Abstract
Several molecular biomarkers have been suggested as predictors of outcome for pediatric ependymomas but deserve further validation in independent case series. We analyzed intracranial ependymomas belonging to a series of 60 patients prospectively treated according to the protocol sponsored by the Italian Association of Pediatric Hematology-Oncology. We used a tissue microarray to analyze nucleolin (NCL), cyclin-dependent kinase inhibitor 2A (CDKN2A), tumor protein 53 (TP53), and epidermal growth factor receptor (EGFR) by immunohistochemistry and by 1q gain by fluorescent in situ hybridization. The mRNA expression levels of EGFR, human telomerase reverse-transcriptase (HTERT), and Prominin 1 (PROM 1)/CD133 were evaluated by quantitative real-time PCR from cases with fresh-frozen tumor material available. Univariate and multivariate analyses of updated clinical data confirmed the prognostic significance of surgery (P < .01) and tumor grading (P < .05) for both relapse-free survival (RFS) and overall survival (OS). Among biomolecular markers, HTERT mRNA expression emerged with the strongest association with OS at multivariate analysis (hazard ratio [HR] = 9.9; P = .011); the 5–year OS was 84% versus 48% in the subgroups with HTERT median value <6 versus ≥6, respectively (P = .005). Five-year RFS was 46% versus 20% in the subgroups with low versus high NCL protein expression, respectively (P = .004), while multivariate Cox analyses gave suggestively high HRs for high versus low NCL (HR = 1.9; P = .090). The other genes tested were not significant at multivariate analyses, and genetic alterations of CDKN2A, TP53, EGFR, and HTERT loci were rare. The PROM1/CD133 cancer stem cell marker was strongly expressed at both RNA and protein levels in a substantial fraction of cases and was suggestively associated with a more indolent form of the disease. We conclude that NCL and HTERT represent the strongest prognostic biomarkers of RFS and OS, respectively, in our ependymoma case series.
Keywords: biomarker, ependymoma, EGFR, HTERT, nucleolin
Ependymomas exhibit heterogeneous clinical courses that cannot be predicted accurately by clinical, pathologic, or molecular markers, with the noticeable exception of extent of surgery. However, several histopathologic features have been investigated in the past as prognostic markers in different case series from multiple studies.1,2 Only recently, molecular profiling of ependymomas revealed candidate prognostic markers that deserve retrospective validation in independent case series preliminary to their application in prospective clinical trials. In particular, 5 molecular markers—epidermal growth factor receptor (EGFR),3 nucleolin (NCL),4 human telomerase reverse-transcriptase (HTERT),3,5 cyclin-dependent kinase inhibitor 2A (CDKN2A) loss,6 and 1q gain6,7—have been proposed as being associated with survival at univariate or multivariate analyses. Here we explored the prognostic value of these molecular markers in an ependymoma cohort treated according to the protocol of the Italian Association of Pediatric Hematology-Oncology (AIEOP).
In particular, contradictory findings have been published on the prognostic value of both EGFR3,4,8 and HTERT protein expression in ependymomas.3,4,9 Such discrepancies may be a result of specificity issues related to the different antibodies used, as well as to inconsistency of immunohistochemistry (IHC) scoring methods. To overcome these issues, we additionally performed analysis of EGFR and HTERT mRNA expression by quantitative real-time PCR (q-PCR). We also analyzed the copy number and mutational status of the genes under study and evaluated the expression of Prominin 1 (PROM1)/CD133 in ependymomas to identify the proportion of cases overexpressing this marker of stemness in relation to primary clinical variables.
Materials and Methods
Patients
The study was carried out on a cohort of 62 children with ependymoma, consecutively treated within the Italian pediatric oncology centers of AIEOP10–12 in the years 1994–2007. The present analysis excluded 2 patients presenting with metastasis. Formalin-fixed paraffin embedded tissue samples from 47 patients and fresh frozen material from 29 cases were available. In all cases, histologic diagnosis and grading according to the 2007 World Health Organization criteria and selection of representative areas were performed by F.G. All parents gave their informed consent prior to the inclusion of their children in the study.
Protein Expression Analyses
Immunohistochemistry
For each case, 3–10 cores, 0.6 mm in diameter, were taken to construct a tissue microarray (TMA) using formalin-fixed paraffin embedded tumor material. IHC was performed by the streptavidin-biotin-immunoperoxidase technique on 5-μm TMA sections. Samples were deparaffinized in xylene and rehydrated through a graded series of ethanol concentrations, then incubated for 15 min in 3% H2O2 in methanol to inhibit endogenous peroxidases. Tissue sections were then incubated in 10% normal rabbit serum to block nonspecific antibody binding, and antigen retrieval was performed in a microwave for 3 × 5 min at full power in sodium citrate buffer (pH 6.0). Anti–pan-EGFR rabbit monoclonal antibody (1:200 diluted, 1902–1, Epitomics), anti-NCL rabbit monoclonal antibody (1:200, clone 4E2, ab 13 541, Abcam), anti-p53 mouse monoclonal antibody (1:100, clone DO1, BD Pharmingen), and anti-p16 mouse monoclonal antibody (1:100, clone 16P07, LabVision) were incubated in humidity chambers for 1 h at room temperature (RT) or overnight at 4°C (anti-EGFR). As negative controls, the primary antibodies were omitted. Tissue sections were incubated with biotinylated secondary immunoglobulin G for 15 min, then incubated with peroxidase-conjugated streptavidin (Ultrateck Sciteck) for 15 min. Peroxidase activity was visualized by immersing sections in 0.03% 3.3′-diaminobenzidine-4HCl (Ultrateck Sciteck) in 0.05 mol/L Tris-HCl buffer at pH 7.6 containing 0.005% H2O2 for 10 min. The tissue sections were rinsed in distilled water, counterstained with hematoxylin, dehydrated with graded ethanol and xylene, and coverslipped. TMA sections were scored blindly to the clinical outcomes, using a brighfield microscope (Nikon) by 2 independent investigators; cases with partially discordant scores were reevaluated jointly until a consensus was reached. For tumors with triplicate cores (80% of cases), the final labeling index (LI) was calculated as the mean of 3 or more cores. The NCL LI was defined as the percentage of immunopositive cells exhibiting nuclear staining in each sample. For tumors exhibiting cellular heterogeneity with more than 3 cores (15% of cases), the mean LI was calculated for each set of triplicates, and the highest LI value was used. NCL immunostaining in tumors was classified as negative, low (<10% of cells exhibiting faint immunostaining), intermediate (10%–40% of cells exhibiting intermediate staining), or high (>40% of cells exhibiting strong staining).4 EGFR membranous and cytoplasmic immunostaining was evaluated on the entire sample at 200× magnification, and protein expression was graded as described:13 strong (moderate to strong continuous membrane and/or cytoplasmic staining in >50% of tumor cells in at least one microscopic field with 400× magnification), weak (faint or partial staining in >50% of tumor cells), negative (faint or partial staining in <50% of tumor cells), or both strong and weak staining being considered as EGFR overexpression. Appropriate controls were included for each antibody.
