Tomato mutants defective in ta-siRNA production have narrow shoestring leaves reminiscent of virus-infected plants. The tomato leaf phenotype is due to deregulated ARF gene expression, but ectopic expression of the same ARF genes in related species fails to recapitulate these developmental defects, analogous to species-specific viral infection symptoms.
Abstract
Interfering with small RNA production is a common strategy of plant viruses. A unique class of small RNAs that require microRNA and short interfering (siRNA) biogenesis for their production is termed trans-acting short interfering RNAs (ta-siRNAs). Tomato (Solanum lycopersicum) wiry mutants represent a class of phenotype that mimics viral infection symptoms, including shoestring leaves that lack leaf blade expansion. Here, we show that four WIRY genes are involved in siRNA biogenesis, and in their corresponding mutants, levels of ta-siRNAs that regulate AUXIN RESPONSE FACTOR3 (ARF3) and ARF4 are reduced, while levels of their target ARFs are elevated. Reducing activity of both ARF3 and ARF4 can rescue the wiry leaf lamina, and increased activity of either can phenocopy wiry leaves. Thus, a failure to negatively regulate these ARFs underlies tomato shoestring leaves. Overexpression of these ARFs in Arabidopsis thaliana, tobacco (Nicotiana tabacum), and potato (Solanum tuberosum) failed to produce wiry leaves, suggesting that the dramatic response in tomato is exceptional. As negative regulation of orthologs of these ARFs by ta-siRNA is common to land plants, we propose that ta-siRNA levels serve as universal sensors for interference with small RNA biogenesis, and changes in their levels direct species-specific responses.
INTRODUCTION
Classical tomato (Solanum lycopersicum) mutants with needle-like leaves were termed “wiry” and were described as having “a virus-like syndrome.” In wiry plants, some leaves have reduced lamina and others are nearly radial, giving the mutant its name. This shoestring disease-mimic phenotype is caused by a single Mendelian mutation that could not be transmitted by grafting or by juice inoculation but was transmitted to progeny as a recessive character (Lesley and Lesley, 1928; Edwardson and Corbett, 1962). Years of collections of single-gene tomato mutations (hosted by the Tomato Genetics Resource Center) and a large-scale screen in a uniform background (Menda et al., 2004) resulted in isolation of several wiry mutations, similar to the original wiry described by Lesley and Lesley (1928). Scanning electron microscopy and anatomical analyses of the original mutant isolate indicated that wiry lamina has normal polarity, but shoestring leaves of the same plant are partially abaxialized (Kim et al., 2003). Thus, unraveling the molecular basis of the wiry syndrome can link patterning processes with the response of plants to pathogen infection. A potential source for such a link was recently offered by the analysis of gene regulation by plant small RNAs that play a role in both patterning processes and host–pathogen interactions (Adenot et al., 2006).
In several different plant species, needle-like leaves lacking lamina expansion are often caused by impaired regulation of organ polarity genes (reviewed in Husbands et al., 2009). Small RNAs are important regulators of some of the organ polarity genes; miR165/6 regulates the adaxial-specifying class III homeodomain-Leu zipper genes, and miR390 is a trigger for the biogenesis of the trans-acting short interfering RNA (ta-siRNA) pathway that regulates two auxin response factors (ARFs), ARF3 and ARF4 (Allen et al., 2005; Fahlgren et al., 2006; Hunter et al., 2006), which stabilize abaxial organ identity in Arabidopsis thaliana (Pekker et al., 2005), tomato, and tobacco (Nicotiana tabacum; Alvarez et al., 2006).
In Arabidopsis, the ta-siRNAs regulating ARF3 and ARF4 (which will be collectively termed here as ARFs) are derived from noncoding genes termed TAS3, collectively, which are cleaved by an ARGONAUTE7 (AGO7)-miR390 complex; the resulting single-stranded cleaved TAS3 RNA is used as a template for the polymerization of double-stranded RNA (dsRNA) by the RNA-DEPENDENT RNA POLYMERASE6 (RDR6) and SUPPRESSOR OF GENE SILENCING3 (SGS3). This dsRNA is then diced by DICER-LIKE4 (DCL4) into phased 21-nucleotide short interfering RNA (siRNA). Two of the resulting ta-siRNAs target ARF3 and ARF4 and have the potential to also target ARF2 (reviewed in Allen and Howell, 2010). The miR390-TAS3-ARFs regulation pathway is highly conserved and can be found in moss, grasses, and dicotyledonous plants (Axtell et al., 2006; Nagasaki et al., 2007; Douglas et al., 2010).
In Arabidopsis, the ta-siRNAs are implicated in regulation of developmental timing as mutants in the pathway have leaf shape defects termed “vegetative phase change” disruptions (Hunter et al., 2003; Xie et al., 2005). In rice (Oryza sativa), mutations in the ta-siRNA biogenesis genes SHOOTLESS2 (SHL2/RDR6), SHL4/SHOOT ORGANIZATION2 (SHO2/AGO7), and SHO1/DCL4 caused absence or abnormal formation of the shoot apical meristem (SAM) (Liu et al., 2007; Nagasaki et al., 2007). The maize (Zea mays) mutant ragged seedling2 (rgd2/ago7) affects medio-lateral expansion but not dorsiventrality of leaves (Douglas et al., 2010), while leafbladeless1 (lbl1/sgs3) has disrupted leaf polarity (Husbands et al., 2009). In Arabidopsis, the ta-siRNAs regulate the mRNA levels of both ARF3 and ARF4 (Peragine et al., 2004; Allen et al., 2005). In rice and maize, however, no ARF4 orthologs are known, but several ARF3 paralogs are present and the levels of at least some are altered in the different mutants that are impaired in ta-siRNA biogenesis (Liu et al., 2007; Nogueira et al., 2007; Douglas et al., 2010).
Notably, small RNAs do not equally regulate ARF3 and ARF4. For example, Axtell et al. (2006) described Arabidopsis small RNAs corresponding to the ARF4 mRNA that originates from the sequence between the two ta-siARF recognition sites as well as from the 3′ terminus and correspond to both sense and antisense strands. In the same study, only a single read of a small RNA that originated from the ARF3 mRNA could be found. Thus, small RNAs that regulate ARF3 and ARF4 but do not equally require the ta-siRNA biogenesis genes potentially exist. These small RNAs may be produced from the ARF transcripts themselves and may not require the same genes for their biogenesis. For example, formation of natural antisense-siRNA shows a differential requirement for RDR6, SGS3, AGO7, and DCL4 (Borsani et al., 2005; Ron et al., 2010), and inverted repeat–derived siRNAs require DCLs but not RDR6 or SGS3 for their biogenesis (Dunoyer et al., 2010) as they are intron-derived siRNAs (Chen et al., 2011). Thus, the ta-siARFs may not be the only small RNAs involved in regulation of ARF3 and ARF4.
Small RNAs are important for antiviral plant defense, and components of small RNA biogenesis or processing are often targets for viral proteins that suppress endogenous defense mechanisms (reviewed in Alvarado and Scholthof, 2009). For example, the RDR6 protein in Nicotiana benthamiana has a role in antiviral defense; tobacco plants with reduced expression of RDR6 were more susceptible to specific viral infection. Also, when Tobacco mosaic virus (TMV)–green fluorescent protein infected these plants, narrow leaves and flowers arose (Qu et al., 2005). The tomato ortholog of SGS3 protein directly interacts with the viral suppressor V2 protein of tomato yellow leaf curl geminivirus, and a mutated version of the V2 protein that could not interact with SGS3 neutralized its ability to suppress RNA silencing (Glick et al., 2008). DCL4 is a component of the antiviral immunity system, and it is the major DICER responsible for the production of virus-derived small RNAs in Arabidopsis (Deleris et al., 2006).
In this study, we show that the four tomato wiry mutants are disrupted in the ta-siRNA biogenesis genes RDR6, SGS3, AGO7, and DCL4. The wiry syndrome results from a failure to negatively regulate ARF3 and/or ARF4, which are differentially misregulated in the different mutants. Small RNA profiles of the wiry mutants revealed complex biogenesis of siRNAs derived from the TAS3 and the ARF4 transcripts. We show that a phenocopy of the wiry syndrome can be stimulated by ectopic expression of ta-siARF–insensitive forms of either ARF3 or ARF4. Surprisingly, such a syndrome cannot be induced in related tobacco and potato (Solanum tuberosum) species, even though ARF3 of Arabidopsis can generate a wiry response in tomato. These results highlight the species-specific modifications that accompany this highly conserved small RNA–based regulatory module.
