Wild-type and active-site defective forms of several Arabidopsis thaliana ARGONAUTE proteins involved in posttranscriptional silencing were used to examine several ARGONAUTE functions, including small RNA binding, interaction with target RNA, slicing or destabilization of target RNA, secondary siRNA formation, and antiviral activity.
Abstract
In RNA-directed silencing pathways, ternary complexes result from small RNA-guided ARGONAUTE (AGO) associating with target transcripts. Target transcripts are often silenced through direct cleavage (slicing), destabilization through slicer-independent turnover mechanisms, and translational repression. Here, wild-type and active-site defective forms of several Arabidopsis thaliana AGO proteins involved in posttranscriptional silencing were used to examine several AGO functions, including small RNA binding, interaction with target RNA, slicing or destabilization of target RNA, secondary small interfering RNA formation, and antiviral activity. Complementation analyses in ago mutant plants revealed that the catalytic residues of AGO1, AGO2, and AGO7 are required to restore the defects of Arabidopsis ago1-25, ago2-1, and zip-1 (AGO7-defective) mutants, respectively. AGO2 had slicer activity in transient assays but could not trigger secondary small interfering RNA biogenesis, and catalytically active AGO2 was necessary for local and systemic antiviral activity against Turnip mosaic virus. Slicer-defective AGOs associated with miRNAs and stabilized AGO-miRNA-target RNA ternary complexes in individual target coimmunoprecipitation assays. In genome-wide AGO-miRNA-target RNA coimmunoprecipitation experiments, slicer-defective AGO1-miRNA associated with target RNA more effectively than did wild-type AGO1-miRNA. These data not only reveal functional roles for AGO1, AGO2, and AGO7 slicer activity, but also indicate an approach to capture ternary complexes more efficiently for genome-wide analyses.
INTRODUCTION
In small RNA-directed RNA silencing pathways, ARGONAUTE (AGO) proteins associate with small RNA to target and silence transcripts (Kim et al., 2009). In plants and animals, interactions between AGO–small RNA complexes and target RNA usually lead to repression of the transcripts through either direct cleavage (slicing) or through other mechanisms, such as target destabilization or translational repression (recently reviewed in Huntzinger and Izaurralde, 2011). However, the relative contribution of each of these mechanisms in target repression has been debated extensively (Eulalio et al., 2008; Filipowicz et al., 2008; Wu and Belasco, 2008). Originally, animal microRNAs (miRNAs) were thought to regulate mRNAs mostly by translational repression because their incomplete base pairing with target RNAs excluded slicing. However, recent work showed that miRNA-mediated (and slicer-independent) mRNA destabilization, mainly through deadenylation of mRNAs, may be a dominant mechanism of transcript regulation by animal miRNAs (Baek et al., 2008; Hendrickson et al., 2009; Guo et al., 2010). mRNA destabilization and translational repression could be mechanistically linked, but whether transcript destabilization occurs preferentially before or immediately after the inactivation of translation is still not clear (Huntzinger and Izaurralde, 2011).
In plants, extensive evidence suggests that miRNAs, with high complementarity to target RNAs, regulate mRNAs through endonucleolytic cleavage (Huntzinger and Izaurralde, 2011). miRNA-guided cleavage products are normally sorted into endogenous RNA degradation pathways, such as 5′-3′ degradation by EXORIBONUCLEASE4, or 3′-5′ degradation by the exosome complex (Souret et al., 2004; Chekanova et al., 2007). However, the fact that slicing occurs broadly on plant miRNA targets does not exclude a role for translational repression, which was initially reported for some of the early characterized target RNAs (Aukerman and Sakai, 2003; Chen, 2004; Gandikota et al., 2007). More recently, (1) the identification of endogenous factors, such as SUO, which is required for miRNA-directed translational repression (Yang et al., 2012); (2) the association of AGO1 with ribosomes in an mRNA-dependent manner (Lanet et al., 2009); and (3) the possibility that miRNA targets may be subject to deadenylation and decapping in plants (Brodersen et al., 2008) has led to the idea that translation repression could be a widespread regulatory mechanism in plants (Brodersen et al., 2008).
AGO proteins have several functional domains, including PAZ, MID, and PIWI domains (Mallory and Vaucheret, 2010). The MID and PAZ domains bind the 5′ monophosphorylated nucleotide and 3′ nucleotide of the guide RNA, respectively. The PIWI domain functions as the ribonucleolytic domain (Song et al., 2004). High-resolution structural analysis of Thermus thermophilus AGO protein suggest a three-step process through which AGO–small RNA complexes bind to and slice target transcripts (Wang et al., 2009). In the nucleation step, the 3′ end of the target RNA is bound at the 5′ end of the guide strand, forming a double helix between the two lobes of the AGO protein. During the propagation step, pivotal movements of the AGO protein permit extension of the double helix and release of the 3′ end of the guide by the PAZ domain. Rotation of the PAZ domain favors the correct positioning of the target RNA cleavage site close to the PIWI domain. Target RNA cleavage occurs at the phosphodiester bond linking nucleotides opposite of positions 10 and 11 of the guide strand and is facilitated by divalent cations (Wang et al., 2009).
The PIWI domain of Arabidopsis thaliana AGOs contains a metal-coordinating triad (Asp-Asp-His [DDH] or Asp-Asp-Asp [DDD]). Mutational analyses revealed that the DDH catalytic motif in AGO1, AGO4, and AGO10 is required for slicer activity in vitro and in vivo (Baumberger and Baulcombe, 2005; Qi et al., 2006; Ji et al., 2011; Zhu et al., 2011). However, AGO10-miRNA complexes do not require slicer activity to exert their function (Zhu et al., 2011).
In addition to directly or indirectly repressing target RNAs, specific AGO–small RNA complexes trigger amplification of secondary small interfering RNA (siRNA) from target transcripts in plants. Trans-acting siRNA (tasiRNA), a class of siRNAs that forms through a highly refined RNA interference mechanism, originates from four families of noncoding (TAS) transcripts after initial miRNA-guided cleavage. TAS1/TAS2 and TAS4 family transcripts are initially targeted and sliced by AGO1-miR173 and AGO1-miR828 complexes, respectively, at a 5′-proximal site (Allen et al., 2005; Yoshikawa et al., 2005; Rajagopalan et al., 2006; Montgomery et al., 2008b). RNA-DEPENDENT RNA POLYMERASE6 (RDR6) uses the 3′ cleavage fragments as templates to produce double-stranded RNA that is processed by DICER-LIKE4 to generate tasiRNAs in register with the miRNA-guided cleavage site (Allen et al., 2005; Dunoyer et al., 2005; Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005; Montgomery et al., 2008b). However, the majority of AGO1-miRNA-target interactions do not lead to efficient siRNA formation, leading to the hypothesis that different AGO-small RNA-target complexes possess distinct properties that lead to recruitment of the RDR6-dependent amplification apparatus. These properties may involve specific AGO1 states that are triggered by either the size of the small RNA or the properties of the precursor from which the small RNA is derived (Chen et al., 2010; Cuperus et al., 2010; Manavella et al., 2012). TAS3a tasiRNAs form through a distinct mechanism involving AGO7, which associates with high specificity with miR390 and directs tasiRNA biogenesis from TAS3 transcripts. AGO7-miR390 complexes function through distinct cleavage and noncleavage modes at two target sites in TAS3 transcripts (Axtell et al., 2006; Montgomery et al., 2008a).
Here, we compared the activities of wild-type and active-site defective forms of several Arabidopsis AGOs. These activities included small RNA binding, interaction with target RNA, slicing or destabilization of target RNA, secondary siRNA formation, and antiviral activity. Arabidopsis AGO2 was identified as an AGO that can target and cleave transcripts but that cannot function in the siRNA amplification pathway. Moreover, AGO2 catalytic residues were essential for antiviral activity in Arabidopsis, as they were required to functionally complement ago2-1 mutants. Catalytic residues of AGO1 and AGO7 were required to complement the morphological and functional defects of Arabidopsis ago1-25 and zip-1 (AGO7-defective) mutants, respectively, supporting the idea that slicer activity is critical for AGO1 and AGO7 in vivo function. Interestingly, both wild-type and active-site defective forms of AGO1, AGO2, AGO7, and AGO10 associated in vivo with miRNAs and/or siRNAs, but target RNAs coimmunoprecipitated more effectively with the active-site defective forms of these AGOs.
RESULTS
To systematically analyze posttranscriptional functions of Arabidopsis AGO1, AGO2, AGO7, and AGO10, constructs encoding proteins with substitutions affecting one or more residues in the catalytic triad of the respective PIWI domains were produced (see Supplemental Figure 1 online). Key residues of the catalytic triad were mutated independently to an Ala, as reported for Arabidopsis AGO1, AGO4, and AGO10 (Baumberger and Baulcombe, 2005; Qi et al., 2006; Zhu et al., 2011) (see Supplemental Figure 1 online). In addition, the third position of the catalytic triad was mutated to an Asp in AGO1 and AGO7 and to a His in AGO2 (see Supplemental Figure 1 online). Wild-type and mutant constructs contained either constitutive (35S) or authentic regulatory sequences for the expression of hemagglutinin (HA)–tagged AGO sequences (see Supplemental Figure 1 online). As AGO2 is involved in antiviral silencing, this will be discussed separately from AGO1, AGO7, and AGO10, which associate with miRNAs that affect developmental processes.
Functional Analysis of Arabidopsis AGO2: Stabilization of Ternary Complexes, Target Slicing and tasiRNA Biogenesis
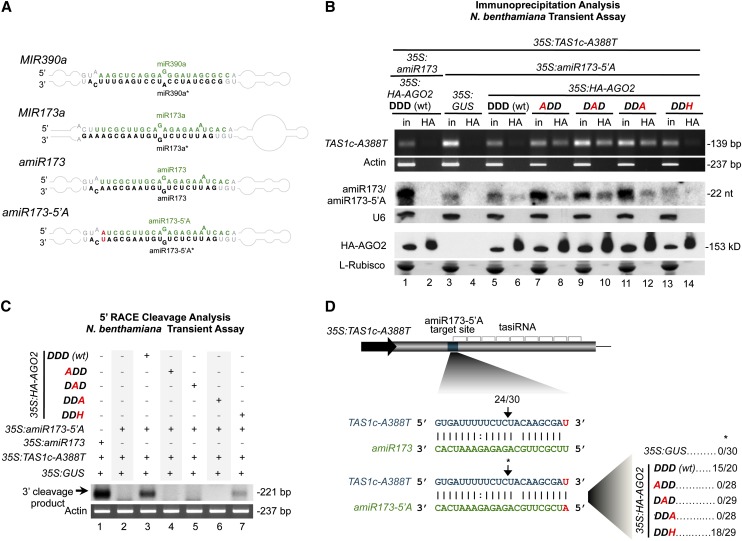
Arabidopsis AGO2 has not been demonstrated as a slicer, although it clearly possesses conserved catalytic triad positions (see Supplemental Figure 1A online). Antiviral and antibacterial defense roles for AGO2 were shown (Harvey et al., 2011; Jaubert et al., 2011; Scholthof et al., 2011; Wang et al., 2011b; Zhang et al., 2011). To test whether or not AGO2 has slicer activity, a modified version of the TAS1c-tasiRNA generating system in Nicotiana benthamiana was used. In the TAS1c-tasiRNA generating system, overexpressed TAS1c transcript is sliced via endogenous AGO1 activity when MIR173 is coexpressed, leading to tasiRNA formation (Montgomery et al., 2008b; Cuperus et al., 2010). Here, we generated a 35S:amiR173-5′A construct expressing, in MIR390 foldback, an artificial form of miR173 containing an AGO2-preferred 5′A (amiR173-5′A) (instead of the AGO1-preferred 5′U of canonical miR173) (Figure 1A), to redirect miR173 association from AGO1 to AGO2. Because the 3′ end of the miR173 target site in TAS1c RNA is an adenosine, we generated a construct (35S:TAS1c-A388T) expressing a modified TAS1c target transcript containing a substitution at the end of the miR173 target site (U instead of A) to maintain full base-pairing with amiR173-5′A.
