The authors developed a comprehensive transcriptome atlas in grapevine by comparing the genes expressed in 54 diverse samples accounting for ∼91% of all known grapevine genes. Using a panel of different statistical techniques, they found that the whole plant undergoes transcriptomic reprogramming, driving it towards maturity.
Abstract
We developed a genome-wide transcriptomic atlas of grapevine (Vitis vinifera) based on 54 samples representing green and woody tissues and organs at different developmental stages as well as specialized tissues such as pollen and senescent leaves. Together, these samples expressed ∼91% of the predicted grapevine genes. Pollen and senescent leaves had unique transcriptomes reflecting their specialized functions and physiological status. However, microarray and RNA-seq analysis grouped all the other samples into two major classes based on maturity rather than organ identity, namely, the vegetative/green and mature/woody categories. This division represents a fundamental transcriptomic reprogramming during the maturation process and was highlighted by three statistical approaches identifying the transcriptional relationships among samples (correlation analysis), putative biomarkers (O2PLS-DA approach), and sets of strongly and consistently expressed genes that define groups (topics) of similar samples (biclustering analysis). Gene coexpression analysis indicated that the mature/woody developmental program results from the reiterative coactivation of pathways that are largely inactive in vegetative/green tissues, often involving the coregulation of clusters of neighboring genes and global regulation based on codon preference. This global transcriptomic reprogramming during maturation has not been observed in herbaceous annual species and may be a defining characteristic of perennial woody plants.
INTRODUCTION
Grapevine (Vitis spp) is the most cultivated fruit crop in the world, covering nearly 7.8 million hectares in 2011 and producing 67.5 million tons of berries (http://www.oiv.int/). The berries are harvested primarily for wine making (68%) but also to provide fresh table grapes (30%), raisins (2%), and minor products, such as grape juice, jelly, ethanol, vinegar, grape seed oil, tartaric acid, and fertilizers. Grape berries contain antioxidants such as polyphenols (e.g., resveratrol) with important health benefits that are valued in the food, cosmetic, and pharmaceutical industries.
Grapevine is a perennial from the family Vitaceae, which includes woody deciduous plants within the basal eudicots (Judd, 1999). It has a biennial reproductive cycle, and its growth characteristics and patterning during development are distinct from annual herbaceous and woody polycarpic plants (Mullins et al., 1992; Carmona et al., 2007; Roubelakis-Angelakis, 2009).
To provide insight into the transcriptional programs controlling the development of different organ systems, we generated a global gene expression atlas for the common grapevine species Vitis vinifera (cv Corvina). Comparable resources for other plant species have been described but none representing perennial woody crops. Functional developmental modules based on expression profiling have been described in Arabidopsis thaliana (Schmid et al., 2005), and dynamic transcriptional profiles representing different cell types and developmental processes have been identified through the analysis of gene expression atlases in rice (Oryza sativa) (Li et al., 2006; Jiao et al., 2009) and barley (Hordeum vulgare) (Druka et al., 2006). A recent atlas of tobacco (Nicotiana tabacum) development based on gene expression profiles from seed to senescence provided new regulatory targets and allowed the manipulation of specific pathways involved in quality control (Edwards et al., 2010). Most recently, whole-plant transcriptome surveys were published for soybean (Glycine max), potato (Solanum tuberosum), tomato (Solanum lycopersicum), and maize (Zea mays) (Aoki et al., 2010; Severin et al., 2010; Massa et al., 2011; Sekhon et al., 2011). Our comprehensive grapevine transcriptome map combined with the complete genome sequence (Jaillon et al., 2007) provides the basis for gene functional analysis on a global scale and elevates grapevine to the status of a model fruit species.
RESULTS AND DISCUSSION
Defining the Grapevine Transcriptome
To study the entire grapevine transcriptome, we collected triplicates of 54 diverse samples representing different vegetative and reproductive organs at various developmental stages. In addition to developing and ripening berries, we included berries that had undergone postharvest withering, a common winemaking process. This represented the only stress condition imposed in our survey (Figure 1; see Supplemental Table 1 online).
Figure 1.
Overview of the V. vinifera cv Corvina Samples Used for Microarray Analysis.
The photographs and diagrams show the shoot/cane organs (A) and berry cluster (B) from clone 48. The exact developmental stages are indicated by the modified E-L classification keys on each picture. Rachis, seed, berry flesh, and skin samples were taken at the stages indicated in (B). Schematic illustrations were modified from Jackson (2000).
The expression of >98% of grapevine genes (http://srs.ebi.ac.uk/) was monitored using the NimbleGen 090918_Vitus_exp_HX12 array. Robust multichip average data were used to evaluate the number of expressed genes, allowing us to identify significant signals representing gene expression and to hypothesize a positive correlation between the number of expressed genes and the degree of bimodal distribution (see Supplemental Figure 1 online).
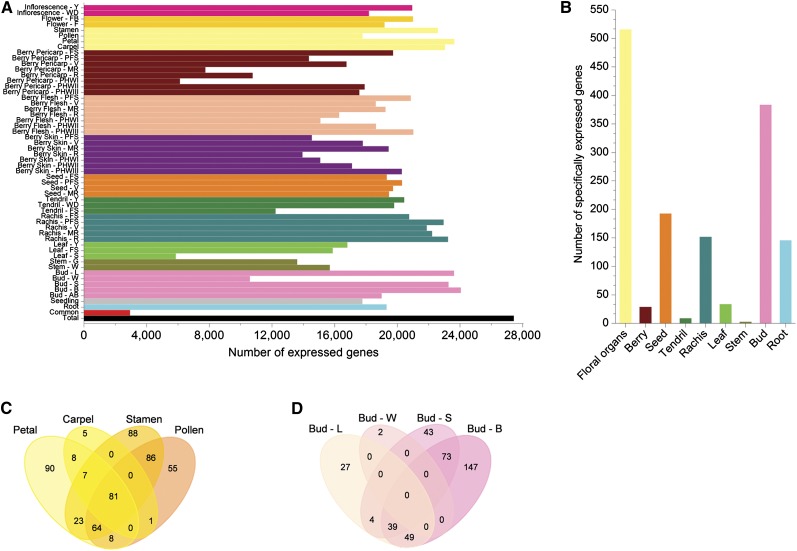
We detected the expression of 27,435 genes in at least one of the 54 samples, representing ∼91% of all genes on the array (Figure 1A; see Supplemental Data Set 1 online). The number of transcripts detected during organ development varied substantially in most of the systems we sampled, fluctuating between 5864 and 24,059 (representing 20 to 81% of all genes on the array). The greatest fluctuations were seen in bud and leaf samples, where more transcripts were detected during active growth and fewer in autumn/winter months when the buds become dormant and the leaves undergo senescence. By contrast, the number of transcripts detected in the seeds remained constant, and there were only minor fluctuations in the number of transcripts detected in the rachis. The distribution of transcripts among grapevine samples, despite their biological complexity, was similar to that previously reported for different rice cell types (Jiao et al., 2009) (see Supplemental Figure 2 online).
To identify and characterize organ-specific genes, we constructed a reduced 38-sample data set, excluding from the analysis samples with redundant organ identity and those collected during senescence and withering (see Supplemental Table 2 online). The floral organs and buds expressed the greatest number of organ-specific transcripts (Figure 2B). Seeds and roots expressed more organ-specific transcripts than leaves, as previously reported (Schmid et al., 2005). Surprisingly, a large number of rachis-specific genes were identified, suggesting this organ is particularly important during grapevine fruiting. By contrast, there were very few genes expressed exclusively in berries, tendrils, or stems.
Figure 2.
Global Gene Expression Patterns in the Different Samples.
(A) Number of genes expressed in each of the 54 samples. Total: number of gene expressed in at least one organ (27,453; ∼93% of all genes on the array). Common: genes expressed in all 54 organs (2948; ∼10% of all genes on the array).
(B) Number of organ-specific genes. Only samples with nonredundant organ identity were analyzed (see Supplemental Table 2 online).
(C) Shared and specific expression profiles of genes expressed in multiple floral organs.
(D) Shared and specific expression profiles of genes expressed at multiple bud developmental stages.
Organ-specific transcripts were analyzed in more detail to identify those expressed in multiple organs (i.e., within the flower) and/or at multiple developmental stages (Figures 2C and 2D; see Supplemental Figure 3 online). Shared expression profiles were more common among different organs than at different developmental stages in the same organ (e.g., no common organ-specific genes were expressed in the developing bud or berry at the different stages we tested). Few organ-specific genes were shared among the different developmental stages of the rachis and seed, but up to 16% of the organ-specific genes expressed in the flower were common to the different floral organs. These data imply that organ identity in the grapevine transcriptome is less important than the developmental stage.
We assessed the function of the organ-specific transcripts and found that bud-specific transcripts were primarily represented by transcription factors, signaling proteins, and transporters (see Supplemental Data Set 2 online). Many of the flower-specific transcripts represented transport functions, including several ABC transporters, some of which may be required to form a cuticular layer in the petals to act as a diffusion barrier (Bessire et al., 2011). Several of the seed-specific transcripts represented transcription factors, including a TT2-like Myb factor (present at the postfruit set [PFS] and veraison stages) required for seed coat proanthocyanidin biosynthesis in Arabidopsis (Baudry et al., 2004). Many of the root-specific transcripts represented secondary metabolic functions, predominantly monolignol and monoterpene biosynthesis. The roots also expressed six transcripts encoding germin-like proteins, which may help to form a defensive barrier during emergence from the soil but are also implicated in symbiosis (Himmelbach et al., 2010). Only a few tendril-specific transcripts were identified, including several related to auxin signaling/responses and one encoding a TT16-like MADS transcription factor that is thought to control organ growth in Arabidopsis (Prasad et al., 2010). Most of the rachis-specific transcripts were identified at the mature stage (Rachis-R). Approximately 30% of these transcripts encoded proteins involved in stress responses, but others were related to transport and signal transduction (e.g., kinases and annexins), indicating that the rachis is not solely a structural organ. Remarkably, more than half of the berry-specific genes we identified do not have an assigned function yet, suggesting that berry development has unique characteristics that are not well understood at the molecular level.