Western blotting
Protein lysates were prepared in radioimmunoprecipitation assay buffer (Sigma) supplemented with protease and phosphatase inhibitors. Fifty μg of cell lysate were loaded on 4%–15% gradient polyacrylamide gel electrophoresis gels (Biorad) and electroblotted onto polyvinylidene difluoride membranes (Amersham Biosciences). Subsequently, membranes were incubated for 1 h atRT in a solution of phosphate buffered saline (PBS) supplemented with 5% nonfat dry milk. For immunodetection, the following primary antibodies were used: anti–total EGFR (rabbit polyclonal 1:1000, Cell Signaling); anti–phosphoTyr1068 EGFR (rabbit polyclonal 1:1000, Cell Signaling); anti–total Akt (rabbit polyclonal 1:1000, Cell Signaling); anti–phosphoSer473 AKT (rabbit polyclonal 1:1000, Cell Signaling); anti-ERK1/2 (clone 137F5 1:1000, Cell Signaling); anti–phosphoT202/Y204 ERK1/2 (clone E10 1:1000, Cell Signaling); anti-CDKN2A/p16 (mouse monoclonal 1:1000, clone 16P07, LabVision); and anti-PROM1/CD133 (rabbit monoclonal clone C24B9 1:1000, Cell Signaling). After overnight incubation at 4°C with the primary antibody, membranes were washed in Tris buffered saline (10 mmol/L Tris-HCl [pH 8.0], 0.15 mol/L NaCl) and 0.05% Tween 20, followed by AlexaFluor680- or IRDye800CW-conjugated goat-antimouse or goat-antirabbit antibodies (Invitrogen or Li-Cor, respectively). An Odyssey infrared imaging system (Li-Cor) was used for detection. Protein loading equivalence was assessed using an anti–glyceraldehyde 3-phosphate dehydrogenase antibody (Sigma).
Gene Expression Analysis by Quantitative Real-time PCR
Frozen tumor tissues containing more than 70% tumor cells, as evaluated by a pathologist from hematoxylin-eosin staining of frozen sections, were included for gene expression analyses. Total RNA was extracted from frozen tumor tissue sections using Trizol reagent (Ambion), and 1 μg RNA was retrotranscribed using Superscript II reverse transcriptase (Gibco) with random primers. Ten nanograms of cDNA were used as template in 20-μL PCR reactions with 1× TaqMan Universal PCR Master Mix (Applera). Relative quantification of gene expression was performed in triplicate using TaqMan Assays on Demand on an ABI Prism 7900HT Sequence Detection System (Applera) by comparative threshold cycle method, using the hypoxanthine phosphoribosyl transferase (HPRT) gene (HPRT predeveloped assay reagent, 4326321E) as an endogenous reference control and normal brain cDNA (Clontech) as calibrator. Assays used were EGFR (Hs01076087_m1, spanning exons 2–3, and Hs00193306_m1, spanning exons 20–21), HTERT (Hs00162669_m1, spanning exons 11–12), and PROM1 (Hs01009261_m1, spanning exons 9–10 that are present in all known isoforms). Included as internal controls were total RNA extracted from CaCo2, CaSki, and G401 cancer cell lines, known to express high levels of PROM1, EGFR, and HTERT, respectively.
DNA Copy Number and Mutational Analyses
Fluorescent in situ hybridization (FISH)
Five-micron sections were cut from TMA blocks for FISH analysis. Slides were deparaffinized in an oven at 60°C for 60 min and incubated in xylene. Slides were then dehydrated in 100% ethanol for 5 min at RT and treated for 20 min at RT in 0.2 mol/L HCl, then with pretreatment solution (1 mol/L sodium thiocyanate) for 30 min at 80°C. Slides were then immersed in pepsin solution (0.65% in protease buffer) for 15–20 min at 37°C, washed twice in buffer for 5 min, and air dried. The 1q25 amplification was detected via a dual-probe hybridization using the SpectrumGreen labeled 1q25 probe and the SpectrumOrange labeled 1q36 probe Visys kit, in accordance with the manufacturer's instructions (Abbott Laboratories). Sections were finally counterstained with 4′-6-diamidino-2-phenylindole (DAPI), coverslipped, and stored in the dark at −20°C prior to analysis. FISH sections were examined with an AxioImager M1 microscope (Carl Zeiss), using a 100× oil immersion objective and analyzed by 2 investigators. Signals were counted for at least 100 tumor cells in each core of a triplicate for each sample. The score was calculated as the ratio between 1q25 and 1q36 signals. A signal ratio between 1 and 2 was considered as 1q25 gain. A signal ratio score >2 was considered as 1q amplification.
Multiplex ligation-dependent probe amplification (MLPA)
Tumor DNA (100 ng) was subjected to DNA copy number analysis using MLPA kits P258-B1, P298-A1, and P171 (MRC-Holland), following manufacturer's instructions and together with normal DNA samples and samples with known gene copy number alterations as controls. Fragment separation was performed on an ABI3130xl genetic analyzer (Applera). Raw data peak pattern evaluation was performed using GeneMapper software (Applera), and Coffalyser software was used for data analysis (MRC-Holland).
Mutational analysis
Exon PCR amplification and direct sequencing were performed to analyze tumor protein 53 (TP53) genes (exons 5–9) and EGFR (exons 18–21). Primers and conditions are available upon request. Sequence reactions were analyzed on an ABI3130xl genetic analyzer (Applera).