Figure 1.
Functional Analysis of AGO2 Catalytic Residues in Ternary Complex Stabilization and Target Transcript Slicing.
(A) Foldbacks of wild-type MIR390a and MIR173a and of artificial miRNAs amiR173 and amiR173-5′A engineered within the MIR390a foldback. Green and black, miRNA guide and miRNA* strands, respectively. Mutagenized positions are shown in red.
(B) Immunoprecipitations with HA-AGO2 proteins. Input and immunoprecipitation fractions from N. benthamiana following coexpression of 35S:amiR173-5′A and 35S:TAS1c-A388T with 35S:GUS, 35S:HA-AGO2-DDD, 35S:HA-AGO2-DAD, 35S:HA-AGO2-DDA, and 35S:HA-AGO2-DDH, as well as coexpression of 35S:amiR173 with 35S:TAS1c-A388T and 35S:HA-AGO2-DDD (wt), were analyzed. Top, ethidium bromide–stained RT-PCR products corresponding to a noncleaved fragment from the TAS1c-A388T target transcript. Middle, amiR173/amiR173-5′A blot is shown as control for HA-AGO2 binding and immunoprecipitation selectivity. Bottom, HA-AGO2 blots. l-Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase) stained membrane was included as input loading and immunoprecipitation control. The catalytic residues are shown in bold, those found in wild-type AGO2 are in black, and those modified are in red. nt, nucleotides.
(C) Ethidium bromide–stained 5′RACE products corresponding to the 3′ cleavage product of TAS1c-A388T targets cleaved by amiR173-5′A/AGO2 complexes. N. benthamiana actin RT-PCR products are shown as control.
(D) Proportion of cloned 5′RACE products corresponding to cleavage within TAS1c-A388T transcripts at the amiR173-5′A targeted site in assays with HA-AGO2 forms.
First, we tested the ability of wild-type and modified AGO2 forms to associate with miRNA (Figure 1B; see Supplemental Figure 1B online). The amiR173-5′A, but not the canonical miR173 expressed from 35S:amiR173 (Cuperus et al., 2010), immunoprecipitated specifically with HA-AGO2-DDD (wt) (for the wild type) (Figure 1B, lanes 6 and 2, respectively). Each modified HA-AGO2 form also associated with amiR173-5′A (Figure 1B, lanes 8, 10, 12, and 14). Second, we analyzed the association between the AGO2 forms and target transcripts. RT-PCR products from TAS1c-A388T target RNA were detected in HA immunoprecipitates containing the HA-AGO2 Ala substitution forms (Figure 1B, lanes 8, 10, and 12) but not in those accumulating HA-AGO2-DDD (wt), HA-AGO2-DDH, or the control vector (Figure 1B, lanes 6, 14, and 4, respectively). This suggests that while each AGO2 form binds miRNA, the altered AGO2 variants associate more effectively with target RNA in ternary complexes.
Third, to test potential AGO2 slicer activity in vivo, 5′ RNA ligase-mediated rapid amplification of cDNA ends (5′RACE) assays were done with each coexpression assay using primers to detect the 3′ product expected after TAS1c-A388T target cleavage. The 3′ cleavage product was detected in assays containing HA-AGO2-DDD (wt) and in the control assay where amiR173 was coexpressed with TAS1c-A388T target (Figure 1C, lanes 3 and 1, respectively). By contrast, no 3′ cleavage fragment was detected in assays containing amiR173-5′A alone or when amiR173-5′A was coexpressed with any of the AGO2 Ala substitution forms (Figure 1C, lanes 2 and 4 to 6). Low levels of cleavage product were detected in assays containing HA-AGO2-DDH (Figure 1C, lane 7). Sequencing confirmed that cleavage guided by amiR173-5′A through exogenous HA-AGO2-DDD (wt) or HA-AGO2-DDH, or by amiR173 most likely through endogenous AGO1, occurred at the canonical site (Figure 1D). No cleavage fragments with ends mapping to the target site were detected through sequencing of samples accumulating the Ala substitution forms (Figure 1D). These results suggest that AGO2 possesses slicer activity.
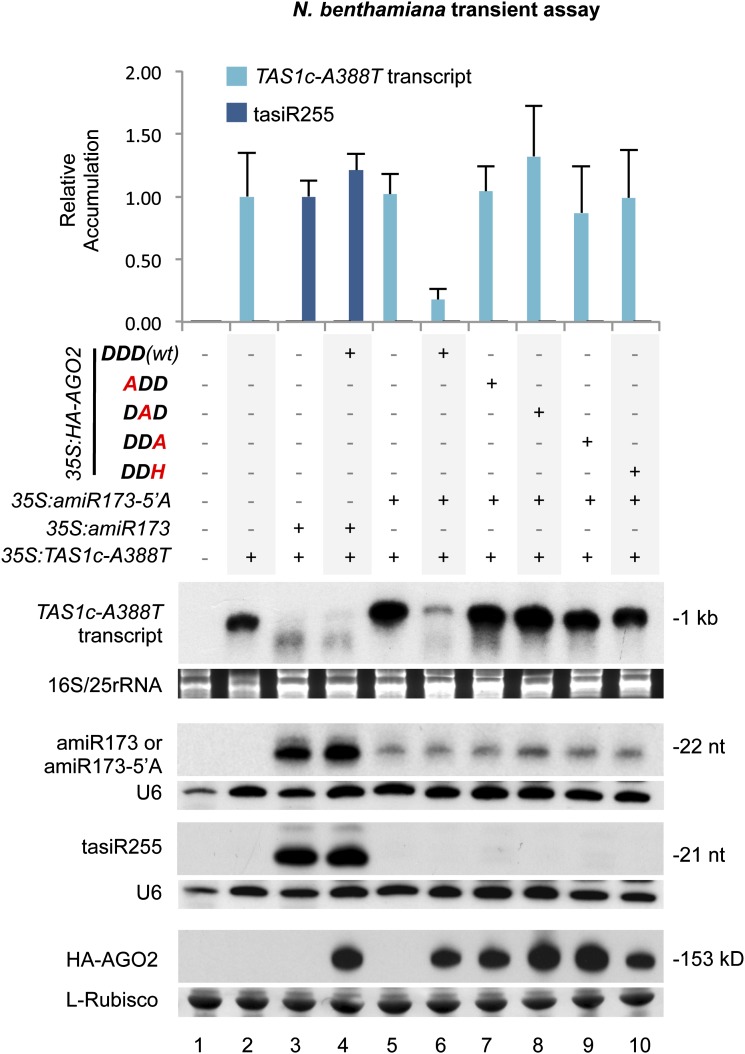
Fourth, we tested AGO2-mediated target transcript destabilization and induction of tasiRNA biogenesis by analyzing TAS1c-A388T target transcript and TAS1c-A388T–derived tasiRNA (tasiR255) accumulation in the presence of the HA-AGO2 forms. We hypothesized that TAS1c-A388T target destabilization could be mediated by both amiR173/AGO1 and amiR173-5′A/AGO2-DDD (wt) complexes. When amiR173 was coexpressed with 35S:TAS1c-A388T, with or without 35S:HA-AGO2-DDD (wt), TAS1c-A388T transcript was destabilized and tasiR255 accumulated to similar levels (Figure 2, lanes 3 and 4). This was likely due to endogenous AGO1 working with amiR173. Target destabilization was not detected in the absence of amiR173 when 35S:TAS1c-A388T was expressed alone, as TAS1c-A388T transcript accumulated to high levels (Figure 2, lane 5). This result indicates that TAS1c-A388T target destabilization was amiR173/AGO1 dependent. When amiR173-5′A was coexpressed with TAS1c-A388T, TAS1c-A388T, target destabilization was not observed (Figure 2, lane 5), indicating that endogenous N. benthamiana AGOs do not function with amiR173-5′A on the target RNA. However, when amiR173-5′A was coexpressed with HA-AGO2-DDD (wt), TAS1c-A388T target RNA was destabilized (Figure 2, lane 6). By contrast, the levels of TAS1c-A388T transcript were high when coexpressed with each of the HA-AGO2 modified forms (Figure 2, lanes 7 to 10) (P < 0.03 for all pairwise t test comparisons). These results indicate that the catalytic residues of AGO2 are required for target transcript destabilization. However, HA-AGO2-DDD (wt)–mediated transcript destabilization did not trigger tasiRNA biogenesis, as no tasiR255 was detected (Figure 2, lane 6). Indeed, tasiR255 was not detected in any of the samples coexpressing any of the HA-AGO2 forms and amiR173-5′A (Figure 2, lanes 7 to 10), suggesting that AGO2 cannot functionally replace AGO1 in the tasiRNA biogenesis pathway.
Figure 2.
Effects of Wild-Type and Modified AGO2 Forms on TAS1c-A388T Transcript Targeting and tasiRNA Biogenesis.
Accumulation of TAS1c-A388T transcript and tasiR255 in N. benthamiana leaves from assays testing AGO2 forms. Constructs were coexpressed as indicated above the blot panels. Top, mean (n = 3) relative TAS1c-A388T transcript (light blue) and tasiR255 (dark blue) levels + sd (lane 2 and lane 3 = 1.0 for TAS1c-A388T transcript and tasiRNA255, respectively). amiR173/amiR173-5′A and HA-AGO2 blots are shown as controls. Only one blot from three biological replicates is shown. U6 and l-Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase) blots, as well as ethidium bromide–stained rRNA gels, are shown as loading controls. nt, nucleotides.
