The combination of metabolomics and transcriptomics on Arabidopsis thaliana lines mutated in 10 steps of the lignin pathway provides insight into monolignol biosynthesis and the metabolic network in which it is embedded. In addition, this work reveals novel pathways and genes associated with lignin biosynthesis.
Abstract
Lignin engineering is an attractive strategy to improve lignocellulosic biomass quality for processing to biofuels and other bio-based products. However, lignin engineering also results in profound metabolic consequences in the plant. We used a systems biology approach to study the plant’s response to lignin perturbations. To this end, inflorescence stems of 20 Arabidopsis thaliana mutants, each mutated in a single gene of the lignin biosynthetic pathway (phenylalanine ammonia-lyase1 [PAL1], PAL2, cinnamate 4-hydroxylase [C4H], 4-coumarate:CoA ligase1 [4CL1], 4CL2, caffeoyl-CoA O-methyltransferase1 [CCoAOMT1], cinnamoyl-CoA reductase1 [CCR1], ferulate 5-hydroxylase [F5H1], caffeic acid O-methyltransferase [COMT], and cinnamyl alcohol dehydrogenase6 [CAD6], two mutant alleles each), were analyzed by transcriptomics and metabolomics. A total of 566 compounds were detected, of which 187 could be tentatively identified based on mass spectrometry fragmentation and many were new for Arabidopsis. Up to 675 genes were differentially expressed in mutants that did not have any obvious visible phenotypes. Comparing the responses of all mutants indicated that c4h, 4cl1, ccoaomt1, and ccr1, mutants that produced less lignin, upregulated the shikimate, methyl-donor, and phenylpropanoid pathways (i.e., the pathways supplying the monolignols). By contrast, f5h1 and comt, mutants that provoked lignin compositional shifts, downregulated the very same pathways. Reductions in the flux to lignin were associated with the accumulation of various classes of 4-O- and 9-O-hexosylated phenylpropanoids. By combining metabolomic and transcriptomic data in a correlation network, system-wide consequences of the perturbations were revealed and genes with a putative role in phenolic metabolism were identified. Together, our data provide insight into lignin biosynthesis and the metabolic network it is embedded in and provide a systems view of the plant’s response to pathway perturbations.
INTRODUCTION
Lignin is an aromatic polymer that is deposited in secondary-thickened cells where it provides strength and impermeability to the wall. In dicot plants, lignin is mainly composed of the monolignols coniferyl and sinapyl alcohol that give rise to the guaiacyl (G) and syringyl (S) units of the lignin polymer, respectively. In addition, a number of other units may be incorporated at lower levels, depending on the species, the genetic background, and environmental conditions (Ralph et al., 2004). The lignin biosynthetic pathway is generally divided in two parts: the general phenylpropanoid pathway from Phe to feruloyl-CoA and the monolignol-specific pathway from feruloyl-CoA to the monolignols (Figure 1). Ten enzymes are involved in the pathway from Phe to the monolignols: phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate:CoA ligase (4CL), hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT), p-coumarate 3-hydroxylase (C3H), caffeoyl-CoA O-methyltransferase (CCoAOMT), cinnamoyl-CoA reductase (CCR), ferulate 5-hydroxylase (F5H), caffeic acid O-methyltransferase (COMT), and cinnamyl alcohol dehydrogenase (CAD) (Figure 1) (Boerjan et al., 2003; Bonawitz and Chapple, 2010). After their biosynthesis, the monolignols are translocated to the cell wall where they are oxidized to radicals that are then coupled in a combinatorial fashion with the formation of various types of chemical bonds of which the ether (8-O-4), resinol (8-8), and coumaran (8-5) bonds are the most prominent ones (Ralph et al., 2004). The ability to obtain plants with altered amounts or structures of lignin by mutant screening (Vermerris et al., 2007) or genetic engineering (Chen and Dixon, 2007) has enabled the properties of plant biomass to be improved for forage digestibility or processing into pulp or fermentable sugars (Pilate et al., 2002; Baucher et al., 2003; Chen and Dixon, 2007). A remarkable insight from this work is that plants with reduced lignin can either grow normally or have dramatic effects on plant growth and development, depending on which gene of the lignin biosynthetic pathway was perturbed. Apparently, plants are sometimes able to cope successfully with a mutation, a phenomenon called phenotypic buffering, whereas in other cases they are not (Fu et al., 2009).
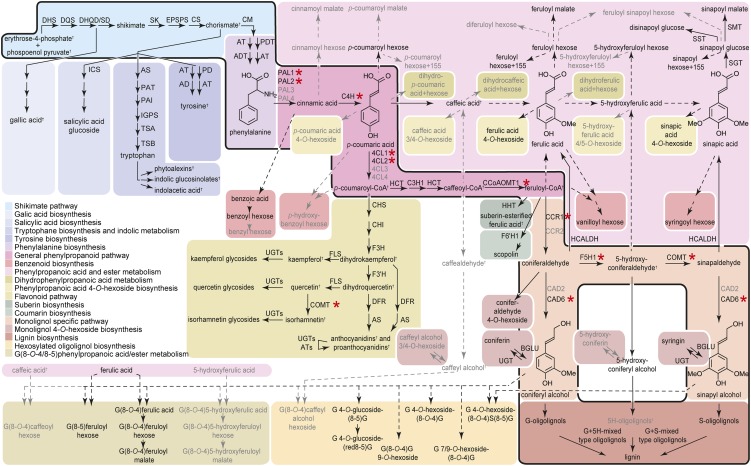
Figure 1.
Metabolic Map of Phenolic Metabolism in the Inflorescence Stem of Arabidopsis.
The main pathways involved in lignin biosynthesis are framed with a bold black border. For enzymes with a red asterisk, mutants were analyzed in this study. For simplicity, proteins corresponding to individual gene family members are given only for the general phenylpropanoid pathway and monolignol-specific pathway. Family members in black were predicted to be involved in developmental lignification (Costa et al., 2003; Goujon et al., 2003a; Raes et al., 2003). Dashed arrows represent suggested pathways. The metabolites that were detectable in wild-type stems are shown in black, whereas those that were detectable only in certain mutants are shown in gray. Expression of all genes was determined via microarrays. The relative abundance of the metabolites was determined via UPLC-MS and/or GC-MS. Metabolites indicated with a cross were not detected or not identified via the used GC-MS and UPLC-MS techniques. This pathway representation was used to map expression and metabolite data of all studied mutant lines and wild-type developmental samples (Figure 5; see Supplemental Figure 5 online). AD, arogenate dehydrogenase; ADT, arogenate dehydratase; AS, anthranilate synthase; AS, anthocyanidin synthase; AT, amino transferase; ATs, acyltransferases; CHI, chalcone isomerase; CHS, chalcone synthase; CM, chorismate mutase; CS, chorismate synthase; DFR, dihydroflavonol 4-reductase; DHS, 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase; DHQD/SD, 3-dehydroquinate dehydratase/shikimate dehydrogenase; DQS, 3-dehydroquinate synthase; EPSPS, 5-enolpyruvylshikimate-3-phosphate synthase; F3H, naringenin 3-dioxygenase; F3′H, flavonoid 3-hydroxylase; F6′H, feruloyl-CoA 6-hydroxylase; FLS, flavonol synthase; HCALDH, hydroxycinnamaldehyde dehydrogenase; ICS, isochorismate synthase; IGPS, indole-3-glycerol phosphate synthase; PAI, phosphoribosylanthranilate isomerase; PAT, phosphoribosylanthranilate transferase; PD, prephenate dehydrogenase; SGT, sinapate 1-glucosyltransferase; SK, shikimate kinase; SMT, sinapoylglucose:malate sinapoyltransferase; SST, sinapoylglucose:sinapoylglucose sinapoylglucosetransferase; TSA, Trp synthase α-subunit; TSB, Trp synthase β-subunit; UGT, UDP-glucosyltransferase. For nomenclature of aromatic molecules, see Supplemental Figure 4 online and Morreel et al. (2010a, 2010b).
In an attempt to understand how plants cope with a genetic defect, we previously showed that altering the expression of genes in the lignin biosynthetic pathway not only results in altered lignification but also in shifts in both primary and secondary metabolism. For example, reductions in lignin in Arabidopsis thaliana pal1 pal2 double mutants, while not leading to abnormalities in overall plant growth, were accompanied by transcript changes of genes involved in phenylpropanoid biosynthesis, carbohydrate metabolism, stress-related pathways, signal transduction, and amino acid metabolism, as studied by cDNA–amplified fragment length polymorphism (Rohde et al., 2004). Whereas all identified phenolic compounds were lower in abundance, nearly all of the detected amino acids accumulated in these mutants. Analogous experiments with CCR-downregulated poplar (Populus tremula × Populus alba) showed an induced stress response and effects on cell wall biosynthetic genes, including the induction of transcripts for PAL and reduction of hemicellulose and pectin biosynthesis. Metabolite analysis showed major increases in phenolic acid glucosides but also shifts in the primary metabolites (e.g., increased levels of maleate, Kreb’s cycle intermediates, and several monosaccharides) (Leplé et al., 2007). Similar effects were observed in CCR-downregulated tobacco (Nicotiana tabacum) where several amino acids accumulated (Dauwe et al., 2007). In CAD-downregulated tobacco, most transcripts of genes involved in phenylpropanoid biosynthesis were lower and transcripts of light- and (nonlignin) cell wall–related genes were higher (Dauwe et al., 2007). Microarray analysis of cad-c cad-d Arabidopsis mutants revealed effects on stress-related pathways and cell wall–related proteins, including lignin, pectin, cellulose, and cell wall–localized proteins (Sibout et al., 2005). Furthermore, a SSH transcript-based comparison of three brown midrib3 (bm3) maize (Zea mays) mutants (mutated in COMT) revealed a feedback on phenylpropanoid and hemicellulose biosynthesis and photosynthesis, and comparisons with bm1 (mutated in CAD) and bm2 (mutated gene unknown) showed a shared response in signaling and regulation (Shi et al., 2006a).
So far, these types of studies have remained fragmentary and disconnected, focusing on a few individual genes in different species, and the transcript and metabolite profiling methods used did not allow the extraction of genome and metabolome-wide conclusions. Hence, the major adjusting and regulatory mechanisms that may exist across the pathway to compensate for either less or modified lignin have remained largely unresolved (Vanholme et al., 2008, 2010a, 2010c). For example, it is unclear whether the flux through the phenylpropanoid pathway is redirected in a systematic way upon blocking particular steps in monolignol synthesis and to what extent feedback systems regulate the pathway. To obtain deeper insight into lignin biosynthesis and the metabolic network it is embedded in, we used a systems biology approach as defined by Ideker et al. (2001) (i.e., the study of the consequences of pathway perturbations, followed by computational analysis of the data). To this end, we systematically analyzed the transcriptome and metabolome of mutants in consecutive steps of the lignin biosynthetic pathway. The spectacular advances in transcriptomics and metabolomics have opened up the possibility of fully exploiting this approach (Oksman-Caldentey and Saito, 2005; Mochida and Shinozaki, 2011), as already illustrated by studies of the consequences of altered expression of five transcription factors involved in glucosinolate biosynthesis in Arabidopsis (Hirai et al., 2007; Malitsky et al., 2008). We chose Arabidopsis as a model because systems biology can be properly performed only in an organism for which thorough basic knowledge exists on the identity and function of genes, proteins, and metabolites. Furthermore, we focused on the inflorescence stem, which is an excellent model for wood formation (Nieminen et al., 2004) as it is rich in fibers and vessels, both cell types undergoing lignification during secondary thickening.
The set of Arabidopsis lines studied were mutated in PAL1 and 2, C4H, 4CL1 and 2, CCoAOMT1, CCR1, F5H1, COMT, and CAD6, genes predicted to be involved in developmental lignification (Boerjan et al., 2003; Costa et al., 2003; Goujon et al., 2003a; Raes et al., 2003; Bonawitz and Chapple, 2010). We show that most of the mutations provoked a strong and organized response at the transcript and metabolite levels, even when these mutants did not have any apparent visible phenotype. Mutants with reduced lignin levels upregulated genes of the pathways that supplied monolignol precursors, whereas mutants with compositional shifts downregulated these pathways. In addition, metabolic profiling showed that perturbations redirected the flux into novel pathways, at the same time revealing metabolic rerouting of accumulating metabolites by 4-O- and 9-O-hexosylation. By combining metabolomic and transcriptomic data in a correlation network, unexpected system-wide consequences of the perturbations were revealed. Furthermore, we explored whether analyzing the response to pathway perturbations can be used to reveal novel genes closely associated with the studied pathway and, thus, to identify prime candidates for genetic improvement strategies. Together, our data provide a systems view of the plants’ response to lignin pathway perturbations.
RESULTS
Collection of Lignin Mutants and Analysis of Growth Dynamics
To study the system-wide consequences of lignin pathway perturbations, a set of Arabidopsis lines was collected, each with a mutation in a single gene of the general phenylpropanoid or monolignol-specific pathway (Figure 1). For 10 genes of the pathway (PAL1, PAL2, C4H, 4CL1, 4CL2, CCoAOMT1, CCR1, F5H1, COMT, and CAD6), we were able to select two independent mutants that were suitable for our approach (Table 1; see Supplemental Figure 1 online). Mutants in C3H and HCT were sublethal and were left out of the analysis. We grew plants under conditions identified to allow the development of a single, strong inflorescence stem with a maximum of secondary cell wall thickening (i.e., shifting plants from short-day to long-day conditions when the average rosette diameter reached ∼6 cm). Ideally, all lines needed to be equally developed at harvest to prevent developmental shifts (previously described for c4h-2, ccr1-3, and ccr1-6; Jones et al., 2001; Mir Derikvand et al., 2008; Schilmiller et al., 2009) from superimposing uninformative molecular changes on the more direct effects of the perturbed lignin pathway. Stem growth rate was slower than the wild type in c4h-2, ccr1-3, ccr1-6, and ccoaomt1-3, while cad6-1 grew faster (Table 1; see Supplemental Figure 2 online). The differences between ccoaomt1-3 or cad6-1 and the wild type were small and, after 44 d in long-day conditions, when all plants ceased growing, only c4h-2, ccr1-3, and ccr1-6 had a significantly reduced final stem height compared with the wild type (Table 1; see Supplemental Figure 2 online). To compensate partially for the developmental delay of these mutant lines, they were sown 2 weeks prior to the other lines for all subsequent analyses.