Cell Cultures
Control cancer cell lines A431, NTERA2, G401, and CaCo2 were purchased from the American Type Culture Collection and maintained in Roswell Park Memorial Institute medium, 10% serum in standard incubators. Cells were harvested at exponential growth for RNA or protein extraction. Tumor-derived cell cultures were established from fresh biopses 1–24 h after surgery, the time spent at RT in stem cell medium (Dulbecco’s modified Eagle's medium F12/B27 supplement, epidermal growth factor [EGF]/basic fibroblast growth factor 20 ng/mL, without serum) for delivery. Biopses were finely aseptically minced, treated briefly (30′–2 h) with Collagenase II (Sigma), and cultured with Amniomax C100 complete medium (Gibco Life Tecnologies). Spectral karyotyping was performed as described.14 Cell cytotoxicity was measured with a sulphorodamine-B assay,15 and drug treatments used EGFR inhibitor 324 674 (Calbiochem) and the phosphatidylinositol 3-kinase inhibitor LY294002 (Sigma). Immunocytochemical analysis was performed on cells grown on Lab-Tek 4-well chamber slides (Nalgene), fixed with PBS 1X paraformaldehyde 3%, sucrose 2%, pH 7.6, permeabilized in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid 20 mM, sucrose 300 mM, NaCl 50 mM, MgCl2 3 mM, Triton X100 0.5%, and blocked in PBS 1X bovine serum albumin 2%, Tween 0.01%. Primary antibodies and dilutions are reported in Supplemental Table 1. Alexa488-conjungated secondary antibodies (Molecular Probes) were used, and nuclei were counterstained with DAPI. Images were acquired with ViewfinderLite software (Pixera) on an Olympus IX70 fluorescence microscope.
Statistical Analyses
The association between 2 dichotomous variables was tested using the Fisher test; the association between a dichotomous and a continuous ordinal variable was tested using the Wilcoxon Mann–Whitney test. Disease outcomes were analyzed in terms of overall survival (OS) and relapse-free survival (RFS). Time was defined as the interval from the date of diagnosis to the date of event, ie, (i) death due to any cause (OS) or (ii) local progression/relapse, local or distant metastases, any joint occurrence of such events, or other malignancy or death with no evidence of disease (NED), whichever came first (RFS). OS and RFS curves were estimated by means of the Kaplan–Meier method; between-group comparisons were performed using the log-rank test. Additional OS and RFS analyses were performed by Cox regression models, the results of which were reported in terms of hazard ratio (HR), the corresponding 95% confidence interval (CI), and the 2-sided Wald test P value. In particular, multivariate models were used to analyze the prognostic effect of clinical variables (tumor site, extent of surgery, histologic grade, radiotherapy administration). For each biological marker independently, we analyses both univariate and multivariate Cox analyses. In the latter, clinical variables were included with the purpose of adjustment; to control for overfitting due to the model's high dimensionality, penalized maximum likelihood estimation methods16 were applied as implemented in the R package Penalized. EGFR, PROM1, and HTERT mRNA expression levels were modeled as continuous variables by means of a nonlinear transformation, ie, 3-knot restricted cubic splines (RCSs),17 whereas the other clinical and biological variables were modeled as categorical by using dummy variables. Concordantly, the values used in the tables presenting the Cox model results were not cutoff values, as in the Kaplan–Meier results, but exact values, ie, the quartiles of the variable distribution. RCS modeling has the advantage of (i) modeling the markers as continuous variables, avoiding their categorization by means of cutoff values, which is instead necessary in order to estimate Kaplan–Meier curves; (ii) allowing the marker's prognostic effect not to be the same in every part of the range; and (iii) taking as reference for HR estimation every value within the range of the variable distribution. Significance level was set at 0.05. All statistical analyses were performed using SAS and R (http://www.r-project.org) software.
Results
Patients
Table 1 shows the characteristics of the 60 children under study. Age range was 4 months to 16 years, with a median of 3 years. Among the 8 children who underwent surgery alone, 1 infant girl died 10 days after surgery from complications. Twenty-one patients of the 36 submitted to radiotherapy were treated according to the first protocol, and 15 according to the second one, as previously outlined.10–12 Forty-one children (68.3%) were rendered without tumor NED after surgery, while 19 (31.7%) were left with tumor residuals. Median follow–up was 94 months (interquartile range: 66–127 months). There were no significant differences in patient and tumor characteristics between the whole study population and the subgroups analyzed by IHC and by molecular markers at the DNA/RNA level.
Table 1.
Patient and tumor characteristics in the whole study population
| Patient Population (N = 60) |
||
|---|---|---|
| n | % | |
| Median age, y (IR) | 3 (2–7.3) | |
| Gender | ||
| Males | 34 | 56.7 |
| Females | 26 | 43.3 |
| Tumor site | ||
| Posterior fossa | 41 | 68.3 |
| Supratentorial areas | 19 | 31.7 |
| Extent of surgery | ||
| Incomplete | 19 | 31.7 |
| Complete | 41 | 68.3 |
| Treatment | ||
| No | 8 | 13.3 |
| CT | 16 | 26.7 |
| RT | 17 | 28.3 |
| CT and RT | 19 | 31.7 |
| Histologic grade | ||
| 2 | 31 | 51.7 |
| 3 | 29 | 48.3 |
Abbreviations: IR, interquartile range; CT, chemotherapy; RT, radiotherapy.
Molecular Markers Analyzed at the Protein Level
Nucleolin
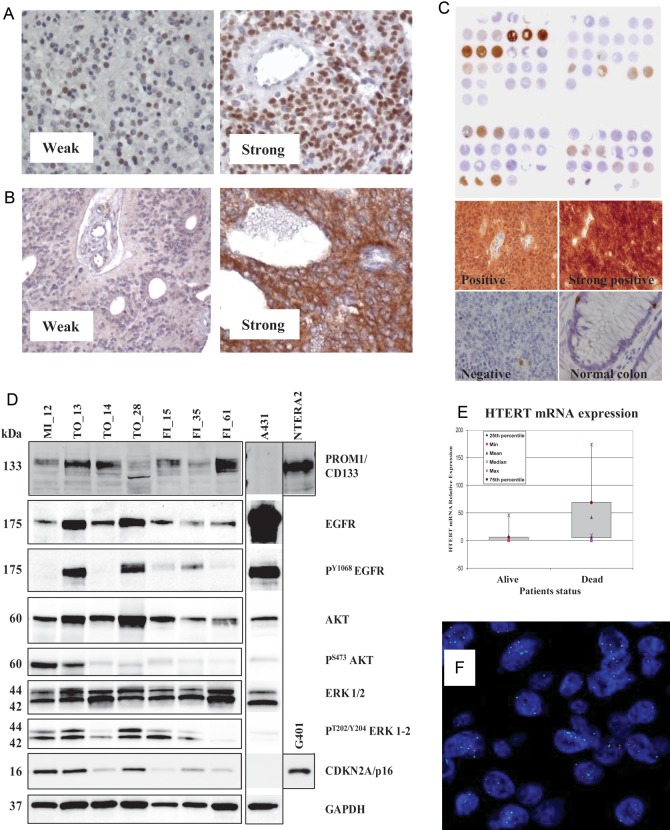
NCL was expressed in 35/47 (74%) examined tumors (Fig. 1A). The intensity of NCL expression was high in 18 patients (38%), intermediate in 2 cases (4%), low in 15 cases (32%), and negative in 12 cases (26%). When cases were sorted into 2 distinct groups with 50% LI used as the cutoff to distinguish between them, no significant association with the clinical factors of resection and age was found, while histologic grading was associated with NCL expression (P = .01).