AGO2 Catalytic Residues Are Required for Antiviral Defense
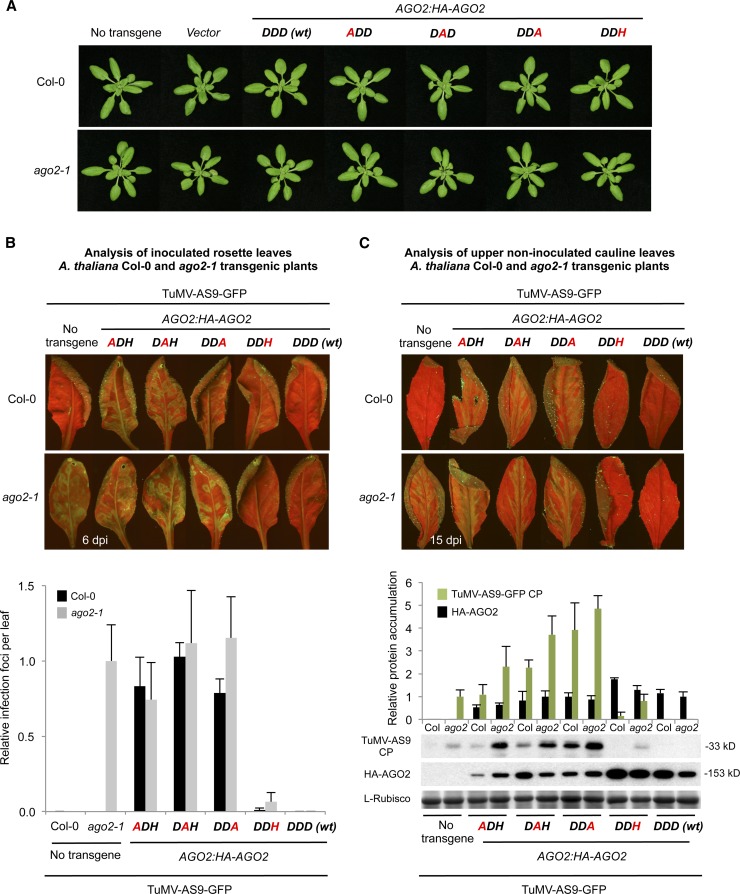
To test the functionality of wild-type and modified AGO2 forms in antiviral defense, genetic complementation assays were done in Arabidopsis ago2-1 mutant plants inoculated with green fluorescent protein (GFP)–expressing forms of Turnip mosaic virus (TuMV). The ago2-1 allele contains a T-DNA insert, but the mutant plants have no obvious morphological phenotype (Lobbes et al., 2006; Harvey et al., 2011). Similarly, neither Columbia-0 (Col-0) nor ago2-1 plants expressing any of the AGO2 constructs had obvious morphological phenotypes (Figure 3A; see Supplemental Figure 2 online). Because wild-type TuMV effectively masks the antiviral effects of RNA silencing responses through the action of a silencing suppressor protein (HC-Pro) (Garcia-Ruiz et al., 2010), AGO2 antiviral functions in Col-0 and ago2-1 nontransgenic and transgenic lines were tested using TuMV-AS9-GFP, an HC-Pro–deficient virus that is sensitive to RNA silencing responses (Garcia-Ruiz et al., 2010). AGO2 function in local and systemic antiviral defense was analyzed in inoculated leaves by detecting GFP fluorescence and counting viral infection foci and in upper noninoculated (cauline) leaves by detecting GFP fluorescence and measuring TuMV coat protein (CP) accumulation.
Figure 3.
Phenotypic and Molecular Analyses of Col-0 and ago-2-1 T3 Transgenic Plants Expressing Wild-Type or Modified AGO2 Forms.
(A) Pictures of 21-d-old Col-0 (top panel) and ago2-1 (bottom panel) T3 transgenic plants.
(B) Analysis of TuMV-GFP-AS9 viral infection in inoculated rosette leaves at 6 d after inoculation. Top, pictures were taken at 6 d after inoculation under UV light. Bottom, the number of infection foci for TuMV-AS9-GFP was expressed relative to those in ago2-1 (12 ± 3 foci per leaf). The graph shows the average and sd for 32 leaves and eight plants per treatment.
(C) Analysis of viral infection in upper noninoculated cauline leaves at 15 d after inoculation. Top, pictures were taken at 15 d after inoculation under UV light. Bottom, accumulation of TuMV-AS9-GFP CP and HA-AGO2 in cauline leaves from ago2-1 and Col-0 transgenic lines at 15 d after inoculation. Mean (n = 4) relative to TuMV-AS9-GFP CP (green) and HA-AGO2 (black) levels + sd [ago2-1 and DDD (wt) = 1.0 for CP and HA-AGO2, respectively]. l-Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase) blot is shown as loading control.
TuMV-AS9-GFP–inoculated leaves of ago2-1 plants expressed high levels of GFP fluorescence, but no fluorescence was detected in inoculated leaves from wild-type Col-0 plants (Figure 3B, top). Cauline leaves of ago2-1 plants, but not of Col-0 plants, displayed GFP fluorescence and accumulated CP (Figure 3C). No GFP fluorescence or infection foci was observed in inoculated or systemic leaves of AGO2:HA-AGO2-DDD (wt) ago2-1 transgenic lines, indicating that the loss of resistance in the ago2-1 mutant was genuinely due to loss of AGO2. By contrast, high levels of GFP fluorescence and a high number of infection foci were detected in inoculated leaves of ago2-1 transgenic plants expressing each of the three AGO2 Ala substitution forms (Figure 3B). High levels of GFP fluorescence and CP were also detected in cauline leaves of these lines (Figure 3C). Finally, low levels of GFP fluorescence and a low number of infection foci were detected in inoculated leaves from ago2-1 plants expressing AGO2:HA-AGO2-DDH (Figure 3B). Cauline leaves of these plants did not show any GFP fluorescence and did not accumulate TuMV-AS9-GFP CP (Figure 3C). Collectively, these results indicate that AGO2 catalytic residues are necessary to block local and systemic TuMV-AS9-GFP infection in Arabidopsis, although the AGO2-DDH variant has partial complementation and antiviral activities.
The Col-0 transgenic plants expressing the AGO2 constructs were also tested for virus susceptibility. No TuMV-AS9-GFP infection foci were observed in inoculated leaves from Col-0 plants expressing AGO2:HA-AGO2-DDD (wt) (Figure 3B, top). Cauline leaves of these plants contained no GFP fluorescence or CP (Figure 3C). By contrast, high levels of GFP fluorescence and a high number of infection foci were detected in inoculated leaves of Col-0 transgenic lines expressing the AGO2 Ala substitution forms (Figure 3B). Cauline leaves of these plants showed high levels of GFP fluorescence and CP (Figure 3C). These results indicate that AGO2 Ala substitution forms interfere with the endogenous AGO2 activity present in Col-0. Low levels of GFP fluorescence and a low number of infection foci were detected in inoculated leaves of Col-0 transgenic plants expressing AGO2:HA-AGO2-DDH (Figure 3B). Cauline leaves of these plants did not display any GFP fluorescence or accumulate CP (Figure 3C). These last observations indicate that AGO2-DDH has little interference activity with endogenous AGO2 activity in Col-0 plants.
AGO1 and AGO7 Catalytic Residues Are Required to Restore Arabidopsis ago1-25 and zip-1 Mutant Defects, Respectively
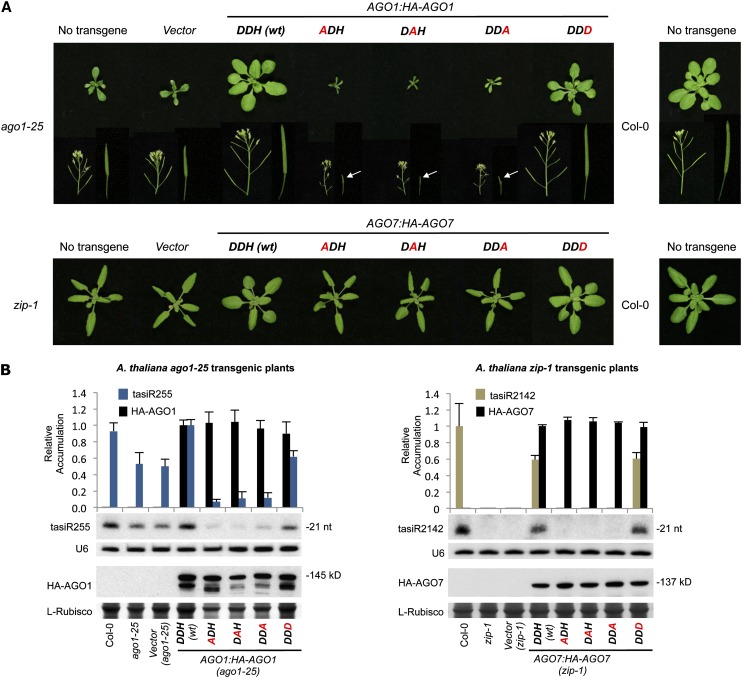
To test functionality of wild-type and mutant AGO1 and AGO7 constructs (see Supplemental Figures 1C and 1D online, respectively), genetic complementation assays in Arabidopsis ago mutants were done. AGO1 constructs were introduced into the Arabidopsis ago1-25 hypomorphic mutant, which grows with a series of vegetative and reproductive defects, such as reduced size, delayed bolting and flowering, and reduced number and size of siliques compared with Col-0 plants (Morel et al., 2002) (Figure 4A, top; see Supplemental Table 1 online). ago1-25 plants have also molecular defects like decreased levels of TAS1 and TAS2 tasiRNAs (Montgomery et al., 2008b). The ago1-25 phenotype was complemented only in the AGO1:HA-AGO1-DDH (wt) and the AGO1:HA-AGO1-DDD transgenic lines (Figure 4A, top; see Supplemental Table 1 online). The three Ala substitution mutant constructs did not complement the ago1-25 phenotype. In fact, plants expressing these constructs had exacerbated developmental defects, as plants were further reduced in size and fertility (Figure 4A, top; see Supplemental Table 1 online). The analysis of TAS1c-dependent tasiRNA (tasiR255) accumulation in the transgenic lines showed that plants transformed with the AGO1:HA-AGO1-DDH (wt) construct accumulated tasiR255 to levels similar to Col-0 plants (1.0 ± 0.07 and 0.93 ± 0.10, respectively) (Figure 4B, left). By contrast, lines expressing any of the Ala substitution mutant constructs had levels of tasiR255 significantly reduced compared with the ago1-25 lines transformed with the empty vector (P < 0.01 for all pairwise t test comparisons). Finally, plants transformed with the AGO1:HA-AGO1-DDD construct accumulated lower levels of tasiR255 than Col-0 and more similar to levels detected in ago1-25 plants transformed with the empty vector (0.61 ± 0.07 and 0.49 ± 0.09, respectively). Collectively, these results suggest that the Ala substitution AGO1 forms interfere with the residual AGO1 activity present in ago1-25. They also suggest that the AGO1-DDD forms have at least partial complementation activity.
Figure 4.
Phenotypic and Molecular Analyses of Arabidopsis ago1-25 and zip-1 Transgenic Plants Expressing Wild-Type or Modified AGO1 and AGO7 Forms, Respectively.
(A) Pictures of 21-d-old ago1-25 (top panel) and zip-1 (bottom panel) T1 transgenic plants. Details of stems containing inflorescences, siliques, or nondeveloped reproductive organs (pointed with a white arrow) are shown for the ago1-25 transgenic lines. The AGO catalytic residues are shown in bold, those found in wild-type AGO1 or AGO7 are in black, and those mutated are in red.