RESULTS
The wiry Syndrome, a Virus Infection Mimic Caused by Mendelian Genes
Tomato leaves comprised a terminal lobed leaflet and one to six pairs of lateral leaflets that initiate basipetally along the margins of leaf primordia and are subsequently separated by a rachis. Usually, the first leaves formed have one to two leaflet pairs, whereas the sixth leaf and subsequent leaves have the full complement of primary and secondary leaflets, each with its own petiole (Figure 1A). As the leaf matures, intercalary leaflets called folioles, having short or no petioles, emerge along the rachis between existing leaflets (Figure 1A). To identify mutants impaired in lamina expansion of leaves, we screened a population of mutagenized tomato lines (Menda et al., 2004) for disrupted leaf growth. In the past, mutants with nearly radial leaves were associated with the shoestring syndrome and the corresponding mutants were named wiry (w) (Lesley and Lesley 1928; Edwardson and Corbett, 1962). Altogether, we isolated 42 independent mutant lines, all were similar to the classical w, and complementation tests showed that these mutants fall into four complementation groups. These groups included new alleles of the original w mutant, new alleles of the previously identified w4 mutant (Clayberg et al., 1966), and alleles of two additional mutants that we named wiry2 (w2) and w3 (see Supplemental Table 1 online for the complete list of alleles). In all mutant lines, complete or partial lamina loss was evident; however, the extent of lamina loss varied considerably between sequentially formed leaves of the same plant and between different mutant lines (Figures 1B to 1D). Overall, the four wiry mutants can be divided into two groups; w, w2, and w4 (Figures 1B and 1C) were indistinguishable, whereas w3 plants (Figure 1D), mutated in the tomato DCL4 (see below), displayed a unique phenotype of spontaneous death with age.
Figure 1.
The Morphological Spectrum of the Wiry Syndrome.
(A) to (D) Heteroblasty (progressing from left to right) of wild-type (WT) tomato leaves (A), a typical strong wiry, w2-1 (B), a weak wiry, w2-3 (C), and a strong w3 allele (D) before the onset of necrosis. In all mutants, the first two to three leaves have five to seven abnormal leaflets and small intercalary folioles, while later-formed leaves lack lamina at different magnitudes.
(E) and (F) Toward flowering, wild-type leaves are not changing (E), while all leaves of strong wiry plants (F) lack or nearly lack leaflets and lamina.
(G) and (H) Scanning electron microscopy images of wild-type leaves (G) showing distinct initiation of trichomes on the abaxial and adaxial sides, whereas initiating needle-like wiry leaves (H) lack adaxial trichomes and the initiation of their abaxial characteristics is delayed.
(I) An 8-week-old w3-1 plant with necrosis throughout its leaves.
(J) Flowers of wild-type and wiry plants. w-1 flower is typical for w, w2, and w4, which are different from w3 flowers.
Bars = 5 cm (A) to (D) and (I) and 2 cm in (E), (F), and (J).
[See online article for color version of this figure.]
In strong alleles of wiry, such as w2-1 and w3-2 (Figures 1B and 1D), more leaflets that lack petioles formed along the rachis of the first three to four leaves. These leaflets were narrow and had entire margins compared with the wild type (cf. Figures 1A with 1B and 1D). In subsequently formed leaves, the number of leaflets decreased, lamina of the developing leaflets diminished, and, occasionally, a trumpet-shaped leaflet was formed (Figure 1B, last leaf). In weak wiry alleles, such as w2-3, the first formed leaves had more leaflets compared with the wild type, and primary leaflets had petiole and secondary leaflets (Figure 1C). As the primary shoot approaches flowering, needle-like leaves developed and leaflet initiation was lost (cf. Figures 1E with 1F). Along the sympodial shoot, a mixture of flat and needle-like leaves formed. The frequency of each class of leaves depended on growth conditions; floral-promoting factors such as high light intensity or a mutation in self pruning favored the formation of strong wiry leaves. Within the leaves, there is also a variation between individual leaflets (see Supplemental Figure 1A online); basal leaflets can be normal, whereas the primary terminal leaflet was often nearly radial (see Supplemental Figure 1B online). The tendency of the terminal leaflet to be nearly radial was also evident in secondary leaflets of weak wiry leaves, where the terminal leaflet was the only one that occasionally lacks lamina (see Supplemental Figures 1B and 1C online). In other lateral leaflets, a radial extension of the midrib was sometimes found, and this extension often had a trumpet-like structure at its tip (see Supplemental Figure 1C online).
Tomato leaf primordia have a distinct rate of trichome development with long trichomes appearing first on the abaxial side at P2 and short club-shaped trichomes appearing at the medial domain of the adaxial side at P3 (Figure 1G; see Supplemental Figure 2A online). By contrast, the shoestring-like leaves of the strong w2-1 line initiated trichomes on the abaxial side at a normal time, whereas their adaxial side lacked clear hallmarks of either abaxial or adaxial features (Figure 1H). As the leaf matured, primarily long abaxial trichomes were found around the needle-like leaves (see Supplemental Figure 2B online). A similar distribution of trichomes characterizes the weak w2-3 allele (see Supplemental Figure 2C online), but in that line, leaflet initiation was maintained, and spacing of leaflets was reduced (see Supplemental Figure 2D online). Thus, regulation of the adaxial and marginal domain of the leaf is a prime target of the WIRY genes.
Overall, the wiry mutants can be divided into two groups: w, w2, and w4 are indistinguishable, and differences between weak and strong alleles of the same complementation group were larger than differences between alleles of a different complementation group (see Supplemental Figures 1D and 1E compared with 1G online). By contrast, w3 plants have similar leaf defects but also accumulate anthocyanin upon germination and as the plant ages, the older leaves develop necrotic lesions and eventually the lamina dries (Figure 1I). With age, the necrosis spreads to the whole plant, eventually causing its death shortly after the formation of the first inflorescence. The appearance of the necrotic lesions varies between the different alleles and is more extreme under intense sunlight. The flowers of w3 mutants are also different from the other wiry mutants. Wild-type tomato flowers have five sepals, five yellow fused petals and stamens, and two to three fused carpels. The flowers of w, w2, and w4 plants are similar with narrow organs that are fused at their base only, whereas w3 flowers, when they develop, have elongated fused sepals enclosing retarded floral organs (Figure 1J). Moreover, w, w2, and w4 mutants are highly similar to each other; in all double mutant combinations that we analyzed, the most extreme phenotype was similar to a strong wiry allele (see Supplemental Figure 1F compared with 1G online). Hence, we hypothesized that w, w2, and w4 are impaired in a common regulatory pathway.
The wiry Mutants Are Impaired in Genes of the ta-siRNA Biogenesis Pathway
Lesley and Lesley (1928) described the wiry syndrome as producing a plant that “looks as though it were suffering from a severe case of the so-called ‘shoestring’ mosaic disease.” In recent years, several studies have linked small RNA biogenesis with antiviral plant defense. For example, the Cucumber mosaic virus (CMV) 2B protein can interact with Arabidopsis AGO1 and inhibit its slicer activity (Zhang et al., 2006). Also, N. benthamiana plants with reduced expression of RDR6 are more susceptible to viral infection (Qu et al., 2005). The very same viruses, CMV and TMV, were also recognized over the years as the cause of the tomato shoestring mosaic disease (Edwardson and Corbett, 1962; Andrade et al., 1981).
The resemblance of the wiry phenotype to virus-infected plants suggested to us that factors participating in the biogenesis of small RNAs might be disrupted in the mutant lines. The four wiry mutants were mapped by crossing mutant lines (as heterozygotes) with the wild tomato relative Solanum pennellii and analysis of marker segregation in F2 families (see Methods). In parallel, we cloned and mapped tomato genes involved in small RNA biogenesis and found that four of them cosegregated with specific wiry mutants. Since many alleles were available for each mutant, we sequenced several lines until several independent alleles were identified: Lesions in three w alleles were found in the tomato RDR6 ortholog (Figure 2A), w2 alleles had lesions in AGO7 (Figure 2B), w4 alleles had lesions in the SGS3 ortholog (Figure 2C), and w3 alleles had lesions in the DCL4 ortholog (Figure 2D). We therefore maintain the syndrome name, wiry, but rename the wiry mutants to permit consistent nomenclature: w- rdr6, w2-ago7, w3-dcl4, and w4-sgs3.