Table 1. Growth and Bolting Characteristics of the 20 Arabidopsis Mutants Used in This Study.
| Growth Characteristics |
|||||
|---|---|---|---|---|---|
| Gene | AGI Code | Mutant Allele | Growth Rate | Final Height | |
| PAL1 | At2g37040 | pal1-2 | (SALK_000357) | – | – |
| pal1-3 | (SALK_022804) | – | – | ||
| PAL2 | At3g53260 | pal2-2 | (SALK_092252) | – | – |
| pal2-3 | (GABI_692H09) | – | – | ||
| C4H | At2g30490 | c4h-2 | (ref3-2) | Reduced** | ±75% of the wild type |
| c4h-3 | (ref3-3) | – | – | ||
| 4CL1 | At1g51680 | 4cl1-1 | (SALK_142526) | – | – |
| 4cl1-2 | (SM_3_27345) | – | – | ||
| 4CL2 | At3g21240 | 4cl2-1 | (SLAT 02_14_04) | – | – |
| 4cl2-3 | (GABI_353A11) | – | – | ||
| CCoAOMT1 | At4g34050 | ccoaomt1-3 | (SALK_151507) | Reduced* | – |
| ccoaomt1-5 | (GABI_007F02) | – | – | ||
| CCR1 | At1g15950 | ccr1-3 | (SALK_123689) | Reduced** | ±20% of the wild type |
| ccr1-6 | (GABI_622C01) | Reduced** | ±65% of the wild type | ||
| F5H1 | At4g36220 | f5h1-2 | (fah1-2) | – | – |
| f5h1-4 | (SALK_063792) | – | – | ||
| COMT | At5g54160 | comt-1 | (SALK_002373) | – | – |
| comt-4 | (SALK_050030) | – | – | ||
| CAD6 | At4g34230 | cad6-1 | (SALK_040062) | Enhanced* | – |
| cad6-4 | (SAIL_776_B06) | – | – | ||
Bolting time and growth are given relative to the wild type; –, no significant difference; **, significant in each of the experiments; *, a tendency, but not significantly different in each of the experiments (see Methods; see Supplemental Figure 2 online for details). AGI, Arabidopsis Genome Initiative.
Deep Phenotyping of Mutants via Transcriptomics and Metabolomics
Replicate samples were taken from 8-, 16-, 24-, and 32-cm-tall wild-type inflorescence stems to evaluate metabolic and transcriptomic shifts during stem development, and these were called WT8cm, WT16cm, WT24cm, and WT32cm, respectively. The mutant lines were harvested in replicate simultaneously with and at the same height as WT24cm with the exceptions of ccr1-6, which was only 19 cm tall, and c4h-2 and ccr1-3, which were harvested simultaneously with WT32cm at 12 and 16 cm tall, respectively. The basal 1 to 9 cm (1 to 6 cm in case of WT8cm) of the inflorescence stems was used for both transcript and metabolite profiling because this region of the stem is relatively enriched in lignifying cells (Nieminen et al., 2004).
Gene expression in the inflorescence stem of the 20 mutant lines and the four wild-type developmental stages was analyzed using Agilent Arabidopsis 3 oligo arrays, harboring 32,221 probes for 25,094 genes (covering 91.53% of the protein coding genes [PCGs] according to the TAIR10 annotated genome). For metabolomics, two different separation techniques were used to cover a wide range of metabolites. Small polar metabolites, such as sugars, sugar alcohols, amino acids, and small organic acids, were determined via a standard gas chromatography–mass spectrometry (GC-MS) method (Roessner et al., 2000; Fernie et al., 2004), whereas phenolic compounds were detected via an established in-house ultrahigh performance liquid chromatography–mass spectrometry (UPLC-MS) method (Morreel et al., 2004, 2010a, 2010b).
The intensity of 26,787 probes (86.14% of the PCGs on the array) was well above background in at least one of the four sampled stages during wild-type stem development. Statistical analysis revealed that the signal of 12,291 probes (46.25% of the PCGs on the array) changed significantly during wild-type stem development, with WT24cm used as the reference sample. In comparison, only 7099 probes (26.91% of the PCGs on the array) were significantly different in at least one of the mutant lines compared with WT24cm. This indicates that the overall change in gene expression provoked by a mutation was more subtle than that caused by progressive development of the inflorescence stem. Remarkably, mutants that had no obvious morphological phenotypes nor developmental shifts (i.e., all mutants except c4h-2, ccr1-3, and ccr1-6) still had up to 675 probes (2.56% of the PCGs on the array) with a signal that was different from the signal in WT24cm (Figure 2A).
Figure 2.
Number of Transcripts and Compounds That Are Significantly Different in Abundance Compared with WT24cm, in Each Mutant Line and in the Wild-Type Developmental Stages.
(A) Number of gene probes that are significantly different as compared with WT24cm. In parentheses: percentage of PCG-representing probes present on the array. WT, the wild type.
(B) Number of compounds detected by GC-MS that accumulate differentially compared with WT24cm. In parentheses: percentage of the total number of compounds detected by GC-MS.
(C) Number of compounds detected by UPLC-MS that accumulate differentially compared with WT24cm. In parentheses: percentage of the total number of compounds detected by UPLC-MS.
Red superscript s and w indicate the stronger and weaker mutant alleles, respectively: substantial residual 4CL2 transcript levels were detected in 4cl2-3 in this study, and the alleles of c4h and ccr1 are described by Schilmiller et al. (2009) and Mir Derikvand et al. (2008).
With GC-MS, 229 compounds were detected in at least one of the lines, 59 of which could be identified by use of the GMD@CSB.DB-library (Kopka et al., 2005) (see Supplemental Data Set 1 online). A compound is defined here as a molecule that elutes at a certain retention time. Because of the derivatization procedure used, some metabolites are detected as multiple compounds (Kopka et al., 2005). The abundance of 86% of compounds varied significantly during wild-type stem development, and the abundance of 93% was significantly altered in at least one of the mutant lines compared with WT24cm (Figure 2B). Notably, the fraction of GC-MS–detected metabolites that differ in abundance due to mutational and developmental effects was larger than the fraction of transcripts that differed in abundance.
Finally, 337 compounds were detected via UPLC-MS (see Supplemental Data Set 1 online), of which 328 were integrated and aligned using the MetAlign software (Lommen, 2009), and nine were manually integrated. A total of 82% of the 328 automatically integrated compounds varied significantly in at least one of the developmental stages compared with WT24cm, whereas 99% varied significantly in at least one of the mutants (Figure 2C). As with the GC-MS–detected metabolites, the fraction of differential UPLC-MS–detected metabolites was larger in the mutant compared with the developmental samples.
Overall, there was a remarkable similarity in the number of differentials between the two mutant alleles of each gene, although this was less pronounced for the transcripts for mutants with a strong and a weak mutant allele. These data underscore the similar molecular response in both mutant alleles of a given gene.
Coordinated Transcript and Metabolite Changes during Inflorescence Stem Development
To have an idea of which biological processes are most prominent over the different stages of wild-type development, the 12,291 probes and 466 metabolites that were significantly differential over developmental time were grouped in clusters (A to F) according to their temporal profiles (Figure 3). Cluster A grouped probes and compounds that were maximal at the WT8cm stage and decreased over development (1703 probes representing 6.57% of the PCGs on the array, 102 GC-MS, and 36 LC-MS compounds; see Supplemental Data Set 2 online). Gene Ontology (GO) analysis via BiNGO (Maere et al., 2005) revealed a significant overrepresentation of the cell cycle and related processes (microtubule biosynthesis, primary cell wall formation, and DNA replication; see Supplemental Data Set 3 online). Furthermore, cluster A contained ∼45% of all compounds detected via GC-MS, including most of the amino acids, indicating that the earlier developmental stages have a high metabolic rate and are enriched in primary metabolism. Cluster A further contained secondary metabolites, such as the flavonol glycosides, and molecules derived from sinapic acid (e.g., sinapoyl malate and sinapic acid O-4 glucoside).
Figure 3.
Relative Gene Expression and Metabolite Abundance over Stem Development.
Cluster A represents the profiles of 1703 probes and 138 compounds that decreased over development. Cluster B shows the profiles of 936 probes and 12 metabolites that had a maximal abundance at the WT16cm stage and are low early and late in development. Cluster C shows the profiles of 1760 transcripts and nine metabolites that had a maximal abundance at the WT24cm stage. Cluster D shows the profiles of 1097 probes and 133 compounds that increased over stem development. Cluster E represents the profile of 1089 probes and 40 metabolites with a minimum at WT16cm but further increased from WT16cm onwards. Cluster F shows the profiles of 840 probes and 22 compounds with a minimum at the WT24cm stage. The colors have no meaning but are used only to visualize the different profiles within each cluster. All probes and metabolites are given in Supplemental Data Set 2 online, whereas GO enrichments of each cluster can be found in Supplemental Data Set 3 online.
Cluster B grouped 936 probes (3.57% of the PCGs on the array) and 12 compounds that had a maximal abundance at the WT16cm stage and of which the abundance was higher in WT24cm compared with WT32cm (see Supplemental Data Set 2 online). GO analysis proved the genes to be enriched in secondary cell wall–related processes, in particular (glucurono)xylan biosynthesis (see Supplemental Data Set 3 online). Cluster C grouped profiles of 1760 probes (6.69% of the PCGs on the array) and nine compounds that increased in abundance up to the WT24cm stage and then decreased (see Supplemental Data Set 2 online) and was enriched in defense response, cell death, and phenylpropanoid biosynthesis genes (see Supplemental Data Set 3 online). PAL1, C4H, 4CL1, 4CL2, HCT, CCR1, CCoAOMT1, F5H1, and COMT were present in this cluster. The occurrence of (glucurono)xylan biosynthesis and lignification in two separate clusters demonstrates the quality of the data set that resolves, in time, two sequential processes of secondary cell wall formation.
Cluster D represented probes and compounds for which the abundance increased over development. It consists of 1097 probes (4.14% of the PCGs on the array) and 14 GC-MS and 119 LC-MS compounds (see Supplemental Data Set 2 online). Genes in response-related processes were enriched, as well as transcription factors and genes involved in auxin transport (see Supplemental Data Set 3 online). This cluster contained virtually all identified oligolignols and the primary metabolite shikimate.
Cluster E grouped profiles that decreased from WT8cm to WT16cm but then increased (1089 probes representing 4.18% of the PCGs on the array and 40 metabolites; see Supplemental Data Set 2 online). Glycolysis-related transcripts were enriched as were categories “translation” and “ribosome” (see Supplemental Data Set 3 online), perhaps reflecting a gradual change from starch respiration in young tissues toward photosynthesis and/or respiration of transported photoassimilates in mature tissues. Consistent with the latter, both Fru-6-P and Glc-6-P were present in cluster E. Finally, cluster F represented 840 probes (3.23% of the PCGs on the array) and 22 compounds that decreased from WT8cm to a minimum in WT24cm then increased again in WT32cm (see Supplemental Data Set 2 online). This cluster was enriched in photosynthesis and related GO processes (see Supplemental Data Set 3 online) and contained the metabolites coniferin and Fru as well as Gln as one of the few amino acids that did not fall into cluster A.
Common System-Wide Responses in Lignin Mutants
To visualize similarities in transcript and metabolite changes among the different mutants, mutant samples were clustered based on their transcript and metabolic profiles (Figure 4). The two mutant alleles of each gene generally clustered together, emphasizing that they provoked a similar response. The only exceptions were the c4h, ccr1, and f5h1 lines in the transcript-based clustering. Notably, c4h-2 and ccr1 mutants clustered with the wild type of a different developmental stage, indicating that the advanced sowing of these mutant lines could not fully compensate for the developmental shift. A difference between the two alleles of both c4h and ccr1 was expected as these lines have a different residual C4H and CCR1 enzymatic activity (Mir Derikvand et al., 2008; Schilmiller et al., 2009). By contrast, the distinction between the two f5h1 lines was unexpected, especially because they nicely cocluster in GC-MS– and UPLC-MS–based clustering. f5h1-2 was similar to comt mutants in the transcript-based clustering, which is in agreement with their respective enzymes catalyzing subsequent reactions in the S-unit–specific branch of monolignol biosynthesis. The f5h1-4 mutant, on the other hand, was very similar to the wild type as both lines appeared to be coclustered based on transcript data. For reasons explained in the Supplemental Text 1 online, f5h1-4 was excluded from all subsequent analyses.
Figure 4.
Sample-Based Clustering of the Mutant Lines and Wild-Type Developmental Series.
(A) Clustering based on the expression of 7099 probes that were significantly different in at least one of the mutant lines.
(B) Clustering based on 212 compounds measured by GC-MS.
(C) Clustering based on 325 compounds measured by UPLC-MS.
Convincingly, mutants of the central part of the phenylpropanoid pathway (c4h-3, 4cl1-1, 4cl1-2, ccoaomt1-3, and ccoaomt1-5) grouped in a separate transcript-based subcluster. In both the GC- and the UPLC-MS–based clustering, this subcluster was further extended with the ccr1 alleles. This indicates that system-wide responses to pathway perturbations are similar in this group of mutants. Surprisingly, based on transcript data, pal1 mutants appeared in the same cluster as f5h1-2 and comt mutants. This is unexpected based on current knowledge of the pathway because PAL1 is the very first enzyme in the general phenylpropanoid pathway, and F5H1 and COMT act on the downstream monolignol-specific pathway. Overall, the clusters obtained from the three different datasets were similar.
For ease of interpretation, the average of the log ratios of the expression/abundance values of the two alleles of pal1, pal2, 4cl1, 4cl2, ccoaomt1, comt, and cad6 were taken for tables and figures, which is sound given the coclustering of the respective alleles (Figure 4). Only for c4h and ccr1 are the data in figures and tables restricted to those of the allele that deviated least from the WT24cm (i.e., c4h-3 and ccr1-6). However, all transcript and metabolic data for both alleles are available at http://www.datadryad.org under accession number doi:10.5061/dryad.jn16g and in Supplemental Data Set 1 online, respectively.
To visualize responses that are shared between the different mutants, the differential transcripts and compounds detected in pal1, pal2, c4h, 4cl1, 4cl2, ccoaomt1, ccr1, f5h1, comt, and cad6 were compared with each other. As deduced from Supplemental Figure 3 online, the number of differential transcripts and metabolites that was shared between multiple mutants was much higher than expected by chance alone. Similarly, the number of transcripts and metabolites differentially in the same direction (up or down) was much higher than expected by chance alone, which indicates that the response in each of the mutants is not random but organized and, thus, biologically relevant.