(B) Accumulation of TAS-dependent tasiRNAs. Left, accumulation of TAS1c-dependent tasiRNA (tasiR255) and AGO1 forms in ago1-25 T1 transgenic lines. Mean (n = 3) relative to tasiR255 (dark blue) and HA-AGO1 (black) levels + sd [DDH (wt) =1.0 for tasiR255 and HA-AGO1]. Right, accumulation of TAS3a-dependent tasiRNA (tasiR2142) and HA-AGO7 forms in zip-1 T1 transgenic lines. Mean (n = 3) relative to tasiR2142 (light brown) and HA-AGO1 (black) levels + sd [Col-0 and DDH (wt) =1.0 for tasiR2142 and HA-AGO7, respectively]. Only one blot from three biological replicates is shown. U6 and l-Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase) blots are shown as loading controls. nt, nucleotides.
AGO1 constructs were also introduced into the wild-type Col-0 background (see Supplemental Figure 3 online). None of these lines had any significant differences in days to bolting or flowering time (see Supplemental Table 1 online). Plants expressing the Ala substitution mutant constructs produced fewer seed, and, in particular, plants expressing the AGO1-DDA forms were significantly shorter (see Supplemental Table 1 online). Levels of tasiR255 in all the Col-0 lines expressing AGO1 forms were slightly reduced compared with the vector-transformed lines (see Supplemental Figure 3B online, left). Taken together, these results suggest that the Ala substitution forms, like in AGO2 Col-0 transgenic lines (Figure 3), interfere with endogenous AGO1 activity (see Supplemental Figures 3A and 3B online, left).
The constructs encoding wild-type or modified AGO7 forms were introduced into the zip-1 mutant, which has an accelerated transition from juvenile to adult vegetative stage (Hunter et al., 2003; Peragine et al., 2004; Yoshikawa et al., 2005; Montgomery et al., 2008a). The zip-1 allele results in a single base pair change that generates an early stop codon, most likely leading to a null allele (Hunter et al., 2003). Moreover, zip-1 plants are impaired in biogenesis of AGO7-dependent tasiRNAs from TAS3 transcripts (Montgomery et al., 2008a). The zip-1 phenotype (accelerated appearance of trichomes in leaves and increased blade length/petiole length ratio) was complemented only in AGO7:HA-AGO7-DDH (wt) and AGO7:HA-AGO7-DDD transgenic lines but not in lines expressing any of the three AGO7 Ala substitution forms (Figure 4A, bottom; see Supplemental Figure 4A online, left). TAS3a-tasiRNAs (tasiR2142) were generated only in the AGO7:HA-AGO7-DDH (wt) and AGO7:HA-AGO7-DDD zip-1 transgenic lines but not in zip-1 lines expressing any of the three Ala substitution forms (Figure 4B, right).
By contrast, when the AGO7 wild-type and mutant constructs were inserted into a Col-0 background, no obvious phenotypes were observed in any of the transgenic lines (see Supplemental Figure 3A, bottom, and Supplemental Figures 4A and 4B, right, online). However, slightly reduced levels of TAS3-dependent tasiR2142 were observed in transgenic lines expressing the AGO7 Ala substitution forms (see Supplemental Figure 3B online, right). This may reflect interference of the Ala substitution AGO7 forms with endogenous AGO7 activity present in Col-0.
In summary, the complementation analyses confirmed that the authentic DDH catalytic triad of AGO1 and AGO7 is required for normal plant development and tasiRNA biogenesis in Arabidopsis. In addition, the presence of an Asp in the third position of AGO1 and AGO7 catalytic triad may be compatible with partial or full AGO activity in AGO1 and AGO7.
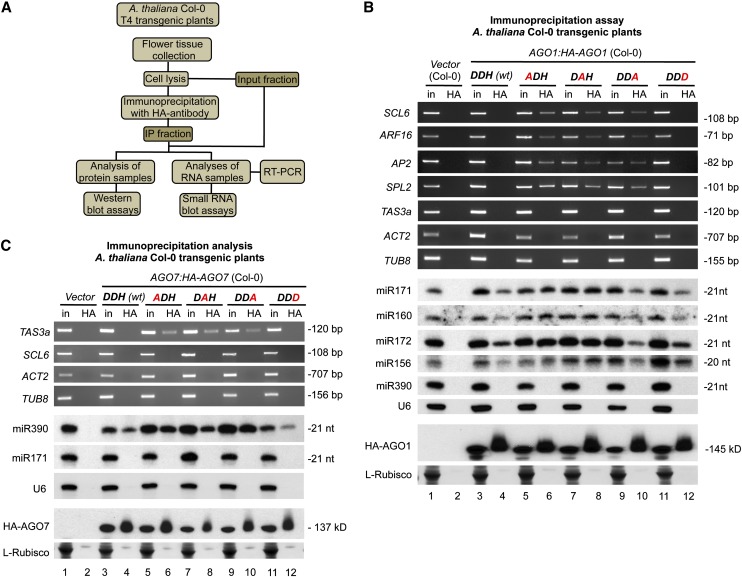
AGO1, AGO7, and AGO10 Active-Site Defective Forms Associate with miRNAs and Stabilize the Interaction with Target Transcripts
Next, we tested the stability of the association between wild-type and modified forms of AGO1 and AGO7 with miRNAs and target transcripts by coimmunoprecipitation analyses in Arabidopsis Col-0 T4 transgenic lines expressing HA-tagged AGO1 or AGO7 forms, respectively. As for AGO2, we used RNA immunoprecipitation followed by RT-PCR detection of fragments from known target transcripts that included the miRNA target site (Figure 5A). HA-AGO1 forms associated to different 5′U miRNAs, such as miR171, miR160, miR169, miR172, and miR156 (Figure 5B), confirming that the substitutions at the catalytic triad did not impair AGO1 miRNA binding activity. Interestingly, RT-PCR products from several known AGO1-miRNA–targeted transcripts, such as SCARECROW-LIKE6 (SCL6), AUXIN RESPONSE FACTOR16 (ARF16), APETALA2 (AP2), and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE2 (SPL2), were detected in HA immunoprecipitates containing the HA-AGO1 Ala substitution forms (Figure 5B, lanes 6, 8, and 10, respectively) but rarely in those containing wild-type HA-AGO1-DDH (wt), HA-AGO1-DDD, or the control vector (Figure 5B, lanes 4, 12, and 2, respectively). As controls, TAS3a transcripts (non-AGO1 but AGO7 target transcripts) and non-miRNA–targeted transcripts, such as ACT2 and TUB8, did not immunoprecipitate with any of the HA-AGO1 proteins (Figure 5B), indicating that the HA-AGO1 immunoprecipitations were selective.
Figure 5.
AGO1 and AGO7 Interactions with miRNAs and Target Transcripts in Arabidopsis Col-0 Transgenic Plants Expressing Wild-Type or Modified AGO1 or AGO7 Forms.
(A) Flowchart of the analytical steps followed to test the association of AGO forms with miRNAs and target transcripts. IP, immunoprecipitation.
(B) Immunoprecipitations with HA-AGO1 forms. Input (in) and immunoprecipitated (HA) fractions from Arabidopsis Col-0 T4 transgenic plants were analyzed. Top, ethidium bromide–stained RT-PCR products corresponding to noncleaved fragments from different known AGO1 target transcripts (SCL6, ARF16, AP2, and SPL2) containing a miRNA target site. Ethidium bromide–stained RT-PCR products corresponding to noncleaved fragments from TAS3a, ACT2, and TUB8 are shown as non-AGO1-targeted controls. Middle, miR171, miR160, miR172, and miR156 blots are shown as controls for HA-AGO1 miRNA association. The miR390 panel shows a HA-AGO1–nonassociated miRNA as an immunoprecipitation control. U6 RNA gel blot was included as input loading and HA-AGO1–nonassociated control. Bottom, HA-AGO1 blots. Other details are as in Figure 1B.
(C) Immunoprecipitations with HA-AGO7 forms. Top, ethidium bromide–stained RT-PCR products corresponding to noncleaved fragments from TAS3a transcripts containing the miR390 cleavable target site. Ethidium bromide–stained RT-PCR products corresponding to noncleaved fragments from SCL6, ACT2, and TUB8 are shown as non-AGO7 targeted controls. Middle, miR390 blot is shown as control for HA-AGO7 miRNA association. The miR171 panel shows a HA-AGO1–nonassociated miRNA as an immunoprecipitation control. Bottom, HA-AGO7 blots. Other details are as in (B).
In plants expressing wild-type and modified HA-AGO7 forms, miR390, but not miR171 or U6 RNA, was immunoprecipitated (Figure 5C, lanes 4, 6, 8, 10, and 12, respectively). RT-PCR products from TAS3a transcripts, including the cleavable miR390 target site at the 3′ end of TAS3a, were detected in the HA immunoprecipitates containing the HA-AGO7 Ala substitution forms (Figure 5C, lanes 6, 8, and 10, respectively) but were essentially absent in those containing wild-type HA-AGO7-DDH, HA-AGO7-DDD or the control vector (Figure 5C, lanes 4, 12, and 2, respectively). As controls, no RT-PCR products corresponding to SCL6 (a non-AGO7 but AGO1 target transcript), ACT2, and TUB8 transcripts were detected in any of the HA immunoprecipitates (Figure 5C).
To further investigate AGO1 function, AGO1-dependent TAS1c tasiRNA formation was tested in the previously described N. benthamiana system. First, the ability of each AGO1 form to associate with miRNAs or siRNAs and to stabilize ternary complexes containing target transcripts was tested. HA-AGO1 forms expressed in N. benthamiana leaves together with TAS1c target RNA and MIR173 associated with miR173 and tasiR255 (see Supplemental Figure 5A online) but not with the AGO1 nonpreferred 5′A TAS1c 3′D2(-) siRNA. RT-PCR products from TAS1c target transcripts containing miR173 target site were detected in the HA immunoprecipitates containing the HA-AGO1 Ala substitution forms (see Supplemental Figure 5A online, lanes 6, 8, and 10, respectively) but rarely in those containing the wild-type HA-AGO1-DDH (wt), HA-AGO1-DDD, or the 35S:GUS (for β-glucuronidase) control (see Supplemental Figure 5A online, lanes 4, 12, and 2, respectively). Similar results were obtained with double and triple AGO1 Ala substitution forms (see Supplemental Figures 1D and 6A online).
Second, the effects of each AGO1 form in target transcript destabilization and tasiRNA biogenesis were tested. In the absence of any exogenously expressed AGO1 protein, TAS1c target transcript was destabilized, and tasiR255 was generated (see Supplemental Figure 5B online, lane 3). TAS1c target transcript was destabilized in samples containing HA-AGO1-DDH (wt) or HA-AGO1-DDD, and tasiR255 accumulated to similar levels than in the control (see Supplemental Figure 5B online, lanes 3, 4, and 8, respectively). By contrast, TAS1c target RNA accumulated to detectable levels in the presence of each of the three Ala substitution forms, and low levels of tasiR255 were generated (see Supplemental Figure 5B online, lanes 5 to 7, respectively). Similar effects were obtained with double and triple AGO1 Ala substitution forms (see Supplemental Figures 1C and 6B online). These results are consistent with interference by the active-site defective AGO1 forms with endogenous AGO1 activity, as previously observed in Arabidopsis transgenic plants (Figure 4B, left; see Supplemental Figure 3B online, left).