Tissue Transcriptome Relationships
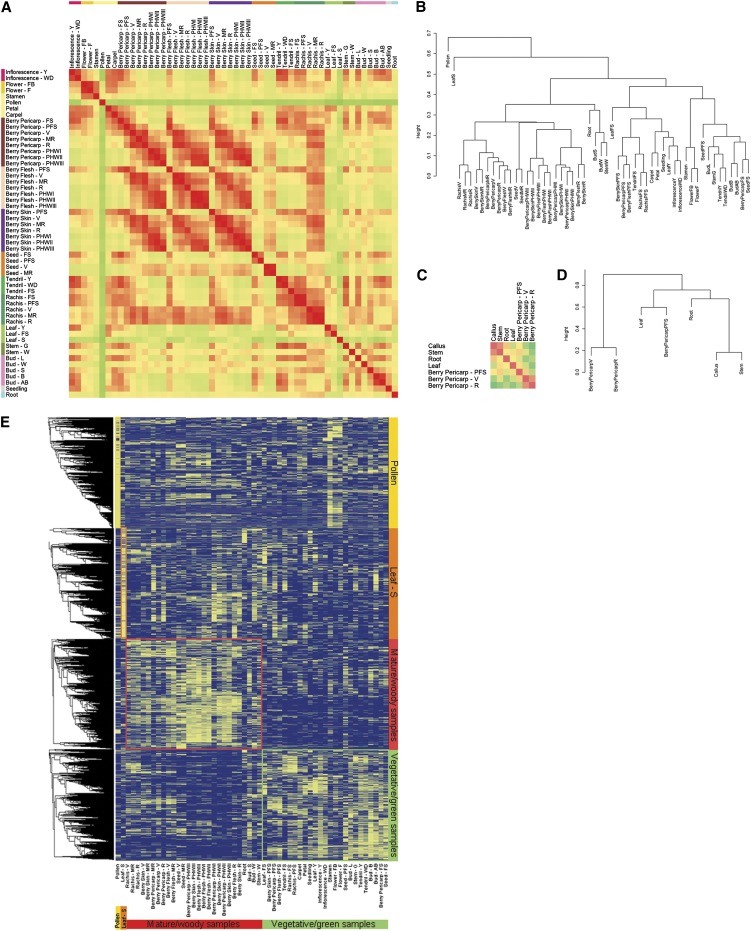
To score the quality of our expression data set, we performed coexpression analysis using selected grapevine genes as queries to identify correlations between genes involved in the same process. We used the closest grapevine homologs of Arabidopsis PSAD1 (photosystem I reaction center subunit II) and LHCII (for light-harvesting complex II), both related to photosynthesis, as well as a regulatory gene (MYBA1) and a structural gene (FLAVANONE 3-HYDROXYLASE1 [F3H1]) from the flavonoid pathway. This identified several photosynthesis-related genes that correlated with PSAD1 and LHCII, and several additional flavonoid pathway genes correlated with MYBA1 and F3H1, with some of them representing known transcriptional hierarchies (see Supplemental Figure 4 online). We generated a Pearson’s distance correlation matrix to compare the transcriptomes from each sample (Figure 3A). This showed a strong correlation among the mature/woody samples and a clear distinction between the mature/woody and vegetative/green samples. The pollen transcriptome was highly distinctive as was the transcriptome of the leaf undergoing senescence, both showing little resemblance to the other samples. The resulting dendrogram showed that samples clustered predominantly in relation to temporal dynamics and that organ identity was less important (Figure 3B; see Supplemental Figure 5A online). Remarkably, this distribution did not depend on the expression levels of the corresponding genes (see Supplemental Figure 5B online). We also noted a separation between ripened berries and vegetative/green tissues when overrepresented berry samples were excluded from the analysis (see Supplemental Figure 5C online). This was confirmed by generating a Pearson’s distance correlation matrix using previously released RNA-seq data mapped onto the 12x Grape Genome, V1 Gene Prediction (Denoeud et al., 2008; Zenoni et al., 2010) (Figures 3C and 3D; see Supplemental Table 3 online). These results confirmed that organ maturity was more important than organ identity in defining a common transcriptome, and the same effects were observed regardless of the analytical method employed and the overrepresentation of particular samples.
Figure 3.
Tissue Transcriptome Relationships.
(A) Correlation matrix of the whole data set. The analysis was performed by comparing the values of the whole transcriptome (29,549 genes) in all 54 samples, using the average expression value of three biological replicates and Pearson’s distance as the metric. Correlation analysis was performed using R software.
(B) Cluster dendrogram of the whole data set. The Pearson’s correlation values were converted into distance coefficients to define the height of the dendrogram.
(C) Correlation matrix for the RNA-seq data set. Reads generated in previous experiments (Denoeud et al., 2008; Zenoni et al., 2010) were remapped on the 12x grapevine genome, V1 gene prediction.
(D) Cluster dendrogram for the RNA-seq data set. Reads generated in previous experiments (Denoeud et al., 2008; Zenoni et al., 2010) were remapped on the 12x grapevine genome, V1 gene prediction.
(E) HCL analysis on the whole 54-sample data set. Pearson’s correlation distance was used as the metric, and TMeV 4.3 software was used to create the transcriptional profiles dendrogram.
The partition between mature/woody and vegetative/green samples was also maintained for gene expression profiles (Figure 3E; see Supplemental Figure 6 online). Hierarchical clustering (HCL) analysis revealed four major groups of genes whose transcriptional profiles defined the mature/woody samples, vegetative/green samples, pollen, and leaves undergoing senescence. The last two samples were typified by their characteristic transcript profiles, validating our hypothesis that these two organs possess highly distinguishable physiological traits based on their unique transcriptomes.
Molecular Biomarkers
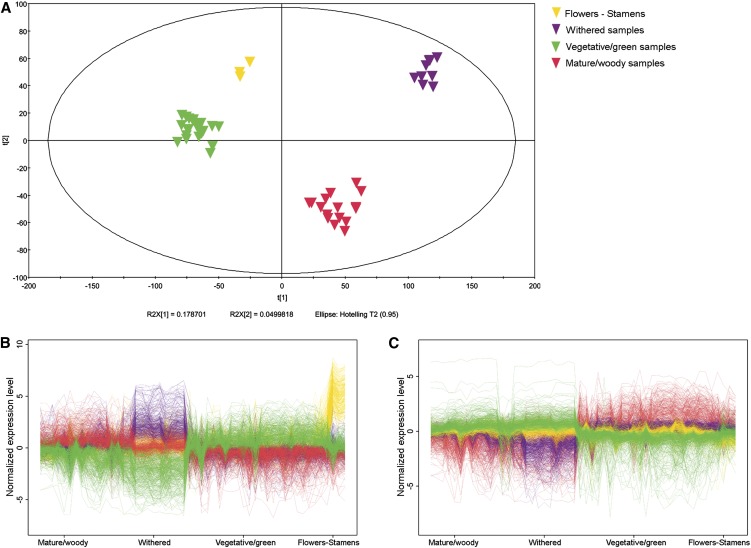
To gain insight into the physiological and molecular factors underlying the separation between samples, we performed principal component analysis (PCA) on the complete data set. We used the first 11 principal components to explain 70.65% of the variability. The second component (11.40%) represented leaves undergoing senescence and the third component (7.99%) represented pollen (see Supplemental Figure 7A online). The relationships among the other samples were investigated in more detail by carrying out a second PCA on the 52-sample reduced data set (without pollen and senescent leaves). The first principal component (19.27%) included four clusters of gene expression profiles (see Supplemental Figure 7B online). We used orthogonal bidirectional projections to latent structures discriminant analysis (O2PLS-DA) (Trygg, 2002) to confirm the PCA data, which verified the four-class distribution: withered berries, mature/woody samples, flowers/stamens, and all the remaining vegetative/green samples (Figure 4A). Samples of berries treated by postharvest withering were clearly separated from the other mature/woody samples, and flowers and stamens were clearly separated from the other vegetative/green samples.
Figure 4.
Global Gene Expression Trends in Grapevine.
(A) Variables and scores scatterplot of the O2PLS-DA model (3 + 5 + 0, UV, R2Y = 0.967, Q2 = 0.868) applied to 52 samples, confirming the separation into four classes sharing similar expression signatures. Components 3 and 5 represent the predictive and orthogonal components identified by the model, whereas 0 represents the background variation (UV = unit variance scaling method).
(B) and (C) The expression profiles of positive (B) and negative (C) putative molecular biomarkers were selected using an S-plot (Wiklund et al., 2008) within the first (positive) and the last (negative) percentile.