Metabolic Maps Reveal Shifts in Phenolic Metabolism upon Pathway Perturbations
The UPLC-MS approach was targeted toward phenolic compounds (i.e., intermediates and products of the perturbed phenylpropanoid and monolignol biosynthetic pathways). Of the 337 detected metabolites, all 128 tentatively structurally identified compounds were phenolic/aromatic; 34 compounds were identified based on their retention time and mass spectral fragmentation in time (MS2) data that were identical to those of standards (see Supplemental Data Set 1 online), and 94 compounds were tentatively identified via MS2 (Morreel et al., 2010b) (see Supplemental Data Set 1 and Supplemental Figure 4 online). The MS2 spectra of 43 tentatively identified compounds that were not published before are listed in Supplemental Figure 4 online, together with the reasoning for their structural elucidation. The 128 tentatively identified phenolic compounds belonged to several metabolic classes, as illustrated in Figure 1.
For each mutation, the altered abundances of metabolites and transcripts involved in phenolic metabolism were visualized by comparison with the control sample WT24cm (Figure 5; see Supplemental Figure 5 and Supplemental Data Sets 1 and 4 online). The metabolic and transcriptomic changes of the wild-type developmental series were similarly compared with WT24cm (see Supplemental Figure 5 and Supplemental Data Sets 1 and 4 online). The response of specific phenolic classes, such as oligolignols, phenylpropanoid 4-O-hexosides, phenylpropanoic acid derivates, benzenoids, and coniferyl alcohol-ferulic acid dimers, and their biosynthetic pathways are described below.
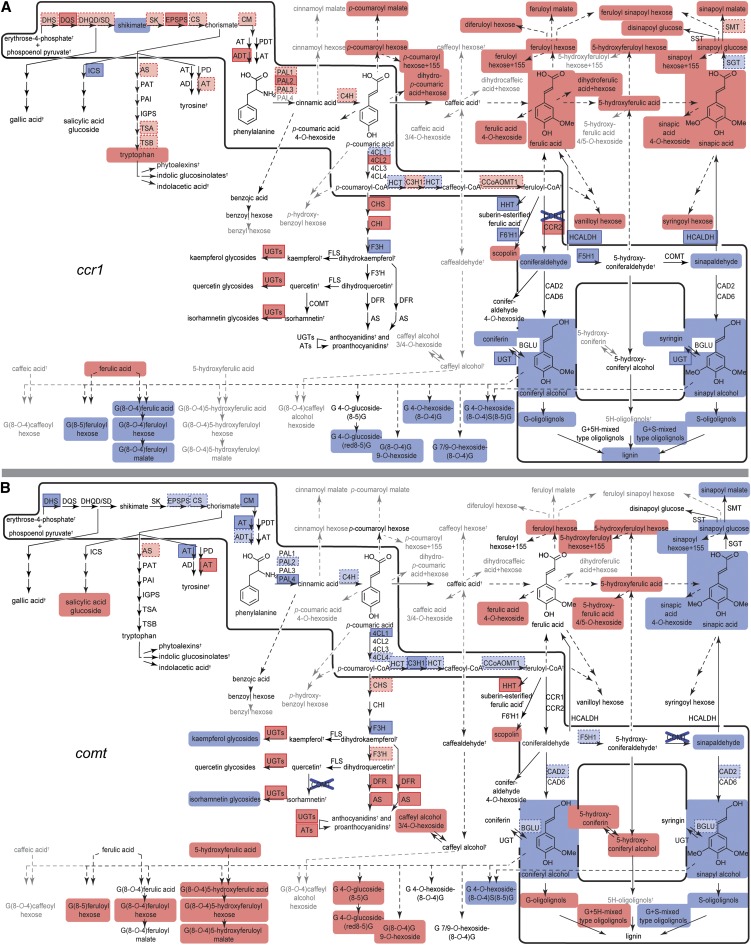
Figure 5.
Metabolic Shifts in Phenolic Metabolism as a Result of a Mutation in the Lignin Biosynthetic Pathway.
The relative increase and decrease of transcript and metabolite abundances in each of the mutants compared with WT24cm was mapped manually on the pathway as discrete features. Differences in metabolites are indicated via round-angled boxes, where red represents a significant increase and blue a significant decrease in abundance. Differences in transcript abundance are indicated with right-angled, framed boxes: significant increases and decreases are visualized via red and blue boxes with solid borders, respectively, whereas tendencies [log2(abundance in the sample/abundance in WT24cm)<−0.3 or >0.3] are visualized via boxes with a pale fill and dashed-line borders. Metabolites that were below detection limit are given in gray. The effects in the ccr1 and comt mutants are in (A) and (B), as indicated. The metabolic shifts in phenolic metabolism of pal1, pal2, c4h, 4cl1, 4cl2, ccoaomt1, f5h1, cad6, and the wild-type developmental stages are given in Supplemental Figure 5 online. For abbreviations, see Figure 1.
Lignin, Oligolignols, and Their Biosynthesis
During the lignification process, monolignols are relocated to the cell wall, where they are oxidized and coupled in a combinatorial fashion to oligolignols and higher molecular weight lignin polymers. Based on their MS2 spectra and retention times by UPLC-MS, 36 compounds found in wild-type stems were identified as oligolignols (di-, tri-, and tetralignols; see Supplemental Data Set 1 online). Oligolignol abundance increased during wild-type development (see Supplemental Figure 5 and Supplemental Data Set 1 online). All oligolignols were less abundant in c4h, 4cl1, ccoaomt1, and ccr1 mutants, whereas f5h1 and comt mutants had reduced amounts of oligolignols that contained S-units (Figure 5; see Supplemental Figure 5 and Supplemental Data Set 1 online). In f5h1 mutants, the reduction in oligolignols with at least one S-unit seemed to be compensated for by an increase in abundance of oligolignols that consist exclusively of G-units, whereas in comt mutants, oligolignols with 5-hydroxyguaiacyl (5H) units [e.g., G(8-O-4)5H] accumulated, derived from coupling of a classical monolignol with 5-hydroxyconiferyl alcohol. To investigate the relationship between oligolignol levels and lignin amount, we measured acetyl bromide lignin in the set of mutant and wild-type plants harvested at the same developmental stage (see Supplemental Figure 6 online). Notably, the relative abundance of the oligolignols in each of the mutants compared with WT24cm largely reflected the relative amount of the lignin polymer (see Supplemental Figure 6 online).
Genes involved in the general phenylpropanoid and the monolignol-specific pathway had maximal expression levels at the 16- to 24-cm stages in the wild type (Figure 6). Remarkably, c4h, 4cl1, ccoaomt1, and ccr1 mutants (i.e., those mutants with reduced lignin levels) showed increased abundance of transcripts of the entire pathway from PAL to CCR (Figure 6; see Supplemental Table 1 online). Increased expression was most prominent in one gene family member of PAL, 4CL, and CCR (i.e., PAL2, 4CL2, and CCR2). Strikingly, the opposite transcriptional response was observed in f5h1 and comt mutants that had normal amounts of lignin but were reduced in oligolignols with S-units (Figure 6; see Supplemental Table 1 online).
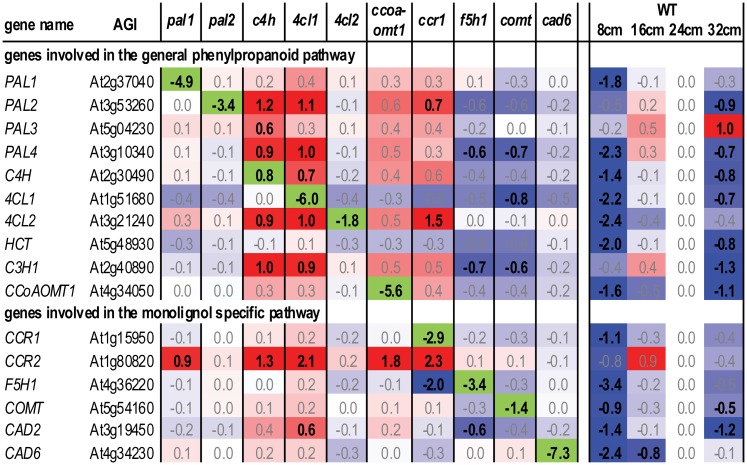
Figure 6.
Transcript Levels of General Phenylpropanoid and Monolignol-Specific Biosynthetic Genes in Each of the Mutants and in the Wild-Type Developmental Series.
Gene names and Arabidopsis Genome Initiative codes were derived from the set of 34 genes described by Raes et al. (2003). Only the genes for which the transcript level was significantly different in at least one of the mutants are given. Full names of the genes are given in Figure 1. Values are log2(abundance in the sample/abundance in WT24cm). In the case of pal1, pal2, 4cl1, 4cl2, ccoaomt1, comt, and cad6, the average of the log ratios of the two alleles was taken. For c4h and ccr1, the values of c4h-2 and ccr1-6 are given, respectively. A red background indicates a higher transcript level in comparison with WT24cm and blue a lower. The color intensity reflects the strength of induction and reduction. A green background represents the transcript level of the mutant gene. Bold values are significantly different from WT24cm. WT, the wild type.
Before polymerization, monolignols are oxidized by laccases and class III peroxidases. Based on the transcript abundance over wild-type development (see Supplemental Figure 7 online), laccases and class III peroxidases could be divided in two sets. The first set had a maximal transcript abundance in WT8cm. This class is potentially involved in the oxidation of monomers for the production of phenylpropanoid dimers that are most abundant at the 8-cm stage (see Supplemental Figure 5 and Supplemental Data Set 1 online). A second set (i.e., LAC2, 4, 11, 16, and 17 and 10 peroxidases) had a maximal transcript abundance in WT16cm or 24cm, the same stages at which lignin biosynthetic genes were also maximally expressed (Figure 6). Most likely, this set of genes contains candidates involved in lignification. Of these, LAC2, LAC11, LAC17, and PER12 responded in the mutants in a similar way as the genes of the phenylpropanoid pathway did in the two groups of mutants (Figure 6; see Supplemental Figure 7 online), making them top candidates for a role in monolignol oxidation. While these data were being prepared for publication, Berthet et al. (2011) provided the first undisputable evidence for a role of LAC17 in lignification, validating our hypothesis.
4-O-Glucosylated Monolignols and Their Metabolism
Coniferin and syringin are thought to be synthesized from their respective aglycone by glucosyltransferases UGT72E2 and UGT72E3. UGT72E2 accepts both coniferyl and sinapyl alcohols as substrate in vitro, whereas UGT72E3 is more specific for sinapyl alcohol (Lim et al., 2005; Lanot et al., 2006). Conversely, β-glucosidases BGLU45 and BGLU46 hydrolyze coniferin and syringin back into their respective aglycones in vitro (Escamilla-Treviño et al., 2006). Over wild-type development, the abundance of coniferin and syringin was highest at the 8-cm stage, but in the mutant samples, they largely followed the abundances of coniferyl and sinapyl alcohol, respectively (see Supplemental Data Set 1 online). Expression of both UGT72E2 and UGT72E3 was reduced in c4h, 4cl1, ccoaomt1, and ccr1 mutants, while BGLU45 and BGLU46 was increased in c4h, 4cl1, and ccoaomt1 mutants (see Supplemental Data Set 4 online). Thus, based on the transcript data, the biosynthesis of coniferin and syringin in the low lignin mutants (i.e., c4h, 4cl1, ccoaomt1, and ccr1) was reduced, while their hydrolysis was increased, which agrees with the observed abundances of coniferin and syringin.
Phenylpropanoic Acid Derivates
Specific phenylpropanoic acids (i.e., cinnamic, p-coumaric, caffeic, ferulic, 5-hydroxyferulic, and sinapic acid) and derivates thereof accumulated in each of the mutants (Figure 5; see Supplemental Figure 5 and Supplemental Data Set 1 online). Most of these metabolites were below the detection limit in the wild-type plants and only found in certain mutants, but for each derivate type, a corresponding ferulic or sinapic acid–derived analog was detectable in the wild type, with the exception of hexose coupled to dihydrogenated phenylpropanoic acid (Figure 1). Strikingly, neither caffeoyl glucose nor caffeoyl malate was detected in ccoaomt1 mutants; instead, the flux in these mutants seemed to be driven toward ferulate esters, flavonol glycosides, and coniferyl alcohol-caffeic acid dimers [i.e., G(8-O-4)caffeoyl hexose] (see Supplemental Figures 5 and 8 and Supplemental Data Set 1 online). This shows that the excess caffeoyl-CoA in ccoaomt1 mutants is metabolized differently from accumulating phenylpropanoids in the other mutants.
The genes involved in sinapate ester biosynthesis in Arabidopsis have been described (Fraser et al., 2007; Sinlapadech et al., 2007). No general response of the sinapate ester pathway was observed in the lignin mutants (see Supplemental Data Set 4 online), showing that this pathway is not responding in an orchestrated way to mutations in the phenylpropanoid pathway.
In addition to the phenylpropanoic acids, four benzenoic hexose esters accumulated in the mutants (i.e., benzoyl, p-hydroxybenzoyl, vanilloyl, and syringoyl hexose) (see Supplemental Figure 5 and Supplemental Data Set 1 online). The exact pathway toward these benzenoids is currently unknown, but these compounds are known to be derived from the corresponding phenylpropanoid acids and/or CoA-esters (Jarvis et al., 2000; Hertweck et al., 2001; Boatright et al., 2004). The accumulation of benzenoids in mutants where the corresponding propanoic acid derivates accumulate proves that part of the excess of phenylpropanoids is metabolized into C6-C1 compounds.
