The authors used a genome-wide approach to identify the DNA sequences associated with the centromeres of potato chromosomes. Five potato centromeres are composed of single- or low-copy sequences, including active genes; six centromeres contain almost exclusively satellite repeats. The results have implications for evolution of repeat-based centromeres from repeatless centromeres.
Abstract
Centromeres in most higher eukaryotes are composed of long arrays of satellite repeats. By contrast, most newly formed centromeres (neocentromeres) do not contain satellite repeats and instead include DNA sequences representative of the genome. An unknown question in centromere evolution is how satellite repeat-based centromeres evolve from neocentromeres. We conducted a genome-wide characterization of sequences associated with CENH3 nucleosomes in potato (Solanum tuberosum). Five potato centromeres (Cen4, Cen6, Cen10, Cen11, and Cen12) consisted primarily of single- or low-copy DNA sequences. No satellite repeats were identified in these five centromeres. At least one transcribed gene was associated with CENH3 nucleosomes. Thus, these five centromeres structurally resemble neocentromeres. By contrast, six potato centromeres (Cen1, Cen2, Cen3, Cen5, Cen7, and Cen8) contained megabase-sized satellite repeat arrays that are unique to individual centromeres. The satellite repeat arrays likely span the entire functional cores of these six centromeres. At least four of the centromeric repeats were amplified from retrotransposon-related sequences and were not detected in Solanum species closely related to potato. The presence of two distinct types of centromeres, coupled with the boom-and-bust cycles of centromeric satellite repeats in Solanum species, suggests that repeat-based centromeres can rapidly evolve from neocentromeres by de novo amplification and insertion of satellite repeats in the CENH3 domains.
INTRODUCTION
The centromere is the chromosomal domain that directs the assembly of the proteinaceous kinetochore, which interacts with spindle microtubules to mediate chromosomal segregation. Centromeric chromatin is defined by the presence of CENH3, a centromere-specific H3 variant. Centromeres in most higher eukaryotic organisms are composed of long arrays of satellite repeats (Henikoff et al., 2001; Jiang et al., 2003). The centromeric satellite repeats are often homogenized throughout the genome; thus, a single repeat dominates all centromeres in most higher eukaryotes. In humans, only the alpha satellite repeats, the primary DNA sequence in all human centromeres, can be used for construction of human artificial chromosomes (Harrington et al., 1997; Ikeno et al., 1998). By contrast, DNA sequences from newly formed centromeres (neocentromeres) are not functional for artificial chromosome formation (Saffery et al., 2001), suggesting that the centromeric satellite repeats are intrinsic for centromere function.
Centromeres may evolve from neocentromeres that emerge in noncentromeric regions (Ventura et al., 2001; Yan et al., 2006; Marshall et al., 2008). Neocentromeres have been repeatedly identified in humans (Marshall et al., 2008). The DNA sequences underlying most human neocentromeres are not distinctly different from the genome average, although neocentromeric DNAs have a relatively higher AT content, ranging from 59.9 to 66.1% compared with the genome average of 59% (Marshall et al., 2008). Most notably, most human neocentromeres do not contain satellite repeats (Marshall et al., 2008). Only one of the ∼100 human neocentromeres landed in a genomic region containing satellite repeats (Hasson et al., 2011). Thus, an intriguing question of centromere evolution is how satellite repeats emerge and invade neocentromeres, eventually resulting in repeat-based centromeres.
Centromeres have been studied in several plant species. Centromere-specific satellite repeats were detected in every chromosome in all plant species analyzed (Jiang et al., 2003). In one extreme case, the centromere of rice chromosome 8 contains only ∼65 kb of the centromeric satellite repeat (CentO), which accounts for <10% of the functional domain of this centromere (Nagaki et al., 2004). We conducted a genome-wide characterization of DNA sequences associated with CENH3 nucleosomes in potato (Solanum tuberosum; 2n = 4x = 48). We discovered centromere-specific satellite repeats in six of the 12 potato centromeres. Surprisingly, five potato centromeres did not include satellite repeats, but contained primarily single- and low-copy sequences, including active genes. In addition, five of the six centromere-specific satellite repeats appeared to have emerged recently in the potato genome because these repeats were not present in closely related Solanum species. Our results suggest that the evolution from a repeatless to a repeat-based centromere is likely completed by a sudden event of de novo amplification and insertion of a satellite repeat in the CENH3 domain rather than a gradual accumulation of repetitive sequences throughout the neocentromere.
RESULTS
Genome-Wide Mapping of DNA Sequences Associated with CENH3 Nucleosomes
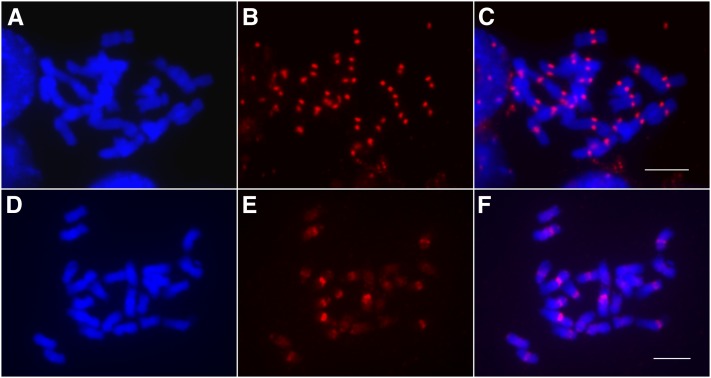
We developed an antibody against the potato CENH3. Immunofluorescence assays showed that the antibody is highly specific to the centromeres of potato chromosomes (Figures 1A to 1C) as well as chromosomes in several wild Solanum species. Chromatin immunoprecipitation (ChIP) was performed using nuclei isolated from leaf tissue of the doubled monoploid potato clone DM1-3 516R44 (hereafter referred to as DM1-3) (2n = 2x = 24), which has been fully sequenced (Xu et al., 2011). Fluorescence in situ hybridization (FISH) using the ChIPed DNA as a probe revealed highly enriched signals in the centromeric regions of multiple DM1-3 chromosomes (Figures 1D to 1F). A ChIPed DNA sample was sequenced using the Illumina Genome Analyzer II platform, generating 43 million 36-bp chromatin immunoprecipitation sequencing (ChIP-seq) reads. Approximately 44% of the reads were mapped to a unique position in the assembled DM1-3 genome (see Methods).
Figure 1.
Immunofluorescence and ChIP-FISH Using Potato CENH3 Antibodies.
(A) Somatic metaphase chromosomes of DM1-3 potato.
(B) Immunofluorescence derived from the anti-CENH3 antibodies.
(C) Image merged from (A) and (B).
(D) Somatic metaphase chromosomes of DM1-3 potato.
(E) FISH signals derived from precipitated DNA isolated from ChIP using anti-CENH3 antibodies.
(F) Image merged from (D) and (E).
Bars = 5 μm.
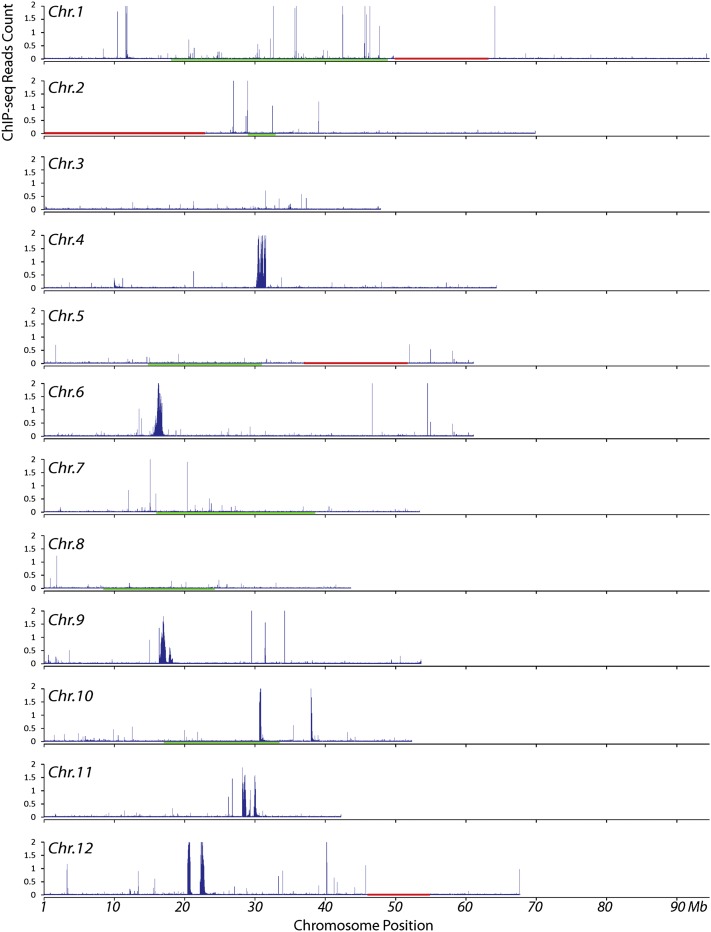
The distribution of unique ChIP-seq reads was displayed in 10-kb windows along the 12 potato chromosomes. Significant sequence enrichment was observed in the centromeres of five potato chromosomes (Cen4, Cen6, Cen9, Cen11, and Cen12) (Figure 2), indicating that the functional cores of these five centromeres contain mainly single and low copy sequences, similar to the centromeres of several rice chromosomes (Yan et al., 2008). The sizes of the CENH3-enriched centromere cores (from the first to the last CENH3 subdomain in each centromere; see below) in the five centromeres were 1313, 1063, 1460, 1930, and 2404 kb, respectively (Figure 2). In Cen10, two CENH3 enriched subdomains, 580 and 360 kb, respectively, were separated by a large chromosomal domain encompassing 6.76 Mb of DNA (Figure 2). The separation of two CENH3 subdomains by a very large chromosomal segment has been reported previously only for dicentric chromosomes. In addition, the two CENH3 subdomains on chromosome 10 span a distance that accounts for more than 10% of the chromosome, which would result in separate immunofluorescence signals on metaphase chromosomes (Zhang et al., 2010). Such separate signals were not observed on potato chromosomes. Thus, this is likely an artifact resulting from sequence misassembly attributable to the whole-genome shotgun approach used to assemble the DM1-3 genome (Xu et al., 2011), which is confirmed by the fact that the 360-kb subdomain is not included in the recombination-suppressed chromosomal domain on the linkage map of this chromosome (Figure 2).