Fig. 1.
Representative examples of IHC stainings of ependymoma samples for (A) nucleolin, (B) EGFR, and (C) CDKN2A/p16 protein expression. (D) Analysis of protein expression by Western blotting from fresh-frozen ependymoma tissues. Equal amounts of protein lysate from cell lines A431, NTERA2, and G401 were loaded as controls. Lanes 2 and 4 correspond to supratentorial ependymomas, while other cases are infratentorial. (E) Differential HTERT mRNA expression in tumor tissue from ependymoma patients who experienced dichotomous outcomes, as detected by qRT-PCR. (F) FISH analysis of an ependymoma sample carrying 1q gain (green signal).
EGFR
In 23/47 samples (49%), EGFR protein expression was detected, intensity of EGFR labeling being strong in 11 samples (23%), weak in 12 samples (26%), and negative in 24 samples (51%) (Fig. 1B). There was no significant correlation between EGFR and NCL staining and no significant association with clinical factors. Notably, Western blot analysis in a subset of fresh-frozen ependymoma samples confirmed EGFR protein expression and identified the presence of Tyr1068-phosphorylated EGFR as well as activated, phospho-Akt and phospho-ERK1/2 mitogen-activated protein kinases (Fig. 1D). Despite the fact that intensity of EGFR staining by IHC in ependymoma tumors was frequently weak and cytoplasmatic, we verified that in ependymoma short-term cell cultures (Supplemental Tables 1 and 2 and Supplemental Figs. 1 and 2), EGFR can localize to the membrane and respond to EGF stimulation, as evidenced by the induction of autophosphorylation events, despite the limited ability to stimulate downstream Akt and ERK1/2 phosphorylation (Supplemental Fig. 3). Furthermore, cell cytotoxicity assay demonstrated a substantial proliferation reduction and morphologic changes of ependymoma cells in response to EGFR-specific small molecule inhibitors (Supplemental Fig. 3).
TP53
No cases presented a positive immunostaining for TP53, suggesting the absence of TP53 mutations associated with protein accumulation. At the DNA level, 3 cases presented single-copy loss of the 17p chromosome arm encompassing the TP53 locus and did not carry TP53 mutations.
CDKN2A/p16
Surprisingly, the p16 tumor suppressor protein was strongly expressed in 7/47 (15%) cases, resembling the expression pattern observed in tumors carrying inactivation of the p16/Rb pathway by viral oncogenic proteins (Fig. 1C). In 6/7 cases, they were supratentorial ependymomas. Furthermore, one additional patient with a supratentorial ependymoma carried a homozygous deletion of the CDKN2A/p16 locus and died of disease at 54 months postdiagnosis. Notably, Western blotting analysis in a subset of fresh-frozen ependymoma samples confirmed aberrantly high p16 protein accumulation in supratentorial samples (Fig. 1D).
Molecular Markers Analyzed at the RNA/DNA Level
EGFR
EGFR mRNA expression was analyzed using 2 different Taqman assays, one spanning exons 2–3 and the other spanning exons 20–21 of the EGFR gene. Both assays displayed very similar quantization of EGFR gene expression (R = 0.91, Supplemental Fig. 4), and results are reported for one assay only. Concurrent analyses of normal brain RNA samples and different pediatric brain tumor samples, as well as relapsing ependymoma samples, confirm the relatively high level of mRNA expression of EGFR in ependymoma, as previously reported by different approaches,8,18 and reveal a lower level of expression in relapses and in spinal ependymomas, compared with primary intracranial ependymomas (Supplemental Fig. 2). Mutational analysis did not identify mutations in the EGFR tyrosine kinase domains, and gene dosage analysis to determine EGFR DNA copy number status using MLPA revealed only one case carrying a low copy amplification, thus confirming recent data on high-throughput copy number analyses that failed to identify recurrent EGFR amplification events in ependymoma.3,18–20
HTERT
HTERT mRNA expression analysis revealed an almost dichotomous low versus high expression pattern. Eleven patients from whom tumor samples were taken expressed high HTERT levels, comparable to those observed in in vitro cancer cell lines carrying HTERT overexpression that were used as internal controls. Notably, among these 11 patients, 9 experienced tumor relapse and 8 died of disease (Fig. 1E). Analysis of HTERT DNA copy number revealed amplification in one case showing a high level of expression of HTERT mRNA.
PROM1/CD133
The stemness cell marker CD133 was expressed at a high level in 20 patients. There was no significant association with clinical features, although in the top quartile of 7 patients expressing particularly high levels of CD133, only 3 died—at 92, 57, and 55 months—the average follow-up time of survivors being 68 months (range, 42–83 mo). Four of the 7 patients were infants; tumor location was supratentorial in 2 and infratentorial in 5 cases. Notably, Western blotting analysis in a subset of fresh-frozen ependymoma samples confirmed variable but consistent PROM1/CD133 protein expression, as compared with the control NTERA2 cell line (Fig. 1D).
Gain of 1q
A gain of 1q chromosomal region, as detected by FISH analysis (Fig. 1F), was observed in 6/47 (13%) cases analyzed, including 1 case of 1q amplification. Three were infant females and 3 were males of 3, 10, and 13 years at diagnosis. There were 4/29 patients <4 years of age, and 2/18 were ≥4 years. Only 1 patient presented a supratentorial tumor, and 4/6 patients died of disease, the survivors being 2 infants with infratentorial ependymomas with follow-ups of 114 and 66 months.
Overall Survival
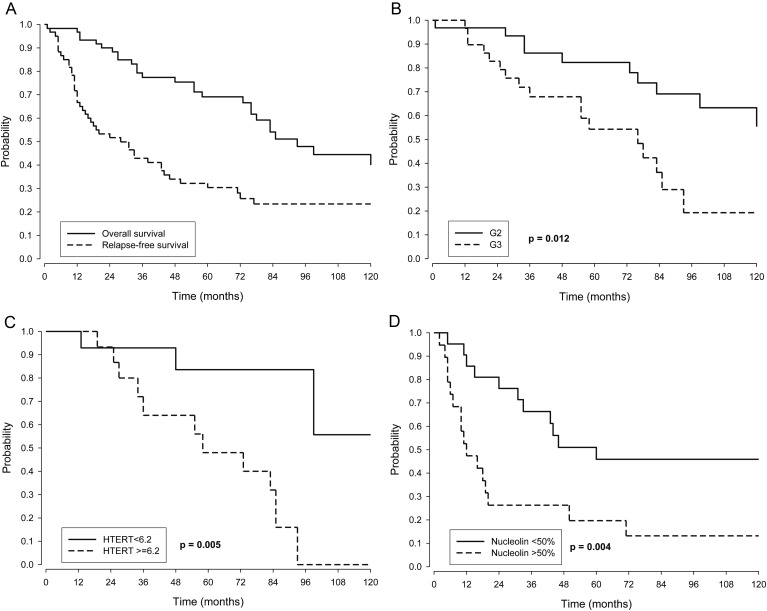
Twenty-eight patients died. Median OS was 93 months. Five- and 10-year OSs were 69.1% (95% CI, 57.8%, 82.6%) and 40.0% (26.7%, 59.9%), respectively (Figure 2A).