Coniferyl Alcohol-Ferulic Acid Dimers
Another class of phenylpropanoic acid-derived metabolites in wild-type Arabidopsis stems comprises coupling products of coniferyl alcohol and ferulic acid [i.e., G(8-O-4)ferulic acid] and different derivatives (Rohde et al., 2004) (Figure 1; see Supplemental Data Set 1 online). Similar to other coniferyl alcohol–derived metabolites, the abundance of coniferyl alcohol-ferulic acid dimers was reduced in c4h, 4cl1, ccoaomt1, and ccr1 mutants and increased in f5h1 and comt mutants. The caffeic acid analog of G(8-O-4)feruloyl hexose [i.e., G(8-O-4)caffeoyl hexose] accumulated in both ccoaomt1 and comt mutants (Figure 5; see Supplemental Figure 5 and Supplemental Data Set 1 online). On the other hand, 5-hydroxyferulic acid analogs of G(8-O-4)feruloyl hexose [e.g., G(8-O-4)5-hydroxyferuloyl hexose] were unique to comt mutants (as all 5-hydroxy substituted phenylpropanoids) (Figure 5; see Supplemental Figure 5 and Supplemental Data Set 1 online). These observations further support the involvement of COMT in the O-3 methylation of caffeoyl-CoA and caffeic acid, even in the presence of CCoAOMT1 and the lack of significant involvement of CCoAOMT1 in the O-5 methylation of 5-hydroxyconiferaldehyde in the presence of COMT (Do et al., 2007).
System-Wide Responses: The Correlation Network
The results described above indicate that perturbation of the lignin biosynthetic pathway has profound effects on the transcript and metabolite levels of this pathway. However, the consequences of the perturbations reach much further than the perturbed pathway itself. A transcript-metabolite correlation network was constructed to visualize these system-wide consequences. In such a network, transcripts and metabolites (nodes) are linked (edges) if the correlation of their abundance was significant. Two nodes that are significantly correlated are likely to have a common in vivo role (e.g., two genes involved in the same metabolic pathway) (Saito et al., 2008; Vandepoele et al., 2009; Lee et al., 2010).
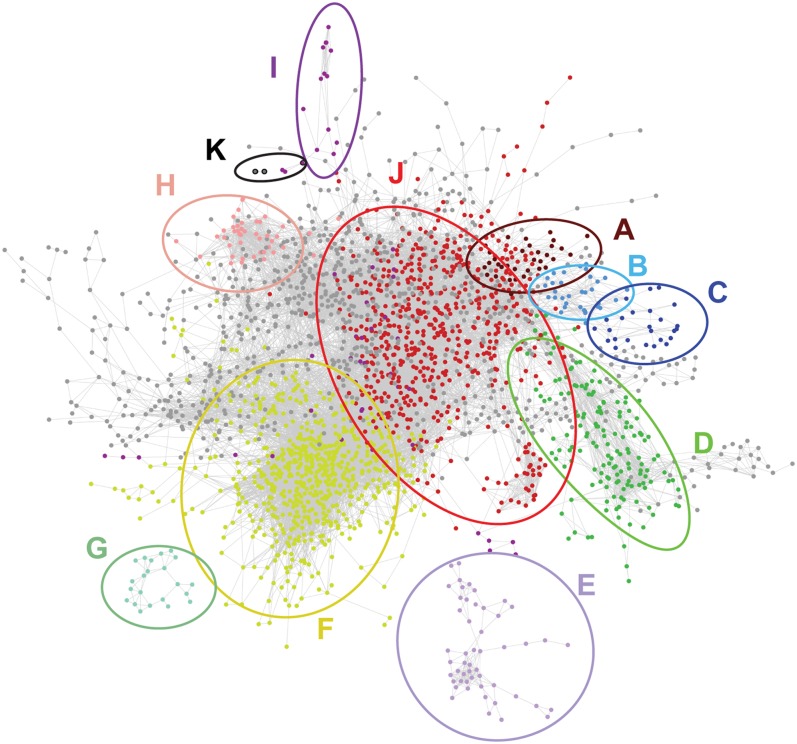
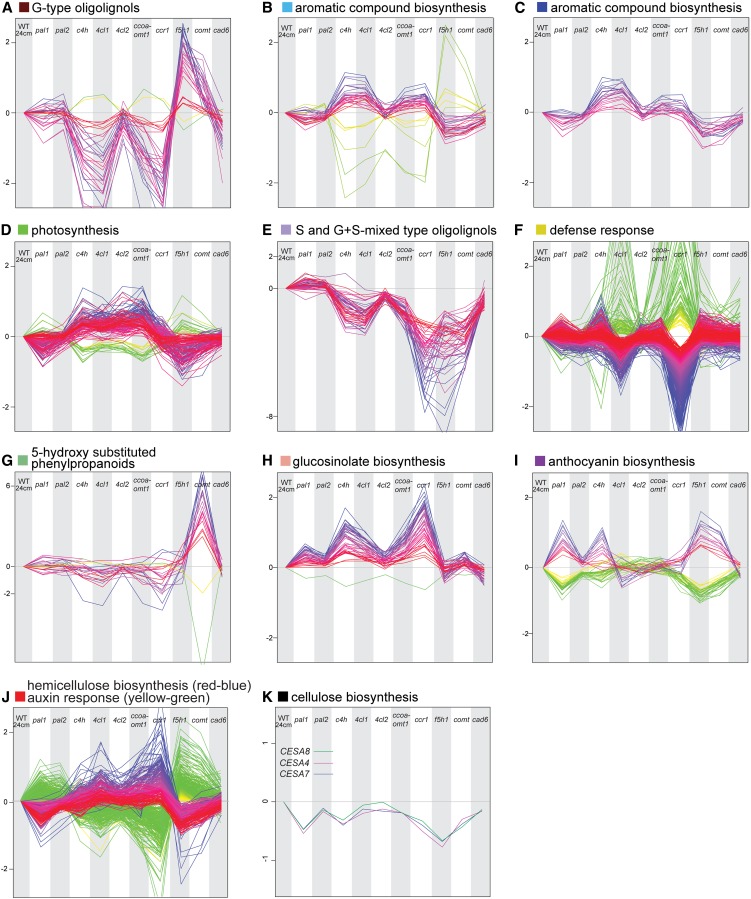
The transcript-metabolite correlation network was based on 3327 probes (12.67% of the PCGs on the array) and 518 compounds that were significantly different in at least one of the mutant lines (see Methods for details). The visualization in Cytoscape (Shannon et al., 2003) revealed that these probes and compounds belonged to different independent networks that consisted of 2778 nodes and 28,206 edges. A total of 2381 of the nodes was connected in one large network with 27,753 edges (Figure 7). To obtain further insight into the substructure of the correlation network, graph-based Heuristic Cluster Chiseling Algorithm (HCCA) clustering was used (Mutwil et al., 2010) to allocate nodes in the network to certain subnetworks (alias clusters), according to their connectivity and, thus, their relative profile (Figures 7 and 8; see Supplemental Data Set 5 online). The enrichment of genes in each of these clusters was determined via BiNGO (Figure 8; see Supplemental Data Set 6 online) (Maere et al., 2005). Two subnetworks enriched in aromatic compound biosynthesis (subnetworks B and C) contained several phenylpropanoid and shikimate pathway genes and genes involved in supplying the methyl-donor S-adenosyl Met (Figure 9). Several genes involved in hemicellulose (mainly glucuronoxylan) biosynthesis were clustered independently (subnetwork J) from phenylpropanoid biosynthetic genes. Cellulose synthase 8 (CESA8, At4g18780), coding for one of the three subunits of the cellulose synthase complex involved in secondary cell wall cellulose biosynthesis, had a profile (subnetwork I) that was different from both hemicellulose and phenylpropanoid biosynthetic genes. Two other genes that are involved in secondary cell wall cellulose biosynthesis (CESA4, At5g44030; CESA7, At5g17420) were correlated with each other (profiles K), but not with any other gene, according to the stringent criteria used to construct the correlation network. Taken together, lignin, cellulose, and hemicelluloses biosynthesis appeared to respond differently to the perturbations, implying that their biosynthetic response to lignin perturbation is largely differentially regulated.
Figure 7.
The Pearson Correlation Network Based on the Abundance Profiles of Transcripts and Metabolites in Mutants and WT24cm.
Each node (dot) represents a probe or a metabolite. Each edge (line) represents a positive or negative correlation between the linked pair of nodes. The network is visualized with the organic layout in Cytoscape (Shannon et al., 2003), so that highly interconnected groups of nodes (that might be considered as subnetworks) are located in the same region. Nodes with the same color belong to the same subnetwork, according to HCCA clustering (Mutwil et al., 2010), and have a significant enrichment for certain GO classes (according to BiNGO) (Maere et al., 2005).
Figure 8.
Abundance Profiles of Transcripts and Metabolites in Subnetworks A to J from the Pearson Correlation Network in Figure 7.
(A) to (J) The color of the squares after the letters A to J is consistent with Figure 7. The two groups of profiles that were negatively correlated with each other, but part of the same subnetwork, are indicated in contrasting colors (i.e., red-blue and yellow-green). The names above the profiles of the nodes within each subnetwork refer to the most significant GO class in case of transcript or to the metabolic class in case of compounds. Unless otherwise indicated, a significant enrichment was found only for the red-blue profiles. WT, the wild type.
(K) The expression profile of genes coding for the three subunits of cellulose synthases involved in secondary cell wall formation.
Figure 9.
List of Genes That Are Coexpressed with Genes Involved in the Phenylpropanoid Biosynthesis, as Derived from the Correlation Network.
Their expressions over wild-type development is given; values and color code are explained in the legend of Figure 6. Full names of the genes are given in Figure 1. WT, the wild type.
Genes involved in photosynthesis were upregulated in c4h, 4cl1, 4cl2, and ccoaomt1 mutants and slightly downregulated in pal1, f5h1-2, and comt mutants (subnetwork D). The transcript levels of genes classified as defense response related by GO annotation were down in 4cl1 and ccr1 mutants but not differential in the other lignin mutants (subnetwork F). Furthermore, transcripts of the glucosinolate biosynthesis genes were more abundant in the low-lignin c4h, 4cl1, ccoaomt1, and ccr1 mutants, whereas transcripts of anthocyanin biosynthesis genes were up in pal1, c4h, f5h1, and comt mutants (subnetworks H and I). Interestingly, subnetwork J was enriched for auxin-response genes; transcripts of many Auxin/Indole-3-Acetic Acid and Auxin Response Factor genes were less abundant in c4h, 4cl1, ccoaomt1, and ccr1 and higher in pal1 and f5h1 (see Supplemental Data Sets 5 and 6 online). In addition, almost all amino acids were represented in the large network and, strikingly, most were higher in abundance in the mutants that had reduced lignin levels (i.e., c4h, 4cl1, ccoaomt1, and ccr1) (see Supplemental Data Set 1 online).
Besides the subnetworks in the large network, some smaller networks were also found, with metabolites and transcripts that were specifically differential in particular mutants. For instance, one small network of 15 nodes contained metabolites that were increased in abundance in the c4h mutants: cinnamic acid, cinnamoyl malate, cinnamoyl Glc, and a number of as yet unknown molecules that were below the detection limit in wild-type plants (data not shown). Furthermore, network G comprised 5-hydroxy substituted phenylpropanoids, which were detected only in comt mutants. Not unexpectedly, the only gene in the network with an opposite profile appeared to be COMT.
Clearly, the impact of a mutation in a gene of the lignin biosynthetic pathway was often more dramatic than expected from the visible phenotype and had more far-reaching consequences than those within the lignin pathway itself.
Low Lignin Mutants Do Not Have More Cellulose
The correlation network described above suggested that cellulose biosynthesis genes were not upregulated in any of the mutants. This was surprising as it has been proposed that a lack of lignin is compensated for by an increase in cellulose (see Discussion). This compensation would require an increase in transcript levels of genes involved in cellulose biosynthesis, a higher translation or passive biosynthesis in which case the protein is not rate limiting. In contrast with what was expected, transcript levels of genes involved in secondary cell wall cellulose biosynthesis were reduced in c4h, 4cl1, ccoaomt1, and ccr1 mutants (Figure 8K). In addition, the reduction was also prominent in pal1, f5h1, and comt mutants that have no reductions in lignin. To investigate further the relationship between lignin and cellulose, we performed cellulose measurements on developing stems of the same set of lignin mutants and demonstrated a significant reduction in cellulose in ccr1, f5h1, and comt mutants but no change in the other mutants (see Supplemental Figure 9 online); similar results were obtained for senesced stems (Rebecca Van Acker, personal communication). Thus, reduced lignin biosynthesis in the low-lignin mutants is not compensated for by increased cellulose biosynthesis.
A Role for TRANSALDOLASE2 in Lignification
Based on the guilt-by-association principle as defined by Saito et al. (2008), our coexpression network provides candidate genes that might encode enzymes active in the general phenylpropanoid or closely related pathways. Indeed, the fact that transcript levels of genes involved in the shikimate and methyl-donor biosynthesis pathways were correlated with transcript levels of genes in the phenylpropanoid pathway provides intrinsic proof that the methodology used is suitable for the selection of novel candidate genes (Figure 9). Therefore, we selected a set of candidate genes from the network that were not yet described to be involved in phenolic biosynthesis (Figure 9). For instance, the two genes annotated as Asp aminotransferases (ASP4, At1g62800; AAT, At2g22250) are likely involved in the aminotransferase step in phenylpropanoid biosynthesis, with Asp as amino group donor and prephenate as amino group acceptor. For AAT, this is in line with its recent in vitro characterization (Graindorge et al., 2010). Two chloroplast-located transporters (PHT4;2, At2g38060; PHT2;1, At3g26570) might be involved in translocating intermediates of the shikimate and Phe biosynthesis (e.g., phospoenolpyruvate, shikimate, and Phe) across the chloroplast membrane (Grace and Logan, 2000; Versaw and Harrison, 2002; Guo et al., 2008). Other genes that were highly coexpressed with known phenylpropanoid genes and for which the homology-based nomenclature suggests a role in phenolic metabolism were annotated as enolase (At1g74030), α/β-hydrolase (At1g19190 and At4g18550), lipase/acylhydrolases (At1g09390 and At1g28610), chalcone-flavanone isomerase (CHI; At5g05270), enoyl-CoA hydratase/crotonase (At1g06550), transaldolase (At5g13420), two dehydrogenases (At5g24760 and At3g19450), and a lysophospholipase (At1g52760). In addition, transcription factors identified here as being coexpressed with genes of the general phenylpropanoid and related pathways are good candidates for regulating these pathways. For example, the coexpressed MYB58 (At1g16490) is already known as a transcriptional activator of lignin biosynthesis in the SND1-mediated transcriptional network regulating secondary cell wall formation (Zhong et al., 2008; Zhou et al., 2009). However, MYB123 (At5g35550) has been described as a master regulator of proanthocyanidin biosynthesis (Debeaujon et al., 2003; Sharma and Dixon, 2005) but our data showing coexpression with genes involved in general phenylpropanoid biosynthesis rather than proanthocyanidin biosynthesis suggest a more general role. For several genes, such as a CCCH-type zinc finger (At1g66810) and WUSCHEL-related Homeobox4 (WOX4; At1g46480), the connection with aromatic metabolism is as yet unclear. Reverse genetics is needed to reveal whether all of these coexpressed genes really play a role in aromatic metabolism.