Figure 2.
Density of Sequence Reads Derived from ChIP with CENH3 Antibodies along Individual Potato Chromosomes.
Read density was represented by the total number of sequence reads in a 10-kb window per base pair mappable region. The x axes show the position on the chromosome. Red horizontal bars mark large physical gaps (>1 Mb) in the pseudomolecules. The red horizontal bar on chromosome 2 represents the unassembled 45S ribosomal gene cluster. The genetic positions of the centromeres of chromosomes 1, 2, 5, 7, 8, and 10 are likely spanned by the green horizontal bars that mark the recombination-suppressed domains on the corresponding linkage maps of these chromosomes (Felcher et al., 2012).
Interspersed CENH3 and H3 Subdomains in Potato Centromeres
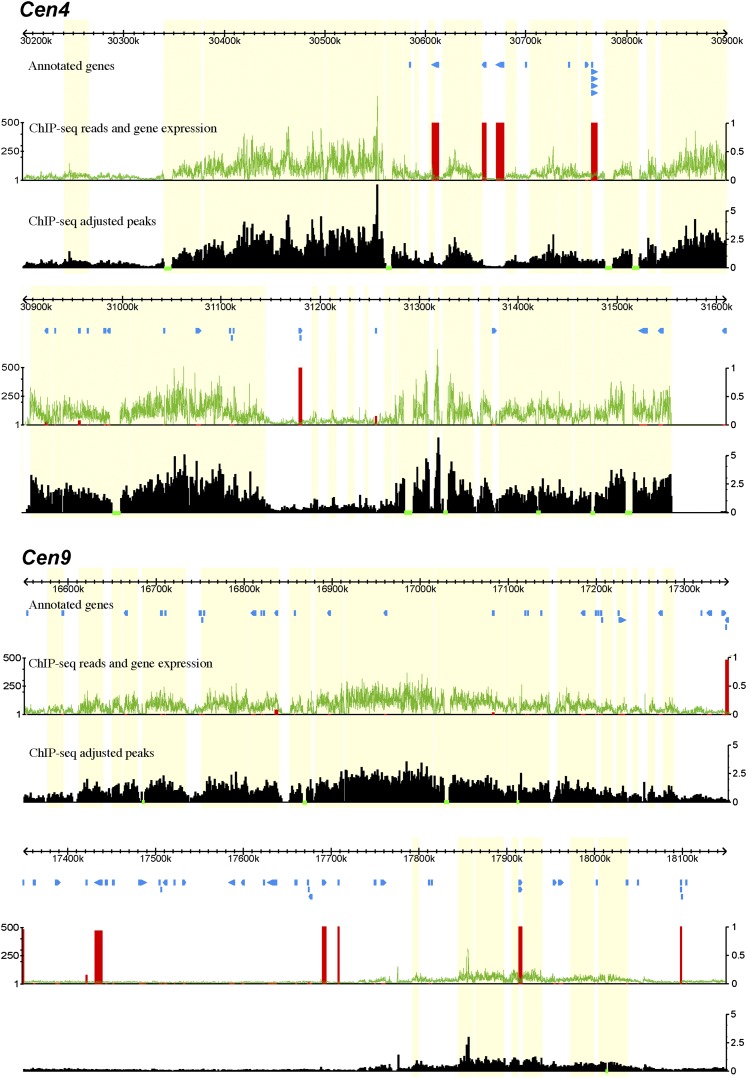
The levels of ChIP-seq enrichment were not uniform across five of the potato centromeres (Cen4, Cen6, Cen9, Cen11, and Cen12) (Figure 3; see Supplemental Figures 1 to 3 online). Each centromere contained several CENH3-enriched subdomains, which were interspersed with subdomains that were not enriched with CENH3. The CENH3-lacking subdomains likely consist of nucleosomes containing H3 (hereafter named as H3 subdomains) (Wu et al., 2011). The structure of intermingled CENH3 and H3 subdomains of potato centromeres is similar to that observed in rice (Oryza sativa) centromeres (Yan et al., 2008).
Figure 3.
Fine Structure of the Functional Cores of Potato Cen4 and Cen9.
The top track in each panel shows the sequence coordinates on the respective potato chromosome. The “annotated genes” track illustrates the positions of annotated genes. The green line in the middle track shows the number of sequence reads derived from ChIP with CENH3 antibodies in 100-bp windows. The vertical red bars represent the percentage of the 32 tissues in which the corresponding gene is expressed (FPKM >0) (1 representing expression in all 32 tissues). The bottom track shows the density of reads in 100-bp windows, adjusted by the length of mappable regions. The horizontal green bars in this track mark the sequencing gap/nonmappable regions. Each green bar region is assigned to an adjacent CENH3 subdomain. All CENH3 subdomains are shaded in yellow.
Cen4 and Cen6 contained large CENH3 subdomains inserted with small H3 subdomains, ranging from 2 to 83 kb (Figure 3; see Supplemental Figure 1 online). Some of the small subdomains were close to the statistical threshold to be annotated as CENH3 or H3 subdomains. Additional ChIP-seq experiments and data with a better signal-to-noise ratio may reveal if these small domains are associated with CENH3 or H3 nucleosome blocks or mixtures of both. Cen9 and Cen11 contained large H3 subdomains, up to 586 kb (Figure 3; see Supplemental Figure 2 online). Centromeres with a fine structure similar to either Cen4/Cen6 or to Cen9/Cen11 were also reported in rice (Yan et al., 2008). Cen12 consisted of two CENH3 subdomains that are separated by a 1.4-Mb H3 subdomain, which is larger than several entire potato centromeres (see Supplemental Figure 3 online). We suspect that sequence misassembly may also contribute to the unusual large size of the H3 subdomain. Thus, we did not use Cen12 for further analysis.
Genes in Potato Centromeres
Annotated gene models were found in each of the four potato centromeres (Cen4, Cen6, Cen9, and Cen11) (Figure 3; see Supplemental Figures 1 and 2 online). We manually examined the annotations and removed gene models with similarity to transposable elements, resulting in a total of 77 genes located within CENH3 subdomains and 98 genes in H3 subdomains in these four centromeres (see Supplemental Data Set 1 online). We then employed the RNA-seq data sets obtained from 32 different tissue types or abiotic/biotic stress treatments of DM1-3 (Xu et al., 2011) to examine the expression of the putative genes within these four centromeres. Interestingly, the majority of these genes (141 of 175) were not expressed (value = 0 fragments per kilobase of exon model per million mapped fragments [FPKM]) in any of the tissues (Figure 3; see Supplemental Figures 1 to 3 online). Only six of the 77 genes (8%) located in CENH3 subdomains showed expression in at least one tissue. By contrast, 28 of the 98 genes (29%) located in the H3 subdomains were expressed at least in one tissue, and 20 of these genes were expressed in more than 14 tissues (see Supplemental Data Set 1 online).
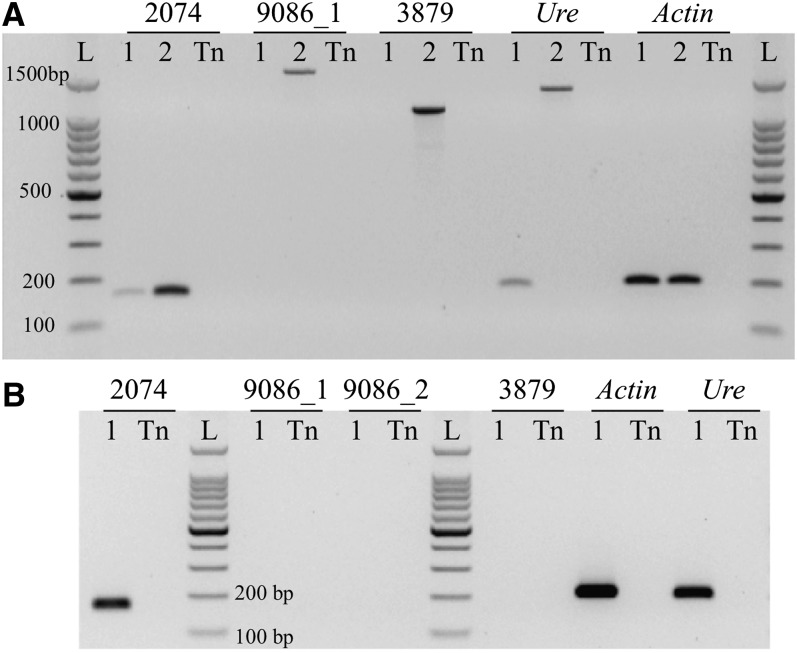
Two of the six putative active genes located within CENH3 subdomains were expressed in leaf tissue based on RNA-seq data. We conducted RT-PCR experiments to confirm the expression of these two genes (PGSC0003DMG400039086 in Cen6 and PGSC0003DMG400012074 in Cen11) in leaves. RNA-seq data suggested that both genes were expressed at low levels in leaves, with FPKM values of 2.354 and 1.685, respectively (see Supplemental Data Set 1 online). RT-PCR confirmed that PGSC0003DMG400012074 was expressed in leaf tissue (Figure 4), although the transcript levels of this gene were approximately one-tenth of the transcript levels derived from the urease gene, a well-characterized potato gene with a low level of transcription (Witte et al., 2005). However, transcripts of PGSC0003DMG400039086 were not detected after 40 PCR cycles, suggesting that this gene is not transcribed in leaves (Figure 4). The association of these two genes with CENH3 nucleosomes was confirmed by quantitative ChIP-PCR analysis. The DNA sequences associated with these two genes were significantly enriched in the ChIPed DNA sample (see Supplemental Figure 4 online).