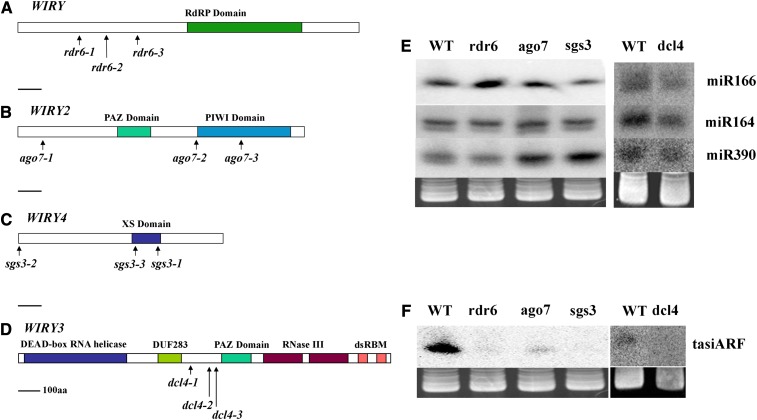
Figure 2.
wiry Mutants Are Impaired in Genes of the ta-siRNA Biogenesis Pathway.
(A) to (D) A scheme of the tomato orthologs of RDR6 (A), AGO7 (B), SGS3 (C), and DCL4 (D) and the lesions identified in the different alleles. aa, amino acids; dsRBM, double-stranded RNA binding motif.
(E) and (F) Different miRNAs, such as miR166, miR164, and miR390, are present in both wild-type (WT) and wiry shoots (E), but the ta-siARF derived from TAS3 could not be detected in any of the four wiry mutants (F).
[See online article for color version of this figure.]
As was shown in Arabidopsis, common to all four genes is their involvement in biogenesis of a specific class of siRNA, the ta-siRNA ( Yoshikawa et al., 2005). Indeed, levels of several microRNAs (miRNAs), such as miR166, miR164, and miR390, were variable among the different mutant lines (Figure 2E), but all lacked the expression of a particular ta-siRNA, ta-siARF (Figure 2F).
ARF3 and ARF4 mRNAs Are Misregulated in the wiry Mutants
The tomato genome contains single orthologs of ARF3 and ARF4 (Wu et al., 2011); both were shown previously to promote and stabilize the abaxial leaf domain (Alvarez et al., 2006). In Arabidopsis, ARF4 RNA is asymmetrically distributed in young leaf primordia, whereas ARF3 RNA can also be detected in the adaxial domain as well as in the SAM (Pekker et al., 2005). In tomato, we detected a similar trend: abaxial expression of ARF4 in leaf primordia and subsequent strong expression in provascular and vascular strands (Figure 3A). Tomato ARF3 distribution was similar in the two sides of leaf primordia, with weaker expression in the SAM (Figure 3B). Thus, tomato ARF3 cannot be considered as a strict abaxial promoting factor. As a marker of tomato leaf polarity, the distribution of a tomato PHB was found to mark the SAM and the adaxial leaf domain (Figure 3C). In agreement with leaf morphology, abaxial markers were widespread in wiry leaves; also, expression of ARF4 was much stronger in ago7-1 apices and was detected in the two sides of the initiating leaf primordia, as well as the SAM (Figure 3D). No change in the expression pattern of ARF3 mRNA could be detected in wiry apices (Figure 3E), whereas PHB mRNA was strongly reduced in leaf primordia (Figure 3F). Thus, the strong wiry shoestring leaves lose morphological and molecular adaxial hallmarks.
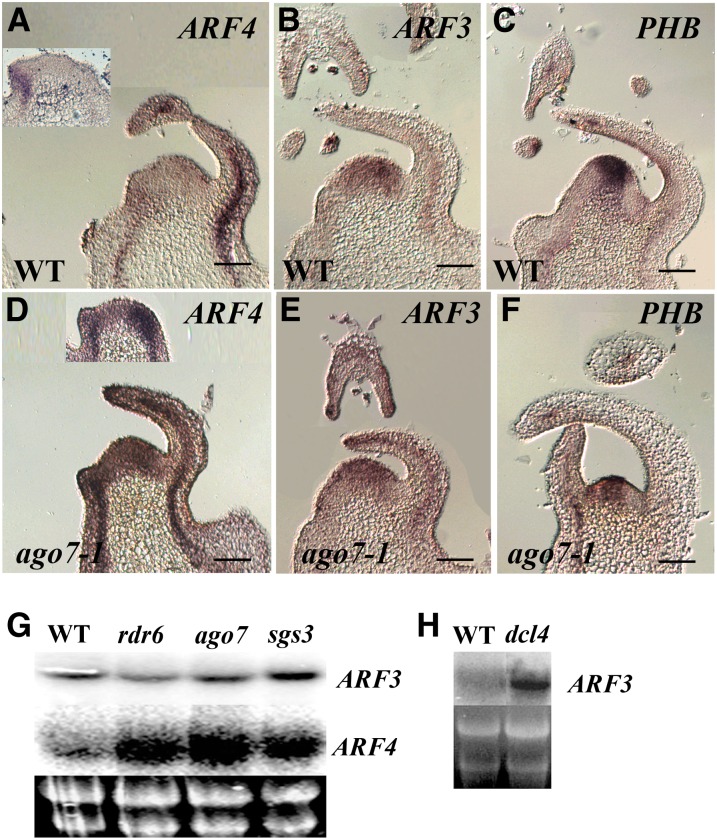
Figure 3.
A Loss of ARF3 and ARF4 Negative Regulation in the wiry Mutants.
(A) to (F) In situ localization of ARF4, ARF3, and PHB transcripts in wild-type (WT) ([A] to [C]) and ago7-1 ([D] to [F]) vegetative apices. ARF4 mRNA is abaxial in leaf primordia and later it is expressed in pro-vascular and vascular strands (A). Its expression is stronger and broader in ago7-1 apices (D). ARF3 mRNA is detected in both sides of leaf primordia, with weaker expression in the SAM (B), and no change is detected in its expression pattern in ago7-1 apices (E). PHB mRNA is adaxial in leaf primordial and in the SAM center (C). It is strongly reduced in ago7-1 vegetative apices but remains adaxial in leaves. Insets in (A) and (D) show sections through P1 leaf primordia. Bars = 50 μm.
(G) and (H) An RNA gel blot of RNA extracted from wild-type and wiry shoots probed with ARF3 and ARF4 cDNAs. Bottom panel is the RNA loaded. Note the high levels of ARF4 mRNA in rdr6, ago7, and sgs3 shoots (G) and the high levels of ARF3 in the dcl4 shoots (H).
[See online article for color version of this figure.]
Both ARF3 and ARF4 transcripts contain two sites that can be targeted by ta-siARF (Alvarez et al., 2006). To determine whether, like Arabidopsis, the tomato ta-siRNA pathway is involved in ARF3 and ARF4 regulation, the mRNA levels of both were examined in shoot apices containing small leaves. High ARF4 mRNA levels were detected in rdr6, ago7, and sgs3 mutants, but levels of ARF3 mRNA in shoots of these mutants were similar to levels found in comparable wild-type shoots (Figure 3G). Notably, however, high levels of ARF3 mRNA were detected in the dcl4 mutant shoots (Figure 3H). Thus, the four WIRY genes are commonly involved in the same siRNA biogenesis pathway, but their targets, ARF3 and ARF4, are not equivalently misregulated in the different mutant backgrounds though identical ta-siARF target sequences are present in the two genes.
Analysis of Small RNAs Derived from the ARF3 and ARF4 Transcripts
To further explore the role of small RNAs in Sl ARF3 and Sl ARF4 regulation, small RNA libraries prepared from apices of wild-type, slrdr6, slago7, and sldcl4 shoots were subjected to Solexa-based deep sequencing (Table 1; see Supplemental Data Set 1 online).
Table 1. General and Specific Reads from the Different Tomato Small RNA Libraries.
| Small RNA | Wild Type | ago7 | dcl4 | rdr6 |
|---|---|---|---|---|
| Number of readsa | 1,801,588 | 1,793,203 | 2,232,298 | 1,807,785 |
| Number of sequencesb | 113,903 | 122,640 | 114,658 | 113,991 |
| Novel sequencesc | – | 21,365 | 27,138 | 22,049 |
| Unique sequencesd | 5,256 | 3,526 | 7,650 | 3,353 |
| miR159 | 42,230 | 23,593 | 38,646 | 26,364 |
| miR164 | 730 | 2,890 | 2,768 | 2,770 |
| miR166 | 17,675 | 27,268 | 16,482 | 10,360 |
| miR390 | 182 | 155 | 251 | 205 |
| ta-siARF | 94 | 0 | 15 | 0 |
Number of reads after adaptor removal and filtering out rRNA and tRNAs.