Putative molecular biomarkers (i.e., transcripts whose presence or absence defines the samples in a given class) were identified by applying four distinct two-class O2PLS-DA models, using in each case the observations from one class as a reference and grouping the other three observations in one unique class (Zamboni et al., 2010). An S-plot (Wiklund et al., 2008) was then used to select putative biomarkers within the first (positive biomarkers) and last (negative biomarkers) percentiles (Figures 4B and 4C; see Supplemental Data Set 3 online). Positive biomarkers representing the flowers and stamens included transcripts corresponding to enzymes in the monoterpenoid and sesquiterpenoid biosynthesis pathways (e.g., enzymes that synthesize germacrene, cadinene, terpineol, pinene and myrcene, which are prominent components of floral scents) (Martin et al., 2010). There were also eight pectinesterase and seven polygalacturonase transcripts encoding cell wall–modifying enzymes involved in flower abscission (van Doorn and Stead, 1997) and pollen tube elongation (Bosch and Hepler, 2005). Notably, a homolog of Arabidopsis galacturonosyltransferase-like 4, which scored the highest p(corr) value, was previously shown to be expressed specifically in Arabidopsis stamens and pollen grains (Kong et al., 2011). Pectinesterase and stilbene synthase transcripts were identified as biomarkers of withered berries, agreeing with previous studies showing that cell wall modification and resveratrol biosynthesis are important aspects of the withering process (Versari et al., 2001; Zamboni et al., 2008). We also identified the Ras GTP binding protein RAN3, which regulates RNA and protein transport through nuclear pores and has previously been identified as a positive biomarker of withering in Corvina berries (Zamboni et al., 2010). Several transcripts encoding stress response, ethylene response, and protein recycling functions were strongly represented in mature/woody samples, along with a population of (predominantly zinc finger) transcription factors, suggesting that significant transcriptional reprogramming is required for the transition to the mature phase. As expected, vegetative/green sample markers were rich in photosynthesis-related transcripts, including those encoding 11 light-harvesting complex subunits, five photosystem reaction center subunits, and the COP-1–interacting protein CIP-7, a positive regulator of light-induced genes (Yamamoto et al., 1998).
Division of Samples into Topics Defined by High-Level Gene Expression
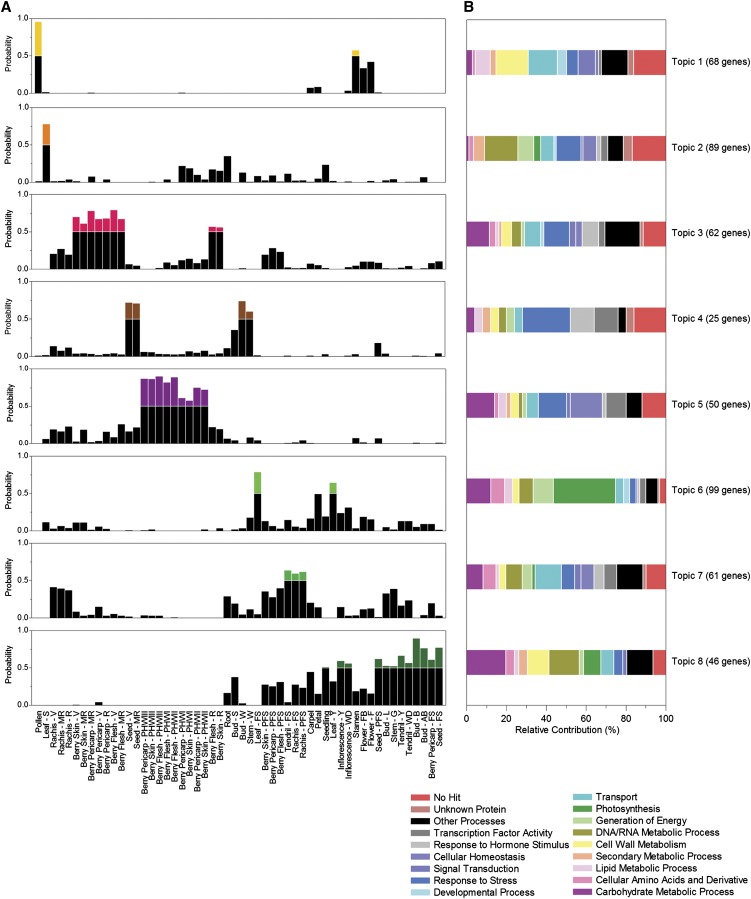
Potential correlations between samples in terms of the magnitude and consistency of gene expression were evaluated using a biclustering analysis method based on a probabilistic topic model called probabilistic latent semantic analysis (PLSA), which allows data sets to be modeled in terms of hidden topics or processes that can reflect underlying meaningful structures (Hofmann, 2001; Joung et al., 2006; Bicego et al., 2010). We applied this method to the entire data set to discover groups of genes sharing compatible expression patterns across subsets of samples (Madeira and Oliveira, 2004; Prelić et al., 2006). The basic idea in the gene expression scenario is that a topic may be roughly intended as a biological process, which can characterize a subset of samples (namely, the samples in which the process is active). At the same time, a topic may be related to the activation of a particular set of genes (namely, the genes related to the particular process). Following this reasoning, the relation between gene expression and samples is said to be mediated through the probabilistic presence of the topics (Joung et al., 2006; Bicego et al., 2010). Penalized likelihood statistical analysis (Bayesian information criterion) (Schwarz, 1978) was used to define the optimal number of topics containing highly correlated samples (see Supplemental Figure 8 online). The eight-topic model confirmed the modulation of the grapevine transcriptome in relation to temporal dynamics, reflecting specific metabolic processes rather than organ identity (Figure 5A). Topic 1 (pollen, stamen, and, with lower probability, whole flower samples) was characterized by the strong expression of genes related to transport, cell wall structure, and lipid metabolism (Figure 5B; see Supplemental Data Set 4 online). The cell wall group included several pectin metabolism genes, the cellulose synthase gene CSLG2 (associated with the inner pollen grain wall or intine), and ECERIFERUM1, whose product is associated with the anther cuticle and the outer pollen grain wall or exine in Arabidopsis, suggesting a protective role during grapevine pollen grain development (Jung et al., 2006). The lipid metabolism group included transcripts for three Gly-Asp-Ser-Leu esterases/acylhydrolases that may regulate changes in lipid composition at the pollen-stigma interface (Updegraff et al., 2009). We also identified a transcription factor homologous to Arabidopsis MYB24, which plays a role in anther development (Matus et al., 2008). Topic 2 (leaves undergoing senescence) was characterized by the strong expression of stress response genes, including those encoding several ribosomal proteins and histones that may control stress-induced gene expression and protein synthesis (Pandey et al., 2008; Falcone Ferreyra et al., 2010), abiotic stress response enzymes, such as stilbene synthase, glutathione S-transferase (oxidative stress), and EARLY LIGHT-INDUCED PROTEIN1 (illumination stress), and pathogen response factors, including metallothionein (Breeze et al., 2011), PATHOGENESIS-RELATED10-like proteins, and two ADP-ribosylation factors (Nomura et al., 2011). Samples from mature/woody samples were distributed over three topics: ripening berries (topic 3), withering berries (topic 5), and veraison and mid-ripening seeds, winter buds, and woody stems (organs related to woody structures or to the dormant state; topic 4). Topics 3 and 5 were characterized by the strong expression of genes related to carbohydrate metabolism (particularly starch and sugar), but remarkably no genes representing secondary metabolism were included. Topics 3 and 5 also included stress response genes relevant to dehydration and/or pathogens, which characterize berry ripening and withering (Davies and Robinson, 2000; Zamboni et al., 2010). Topic 5 also included the high-level expression of polyubiquitin, protease, and proteasome subunit genes, representing the transcriptional control of protein degradation and recycling during withering, where dehydration and sugar concentration lead to significant physiological changes. Topic 4 was represented by a small number of genes, mainly encoding stress and hormone response proteins, such as metallothionein and dehydrin, an ABA-INDUCED WHEAT PLASMA MEMBRANE-19 protein homolog that could mediate ABA-induced freezing tolerance, and the dormancy regulator DRM1. Topic 4 also contained an AtMYB73 homolog, which is related to cold acclimation in Arabidopsis (Jung et al., 2008).
Figure 5.
Biclustering Analysis with the PLSA Algorithm.
(A) Samples were divided into eight topics defined by high-level gene expression.
(B) Functional category distribution of topic-specific transcripts. The V1 version of the 12x draft annotation of the grapevine genome allows the identification of ∼70% of genes. This was manually verified and transcripts were grouped into the 18 most represented functional categories, based on Plant GO Slim biological processes classification.
Samples from vegetative/green samples were also distributed over three topics: green leaves (topic 6), rachis and tendrils at fruit set and rachis at postfruit set (topic 7), and young green tissues (topic 8). Topic 6 was characterized by the high-level expression of genes related to photosynthesis and glycolysis, as expected for a grouping of young and mature leaves and (as minor contributors) petals, including those encoding several apoproteins of the light-harvesting complex associated with photosystem II (Lhcb) and a homolog of the Arabidopsis circadian clock Myb transcription factor CCA1, supporting its role in the regulation of Lhcb expression and its close association with circadian rhythms in the grapevine leaf (Wang and Tobin, 1998). Topic 7 grouped the first two rachis stages and the last tendril stage, confirming the ontogenic relationship between these two organs, which are peculiar to grapevine. The three last rachis stages, the berry pericarp, skin, and flesh at PFS, the green stem, and root samples were also represented (albeit with a lower probability) in this topic. All these organs are characterized by reaching their final shape and size and by a forthcoming metabolic shift to the mature phase. Many of the strongly expressed genes included in this topic are involved in transport and stress responses, including at least four encoding aquaporins that regulate the movement of water across membranes. This is consistent with the translocation activity of most of the organs represented in this topic (Shatil-Cohen et al., 2011). Transcripts representing the grape homologs of the Arabidopsis transcription factors ETHYLENE INSENSITIVE3 (EIN3) and JASMONATE-ZIM DOMAIN1 (JAZ1), which may integrate ethylene and jasmonate signaling during development (Zhu et al., 2011), were also strongly expressed in this topic. The JAZ1 homolog (but not the EIN3 homolog) was also strongly expressed in topic 6, perhaps reflecting a role in the repression of epidermal differentiation as previously established in Arabidopsis (Qi et al., 2011). Topic 8 included the two inflorescence stages, seeds at fruit set and postfruit set, berry pericarp at fruit set, latent bud, bud at burst, bud after burst, green stem, and young and well-developed tendrils. These growing organs were characterized by the high-level expression of genes involved in growth (e.g., carbohydrate and cell wall metabolism, photosynthesis, and ribosomal activity). The protection of such developing organs is underlined by the strong expression of genes encoding flavanone-3-hydroxylase and leucoanthocyanidin dioxygenase, which contribute to the accumulation of flavonoid compounds that protect plants against UV radiation.