Figure 4.
RT-PCR Analysis of Two Potato Genes Located in the CENH3 Subdomains.
All RT-PCR experiments were conducted using young leaves of DM1-3.
(A) Cycle-limited RT-PCR analysis of PGSC0003DMG400012074 (2074) and PGSC0003DMG400039086 (primer pair 9086_1; see Supplemental Table 3 online). 1, DM1-3 cDNA; 2, DM1-3 genomic DNA; Tn, technical negative control (water); L, DNA ladder. PGSC0003DMG400043879 (3879) is an untranscribed gene located in Cen12 and is used as a negative control. The expression of the Urease gene (Ure) and Actin gene (Actin) was used for comparison.
(B) Gel electrophoresis analysis of the expression of the genes as in (A) after 40 PCR cycles. No amplicons were obtained for PGSC0003DMG400039086 (both primer pair 9086_1 and primer pair 9086_2) and PGSC0003DMG400043879 (3879).
Identification of Centromere-Specific Satellite Repeats in Potato
Megabase-sized satellite repeat arrays are common to centromeres in both animal and plant species (Henikoff et al., 2001; Jiang et al., 2003). However, centromeric satellite repeats are frequently absent in assembled genome sequences due to technical barriers in de novo assembly of highly repetitive sequences. Even if centromeric satellite repeats were represented within the assembled genome sequence, ChIP-seq reads derived from these repeats will not be included in the mapping process due to the requirement of the reads to align uniquely (see Methods). ChIP-seq sequence enrichment, which is measured by alignment of ChIP-seq reads to the assembled potato genome, was not observed in six of the 12 potato centromeres (Cen1, Cen2, Cen3, Cen5, Cen7, and Cen8) (Figure 2). We hypothesized that these six centromeres contain long arrays of satellite repeats, which is supported by the fact that approximately half of the DM1-3 chromosomes showed significantly enhanced centromeric FISH signals using ChIPed DNA as a probe (Figure 1). Such enhanced signals were likely derived from highly repetitive centromeric repeats because these repeats would be enriched in the ChIPed DNA.
We designed a two-step procedure, which combined whole-genome shotgun (WGS) sequencing reads with CENH3 ChIP-seq data, to identify satellite repeats associated with potato centromeres. First, we used a similarity-based sequence clustering approach for de novo identification of repetitive sequences (Macas et al., 2007; Novák et al., 2010) using a set of 1.24 million 454 WGS reads generated from DM1-3 (Torres et al., 2011). This analysis resulted in repeat clusters representing different repeat families in the DM1-3 genome. The sequence proportion (%) of each repeat family was estimated based on the number of 454 sequence reads associated with individual clusters (Table 1). In the second step, we mapped the CENH3 ChIP-seq reads to the repeat clusters. We then calculated ratios of ChIP-seq reads to 454 reads associated with each cluster (Table 1). This ratio was indicative of the level of enrichment of individual repeats in potato centromeres (see Supplemental Figure 5 online). Potential satellite repeats were then identified based on their reconstructed consensus sequences and structure of the cluster graphs (Novák et al., 2010).
Table 1. Characteristics of Sequence Clusters Containing Putative Satellite Repeats.
| Proportion (%) |
Ratio (ChIP-seq/WGS) | Probe (GenBank Acc. No.)b | |||||
|---|---|---|---|---|---|---|---|
| Clustera | WGSc | ChIP-seqd | Repeat | Monomer (bp) | Chromosomal Locations | ||
| 49 | 0.100 | 2.780 | 27.80 | St49 | 2754 | 1335 (JQ731639) | Centromere 5 |
| 57 | 0.080 | 1.587 | 19.74 | St57 | 1924 | 1338 (JQ731642) | Centromere 7 |
| 24 | 0.268 | 3.124 | 11.68 | St24 | 979 | 1336 (JQ731640) | Centromere 1 |
| 3 | 2.272 | 13.730 | 6.04 | ||||
| St3-58 | 2957 | 1331 (JQ731637) | Centromere 2 | ||||
| St3-238 | 3814 | 1333 (JQ731638) | Centromere 8 | ||||
| St3-294 | (5390)e | 1340 (JQ731643) | Centromeres 3/9 | ||||
| 18 | 0.310 | 1.459 | 4.70 | St18 | 1180 | 1337 (JQ731641) | Centromere 9 |
Clusters generated by analysis of whole-genome 454 sequencing data.
Clones of genomic fragments obtained by PCR with primers based on predicted consensus sequences.
Whole-genome sequencing (454 reads).
CENH3 ChIP-seq data (Illumina reads).
Estimated from predicted sequence but not verified by PCR (only a partial fragment cloned).
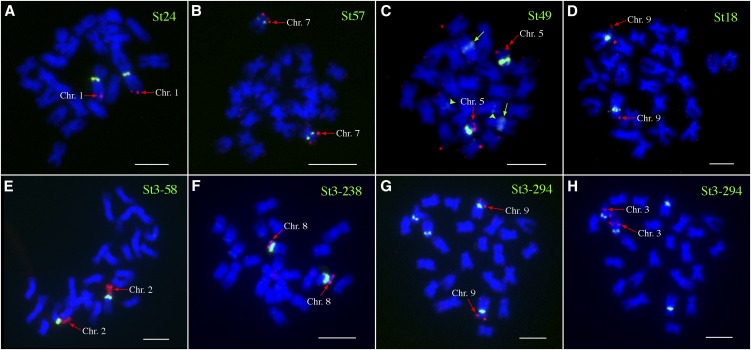
Three clusters showed >10-fold enrichment in ChIP-seq data, representing repeats St49, St57, and St24 (Table 1; see Supplemental Figure 5 online). Tandem arrangement of these three repeats in the genome was confirmed using PCR with primers directed outward from the reconstructed consensus sequences and by sequencing the cloned PCR products. FISH using the cloned probes revealed that St24 is highly specific to Cen1 (Figure 5A). No cross-hybridization was observed on other potato chromosomes. St57 hybridized to Cen7 with very weak noncentromeric signals observed on other potato chromosomes (Figure 5B). St49 generated strong FISH signals in Cen5. Weak hybridization signals from St49 were also observed in the interstitial regions of two other chromosomes (Figure 5C).
Figure 5.
FISH Mapping of Centromeric Repeats in DM1-3 Potato.
(A) Repeat St24 was mapped to Cen1 together with BAC clone 96H03, which is specific to 1L.
(B) Repeat St57 was mapped to Cen7 together with BAC clone 186I02, which is specific to 7S.
(C) Repeat FISH St49 was mapped to Cen5 together with BAC clone 44A21, which is specific to 5S. Two green arrows point to the second signals that are much weaker than the Cen5-specific signals. The two green arrowheads point to the third signals that were very weak but consistently observed.
(D) Repeat St18 was mapped to Cen9 together with BAC clone 135I22, which is specific to 9S.
(E) Repeat St3-58 was mapped to Cen2 together with the 45S rDNA probe, which is specific to 2S.
(F) Repeat St3-238 was mapped to Cen8 together with BAC clone 122L16, which is specific to 8S.
(G) Repeat St3-294 was mapped to the centromeres of two pairs of chromosomes. The first pair of chromosomes were identified to be chromosome 9 using BAC clone 135I22, which is specific to 9S.
(H) The second pair of chromosomes hybridized to St3-294 were identified to be chromosomes 3 using BAC clone 79E02, which is specific to 3L.
Bars = 5 μm.
Several additional clusters representing relatively abundant repeat families showed the ChIP-seq enrichment ratio ranging from approximately four to approximately nine (see Supplemental Figure 5 online). However, with the exception of clusters 18 and 3, these clusters lacked tandem arrangement and generated noncentromeric or dispersed hybridization patterns. Cluster 18 included a tandem repeat designated St18 that showed strong hybridization to Cen9 with very weak hybridization to other chromosomal locations (Figure 5D). Cluster 3 included a complex group of related sequences, some of which showed similarities to the gag-pol regions of Ty3/gypsy retroelements, while other reads formed ring-like structures within the cluster graph, indicating their tandem arrangement (Novák et al., 2010). Three subclusters of Cluster 3 representing potential satellite repeats were designated as St3-58, St3-238, and St3-294 and were investigated further. Using PCR and cloning experiments, these subclusters were proven to be tandemly organized with the exception of St3-294 where only a partial sequence was cloned due to its extremely long predicted monomer (5.4 kb) (Table 1). FISH experiments with St3-58 and St3-238 probes produced strong hybridization signals on Cen2 and Cen8, respectively (Figures 5E and 5F), and St3-294 hybridized to both Cen9 and Cen3 (Figures 5G and 5H).