Fig. 2.
(A) Kaplan–Meier OS and RFS curves in the whole series. (B and C) Kaplan–Meier OS curves according to (B) histologic grade 2 (G2) and grade 3 (G3) and (C) HTERT mRNA expression. (D) Kaplan–Meier RFS curves according to nucleolin protein expression.
Among the clinical variables studied (tumor site, histologic grade, extent of surgery, and radiotherapy as upfront), extent of surgery was significant at univariate analysis by Kaplan–Meier survival estimation, with 10-year OS of 49.5% (95% CI, 33.1%, 73.9%) for patients with complete surgery and 15.6% (3.2%, 76.3%) for patients with incomplete surgery (P = .013). Grade too was statistically significant, with 10-year OS of 55.4% (36.8%, 83.4%) for children with grade 2 tumors and 19.3% (6.5%, 57.1%) for those with grade 3 tumors (P = .012; Fig. 2B). At multivariate Cox regression analysis (Table 2, left), both extent of surgery and histologic grade confirmed their significant association with OS.
Table 2.
Multivariate Cox analysis of clinical factors’ prognostic effect on OS and RFS
| OS |
RFS |
|||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P (2–sided Wald test) | HR | 95% CI | P (2–sided Wald test) | |
| Tumor site | 0.861 | 0.721 | ||||
| PF vs ST | 1.1 | (0.4, 2.7) | 1.1 | (0.6, 2.3) | ||
| Extent of surgery | 0.006 | 0.001 | ||||
| Incomplete vs complete | 3.2 | (1.4, 7.2) | 3.1 | (1.5, 6.1) | ||
| Histologic grade (G) | 0.015 | 0.023 | ||||
| G3 vs G2 | 2.8 | (1.2, 6.3) | 2.1 | (1.1, 3.9) | ||
| RT | 0.408 | 0.006 | ||||
| No vs yes | 1.5 | (0.6, 3.5) | 2.6 | (1.3, 5.2) | ||
Abbreviations: PF, posterior fossa; ST, supratentorial areas; RT, radiotherapy.
As regards biological variables, Table 3 (left) shows Kaplan–Meier OS estimates, and Table 4 shows the univariate and multivariate analyses by Cox regression model. HTERT was the only statistically significant factor for OS at both univariate and at multivariable analyses: 5- and 10–year OSs were 83.6% and 48% and 55.7% and 0%, respectively, at HTERT median value cutoff (P = .005; Table 3, left; Fig. 2C), while HR for HTERT values of 33.5 versus 1.2 was 9.9 (P = .011; Table 4) in the multivariable Cox model.
Table 3.
Kaplan–Meier OS and RFS analysis for biological factors
| OS |
RFS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5-y | 95% CI | 10-y | 95% CI | P | 5-y | 95% CI | 10-y | 95% CI | P | |
| 1q gain (FISH) | 0.554 | 0.882 | ||||||||
| No | 63.0% | (46.8, 84.9%) | 39.3% | (20.8, 74.4%) | 26.2% | (14.1, 48.6%) | 21.0% | (9.8, 44.7%) | ||
| Yes | 66.7% | (37.9, 100%) | 22.2% | (4.1, 100%) | 16.7% | (2.8, 99.7%) | 16.7% | (2.8, 99.7%) | ||
| Nucleolin (IHC, % positive cells) | 0.122 | 0.004 | ||||||||
| ≤05% | 89.7% | (77.2, 100%) | 52.9% | (31.5, 88.7%) | 45.9% | (28.6, 73.8%) | 45.9% | (28.6, 73.8%) | ||
| >50% | 49.6% | (30.9, 79.7%) | 42.5% | (24.2, 74.6%) | 19.7% | (7.7, 50.6%) | 13.2% | (3.8, 45.3%) | ||
| EGFR (IHC) | 0.338 | 0.025 | ||||||||
| Strong | 72.7% | (50.6, 100%) | 62.3% | (38.9, 100%) | 63.6% | (40.7, 99.5%) | 53.0% | (29.9, 94.0%) | ||
| Low | 72.0% | (57.1, 90.8%) | 34.7% | (18.6, 64.5%) | 21.9% | (11.0, 43.6%) | 17.5% | (7.8, 39.6%) | ||
| EGFR (mRNA)* | 0.244 | 0.517 | ||||||||
| <10.1 | 75.4% | (54.4, 100%) | 25.9% | (8.1, 82.7%) | 32.0% | (15.0, 68.4%) | 10.7% | (1.8, 62.7%) | ||
| ≥1.01 | 53.6% | (31.9, 90.1%) | 40.2% | (18.6, 86.6%) | 21.4% | (7.9, 58.4%) | 21.4% | (7.9, 58.4%) | ||
| PROM1 (mRNA)* | 0.858 | 0.375 | ||||||||
| <53.5 | 78.0% | (58.6, 100%) | 23.4% | (4.8, 100%) | 17.8% | (5.6, 56.7%) | 17.8% | (5.6, 56.7%) | ||
| ≥5.35 | 55.0% | (33.7, 89.7%) | 18.3% | (3.4, 97.7%) | 35.7% | (17.7, 72.1%) | 17.9% | (5.4, 59.6%) | ||
| HTERT (mRNA)* | 0.005 | 0.223 | ||||||||
| <6.2 | 83.6% | (64.9, 100%) | 55.7% | (24.1, 100%) | 35.7% | (17.7, 72.1%) | 17.9% | (3.8, 84.5%) | ||
| ≥2.6 | 48.0% | (27.3, 84.5%) | 0% | – | 20.0% | (7.3, 55.0%) | 10.0% | (1.8, 55.6%) | ||
*The cutoff value is the median of the variable distribution.
Table 4.