Sequences are counted only when present in at least five independent reads.
Sequences not present in the wild-type library.
Sequences present only in this library.
–, no novel sequences.
The different small RNA libraries were prepared from shoot apices containing primordial leaves and flowers. These libraries contained both miRNAs common to all tested flowering plants as well as tomato-specific miRNAs (see Supplemental Table 2 online). Some of these miRNAs, such as miR164, were more prevalent in libraries of wiry apices compared with the wild type, likely reflecting their novel phenotype. Other miRNAs, such as miR166, were more abundant in the ago7 apices but less abundant in the rdr6 apices, suggesting a genotypic (i.e., phenotype-independent) cause (Table 1). A couple hundred miR390 molecules corresponding to two nearly identical types were detected (Table 1; see Supplemental Table 2 online), but neither miR173 nor miR828, which are implicated in ta-siRNA biogenesis in Arabidopsis, were identified. The ta-siARFs, products of processing of TAS3 genes, were found in the wild type but were absent in the libraries of rdr6 and ago7 and were dramatically reduced in the dcl4 mutant lines. These results are consistent with the initial RNA gel blot analysis (Figure 2F). Further description of the small RNA libraries can be found in Supplemental Table 2 and Supplemental Figure 3 online.
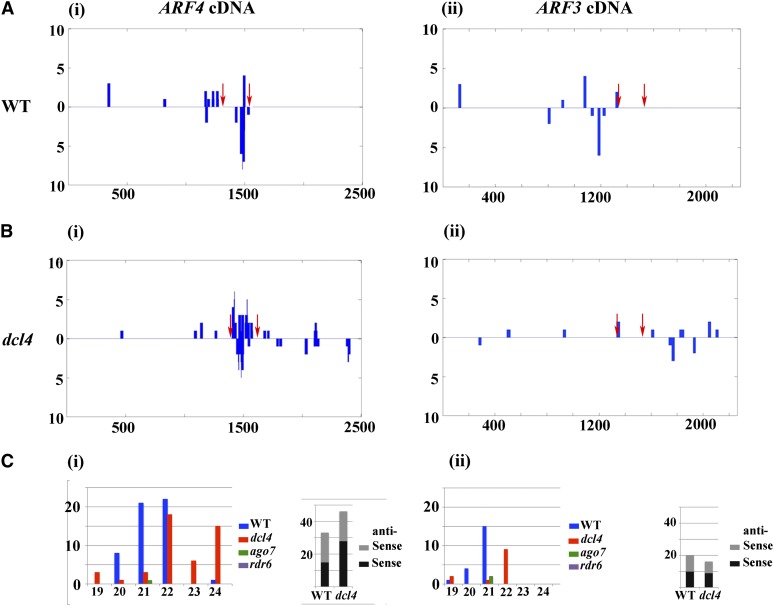
We next surveyed the libraries for small RNAs that are derived specifically from the two ARF genes (Figure 4; see Supplemental Data Set 2 online). In wild-type apices, 52 reads of 20 different sequences derived from ARF4 mRNA were found; the majority originated from the 186 bp flanked by the two ta-siARF binding sites (Figure 4A). Most of these reads originate from the antisense orientation of ARF4 and are 21 and 22 nucleotides long (Figure 4C). Twenty reads of 21 nucleotides from the ARF3 mRNA were found, and these originated from all parts of the RNA but not from the sequence flanked by the two ta-siARF binding sites (Figure 4A). All of these small RNAs were absent in the rdr6 and ago7 apices. By contrast, more reads and sequences derived from the 186-bp ARF4 region flanked by the two ta-siARF binding sites were found in the dcl4 apices (Figure 4B). Half of these sequences originated from the antisense orientation and their size shifted to 22 and 24 nucleotides (Figure 4C). Such a shift suggests that in the absence of DCL4, other DCL enzymes are now processing a dsRNA corresponding to the ARF4 gene (Gasciolli et al., 2005). In addition, a set of small RNAs derived from the 3′ end of ARF4 were also found in dcl4 libraries (Figure 4B). By contrast, very few reads originating from ARF3 could be detected either in wild-type or dcl4 apices (15 compared with 10; Figures 4B and 4C). Thus, a specific type of small RNAs, ones that can be formed in the absence of DCL4, can target ARF4, but at the same time, cannot target ARF3.
Figure 4.
Small RNAs Derived from the ARF4 and ARF3 Transcripts.
(A) In the wild type (WT), ARF4-derived siRNAs (i) originate mainly from the sequence flanked by the two ta-siARF recognition sites (red arrows), and ARF3-derived siRNAs (ii) are present in low numbers and originate from all parts of the mRNA.
(B) More ARF4-derived siRNAs were found in dcl4 apices (i) as well as siRNAs from the gene’s 3′ end but not from ARF3 (ii).
(C) The ARF4-derived siRNAs (i) are 20 to 22 nucleotides in the wild type and primarily 22 to 24 nucleotides in dcl4 but the ARF3-derived siRNAs (ii) are 20 to 21 nucleotides in the wild type and primarily 22 nucleotides in dcl4.
In (A) and (B), the x axis marks the position along the gene, but in (C), it marks the small RNA size. The y axis marks the numbers of reads.
Novel TAS3-Derived Small RNAs Are Made in the Absence of DCL4
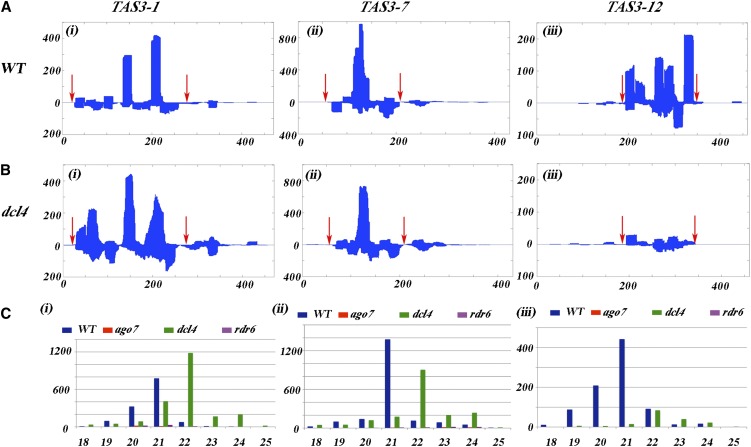
Many of the ARF4-derived small RNAs originate from the region flanked by the two ta-siARF target sites. Formation of such secondary siRNAs is consistent with the two-hit model suggested before (Axtell et al., 2006). By this model, a single-stranded RNA that is targeted twice by an RNA-induced silencing complex complex can form a template for RNA Dependent RNA Polymerase for production of dsRNA. Such templates are the TAS genes themselves, and we therefore surveyed the tomato genome for TAS3 genes. Based on homology with other species, three ta-siARF producing loci were found in the tomato genome and are named here after their chromosomal origin TAS3-1, TAS3-7, and TAS3-12. ESTs for TAS3-1 and TAS3-12 are present in public databases, and all three have two miR390 recognition sites (Figure 5; see Supplemental Figure 4 online), but only TAS3-1 produces two ta-siARFs, as do the Arabidopsis TAS3 genes, whereas TAS3-7 and TAS3-12 produce only one ta-siARF each. The three genes have a region of 135 to 150 bp from which the majority of the small RNAs are derived. This region starts and ends with a miR390 recognition site (Figure 5; see Supplemental Figure 4A online). Analysis of small RNAs derived from TAS3 shows that hundreds of reads in the wild-type library are derived from each of the three loci (Figure 5A). These reads correspond to more than 50 different sequences and are derived from the sense and antisense strands of each gene (Figure 5C).
Figure 5.
Normal and DCL4-Independent Production of TAS3 siRNAs.
(A) Wild-type (WT) small RNAs derived from the TAS3-1 (i), TAS3-7 (ii), and TAS3-12 (iii) genes. Most small RNAs are derived from the fragment flanked by the miR390 recognition sites (red arrows).
(B) In the dcl4 library, small RNAs from TAS3-1 (i) and TAS3-7 (ii) are present, but small RNAs from TAS3-12 are not present (iii).
