Plant cells communicate by movement of signaling agents through cytoplasmic bridges, termed plasmodesmata (PD). In this study, we characterize two members of the Germin-like protein family that are located within PD. PDGLP1/2 overexpression phenotypes had a reduction in primary root meristem size, likely due to their inability to form functional PD complexes.
Abstract
In plants, a population of non-cell-autonomous proteins (NCAPs), including numerous transcription factors, move cell to cell through plasmodesmata (PD). In many cases, the intercellular trafficking of these NCAPs is regulated by their interaction with specific PD components. To gain further insight into the functions of this NCAP pathway, coimmunoprecipitation experiments were performed on a tobacco (Nicotiana tabacum) plasmodesmal-enriched cell wall protein preparation using as bait the NCAP, pumpkin (Cucurbita maxima) PHLOEM PROTEIN16 (Cm-PP16). A Cm-PP16 interaction partner, Nt-PLASMODESMAL GERMIN-LIKE PROTEIN1 (Nt-PDGLP1) was identified and shown to be a PD-located component. Arabidopsis thaliana putative orthologs, PDGLP1 and PDGLP2, were identified; expression studies indicated that, postgermination, these proteins were preferentially expressed in the root system. The PDGLP1 signal peptide was shown to function in localization to the PD by a novel mechanism involving the endoplasmic reticulum-Golgi secretory pathway. Overexpression of various tagged versions altered root meristem function, leading to reduced primary root but enhanced lateral root growth. This effect on root growth was corrected with an inability of these chimeric proteins to form stable PD-localized complexes. PDGLP1 and PDGLP2 appear to be involved in regulating primary root growth by controlling phloem-mediated allocation of resources between the primary and lateral root meristems.
INTRODUCTION
In plants, intercellular communication is achieved by means of non-cell-autonomous signaling molecules that move either across the intervening cell wall or through the specialized plasma membrane–lined cylindrical cytoplasmic channels, termed plasmodesmata (PD) (Robards and Lucas, 1990; Lucas and Lee, 2004; Maule, 2008; Lucas et al., 2009; Xu and Jackson, 2010). Trafficking through PD can occur through microchannels formed by proteins embedded in both the plasma membrane and a centrally located appressed form of the endoplasmic reticulum (ER) that establishes an endomembrane continuum between neighboring plant cells. This PD-established cytoplasmic pathway also allows for the supply of metabolites and mineral nutrients required for heterotrophic growth. In addition, PD mediate the cell-to-cell trafficking of information macromolecules, including proteins (Lucas et al., 1995; Helariutta et al., 2000; Sessions et al., 2000; Kurata et al., 2005a) and various forms of RNA (Jorgensen et al., 1998; Xoconostle-Cázares et al., 1999; Kim et al., 2001; Haywood et al., 2005; Banerjee et al., 2006; Wright et al., 2007; Martin et al., 2009).
The maize (Zea mays) transcription factor KNOTTED1 (KN1) (Vollbrecht et al., 1991) that functions in the meristem to regulate cell fate was the first plant protein identified as having the capacity to traffic through PD (Lucas et al., 1995). Subsequently, numerous non-cell-autonomous plant transcription factors have been characterized (Kurata et al., 2005b; Wu and Gallagher, 2011). Interestingly, KN1 can also bind and transport its own mRNA through PD (Lucas et al., 1995; Kim et al., 2005), and, consistent with discoveries based on viral movement proteins (MPs) facilitating the trafficking of their infectious RNA/DNA (Lucas, 2006), an ever increasing population of RNA molecules has been shown to move locally and over long distances (Kehr and Buhtz, 2008). Movement of the gene silencing signal, presumably in the form of a small RNA, also occurs through PD (Yoo et al., 2004; Dunoyer et al., 2005; Kurata et al., 2005b; Carlsbecker et al., 2010; Chitwood and Timmermans, 2010; Molnar et al., 2010).