As a proof of principle, we investigated the knockout mutant of the candidate gene TRANSALDOLASE2 (At5g13420), which we called tra2. The senesced inflorescence stem of tra2 had reduced acetyl bromide lignin levels and an increased S/G ratio (see Supplemental Figure 10 online). In addition, phenolic profiling of developing inflorescence stems showed that the levels of oligolignols were reduced (see Supplemental Figure 10 online). These data suggest a role for TRA2 in (the flux toward) phenolic metabolism, thus supporting the value of the correlation-network approach to discovering genes involved in lignification.
DISCUSSION
In this article, we investigated the system-wide consequences of lignin pathway perturbations in Arabidopsis to obtain a deeper insight into how the lignin biosynthetic pathway is regulated, how it integrates with other biochemical pathways and processes, and how it copes with perturbations. At the same time, our objective was to use this approach to discover new pathways and genes closely connected to lignin biosynthesis. To this end, we analyzed two independent mutant alleles in 10 of the 12 genes involved in developmental lignification in Arabidopsis. Most of the mutations in the pathway did not result in strong morphological or developmental phenotypes. Only hct and c3h1 mutants had to be eliminated from the analyses because all currently available mutant lines are sublethal and unable to develop a significant inflorescence stem.
Although no developmental phenotypes were observed for most of the analyzed mutants, large effects manifested at the transcriptome and metabolome levels. These effects were organized responses of the plant to the pathway perturbations; the two mutant alleles grouped together when the transcriptome and metabolome data sets were hierarchically clustered. Even more interesting was that the system-wide effects were often similar in different mutants, indicating common responses in subsets of the mutants. Those responses and their possible causes are explored in more detail below.
Shikimate, Phenylpropanoid, and Methyl Donor Pathways Are Tightly Coregulated
Previous studies have demonstrated that perturbation of the lignin biosynthetic pathway affects the expression level of other genes on the same pathway (Rohde et al., 2004; Sibout et al., 2005; Shi et al., 2006a; Dauwe et al., 2007; Leplé et al., 2007; Vanholme et al., 2010c). Consequently, feedback and potentially feed-forward regulatory mechanisms must operate. By comparing such responses for mutants in consecutive steps of the same pathway, this study allowed general response mechanisms to be deduced and correlated with metabolic changes. A striking observation was that c4h, 4cl1, ccoaomt1, and ccr1 mutants all had increased transcript levels of several genes of the shikimate, phenylpropanoid, and methyl donor pathways (Figures 6 to 9), in agreement with the role of these pathways in supplying phenylpropanoid and methyl-groups for lignification. Thus, although the shikimate and methyl donor pathways are involved in the biosynthesis of many more metabolites than just monolignols, members of these pathways appear to respond to the same signal, allowing coordinated expression, and illustrating the integration of phenylpropanoid and monolignol synthesis.
What could be the causal reason for the upregulation of transcript levels in these pathways? Our data show that c4h, 4cl1, ccoaomt1, and ccr1 mutants are characterized by reduced levels of monolignol-derived compounds, such as monolignol glucosides and oligolignols. On the other hand, the transcript levels of the pathways that were upregulated in these mutants were reduced in f5h1 and comt mutants, which are characterized by shifts from S to 5H and/or G containing compounds and oligolignols. These observations suggest that the abundance of one or more G-type molecules (e.g., coniferyl alcohol, coniferin, G-type oligomers, or G-type lignin) may regulate the transcript levels of genes from their own biosynthetic pathway (Figure 10). In our model, low levels of G-type molecules result in higher transcript levels of the phenylpropanoid pathway, and high levels result in reduced transcript levels in this pathway. This hypothesis is further strongly supported by increased transcript levels of lignin-related genes in F5H1 overexpressing (C4H:F5H1) and comt C4H:F5H1 Arabidopsis lines where G-type metabolites are replaced by S- and 5H-type metabolites, respectively (Vanholme et al., 2010c), by the increased transcript levels in CCR-deficient tobacco and poplar (Dauwe et al., 2007; Leplé et al., 2007), and by the reduced transcript levels of lignin-related genes in the bm3 (i.e., comt) maize mutants (Shi et al., 2006a). As summarized in Figure 10, in all these cases, it is the level of coniferyl alcohol and other G-type compounds that is inversely correlated to the transcript levels of the shikimate and phenylpropanoid pathway genes. In addition to coniferyl alcohol, the most obvious candidate molecule for such a control mechanism is coniferin, which correlates in abundance with that of coniferyl alcohol in our dataset. Dilignols (or hexosylated dilignols) might also have such function. Further research is needed to test this hypothesis.
Figure 10.
Schematic Overview of Transcriptional Regulation in Different Arabidopsis Lines with Altered Monolignol Biosynthesis.
Black arrows depict the metabolic conversions. Gray dotted lines denote feedback regulation, and a blue cross or red arrow represents knockout or overexpression of genes, respectively. Transcript abundances of genes are represented by framed boxes, whereas monolignol and lignin abundance is represented by unframed ellipses. As for gene expression, red and blue point toward higher and lower abundances, respectively. White stands for no difference, and gray is unknown. Data for C4H:F5H1 and comt C4H:F5H1 are from Vanholme et al. (2010c).
Alternatively, or additionally, the altered structure of the cell wall itself might be the causal signal that initiates transcriptional feedback, as proposed before (Ellis et al., 2002; Zhong et al., 2002; Caño-Delgado et al., 2003; Dauwe et al., 2007; Vanholme et al., 2010c). This hypothesis would be conceivable for the series of c4h, ccoaomt1, ccr1, and comt C4H:F5H mutants that have lower lignin content and for which alterations in cell wall structure have already been demonstrated (Jones et al., 2001; Do et al., 2007; Mir Derikvand et al., 2008; Schilmiller et al., 2009). However, it does not explain why f5h1 and comt mutants with normal lignin levels downregulate the phenylpropanoid pathway. This would imply that the sensing mechanism is able to differentiate between different lignin compositions, which we believe is a less attractive hypothesis than the soluble G-type compound as a regulator of the phenylpropanoid pathway.
Specific Gene Family Members Respond to the Lignin Pathway Perturbations
Notably, genes that are coexpressed with phenylpropanoid biosynthesis genes in the mutant samples are not necessarily coexpressed with phenylpropanoid biosynthesis genes during wild-type development (Figures 6 and 9). Evidence from external expression data sets of wild-type Arabidopsis (Schmid et al., 2005; Mutwil et al., 2008) demonstrates that several genes expressed upon pathway perturbations are relatively weakly expressed in inflorescence stems compared with other organs. For instance, α/β hydrolase (At4g18550) is specifically expressed in petals, whereas two genes that are annotated as unknown (At5g42690 and At2g14520) are normally specifically expressed in roots, according to the AtGenExpress tissue atlas (Schmid et al., 2005; Mutwil et al., 2008). This suggests that these three genes are recruited into phenylpropanoid biosynthesis in response to pathway perturbations but that they have other roles in wild-type plants under normal conditions. This is also seen within phenylpropanoid biosynthesis itself, specifically with PAL2, 4CL2, and CCR2 (Figure 6). Whereas PAL1, 4CL1, and CCR1 are prominently expressed during developmental lignin biosynthesis, these gene family members appear to respond less (or not at all) to pathway perturbations. PAL2, 4CL2, and CCR2, on the other hand, are prominently upregulated in the low-lignin mutants. Strikingly, pal2 and 4cl2 mutants neither resulted in reduced lignin levels nor did they affect transcript levels of the phenylpropanoid biosynthetic genes. Thus, likely, PAL2, 4CL2, and CCR2 are upregulated in an attempt to compensate for the reduced lignin levels in c4h, 4cl1, ccoaomt1, and ccr1 mutants. A similar situation is observed for the laccases LAC4/IRX12 and LAC17. Both are involved in lignin polymerization in the Arabidopsis stem (Brown et al., 2005; Berthet et al., 2011), but only LAC17, not LAC4, had a tendency to be upregulated in some of the lignin-deficient mutants (see Supplemental Figure 7 online).
Coniferin and Syringin as Storage Metabolites
The biological role of 4-O-glucosylated coniferyl and sinapyl alcohol (coniferin and syringin, respectively) is not yet understood (Escamilla-Treviño et al., 2006; Lanot et al., 2006; Vanholme et al., 2010b). According to current models, they might function either as storage or as transport forms (or both) of their corresponding aglycones, although no experimental proof is available (Samuels et al., 2002; Kaneda et al., 2008; Liu et al., 2011). The transcript data suggest that the biosynthesis of coniferin and syringin competes with the biosynthesis of lignin (i.e., their biosynthesis via UGT72E2 and UGT72E3 is disfavored at the moment that there is a need for lignin biosynthesis) (see Supplemental Data Set 4 online). This observation is in agreement with the hypothesis that coniferin and syringin are storage forms of their aglycones and contradicts the idea that they are needed for transport (in which case the UGT72Es would be expected to be upregulated).
New Candidate Genes Involved in Monolignol Biosynthesis and Coupling
We used our coexpression analysis to identify genes that might have as yet unappreciated roles in phenylpropanoid or related metabolism (Figure 9). As a proof of principle, we investigated a T-DNA insertion mutant of one of these genes, TRA2. Arabidopsis has two transaldolase isoforms that share low sequence similarity (Caillau and Paul Quick, 2005). Both transaldolases catalyze a plastid-localized reaction of the nonoxidative branch of the oxidative pentose phosphate pathway, which gives rise to erythrose-4-phosphate, a precursor of the shikimate pathway (Kruger and von Schaewen, 2003). The oxidative pentose phosphate pathway has a central role in both primary and secondary metabolism (Caillau and Paul Quick, 2005) that has been illustrated by the effects on photosynthesis, sugar levels, and phenolic compounds provoked by disturbing the flux through this pathway in tobacco (Henkes et al., 2001).
Strikingly, although this enzyme acts at an early stage in the supply of shikimate pathway precursors, lignin amount and oligolignol levels were reduced in tra2, whereas no growth abnormalities were seen (see Supplemental Figure 10 online). Although further research is needed to determine the exact role of TRA2, these initial results prove a role for this gene in influencing the flux toward lignin and illustrate the power of our approach for identifying novel players in lignification and phenolic metabolism.
Pathways for Overaccumulating Phenylpropanoids
The accumulation of free phenylpropanoic acids and their derivates (4-O-etherified to hexose and 9-O-esterified to hexose and malic acid) in c4h, 4cl1, ccoaomt1, ccr1, f5h1, comt, and cad6 mutants reveals common biological mechanisms to cope with the accumulation of phenylpropanoids upon pathway perturbations (Figures 5, 7, and 8; see Supplemental Figure 5 and Supplemental Data Set 1 online). Hexose adducts of xenobiotics are generally considered as being detoxification products (Coleman et al., 1997). Our data suggest that in addition to hexosylation of xenobiotics, 4-O- and 9-O-hexosylation of accumulating phenylpropanoids represent a detoxification strategy. Transgenic Arabidopsis that upregulated F5H in a comt mutant background overproduced 5-hydroxyphenylpropanoids, some of which were also 4-O- and 9-O-hexosylated, or hexosylated at both 4-O and 9-O positions (Vanholme et al., 2010c). The absence of a general increase in expression of genes that might catalyze the 9-O-esterification or 4-O-addition (i.e., SGTs and UGT72s) in c4h, 4cl1, ccoaomt1, ccr1, f5h1, comt, and cad6 mutants compared with the wild type (see Supplemental Data Set 4 online) implies that biosynthesis of 9-O-hexoside and 4-O-glucoside phenylpropanoic acids may occur via a so-called passive metabolic route (Vanholme et al., 2010c). Alternatively, hexosylation of accumulating phenylpropanoic acids is regulated posttranscriptionally or might occur via other UDP-glucosyltransferases.
Alternative Biosynthetic Routes to Ferulic Acid in c4h, 4cl1, ccoaomt1, and ccr1 Mutants
Ferulic acid and several ferulic acid derivatives accumulated in c4h, 4cl1, ccoaomt1, and ccr1 mutants, while in the same mutants, the transcript level of hydroxycinnamaldehyde dehydrogenase (HCALDH, At3g24508) was reduced or not differential and the level of coniferaldehyde was reduced (Figure 5; see Supplemental Figure 5 and Supplemental Data Sets 1 and 4 online). This observation contradicts the current model of the pathway in the wild type (Nair et al., 2004) and indicates the existence of HCALDH-independent routes toward ferulic acid. A first alternative route might take place in the ccr1 mutant. The most evident route toward ferulic acid is a one-step reaction from feruloyl-CoA (the CCR1 substrate), possibly committed via a thioesterase (Dauwe et al., 2007; Mir Derikvand et al., 2008), as the nonenzymatic autohydrolysis of thioesters is slow at neutral pH (Bracher et al., 2011).
A second alternative route toward ferulic acid must occur in 4cl1 and ccoaomt1 mutants. Here, the biosynthesis of ferulic acid most likely occurs via caffeic acid. In the case of the ccoaomt1 mutant, caffeic acid might be formed from caffeoyl-CoA via a thioesterase, a biosynthetic route that was also suggested to occur in CCoAOMT-downregulated alfalfa (Medicago sativa) and poplar (Meyermans et al., 2000; Guo et al., 2001; Chen et al., 2006). In the case of the 4cl1 mutant, caffeic acid might be directly derived from p-coumaric acid; although p-coumaric acid is a poor substrate for C3H1 in Arabidopsis, and not a substrate in poplar (Schoch et al., 2001; Franke et al., 2002; Chen et al., 2011), it has recently been shown that the poplar C4H and C3H form a complex and that this complex is able to convert p-coumaric acid to caffeic acid (Chen et al., 2011). In both cases, COMT is able to methylate caffeic acid to form ferulic acid (Goujon et al., 2003b; Do et al., 2007).
The accumulation of ferulic acid and feruloyl hexose in c4h mutants was most surprising and cannot be properly explained at this moment. Since these are not knockout mutants (Schilmiller et al., 2009), perhaps the residual flux that passes C4H is predominantly used for the biosynthesis of ferulic acid metabolites, instead of monolignols. However, the low lignin levels in c4h mutants would be expected to signal a higher demand for monolignols. Alternatively, ferulic acid might be synthesized via an alternative route, without the need for C4H activity. Metabolic flux analyses are needed to reveal such a pathway, if it exists.