Gene Coexpression Dynamics Contribute to the Division between Green/Vegetative and Mature/Woody Samples
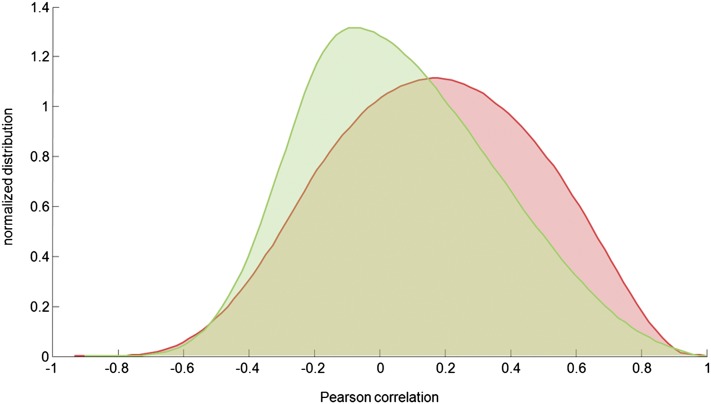
We studied the transcriptomic behavior of clustered samples in more detail by analyzing the coexpression of genes previously identified by HLC analysis as typical representatives of vegetative/green or mature/woody samples (Figure 3E). We looked at the correlation among gene pairs from these selected groups independently (see Supplemental Data Set 5 online). Transcriptome correlation analysis in vegetative/green samples revealed genes potentially involved in diverse processes, such as photosynthesis, secondary metabolism, and hormone signaling. A clear example of genes from the same pathway with a high degree of gene pair correlation is provided by two linalool synthases and three 1,8-cineole synthases from the plastidial 2-methyl-d-erythritol-4-phosphate pathway (Bohlmann et al., 1998; Emanuelli et al., 2010). In mature/woody samples, transcriptome correlation revealed several genes potentially involved in defense/stress responses, lipid metabolism, and cell wall assembly. For example, the dehydration-responsive protein RD22 was highly correlated with many late embryogenesis abundant proteins, which protect tissues from water loss (Hanana et al., 2008; Olvera-Carrillo et al., 2010). The expression profiles of mature/woody genes in the mature/woody samples were evidently more correlated than those of green/vegetative genes in green/vegetative samples (Figure 6). Surprisingly, the most correlated gene pairs in vegetative/green samples (>99 percentile) showed a higher correlation in the mature/woody samples sub–data set than the converse analysis in which the most correlated mature/woody gene pairs were investigated in the vegetative/green samples sub–data set (see Supplemental Figure 9 online). Furthermore, the 1000 best-correlated gene pairs in mature/woody samples represented only 105 single genes, whereas those in green/vegetative samples represented 163 single genes, indicating that individual mature/woody genes participate on average in more gene pairs to establish tightly correlated groups or small networks (see Supplemental Figure 10 online). This suggests that the onset of the mature/woody developmental program is characterized by the coexpression of a few genes belonging to the same metabolic pathways.
Figure 6.
Coexpression Distribution among Green/Vegetative Samples and Ripe/Woody Samples.
Pairwise gene correlation analysis was computed by calculating the Pearson’s correlation for each gene pair in both specific subsets of organs. Curve distributions are represented by the areas under the curves normalized to 1. Green curve, green/vegetative samples; red curve, ripe/woody samples.
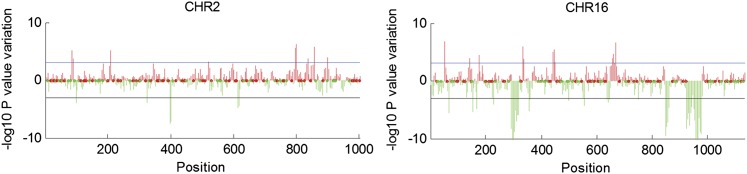
The chromosomal locus of a gene influences its transcription in higher eukaryotes (Williams and Bowles, 2004; Weber and Hurst, 2011), so we integrated the pairwise correlation analysis with a sliding-window analysis of coexpressed neighboring genes. This identified several chromosome regions containing neighboring genes coexpressed at a higher frequency (over a threshold P value) than would be expected by chance (see Supplemental Figures 11A and 12 and Supplemental Data Set 6 online). Most of these regions contained duplicated genes, as previously reported in other eukaryotes (Williams and Bowles, 2004; Weber and Hurst, 2011). A remarkable example is provided by cluster 34 on chromosome 16 (chr16-clA34), which includes 35 stilbene synthase genes. Some groups of coexpressed neighboring genes identified during the whole data set analysis were found to be coexpressed in a particular subset of samples following a more detailed analysis (e.g., chr3-clA5 in withered berries, roots, and seeds and chr10-clA18 in green buds and other vegetative samples). To determine whether vegetative/green or mature/woody samples could be characterized specifically by the coexpression of neighboring genes, we analyzed changes in coexpression between the two groups of samples (see Supplemental Figures 11B, 11C, and 13 online). Significant coexpression peaks found on chromosome 2 during the whole data set analysis were shown to be confined predominantly to mature/woody samples, such as cluster chr2-clMW5, which contained R2R3 Myb family genes involved in the control of anthocyanin synthesis (Matus et al., 2008). Conversely, coexpression peak chr2-clVG5 contained thaumatin and osmotin genes that are likely to be involved in defense responses during vegetative growth (de Freitas et al., 2011a, 2011b) (Figure 7; see Supplemental Data Sets 7 and 8 on line). Several Phe ammonia lyase (PAL) genes were clustered on chromosome 16, one group coexpressed in mature/woody organs, and another in vegetative/green samples, suggesting phenylpropanoid-derived compounds are abundant in both types of samples. The presence of multiple segmental duplications in this region could explain the divergence of PAL gene expression profiles within the cluster (Giannuzzi et al., 2011). The coexpression of neighboring genes with apparently uncorrelated functions was observed in both vegetative/green and mature/woody samples, which contrasts with the coexpression analysis data covering the entire data set. This may suggest a partnership between genes in the same cluster and a given developmental process, despite their apparently unrelated molecular or cellular functions, and may be related to epigenetic regulatory mechanisms that exert their effects over genes in the same chromatin domain and recruit them into coregulated pathways.
Figure 7.
Sliding-Window Analysis of Coexpression along Grapevine Chromosomes 2 and 16.
Red and green lines correspond to positions on the chromosome where coexpression is specific for nonvegetative samples (positive variation) and vegetative samples (negative variation), respectively (see Supplemental Methods 1 online for further details on sliding-window analysis).
Vegetative/Green and Mature/Woody Samples Have Different Codon Usage Preferences
Cellular tRNA pools can be highly dynamic, helping to tune protein synthesis to meet specific physiological or developmental requirements (Najafabadi et al., 2009). The analysis of mutual information (MI) between codon usage and 60 transcriptional clusters revealed the nonrandom use of many codons in genes with the same expression profiles (see Supplemental Figure 14 online). This suggested that tRNA availability may contribute to the regulation of gene expression in grapevine leading to the hypothesis that transcriptomic differences between vegetative/green and mature/woody clusters may be associated with differences in tRNA availability. Indeed, we found that genes belonging to the green/vegetative and mature/woody clusters (Figure 3E) have significantly (P < 0.0001) different codon usage preferences (see Supplemental Table 4 online).
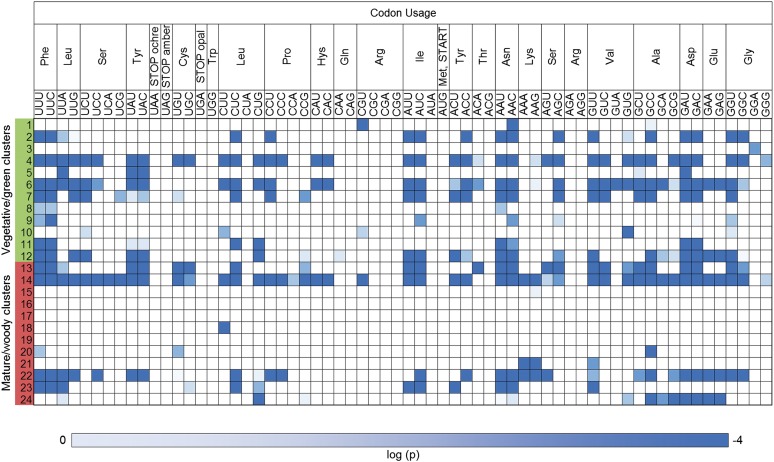
To gain more insight into the expression profiles that contribute most to the codon usage diversity, we grouped vegetative/green and mature/woody samples into 12 clusters and tested coclustering genes for their codon usage preferences. Genes expressed in either the vegetative/green or mature/woody samples were compositionally diverse, confirming an association between transcriptional and compositional clustering (Figure 8; see Supplemental Figure 15 online). This means that grapevine genes defining vegetative/green and mature/woody samples not only have distinct expression profiles but also different codon usage preferences and implies that a typical green/vegetative gene is disadvantaged if expressed in mature/woody samples and vice versa. Clusters with the most significant preferential codon usage often represented specific developmental phases in certain samples (e.g., rachis and tendril, cluster 4; berry withering, clusters 13 and 22; and seeds, cluster 24).