A similar transient assay approach was used to study the role of AGO10 catalytic residues in ternary complex stabilization, target transcript destabilization, and tasiRNA biogenesis. Both HA-AGO10-DDH (wt) and HA-AGO10-DAH coimmunoprecipitated with miR173 (see Supplemental Figures 1E and 5C online, lane 6), but RT-PCR fragments from TAS1c transcripts were detected only in HA immunoprecipitates containing the AGO10 Ala substitution form. As an immunoprecipitation control, 35S:MIR390a was coexpressed with 35S:HA-AGO10-DDH (wt) and 35S:TAS1c but failed to associate with HA-AGO10-DDH (wt) (see Supplemental Figure 5C online, lane 8). In addition, the effects of the HA-AGO10 forms on TAS1c target transcript destabilization and TAS1c-dependent tasiRNA biogenesis were tested. In the absence of any exogenous AGO, TAS1c target transcript was destabilized and tasiR255 formed in a miR173-dependent manner (see Supplemental Figure 5D online, lanes 2 and 3, respectively). However, TAS1c transcript accumulation was significantly higher in the presence of HA-AGO10-DAH relative to wild-type HA-AGO10-DDH (wt) (see Supplemental Figure 5D online, lanes 4 and 5, respectively; P = 0.01, t test). Finally, 35S:MIR390a was coexpressed with 35S:TAS1c and 35S:HA-AGO10-DDH (wt) as an additional control. In this case, TAS1c target RNA levels were similar to those in the control sample and no tasiRNAs were formed (see Supplemental Figure 5D online, lanes 2 and 6, respectively).
In summary, these results indicate that mutations affecting AGO1, AGO7, and AGO10 catalytic triad residues do not disrupt association with miRNAs or siRNAs. However, AGO1, AGO7, and AGO10 Ala substitution forms associated more stably in ternary complexes containing targeted transcripts.
Genome-Wide Profiling of miRNAs and Target RNAs in Catalytically Active or Inactive AGO1 Complexes
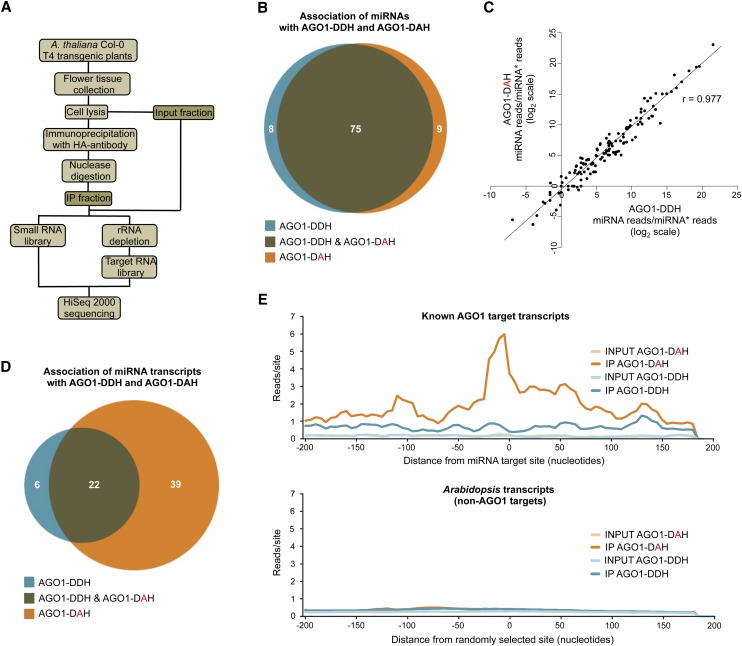
To determine in a genome-wide manner if slicer-defective AGO1 forms associated with miRNAs similarly to wild-type AGO1 and if they could be used for global profiling of target RNAs, small RNA and target RNA libraries from immunoprecipitates obtained from Arabidopsis Col-0 T4 transgenic plants expressing either catalytically active or inactive AGO1 were analyzed (Figure 6A). RNAs were nuclease digested during the immunoprecipitation to enrich immunoprecipitates in AGO1-protected target RNAs (Figure 6A). Libraries were also prepared using immunoprecipitates from transgenic plants containing an empty vector.
Figure 6.
Sequencing Analysis of miRNAs and Target RNAs Recovered from Catalytically Active or Inactive AGO1 Complexes.
(A) Flowchart of analytical steps to test immunoprecipitation of AGO1 forms with miRNAs and target transcripts.
(B) Venn diagram showing the association of miRNAs with distinct AGO1 forms. The number of miRNAs present in each category is shown.
(C) Scatterplot of the miRNA/miRNA* ratio from AGO1-DDH and AGO1-DAH for all known miRNAs in the immunoprecipitated fractions. The Pearson correlation coefficient (r) is shown.
(D) Venn diagram showing the association of known AGO1 target sites with distinct AGO1 forms. The number of miRNA target sites present in each category is shown.
(E) Distribution of transcript reads relative to miRNA target sites. Reads from AGO1-DDH and AGO1-DAH input or immunoprecipitated fractions from miRNA target sites (top) or from randomly selected sites (bottom) are plotted by the scrolling window method (20-nucleotide windows, 5-nucleotide scroll). Plots are centered on miRNA target sites (top) or randomly selected sites (bottom) and include 200 nucleotides upstream and downstream.
First, sequence analysis of miRNAs showed that a large subset of miRNAs (75) was enriched at least fourfold in immunoprecipitates containing either catalytically active or inactive AGO1 (Figure 6B). A small subset of miRNAs was enriched only in AGO1-DDH or AGO1-DAH immunoprecipitates (8 and 9, respectively) (Figure 6B). These results suggest that the inactivation of AGO1 slicer activity does not affect AGO1 miRNA binding specificity. Because AGO1 slicer activity could be required for passenger strand removal (Matranga et al., 2005; Iki et al., 2010) we analyzed the effect of AGO1 slicer activity on miRNA maturation by comparing the ratio between the reads corresponding to guide strands (miRNA reads) and passenger strands (miRNA* reads) from AGO1-DDH and AGO1-DAH immunoprecipitates (Figure 6C). If AGO1 slicer activity is required for guide strand maturation, miRNA/miRNA* ratios between AGO1-DAH and AGO1-DDH immunoprecipitates should decrease. However, these ratios were highly correlated (Pearson correlation coefficient = 0.977) (Figure 6C), suggesting that the inactivation of AGO1 catalytic activity does not affect miRNA maturation as previously proposed (Iki et al., 2010).
Second, transcript reads within a 401-nucleotide region flanking known miRNA-guided AGO1 target sites were analyzed for enrichment in immunoprecipitates compared with input RNA fractions. Thirty-nine target sites were enriched at least fourfold only in the AGO1-DAH immunoprecipitates. Twenty-two sites were enriched in immunoprecipitates containing both catalytically active and inactive AGO1, and six sites were enriched only in the catalytically active AGO1-DDH immunoprecipitates (Figure 6D). Although reads corresponding to RNA fragments from AGO1-DDH immunoprecipitates were enriched in target site regions relative to input RNA fractions, reads from AGO1-DAH immunoprecipitates were enriched up to approximately sevenfold more on average in the immediate target site vicinity compared with AGO1-DDH immunoprecipitates (Figure 6E, top). This pattern was not observed for 100 sets of randomly selected sites on non-AGO1 target transcripts (Figure 6E, bottom). Alternatively, target site enrichment could result from the increased abundance of miRNA targets in samples containing AGO1-DAH. However, miRNA target transcripts were on average 1.1 times more abundant in AGO1-DAH input samples relative to AGO1-DDH input samples, but this difference was not significant (P = 0.35, Welch two-sample t test). Therefore, the genome-wide data support the idea that AGO1 slicer-defective forms associate more stably with target RNAs in ternary complexes.
DISCUSSION
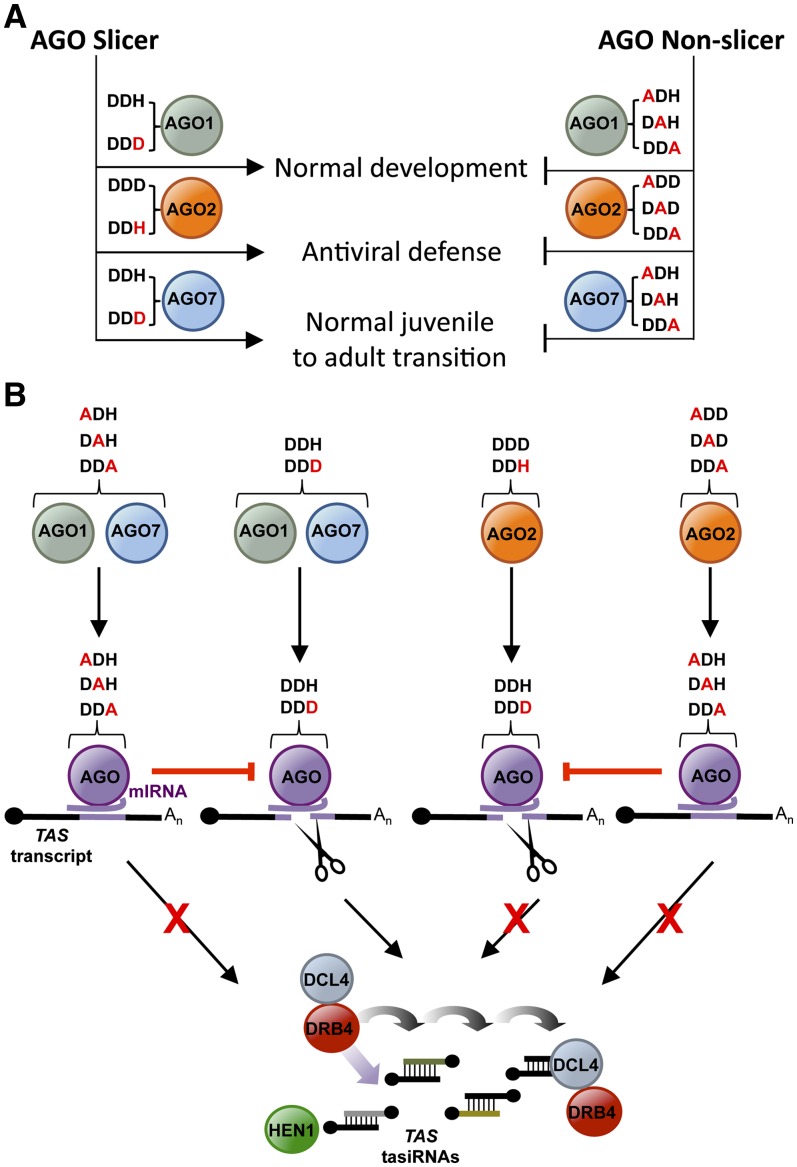
Comparative analyses of the activities of catalytically active or inactive AGO forms were done to understand the relevance of AGO slicer function in small RNA binding, target transcript interaction, target slicing or destabilization, secondary siRNA biogenesis, and antiviral defense (for a summary of the results, see Figure 7). The catalytic residues of AGO1, AGO2, and AGO7 were found to be essential for phenotypic and/or functional complementation of Arabidopsis ago1-25, ago2-1, and zip-1 defects (Figure 7A). The AGO1 and AGO7 catalytic residues were required for normal plant development and tasiRNA biogenesis. Complementation assays in ago2-1 plants with different AGO2 forms demonstrated the requirement of AGO2 catalytic residues for antiviral activity against TuMV-AS9-GFP, reinforcing previous observations that link AGO2 with antiviral defense in plants (Harvey et al., 2011; Jaubert et al., 2011; Scholthof et al., 2011; Wang et al., 2011b; Zhang et al., 2011). AGO2 may function to catalyze cleavage of viral RNA, although direct evidence for AGO2 function on viral RNA remains to be demonstrated.