Lignin Mutants Do Not Compensate for Low Lignin by Increased Cellulose Biosynthesis
The potential crosstalk between the biosynthesis of the different cell wall polymers is a hot topic of discussion. Based on the concomitant increase in cellulose and decrease in lignin in 4CL downregulated poplar trees, it was proposed that a lack of lignin was compensated for by an increase in cellulose (Hu et al., 1999; Li et al., 2003). Consistent with this hypothesis, COMT-downregulated poplar trees with reduced lignin levels were also increased in cellulose (Jouanin et al., 2000).
Lignin amount and oligolignol levels were severely reduced in c4h, 4cl1, ccoaomt1, and ccr1 mutants (Figure 5; see Supplemental Data Set 1 and Supplemental Figures 5 and 6 online), but in these same mutants, transcript levels of genes involved in secondary cell wall cellulose biosynthesis were reduced (Figure 8K). Importantly, cell wall analysis of inflorescence stems proved that cellulose amounts were significantly reduced in ccr1, f5h1, and comt mutants, and there was no increase in cellulose amounts in any of the lignin mutants analyzed (see Supplemental Figure 9 online). Thus, our data do not support the idea that reduced lignin biosynthesis in c4h, 4cl1, ccoaomt1, and ccr1 mutants might be compensated for by increased cellulose biosynthesis, at least not in Arabidopsis.
System-Wide Effects in Lignin Mutants
The correlation network revealed the system-wide effects of the mutations and prompts some striking observations that cannot be fully explained with current knowledge. One observation was that the transcript levels of genes involved in glucosinolate biosynthesis were increased in the very same mutants in which those of lignin biosynthesis were increased (i.e., c4h, 4cl1, ccoaomt1, and ccr1), suggesting that both pathways respond (at least partly) to a common signal (Figure 8H). This could be a G-type metabolite or a signal derived from aberrant cell walls as during herbivore attack when glucosinolates are induced for chemical defense (Raybould and Moyes, 2001; Malitsky et al., 2008).
Similarly, almost all detected amino acids were higher in abundance in the same set of low-lignin mutants (i.e., c4h, 4cl1, ccoaomt1, and ccr1) but were unaltered in the mutants with a shift in lignin composition (f5h1 and comt) (see Supplemental Data Set 1 online). This suggests another response to G-type metabolites or to the aberrant cell wall of lignin mutants. However, the biological role for this accumulation remains speculative.
Another intriguing observation is the higher expression of nuclear genes encoding chloroplast-localized proteins involved in photosynthesis in c4h, 4cl1, 4cl2, and ccoaomt1 mutants compared with WT24cm (Figure 8D). These observations are indicative of enhanced carbon assimilation in these mutants. The effect on photosynthesis gene expression might arise from increased shikimate biosynthesis in the chloroplast, but this explanation could not hold for the complete set of low-lignin mutants. Remarkably, 45% of differentially expressed genes in the 4cl2 mutant were related to processes in the chloroplast, and the mutant has neither an obvious lignin phenotype nor differences in the phenolic pool. This suggests that 4CL2 does not have a clear role in stem lignin biosynthesis but rather has a role in chloroplast-related processes.
Curiously, expression levels of genes involved in the defense response were significantly lower in 4cl1 and ccr1 mutants (Figure 8F). Among others, 25 nucleotide binding site and Leu-rich repeat proteins and 14 receptor-like proteins and protein kinases were downregulated in these mutants (see Supplemental Data Set 5 online). By contrast, transcripts for glucosinolate metabolism were more abundant in both 4cl1 and ccr1 mutants than in the wild type, as discussed above. Thus, 4cl1 and ccr1 downregulate a specific part of the defense response. These observations may add small pieces to the complex puzzle of why some lignin mutants develop growth abnormalities whereas other mutants do not.
Conclusion
We have shown that the system-wide analysis of perturbations in consecutive steps of a metabolic pathway, using a combination of transcriptomics, metabolomics, and MS2 discovery of new metabolites provides insight into the regulatory circuit of the perturbed pathway. Furthermore, the approach discloses branching pathways, suggests the existence of alternative pathways, and provides candidate genes involved in them. In addition, our data show how plants respond as a biological system to these perturbations. Because most of the mutants do not have an obvious visible phenotype, the system-wide responses observed at the transcriptome and metabolome level likely result from the plant’s attempt to compensate for the mutation, a phenomenon known as phenotypic buffering (Fu et al., 2009). Understanding these buffering mechanisms may help in designing plants with low lignin yet normal development in the transition toward a bio-based economy.
METHODS
Plant Lines
c4h-2 (ref3-2), c4h-3 (ref3-3), and f5h1-2 (fah1-2) are ethyl methanesulfonate–generated mutants (Chapple et al., 1992; Meyer et al., 1996; Ruegger et al., 1999; Ruegger and Chapple, 2001; Schilmiller et al., 2009). The following T-DNA insertional mutants from SALK (Alonso et al., 2003), GABI-Kat (Rosso et al., 2003), SM/SLAT (Tissier et al., 1999), and SAIL collections (Sessions et al., 2002) were ordered via the European Arabidopsis Stock Centre: pal1-2 and -3, pal2-2 and -3, 4cl1-1 and -2, 4cl2-1, ccoaomt1-3, comt-1 and -4, f5h1-4, cad6-1 and -4, ccr1-6, and tra2 (SALK_094724). The mutants 4cl2-3 and ccoaomt1-5 were delivered as homozygotes by the GABI-Kat project. ccoaomt1-3, ccr1-3 (ccr1s), and ccr1-6 (ccr1g) were provided by Lise Jouanin as homozygous seed stocks (Mir Derikvand et al., 2008; Ruel et al., 2009). Primer sequences for genotyping are given in Supplemental Table 2 online. In case of c4h-2, segregating seed stocks of a heterozygous parental line were kept due to sterility of the homozygous plant.
Growth Conditions
Twenty independent mutant lines and wild-type plants were germinated on soil (Saniflor ref. 252020) supplemented with 10% (volume) vermiculite. Short-day conditions were 9 h light/15 h dark, and long-day conditions were 16 h light/8 h dark.
Biological Material
Inflorescence stems were flash-frozen in liquid nitrogen. The basal 1 to 9 cm of individual inflorescences was ground in 2-mL Eppendorf tubes using a Retsch mill (20 Hz, 5-mm bead). For metabolite profiling, 10 to 13 stems of each genotype/developmental stage were used individually. For microarray analysis, aliquots of a minimum of eight different samples of the same genotype/developmental stage were pooled to obtain one biological sample. For each genotype/developmental stage, two biological replicates were prepared for microarray analysis.
Metabolite Extraction
For each mutant and wild-type developmental stage, ground plant material was individually extracted with 500 μL methanol (15 min, 70°C, 1000 rpm), containing internal standards (o-anisic acid, 2.4 mg/L; 4-hydroxyphenyl-2-butanon, 6 mg/L; methyl-5-acetylsalicylate, 10 mg/L; methyl nonadecanoate, 16 mg/L). The cooled extract was loaded on a 60-mg Varian Bond Elut Plexa (polystyrene-divinylbenzene) column, pre-equilibrated with methanol, and subsequently washed with 400 μL methanol. The combined phases were split into 200 μL for GC-MS analysis and 700 μL for UPLC-MS analysis. Both fractions were freeze-dried.
UPLC-MS Analysis
Samples were redissolved in 80 μL water and transferred to an injection vial. Fifteen microliters of each sample was injected on a Waters Acquity UPLC system equipped with an Acquity UPLC BEH C18 (2.1 × 100 mm, 1.7 µm) column. A gradient of two buffers was used: buffer A (100/1/0.1 water/acetonitrile/ammonium acetate [2 M], pH 5), buffer B (100/1/0.1 acetonitrile/water/ammonium acetate [2 M], pH 5); 95% A for 0.1 min decreased to 55% A in 36.9 min (200 µL/min, column temperature 40°C). A UV/Vis absorption spectrum was measured (190 to 600 nm). Atmospheric pressure chemical ionization, in the negative ionization mode, was used to couple UPLC with the ion trap mass spectrometry instrument (LCQ Classic; ThermoQuest; vaporizer temperature 450°C, capillary temperature 150°C, source current 5 mA, sheath gas flow 21, auxiliary gas flow 3, mass range 135 to 1000 atomic mass units).
Peaks were integrated and aligned using MetAlign software (http://www.pri.wur.nl/UK/products/MetAlign; Lommen (2009)), with the following parameters: retention begin = 1, retention end = 2500, max amplitude = 109, peak slope factor = 1.0, peak threshold factor = 1.5, average peak width at half height = 10, no scaling, begin of first region = 0 (max shift 40), end of first region = 2800 (max shift 60), no prealign processing. In Excel, peaks that were not present in all biological replicates of at least one single mutant were removed. Also, the front signal (<3 min) was removed. Based on retention time and the intensity, an in-house R script grouped peaks that were derived from the same compound. The final list of compounds was manually correct.
For phenolic profiling of the tra2 mutant and the targeted analysis of caffeoyl Glc and caffeoyl malate, similar buffers were used as above, but with 0.1 formic acid (pH 2) instead of ammonium acetate. Separation was done as above on a Waters Acquity UPLC system, but coupled to a Synapt Q-Tof (Waters).
For accurate mass data, one sample of each genotype and developmental stage was injected and analyzed via UPLC-Fourier transform-ion cyclotron-MS (LTQ FT Ultra; Thermo Electron) with electrospray ionization (ESI) in the negative mode according to Morreel et al. (2010a), but with 0.1 acetic acid, pH 4, in buffers A and B instead of formic acid.
GC-MS Analysis
A procedure adapted from Roessner et al. (2000) was used. Samples were successively derivatized by methoxyamine hydrochloride (Meox) and N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA): 40 μL Meox solution (48 mg Meox in 2.4 mL pyridine) was added to the dried extracts, and the samples were incubated for 1.5 h at 30°C. Forty-five microliters of MSTFA mix (2400 μL MSTFA + 300 μL alkane mixture) was added, and the samples were incubated for another 0.5 h at 37°C. One microliter was splitless injected in a HP6890 gas chromatograph. Inlet temperature was 230°C. A constant He flow (1 mL/min) was used on the Varian FactorFour capillary column VF-5ms (5% phenyl and 95% dimethylpolysiloxane). Oven temperature was 70°C for the first 5 min, increased with 5°C/min to 325°C and kept for 1 min. The HP5973 quadrupole mass detector coupled to the gas chromatograph performed electron impact ionization (60 to 600 atomic mass units; 7.8 to 68.8 min).
Integration and alignment were performed using the R xcms package (Smith et al., 2006) with following parameters for the integration: FWHM = 3, max = 300, snthresh = 3, step = 0.1, steps = 2, mzdiff = 0.5; and for the alignment: bw = 2, minfac = 0.3, max = 300. Peakfilling was used. In Excel, peaks were manually curated. Peaks were manually annotated by use of the GMD@CSB.DB library from the Max Planck Institute for Plant Physiology (Golm, Germany) (Stein, 1999; Kopka et al., 2005) and AMDIS software (Davies, 1998).
RNA Isolation
Total RNA was prepared from 100 mg ground plant material (see above) using the mirVanaTM miRNA isolation kit (Ambion), recovering a fraction of microRNA and a fraction of total RNA (200-bp cutoff).
Microarray Design
Two-color full-genome Agilent Arabidopsis 3 oligo arrays were used. These contain 32,221 unique probes, representing 25,094 genes. The MicroArray Quality Control consortium reported several times the good quality of the Agilent two-color arrays (Kuo et al., 2006; Patterson et al., 2006; Shi et al., 2006b). The transcripts of two independent biological replicates (pools; see above) of 20 mutants and four wild-type developmental stages were measured. Each pool was measured twice (once Cy3 and once Cy5 labeled), except for WT24cm and WT32cm, which were measured six times. The samples were hybridized in two sets, both containing the two biological replicates of WT24cm, WT32cm, and of one mutant allele for each gene of interest. The first set was further extended with the replicates of WT8cm and WT16cm (see Supplemental Figure 11 online). Preparation and labeling of the samples, hybridization, washing, scanning, and feature extraction of Agilent 60-mer oligonucleotide microarrays was conducted at the VIB microarray facility (http://www.microarrays.be) and conducted according to the manufacturer (Agilent).
Growth Statistics
The height of plants in the eight trays was measured daily. Two replicates of each mutant and 10 replicates of the wild type were grown in each tray. Data were analyzed with longitudinal statistics in R (version 2.6.2.). Data from 4 until 12 d were used to detect lines with different growth rate. For the covariance, an autoregressive structure with heterogeneity was chosen based on the Akaike information criterion: gls(height∼time+line+time:line, correlation=corAR1(form=∼1|line), weights=varIdent(form=∼1|T), data=data, na.action=na.exclude). Data from 27 until 44 d were used to detect lines with different final height. Again, based on Akaike information criterion, an autoregressive structure fitted best, and the following command was used: gls(height∼time+line, correlation=corAR1(form=∼1|line), data=data, na.action=na.exclude). The Benjamini and Hochberg multiple test correction was calculated with the function mt.rawp2adjp(proc=”BH”) in the package multtest.