Figure 8.
Mutual Information of Synonymous Codon Usage in Grapevine Gene Coexpression Clusters.
Each row represents a coexpression cluster, whereas each column represents a synonymous codon. Significant mutual information is shown in blue (P ≤ 10−4).
Summary and Conclusions
We constructed a genome-wide transcriptomic atlas of a woody fruit crop, using grapevine as a model because it is the most widely cultivated fruit crop in the world. We analyzed gene expression profiles in 54 diverse organ/tissue samples using a comprehensive grapevine genome microarray and detected the expression of ∼91% of the predicted genes from the latest 12x grapevine genome annotation in at least one sample. The remaining genes are probably expressed uniquely under conditions that were not evaluated in our survey (i.e., different forms of biotic and abiotic stress) (see Supplemental Data Set 1 and Supplemental Figure 16 online). Microarray analysis revealed that samples with unique characteristics (such as pollen grains and leaves undergoing senescence) were clearly distinguishable at the transcriptomic level from all other samples, which grouped more according to their maturity and developmental stage than their organ or tissue identity, as also supported by the in silico analysis of RNA-seq data. Previous studies have focused mostly on berry development and ripening (Zamboni et al., 2010; Zenoni et al., 2010; Tornielli et al., 2012), but our transcriptomic atlas presents a comprehensive grapevine transcriptome.
The fundamental reprogramming of the transcriptome during maturation was highlighted by all three statistical approaches we used to mine our microarray data. These different methods also allowed us to identify the transcriptional relationships among samples (Pearson’s correlation distance approach), putative biomarkers (O2PLS-DA approach), and sets of strongly and consistently expressed genes that define groups (topics) of similar samples (biclustering analysis based on a topic model approach).
Coexpression analysis provided further insight into the dynamic reprogramming of the transcriptome during maturation by revealing specific characteristics that defined vegetative/green and mature/woody samples. The shift to the mature/woody developmental program results from the reiterative coactivation of particular pathways that are inactive or minimally active in vegetative/green samples, whereas some pathways that are active in vegetative/green samples remain at least partially active after maturation. In many cases, the coexpression of genes and, indeed, pathways involved in the maturation process involved the coregulation of neighboring genes in clusters as well as global regulation based on codon usage preference. This peculiar behavior of the grapevine transcriptome might be shared with other perennial woody plants, but it has not been reported previously in the transcriptomes of herbaceous annual species.
The grapevine genome sequence revealed several examples of expanding gene families (Jaillon et al., 2007; Velasco et al., 2007; Matus et al., 2008), and some of which may have an impact on ripe berry quality and the organoleptic properties of wine. Our gene expression atlas provides further insight into the molecular mechanisms underlying berry development, particularly the biclustering topic model analysis that identified both structural and regulatory genes that are potentially the key players defining groups of organs with similar developmental and metabolic features. Many genes that define the ripe berry topic currently have no known function and therefore are important targets for functional annotation to increase our knowledge of the processes that control berry ripening.
Combined with the complete grapevine genome sequence, our comprehensive transcriptome atlas elevates grapevine to the status of a model fruit tree species, facilitating large-scale investigations of gene function in the future. Our gene expression survey could be used to infer the specific metabolic processes and cellular structures within each of the samples, as recently reported in tomato (Matas et al., 2011). The transcriptome atlas will also support vineyard management by providing the means to pinpoint molecular changes that affect yield, quality, environmental responses, and molecular factors that underlie the phenotypic plasticity of different grapevine varieties during cultivation.
METHODS
Vineyard Features
Grapevine (Vitis vinifera cv Corvina, clone 48) samples were collected from a 7-year-old vineyard (45° 27′ 17′′ N, 11° 03′ 14′′ E, Montorio, Verona Province, Italy) during the 2008/2009 growing seasons at the same time of day (∼9:30 am). The vineyard was 130 m above sea level, and the soil comprised 36% sand, 36% clay, and 28% silt. The replacement cane Guyot rows were north–south oriented, and 41B was used as the rootstock.
Sample Collection
We collected 54 grapevine samples (bud, inflorescence, tendril, leaf, stem, root, developing berry, withering berry, seed, rachis, anther, carpel, petal, pollen, and seedling) covering most organs at several developmental stages (see Supplemental Table 1 online). Three biological replicates were taken for each sample. Buds were collected at five developmental stages, the first corresponding to the first-season latent bud (E-L 23), the second representing the winter dormant bud (E-L 1), the third corresponding to the bud-swelling stage (E-L 2), the fourth representing the initial bud burst, showing a green tip (E-L 4), and the last representing bud burst, when a rosette of leaf tips is visible (E-L 5). Inflorescences were collected at two developmental stages, the first representing the young inflorescence with single flowers in compact groups (E-L 14) and the second representing a well-developed inflorescence with separated flowers (E-L 17). Flowers were collected at the beginning of flowering (10% of caps off; E-L 20) and at the 50% caps off stage, which is considered the flowering phase (E-L 23). Floral organs were collected from undisclosed flowers collected at two time points corresponding to E-L 20 (10% caps off, 16 to 18 leaves) and E-L 23 (50% caps off, 17 to 20 leaves). A pool of these two developmental stages was created for each sample of petal, anther, and carpel. Pollen was collected from opened flowers (>50% caps off, E-L 25). Tendrils are slender structures with the same developmental origin as the inflorescence. They grow opposite the leaf at each node, except for the first two to three supporting leaves at the base of the shoot. Tendrils were collected at three developmental stages, the first corresponding to a pool of tendrils collected when the shoot bears seven separated leaves (E-L 14), the second corresponding to a pool of well-developed tendrils collected when the shoot bears 12 separated leaves (E-L 17), and the last corresponding to a pool of mature-coiled tendrils collected at fruit set (berry diameter ∼4 mm; E-L 29). Leaves were collected at three developmental stages, the first representing a pool of young light-green leaves starting from the second from the tip, when the shoot bears approximately five well-separated leaves (E-L 14), the second corresponding to mature leaves collected when the berry size was ∼4 mm diameter (E-L 29), and the third representing leaves undergoing senescence collected before the beginning of leaf fall (E-L 43). Berries (pericarp) were sampled at five developmental time points by freezing whole berries and removing the seeds. The first stage (15 d after flowering [DAF]; E-L 29) corresponds to the fruit set, when young berries are enlarging (>3 mm diameter); the second stage (35 DAF; E-L 32) is the PFS, when berries (>7 mm diameter) start touching; the third stage (70 DAF; E-L 35) is the veraison, when berries begin to change color and enlarge (10.4° Brix); the fourth stage (84 DAF; E-L 36) corresponds to the mid-ripening stage (15.5° Brix); and the final stage (115 DAF; E-L 38) represents complete ripening (20.0° Brix). The sugar content (mean Brix degree value) was recorded at each time point using a PR-32 bench refractometer (Atago Co.). Starting from the PFS stage, berries were further dissected into skin and flesh tissues. After harvest, clusters were placed for three months in single layers in a naturally ventilated room with no automated temperature or humidity control. Withered berries were sampled each month, and weight percentages of the withering samples were compared with the weight of the ripening berries (E-L 38). The sugar content was recorded as above. At the first withering stage (WI), berry weight was 76.4% the ripe value and the sugar content was 24.5° Brix. The second stage (WII) was characterized by 69.7% berry weight and 25.9° Brix, and the last stage (WIII) was characterized by 67.3% berry weight and 26.7° Brix. At each time point, berries were further dissected into skin and flesh tissues. Seeds were collected at the first four stages of berry development, corresponding to E-L 29, E-L 32, E-L 35, and E-L 36. The rachis is the main inflorescence axis of the grape berry cluster, and rachis samples were collected along with the berry samples. Stems were collected at two developmental stages, the first representing a pool of stems collected starting from the second node from the tip (E-L 14) and the second representing a pool of woody stems (cane) collected at E-L 43. Corvina roots were collected from in vitro cuttings. The growth medium (HB) was prepared as described by Blaich (1977). Developing young roots were pooled to create three biological replicates. Ripened seeds were stored a 4°C for at least 2 weeks and then planted in soil under normal greenhouse conditions. Seedlings were collected after 2 months to create three pools at three different developmental stages. Cotyledons were still closed in the first stage, just opened in the second stage, and wide open at the third stage.