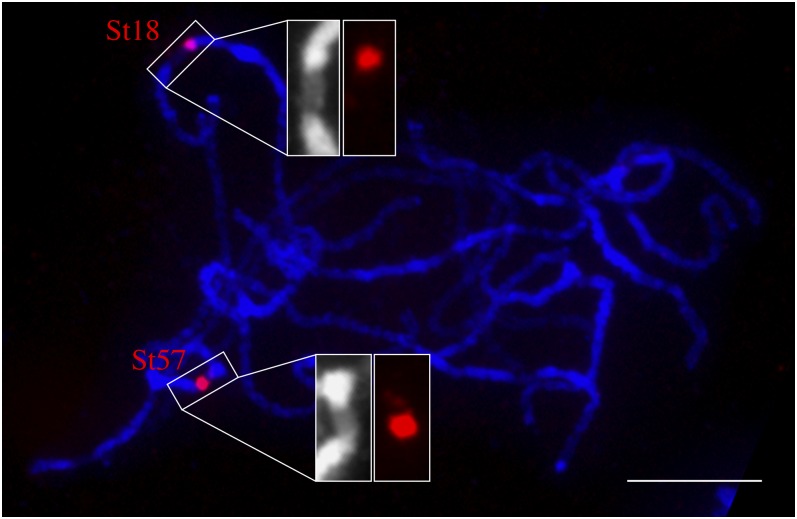
In summary, we identified satellite repeats associated with each of the six centromeres that were not enriched with unique ChIP-seq reads. Thus, the functional domains of these six centromeres are likely composed mainly or exclusively of these satellite repeats. The only exception is Cen9, which was enriched with ChIP-seq reads but also contained two centromere-specific repeats (St18 and St3-294) (Figure 5d). We examined the location of St18 on meiotic pachytene chromosomes of DM1-3. FISH signals from St18 mapped to the edge of the primary constriction of chromosome 9. By contrast, the entire St57 array mapped within the primary constriction of chromosome 7 (Figure 6). These results showed that only a fraction of the St18 repeat array is likely associated with CENH3 nucleosomes, which is correlated with its relatively lower ChIP-seq sequence proportion ratio (4.7) compared with the ratio of St57 (19.7).
Figure 6.
Locations of St18 and St57 on the Pachytene Chromosomes of DM1-3 Potato.
St18 is located at the edge of the primary constriction of chromosome 9. FISH signal from St57 is almost completely located within the primary constriction of chromosome 7. Chromosomes were stained by DAPI. Note: The primary constriction of the pachytene chromosomes can be readily identified based on their distinctly lower level of staining compared with the brightly stained pericentromeric heterochromatin. Bar = 10 μm.
Potato Centromeric Satellite Repeats Are Organized as Megabase-Sized Tandem Arrays
Fiber-FISH was conducted to reveal the organization of the six centromeric repeats (St18, St24, St49, St57, St3-58, and St3-238) in the DM1-3 genome. Long contiguous fiber-FISH signals were observed from all six probes (see Supplemental Figure 6 online), confirming that these repeats are organized as long tandem arrays. The St57 array spanned 283.4 ± 42 μm (n = 24), representing an average of 909.7 ± 134.8 kb of each array (see Supplemental Table 1 online). The sizes of the fiber-FISH signals from the other five probes were >1000 kb. Because DNA molecules >1000 kb tend to break during DNA fiber preparation, it is difficult to measure DNA loci that are multiple megabases in size. We obtained a minimum of five high-quality fiber-FISH signals from each probe. Measurements from these signals suggested that the St3-58 array was ∼1500 kb. The other four satellite repeat arrays were >2000 kb (see Supplemental Table 1 online). Thus, these repeats can potentially span the entire CENH3 domains of their respective centromeres based on the fact that the sizes of the CENH3 domains of Cen4, Cen6, Cen9, and Cen11 are between 1063 and 1930 kb.
Origin of Potato Centromeric Satellites Repeats
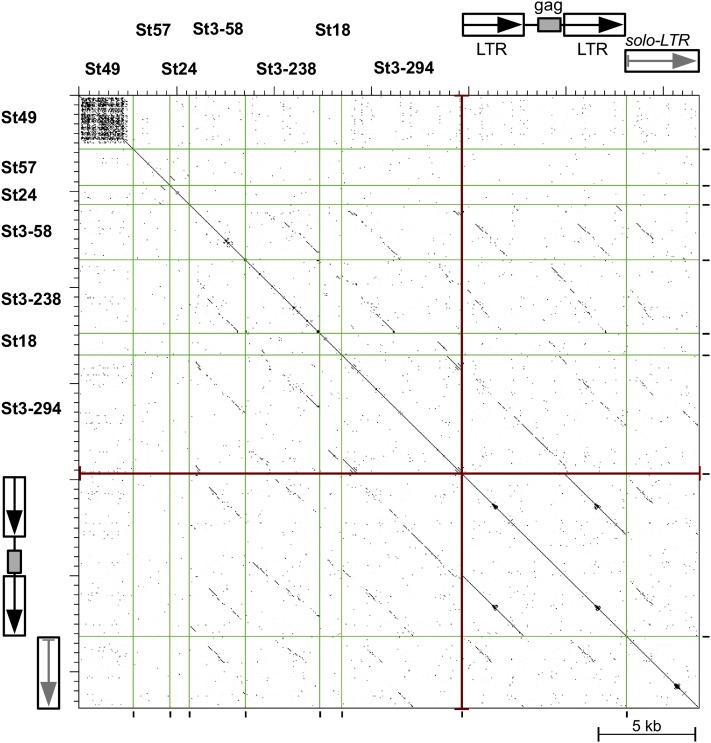
A striking feature of all identified centromeric satellites was their long monomer sizes, ranging from 979 bp in St24 to 5.4 kb in St3-294, which is far longer than the most frequent monomer sizes associated with known plant satellite repeats (135 to 195 and 315 to 375 bp) (Macas et al., 2002). Based on their sequence similarity, the potato centromeric satellites could be divided into three groups suggestive of origins from different sequences (Figure 7). Repeats St57 and St24, which represent the first group, shared a short (∼200 bp) region of similarity; otherwise, they differed from each other as well as from all other centromeric repeats (Figure 7). St49, representing the second group, showed no similarity to other centromeric repeats but had partial similarity to three other repeat families in the DM1-3 genome, all of them being putative satellites but none showing substantial enrichment in the CENH3 ChIP-seq reads (clusters 21, 45, and 66). Due to the presence of multiple short A/T-rich and telomere-like motifs in their sequences, they appear to be members of a broader group of Solanum repeats that also includes the previously reported telomere-similar centromeric satellite repeats in Solanum bulbocastanum (Tek and Jiang, 2004).
Figure 7.
Dot-Plot Similarity Comparison of Potato Centromeric Satellite Sequences and Selected Retrotransposon-Like Sequences.
Individual sequences are separated by vertical lines and their similarities exceeding 60% over a 100-bp sliding window are displayed as black dots or diagonal lines. The retrotransposon-like sequences with similarities to St3 and St18 satellites are represented by nonautonomous LTR retroelement (NA-RE) and related solo-LTR sequence, both identified in potato BAC clone BA251C18 (GenBank accession number GU906238, positions 23518-24812/28493-35636 and 24813-28492, respectively).
[See online article for color version of this figure.]
The third and largest group included St18 and the three satellites derived from cluster 3, and these four repeats shared partial sequence similarities. Moreover, a number of similarities were also detected in long terminal repeat (LTR) retroelement-like sequences present in the potato genomic BAC clones. These repeats showed the highest similarities to a group of nonautonomous LTR retrotransposons and related solo-LTR sequences shown on Figure 7. The depicted element consists of a truncated internal region containing a gag-coding domain but lacking the rest of the gag-pol region, surrounded by two LTRs. Comparison of the inferred GAG protein sequence to those of other plant retrotransposons suggested classification of this element as a member of Chromovirus clade of Ty3/gypsy elements (Gorinsek et al., 2004). The satellite repeats originated either from its LTR sequence (St3-58, St3-238, or St18) or included nearly the entire element (St3-294). This finding is in line with the observed clustering of these repeats with the LTR retrotransposon sequences that formed cluster 3.
Evolution of the Centromeric Satellite Repeats in Solanum Species
We selected a set of diploid Solanum species (Figure 8) to study the evolution of the centromeric satellite repeats. Each species was assigned to a different genome type (A, B, P, and E) based mainly on traditional chromosome pairing studies (Matsubayashi, 1991; Gavrilenko, 2007). Although the exact divergence between potato and each of these species is not known, the genus Solanum diverged from its closest related genus ∼12 million years ago (Wikström et al., 2001); tomato (Solanum lycopersicum) and potato may have diverged seven million years ago (Nesbitt and Tanksley, 2002). Solanum verrucosum is most closely related to potato and was proposed as the progenitor of cultivated potato (Hawkes, 1990).
Figure 8.
Phylogenetic Relationships of the Solanum Species Used in Evolutionary Study of the Potato Centromere-Specific Satellite Repeats.
The bootstrap values were based on plastid DNA analyses of Spooner et al. (1993), Spooner and Castillo (1997), and Castillo and Spooner (1997).