Microarray Analysis: Linear Mixed Model
For each array, standard Pearson correlations were calculated between background log2 ratios and foreground log2 ratios (Scharpf et al., 2007). Subsequently, Loess normalization was performed on log2 transformed signals using the SAS Loess procedure (version 9.1.2). Loess normalized expression data were further analyzed using two interconnected analysis of variance models according to Wolfinger et al. (2001). The SAS proc mixed procedure was used to fit the parameters. The normalization model was:
where ygijklm is the log2 of the Loess normalized intensity measurements of probe g, array i, dye j, variety k, biological replicate l, and loop m. μ represents an overall mean value, L is the (fixed) main loop effect, A is the (random) effect for arrays, nested within loop, and r is the error. Ai(Lm) and rgijklm are assumed to be normally distributed random variables and to be independent both across their indices and with each other. A random interaction effect between dye and array and a fixed main dye effect were left out of the model after testing the significance with a likelihood ratio test and an F test, respectively. The model adopted at the probe level was:
where μ′ represents the mean value per probe, L the probe-specific loop effect, A the probe-specific array effects reflecting the spot-to-spot variability inherent in spotted microarray data, D the probe-specific dye effects, V the probe-specific variety effects, nested under loop, and B the probe-specific replicate effects, nested under variety. Ai(Lm), Bl(Vk), and εijklm are assumed to be normally distributed random variables and to be independent both across their indices and with each other. Restricted maximum likelihood was used to estimate the variance components. For the probe model, type III F tests and P values were calculated for the variety terms. The Kenward-Roger method (Kenward and Roger, 1997), recommended by Verbeke and Molenberghs (2009) when sample sizes are small at each level of the data set, was used to calculate denominator degrees of freedom for these approximate F tests. False discovery rate (FDR)–adjusted P values, described by Benjamini and Hochberg (1995), were calculated. The estimates of primary interest are those of the variety [Vk(Lm)] effects. Differences of the estimates between all mutant lines and WT24cm for the variety effects along with mixed-model based t tests and P values were calculated for those probes that had a significant FDR-adjusted P value for the F tests (fdr≤0.01). Given the design (see Supplemental Figure 11 online), there is a confounding effect between loop and variety. For WT24cm and WT32cm, two estimates were obtained, one for each loop. Lines from loop 1 were compared with WT24cm from loop 1, and lines from loop 2 were compared with WT24cm from loop 2.
On those probes that had a significant FDR-adjusted P value for the F tests (FDR ≤ 0.01) the balanced decision strategy (Moerkerke and Goetghebeur, 2006) was conducted. A target alternative of 1 was chosen for each probe. A probe signal was considered to be significantly differential between a mutant line and WT24cm when the R-ratio was >1. A weight ratio of 10 was used. When taking the average of two mutant lines, significances were calculated as follows: all significantly differential probe signals from one line that had a difference in least square means estimates of at least 0.3 in the same direction in the other line were retained as significant differential and vice versa. The choice of −0.3 and +0.3 corresponds with the 90th percentile of the absolute values of all differences in least square means estimates.
GC-MS and UPLC-MS Analysis: Linear Mixed Model
For both GC-MS and UPLC-MS, peaks were normalized to dry weight. The following normalization model was adopted on the combined data set of GC-MS and UPLC-MS:
where yijkl is the base-2 logarithm of the normalized peak area of metabolite l for run i, number in the run j, and mutant/wild-type developmental stage (here called variety) k. μ represents an overall mean value, R is the main run effect, N is the effect for the location in the run, nested within run, and ε the error term. Ri, Nj(Ri), and rijkl are assumed to be normally distributed random variables with zero means and to be independent both across their indices and with each other. The metabolite model is described as follows:
where rijk represents the conditional residual from the previous model, μ′ the mean value per metabolite, R the metabolite-specific run effect, Loc the metabolite-specific effect for the location in the run, nested within run, and V the metabolite-specific variety effects. Loc is an interval variable created from position within the run (positions 1 to 9, 10 to 18, 19 to 27, and 28 to 36) to avoid overparameterization of the model. Ri, Locj(Ri), and γijkl are assumed to be normally distributed random variables with zero means and to be independent both across their indices and with each other. They have different variances across the metabolite index l. Residual analysis showed many outliers. This was due to incorrect peak identification of the Metalign software, resulting in erroneous zero values. Therefore, all observations with a residual value less than −4 were set to zero and subsequently estimated with the k-nearest neighbor procedure with k = 10 (R package impute.knn). Further analysis, including multiple testing correction, calculation of the differences of the least-squares means for the variety effects, and balance test, was performed as for the microarray analysis.
Hierarchical Clustering of Samples
Hierarchical clustering was performed with Ward’s method using the dist and agnes function from the R cluster package. The distance metric used was the Euclidean distance. As input data, the differences in estimates between all lines and WT24cm for the significantly differential probes and metabolites were used.
Probe and Metabolite Clustering on Wild-Type Profiles
Clusters based on the expression in wild-type developmental stages were generated in Excel according to the formulas given in Supplemental Data Set 2 online.
Correlation Network
Because the correlation of profiles was often biased by extremely high or low expression/abundance values in a few mutants with a clear developmental effect (such as c4h-2 and ccr1-3), these samples were left out from the analysis. Despite its aberrant development, the weaker ccr1-6 allele was retained in the sample set to keep information of at least one ccr1 mutant. As the number of samples for c4h and ccr1 mutants was now reduced to one, and since bias toward certain samples during the correlation analysis needs to be avoided, data from the two alleles of all other mutants were merged by taking the average value of each transcript and each metabolite of both alleles. For pal1, pal2, 4cl1, 4cl2, ccoaomt1, comt, and cad6 mutants, this was justified by their similarity, as demonstrated by the hierarchical clustering of samples (Figure 4). As explained in the Supplemental Text 1 online, f5h1-4 was left out. This selection procedure resulted in a set of 11 samples: the average of both mutant alleles of pal1, pal2, 4cl1, 4cl2, ccoaomt1, comt, and cad6, as well as c4h-3, ccr1-6, f5h1-2, and the reference sample WT24cm.
The correlation network was constructed with transcripts and metabolites that were significantly different in at least one of the averaged values of the two mutant alleles of each gene compared with WT24cm, with the exception of transcripts of the f5h1 mutant, which were used if they were significantly different in f5h1-2 (in total 3327 probes, 196 GC-MS, and 324 UPLC-MS compounds). For those genes that were mutated, the expression in the corresponding mutant was replaced via the k-nearest neighbor procedure with k = 10 (R package impute.knn), which led to 10 extra imputed probes. A relevance network was constructed using a statistically sound two-stage coexpression detection algorithm (Zhu et al., 2005). The following parameters were used: FDR α = 0.05 and minimal acceptable strength 0.7. Two measures of association were considered: the Pearson correlation and the Kendall’s tau coefficient. Calculations were done with the R GeneNT package. The complete network was visualized with Cytoscape 2.6.1 (organic layout) (http://www.cytoscape.org/; Shannon et al., 2003) and is provided as Supplemental Data Set 7 online. Subnetworks were generated using HCCA clustering (Mutwil et al., 2010). For BiNGO analysis (Maere et al., 2005), each subcluster was split in two sets (if appropriate) according to their relative expression profile (positively and negatively correlated nodes, respectively). A corrected P value cutoff of 0.01 was used for enrichment, and the top 11 subclusters are presented in Figures 6 and 7.
Cell Wall Analysis
The acetyl bromide protocol published by Dence (1992), optimized for small sample quantities of Arabidopsis thaliana, was used for lignin quantification. Cellulose amount was determined via the sulfuric acid-phenol colorimetric method based on the methods of DuBois et al. (1956) and Masuko et al. (2005).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the accession numbers listed in Table 1. Microarray data are available in *.xlsx format at http://www.datadryad.org. under accession number doi:10.5061/dryad.jn16g and in the http://www.ebi.ac.uk/arrayexpress/ database under accession number E-MTAB-1284.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Representation of the 20 Mutant Genes Used in This Study.
Supplemental Figure 2. Growth Characteristics of the 20 Arabidopsis Lines Given in Table 1.
Supplemental Figure 3. Number of Transcripts, Compounds Detected by GC-MS, and Compounds Detected by UPLC-MS That Are Differential in One or Multiple Mutants.
Supplemental Figure 4. MS2 Spectra and the Reasoning for the (Tentative) Structural Identification of 34 Metabolites of Which the MS2 Spectra Were Not Published Before.
Supplemental Figure 5. Metabolic Shifts in Phenolic Metabolism of the Lignin Mutants and the Wild-Type Developmental Stages.
Supplemental Figure 6. Lignin Amount in the Inflorescence Stem of the Wild Type and Mutants of the Lignin Biosynthetic Pathway.
Supplemental Figure 7. Transcript Levels of Genes Coding for Laccases and Class III Peroxidases.
Supplemental Figure 8. Targeted Analysis of Caffeoyl Glucose and Caffeoyl Malate in ccoaomt1 Mutants.
Supplemental Figure 9. Cellulose Amount in the Inflorescence Stem of the Wild Type and Mutants of the Lignin Biosynthetic Pathway.
Supplemental Figure 10. Reverse Genetics Suggest a Role for TRANSALDOLASE2 (At5g13420) in Lignification.
Supplemental Figure 11. Microarray Design.
Supplemental Table 1. GSEA on the Pathway from PAL to CCR.
Supplemental Table 2. Sequence of Primers Used for Genotyping the Mutants.
Supplemental Text 1. The Aberrant Transcriptional Response of f5h1-4 Mutants.
Supplemental Data Set 1. Abundance of All Molecules Detected by GC- and UPLC-MS in Each of the Lignin Mutants and Wild-Type Developmental Stages, Relative to WT24cm.
Supplemental Data Set 2. Wild-Type Developmental Clusters.
Supplemental Data Set 3. Gene Ontology Analysis of Wild-Type Developmental Clusters.
Supplemental Data Set 4. Transcripts for Phenolic Metabolism in Each of the Mutants and Wild-Type Developmental Stages Compared with WT24cm.
Supplemental Data Set 5. Subnetworks in the Correlation Network of Transcripts and Metabolites in the Mutant Samples.
Supplemental Data Set 6. Gene Ontology Analysis of Subnetworks in the Correlation Network of Transcripts and Metabolites in the Mutant Samples.
Supplemental Data Set 7. The Correlation Network of Genes and Metabolites as a Cytoscape-Compatible .csv File.
Acknowledgments
We thank Clint Chapple for providing the fah1-2, c4h-2 (ref3-2), and c4h-3 (ref3-3) lines, Lise Jouanin for providing ccoaomt1-3, ccr1-3, and ccr1-6 lines, Bart Ivens for technical assistance, Martine De Cock and Annick Bleys for help in preparing the article, and Eric Messens for useful discussions. We acknowledge partial funding through the Research Foundation-Flanders (Grant G.0352.05N), the European Commission’s Directorate General for Research within the 7th Framework Program (FP7/ 2007-2013) under Grant 211982 (RENEWALL), the Hercules program of Ghent University for the Synapt Q-Tof (Grant AUGE/014), the “Bijzondere Onderzoeksfonds-Zware Apparatuur” of Ghent University for the Fourier transform-ion cyclotron-MS instrument (174PZA05), and the Multidisciplinary Research Partnership ‘Biotechnology for a Sustainable Economy’(01MRB510W) of Ghent University. R.V. is indebted to the Agency for Innovation by Science and Technology for a predoctoral fellowship and the Research Foundation-Flanders for a postdoctoral fellowship.
AUTHOR CONTRIBUTIONS
R.V., A.R., and W.B. designed the research. R.V., V.S., and L.S. performed research. R.V., K.M., G.G., and J.H.C. contributed analytic tools. V.S. contributed statistical analyses. R.V., B.V., K.M., and W.B. analyzed the data. R.V., B.V., C.H., A.R., K.M., and W.B. wrote the article.
Glossary
- PCG
protein coding gene
- GC-MS
gas chromatography–mass spectrometry
- UPLC-MS
ultrahigh performance liquid chromatography–mass spectrometry
- GO
Gene Ontology
- MS2
to be defined
- HCCA
to be defined
- Meox
methoxyamine hydrochloride
- MSTFA
N-methyl-N-(trimethylsilyl)trifluoroacetamide
- FDR
false discovery rate
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baucher M., Halpin C., Petit-Conil M., Boerjan W. (2003). Lignin: Genetic engineering and impact on pulping. Crit. Rev. Biochem. Mol. Biol. 38: 305–350 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 57: 289–300 [Google Scholar]
- Berthet S., Demont-Caulet N., Pollet B., Bidzinski P., Cézard L., Le Bris P., Borrega N., Hervé J., Blondet E., Balzergue S., Lapierre C., Jouanin L. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright J., Negre F., Chen X., Kish C.M., Wood B., Peel G., Orlova I., Gang D., Rhodes D., Dudareva N. (2004). Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 135: 1993–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonawitz N.D., Chapple C. (2010). The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annu. Rev. Genet. 44: 337–363 [DOI] [PubMed] [Google Scholar]
- Bracher P.J., Snyder P.W., Bohall B.R., Whitesides G.M. (2011). The relative rates of thiol-thioester exchange and hydrolysis for alkyl and aryl thioalkanoates in water. Orig. Life Evol. Biosph. 41: 399–412 [DOI] [PubMed] [Google Scholar]
- Brown D.M., Zeef L.A.H., Ellis J., Goodacre R., Turner S.R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillau M., Paul Quick W. (2005). New insights into plant transaldolase. Plant J. 43: 1–16 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A., Penfield S., Smith C., Catley M., Bevan M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34: 351–362 [DOI] [PubMed] [Google Scholar]
- Chapple C.C.S., Vogt T., Ellis B.E., Somerville C.R. (1992). An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Dixon R.A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chen F., Srinivasa Reddy M.S., Temple S., Jackson L., Shadle G., Dixon R.A. (2006). Multi-site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.). Plant J. 48: 113–124 [DOI] [PubMed] [Google Scholar]
- Chen H.-C., Li Q., Shuford C.M., Liu J., Muddiman D.C., Sederoff R.R., Chiang V.L. (2011). Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl. Acad. Sci. USA 108: 21253–21258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J.O.D., Blake-Kalff M.M.A., Davies T.G.E. (1997). Detoxification of xenobiotics by plants: Chemical modification and vacuolar compartmentation. Trends Plant Sci. 2: 144–151 [Google Scholar]
- Costa M.A., Collins R.E., Anterola A.M., Cochrane F.C., Davin L.B., Lewis N.G. (2003). An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof. Phytochemistry 64: 1097–1112 [DOI] [PubMed] [Google Scholar]
- Dauwe R., et al. (2007). Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J. 52: 263–285 [DOI] [PubMed] [Google Scholar]
- Davies A.N. (1998). The new automated mass spectrometry deconvolution and identification system (AMDIS). Spectrosc. Eur. 10: 24–27 [Google Scholar]
- Debeaujon I., Nesi N., Perez P., Devic M., Grandjean O., Caboche M., Lepiniec L. (2003). Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dence C.W. (1992). The determination of lignin. In Methods in Lignin Chemistry, S.Y. Lin and C.W. Dence, eds (Berlin: Springer-Verlag), pp. 33–61.