RNA Extraction
For most samples, ∼100 mg of tissue was ground under liquid nitrogen, and total RNA was extracted using the Spectrum Plant Total RNA kit (Sigma-Aldrich) following the manufacturer’s protocol. For berry flesh, senescing leaves, and woody stems, ∼400 mg of ground material was required, and for berry pericarp and skin, seed, rachis, root, and latent and winter buds, ∼200 mg of ground material was required. We precipitated the total RNA from winter buds, seeds, woody stems, and rachis at veraison and mid-ripening with LiCl to remove contaminants that absorbed at 230 nm. LiCl was mixed with total RNA to a final concentration of 2.5 M, incubated overnight at 4°C, and centrifuged at 13,000g, and the pellet was washed with 70% ethanol before resuspending in water. RNA quality and quantity were determined using a Nanodrop 2000 spectrophotometer (Thermo Scientific) and a Bioanalyzer Chip RNA 7500 series II (Agilent).
Microarray Hybridization and Data Extraction
We performed cDNA synthesis, labeling, hybridization, and washing steps according to the NimbleGen Arrays User’s Guide (version 3.2). Each sample was hybridized to a NimbleGen microarray 090818 Vitis exp HX12 (Roche, NimbleGen), which contains probes targeted to 29,549 predicted grapevine genes (http://ddlab.sci.univr.it/FunctionalGenomics/), representing ∼98.6% of the genes predicted from the V1 annotation of the 12x grapevine genome (http://srs.ebi.ac.uk/) and 19,091 random probes as negative controls. Each microarray was scanned using an Axon GenePix 4400A (Molecular Devices) at 532 nm (Cy3 absorption peak) and GenePix Pro7 software (Molecular Devices) according to the manufacturer’s instructions. Images were analyzed using NimbleScan v2.5 software (Roche), which produces pair files containing the raw signal intensity data for each probe and calls files with normalized expression data derived from the average of the intensities of the four probes for each gene. All microarray expression data are available in the Gene Expression Omnibus under the series entry GSE36128 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=lfcrxesyciqgsjoandacc=GSE36128).
Statistical Evaluation of Gene Expression and Tissue Specificity
To find the threshold expression level, which defines a gene as “expressed” or “nonexpressed,” we computed the log2 data and estimated the control group probability density for the 19,091 random probes in each experiment using a normal kernel smoothing method with the threshold P = 0.05 and averaged the biological replicates only if at least two (out of three) expression values exceeded the threshold. For each group of samples (see Supplemental Table 2 online), we defined a transcript as tissue specific if its mean expression value exceeded the threshold in at least one sample form a given organ.
Functional Category Distribution and GO Enrichment Analysis
All transcripts were annotated against the V1 version of the 12x draft annotation of the grapevine genome (http://genomes.cribi.unipd.it/DATA/), allowing 70% of the genes to be identified. This was verified manually and integrated using gene ontology (GO) classifications. Transcripts were then grouped into the 16 most represented functional categories (GO:0008150, other processes; GO:0051090, transcription factor activity; GO:0009725, response to hormone stimulus; GO:0019725, cellular homeostasis; GO:0007165, signal transduction; GO:0006950, response to stress; GO:0032502, developmental process; GO:0006810, transport; GO:0015979, photosynthesis; GO:0006091, generation of energy; GO:0090304, DNA/RNA metabolic process; GO:0044036, cell wall metabolism; GO:0019748, secondary metabolic process; GO:0006629, lipid metabolic process; GO:0006520, cellular amino acids and derivative metabolic process; GO:0005975, carbohydrate metabolic process), based on GO biological processes. Genes with unknown functions or with a “no hit” annotation were also included. The distribution of functional categories is represented in a histogram showing the percentage of the genes in each topic (Figure 6B).
GO annotation analysis was applied to gene clusters and organ-specific genes using the BiNGO 2.3 plug-in tool in Cytoscape version 2.6 with PlantGOslim categories, as described by Maere et al. (2005). Overrepresented PlantGOslim categories were identified using a hypergeometric test with a significance threshold of 0.05 for gene clusters and of 0.5 for organ-specific genes, after Benjamini and Hochberg false discovery rate correction (Klipper-Aurbach et al., 1995).
Estimation of Bimodal Distribution
For each sample experiment, we first averaged the replicate genome-wide data and estimated the probability distribution using a normal kernel smoothing method. We then calculated the mean and sd by optimally fitting the data to a unimodal normal distribution. Finally, we computed the mean square of the difference between the estimated distribution and the normal unimodal distribution with the estimated mean and variance. The mean square is a measure of the “distance” of data from a unimodal normal distribution. We then ordered these error data according to the number of expressed genes in each organ and found a positive trend (see Supplemental Figure 1 online).
Correlation Analysis
A correlation matrix was prepared using R software and Pearson’s correlation coefficient as the statistical metric to compare the values of the whole transcriptome (29,549 genes) in all 54 samples, using the average expression value of the three biological replicates. Correlation values were converted into distance coefficients to define the height scale of the dendrogram. FPKM (fragments per kilobase per million of reads mapped) values were used to create the correlation matrix and the cluster dendrogram from the RNA-seq data set. MATLAB scripts were used to analyze the correlation among samples at different statistical metrics (euclidean, spearman rank, cityblock, and cosine) and at three expression levels (top 20%, between 20% and 80%, and bottom 20%).
Remapping Reads on the 12x Grapevine Genome Prediction
Illumina sequences derived from poly(A+) RNA isolated from four Pinot noir tissues (in vitro–cultivated juvenile leaf, juvenile stem, juvenile root, and embryonic callus) and three developmental stages of Corvina berry pericarp (PFS, veraison, and ripening) were previously generated using the Solexa/Illumina technology (Denoeud et al., 2008) and Illumina genome analyzer II (Zenoni et al., 2010) platforms, respectively. Sequence alignments were generated with TopHat version 1.0.14 (Trapnell et al., 2009) (see Supplemental Data Set 2 online). The V. vinifera RefSeq sequences were based on the 12-fold PN40024 genome newer Version 1 (http://srs.ebi.ac.uk/). Gene expression was evaluated using Cufflinks software (version 0.9.2; http://cufflinks.cbcb.umd.edu/) (Trapnell et al., 2010). Briefly, Cufflinks uses the alignment information at each gene locus to assign multimapping reads to a specific locus using a maximum likelihood estimation. On the basis of the relative abundance of fragments (defined as a single read in single-end experiments or as two reads from the same cDNA in paired-end experiments), the software is able to compute the normalized expression measure as FPKM. The number of reads falling in a given gene locus can be estimated from the FPKM value as follows: n = FPKM × L × NTot × 10−9, where n = number of mapping reads at a given gene locus, L = estimated length (bp) of the gene locus, NTot = number of total mapping reads, and FPKM = gene locus FPKM value.
PCA, O2PLS, and Putative Marker Genes
PCA was performed using SIMCA P+ (Umetrics). O2PLS-DA was used to integrate the PCA data and reduce experimental variability. The latent structures of the joint X-Y correlated variation were used to identify small groups of correlated variables belonging to the two different blocks by evaluating the similarity between each variable and the predictive latent components of the X-Y O2PLS model by means of their correlation. The significance threshold for the similarity was set using a permutation test, and data integration was performed on each small group of X-Y variables with significant correlation. O2PLS-DA allowed the identification of latent variables yielding a parsimonious and efficient representation of the process. To define the number of latent components for our O2PLS-DA models, we applied partial cross-validation and a permutation test to reveal overfitting. Multivariate data analysis was performed using SIMCA P+ (Umetrics). Putative biomarker transcripts were selected from the class-specific S-plots within the first (positive biomarkers) and the last (negative biomarkers) percentile (Wiklund et al., 2008). Gene expression values from the 52-sample data set of each group were log2 transformed and normalized. Expression profiles were plotted in two different graphs describing the peculiar trends of positive and negative biomarker genes (R software).
Hierarchical Clustering
Cluster analysis was performed by the k-means method with Pearson’s correlation distance (TMeV 4.3; http://www.tm4.org/mev) on the 54-sample data set. HCL was performed on each cluster to represent gene relationships in dendrograms (TMeV), with Pearson’s correlation distance as the metric. An entire HCL representation was created by joining the four groups. Supplemental Data Set 1 online provides information about the membership of different clusters.
Biclustering Analysis with the PLSA Algorithm
Biclustering analysis aims to discover groups of genes sharing compatible expression patterns across subsets of samples (Madeira and Oliveira, 2004; Prelić et al., 2006). We used a technique (Joung et al., 2006; Bicego et al., 2010) that employs PLSA, which allows data sets to be modeled in terms of hidden topics or processes that can reflect underlying meaningful structures. The basic idea in the gene expression scenario is that a topic may be roughly intended as a biological process, which can characterize a subset of samples (namely, the samples where the process is active). At the same time, a topic may induce the activation of a particular set of genes (namely, the genes related to the particular process). Following this reasoning, it can be said that the relation between gene expression and samples is mediated through the probabilistic presence of the topics (Joung et al., 2006; Bicego et al., 2010). Given the expression matrix, the relation topics/samples and genes/topics were learned using the expectation maximization algorithm (Hofmann, 2001). To avoid local minima, we performed 20 different training scenarios starting from different random initializations and retained the best model. The number of topics (representing the free parameter of the model) was set using the classic Bayesian information criterion, a penalized likelihood criterion (Schwarz, 1978), and training the model with two to 30 topics (see Supplemental Figure 8 online). The first type of information (relation topic/samples) is completely encoded in the probability distribution p(z|d), representing the probability of finding the topic “z” in the sample “d.” The second type of information (relation topic/genes) was inferred by selecting the 500 highest entries of the p(w|z) matrix, which describes the probability of the gene “w” given the topic “z,” namely, the level of presence of such gene in such topic. Subsequently, for every topic, the selected genes were grouped by their functional category.