Cox OS analysis of biological variables’ prognostic effect
| Univariate Analysis |
Multivariate Analysis |
|||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P (2–sided Wald test) | HR | 95% CI | P (2–sided Wald test) | |
| 1q gain (FISH) | 0.554 | 0.202 | ||||
| Yes vs no | 1.4 | (0.5, 4.4) | 2.0 | (0.7, 5.8) | ||
| Nucleolin (IHC, % positive cells) | 0.130 | 0.539 | ||||
| >50% vs ≤50% | 2.1 | (0.8, 5.6) | 1.3 | (0.6, 2.6) | ||
| EGFR (IHC) | 0.340 | 0.714 | ||||
| Low vs strong | 1.7 | (0.6, 5.2) | 1.2 | (0.5, 2.5) | ||
| EGFR (mRNA)* | 0.328 | 0.377 | ||||
| 10.1 vs 5.1 | 1.6 | (0.9, 2.8) | 1.4 | (0.8, 2.4) | ||
| 32.7 vs 5.1 | 3.2 | (0.7, 14.6) | 2.6 | (0.7, 9.8) | ||
| PROM1 (mRNA)* | 0.993 | 0.967 | ||||
| 53.5 vs 26.8 | 1.0 | (0.5, 1.9) | 0.9 | (0.5, 1.9) | ||
| 80.9 vs 26.8 | 1.0 | (0.4, 2.3) | 0.9 | (0.4, 2.2) | ||
| HTERT (mRNA)* | 0.024 | 0.011 | ||||
| 6.2 vs 1.2 | 1.5 | (1.1, 2.1) | 1.7 | (1.2, 2.3) | ||
| 33.5 vs 1.2 | 7.1 | (1.7, 29.2) | 9.9 | (2.2, 44.3) | ||
*The 3 values are the quartiles of the variable distribution.
Relapse-free Survival
Forty-four patients relapsed. Median RFS was 28 months, and median time to death after relapse was 30 months. Five- and 10-year RFSs were 30.4% (95% CI, 20.6%, 44.9%) and 23.4% (14.3%, 38.2%), respectively (Fig. 2A). All tested clinical variables but tumor site were significantly associated with RFS. In particular, 10–year RFSs were as follows: extent of surgery, 35.0% (22.7%, 54.0%) for patients with complete surgery and 0% for patients with incomplete surgery (P = .012); histologic grade, 32.7% (19.2%, 55.5%) for patients having grade 2 tumors and 12.8% (4.2%, 38.8%) for those with grade 3 tumors (P = .021); use of radiation therapy after surgery, 15.6% (6.0%, 41.0%) for patients not submitted to radiation and 28.5% (16.2%, 50.0%) for patients submitted to radiation (P = .011). At multivariable Cox analysis (Table 2, right) all 3 factors remained statistically significant.
Kaplan–Meier estimates (Table 3, right) showed a significant association of EGFR and NCL with RFS. Univariate and multivariate analyses by Cox regression model (Table 5) showed that NCL protein expression was significantly associated with RFS at univariate analysis. In particular, 5- and 10-year RFSs were 45.9% and 19.7% and 45.9% and 13.2%, respectively, for those children with low and high NCL expression (P = .004; Fig. 2D), and NCL was the only suggestive marker at multivariate Cox model analysis (P = .090; Table 5). The RFS Kaplan–Meier curves stratified by anatomic site showed that the NCL prognostic effect held in the subgroups of posterior fossa (10-year RFS estimates: 28.6% [95% CI, 12.5, 65.4] vs 16.7% [5.0, 56.1], P = .221) and supratentorial (10-year RFS estimates: 85.7% [95% CI, 63.3, 100] vs 0%, P = .0004), with a major difference in the latter. However, due to the small size of the subgroups, the estimates were imprecise, which also hampered a rigorous multivariable subgroup analysis. This issue would necessitate testing in larger series.
Table 5.
Cox RFS analysis of biological variables’ prognostic effect
| Univariate Analysis |
Multivariate Analysis* |
|||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P (2–sided Wald test) | HR | 95% CI | P (2–sided Wald test) | |
| 1q gain (FISH) | 0.829 | 0.171 | ||||
| Yes vs no | 1.1 | (0.4, 3.0) | 2.0 | (0.7, 5.4) | ||
| Nucleolin (IHC, % positive cells) | 0.006 | 0.090 | ||||
| >50% vs ≤50% | 3.0 | (1.4, 6.6) | 1.9 | (0.9, 3.8) | ||
| EGFR (IHC) | 0.298 | 0.175 | ||||
| Low vs strong | 2.9 | (1.1, 7.7) | 1.7 | (0.8, 3.5) | ||
| EGFR (mRNA)* | 0.422 | 0.396 | ||||
| 10.1 vs 5.1 | 1.3 | (0.9, 1.9) | 1.3 | (0.9, 1.9) | ||
| 32.7 vs 5.1 | 1.6 | (0.7, 4.2) | 1.8 | (0.7, 4.7) | ||
| PROM1 (mRNA)* | 0.496 | 0.628 | ||||
| 53.5 vs 26.8 | 1.1 | (0.7, 1.9) | 1.1 | (0.6, 1.9) | ||
| 80.9 vs 26.8 | 1.1 | (0.6, 2.2) | 1.0 | (0.5, 2.1) | ||
| HTERT (mRNA)* | 0.292 | 0.337 | ||||
| 6.1 vs 1.2 | 1.2 | (0.9, 1.5) | 1.2 | (0.9, 1.5) | ||
| 33.5 vs 1.2 | 2.1 | (0.9, 5.7) | 2.0 | (0.8, 5.4) | ||
*The 3 values are the quartiles of the variable distribution.
Discussion
Univariate and multivariate analyses of updated clinical data confirmed our previous report about the prognostic significance of extent/completeness of surgery and tumor grade for both RFS and OS.10 In addition, the use of radiation therapy appeared to be a significant prognostic parameter at multivariate analysis for RFS, thus outlining the importance of radiation therapy as a major step forward in the management of childhood ependymoma.10,11,21 Some children who were not submitted to irradiation at first diagnosis, mostly because they were <3 years of age, showed good response to rescue radiotherapy at first relapse, although our recent data clearly show that this approach achieves poorer results compared with firstline irradiation.12
Analysis of gene expression by qRT-PCR is a robust, easy to perform, and straightforward approach already used in several clinical applications,22–25 thanks to the very high specificity and sensitivity of the assay. This approach may avoid the limitations of IHC in those instances in which no robust antibodies are available and is particularly informative when the level of expression of the target gene from normal cells or from negative cancer cells is negligible, as is the case of the molecular markers under investigation, HTERT and EGFR.26–28 HTERT sustains normal cell lifespan and proliferation,29 while cancer cells frequently achieve immortalization by aberrant activation of telomerase expression,30 and HTERT overexpression has been proposed as a prognostic marker and therapeutic target in multiple tumor types.31,32 Ependymomas are characterized by heterogeneous clinical courses that cannot be predicted accurately by clinical, pathologic, or molecular markers, hence the necessity to advance our knowledge of this disease. In the present work, we explored in AIEOP ependymoma series the prognostic impact of previously reported molecular markers, by means of IHC and qRT-PCR. Our data show that elevated HTERT gene mRNA expression and NCL protein expression are promising prognostic markers for worse OS and RFS, respectively, worth testing additionally in larger and prospective cohorts. Recently, several reports showed genetic heterogeneity of ependymoma in relation to tumor location and patients' age, and we recognize that subgroup analyses of molecular-marker status taking into account this evidence may provide useful clinical information for future trials. Unfortunately, most of the cohorts used to retrospectively validate prognostic molecular markers so far,3–7 including our own, suffer limitations in sample size, patients' age, or tumor location distribution, or are even not trial based. Therefore, robust subgroup analyses in uniform trial cohorts are needed and will be possible only in the framework of upcoming, properly sized, multinational clinical trials.