- Do C.-T., Pollet B., Thévenin J., Sibout R., Denoue D., Barrière Y., Lapierre C., Jouanin L. (2007). Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226: 1117–1129 [DOI] [PubMed] [Google Scholar]
- DuBois M., Gilles K.A., Hamilton J.K., Rebers P.A., Smith F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28: 350–356 [Google Scholar]
- Ellis C., Karafyllidis I., Wasternack C., Turner J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla-Treviño L.L., Chen W., Card M.L., Shih M.-C., Cheng C.-L., Poulton J.E. (2006). Arabidopsis thaliana β-glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides. Phytochemistry 67: 1651–1660 [DOI] [PubMed] [Google Scholar]
- Fernie A.R., Trethewey R.N., Krotzky A.J., Willmitzer L. (2004). Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5: 763–769 [DOI] [PubMed] [Google Scholar]
- Franke R., Humphreys J.M., Hemm M.R., Denault J.W., Ruegger M.O., Cusumano J.C., Chapple C. (2002). The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 30: 33–45 [DOI] [PubMed] [Google Scholar]
- Fraser C.M., Thompson M.G., Shirley A.M., Ralph J., Schoenherr J.A., Sinlapadech T., Hall M.C., Chapple C. (2007). Related Arabidopsis serine carboxypeptidase-like sinapoylglucose acyltransferases display distinct but overlapping substrate specificities. Plant Physiol. 144: 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., et al. (2009). System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat. Genet. 41: 166–167 [DOI] [PubMed] [Google Scholar]
- Goujon T., Sibout R., Eudes A., MacKay J., Joulanin L. (2003a). Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiol. Biochem. 41: 677–687 [Google Scholar]
- Goujon T., Sibout R., Pollet B., Maba B., Nussaume L., Bechtold N., Lu F., Ralph J., Mila I., Barrière Y., Lapierre C., Jouanin L. (2003b). A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Mol. Biol. 51: 973–989 [DOI] [PubMed] [Google Scholar]
- Grace S.C., Logan B.A. (2000). Energy dissipation and radical scavenging by the plant phenylpropanoid pathway. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graindorge M., Giustini C., Jacomin A.C., Kraut A., Curien G., Matringe M. (2010). Identification of a plant gene encoding glutamate/aspartate-prephenate aminotransferase: The last homeless enzyme of aromatic amino acids biosynthesis. FEBS Lett. 584: 4357–4360 [DOI] [PubMed] [Google Scholar]
- Guo B., Jin Y., Wussler C., Blancaflor E.B., Motes C.M., Versaw W.K. (2008). Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 177: 889–898 [DOI] [PubMed] [Google Scholar]
- Guo D., Chen F., Inoue K., Blount J.W., Dixon R.A. (2001). Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa. impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13: 73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkes S., Sonnewald U., Badur R., Flachmann R., Stitt M. (2001). A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 13: 535–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertweck C., Jarvis A.P., Xiang L., Moore B.S., Oldham N.J. (2001). A mechanism of benzoic acid biosynthesis in plants and bacteria that mirrors fatty acid β-oxidation. ChemBioChem 2: 784–786 [DOI] [PubMed] [Google Scholar]
- Hirai M.Y., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 104: 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.-J., Harding S.A., Lung J., Popko J.L., Ralph J., Stokke D.D., Tsai C.-J., Chiang V.L. (1999). Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat. Biotechnol. 17: 808–812 [DOI] [PubMed] [Google Scholar]
- Ideker T., Galitski T., Hood L. (2001). A new approach to decoding life: Systems biology. Annu. Rev. Genomics Hum. Genet. 2: 343–372 [DOI] [PubMed] [Google Scholar]
- Jarvis A.P., Schaaf O., Oldham N.J. (2000). 3-Hydroxy-3-phenylpropanoic acid is an intermediate in the biosynthesis of benzoic acid and salicylic acid but benzaldehyde is not. Planta 212: 119–126 [DOI] [PubMed] [Google Scholar]
- Jones L., Ennos A.R., Turner S.R. (2001). Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26: 205–216 [DOI] [PubMed] [Google Scholar]
- Jouanin L., Goujon T., de Nadaï V., Martin M.-T., Mila I., Vallet C., Pollet B., Yoshinaga A., Chabbert B., Petit-Conil M., Lapierre C. (2000). Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol. 123: 1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M., Rensing K.H., Wong J.C.T., Banno B., Mansfield S.D., Samuels A.L. (2008). Tracking monolignols during wood development in lodgepole pine. Plant Physiol. 147: 1750–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenward M.G., Roger J.H. (1997). Small sample inference for fixed effects from restricted maximum likelihood. Biometrics 53: 983–997 [PubMed] [Google Scholar]
- Kopka J., et al. (2005). GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kruger N.J., von Schaewen A. (2003). The oxidative pentose phosphate pathway: Structure and organisation. Curr. Opin. Plant Biol. 6: 236–246 [DOI] [PubMed] [Google Scholar]
- Kuo W.P., et al. (2006). A sequence-oriented comparison of gene expression measurements across different hybridization-based technologies. Nat. Biotechnol. 24: 832–840 [DOI] [PubMed] [Google Scholar]
- Lanot A., Hodge D., Jackson R.G., George G.L., Elias L., Lim E.-K., Vaistij F.E., Bowles D.J. (2006). The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 48: 286–295 [DOI] [PubMed] [Google Scholar]
- Lee I., Ambaru B., Thakkar P., Marcotte E.M., Rhee S.Y. (2010). Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat. Biotechnol. 28: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplé J.-C., et al. (2007). Downregulation of cinnamoyl-coenzyme A reductase in poplar: Multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhou Y., Cheng X., Sun J., Marita J.M., Ralph J., Chiang V.L. (2003). Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. USA 100: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E.-K., Jackson R.G., Bowles D.J. (2005). Identification and characterisation of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS Lett. 579: 2802–2806 [DOI] [PubMed] [Google Scholar]
- Liu C.-J., Miao Y.-C., Zhang K.-W. (2011). Sequestration and transport of lignin monomeric precursors. Molecules 16: 710–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommen A. (2009). MetAlign: Interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal. Chem. 81: 3079–3086 [DOI] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Malitsky S., Blum E., Less H., Venger I., Elbaz M., Morin S., Eshed Y., Aharoni A. (2008). The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol. 148: 2021–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuko T., Minami A., Iwasaki N., Majima T., Nishimura S.-I., Lee Y.C. (2005). Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 339: 69–72 [DOI] [PubMed] [Google Scholar]
- Meyer K., Cusumano J.C., Somerville C., Chapple C.C.S. (1996). Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc. Natl. Acad. Sci. USA 93: 6869–6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyermans H., et al. (2000). Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J. Biol. Chem. 275: 36899–36909 [DOI] [PubMed] [Google Scholar]
- Mir Derikvand M., Sierra J.B., Ruel K., Pollet B., Do C.-T., Thévenin J., Buffard D., Jouanin L., Lapierre C. (2008). Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227: 943–956 [DOI] [PubMed] [Google Scholar]
- Mochida K., Shinozaki K. (2011). Advances in omics and bioinformatics tools for systems analyses of plant functions. Plant Cell Physiol. 52: 2017–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerkerke B., Goetghebeur E. (2006). Selecting “significant” differentially expressed genes from the combined perspective of the null and the alternative. J. Comput. Biol. 13: 1513–1531 [DOI] [PubMed] [Google Scholar]
- Morreel K., Dima O., Kim H., Lu F., Niculaes C., Vanholme R., Dauwe R., Goeminne G., Inzé D., Messens E., Ralph J., Boerjan W. (2010b). Mass spectrometry-based sequencing of lignin oligomers. Plant Physiol. 153: 1464–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K., Kim H., Lu F., Dima O., Akiyama T., Vanholme R., Niculaes C., Goeminne G., Inzé D., Messens E., Ralph J., Boerjan W. (2010a). Mass spectrometry-based fragmentation as an identification tool in lignomics. Anal. Chem. 82: 8095–8105 [DOI] [PubMed] [Google Scholar]
- Morreel K., Ralph J., Kim H., Lu F., Goeminne G., Ralph S., Messens E., Boerjan W. (2004). Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol. 136: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M., Øbro J., Willats W.G.T., Persson S. (2008). GeneCAT—Novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res. 36: W320–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M., Usadel B., Schütte M., Loraine A., Ebenhöh O., Persson S. (2010). Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol. 152: 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair R.B., Bastress K.L., Ruegger M.O., Denault J.W., Chapple C. (2004). The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen K.M., Kauppinen L., Helariutta Y. (2004). A weed for wood? Arabidopsis as a genetic model for xylem development. Plant Physiol. 135: 653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksman-Caldentey K.-M., Saito K. (2005). Integrating genomics and metabolomics for engineering plant metabolic pathways. Curr. Opin. Biotechnol. 16: 174–179 [DOI] [PubMed] [Google Scholar]
- Patterson T.A., et al. (2006). Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat. Biotechnol. 24: 1140–1150 [DOI] [PubMed] [Google Scholar]
- Pilate G., et al. (2002). Field and pulping performances of transgenic trees with altered lignification. Nat. Biotechnol. 20: 607–612 [DOI] [PubMed] [Google Scholar]
- Raes J., Rohde A., Christensen J.H., Van de Peer Y., Boerjan W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J., Lundquist K., Brunow G., Lu F., Kim H., Schatz P.F., Marita J.M., Hatfield R.D., Ralph S.A., Christensen J.H., Boerjan W. (2004). Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem. Rev. 3: 29–60 [Google Scholar]
- Raybould A.F., Moyes C.L. (2001). The ecological genetics of aliphatic glucosinolates. Heredity (Edinb) 87: 383–391 [DOI] [PubMed] [Google Scholar]
- Roessner U., Wagner C., Kopka J., Trethewey R.N., Willmitzer L. (2000). Technical advance: Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Rohde A., et al. (2004). Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Ruegger M., Chapple C. (2001). Mutations that reduce sinapoylmalate accumulation in Arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics 159: 1741–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M., Meyer K., Cusumano J.C., Chapple C. (1999). Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis. Plant Physiol. 119: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel K., Berrio-Sierra J., Derikvand M.M., Pollet B., Thévenin J., Lapierre C., Jouanin L., Joseleau J.-P. (2009). Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol. 184: 99–113 [DOI] [PubMed] [Google Scholar]
- Saito K., Hirai M.Y., Yonekura-Sakakibara K. (2008). Decoding genes with coexpression networks and metabolomics - ‘Majority report by precogs’. Trends Plant Sci. 13: 36–43 [DOI] [PubMed] [Google Scholar]
- Samuels A.L., Rensing K.H., Douglas C.J., Mansfield S.D., Dharmawardhana D.P., Ellis B.E. (2002). Cellular machinery of wood production: Differentiation of secondary xylem in Pinus contorta var. latifolia. Planta 216: 72–82 [DOI] [PubMed] [Google Scholar]
- Scharpf R.B., Iacobuzio-Donahue C.A., Sneddon J.B., Parmigiani G. (2007). When should one subtract background fluorescence in 2-color microarrays? Biostatistics 8: 695–707 [DOI] [PubMed] [Google Scholar]
- Schilmiller A.L., Stout J., Weng J.-K., Humphreys J., Ruegger M.O., Chapple C. (2009). Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 60: 771–782 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schoch G., Goepfert S., Morant M., Hehn A., Meyer D., Ullmann P., Werck-Reichhart D. (2001). CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 276: 36566–36574 [DOI] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.B., Dixon R.A. (2005). Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana. Plant J. 44: 62–75 [DOI] [PubMed] [Google Scholar]
- Shi C., Koch G., Ouzunova M., Wenzel G., Zein I., Lübberstedt T. (2006a). Comparison of maize brown-midrib isogenic lines by cellular UV-microspectrophotometry and comparative transcript profiling. Plant Mol. Biol. 62: 697–714 [DOI] [PubMed] [Google Scholar]
- Shi L., et al. MAQC Consortium (2006b). The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 24: 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R., Eudes A., Mouille G., Pollet B., Lapierre C., Jouanin L., Séguin A. (2005). CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinlapadech T., Stout J., Ruegger M.O., Deak M., Chapple C. (2007). The hyper-fluorescent trichome phenotype of the brt1 mutant of Arabidopsis is the result of a defect in a sinapic acid: UDPG glucosyltransferase. Plant J. 49: 655–668 [DOI] [PubMed] [Google Scholar]
- Smith C.A., Want E.J., O’Maille G., Abagyan R., Siuzdak G. (2006). XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78: 779–787 [DOI] [PubMed] [Google Scholar]
- Stein S.E. (1999). An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 10: 770–781 [Google Scholar]
- Tissier A.F., Marillonnet S., Klimyuk V., Patel K., Torres M.A., Murphy G., Jones J.D.G. (1999). Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Quimbaya M., Casneuf T., De Veylder L., Van de Peer Y. (2009). Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 150: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. (2010b). Lignin biosynthesis and structure. Plant Physiol. 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Morreel K., Ralph J., Boerjan W. (2008). Lignin engineering. Curr. Opin. Plant Biol. 11: 278–285 [DOI] [PubMed] [Google Scholar]
- Vanholme R., et al. (2010c). Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J. 64: 885–897 [DOI] [PubMed] [Google Scholar]
- Vanholme R., Van Acker R., Boerjan W. (2010a). Potential of Arabidopsis systems biology to advance the biofuel field. Trends Biotechnol. 28: 543–547 [DOI] [PubMed] [Google Scholar]
- Verbeke G., Molenberghs G. (2009). Linear Mixed Models for Longitudinal Data. (New York: Springer-Verlag). [Google Scholar]
- Vermerris W., Saballos A., Ejeta G., Mosier N.S., Ladisch M.R., Carpita N.C. (2007). Molecular breeding to enhance ethanol production from corn and sorghum stover. Crop Sci. 47: S142–S153 [Google Scholar]
- Versaw W.K., Harrison M.J. (2002). A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14: 1751–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfinger R.D., Gibson G., Wolfinger E.D., Bennett L., Hamadeh H., Bushel P., Afshari C., Paules R.S. (2001). Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8: 625–637 [DOI] [PubMed] [Google Scholar]
- Zhong R., Kays S.J., Schroeder B.P., Ye Z.-H. (2002). Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14: 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., Zhou J., McCarthy R.L., Ye Z.-H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Lee C., Zhong R., Ye Z.-H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Hero A.O., Qin Z.S., Swaroop A. (2005). High throughput screening of co-expressed gene pairs with controlled false discovery rate (FDR) and minimum acceptable strength (MAS). J. Comput. Biol. 12: 1029–1045 [DOI] [PubMed] [Google Scholar]