Coexpression Analysis
Coexpression analysis of the whole data set was performed as sanity test to score the quality of the expression data with a small number of selected genes as queries, using the Pearson correlation distance (CorTo; http://www.usadellab.org/cms/index.php?page=corto).
Pairwise Gene Correlation Analysis
We averaged replicate genome-wide data and computed the Pearson correlation for each gene pair of a specific group of genes, using data relative to a specific group of samples. This was achieved by computing four pairwise gene correlation analyses: mature/woody cluster genes over mature/woody samples, mature/woody cluster genes over vegetative/green samples, vegetative/green cluster genes over vegetative/green samples, and vegetative/green cluster genes over mature/woody samples.
Sliding-Window Analysis of Chromosomal Coexpression
As previously described (Williams and Bowles, 2004), we averaged replicate genome-wide data and computed the mean Pearson's correlation coefficient (R) of all possible pairs of neighboring genes for each group over a sliding window of size 10 to give a measure of similarity in expression profiles. We therefore assessed 45 different correlations, and the mean R was used as a measure of the level of coexpression for each particular block. These mean R values may be interpreted as the degree of coexpression for each chromosomal region of 10 genes. Neighboring genes were defined as genes that were immediately adjacent in the grapevine genome. The mean R calculated from the real data set was then compared with the mean R calculated from 10,000 data sets, in which the order of both the genes and experiments were randomized. The distance between genes was defined as the distance in base pairs on either strand between the last coding position of the first gene and the first coding position of the second. In the case of gene families, the specificity of the probe set for each single gene was assessed to exclude the possibility of cross-hybridization signals and misleading coexpression results.
Codon Usage Preference Analysis
MI between codon usage and expression profile was calculated by comparing variable γ (i.e., the normalized genic frequency of each codon) and cluster α (a list of genes assigned to a given cluster) to determine any nonrandom distribution (Elemento et al., 2007; Najafabadi et al., 2009). The number of bins was set to five and gene cluster assignments were shuffled 104 times for the assessment of MI significance. The normalized frequency of a synonymous codon in a given gene was calculated as the usage of that codon divided by the usage of the corresponding amino acid in the same gene product. This statistic was calculated only when the corresponding amino acid was present more than five times the degeneracy of the encoded amino acid. Gene clusters were defined by the k-means method with Pearson's correlation distance (TMeV 4.3; http://www.tm4.org/mev). The MI-RSCU package of the ICodPack suite was used to calculate the mutual information of each codon. More information can be found in Supplemental Methods 1 online. The codon usage diversity between genes belonging to the green/vegetative and mature/woody was calculated using the PIRSCU script (Najafabadi et al., 2009). In brief, the normalized frequency of each codon in each gene was calculated as the usage of that codon divided by the usage of the amino acid it codes for. The distance (d) of a pair of genes was calculated as the absolute value of the difference between the normalized codon usage frequencies in the two genes. The distance of all gene pairs was calculated and gene pairs were sorted according to their d values. Then, the sorted gene pairs were divided into 50 several equally populated bins and for each bin the likelihood of being in the same cluster was calculated as by Najafabadi et al. (2009). Pearson correlation coefficient between minimum d value for each bin and the L values associated with that bin were calculated. The significance of Pearson correlation coefficient was estimated by randomly shuffling gene cluster assignments 104 times, each time repeating the calculations and comparing with the original correlation coefficient.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Plot Showing the Relationship between the Number of Expressed Genes in Each Organ and the Bimodal Character of the Expression Distribution.
Supplemental Figure 2. Distribution of Genes Expressed among Different Grapevine Tissues.
Supplemental Figure 3. Venn Diagrams Showing the Distribution of Organ-Specific Genes among Seeds, Berries, and Rachis Developmental Stages.
Supplemental Figure 4. Coexpression Analysis on Selected Genes Belonging to Flavonoid Pathway and Photosynthesis.
Supplemental Figure 5. Cluster Dendrograms Obtained with Different Distance Metrics Converted into Distance Coefficients, with Different Gene Expression Level Subsets and on a Reduced-Sample Data Set.
Supplemental Figure 6. Enriched GO Terms for Genes in the Four Clusters Defined by HCL Analysis.
Supplemental Figure 7. PCA Analysis Using Simca-P+ 12.0 (Umetrics).
Supplemental Figure 8. Plot Representing the Penalized Likelihood Approach (Bayesian Information Criterion).
Supplemental Figure 9. Coexpression Distribution Profiles.
Supplemental Figure 10. Networks of the 1000 Most Correlated Gene Pairs.
Supplemental Figure 11. Sliding-Window Analysis of Coexpression along Grapevine Chromosomes.
Supplemental Figure 12. Gene Cluster Correlation Matrices.
Supplemental Figure 13. Sliding-Window Analysis of Coexpression along Chromosomes: Vegetative and Nonvegetative Differences/Loadings.
Supplemental Figure 14. The Significance of Correlation between Codon Usage and 60 Clusters of Genes According to Expression Profile.
Supplemental Figure 15. Heat Map Summarizing the Expression Profiles of 24 Gene Clusters (12 Representing Vegetative/Green and 12 Representing Mature/Woody Genes) Analyzed for Preferential Codon Usage.
Supplemental Figure 16. Enriched GO Terms in Nonexpressed Genes.
Supplemental Table 1. Description of Samples of Vitis vinifera cv Corvina Used for Microarray Analysis.
Supplemental Table 2. Description of Groups of Organs Considered in Organ-Specific Gene Expression Analysis.
Supplemental Table 3. Summary of Reads Number in RNA-seq Experiments.
Supplemental Table 4. Preference in Codon Usage of Vegetative/Green and Mature/Woody Genes.
Supplemental Data Set 1. Transcriptome Description in Grapevine Atlas.
Supplemental Data Set 2. Organ-Specific Transcripts.
Supplemental Data Set 3. Molecular Biomarkers.
Supplemental Data Set 4. PLSA Topic-Specific Transcripts.
Supplemental Data Set 5. List of the 1000 Most Correlated Gene Pairs in Vegetative/Green and Mature/Woody Samples.
Supplemental Data Set 6. List of Coexpressed Genes along Grapevine Chromosomes in All Samples.
Supplemental Data Set 7. List of Coexpressed Genes along Grapevine Chromosomes in Mature/Woody Samples.
Supplemental Data Set 8. List of Coexpressed Genes along Grapevine Chromosomes in Vegetative/Green Samples.
Acknowledgments
We thank Giorgio Pasqua for providing plant material and for his support with grapevine genomics. We thank Pietro Lovato for his support in performing PLSA analysis. This work was supported by the Completamento del Centro di Genomica Funzionale Vegetale project funded by the CARIVERONA Bank Foundation, by the Valorizzazione dei Principali Vitigni Autoctoni Italiani e dei loro Terroir (Vigneto) project funded by the Italian Ministry of Agricultural and Forestry Policies, by Valorizzazione della tipicità dei vitigni autoctoni e dei vini veneti (Valvive) funded by Regione Veneto, and by Joint Project 2011 between Pasqua Vigneti e Cantine SpA and the Biotechnology Department of University of Verona.
AUTHOR CONTRIBUTIONS
M.P. designed the experiment and supervised the project. M.F. performed RNA extraction, hybridization, and microarray data analysis. M.F., S.D.S., S.Z., G.B.T., and M.P. interpreted the data and wrote the article. S.Z., A.Z., G.B.T., and M.F. performed plant material collection. L.F., A.P., M.B., S.D.S., G.B.T., V.M., and A.Z. developed and/or applied statistical tools. L.V. performed RNA-seq data analysis. A.F. and M.D. optimized the microarray platform.