(C) Size distribution of small RNAs derived from the TAS3 genes. In the wild type, most are 21 nucleotides, but in the dcl4, they are shifted to 22 nucleotides.
In (A) and (B), the x axis marks the position along the gene, but in (C), it marks the small RNA size. The y axis marks the numbers of reads.
Comparing the different libraries for small RNAs derived from the TAS3 genes shows that in the ago7 and rdr6 libraries, the number of reads was reduced to near zero (Figure 5C). However, in the dcl4 apices, there were more reads of sequences from the TAS3-1 gene than were in the wild-type apices (cf. Figure 5A with 5B). These reads came from sequences of different position and different sizes than present in the wild type (Figure 5C), and a similar pattern was found for small RNAs that originate from the TAS3-7 gene. By contrast, the number of small RNAs from TAS3-12 gene was reduced in dcl4 (cf. Figure 5A with 5B).
In the wild type, the majority of the TAS3-derived small RNAs were 21 nucleotides long, but in the dcl4 apices, most were 22 nucleotides (Figure 5C). In the absence of Arabidopsis DCL4, some RDR6-dependent siRNAs were produced by DCL2 and DCL3 (Gasciolli et al., 2005). Based on the increase of the 22-nucleotide small RNA class in the dcl4 apices, we suggest that another DCL is compensating for the absence of DCL4 in tomato. Notably, in the Arabidopsis dcl4 mutant, TAS3-derived small RNAs are also formed, but their phasing is lost (Howell et al., 2007). A loss of phasing and production of 22-nucleotide small RNAs may alter the nature/efficiency of the TAS3-derived RNAs and, in particular, the formation of the conserved ta-siARFs. Indeed, there are no ta-siARF small RNAs in the rdr6 and ago7 apices (Table 1), and while there are some TAS3-derived sequences in the dcl4 apices (Figure 5B), the total number of ta-siARF–like sequences is strongly reduced (Table 2; see Supplemental Figure 4B online).
Table 2. Prevalence of ta-siARFs in Wild-Type and wiry Apices.
| Library | Loci of Origin | No. of Sequences | No. of Reads | Length |
|---|---|---|---|---|
| Wild type | TAS3-1, 7, 12 | 5, 3, 10 | 20, 8, 66 | 19–23 |
| rdr6 | TAS3-1, 7, 12 | 0, 0, 0 | 0, 0, 0 | – |
| ago7 | TAS3-1, 7, 12 | 0, 0, 0 | 0, 0, 0 | – |
| dcl4 | TAS3-1, 7, 12 | 5, 2, 0 | 7, 8, 0 | 22–23 |
–, no reads in these samples.
ta-siRNA–Insensitive ARF Forms Can Stimulate a wiry Syndrome
As noted above, the dcl4 mutation, where high levels of ARF3 are found, is distinct from the other wiry mutants (Figure 1). To examine whether differences in regulation of the two ARFs can underlie the difference between the two wiry classes, we overexpressed the two genes. 35S:ARF4 plants are largely indistinguishable from the wild type, whereas the lamina of 35S:ARF3 leaves is slightly narrower and curved downward with leaflets having slightly deeper serrations than the wild type (Figures 6A to 6C).
Figure 6.
Loss of ARF3 and ARF4 Regulation Is the Basis of the wiry Syndrome.
(A) to (C) Leaf 7 of wild-type (WT) (A), 35S:ARF3 (B), and 35S:ARF4 (C) plants.
(D) A scheme of the ARF genes and construction of their ta-siRNA–insensitive forms by introduction of silent mutations into the two ta-siARF recognition sites.
(E) and (F) Overexpressing the ta-siARF–insensitive form (Slm) of ARF3 (E) or ARF4 (F) in the wild type resulted in a strong wiry phenotype.
(G) Expressing 35S:ARF4 in the weak ago7-3 mutant resulted in a very strong wiry phenotype.
(H) to (J) Rescue of ago7-3 by downregulation of both ARF3 and ARF4 mRNA using an artificial miRNA; compare ago7-3 (H), pFIL>>amiR-ARF (I), and ago7-3 pFIL>>amiR-ARF (J).
Bars = 2 cm.
[See online article for color version of this figure.]
If ta-siRNA regulation imposes an important negative regulation of the two ARFs, bypassing such activities would have a strong impact on ARF overexpression. To examine that inference, the two conserved 21-bp ta-siARF recognition sites (Alvarez et al., 2006) were sequentially modified; silent mutations were introduced into each of the two ta-siARF recognition sites (termed A and B) to generate mA and mB forms of ARF3 and ARF4 (Figure 6D). The four mutant cDNAs were expressed in plants under the 35S promoter and in no case stimulated wiry leaves. In fact, their effects were only mildly stronger than the effects of overexpressing the native forms of ARF3 or ARF4 (see Supplemental Figures 5A to 5D online).
In sharp contrast, plants expressing cDNAs with 35S:mARF3 and 35S:mARF4 that had silent mutations in both of the two ta-siARF binding sites (mAB) had minute needle-like leaves and could not form roots in the tissue culture medium used for their regeneration (see Supplemental Figures 5E and 5F online). To bypass the rooting barrier, a transactivation approach was employed, and several lines with ta-siRNA–insensitive ARFs driven by an OP promoter were generated. These responder lines were crossed with the pFIL:LhG4 driver that, unlike in Arabidopsis plants, directs expression in both sides of leaf primordia (Lifschitz et al., 2006). The resulting pFIL>>mARF3 and pFIL>>mARF4 plants had a range of phenotypes resembling intermediate and strong wiry alleles (Figures 6E and 6F; compare with Supplemental Figures 5G and 5H online). In general, the effects of mARF3 were marginally stronger, though mARF4 lines that manifest a strong wiry phenotype were obtained as well. Significantly, overexpression of ARF3 was similar to mutations in RDR6 or AGO7 but not in DCL4; no necrotic lesions ever developed on the shoestring leaves, and floral organs were narrow and unfused. We therefore attribute the unique defects in dcl4 lines to a failure to regulate other targets. Given the large number of small RNAs that are specifically altered in that background (Table 1; see Supplemental Figure 3C online), it is difficult to predict which genes may account for these defects.
In support of a role of ta-siRNA regulation in restriction of ARF activities, expressing either 35S:ARF4 or 35S:ARF3 in the background of the weak ago7-3 also had dramatic effects; all leaves starting from leaf 1 became nearly radial, short, and devoid of lamina (Figure 6G; see Supplemental Figure 5I online).
A Failure to Negatively Regulate ARF3 and ARF4 Underlies the wiry Phenotype
If a failure to negatively regulate ARF3 and/or ARF4 by ta-siARFs is the prime cause of the wiry syndrome, than reduced activity of the two genes should rescue the wiry lamina phenotype. Knocking out both genes in tomato was accomplished by the expression of an artificial miRNA directed against the same sequence targeted by ta-siARF (amiR-ARF; Alvarez et al., 2006). However, 35S:amiR-ARF plants are sterile; therefore, both pFIL:LhG4 and OP:amiR-ARF were introduced into ago7-3. RNA gel blot analysis showed a significant reduction in the levels of the ARF genes in plants expressing the miRNA directed against them (see Supplemental Figure 6 online). Significantly, pFIL>>amiR-ARF and pFIL>>amiR-ARF ago7-3 were indistinguishable (Figures 6I and 6J), and the mutant background was therefore confirmed by sequencing. Our results suggest therefore that both ARF3 and ARF4 are regulated in vivo by the ta-siARF, both can induce a wiry phenotype when this regulation is bypassed, and a failure to negatively regulate these ARFs is the prime source of the wiry syndrome.