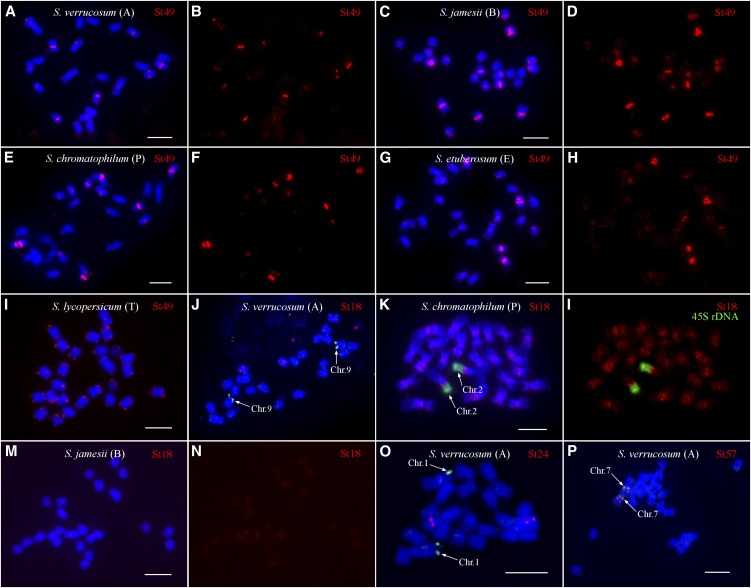
St49 was detected in all species analyzed. The FISH patterns on metaphase chromosomes were similar in all species except for tomato. Hybridization signals were observed in the centromeric or pericentromeric regions of most chromosomes (Table 2, Figures 9A to 9H), and only a few chromosomes in S. verrucosum and Solanum chromatophilum did not show unambiguous signals. Most FISH signals were difficult to score as centromeric or pericentromeric due to low FISH resolution on metaphase chromosomes. However, some FISH signals were clearly located in pericentromeric regions. Massive St49 signals were observed in centromeric/pericentromeric regions of several Solanum jamesii chromosomes (Figure 9C). Interestingly, St49 hybridized exclusively to the telomeric regions of all tomato chromosomes (Figure 9I), consistent with its sequence similarity (77% sequence identity over 854 bp) to the previously reported telomere-similar centromeric repeats in S. bulbocastanum (Tek and Jiang, 2004). Weak telomeric signals were also observed on chromosomes of Solanum etuberosum (Figure 9G), which is phylogenetically more closely related to tomato than to potato (Figure 8). These results show that St49 is an ancient repeat and derived from a telomere-similar sequence.
Table 2. Summary of FISH Analysis of Centromere-Specific Satellite Repeats in Six Diploid Solanum Species.
| Repeat | DM1-3 S. tuberosum (A Genome) | S. verrucosum (A Genome) | S. jamesii (B Genome) | S. chromatophilum (P Genome) | S. lycopersicum (T Genome) | S. etuberosum, S. palustre (E Genome)a |
|---|---|---|---|---|---|---|
| St49 | One location to Cen5, another noncentromeric location on the second chromosome | Strong centromeric and/or pericentromeric signals on multiple chromosomes | Strong centromeric and/or pericentromeric signals on multiple chromosomes | Strong centromeric and/or pericentromeric signals on multiple chromosomes | Telomeric signals on all chromosomes | Strong centromeric and/or pericentromeric signals on multiple chromosomes and weak telomeric signals on all chromosomes |
| St18 | Specific to Cen9 | Single centromeric location, not Cen9 | No hybridizationb | Dispersed signals at all chromosomes, enhanced centromeric/pericentromeric signals at some chromosomes (Figure 9l) | No hybridization | No hybridization |
| St24 | Specific to Cen1 | Single centromeric location, not Cen1 | No hybridization | No hybridization | No hybridization | No hybridization |
| St57 | Specific to Cen7 | Specific to Cen7 | No hybridization | No hybridization | No hybridization | No hybridization |
| St3-58 | Specific to Cen2 | No hybridization | No hybridization | No hybridization | No hybridization | No hybridization |
| St3-238 | Specific to Cen8 | No hybridization | No hybridization | Dispersed signals at all chromosomes, enhanced centromeric/pericentromeric signals at some chromosomes | No hybridization | No hybridization |
| St3-294c | Specific to Cen3 | No hybridization | No hybridization | Dispersed signals at all chromosomes, enhanced centromeric/pericentromeric signals at some chromosomes | No hybridization | No hybridization |
Both S. etuberosum and S. palustre contain the E genome. The FISH results from these two species were identical.
“No hybridization” refers to all FISH signal patterns with very weak and dispersed hybridization or patterns that are inconsistent and are not specific to centromeric regions. See one example in Figure 9N.
A St3-294 probe that excludes St18-related sequences was used in FISH analysis.
Figure 9.
FISH Mapping of Potato Centromere-Specific Satellite Repeats in Different Solanum Species.
(A) FISH of St49 on metaphase chromosomes of S. verrucosum.
(B) Digitally separated FISH signals from (A).
(C) FISH of St49 on metaphase chromosomes of S. jamesii.
(D) Digitally separated FISH signals from (C).
(E) FISH of St49 on metaphase chromosomes of S. chromatophilum.
(F) Digitally separated FISH signals from (E).
(G) FISH of St49 on metaphase chromosomes of S. etuberosum.
(H) Digitally separated FISH signals from (G).
(I) FISH of St49 on metaphase chromosomes of tomato (S. lycopersicum).
(J) FISH of St18 on metaphase chromosomes of S. verrucosum. St18 is not located on chromosome 9, which is identified by BAC 135I22 (arrows).
(K) FISH of St18 on metaphase chromosomes of S. chromatophilum. Chromosome 2 is identified by the FISH signals from 45S rDNA (arrows).
(L) Digitally separated FISH signals from (K).
(M) FISH of St18 on metaphase chromosomes of S. jamesii.
(N) Digitally separated FISH signals from (M).
(O) FISH of St24 on metaphase chromosomes of S. verrucosum. St24 is not located on chromosome 1, which is identified by BAC 96H03 (arrows).
(P) FISH of St57 on metaphase chromosomes of S. verrucosum. St57 is colocalized on chromosome 7, which is identified by BAC 186I02 (arrows).
Letters in parentheses represent the genome of the Solanum species. Bars = 5 μm.
St18, associated with Cen9 in DM1-3 (Figure 5D), hybridized to the centromeric region of single pair of chromosomes in S. verrucosum. However, the St18-associated S. verrucosum chromosome is not chromosome 9 (Figure 9J). St18 hybridized to broad regions of all S. chromatophilum chromosomes (Figures 9K and 9L). Enhanced centromeric/pericentromeric signals were observed in several chromosomes, including chromosome 2, which can be identified by its association with the 45S rRNA genes (45S rDNA) (Figures 9K and 9L). St18 produced similar, but much weaker, FISH signals on S. jamesii chromosomes (Figures 9M and 9N). We observed either no or very weak and dispersed FISH signals on chromosomes in other Solanum species (Table 2).
St24, associated with Cen1 in DM1-3 (Figure 5A), hybridized to the centromeric region of a single pair of chromosomes of S. verrucosum. However, the St24 signals were not located on chromosome 1 in S. verrucosum (Figure 9O). St24 did not generate any FISH signals on chromosomes in other Solanum species.
St57 is associated with Cen7 in DM1-3 (Figure 5B) and hybridized also to Cen7 in S. verrucosum (Figure 9P). St57 generated either no or very weak and dispersed FISH signals in other Solanum species.
St3-58, St3-238, and St3-294 generated very weak and dispersed FISH signals in all Solanum species analyzed (Table 2). No distinct centromeric signals were observed in any species, including S. verrucosum (Table 2).
In summary, among six centromere-specific satellite repeats identified in cultivated potato, only St49 represents an ancient repeat and was detected in all Solanum species analyzed. The other five repeats were either detected only in S. verrucosum, which is most closely related potato or absent in all Solanum species analyzed. Thus, these five repeats were amplified very recently in the potato genome. St18 and St24 were mapped to different chromosomes in S. verrucosum in comparison to potato. It is possible that these two repeats were independently amplified in the two species. Alternatively, structural rearrangements of the chromosomes in the two species may result in the relocations of the centromeric satellite or the BAC markers on different chromosomes.
DISCUSSION
Genes in Centromeres
Neocentromeres often emerge in gene-poor regions (Cardone et al., 2006; Ventura et al., 2007; Lomiento et al., 2008). This is correlated with the fact that centromeric chromatin appears to be incompatible with transcription (Allshire et al., 1995). Indeed, neocentromere formation can cause reduction of transcription or silencing of the underlying genes (Ishii et al., 2008; Ketel et al., 2009). In the nematode Caenorhabditis elegans, which has holocentric chromosomes, genes transcribed in embryos are refractory to CENH3 incorporation, whereas silent genes in embryos are permissive to CENH3 incorporation (Gassmann et al., 2012). Active genes were found in several rice centromeres (Nagaki et al., 2004; Yan et al., 2006, 2008). However, these genes are all located within the H3 subdomains embedded in the centromeric cores (Yan et al., 2008). Thus, the genes in rice centromeres are associated with H3 nucleosomes and show similar histone H3 modification patterns to those located outside of the centromeres (Wu et al., 2011). Most active genes (based on RNA-seq annotation) in potato centromeres were mapped within H3 subdomains. We demonstrate, however, that one transcribed gene, PGSC0003DMG400012074 in Cen11, is associated with CENH3 nucleosomes. Thus, CENH3 nucleosomes are not completely incompatible with transcription in potato. It will be interesting to explore if PGSC0003DMG400012074 is an essential gene and whether its relatively low level of expression is compatible with its biological functions.
Origin of Centromeric Satellite Repeats
Centromeres in most higher eukaryotes are composed of long arrays of satellite repeats. Such satellite repeat arrays can occupy the entire CENH3 domain in the centromere and can extend into the pericentromeric regions (Shibata and Murata, 2004; Houben et al., 2007). Centromeric satellite repeats evolve rapidly and different repeats can emerge in centromeres from closely related species (Lee et al., 2005). However, the origin of centromeric satellite repeats has been elusive. We demonstrate that at least three centromere-specific satellite repeats, St3-58, St3-238, and St3-294, emerged in potato since its divergence from its closest relative S. verrucosum. Interestingly, all three repeats showed sequence similarity with a retrotransposon. In addition, St18, which is present in S. verrucosum but on a different chromosome, is also related to retrotransposon sequences.