Although the presence of a single patient carrying a CDKN2A homozygous deletion in our series prevented prognostic evaluation of this marker, combined IHC analysis of the CDKN2A/p16 protein showed frequent p16 protein accumulation in supratentorial ependymomas, suggesting that the p16/Rb pathway is directly inactivated in most supratentorial tumors, either by loss of p16 or by downstream events. The genetic or environmental nature of the causative event leading to p16 accumulation in these tumors remains to be determined, but it is tempting to speculate that it may relate to the presence of polyomavirus infections previously described in ependymoma.33,34
Gain of 1q did not reveal any prognostic significance. This may in part be due to the relatively low frequency of 1q gain in our case series (13%). It should be noted that although gain of 1q is reported at variable frequencies in older studies, case series analyzed more recently by high-throughput copy number analyses, which provide a robust evaluation of 1q status thanks to dozens of 1q loci interrogated simultaneously for copy number estimation, concordantly displayed a frequency of a large 1q gain of 10%.6,18–20
For the first time, we provide evidence of elevated mRNA expression of PROM1/CD133—a stemness marker frequently expressed in the cancer-initiating cells of many tumor types35—in a high proportion of ependymoma tumor tissue samples. Expression of PROM1 did not significantly impact patient survival, but, notably, patients displaying peculiarly elevated expression of PROM1 were characterized by prolonged survival despite the occurrence of tumor relapse, suggesting the presence of an indolent form of disease, although such an issue deserves further investigation. Expression of PROM1 was recently described in ependymoma-derived cancer stem cell lines.36,37
EGFR protein expression did not impact OS and RFS at univariate and multivariate Cox analyses. Surprisingly, there was a trend for high EGFR protein expression to display a better prognosis by means of the Kaplan–Meier method. In addition, we showed that even at the mRNA expression level, EGFR failed to demonstrate any significant association with survival, further supporting a lack of prognostic value for this molecular marker. Analysis of ependymoma cell cultures confirmed substantial EGFR protein expression by tumor cells and verified their ability to respond to EGF stimulation by activating EGFR phosphorylation events and in part signaling to Akt. Inhibition of EGFR with a specific small molecule inhibitor demonstrated mainly cytostatic effects accompanied by profound morphologic changes resembling senescence, similar to what was observed in recent reports.36,37 Therefore, despite the established expression of EGFR in ependymoma, several observations—including contradictory results as a prognostic marker from the literature, an expression pattern that is mainly intracytoplasmic, and the lack of recurrent mutation events3,18–20—indicate that the biological significance of EGFR in pediatric intracranial ependymoma needs to be further ascertained.
Supplementary Material
Funding
This work was funded by the Associazione Italiana per la Ricerca sul Cancro, the Fondazione Italiana per la Lotta al Neuroblastoma (Progetto Pensiero), and the Associazione Bianca Garavaglia Onlus. Their support is gratefully acknowledged. The funding agencies had no role in the study.
Supplementary Material
Acknowledgments
P. M. and F. R. B. contributed equally to the study. Symbols in affiliations used to denote correct affiliations for authors with the same initials.
Conflict of interest statement. None declared.
References
- 1.Massimino M, Buttarelli FR, Antonelli M, Gandola L, Modena P, Giangaspero F. Intracranial ependymoma: factors affecting outcome. Future Oncol. 2009;5(2):207–216. doi: 10.2217/14796694.5.2.207. [DOI] [PubMed] [Google Scholar]
- 2.Kuncova K, Janda A, Kasal P, Zamecnik J. Immunohistochemical prognostic markers in intracranial ependymomas: systematic review and meta-analysis. Pathol Oncol Res. 2009;15(4):605–614. doi: 10.1007/s12253-009-9160-2. [DOI] [PubMed] [Google Scholar]
- 3.Mendrzyk F, Korshunov A, Benner A, et al. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res. 2006;12(7 Pt 1):2070–2079. doi: 10.1158/1078-0432.CCR-05-2363. [DOI] [PubMed] [Google Scholar]
- 4.Ridley L, Rahman R, Brundler MA, et al. Multifactorial analysis of predictors of outcome in pediatric intracranial ependymoma. Neuro Oncol. 2008;10(5):675–689. doi: 10.1215/15228517-2008-036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabori U, Ma J, Carter M, et al. Human telomere reverse transcriptase expression predicts progression and survival in pediatric intracranial ependymoma. J Clin Oncol. 2006;24(10):1522–1528. doi: 10.1200/JCO.2005.04.2127. [DOI] [PubMed] [Google Scholar]
- 6.Korshunov A, Witt H, Hielscher T, et al. Molecular staging of intracranial ependymoma in children and adults. J Clin Oncol. 2010;28(19):3182–3190. doi: 10.1200/JCO.2009.27.3359. [DOI] [PubMed] [Google Scholar]
- 7.Carter M, Nicholson J, Ross F, et al. Genetic abnormalities detected in ependymomas by comparative genomic hybridisation. Br J Cancer. 2002;86(6):929–939. doi: 10.1038/sj.bjc.6600180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbertson RJ, Bentley L, Hernan R, et al. ERBB receptor signaling promotes ependymoma cell proliferation and represents a potential novel therapeutic target for this disease. Clin Cancer Res. 2002;8(10):3054–3064. [PubMed] [Google Scholar]
- 9.Tabori U, Wong V, Ma J, et al. Telomere maintenance and dysfunction predict recurrence in paediatric ependymoma. Br J Cancer. 2008;99(7):1129–1135. doi: 10.1038/sj.bjc.6604652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massimino M, Gandola L, Giangaspero F, et al. Hyperfractionated radiotherapy and chemotherapy for childhood ependymoma: final results of the first prospective AIEOP (Associazione Italiana di Ematologia-Oncologia Pediatrica) study. Int J Radiat Oncol Biol Phys. 2004;58(5):1336–1345. doi: 10.1016/j.ijrobp.2003.08.030. [DOI] [PubMed] [Google Scholar]