The Leaves of Different Species Respond Differentially to Ectopic ARF Expression
The potential of the tomato ARF genes to induce wiry leaves is in a sharp contrast to the reported effects of these genes in Arabidopsis, where overexpression of ta-siRNA–insensitive forms of ARF3 stimulated the formation of trichomes on the abaxial side of early formed leaves and altered leaf growth (Hunter et al., 2006). However, in no case did its overexpression interfere with the formation of a flat lamina. It is therefore possible that tomato ARF genes have a different function than their Arabidopsis counterparts. To investigate this, we introduced the ta-siARF–insensitive form of tomato ARF3 into Arabidopsis. Conversely, we also introduced a ta-siARF–insensitive form of Arabidopsis ARF3 into tomato. The effects of overexpressing the tomato gene in Arabidopsis were very similar to the effects of overexpressing the gene native to that species; slow-growing plants that showed abaxial trichomes on early formed leaves (leaf 2-3 compared with leaf 4-5 in the wild type) and leaves that were shorter and proportionally broader than the wild type (cf. Figure 7A with 7B and 7C; see Supplemental Figures 7A to 7C online). Likewise, tomato plants overexpressing the Arabidopsis ARF3 showed the exact same effects as the plants expressing the tomato gene: miniature shoots with radial leaves that could not form roots (see Supplemental Figure 7D online). Weaker lines that were recovered from the tissue culture were highly similar to wiry plants (Figure 7D). We therefore conclude that products of the genes from the two species have comparable activity and that differences between the highly malformed wiry plants and the nearly normal ago7/zippy mutants represent differential response of the leaves of the two species to the same ARF misexpression.
Figure 7.
Species-Specific Responses to ARF3 Activities.
(A) to (C) Arabidopsis plants (A) overexpressing a ta-siARF–insensitive (m) ARF3 of either Arabidopsis (B) or tomato (C) origin have bifacial flat leaves. WT, the wild type.
(D) Tomato plant overexpressing Arabidopsis mARF3.
(E) to (H) Comparison of wild-type (i) and 35S:Sl-mARF3 (ii) tobacco plants. Tobacco leaves (E) are mildly modified, whereas flowers (F) are smaller and the tips of the petals are narrow. N. benthamiana leaves (G) are narrow and curled down, whereas flowers (H) are smaller and the petals are narrow and separated.
(I) and (J) Leaves of normal (I) and 35S:At-mARF3 (J) potato plants.
Bars = 2 cm.
[See online article for color version of this figure.]
To determine whether the response to altered levels of ARFs is dependent on phylogenetic origin (Solanaceae versus Brassicaceae), or perhaps on leaf structure (simple versus compound), we expressed the ta-siARF–insensitive forms of tomato ARF in tobacco (Nicotiana tabacum) and N. benthamiana, simple-leaved Solanaceae species. In both plants, expression of 35S:mARF3 or 35S:mARF4 caused malformed leaves, but lamina loss was never observed (Figures 7E and 7G; see Supplemental Figures 7E to 7H online). In both, the flowers were smaller and the distal tip of the petals was narrow (Figure 7F; see Supplemental Figure 7F online), but in N. benthamiana, the petals were largely unfused (Figure 7H). We next examined the response of potato, a compound-leaved Solanaceae species, to ARF misexpression. The response of potato plants to 35S:mARF3 was weak, and given the high similarity of the tomato and potato genes, we were concerned about potential cosuppression. To bypass such a potential problem, we expressed in potato the ta-siARF–insensitive Arabidopsis ARF genes. In the wild type, the first leaves have one to three leaflets, and as the plant ages, the number of leaflets increases (Figure 7I). The 35S:At-mARF3 and 35S:At-mARF4 potato plants displayed slightly malformed epinastic leaves but all had expanded lamina (Figure 7J; see Supplemental Figures 7I and 7J online). Our results imply therefore that the developmental responses to ARF expression are species specific and do not depend on leaf shape or family origin.
DISCUSSION
The Molecular Basis of a Disease Mimic Syndrome
In this study, four tomato mutants with highly malformed shoots were analyzed. In all mutants, leaf shape ranged from flat lamina in the first two to three leaves to nearly radial leaflets or needle-like leaves that lack lamina and leaflets altogether in later formed leaves. These effects are collectively termed wiry, to reflect the nature of the extreme mutant leaves. All four mutants are impaired in various components of siRNA biogenesis, and in all mutant lines, specific ARF genes are misregulated. Expression of ta-siARF–insensitive versions of these ARFs, either ARF3 or ARF4, can mimic the wiry syndrome, and expression of an amiR-ARF that targets both genes can rescue wiry plants.
The analogy between the wiry mutants and viral infected plants was first made by Lesley and Lesley (1928) upon analysis of the first wiry mutant line. By their analysis, this single Mendelian mutation was mimicking the shoestring mosaic disease, a phenotype commonly seen in tomato plants infected by what today is known as CMV, Tomato mosaic virus, and more commonly, by their combination with other viruses (Edwardson and Corbett, 1962; Andrade et al., 1981). Here, we showed that wiry mutants are impaired in enzymes known to be involved in an antiviral defense mechanism as well as gene silencing (reviewed in Alvarado and Scholthof, 2009). The RDR6 and the DCL4 RNA processing pathways of Arabidopsis are targeted by various viral suppressor proteins, and, in agreement, expression of these viral suppressors lead to similar defects as found in the rdr6 or dcl4 mutants (Deleris et al., 2006; Moissiard et al., 2007). By analogy, we suggest that the molecular basis of the Mendelian wiry syndrome and of that of viral-infected tomato plants displaying shoestring symptoms are similar; in both cases, specific classes of small RNAs fail to accumulate. As shown here, when ta-siARFs fail to accumulate in tomato, misexpression of ARF3 and/or ARF4 leads to needle-like leaves. Notably, only few tomato viral diseases result in the formation of shoestring leaves. Indeed, many mechanisms direct plant responses to infection: the nature of the viral suppressor proteins (Díaz-Pendón and Ding, 2008), the spatial distribution of virus particles in the plant, the plant’s physiological status, and in many cases, the coinfection by other viruses.
Conservation and Divergence of ARF Regulation by ta-siRNA
In land plants, the regulatory pathway consisting of miR390, the TAS3-derived ta-siARF and its ARF targets, is highly conserved (Axtell et al., 2006), and as we show, tomato is not exceptional. However, there are also species-specific modifications; it was suggested that Arabidopsis AGO7 is dedicated to miR390-regulated processes (Montgomery et al., 2008), but numerous tomato small RNAs require AGO7 for their biogenesis. In fact, the fraction of small RNAs that commonly require the biogenesis enzymes tested (DCL4, RDR6, and AGO7) is only 4% of the small RNA types found in the wild type, while small RNAs that require only one or two of these enzymes for their biogenesis constitute 31% (see Supplemental Figure 3 online). Thus, a diverse involvement of each of these enzymes in various small RNA biogenesis pathways is implicated.
The sources and the functions of the tomato small RNAs that require only AGO7 and what causes the shift from 21 to 22 nucleotides in rdr6 libraries (see Supplemental Figure 3 online) are presently unknown. However, this phenomenon illustrates the complexity of tomato small RNA biogenesis and our limited appreciation of its significance. The DCL4 enzyme has a significant role in dicing dsRNA into 21-nucleotide sRNAs that can trigger silencing of matching targets, be it in cis or in trans (Dunoyer et al., 2005; Yoshikawa et al., 2005). Small RNAs derived from TAS3 and ARF4 have different sizes (Figures 4 and 5), suggesting that different dicing enzymes cleave that dsRNA derived from these genes. Similarly, Dunoyer et al. (2010) showed that two inverted repeat loci in Arabidopsis could be cleaved into 21-, 22-, and 24-nucleotide small RNAs. In agreement, in the absence of DCL4, other DCL proteins can compensate for its loss and process dsRNA to produce mainly 22- or 24-nucleotide instead of 21-nucleotide sRNAs (Gasciolli et al., 2005). Such flexibility and complexity in small RNA production might give an advantage in silencing RNA of endogenous but primarily exogenous origin. Viruses can rapidly change and in return host plants need flexible and evolving counteracting machinery. One plausible strategy to achieve such machinery is by nonspecific interaction of dsRNA with DCL proteins, giving the opportunity to substitute for a particular DCL protein that may be targeted for suppression upon viral infection.
Conservation and Divergence of Leaf Form Regulation by ta-siARF Targets
The ta-siARF and its ARF target is one of the most conserved small RNA-target pairs in the plant kingdom (Axtell and Bowman, 2008). By contrast, the phenotypes of mutants impaired in ta-siRNA biogenesis are different in different species. The dramatic phenotype of tomato wiry is in a sharp contrast with other eudicots; in Arabidopsis, the ago7/zippy, rdr6, and sgs3 mutants have normal blade growth and leaf polarity (Hunter et al., 2003; Peragine et al., 2004; Xie et al., 2005) as do tobacco plants with reduced RDR6 expression (Qu et al., 2005). By contrast, some monocots are more similar to tomato where ta-siRNA biogenesis mutants have narrow leaves or even arrested meristem development shortly after zygote formation (Liu et al., 2007; Nagasaki et al., 2007; Nogueira et al., 2007; Douglas et al., 2010). It could be speculated that tomato leaves are unique, as they are compound and respond differently as was shown for KNOX misexpression (Hareven et al., 1996). However, even potato, a closely related species with complex leaves, does not produce wiry leaves upon overexpression of ta-siARF–insensitive forms of the ARFs. Thus, as in other cases, it is important to compare gene function among different species (Efroni et al., 2010) to reach conclusions regarding general or specialized gene functions.