Figure 7.
Models Summarizing the Functional Analyses of Wild-Type and Modified Forms of AGO1, AGO2, and AGO7.
(A) Effects of the different AGO forms on key biological processes.
(B) Effects of the different AGO forms on miRNA association, target transcript interaction, ternary complex stability, and tasiRNA biogenesis resulting from the interaction of AGO1, AGO2, and AGO7 with TAS transcripts.
The interference effect of AGO Ala substitution forms with residual endogenous AGO1 activity in the ago1-25 hypomorphic mutant was evident by the increased severity of developmental and fertility defects. As AGO1 is known to associate with miRNAs that repress target mRNAs involved in developmental pathways, these results are consistent with a global impact of AGO1 slicer activity on normal development in Arabidopsis. Interference with endogenous functions was also observed in Col-0 plants expressing AGO2 Ala substitution forms, as these plants lost resistance to TuMV-AS9-GFP. These two examples of interference may reflect the possibility that the defective AGO proteins compete with functional AGO proteins for specific factors. In particular, AGO slicer-deficient forms act as dominant-negative repressors of catalytically active endogenous AGOs, perhaps by sequestering miRNAs or miRNA-target RNA complexes. Complementation assays in Arabidopsis with AGO4 and AGO10 wild-type and Ala substitution forms showed that these particular AGOs also have slicer-independent functions. In the case of AGO4, slicer activity was not required for non-CpG methylation and silencing at the SUPERMAN (SUP) locus, as the SUP phenotype (increased no. of stamens and incompletely fused carpels) in clk-3/ago4-1 plants was recovered by either AGO4 wild-type or catalytic mutant forms (Qi et al., 2006). In the case of AGO10, both wild-type and Ala substitution forms rescued the ago10 pinhead-2 mutant phenotype in Arabidopsis Landsberg erecta ecotype and showed restored HD-ZIP III family expression, suggesting that the slicer activity of AGO10 is not required to exert its function during development (Zhu et al., 2011).
Several of the results presented here point toward Arabidopsis AGO2 functioning as a slicer. First, miR173-5′A-guided TAS1c-A388T target RNA 3′ cleavage products were detected by 5′RACE when wild-type AGO2 was expressed but not in the presence of any of the AGO2 Ala substitution forms. Second, TAS1c-A388T target RNAs in transient assays are destabilized in the presence of wild-type AGO2 but not when AGO2 Ala substitution forms are expressed. Third, TAS1c-A388T target transcripts were detected exclusively in immunoprecipitates containing miR173-5′A/AGO2 Ala substitution complexes. Fourth, TuMV-AS9-GFP resistance in Arabidopsis requires the presence of an AGO2 form with an active catalytic motif, as ago2-1 transgenic lines expressing AGO2 Ala substitution forms are susceptible to this virus.
The finding that AGO2 cannot substitute for AGO1 in triggering tasiRNA formation from TAS1c transcripts adds more insight into the requirements needed for secondary siRNA formation in plants. The inhibitory effects of AGO1 catalytic mutants on TAS1c transcript cleavage and, subsequently, on tasiRNA biogenesis, reinforce the idea that cleavage of TAS1c transcripts is necessary to induce tasiRNA formation. However, the fact that AGO2-mediated TAS1c-A388T transcript cleavage does not lead to tasiRNA biogenesis supports that the cleavage event is necessary but not sufficient to trigger formation of tasiRNA (Chen et al., 2010; Cuperus et al., 2010). This reinforces the idea that AGO1 has a specialized function to recruit RDR6 complexes to some target transcripts (Montgomery et al., 2008b; Cuperus et al., 2010). Previous reports suggested that AGO1 complexes associating with a 22-nucleotide miRNA derived from foldbacks with MIRNA/MIRNA* duplex asymmetry (such as the MIR173 foldback) adopt a state that leads to recruitment of RDR6 complexes to the sliced target (Chen et al., 2010; Cuperus et al., 2010). Recently, it was proposed that the base pair asymmetry within the miRNA duplex is the key trigger that transmits a signal to AGO1 to adopt the RDR6-recruiting state (Manavella et al., 2012). Here, AGO2 association with the 22-nucleotide miR173-5′A, originated from a precursor with an asymmetric foldback (Figure 1A), leads to TAS1c-A388T transcript cleavage but does not induce tasiRNA formation. Therefore, recruitment of RDR6 complexes may depend not only on the size of the guide miRNA or on the asymmetry of a miRNA duplex but also on the ability of a specific AGO protein to adopt the trigger state. However, the possibilities that the levels of accumulated miR173-5′A are not sufficient for triggering secondary siRNA biogenesis or that AGO2 may be producing secondary siRNAs that are somehow unstable cannot be ruled out.
Genome-wide identification of small RNAs that interact specifically with several Arabidopsis AGOs has been achieved by immunoprecipitation of tagged AGO proteins followed by high-throughput sequencing of bound small RNAs (Mi et al., 2008; Montgomery et al., 2008a; Cuperus et al., 2010; Havecker et al., 2010; Zhu et al., 2011). In several animal systems, identification of AGO targets by immunoprecipitation followed by high-throughput sequencing was achieved by stabilizing AGO-target transcript complexes with a UV cross-linking treatment prior to the immunoprecipitation (Chi et al., 2009; Zisoulis et al., 2010; Leung et al., 2011). In Arabidopsis, similar approaches have not yet been reported. We suggest that the recovery and analysis of Arabidopsis ternary complexes may be complicated by extensive target slicing or by accelerated target destabilization. In this work, slicer-deficient AGO forms were shown to associate with miRNAs and stabilize AGO-miRNA-target interactions, as targeted but nonsliced transcripts were detected in HA-AGO immunoprecipitates far more easily than those containing AGO wild-type forms. Moreover, epitope-tagged AGO catalytic mutant proteins expressed under native promoters may serve to identify targets more efficiently in a genome-wide surveys, as shown here for AGO1. This may be particularly important for the discovery of new AGO targets. However, if the slicing-dependent model for passenger strand clearance from siRNA duplexes is valid in Arabidopsis (Matranga et al., 2005), the identification of siRNA targets with slicer-deficient AGOs could be compromised. This possibility is not yet resolved, although tasiR255 was shown here to associate with single, double, and triple AGO1 Ala substitution forms.
Finally, comparative analyses of those target transcripts associating with wild-type or catalytic mutant AGOs might help identify which pools of transcripts are regulated in a slicing-dependent manner and which are not. This may help clarify the specific contribution of RNA cleavage versus translational repression in the genomic regulation of AGO targets in plants and might contribute to the discovery of new mechanisms of regulation of target transcripts by AGO complexes.
METHODS
Plant Materials and Growth Conditions
ago1-25, ago2-1, and zip-1 alleles were previously described (Morel et al., 2002; Hunter et al., 2003; Lobbes et al., 2006).
Nicotiana benthamiana and Arabidopsis thaliana plants were grown in standard greenhouse conditions with supplemental light on a 16-h-light/8-h-dark cycle. For the phenotypic analyses, transgenic plants were grown in a growth chamber with 22°C constant temperature and long-day conditions (16 h light/8 h dark cycle). Arabidopsis plants were genetically transformed using the floral dip method with Agrobacterium tumefaciens GV3101 strain (Clough and Bent, 1998). Transgenic plants were grown on plates containing Murashige and Skoog medium and hygromycin (50 μg/mL) for 10 d before being transferred to soil.
Transgene Constructs
35S:GUS, 35S:TAS1c, 35S:MIR173, 35S:MIR390a, 35S:amiR173, 35S:HA-AGO1-DDH (wt), 35S:HA-AGO2-DDD (wt), AGO2:HA-AGO2-DDD (wt), and AGO7:HA-AGO7-DDH (wt) constructs were described previously (Montgomery et al., 2008a; Cuperus et al., 2010).
MIR390a-derived artificial amiR173-5′A construct (35S:amiR173-5′A) was designed by ligating overlapping oligonucleotides into a pMDC32-derived vector containing ∼200 bp upstream and downstream of the MIR390a foldback as previously described (Cuperus et al., 2010).
To generate 35S:HA-AGO10-DDH (wt), a sequence coding for a triple HA epitope tag was fused by PCR amplification to the AGO10 open reading frame using Col-0 cDNA and the primers 3xHA-AGO10-F and AGO10-R (see Supplemental Table 2 online). The resulting DNA fragment was cloned into pENTR/D-TOPO (Invitrogen). AGO10 3′ regulatory sequence was PCR amplified from genomic DNA with AGO10-3pUTR-F2 and AGO10-3pUTR-R2 primers (see Supplemental Table 2 online) and introduced by ligation into an AscI site directly after the AGO10 stop codon within the pENTR vector, generating the pENTR-HA-AGO10-DDH (wt) plasmid. The HA-AGO10-DDH (wt) cassette was recombined into the plant transformation vector pMDC32 containing the 35S promoter sequence (Curtis and Grossniklaus, 2003).
35S:TAS1c-A388T was obtained by PCR mutagenesis using 35S:TAS1c as template and mutagenic oligos TAS1c-A388T-F and TAS1c-A388T-R (see Supplemental Table 2 online).
35S:HA-AGO1, 35S:HA-AGO2, and 35S:HA-AGO10 active-site mutant clones were obtained by mutagenesis PCR reactions using pENTR:HA-AGO1-DDH (wt), pENTR:HA-AGO2-DDD (wt), and pENTR:HA-AGO10-DDH (wt) clones as templates, respectively, and AGO1, AGO2, or AGO10 mutagenic oligos listed in Supplemental Table 2 online.