Retrotransposons appear to be a major resource for the origin of new centromeric satellite repeats in potato. Sudden amplification of a long array of a satellite repeat, named Sobo, from a retrotransposon was previously reported in a wild potato species S. bulbocastanum (Tek and Jiang, 2004). The 4.7-kb monomer of the Sobo repeat is a retrotransposon-related sequence and was amplified into a single array spanning ∼360 kb (Tek et al., 2005). The monomers of this repeat share >99% sequence identity, suggesting that Sobo was likely derived from rolling circle replication of an extrachromosomal circular DNA (eccDNA) followed by reinsertion into the S. bulbocastanum genome (Tek et al., 2005). Satellite repeats can also be amplified from a small portion of a retrotransposon (Langdon et al., 2000; Macas et al., 2009). Most interestingly, a recent report demonstrated that several centromere-specific satellite repeats in chicken showed partial sequence similarity to a retrotransposon (Shang et al., 2010). The monomer sizes of the chicken centromeric satellite repeats ranged from 0.7 to 3.2 kb, similar to the potato centromeric satellite repeats. Thus, amplified DNA from retrotransposons may be a common source for centromeric DNA in different higher eukaryotes.
Evolution of Repeat-Based Centromeres
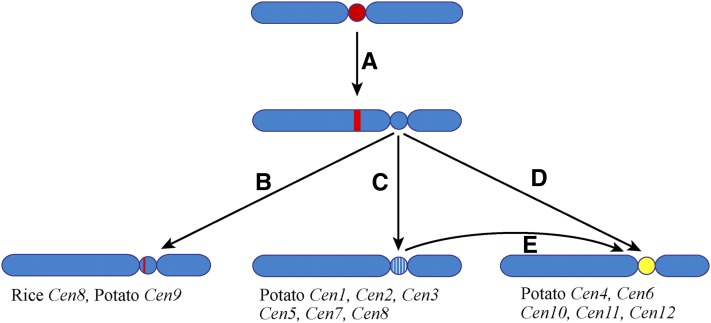
If centromeres evolve from newly formed centromeres with a typical genomic structure similar to human neocentromeres, then most of these new centromeres may be short lived because centromeres analyzed in most higher eukaryotes to date are composed of long arrays of satellite repeats (Figure 10). Only a few centromeres have an intermediate genomic structure between human neocentromeres and repeat-based centromeres. The centromere of rice chromosome 8 (Cen8) spans an ∼750-kb CENH3 domain, which includes only ∼65 kb of the 155-bp rice centromeric satellite repeat (Nagaki et al., 2004; Wu et al., 2009) (Figure 10). The rest of the DNA sequences within the CENH3 domain of Cen8 are similar to the average rice genome sequences, including actively transcribed genes (Nagaki et al., 2004; Yan et al., 2005). Interestingly, we were unable to identify satellite repeats in five of the 12 potato centromeres. The DNA sequences within these five potato centromeres were not distinctly different from the flanking sequences in the pericentromeric regions. The DNA sequences associated with these five centromeres appear to be evolving similarly as typical intergenic sequences by accumulation of DNA mutation and transposon insertions (Figure 10).
Figure 10.
A Model of Centromere Evolution.
(A) A neocentromere activation event may result in the repositioning of the centromere.
(B) The evolutionarily new centromere acquired a satellite repeat array during evolution. The satellite repeat may be derived from other centromeres, such as rice Cen8, or a new repeat, such as potato Cen9. The satellite repeat array in the evolutionarily new centromere may expand and eventually occupy the entire centromere.
(C) The evolutionarily new centromere may survive for several million years without satellite repeat invasion. Such evolutionarily new centromeres will slowly evolve by accumulating DNA mutations and transposable elements (white lines).
(D) and (E) A de novo DNA amplification of a satellite repeat, possibly based on an eccDNA-mediated mechanism, and insertion of the repeat (yellow) in the CENH3 domain can turn an evolutionarily new centromere into a repeat-based centromere.
The presence of the two distinct types of centromeres in potato suggests that the evolution from neocentromeres to repeat-based centromeres is a sudden rather than a gradual process. A de novo amplification and insertion of a megabase-sized satellite repeat array can dramatically turn a repeatless centromere into a repeat-based centromere, although it can also be achieved by an insertion of a relatively short array followed by rapid expansion of the array. De novo amplification of satellite repeats, possibly via eccDNA-based mechanisms, may occur constantly during genome evolution, such as the Sobo repeat in S. bulbocastanum (Tek et al., 2005). However, most of such newly amplified satellite repeats will not be fixed in the population because these repeats will be unlikely to impact the fitness of the organism. By contrast, a new satellite repeat inserted into a centromere will have a better chance to be fixed, as satellite repeats are likely favorable for organizing CENH3-associated nucleosome arrays. CENH3 nucleosomes are conformationally more rigid than H3-associated nucleosomes (Black et al., 2007). This unique physical characteristic of CENH3 nucleosome arrays may help to orient and distribute the forces from microtubule binding and pulling during anaphases. Alternatively, insertion of a satellite repeat array will result in the expansion of CENH3 domains of the neocentromeres, which are usually smaller than normal centromeres. A centromere with an expanded CENH3 domain will be favorably transmitted due to competition in female meiosis (Fishman and Saunders, 2008; Malik and Henikoff, 2009).
Centromeric satellite repeats are often homogenized in the entire genome. Thus, in most higher eukaryotes, a single type of satellite repeat dominates all centromeres. Potato and chicken are among the rare examples in which a species contains multiple centromeric satellite repeats (Shang et al., 2010). The mechanism of genome-wide homogenization of a single centromeric repeat is not known. Different satellite repeats may have different levels of fitness for organizing CENH3 nucleosomes, which explains the fact that the most common monomer sizes of the centromeric satellite repeats are 150 to 180 bp. We hypothesize that movement of such a favorable satellite repeat from one centromere to another centromere can also be achieved by eccDNA-based systems. Tens of thousands of short eccDNAs (<400 bp) have recently been reported in mammalian species (Shibata et al., 2012). Thus, eccDNAs are likely more widely present in higher eukaryotes than what we previously understood. In addition, satellite repeats are prone to eccDNA formation possibly via intrachromosomal homologous recombination, which has recently been demonstrated in both animal and plant species (Cohen et al., 2006; Cohen et al., 2008; Navrátilová et al., 2008). Rolling circle replication of eccDNA and reinsertion into the genome may eventually spread a single satellite repeat to all centromeres. Interestingly, none of the potato and chicken centromeric repeats resemble the classical centromeric satellites with monomeric sizes of 150 to 180 bp. Thus, these repeats may not represent the most favorable repeat to be fixed for centromeres.
METHODS
Plant Materials
DM1-3 516R44 (DM1-3), a homozygous doubled monoploid (2n = 2x = 24) clone developed from a diploid potato species Solanum phureja, was used for ChIP and cytogenetic studies. Six wild Solanum species, including Solanum verrucosum (A genome, PI 275260), Solanum jamesii (B genome, PI 620869), Solanum chromatophilum (P genome, PI 365339), tomato (Solanum lycopersicum cv MicroTom) (T genome), Solanum etuberosum (E genome, PI 558288), and Solanum palustre (E genome, PI 558245), were used for FISH mapping of the centromeric repeats identified in DM1-3. All these species are diploids with a chromosome number of 24. Seeds of all Solanum species, with the exception of the tomato cultivar MicroTom, were obtained from the USDA/Agricultural Research Service Potato Introduction Station, Sturgeon Bay, WI.
ChIP, ChIP-seq, and Quantitative ChIP-PCR
The potato (Solanum tuberosum) CENH3 antibody is a rabbit polyclonal antiserum and was raised against the peptide acetyl-RTKHLAKRSRTKPSVAC-amide. ChIP was performed as previously described (Nagaki et al., 2003) with only minor modifications. Approximately 10 g of fresh leaf tissue was collected from young potato DM1-3 plants grown in the greenhouse. Nuclei extracted from leaf tissue were digested with micrococcal nuclease (Sigma-Aldrich). After two rounds of centrifugation at 13,000 rpm for 10 min at 4°C, the digested chromatin was used for ChIP experiments using the potato CENH3 antibody. Approximately 30 ng of ChIP DNA was used for high-throughput sequencing library preparation using the ChIP-seq protocol provided by Illumina, including repairing the ends of DNA fragments, poly(A) tailing of the 3′ ends, ligation of paired-end adapters, fractionation of 150- to 300-bp adapter-ligated DNA using 2% agarose gel, and enrichment of sized adapter-modified DNA fragments by PCR. The enriched DNA sample was sequenced using Illumina Genome Analyzer II generating 36-bp sequence reads.
ChIP-qPCR was performed to confirm the relative enrichment of specific sequences within anti-CENH3 precipitated DNA relative to the DNA sample prepared from mock (normal IgG) immunoprecipitation reaction. Targeting DNA fragments used for quantitative PCR signal quantification were designed to be between 100 to 120 bp in length, to ensure that the amplified size range fit within individual mononucleosomes. We used the actin gene, which is located outside of the centromeres, as a negative control to normalize the enrichment of each positive amplicon. We calculated the difference (ΔCT) in the PCR cycle threshold (CT) to determine the relative enrichment of each amplicon as previously described (Yan et al., 2005).
Mapping of ChIP-seq Reads to the Potato Genome
Sequence reads generated from ChIP-seq were aligned to the recently released potato genome sequence map derived from the DM1-3 clone (PGSC_DM_v3_2.1.10_pseudomolecule downloaded from http://potatogenomics.plantbiology.msu.edu/) using the MAQ alignment program (Li et al., 2008). We allowed 1-bp mismatch between each sequence read (36 bp) and the reference genome; only reads that mapped to a unique position of the potato genome were retained for further analysis. To map the CENH3 enrichment along each chromosome in an unbiased approach, we first identified all uniquely mappable regions in the DM1-3 genome. We generated 36-bp reads starting from every base pair of the potato genome and mapped the reads to the genome, retaining the reads that mapped to a unique position. The genomic position of the starting nucleotide of a unique read was considered as a uniquely mappable region. We then divided each chromosome into 10-kb windows and calculated the unique read number per base pair mappable region in each window. Thus, read density equals the number of unique reads in a 10-kb window per the length of mappable region in the same window.
