) Luminal B; (d) ERBB2; and (e) Basal subtype. The p values shown were obtained from a long-rank test among three groups.
Discussion
There is a large amount of evidences showing that FBXW7 is a tumor suppressor gene in human cancer (review see 20). Searching distribution of somatic mutations in FBXW7 in Catalogue of Somatic Mutation In Cancer (COSMIC) (http://www.sanger.ac.uk/genetics/CGP/cosmic/), we found that there is about 1% of FBXW7 point mutation in breast cancer. However deletions of chromosome 4q31, on which FBXW7 is located, are found in more than 30% of breast cancer cell lines and primary cancers (37, 30). Since loss of only one copy of FBXW7 can have a substantial effect on tumor development, it is likely that the overall impact of this gene in human breast cancer is greater. Consistent with these findings, this study showed that the FBXW7 expression levels were significantly reduced in breast cancer compared to the normal breast tissues in both datasets. Intriguingly, luminal A tumors displayed the lowest average level of FBXW7 expression whereas normal-like tumors displayed the highest average level of FBXW7 expression.
In this meta-analysis study, we used the public transcriptional profiles to evaluate the prognostic value of FBXW7 expression in breast cancer. We found that the lower levels of FBXW7 were significantly associated with shorter DFS, particularly in patients with ER-negative and basal subtype tumors. With no effect was seen in patients with ER-positive and other subtype tumors, we conclude that the association between FBXW7 expression and DFS in the complete patient sample set was driven by the ER-negative and basal subtype tumors. The recent study by Ibusuki et al (21) found no significant difference in DFS between the patients with low and high FBXW7 expression, which is possibly due to that very small proportion of tumors was ER-negative in the study.
On the other hand, there is no effect of FBXW7 expression on OS in the complete patient sample set. With stratification of patients according to ER status, we found that higher levels of FBXW7 expression significantly increased OS time in the patients with ER-negative, but not ER-positive tumors. Surprisingly, with stratification of patients according to molecular subtypes, we found the opposite effect of FBXW7 mRNA levels on OS. In normal-like subtype, the patients in the high FBXW7 expression group had a significantly poorer OS than those in the low FBXW7 expression group. In ERBB2 and basal subtype, the patients in the high FBXW7 expression group had a significantly better OS than those in the low FBXW7 expression group. The different effects of FBXW7 on OS in the breast cancer subtypes could reflect the different progression pathways. There is a recent study showing that hypermethylation of FBXW7β (reduced expression of FBXW7β) is related to favorable OS (38). The additional studies are needed to get insight into how FBXW7 functions differently in these different subtypes.
However, our results should be interpreted with caution since there are some limitations of this meta-analysis. Major limitation was unable to explore the potential for confounding by various clinical factors, such as age, disease stage, different treatment regimes. The other limitation is that published articles often lack sufficient information to allow adequate assessment of the quality of the study, which subsequently influences the confident level of meta-analysis. Finally, the studies included in this meta-analysis were from different populations, it is possible that demographic factors can confound our results.
Conclusion
The meta-analysis of transcriptional profiles showed that FBXW7 expression may be useful as a prognostic factor for patients with ER-negative and basal subtype tumors.
Acknowledgments
We thank Dr. Erik Fredlund (University of California, San Francisco) for providing us the microarray data and clinic information of Dataset 1 to 8. GW is supported by National Natural Science Foundation of China No. 81172528, and Doctoral Fund of Ministry of Education of China No. 20110131110035. JHM is supported by the National Institutes of Health, National Cancer Institute grant R01 CA116481, the Low Dose Scientific Focus Area, Office of Biological & Environmental Research, US Department of Energy (DE-AC02–05CH11231), and Laboratory Directed Research & Development Program (LDRD).
Footnotes
Conflict of interest
The authors have no conflict of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378:1461–84. doi: 10.1016/S0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- 3.Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol. 2004;14:1852–7. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 4.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. Embo J. 2004;23:2116–25. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–7. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 7.Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, et al. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 8.Oberg C, Li J, Pauley A, Wolf E, Gurney M, et al. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem. 2001;276(38):35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- 9.Gupta-Rossi N, Le Bail O, Gonen H, et al. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276(37):34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- 10.Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, et al. Fbxw7/Cdc4 is a p53-dependent haploinsufficient tumour suppressor gene. Nature. 2004;432:775–9. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 11.Fujii Y, Yada M, Nishiyama M, Kamura T, Takahashi H, et al. Fbxw7 contributes to tumor suppression by targeting multiple proteins for ubiquitin-dependent degradation. Cancer Sci. 2006;97:729–36. doi: 10.1111/j.1349-7006.2006.00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon YW, Kim IJ, Wu D, Lu J, Stock WA, Jr, et al. Pten regulates Aurora-A and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol Cancer Res. 2012;10:834–844. doi: 10.1158/1541-7786.MCR-12-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao JH, Kim IJ, Wu D, Climent J, Kang HC, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu L, Kim YA, Wang X, Wu X, Yue P, et al. Perifosine inhibits mammalian target of rapamycin signaling through facilitating degradation of major components in the mTOR axis and induces autophagy. Cancer Res. 2009;69:8967–76. doi: 10.1158/0008-5472.CAN-09-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu N, Li H, Li S, Shen M, Xiao N, et al. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem. 2010;285:18858–67. doi: 10.1074/jbc.M109.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao D, Zheng HQ, Zhou Z, Chen C. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010;70:4728–38. doi: 10.1158/0008-5472.CAN-10-0040. [DOI] [PubMed] [Google Scholar]
- 17.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–9. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–4. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 19.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 20.Cheng Y, Li G. Role of the ubiquitin ligase Fbw7 in cancer progression. Cancer Metastasis Rev. 2012;31:75–87. doi: 10.1007/s10555-011-9330-z. [DOI] [PubMed] [Google Scholar]
- 21.Ibusuki M, Yamamoto Y, Shinriki S, Ando Y, Iwase H. Reduced expression of ubiquitin ligase FBXW7 mRNA is associated with poor prognosis in breast cancer patients. Cancer Sci. 2011;102(2):439–45. doi: 10.1111/j.1349-7006.2010.01801.x. [DOI] [PubMed] [Google Scholar]
- 22.Pawitan Y, Bjöhle J, Amler L, Borg AL, Egyhazi S, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loi S, Haibe-Kains B, Desmedt C, Lallemand F, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–46. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 25.Miller LD, Smeds J, George J, Vega VB, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–5. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, et al. TRANSBIG Consortium: Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt M, Böhm D, von Törne C, Steiner E, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–13. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Sieuwerts AM, McGreevy M, Casey G, et al. The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res Treat. 2009;116:303–9. doi: 10.1007/s10549-008-0183-2. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 30.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 31.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 32.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–50. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson AL, Wang ZC, De Nicolo A, Lu X, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Climent J, Perez-Losada J, Quigley DA, Kim IJ, Delrosario R, Jen KY, Bosch A, Lluch A, Mao JH, Balmain A. Deletion of the hPER3 gene on chromosome 1p36 in recurrent ER-positive breast cancer. Journal of Clinical Oncology. 2010;28:3770–3778. doi: 10.1200/JCO.2009.27.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akhoondi S, Lindström L, Widschwendter M, Corcoran M, Bergh J, et al. Inactivation of FBXW7/hCDC4-β expression by promoter hypermethylation is associated with favorable prognosis in primary breast cancer. Breast Cancer Res. 2010;12(6):R105. doi: 10.1186/bcr2788. [DOI] [PMC free article] [PubMed] [Google Scholar]