To generate AGO1:HA-AGO1-DDH (wt), AGO1 coding sequence was PCR amplified from Col-0 cDNA in frame with an N-terminal triple HA epitope sequence using Pfu Ultra polymerase (Stratagene) and the primers 3xHA-AGO1-F and AGO1-R (see Supplemental Table 2 online). The resulting DNA fragment was cloned into the pENTR D-TOPO vector (Invitrogen). AGO1 5′ regulatory sequence was PCR amplified from Col-0 genomic DNA using AGO1-Prom-F and AGO1-Prom-NotI-R, cloned into the pENTR D-TOPO vector, digested from the resulting plasmid with NotI, and ligated into the NotI site within the plasmid containing HA-AGO1 coding sequence. AGO1 3′ regulatory sequence was PCR amplified from Col-0 genomic DNA using AGO1-3′UTR-AscI-F and AGO1-3′UTR-R, cloned into the pENTR D-TOPO vector, digested from the resulting plasmid with AscI, and ligated into the AscI site within the plasmid containing HA-AGO1 coding and 5′ regulatory sequences to generate pENTR-AGO1:HA-AGO1-DDH (wt). Finally, the AGO1:HA-AGO1-DDH (wt) cassette was cloned by recombination into pMDC99, a Gateway-compatible plant transformation vector (Curtis and Grossniklaus, 2003).
AGO1:HA-AGO1, AGO2:HA-AGO2, and AGO7:HA-AGO7 active-site mutant clones were obtained by mutagenesis PCR reactions using pENTR-pAGO1:HA-AGO1-DDH (wt), pENTR-AGO2:HA-AGO2, and pENTR-AGO7:HA-AGO7-DDH (wt) (Montgomery et al., 2008a) as templates, respectively, and AGO1, AGO2, and AGO7 mutagenic oligos listed in Supplemental Table 2 online.
Transient Expression Assays
Transient expression assays in N. benthamiana leaves were done as described (Llave et al., 2002; Cuperus et al., 2010) with Agrobacterium GV3101.
Virus Infection Assays
Viruses and infection assays in Arabidopsis were as described (Garcia-Ruiz et al., 2010).
RNA Gel Blot Assays
Total RNA from N. benthamiana was isolated using TRIzol reagent (Invitrogen) as described (Cuperus et al., 2010). Triplicate samples from pools of N. benthamiana infiltrated leaves were analyzed. RNA gel blot assays were done as described (Montgomery et al., 2008b; Cuperus et al., 2010). Oligonucleotides used as probes for small RNA gel blots are listed in Supplemental Table 3 online.
5′RACE
5′ RNA ligase-mediated RACE was done as described (Montgomery et al., 2008b) with the Generacer kit (Invitrogen) TAS1c-707-5′RACE and TAS1c-573-5′RACE primers (see Supplemental Table 2 online). 5′RACE products were gel purified using MinElute gel extraction kit (Qiagen), cloned in TOPO TA (Invitrogen), introduced into Escherichia coli DH10B, screened for inserts, and sequenced.
RNA Immunoprecipitation Followed by RT-PCR
Two Agrobacterium-infiltrated N. benthamiana leaves or 1 g of inflorescence tissue from transgenic T4 Arabidopsis plants (4 to 6 weeks old, flower stages 1 to 12) were ground to a fine powder with a mortar and pestle in liquid nitrogen and homogenized in 12.5 mL/g lysis buffer (50 mM Tris-HCl, pH 7.4, 2.5 mM MgCl2, 100 mM KCl, 0.1% Nonidet P-40, 1 μg/mL leupeptin, 1 μg/mL aprotonin, 0.5 mM phenylmethylsulfonyl fluoride, one tablet of Complete proteinase inhibitor tablet [Roche], and 50 units/mL RNase OUT [Invitrogen]). Cell debris was pelleted by centrifugation for 5 min at 12,000 rcf at 4°C. Clarified lysates were incubated with 4 μg/mL of 12CA5 anti-HA antibody (Roche) for 15 min at 4°C and then with 100 μL of Protein-A agarose (Roche) per milliliter for 30 min at 4°C. Beads were washed six times for 10 min with lysis buffer at 4°C and then divided for protein and RNA analysis. RNAs were recovered by incubating the beads in 0.5 volumes of proteinase K buffer (0.1 M Tris-HCl, pH 7.4, 10 mM EDTA, 300 mM NaCl, 2% SDS, and 1 μg/μL proteinase K [Roche]) for 15 min at 65°C, extraction with saturated phenol, phenol:chloroform:isoamyl alcohol and chloroform, and ethanol precipitation. For small RNA gel blot assays, 2 to 5 μg of total RNA was used for the input fraction, and 20 to 40% of the RNA immunoprecipitate was used for the immunoprecipitation (HA) fraction. For protein blot assays, 10 μL of clarified eluate was loaded for the input fraction, and 3% of the immunoprecipitated beads was used for the immunoprecipitation (HA) fraction. HA-AGOs were detected by immunoblotting and chemiluminescence using anti-HA peroxidase 3F10 antibody (Roche) at a 1:1000 dilution and Western Lighting plus-ECL substrate (Perkin-Elmer,).
RT-PCR amplifications were done using those RNA samples used for RNA gel blots. Briefly, cDNA was obtained from 2 to 4 μg of total RNA or 25% of the RNA sample resulting from the immunoprecipitation, using the Superscript III system (Invitrogen). PCR to amplify fragments including the miRNA cleavage site from Arabidopsis AP2, ARF16, SCL6, SPL2, TAS1c, and TAS3a transcripts was done with the following oligo pairs: AP2-CS-F/AP2-CS-R, ARF16-CS-F/ARF16-CS-R, SCL6-CS-F/SCL6-CS-R, TAS1c-CS-F/TAS1c-CS-R, and TAS3a-CS-F/TAS3a-CS-R, respectively. Oligo pair ACT-benth-F/ACT-benth-R was used to amplify a fragment of N. benthamiana actin transcript, and oligo pairs ACT2-F/ACT2-R and TUB8-F/TUB8-R were used to amplify a fragment of Arabidopsis ACT2 and TUB8 transcripts, respectively. The sequences of all these oligos are listed in Supplemental Table 2 online.
RNA Immunoprecipitation for High-Throughput Sequencing Analysis
RNA immunoprecipitated from 2 g of inflorescence tissue from transgenic T4 Arabidopsis plants (4 to 6 weeks old, flower stages 1 to 12) was used to generate either small RNA or transcript libraries. The immunoprecipitation protocol was essentially as described above but with the following modifications. Clarified lysates were filtered with sterile Miracloth paper (Calbiochem) and incubated with 16 μg/mL of 12CA5 anti-HA antibody (Roche) for 30 min at 4°C; after six washes, beads were incubated at 22°C for 30 min in a Thermomixer R (Eppendorf) with intermittent shaking (1200 rpm for 15 s every 2 min), in an equal volume of Nuclease P1 Digestion Buffer (lysis buffer supplemented with 0.125 mM ZnCl2) including 0.5 ng/μL of nuclease P1 (Sigma-Aldrich). Beads were pelleted by centrifugation for 1 min at 756 rcf at 4°C and the supernatant removed. Beads were then washed two times at 4°C for 2 min in P1 Digestion STOP buffer (lysis buffer supplemented with 50 mM EDTA, pH 8.0). Finally, beads were washed once at 4°C for 2 min in lysis buffer prior to proteinase K treatment.
Preparation of Small RNA Libraries
Ten microliters (or 100%) of the immunoprecipitated RNA was mixed with 15 pmol of Cloning Linker 1 (IDT; see Supplemental Table 2 online) and incubated at 70°C for 2 min. The mixture was cooled on ice for 2 min before adding 1.5 μL of 10× T4 RNA ligase reaction buffer (NEB), 40 units of RNase OUT), and 10 units of T4 RNA Ligase 2 truncated K227Q (NEB). Ligation reactions were incubated overnight at 16°C. Digestion of unligated Cloning Linker 1 was done by first adding 20 units of deadenylase (NEB) to the overnight ligation and incubating at 30°C for 15 min, then adding 10 units of Exonuclease VII (Affymetrix) and incubating at 37°C for 15 min. Next, 15 pmol of RNA 5′ adapter (see Supplemental Table 2 online) was denatured at 70°C for 2 min followed by transfer to ice for at least 2 min. Denatured RNA 5′ adapter was added to the ligation mixture with 0.48 mM of ATP and five units of T4 RNA Ligase 1 (Ambion), and the resulting mixture was incubated at 28°C for 1 h, then placed on ice. For cDNA synthesis, 5 µM of RT primer (see Supplemental Table 2 online) was used with the SuperScript III first-strand synthesis system (Invitrogen). After cooling the cDNA synthesis reactions on ice, two units of E. coli RNase H (Invitrogen) were added, and the reaction was incubated at 37°C for 20 min. Linear PCR reactions were done with half of the cDNA and in the presence of 1× Phusion HF buffer, 0.25 µM of P5 primer (see Supplemental Table 2 online), 0.25 µM of P7-modban (see Supplemental Table 2 online), 0.1 mM deoxynucleotide triphosphate mix, and 1 unit of Phusion Polymerase (Thermo Scientific). The following PCR conditions were used: 98°C for 30 s, 17 cycles of 98°C for 10 s, 60°C for 30 s, 72°C for 15 s, and 72°C for 5 min. The Qiagen PCR purification kit was used to clean up the PCR products before loading samples onto a 6% native polyacrylamide gel. The desired amplicons (between ∼100 and 108 bp) were recovered from the gel using electrotransfer onto DE81 paper as previously described (Fahlgren et al., 2009). DNA concentrations were quantified using the Qubit HS assay kit (Invitrogen) and submitted for sequencing on a HiSeq 2000 sequencer (Illumina).
miRNA Sequencing Analysis
Sequencing reads from small RNA libraries (immunoprecipitated from plants containing AGO1-DDH, AGO1-DAH, or empty vector) were computationally processed to remove 3′ adaptor sequences, and sequences were consolidated to generate read counts per unique small RNA. Unique small RNAs were mapped to the Arabidopsis genome (TAIR10) using Bowtie v0.12.8 (Langmead et al., 2009) with settings allowing for perfect matches only (bowtie –f –v 0 –a –S –p 10). Libraries were normalized for sequencing depth differences by randomly sampling genome-mapped small RNA read counts from each library to match the proportion of mapped reads from the smallest library, relative to the larger libraries, resulting in ∼20 million reads per sample (see Supplemental Data Set 1A online). Reads per miRNA and miRNA* were determined for each sample using a list of annotated mature Arabidopsis miRNA and miRNA* sequences (see Supplemental Data Set 1B online). Immunoprecipitate enrichment was calculated for each mature miRNA as log2(IP reads + 1/vector reads+ 1) for AGO1-DDH and AGO1-DAH samples. A Venn diagram was generated for mature miRNA with 10 or more reads in at least one library (immunoprecipitates of AGO1-DDH, AGO1-DAH, and empty vector) that were enriched fourfold or greater in one or both AGO1-DDH and AGO1-DAH samples, relative to the empty vector sample, using the Venn Diagram Generator (http://jura.wi.mit.edu/bioc/tools/venn.php; Figure 6B; see Supplemental Data Set 1B online).