Glossary
- HCL
hierarchical clustering
- PCA
principal component analysis
- O2PLS-DA
orthogonal bidirectional projections to latent structures discriminant analysis
- PLSA
probabilistic latent semantic analysis
- PFS
postfruit set
- MI
mutual information
- DAF
days after flowering
- GO
gene ontology
- FPKM
fragments per kilobase per million of reads mapped
References
- Aoki K., et al. (2010). Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics 11: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39: 366–380 [DOI] [PubMed] [Google Scholar]
- Bessire M., Borel S., Fabre G., Carraça L., Efremova N., Yephremov A., Cao Y., Jetter R., Jacquat A.C., Métraux J.P., Nawrath C. (2011). A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 23: 1958–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicego M., Lovato P., Ferrarini A., Delledonne M. (2010). Biclustering of expression microarray data with topic models. In International Conference on Pattern Recognition 2010, pp. 2728–2731
- Blaich R. (1977). Attempts at artificial mycorrhiza formation in Vitis riparia. Vitis 16: 32–37 [Google Scholar]
- Bohlmann J., Meyer-Gauen G., Croteau R. (1998). Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 95: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M., Hepler P.K. (2005). Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 17: 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona M.J., Calonje M., Martínez-Zapater J.M. (2007). The FT/TFL1 gene family in grapevine. Plant Mol. Biol. 63: 637–650 [DOI] [PubMed] [Google Scholar]
- Davies C., Robinson S.P. (2000). Differential screening indicates a dramatic change in mRNA profiles during grape berry ripening. Cloning and characterization of cDNAs encoding putative cell wall and stress response proteins. Plant Physiol. 122: 803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Freitas C.D., Lopes J.L., Beltramini L.M., de Oliveira R.S., Oliveira J.T., Ramos M.V. (2011a). Osmotin from Calotropis procera latex: New insights into structure and antifungal properties. Biochim. Biophys. Acta 1808: 2501–2507 [DOI] [PubMed] [Google Scholar]
- de Freitas C.D., Nogueira F.C., Vasconcelos I.M., Oliveira J.T., Domont G.B., Ramos M.V. (2011b). Osmotin purified from the latex of Calotropis procera: Biochemical characterization, biological activity and role in plant defense. Plant Physiol. Biochem. 49: 738–743 [DOI] [PubMed] [Google Scholar]
- Denoeud F., Aury J.M., Da Silva C., Noel B., Rogier O., Delledonne M., Morgante M., Valle G., Wincker P., Scarpelli C., Jaillon O., Artiguenave F. (2008). Annotating genomes with massive-scale RNA sequencing. Genome Biol. 9: R175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka A., et al. (2006). An atlas of gene expression from seed to seed through barley development. Funct. Integr. Genomics 6: 202–211 [DOI] [PubMed] [Google Scholar]
- Edwards K.D., Bombarely A., Story G.W., Allen F., Mueller L.A., Coates S.A., Jones L. (2010). TobEA: An atlas of tobacco gene expression from seed to senescence. BMC Genomics 11: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O., Slonim N., Tavazoie S. (2007). A universal framework for regulatory element discovery across all genomes and data types. Mol. Cell 28: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelli F., Battilana J., Costantini L., Le Cunff L., Boursiquot J.M., This P., Grando M.S. (2010). A candidate gene association study on muscat flavor in grapevine (Vitis vinifera L.). BMC Plant Biol. 10: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone Ferreyra M.L., Pezza A., Biarc J., Burlingame A.L., Casati P. (2010). Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol. 153: 1878–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannuzzi G., D’Addabbo P., Gasparro M., Martinelli M., Carelli F.N., Antonacci D., Ventura M. (2011). Analysis of high-identity segmental duplications in the grapevine genome. BMC Genomics 12: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanana M., Deluc L., Fouquet R., Daldoul S., Léon C., Barrieu F., Ghorbel A., Mliki A., Hamdi S. (2008). [Identification and characterization of “rd22” dehydration responsive gene in grapevine (Vitis vinifera L.)]. C. R. Biol. 331: 569–578 [DOI] [PubMed] [Google Scholar]
- Himmelbach A., Liu L., Zierold U., Altschmied L., Maucher H., Beier F., Müller D., Hensel G., Heise A., Schützendübel A., Kumlehn J., Schweizer P. (2010). Promoters of the barley germin-like GER4 gene cluster enable strong transgene expression in response to pathogen attack. Plant Cell 22: 937–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann T. (2001). Unsupervised learning by probabilistic latent semantic analysis. Mach. Learn. 42: 177–196 [Google Scholar]
- Jackson R.S. (2000). Wine Science: Principles, Practice, Perception, 2nd ed. (San Diego, CA: Academic Press) [Google Scholar]
- Jaillon O., et al. French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jiao Y., et al. (2009). A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat. Genet. 41: 258–263 [DOI] [PubMed] [Google Scholar]
- Joung J.G., Shin D., Seong R.H., Zhang B.T. (2006). Identification of regulatory modules by co-clustering latent variable models: Stem cell differentiation. Bioinformatics 22: 2005–2011 [DOI] [PubMed] [Google Scholar]
- Judd W.S. (1999). Plant Systematics: A Phylogenetic Approach. (Sunderland, MA: Sinauer Associates) [Google Scholar]
- Jung C., Seo J.S., Han S.W., Koo Y.J., Kim C.H., Song S.I., Nahm B.H., Choi Y.D., Cheong J.J. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K.H., Han M.J., Lee D.Y., Lee Y.S., Schreiber L., Franke R., Faust A., Yephremov A., Saedler H., Kim Y.W., Hwang I., An G. (2006). Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18: 3015–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipper-Aurbach Y., Wasserman M., Braunspiegel-Weintrob N., Borstein D., Peleg S., Assa S., Karp M., Benjamini Y., Hochberg Y., Laron Z. (1995). Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med. Hypotheses 45: 486–490 [DOI] [PubMed] [Google Scholar]
- Kong Y., Zhou G., Yin Y., Xu Y., Pattathil S., Hahn M.G. (2011). Molecular analysis of a family of Arabidopsis genes related to galacturonosyltransferases. Plant Physiol. 155: 1791–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang X., Stolc V., Li X., Zhang D., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Deng X.W. (2006). Genome-wide transcription analyses in rice using tiling microarrays. Nat. Genet. 38: 124–129 [DOI] [PubMed] [Google Scholar]
- Madeira S.C., Oliveira A.L. (2004). Biclustering algorithms for biological data analysis: A survey. IEEE/ACM Trans. Comput. Biol. Bioinform. 1: 24–45
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Martin D.M., Aubourg S., Schouwey M.B., Daviet L., Schalk M., Toub O., Lund S.T., Bohlmann J. (2010). Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 10: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa A.N., Childs K.L., Lin H., Bryan G.J., Giuliano G., Buell C.R. (2011). The transcriptome of the reference potato genome Solanum tuberosum Group Phureja clone DM1-3 516R44. PLoS ONE 6: e26801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matas A.J., et al. (2011). Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 23: 3893–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus J.T., Aquea F., Arce-Johnson P. (2008). Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins M.G., Bouquet A., Williams L.E. (1992). Biology of the Grapevine. (Cambridge, UK: Cambridge University Press) [Google Scholar]
- Najafabadi H.S., Goodarzi H., Salavati R. (2009). Universal function-specificity of codon usage. Nucleic Acids Res. 37: 7014–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Mecey C., Lee Y.N., Imboden L.A., Chang J.H., He S.Y. (2011). Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 10774–10779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvera-Carrillo Y., Campos F., Reyes J.L., Garciarrubio A., Covarrubias A.A. (2010). Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 154: 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Chakraborty S., Datta A., Chakraborty N. (2008). Proteomics approach to identify dehydration responsive nuclear proteins from chickpea (Cicer arietinum L.). Mol. Cell. Proteomics 7: 88–107 [DOI] [PubMed] [Google Scholar]
- Prasad K., Zhang X., Tobón E., Ambrose B.A. (2010). The Arabidopsis B-sister MADS-box protein, GORDITA, represses fruit growth and contributes to integument development. Plant J. 62: 203–214 [DOI] [PubMed] [Google Scholar]
- Prelić A., Bleuler S., Zimmermann P., Wille A., Bühlmann P., Gruissem W., Hennig L., Thiele L., Zitzler E. (2006). A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics 22: 1122–1129 [DOI] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubelakis-Angelakis K.A. (2009). Grapevine Molecular Physiology and Biotechnology. (Dordrecht, The Netherlands: Springer Science+Business Media B.V.) [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schwarz G. (1978). Estimating the dimension of a model. Ann. Stat. 6: 461–464 [Google Scholar]
- Sekhon R.S., Lin H., Childs K.L., Hansey C.N., Buell C.R., de Leon N., Kaeppler S.M. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Severin A.J., et al. (2010). RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatil-Cohen A., Attia Z., Moshelion M. (2011). Bundle-sheath cell regulation of xylem-mesophyll water transport via aquaporins under drought stress: a target of xylem-borne ABA? Plant J. 67: 72–80 [DOI] [PubMed] [Google Scholar]
- Tornielli G.B., Zamboni A., Zenoni S., Delledonne M., Pezzotti M. (2012). Transcriptomics and metabolomics for the analysis of grape berry development. In The Biochemestry of Grape Berry, H. Geros, M.M. Chavez, and S. Delrot, eds (Sharjan, United Arab Emirates: Bentham Science Publishers), pp. 218–240 [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trygg J. (2002). O2-PLS for qualitative and quantitative analysis in multivariate calibration. J. Chemometr. 16: 283–293 [Google Scholar]
- Updegraff E.P., Zhao F., Preuss D. (2009). The extracellular lipase EXL4 is required for efficient hydration of Arabidopsis pollen. Sex. Plant Reprod. 22: 197–204 [DOI] [PubMed] [Google Scholar]
- van Doorn W.G., Stead A.D. (1997). Abscission of flowers and floral parts. J. Exp. Bot. 48: 821–837 [Google Scholar]
- Velasco R., et al. (2007). A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2: e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versari A., Parpinello G.P., Tornielli G.B., Ferrarini R., Giulivo C. (2001). Stilbene compounds and stilbene synthase expression during ripening, wilting, and UV treatment in grape cv. Corvina. J. Agric. Food Chem. 49: 5531–5536 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Tobin E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Weber C.C., Hurst L.D. (2011). Support for multiple classes of local expression clusters in Drosophila melanogaster, but no evidence for gene order conservation. Genome Biol. 12: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklund S., Johansson E., Sjöström L., Mellerowicz E.J., Edlund U., Shockcor J.P., Gottfries J., Moritz T., Trygg J. (2008). Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 80: 115–122 [DOI] [PubMed] [Google Scholar]
- Williams E.J., Bowles D.J. (2004). Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 14: 1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y.Y., Matsui M., Ang L.H., Deng X.W. (1998). Role of a COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell 10: 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni A., et al. (2010). Identification of putative stage-specific grapevine berry biomarkers and omics data integration into networks. Plant Physiol. 154: 1439–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni A., Minoia L., Ferrarini A., Tornielli G.B., Zago E., Delledonne M., Pezzotti M. (2008). Molecular analysis of post-harvest withering in grape by AFLP transcriptional profiling. J. Exp. Bot. 59: 4145–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S., Ferrarini A., Giacomelli E., Xumerle L., Fasoli M., Malerba G., Bellin D., Pezzotti M., Delledonne M. (2010). Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol. 152: 1787–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]