We used SICER (version 1.03) to identify the CENH3 subdomains in each potato centromere. The software was optimized for diffuse, variable-length regions spanning from several nucleosomes to large domains (Zang et al., 2009). We used 1-kb windows, which required the P value of a CENH3 subdomain to be <0.0001, and allowed 1-kb gaps in the defined CENH3 subdomains. If a sequence gap or a nonmappable region in the core of the centromeres was located adjacent to a defined CENH3 subdomain(s), this region was then arbitrarily assigned to a CENH3 subdomain. Few genes spanned a transition zone between a CENH3 and a H3 subdomain. A gene was considered to be located within the CENH3/H3 subdomain if >50% of the sequence of this gene was located within this specific subdomain. The reference genome of potato (PGSC_DM_v3_2.1.10_pseudomolecules), gene annotation (PGSC_DM_v3.4_gene.fasta), and gene expression value based on RNA-seq data from 40 DM1-3 libraries (32 different tissues/treatments) were downloaded from http://potatogenomics.plantbiology.msu.edu/index.html (Xu et al., 2011). The expression data of libraries derived from the same tissue/treatment showed high correlations (Pearson correlation coefficient >0.96). Thus, we averaged the FPKM values of libraries from same tissue/treatment and used the expression data from each of the 32 tissue/treatment in our analysis.
Repeat Identification and Characterization
Similarity-based clustering and repeat identification in a set of 1,238,463 WGS 454 sequences derived from DM1-3 was performed as previously described (Torres et al., 2011). Investigation of cluster graphs was performed using the SeqGrapheR program (Novák et al., 2010). Cloning of selected satellite repeats was done using PCR primers designed according to their reconstructed consensus sequences (see Supplemental Table 2 online). To identify repeats associated with CenH3 nucleosomes, a set of 10 million randomly sampled ChIP-seq reads was mapped to 454 read clusters based on their sequence similarities detected using PatMaN program (Prüfer et al., 2008), allowing for maximum of three mismatches including two gaps (the gaps were allowed in order to compensate for homopolymer sequencing errors in the 454 reads). Each ChIP-seq read was mapped to a maximum of one cluster, based on its best similarity detected among 454 reads.
FISH, Fiber-FISH, and Immunofluorescence
Preparation of mitotic and meiotic chromosomes, FISH, and fiber-FISH were performed following published protocols (Jackson et al., 1998; Dong et al., 2000; Lou et al., 2010). DNA probes for each centromere-specific satellite repeat were amplified by PCR from the DM1-3 genomic DNA. Primers were designed from bioinformatically extracted repeat cluster (see Supplemental Table 2 online). The amplified DNAs were labeled with either biotin-16-UTP or digoxigenin-11-dUTP (Roche Diagnostics) using a standard nick translation reaction. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) in Vectashield antifade solution (Vector Laboratories). The FISH images were processed with Meta Imaging Series 7.5 software. The final contrast of the images was processed using Adobe Photoshop CS3 software. The cytological measurements of the fiber-FISH signals were converted into kilobases using a 3.21-kb/μm conversion rate (Cheng et al., 2002).
Root tips harvested from plants were fixed in 4% (w/v) paraformaldehyde for 15 min at room temperature. The root tips were washed with 1× PBS three times, each for 5 min, and were squashed on glass slides. After removal of the cover slip, the slides were dehydrated using ethanol (70, 90, and 100%) and then incubated in a humid chamber at 37°C for overnight with the rabbit primary sera antibody against potato CENH3 diluted 1:500 in TNB buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, and 0.5% blocking reagent). After three rounds of washing in 1× PBS, the slides were incubated with Cy3-conjugated goat anti-rabbit antibody (1:1000) at 37°C for 1 h. After three rounds of washing in 1× PBS, the slides dried at room temperature and the chromosomes were counterstained with DAPI.
Expression of Centromeric Genes
Expression of the potato genes PGSC0003DMG400012074 and PGSC0003DMG400039086 was quantified by real-time PCR. An untranscribed gene, PGSC0003DMG400043879, located in Cen12, was used as a negative control. RNA was extracted from pooled leaf tissue of three young DM1-3 plants (full expanded terminal leaflets from the top of the plants) using the Qiagen Plant RNeasy kit with the on-column DNase digestion according to the manufacturer’s instructions. RNA was again treated with Turbo DNA-Free (Ambion/Applied Biosystems). RNA quality/quantity and integrity were evaluated using Nanodrop absorbance and agarose gel electrophoresis, respectively. Super Script III reverse transcriptase (Invitrogen) and oligo(dT) primers were used to generate the first-strand cDNA. A control without reverse transcriptase was used to confirm that the RNA was free of any DNA contamination. Primers and genes used in this study are listed in Supplemental Table 3 online. All primers were first tested on both genomic DNA and cDNA by regular PCR, with 37 cycles of heat denaturation at 95°C for 20 s, annealing at 60°C for 20 s (63°C for PGSC0003DMG400043879), and extension at 72°C for 30 s after an initial heat denaturation at 95°C for 5 min. For cycle-limited PCR, the number of cycles was reduced to 33. Amplification products were analyzed by agarose electrophoresis and ethidium bromide staining. All RT-PCR reactions and subsequent amplicon melting curves were performed in triplicate using Dynamo SYBR Green master on the MJ Research Opticon 2. Normalized expression level was calculated using the comparative Ct method and the reference gene actin 97, according to the equation 2−ΔCT where ΔCT = CT (target gene) − CT (actin 97). The normalized expression level of the gene urease was used for comparison because urease is a single-copy gene in potato with a low level of mRNA accumulation (Witte et al., 2005).
Accession Numbers
Sequence data for the centromeric satellite repeat sequences can be found in the GenBank data library under accession numbers JQ731637 to JQ731643.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Fine Structure of the Functional Core of Potato Cen6.
Supplemental Figure 2. Fine Structure of the Functional Core of Potato Cen11.
Supplemental Figure 3. Fine Structure of the Functional Core of Potato Cen12.
Supplemental Figure 4. ChIP-qPCR Confirmation of Potato Genes Associated with CENH3 Nucleosomes.
Supplemental Figure 5. Proportion of Repeat Families in WGS-454 and CENH3 ChIP-seq Data.
Supplemental Figure 6. Representative Fiber-FISH Images from Six Centromere-Specific Satellite Repeats.
Supplemental Table 1. Measurements of Fiber-FISH Signals from Six Centromeric Repeats.
Supplemental Table 2. PCR Primers Used for Cloning Potato Satellite Repeats.
Supplemental Table 3. Primer Set Used to Analyze Putative Active Genes Located in the CENH3 Domain.
Supplemental Data Set 1. Genes and Their Associated RNA-seq Data in Four Potato Centromeres.
Acknowledgments
We thank David Spooner for constructing the phylogenetic tree in Figure 8. This work was supported by Grants DBI-0604907 and DBI-0834044 from the National Science Foundation to C.R.B., by Grants DBI-0922703 and DBI-0923640 and Hatch funds to J.J., by Grants LH11058 and AVOZ50510513 from the Ministry of Education, Youth, and Sport and from the Academy of Sciences of the Czech Republic to J.M., and by a fellowship from the Brazilian Ministry of Education to G.A.T.
AUTHOR CONTRIBUTIONS
J.J. and J.M. designed the research. Z.G., A.K., G.A.T., K.W., M.I., W.Z., and P.N. performed the research. Y.W., P.N., J.M., and J.J. analyzed the data. J.J., J.M., and C.R.B. wrote the article. J.M. and J.J. are joint senior authors who contributed to this project equally.
Glossary
- ChIP
chromatin immunoprecipitation
- FISH
fluorescence in situ hybridization
- ChIP-seq
chromatin immunoprecipitation sequencing (ChIP-seq)
- FPKM
fragments per kilobase of exon model per million mapped fragments
- WGS
whole-genome shotgun
- LTR
long terminal repeat
- eccDNA
extrachromosomal circular DNA
- DAPI
4′,6-diamidino-2-phenylindole
References
- Allshire R.C., Nimmo E.R., Ekwall K., Javerzat J.P., Cranston G. (1995). Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9: 218–233 [DOI] [PubMed] [Google Scholar]
- Black B.E., Brock M.A., Bédard S., Woods V.L., Jr, Cleveland D.W. (2007). An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 104: 5008–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone M.F., et al. (2006). Independent centromere formation in a capricious, gene-free domain of chromosome 13q21 in Old World monkeys and pigs. Genome Biol. 7: R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo R.O., Spooner D.M. (1997). Phylogenetic relationships of wild potatoes, Solanum series Conicibaccata (sect. Petota). Syst. Bot. 22: 45–83 [Google Scholar]
- Cheng Z.K., Buell C.R., Wing R.A., Jiang J. (2002). Resolution of fluorescence in-situ hybridization mapping on rice mitotic prometaphase chromosomes, meiotic pachytene chromosomes and extended DNA fibers. Chromosome Res. 10: 379–387 [DOI] [PubMed] [Google Scholar]
- Cohen S., Houben A., Segal D. (2008). Extrachromosomal circular DNA derived from tandemly repeated genomic sequences in plants. Plant J. 53: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Cohen Z., Bacharach E., Lavi S. (2006). Mouse major satellite DNA is prone to eccDNA formation via DNA Ligase IV-dependent pathway. Oncogene 25: 4515–4524 [DOI] [PubMed] [Google Scholar]
- Dong F.G., Song J.Q., Naess S.K., Helgeson J.P., Gebhardt C., Jiang J.M. (2000). Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor. Appl. Genet. 101: 1001–1007 [Google Scholar]
- Felcher K.J., Coombs J.J., Massa A.N., Hansey C.N., Hamilton J.P., Veilleux R.E., Buell C.R., Douches D.S. (2012). Integration of two diploid potato linkage maps with the potato genome sequence. PLoS ONE 7: e36347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L., Saunders A. (2008). Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322: 1559–1562 [DOI] [PubMed] [Google Scholar]
- Gassmann R., et al. (2012). An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature 484: 534–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilenko T. (2007). Potato cytogenetics. In Potato Biology and Biotechnology: Advances and Perspectives, D. Vreugdenhil, J. Bradshaw, C. Gebhardt, F. Govers, D.K.L. MacKerron, M.A. Taylor, and H.A. Ross, eds (Amsterdam: Elsevier), pp. 203–216.