- 11.Massimino M, Giangaspero F, Garre ML, et al. Salvage treatment for childhood ependymoma after surgery only: pitfalls of omitting “at once” adjuvant treatment. Int J Radiat Oncol Biol Phys. 2006;65(5):1440–1445. doi: 10.1016/j.ijrobp.2006.03.052. [DOI] [PubMed] [Google Scholar]
- 12.Massimino M, Gandola L, Barra S, et al. Infant ependymoma in a 10-year AIEOP (Associazione Italiana Ematologia Oncologia Pediatrica) experience with omitted or deferred radiotherapy. Int J Radiat Oncol Biol Phys. 2011;80(3):807–14. doi: 10.1016/j.ijrobp.2010.02.048. [DOI] [PubMed] [Google Scholar]
- 13.Miyanaga T, Hirato J, Nakazato Y. Amplification of the epidermal growth factor receptor gene in glioblastoma: an analysis of the relationship between genotype and phenotype by CISH method. Neuropathology. 2008;28(2):116–126. doi: 10.1111/j.1440-1789.2007.00853.x. [DOI] [PubMed] [Google Scholar]
- 14.Lualdi E, Modena P, Debiec-Rychter M, et al. Molecular cytogenetic characterization of proximal-type epithelioid sarcoma. Genes Chromo Cancer. 2004;41(3):283–290. doi: 10.1002/gcc.20086. [DOI] [PubMed] [Google Scholar]
- 15.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1(3):1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 16.Moons KG, Donders AR, Steyerberg EW, Harrell FE. Penalized maximum likelihood estimation to directly adjust diagnostic and prognostic prediction models for overoptimism: a clinical example. J Clin Epidemiol. 2004;57(12):1262–1270. doi: 10.1016/j.jclinepi.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Durrleman S, Simon R. Flexible regression models with cubic splines. Stat Med. 1989;8(5):551–561. doi: 10.1002/sim.4780080504. [DOI] [PubMed] [Google Scholar]
- 18.Modena P, Lualdi E, Facchinetti F, et al. Identification of tumor-specific molecular signatures in intracranial ependymoma and association with clinical characteristics. J Clin Oncol. 2006;24(33):5223–5233. doi: 10.1200/JCO.2006.06.3701. [DOI] [PubMed] [Google Scholar]
- 19.Taylor MD, Poppleton H, Fuller C, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8(4):323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Johnson RA, Wright KD, Poppleton H, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466(7306):632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massimino M, Gandola L, Garre ML, et al. Do we really need class 1 evidence results to give adjuvant radiation therapy to childhood intracranial ependymomas? Childs Nerv Syst. 2009;25(6):641–2. doi: 10.1007/s00381-009-0847-1. author reply 643–4. [DOI] [PubMed] [Google Scholar]
- 22.Bustin SA, Mueller R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin Sci (Lond) 2005;109(4):365–379. doi: 10.1042/CS20050086. [DOI] [PubMed] [Google Scholar]
- 23.Skrzypski M. Quantitative reverse transcriptase real-time polymerase chain reaction (qRT-PCR) in translational oncology: lung cancer perspective. Lung Cancer. 2008;59(2):147–154. doi: 10.1016/j.lungcan.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Bustin SA, Mueller R. Real-time reverse transcription PCR and the detection of occult disease in colorectal cancer. Mol Aspects Med. 2006;27(2–3):192–223. doi: 10.1016/j.mam.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Gabert J, Beillard E, van der Velden VH, et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—a Europe Against Cancer program. Leukemia. 2003;17(12):2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- 26.Wu YL, Dudognon C, Nguyen E, et al. Immunodetection of human telomerase reverse-transcriptase (hTERT) re-appraised: nucleolin and telomerase cross paths. J Cell Sci. 2006;119(Pt 13):2797–2806. doi: 10.1242/jcs.03001. [DOI] [PubMed] [Google Scholar]
- 27.Dei Tos AP, Ellis I. Assessing epidermal growth factor receptor expression in tumours: what is the value of current test methods? Eur J Cancer. 2005;41(10):1383–1392. doi: 10.1016/j.ejca.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Fruehauf J. EGFR function and detection in cancer therapy. J Exp Ther Oncol. 2006;5(3):231–246. [PubMed] [Google Scholar]
- 29.Masutomi K, Yu EY, Khurts S, et al. Telomerase maintains telomere structure in normal human cells. Cell. 2003;114(2):241–253. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- 30.Janknecht R. On the road to immortality: HTERT upregulation in cancer cells. FEBS Lett. 2004;564(1–2):9–13. doi: 10.1016/S0014-5793(04)00356-4. [DOI] [PubMed] [Google Scholar]
- 31.Kelland LR. Overcoming the immortality of tumour cells by telomere and telomerase based cancer therapeutics—current status and future prospects. Eur J Cancer. 2005;41(7):971–979. doi: 10.1016/j.ejca.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 32.Liu JP, Chen W, Schwarer AP, Li H. Telomerase in cancer immunotherapy. Biochim Biophys Acta. 2010;1805(1):35–42. doi: 10.1016/j.bbcan.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Croul S, Otte J, Khalili K. Brain tumors and polyomaviruses. J Neurovirol. 2003;9(2):173–182. doi: 10.1080/13550280390194055. [DOI] [PubMed] [Google Scholar]
- 34.Bergsagel DJ, Finegold MJ, Butel JS, Kupsky WJ, Garcea RL. DNA sequences similar to those of simian virus 40 in ependymomas and choroid plexus tumors of childhood. N Engl J Med. 1992;326(15):988–993. doi: 10.1056/NEJM199204093261504. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, Wu PY. CD133 as a marker for cancer stem cells: progresses and concerns. Stem Cells Dev. 2009;18(8):1127–1134. doi: 10.1089/scd.2008.0338. [DOI] [PubMed] [Google Scholar]
- 36.Servidei T, Meco D, Trivieri N, et al. Effects of epidermal growth factor receptor blockade on ependymoma stem cells in vitro and in orthotopic mouse models. Int J Cancer. 2012;131(5):E791–803. doi: 10.1002/ijc.27377. [DOI] [PubMed] [Google Scholar]
- 37.Guan S, Shen R, Lafortune T, et al. Establishment and characterization of clinically relevant models of ependymoma: A true challenge for targeted therapy. Neuro Oncol. 2011;13(7):748–758. doi: 10.1093/neuonc/nor037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.