On the basis of the different cases where loss of ta-siRNA was studied, we conclude that regulation of the ARF genes by this mechanism cannot be used for a highly conserved universal developmental pathway. Instead, we suggest that while these ARFs have a conserved developmental role in stabilizing leaf polarity (Pekker et al., 2005; Alvarez et al., 2006), their regulation by small RNAs has an important physiological function that is implemented in a species-specific manner.
A Suggested Role for the Highly Conserved ta-siRNA-ARF Regulatory Module
ta-siRNA is the only known class of small RNAs that requires miRNA and siRNA biogenesis for its production, and genes related to ARF3 are the only known targets of ta-siRNA that are conserved among land plants. Therefore, levels of ARF3 genes are universal sensors of small RNA biogenesis, and when this process is challenged, their levels change. ARF3 and ARF4 act as negative regulators of auxin signaling by competing with positive regulators for the binding of AuxRE in auxin-regulated promoters (Vernoux et al., 2011). Together, plants use deregulation of ARF3 by loss of ta-siRNA to ameliorate auxin responses upon interference with small RNA biogenesis. What is the advantage of such a response? And why would a pathway dedicated to RNA silencing target developmental genes as well?
Many plant viruses have suppressor proteins that interfere with either siRNA or miRNA biogenesis (reviewed in Ding and Voinnet, 2007). Jay et al. (2011) suggested that plant viruses suppresses antiviral defense mechanisms as a means to exploit plant development and hormonal pathways for optimization of endogenous conditions for their own replication and do not alter them merely as a defense mechanism. For example, Padmanabhan et al. (2008) suggested that TMV replicase protein interacts with several auxin/indole-3-acetic acid proteins and changes their nuclear localization to direct a cellular environment more compatible for viral replication. Likewise, evidence indicates that pathogen infection, bacteria and fungi alike, can result in changes of cellular auxin and altered expression of auxin signaling genes (reviewed in Bari and Jones, 2009). It is possible that in plant evolutionary history, viruses triggered auxin signals to modify the cellular environment in their favor (Jay et al., 2011). Plants, in response, used the small RNA sensory mechanism to balance auxin signals through orthologs of ARF3 genes. Thus, while the expression of genes involved in ta-siRNA biogenesis, and in particular, that of AGO7 is restricted in Arabidopsis (Chitwood et al., 2009), its activity is required to restrict the accumulation of leaf infected green fluorescent protein RNA particles (Qu et al., 2008).
Evidently, most viral infections do not result in disease symptoms, as the plant is able to balance the virus levels and their effect on the plant cells. However, in cases such as CMV or Tomato mosaic virus infection in tomato, the plant lose its ability to keep virus levels in check and to maintain the balance between small RNA levels and auxin signals. As a result, ARF mRNA levels are upregulated and species-specific disease symptoms, analogous to species-specific hormonal responses, appear.
METHODS
Plant Material and Genetics
Tomato (Solanum lycopersicum) plants (cv M82) were grown in greenhouse conditions with temperatures ranging between 18 and 25°C. All mutants were described by Menda et al. (2004). Allelism tests and combination of wiry mutants with transgenic lines were performed with fertile sibs. To map the w, w2, w3, and w4 mutants, heterozygous plants were crossed with Solanum pennellii, and F2 plants showing the wiry phenotype were used. The w mutant cosegregated with TG182 (chromosome 4), w2 with TG24 (chromosome 1), w3 with DCL4 (chromosome 7), and w4 with TG2 (chromosome 4). Genes involved in small RNA biogenesis were mapped, and ones tightly linked with the wiry mutants were sequenced in the corresponding mutant lines.
RNA Isolation and Analysis
Total RNA was extracted using TRI Reagent (Sigma-Aldrich) according to the manufacturer’s instructions. High molecular weight RNA was normalized by spectrophotometry to 20 µg/lane. Radiolabeled probes for RNA gel blots of mRNAs were made by random priming reactions. Probes consisted of the full-length cDNAs (see Supplemental Table 3 online for list of primers). Equivalent loading of samples was monitored by detection of 28S and 18S RNA in all gels prior to blot transfer.
Low molecular weight RNA was purified using TRI Reagent, resolved on a 17% polyacrylamide-urea gel, transferred to a Zeta-Probe GT membrane (Bio-Rad), and probed with a P32 end-labeled oligonucleotides, complementary to the mature miRNAs and ta-siARF (5′-AAGAACTGGAACGTTCTGGAA-3′) where an LNA probe was used. Equivalent loading of samples was monitored by 5S RNA levels prior to RNA gel blotting.
In Situ Hybridization
Tissue preparation, histological analyses, tissue clearing, and in situ hybridization were performed according to Pekker et al. (2005). ARF cDNAs were amplified by PCR and cloned into the pDRIVE vector (Qiagen PCR cloning kit). ARF3, ARF4, and PHB (Solyc02g024070) probes were generated by linearizing the described cDNA-containing plasmids and synthesizing digoxigenin-labeled antisense RNA using T7 RNA polymerase.
Cloning and Plant Transformation
cDNAs of the tomato and Arabidopsis thaliana genes (see Supplemental Table 3 online for primers used) were transformed into a transactivation (10OP-BJ36) or 35S direct fusion (ART7) vectors. Assembly PCR was used to construct the mutated forms of ARF3 and ARF4. FILpro:LhG4 and OP:amiR-ARF were described earlier (Alvarez et al., 2006). All constructs were subcloned into the pART27 binary vector (Eshed et al., 2001) and were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. Cotyledon transformation in tomato and leaf disc transformation in tobacco (Nicotiana tabacum) and potato (Solanum tuberosum cv Desiree) were performed according to McCormick (1991), Beaujean et al. (1998), and Horsch et al. (1985), respectively. Phenotypic analyses were performed with selected OP:GENE responder lines that were crossed to promoter:LhG4 driver lines. Several independent responder lines were crossed to the FILpro driver, and a representative line was selected for further analysis. To generate FILpro>>OP in a wiry mutant background, OP:GENE and FILpro:LhG4 plants were separately crossed to plants heterozygous for the wiry mutant. Resulting F1 plants were characterized by segregation and when necessary, validated by sequencing.
High-Throughput Sequencing of Tomato Small RNAs
Since ARF3 and ARF4 have an important role in early stages of leaf initiation (Pekker et al., 2005), we used for our analyses RNA collected from shoot apices including three to five primordial leaves. The first two leaves were removed since in all mutants those leaves have expanded lamina. Production of small RNA libraries was done using the Illumina Small RNA sample prep kit (FC-102-1009) according to the manufacturer’s protocol.
Approximately 26 million reads were obtained from high-throughput sequencing of small RNAs from the four libraries. Sequence adaptors (Illumina Small RNA version 1.5) were identified using crossmatch-minmatch 10 -minscore 10 (http://www.incogen.com/public_documents/vibe/details/crossmatch.html). Adaptor sequences were then removed and reads of 18 to 26 nucleotides were selected. Within each library, the unique sequences were reported as tags containing sequence information and frequency. The tags were compared with a database of ribosomal and tRNAs. The ribosomal sequences were obtained by combining Rfam-rRNA sequences and Arabidopsis rRNAs. Rfam was used also to filter the tRNA sequences. The search of ribosomal and tRNAs was performed using NCBI-BLASTN (Altschul et al., 1997).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: tomato sequences: TA3-1, JX047545; TA3-7, JX047546; TA3-12, JX047547; RDR6, Solyc04g014870/JX047548; AGO7, Solyc01g010970/JX047549; DCL4, Solyc07g005030/JX047550; SGS3, Solyc04g02530/JX047551; ARF3, Solyc02g077560; ARF4, Solyc11g069190; Arabidopsis sequences: ARF3, At2G33860; ARF4, At5G60450.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Shoots of Weak and Strong wiry Plants.
Supplemental Figure 2. Scanning Electron Microscopy Images of wiry Mutant Apices.
Supplemental Figure 3. Properties of Wild-Type and wiry Small RNA Libraries.