- Gorinsek B., Gubensek F., Kordis D. (2004). Evolutionary genomics of chromoviruses in eukaryotes. Mol. Biol. Evol. 21: 781–798 [DOI] [PubMed] [Google Scholar]
- Harrington J.J., Van Bokkelen G., Mays R.W., Gustashaw K., Willard H.F. (1997). Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 15: 345–355 [DOI] [PubMed] [Google Scholar]
- Hasson D., Alonso A., Cheung F., Tepperberg J.H., Papenhausen P.R., Engelen J.J.M., Warburton P.E. (2011). Formation of novel CENP-A domains on tandem repetitive DNA and across chromosome breakpoints on human chromosome 8q21 neocentromeres. Chromosoma 120: 621–632 [DOI] [PubMed] [Google Scholar]
- Hawkes J.G. (1990). The Potato: Evolution, Biodiversity and Genetic Resources. (Washington, DC: Smithsonian Institution Press).
- Henikoff S., Ahmad K., Malik H.S. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Houben A., Schroeder-Reiter E., Nagaki K., Nasuda S., Wanner G., Murata M., Endo T.R. (2007). CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma 116: 275–283 [DOI] [PubMed] [Google Scholar]
- Ikeno M., Grimes B., Okazaki T., Nakano M., Saitoh K., Hoshino H., McGill N.I., Cooke H., Masumoto H. (1998). Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 16: 431–439 [DOI] [PubMed] [Google Scholar]
- Ishii K., Ogiyama Y., Chikashige Y., Soejima S., Masuda F., Kakuma T., Hiraoka Y., Takahashi K. (2008). Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321: 1088–1091 [DOI] [PubMed] [Google Scholar]
- Jackson S.A., Wang M.L., Goodman H.M., Jiang J.M. (1998). Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome 41: 566–572 [PubMed] [Google Scholar]
- Jiang J.M., Birchler J.A., Parrott W.A., Dawe R.K. (2003). A molecular view of plant centromeres. Trends Plant Sci. 8: 570–575 [DOI] [PubMed] [Google Scholar]
- Ketel C., Wang H.S.W., McClellan M., Bouchonville K., Selmecki A., Lahav T., Gerami-Nejad M., Berman J. (2009). Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 5: e1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon T., Seago C., Jones R.N., Ougham H., Thomas H., Forster J.W., Jenkins G. (2000). De novo evolution of satellite DNA on the rye B chromosome. Genetics 154: 869–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.R., Zhang W.L., Langdon T., Jin W.W., Yan H.H., Cheng Z.K., Jiang J.M. (2005). Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. USA 102: 11793–11798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R. (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomiento M., Jiang Z.S., D’Addabbo P., Eichler E.E., Rocchi M. (2008). Evolutionary-new centromeres preferentially emerge within gene deserts. Genome Biol. 9: R173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Q.F., Iovene M., Spooner D.M., Buell C.R., Jiang J.M. (2010). Evolution of chromosome 6 of Solanum species revealed by comparative fluorescence in situ hybridization mapping. Chromosoma 119: 435–442 [DOI] [PubMed] [Google Scholar]
- Macas J., Koblízková A., Navrátilová A., Neumann P. (2009). Hypervariable 3′ UTR region of plant LTR-retrotransposons as a source of novel satellite repeats. Gene 448: 198–206 [DOI] [PubMed] [Google Scholar]
- Macas J., Mészáros T., Nouzová M. (2002). PlantSat: A specialized database for plant satellite repeats. Bioinformatics 18: 28–35 [DOI] [PubMed] [Google Scholar]
- Macas J., Neumann P., Navrátilová A. (2007). Repetitive DNA in the pea (Pisum sativum L.) genome: Comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics 8: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H.S., Henikoff S. (2009). Major evolutionary transitions in centromere complexity. Cell 138: 1067–1082 [DOI] [PubMed] [Google Scholar]
- Marshall O.J., Chueh A.C., Wong L.H., Choo K.H.A. (2008). Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 82: 261–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi M. (1991). Phylogenetic relationships in the potato and its related species. In Chromosome Engineering in Plants: Genetics, Breeding, Evolution, T. Tsuchiya and P. Gupta, eds (Amsterdam: Elsevier), pp. 93–118.
- Nagaki K., Cheng Z.K., Ouyang S., Talbert P.B., Kim M., Jones K.M., Henikoff S., Buell C.R., Jiang J.M. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36: 138–145 [DOI] [PubMed] [Google Scholar]
- Nagaki K., Talbert P.B., Zhong C.X., Dawe R.K., Henikoff S., Jiang J.M. (2003). Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163: 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navrátilová A., Koblízková A., Macas J. (2008). Survey of extrachromosomal circular DNA derived from plant satellite repeats. BMC Plant Biol. 8: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt T.C., Tanksley S.D. (2002). Comparative sequencing in the genus Lycopersicon. Implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics 162: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P., Neumann P., Macas J. (2010). Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K., Stenzel U., Dannemann M., Green R.E., Lachmann M., Kelso J. (2008). PatMaN: Rapid alignment of short sequences to large databases. Bioinformatics 24: 1530–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffery R., Wong L.H., Irvine D.V., Bateman M.A., Griffiths B., Cutts S.M., Cancilla M.R., Cendron A.C., Stafford A.J., Choo K.H.A. (2001). Construction of neocentromere-based human minichromosomes by telomere-associated chromosomal truncation. Proc. Natl. Acad. Sci. USA 98: 5705–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W.H., Hori T., Toyoda A., Kato J., Popendorf K., Sakakibara Y., Fujiyama A., Fukagawa T. (2010). Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 20: 1219–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y., Kumar P., Layer R., Willcox S., Gagan J.R., Griffith J.D., Dutta A. (2012). Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science 336: 82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata F., Murata M. (2004). Differential localization of the centromere-specific proteins in the major centromeric satellite of Arabidopsis thaliana. J. Cell Sci. 117: 2963–2970 [DOI] [PubMed] [Google Scholar]
- Spooner D.M., Anderson G.J., Jansen R.K. (1993). Chloroplast DNA evidence for the interrelationships of tomatoes, potatoes, and pepinos (Solanaceae). Am. J. Bot. 80: 676–688 [Google Scholar]
- Spooner D.M., Castillo R. (1997). Reexamination of series relationships of South American wild potatoes (Solanaceae: Solanum sect. Petota): Evidence from chloroplast DNA restriction site variation. Am. J. Bot. 84: 671–685 [PubMed] [Google Scholar]
- Tek A.L., Jiang J. (2004). The centromeric regions of potato chromosomes contain megabase-sized tandem arrays of telomere-similar sequence. Chromosoma 113: 77–83 [DOI] [PubMed] [Google Scholar]
- Tek A.L., Song J.Q., Macas J., Jiang J. (2005). Sobo, a recently amplified satellite repeat of potato, and its implications for the origin of tandemly repeated sequences. Genetics 170: 1231–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., et al. Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195 [DOI] [PubMed] [Google Scholar]
- Torres G.A., Gong Z.Y., Iovene M., Hirsch C.D., Buell C.R., Bryan G.J., Novák P., Macas J., Jiang J.M. (2011). Organization and evolution of subtelomeric satellite repeats in the potato genome. G3 1: 85–92. [DOI] [PMC free article] [PubMed]
- Ventura M., Antonacci F., Cardone M.F., Stanyon R., D’Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M. (2007). Evolutionary formation of new centromeres in macaque. Science 316: 243–246 [DOI] [PubMed] [Google Scholar]
- Ventura M., Archidiacono N., Rocchi M. (2001). Centromere emergence in evolution. Genome Res. 11: 595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikström N., Savolainen V., Chase M.W. (2001). Evolution of the angiosperms: Calibrating the family tree. Proc. Biol. Sci. 268: 2211–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte C.P., Tiller S., Isidore E., Davies H.V., Taylor M.A. (2005). Analysis of two alleles of the urease gene from potato: Polymorphisms, expression, and extensive alternative splicing of the corresponding mRNA. J. Exp. Bot. 56: 91–99 [DOI] [PubMed] [Google Scholar]
- Wu J.Z., et al. (2009). Comparative analysis of complete orthologous centromeres from two subspecies of rice reveals rapid variation of centromere organization and structure. Plant J. 60: 805–819 [DOI] [PubMed] [Google Scholar]
- Wu Y.F., Kikuchi S., Yan H.H., Zhang W.L., Rosenbaum H., Iniguez A.L., Jiang J.M. (2011). Euchromatic subdomains in rice centromeres are associated with genes and transcription. Plant Cell 23: 4054–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.H., et al. (2006). Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell 18: 2123–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.H., Jin W.W., Nagaki K., Tian S., Ouyang S., Buell C.R., Talbert P.B., Henikoff S., Jiang J.M. (2005). Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell 17: 3227–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.H., Talbert P.B., Lee H.R., Jett J., Henikoff S., Chen F., Jiang J.M. (2008). Intergenic locations of rice centromeric chromatin. PLoS Biol. 6: e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W. (2009). A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25: 1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.L., Friebe B., Gill B.S., Jiang J.M. (2010). Centromere inactivation and epigenetic modifications of a plant chromosome with three functional centromeres. Chromosoma 119: 553–563 [DOI] [PubMed] [Google Scholar]