Strand-Specific Target Transcript Library Preparation
Ten micrograms of total RNA or 10 μL (100%) of the immunoprecipitated RNA was treated with TURBO DNAase I DNA-free (Ambion). Samples were depleted of rRNAs by treatment with Ribominus plant kit for RNA-Seq (Invitrogen) according to manufacturer’s instructions. cDNA synthesis and strand-specific target transcript libraries from both input RNA and immunoprecipitated RNA were made as described (Wang et al., 2011a), with the following modifications. Immunoprecipitated RNAs were not fragmented with metal ions prior to library construction, and 16 cycles were used in the linear PCR reaction. DNA adaptors 1 and 2 were annealed to generate the Y-shape adaptors, and oligonucleotides PE-F and PE-R were used in the linear PCR (see Supplemental Table 2 online). DNA amplicons were analyzed with a Bioanalyzer (DNA HS kit; Agilent), quantified using the Qubit HS Assay Kit (Invitrogen), and sequenced on a HiSeq 2000 sequencer (Illumina).
Transcript Sequencing Analysis
Sequencing reads from transcript libraries (input or immunoprecipitate samples containing AGO1-DDH or AGO1-DAH, two replicates each) were consolidated to generate read counts per unique sequence, and unique sequences were mapped to Arabidopsis transcripts (TAIR10) using Bowtie (Langmead et al., 2009) with the settings described above. Aligned sequences were filtered to remove reads that mapped to rRNA, tRNA, small nuclear RNA, small nucleolar RNA, and transposable elements and to remove reads that mapped to transcripts from multiple genes or were derived from the transcript antisense strand. Libraries were normalized for sequencing depth differences by randomly sampling as described above, resulting in ∼1 million reads per sample (see Supplemental Data Set 1A online). Known miRNA target site regions on Arabidopsis transcripts were used for the analyses presented, excluding TAS3 family transcripts, which are targeted by AGO7-miR390 complexes (Montgomery et al., 2008a) (see Supplemental Data Set 1C online). Transcript reads were averaged between replicates, and reads that mapped within 200 nucleotides upstream or downstream of known target sites were used for analysis (see Supplemental Data Set 1D online). Target sites were represented as a single nucleotide from the center of the target site. Immunoprecipitate enrichment was calculated for each target site as log2(IP average reads + 1/input average reads + 1) for AGO1-DDH and AGO1-DAH samples. A Venn diagram was generated, as above, for target sites with four or more average reads in at least one library (inputs or immunoprecipitates of AGO1-DDH and AGO1-DAH) that were enriched fourfold or greater in one or both AGO1-DDH and AGO1-DAH samples, relative to input samples (Figure 6D; see Supplemental Data Set 1D online). The distribution of reads across target site regions was plotted by the scrolling window method (20-nucleotide windows, five-nucleotide scroll). Windows were positioned relative to the miRNA target site, and windows at the same relative distance were averaged across all target site regions analyzed (Figure 6E, top). For comparison to non-AGO1 target transcripts, sets of randomly selected transcript regions were generated. Each selected set contained 260 transcripts that were sampled such that the transcript expression distribution approximately matched the expression range of known miRNA target transcripts. Scrolling-window read counts from randomly selected sites for each random transcript set were calculated as for authentic target regions and were averaged across 100 transcript sets (Figure 6E, bottom).
Accession Number
Sequence data from this article can be found in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE40259.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. AGO1, AGO2, AGO7, and AGO10 Wild-Type and Mutant Constructs.
Supplemental Figure 2. Accumulation of HA-AGO2 Forms in Arabidopsis Col-0 and ago2-1 T3 Transgenic Lines Expressing Different AGO2 Forms.
Supplemental Figure 3. Phenotypic and Molecular Analyses of Arabidopsis Col-0 Transgenic Plants Expressing Wild-Type or Active-Site Substitution AGO1 and AGO7 Forms, Respectively.
Supplemental Figure 4. Phenotypic Analyses of Arabidopsis zip-1 and Col-0 T1 Transgenic Plants Expressing Wild-Type or Modified AGO7 Forms.
Supplemental Figure 5. AGO1 and AGO10 Interactions with Small RNAs and Target Transcripts in N. benthamiana Transient Assays Overexpressing Wild-Type or Modified AGO Forms.
Supplemental Figure 6. AGO1 Interactions with Small RNA and Target Transcripts in N. benthamiana Transient Assays Overexpressing AGO1 Wild-Type, Double, or Triple Active-Site Substitution Forms Together with miR173 and TAS1c.
Supplemental Table 1. Phenotypic Analyses of Arabidopsis ago1-25 and Col-0 Transgenic Plants Expressing Wild-Type and Modified AGO1 Constructs.
Supplemental Table 2. Oligonucleotides Used.
Supplemental Table 3. Probes for Small RNA Gel Blot.
Supplemental Data Set 1A. Summary of High-Throughput RNA Sequencing Libraries.
Supplemental Data Set 1B. miRNA Sequencing Data from AGO1-DDH and AGO1-DAH Immunoprecipitates.
Supplemental Data Set 1C. AGO1-miRNA Target Sites.
Supplemental Data Set 1D. miRNA Target Region Sequencing Data from AGO1-DDH and AGO1-DAH Immunoprecipitate and Input Samples.
Acknowledgments
We thank Goretti Nguyen for excellent technical assistance. A.C. was supported by a postdoctoral fellowship from the Ministerio de Ciencia e Innovacion (BMC-2008-0188). H.G.-R. was the recipient of a Helen Hay Whitney Postdoctoral fellowship (F-972). This work was supported by grants from the National Science Foundation (MCB-1231726), the National Institutes of Health (AI043288), and Monsanto Corporation.
AUTHOR CONTRIBUTIONS
J.C.C. and A.C. conceived the project. A.C. designed research. A.C., H.G.-R, K.B.G, T.A.M., T.N., and J.T.C. performed research. A.C., N.F., K.B.G., and J.C.C. analyzed the data. A.C., N.F., and J.C.C. wrote the article.
Glossary
- miRNA
microRNA
- tasiRNA
trans-acting small interfering RNA
- siRNA
small interfering RNA
- HA
hemagglutinin
- 5′RACE
5′ rapid amplification of cDNA ends
- GFP
green fluorescent protein
- TuMV
Turnip mosaic virus
- Col-0
Columbia-0
- CP
coat protein
References
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. (2008). The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Chekanova J.A., et al. (2007). Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131: 1340–1353 [DOI] [PubMed] [Google Scholar]
- Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi S.W., Zang J.B., Mele A., Darnell R.B. (2009). Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Himber C., Voinnet O. (2005). DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 37: 1356–1360 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. (2008). Getting to the root of miRNA-mediated gene silencing. Cell 132: 9–14 [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Sullivan C.M., Kasschau K.D., Chapman E.J., Cumbie J.S., Montgomery T.A., Gilbert S.D., Dasenko M., Backman T.W., Givan S.A., Carrington J.C. (2009). Computational and analytical framework for small RNA profiling by high-throughput sequencing. RNA 15: 992–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Gandikota M., Birkenbihl R.P., Höhmann S., Cardon G.H., Saedler H., Huijser P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49: 683–693 [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz H., Takeda A., Chapman E.J., Sullivan C.M., Fahlgren N., Brempelis K.J., Carrington J.C. (2010). Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell 22: 481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey J.J., Lewsey M.G., Patel K., Westwood J., Heimstädt S., Carr J.P., Baulcombe D.C. (2011). An antiviral defense role of AGO2 in plants. PLoS ONE 6: e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker E.R., Wallbridge L.M., Hardcastle T.J., Bush M.S., Kelly K.A., Dunn R.M., Schwach F., Doonan J.H., Baulcombe D.C. (2010). The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson D.G., Hogan D.J., McCullough H.L., Myers J.W., Herschlag D., Ferrell J.E., Brown P.O. (2009). Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 7: e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C., Sun H., Poethig R.S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13: 1734–1739 [DOI] [PubMed] [Google Scholar]
- Huntzinger E., Izaurralde E. (2011). Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Nishikiori M., Jaudal M.C., Matsumoto-Yokoyama E., Mitsuhara I., Meshi T., Ishikawa M. (2010). In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell 39: 282–291 [DOI] [PubMed] [Google Scholar]
- Jaubert M.J., Bhattacharjee S., Mello A.F., Perry K.L., Moffett P. (2011). ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 156: 1556–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L., et al. (2011). ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 7: e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Han J., Siomi M.C. (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Lanet E., Delannoy E., Sormani R., Floris M., Brodersen P., Crété P., Voinnet O., Robaglia C. (2009). Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., Young A.G., Bhutkar A., Zheng G.X., Bosson A.D., Nielsen C.B., Sharp P.A. (2011). Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat. Struct. Mol. Biol. 18: 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Lobbes D., Rallapalli G., Schmidt D.D., Martin C., Clarke J. (2006). SERRATE: A new player on the plant microRNA scene. EMBO Rep. 7: 1052–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A., Vaucheret H. (2010). Form, function, and regulation of ARGONAUTE proteins. Plant Cell 22: 3879–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P.A., Koenig D., Weigel D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 109: 2461–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D. (2005). Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620 [DOI] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008a). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Montgomery T.A., Yoo S.J., Fahlgren N., Gilbert S.D., Howell M.D., Sullivan C.M., Alexander A., Nguyen G., Allen E., Ahn J.H., Carrington J.C. (2008b). AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl. Acad. Sci. USA 105: 20055–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel J.B., Godon C., Mourrain P., Béclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., He X., Wang X.J., Kohany O., Jurka J., Hannon G.J. (2006). Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof H.B., Alvarado V.Y., Vega-Arreguin J.C., Ciomperlik J., Odokonyero D., Brosseau C., Jaubert M., Zamora A., Moffett P. (2011). Identification of an ARGONAUTE for antiviral RNA silencing in Nicotiana benthamiana. Plant Physiol. 156: 1548–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Souret F.F., Kastenmayer J.P., Green P.J. (2004). AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15: 173–183 [DOI] [PubMed] [Google Scholar]
- Wang L., Si Y., Dedow L.K., Shao Y., Liu P., Brutnell T.P. (2011a). A low-cost library construction protocol and data analysis pipeline for Illumina-based strand-specific multiplex RNA-seq. PLoS ONE 6: e26426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.B., Jovel J., Udomporn P., Wang Y., Wu Q., Li W.X., Gasciolli V., Vaucheret H., Ding S.W. (2011b). The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23: 1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J. (2009). Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461: 754–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Belasco J.G. (2008). Let me count the ways: Mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell 29: 1–7 [DOI] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wu G., Poethig R.S. (2012). Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhao H., Gao S., Wang W.C., Katiyar-Agarwal S., Huang H.D., Raikhel N., Jin H. (2011). Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(∗)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol. Cell 42: 356–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Hu F., Wang R., Zhou X., Sze S.H., Liou L.W., Barefoot A., Dickman M., Zhang X. (2011). Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis D.G., Lovci M.T., Wilbert M.L., Hutt K.R., Liang T.Y., Pasquinelli A.E., Yeo G.W. (2010). Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 17: 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]