Supplemental Figure 4. Processing of the Tomato TAS3 Genes.
Supplemental Figure 5. The Two ta-siARF Binding Sites Are Functional.
Supplemental Figure 6. ARF Levels Are Reduced by miR-ARF Expression.
Supplemental Figure 7. Species-Specific Responses to ARF3 or ARF4 Expression.
Supplemental Table 1. wiry Alleles Characterized in This Work.
Supplemental Table 2. Prevalence of Selected Small RNAs in Libraries of Wild-Type and Mutant Shoots.
Supplemental Table 3. Primers for PCR-Mediated Cloning.
Supplemental Data Set 1. Tomato Small RNA Sequences.
Supplemental Data Set 2. Small RNA Sequences from the TAS3, ARF3, and ARF4 Genes.
Acknowledgments
We thank Eyal Arazi and Guy Gafny for their dedicated work. We thank members of the Eshed lab, John Bowman, and Eliezer Lifschitz for comments and discussions and Dena Leshkowitz for help with analysis of small RNA libraries. This work was made possible with funding from Research Grant 1294-10 from Israel Science Foundation and from MINERVA to Y.E. Y.E. is an incumbent of the Mimran Family Professorial Chair.
AUTHOR CONTRIBUTIONS
T.Y., I.P., J.P.A., and Y.E. designed the research. T.Y., I.P., D.P., A.P., M.S., G.W., J.P.A., Z.A., and Y.E. performed the experiments and analyzed the “wet” data. T.Y. and G.F. characterized the small RNA transcriptome. T.Y. and Y.E. wrote the article.
Glossary
- ta-siRNA
trans-acting short interfering RNA
- dsRNA
double-stranded RNA
- siRNA
short interfering RNA
- SAM
shoot apical meristem
- TMV
Tobacco mosaic virus
- CMV
Cucumber mosaic virus
- ta-siARF
to be defined
- miRNA
microRNA
References
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouché N., Gasciolli V., Vaucheret H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Allen E., Howell M.D. (2010). miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin. Cell Dev. Biol. 21: 798–804 [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado V., Scholthof H.B. (2009). Plant responses against invasive nucleic acids: RNA silencing and its suppression by plant viral pathogens. Semin. Cell Dev. Biol. 20: 1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J.P., Pekker I., Goldshmidt A., Blum E., Amsellem Z., Eshed Y. (2006). Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade O., Latrre B.A., Escaffi O. (1981). Tomato mosaic virus associated with shoestring symptom in Chilean tomatoes. Plant Disease 65: 761–762.
- Axtell M.J., Bowman J.L. (2008). Evolution of plant microRNAs and their targets. Trends Plant Sci. 13: 343–349 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Bari R., Jones J.D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Beaujean A., Sangwan R.S., Hodges M., Sangwan-Norreel B.S. (1998). Effect of ploidy and homozygosity on transgene expression in primary tobacco transformants and their androgenetic progenies. Mol. Gen. Genet. 260: 362–371 [DOI] [PubMed] [Google Scholar]
- Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Meng Y., Yuan C., Bai L., Huang D., Lv S., Wu P., Chen L.L., Chen M. (2011). Plant siRNAs from introns mediate DNA methylation of host genes. RNA 17: 1012–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Nogueira F.T., Howell M.D., Montgomery T.A., Carrington J.C., Timmermans M.C. (2009). Pattern formation via small RNA mobility. Genes Dev. 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayberg C.D., Butler L., Rick C.M., Robinson R.W. (1966). Third list of known genes in the tomato. J. Hered. 57: 189–196 [Google Scholar]
- Deleris A., Gallego-Bartolome J., Bao J., Kasschau K.D., Carrington J.C., Voinnet O. (2006). Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71 [DOI] [PubMed] [Google Scholar]
- Díaz-Pendón J.A., Ding S.W. (2008). Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46: 303–326 [DOI] [PubMed] [Google Scholar]
- Ding S.W., Voinnet O. (2007). Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas R.N., Wiley D., Sarkar A., Springer N., Timmermans M.C., Scanlon M.J. (2010). ragged seedling2 encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell 22: 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Brosnan C.A., Schott G., Wang Y., Jay F., Alioua A., Himber C., Voinnet O. (2010). An endogenous, systemic RNAi pathway in plants. EMBO J. 29: 1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dunoyer P., Himber C., Voinnet O. (2005). DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 37: 1356–1360 [DOI] [PubMed] [Google Scholar]
- Edwardson J.R., Corbett M.K. (1962). A virus-like syndrome in tomato caused by a mutation. Am. J. Bot. 49: 571–575 [Google Scholar]
- Efroni I., Eshed Y., Lifschitz E. (2010). Morphogenesis of simple and compound leaves: A critical review. Plant Cell 22: 1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y., Baum S.F., Bowman J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99: 199–209 [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Glick E., Zrachya A., Levy Y., Mett A., Gidoni D., Belausov E., Citovsky V., Gafni Y. (2008). Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc. Natl. Acad. Sci. USA 105: 157–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hareven D., Gutfinger T., Parnis A., Eshed Y., Lifschitz E. (1996). The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell 84: 735–744 [DOI] [PubMed] [Google Scholar]
- Horsch R.B., Fry J.E., Hoffmann N.L., Eichholtz D., Rogers S.G., Fraley R.T. (1985). A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C., Sun H., Poethig R.S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13: 1734–1739 [DOI] [PubMed] [Google Scholar]
- Hunter C., Willmann M.R., Wu G., Yoshikawa M., de la Luz Gutiérrez-Nava M., Poethig S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands A.Y., Chitwood D.H., Plavskin Y., Timmermans M.C. (2009). Signals and prepatterns: New insights into organ polarity in plants. Genes Dev. 23: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay F., Wang Y., Yu A., Taconnat L., Pelletier S., Colot V., Renou J.P., Voinnet O. (2011). Misregulation of AUXIN RESPONSE FACTOR 8 underlies the developmental abnormalities caused by three distinct viral silencing suppressors in Arabidopsis. PLoS Pathog. 7: e1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Pham T., Hamidi A., McCormick S., Kuzoff R.K., Sinha N. (2003). Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX genes in generating compound leaves. Development 130: 4405–4415 [DOI] [PubMed] [Google Scholar]
- Lesley J.W., Lesley M.M. (1928). The “wiry” tomato. A recessive mutant form resembling a plant affected with mosaic disease. J. Hered. 8: 337–344 [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J.P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., et al. (2007). Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell 19: 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S. (1991). Transformation of tomato with Agrobacterium tumefaciens In Plant Tissue Culture Manual, Vol. B6, K. Lindsey, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–9.
- Menda N., Semel Y., Peled D., Eshed Y., Zamir D. (2004). In silico screening of a saturated mutation library of tomato. Plant J. 38: 861–872 [DOI] [PubMed] [Google Scholar]
- Moissiard G., Parizotto E.A., Himber C., Voinnet O. (2007). Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA 13: 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T.A., Yoo S.J., Fahlgren N., Gilbert S.D., Howell M.D., Sullivan C.M., Alexander A., Nguyen G., Allen E., Ahn J.H., Carrington J.C. (2008). AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl. Acad. Sci. USA 105: 20055–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H., Itoh J., Hayashi K., Hibara K., Satoh-Nagasawa N., Nosaka M., Mukouhata M., Ashikari M., Kitano H., Matsuoka M., Nagato Y., Sato Y. (2007). The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 104: 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira F.T., Madi S., Chitwood D.H., Juarez M.T., Timmermans M.C. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan M.S., Kramer S.R., Wang X., Culver J.N. (2008). Tobacco mosaic virus replicase-auxin/indole acetic acid protein interactions: Reprogramming the auxin response pathway to enhance virus infection. J. Virol. 82: 2477–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I., Alvarez J.P., Eshed Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F., Ye X., Hou G., Sato S., Clemente T.E., Morris T.J. (2005). RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 79: 15209–15217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F., Ye X., Morris T.J. (2008). Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. USA 105: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M., Alandete Saez M., Eshed Williams L., Fletcher J.C., McCormick S. (2010). Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 24: 1010–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Wang F., Cheng L., Kong F., Peng Z., Liu S., Yu X., Lu G. (2011). Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep. 30: 2059–2073 [DOI] [PubMed] [Google Scholar]
- Vernoux T., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yuan Y.R., Pei Y., Lin S.S., Tuschl T., Patel D.J., Chua N.H. (2006). Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]