The capacity of a protein for cell-to-cell movement has been shown to depend upon a number of factors. Some non-cell-autonomous proteins (NCAPs) can interact with components of the PD to induce a significant increase in the microchannel size exclusion limit (SEL) from a value of ∼800 to 1200 D to values on the order of 15 to 40 kD, depending on the specific NCAP (Wolf et al., 1989; Noueiry et al., 1994; Lucas et al., 1995; Xoconostle-Cázares et al., 1999; Taoka et al., 2007). In this dilated state, NCAPs, like KN1, appear to undergo partial unfolding to pass through the PD microchannels (Kragler et al., 1998; Xu et al., 2011). Motifs on some NCAPs that are required for this increase in PD SEL have been identified, and engineered cell-autonomous proteins carrying such motifs are able to move cell to cell (Ishiwatari et al., 1998; Aoki et al., 2002; Kim et al., 2005; Taoka et al., 2007). Thus, for this class of NCAPs, molecular size/shape does not limit their ability to move into neighboring cells. For other NCAPs that lack this capacity for PD microchannel dilation, movement appears to be regulated by their basic size/shape (Oparka et al., 1999; Crawford and Zambryski, 2000).
The intercellular trafficking of both endogenous NCAPs and viral MPs can be further regulated by posttranslational modifications, such as phosphorylation and glycosylation. For example, mutations of specific C-terminal phospho-residues on the Tobacco mosaic virus (TMV) MP can block both cell-to-cell and long-distance viral movement (Waigmann et al., 2000). Another example is provided by studies on the pumpkin (Cucurbita maxima) phloem NCAP, PHLOEM PROTEIN16 (Cm-PP16), an endogenous equivalent to viral MPs (Xoconostle-Cázares et al., 1999). In this case, Cm-PP16 must be phosphorylated and glycosylated to interact and bind with the NCAPP1 receptor (Lee et al., 2003) for delivery to the PD orifice (Taoka et al., 2007). Interestingly, the NCAPP1 receptor must similarly be both phosphorylated and glycosylated for effective binding to Cm-PP16.
A comprehensive understanding of the role played by this PD-facilitated NCAP pathway, in contributing to the control of developmental and physiological processes, requires isolation and characterization of the proteins, within the PD, that participate in these trafficking steps. Progress toward this goal has been recently accomplished through a combination of biochemical methods to enrich for putative PD proteins and advanced proteomics methods (Lee et al., 2003, 2005; Sagi et al., 2005; Taoka et al., 2007; Fernandez-Calvino et al., 2011; Jo et al., 2011). Subsequent studies on a few select candidate proteins, contained within these PD proteomes, have offered insight into mechanisms used for targeting to PD (Thomas et al., 2008; Simpson et al., 2009). However, in general, the functions of the majority of these proteins remain to be elucidated.
In this study, we used the phloem NCAP Cm-PP16 as bait in coimmunoprecipitation (co-IP) experiments performed on a tobacco (Nicotiana tabacum) plasmodesmal-enriched cell wall protein (PECP) preparation to identify PD-located proteins that are specifically involved in NCAP trafficking. These co-IP studies yielded six putative tobacco PD proteins, and, in this study, we characterize two Arabidopsis thaliana orthologs, PLASMODESMAL GERMIN-LIKE PROTEIN 1 (PDGLP1) and PDGLP2.
RESULTS
Identification of PDGLP1 and PDGLP2
Our Cm PP16-PECP co-IP experiments identified six interacting proteins (see Supplemental Table 1 online), including a tobacco GERMIN-LIKE PROTEIN (GLP), as candidate PD proteins. Transgenic tobacco leaves expressing either TMV MP-GFP (for green fluorescent protein) or Cucumber mosaic virus (CMV) MP-GFP, both established PD markers (Blackman et al., 1998; Waigmann et al., 2004), were used for Nt GLP-RFP (for red fluorescent protein) transient expression studies to test for protein subcellular localization. Colocalization between Nt GLP-RFP, TMV MP-GFP, and CMV MP-GFP (see Supplemental Figure 1A online) provided support for the hypothesis that Nt GLP-RFP is localized to PD; thus, this member of the GLP family was designated as Nt-PDGLP1. A bioinformatics analysis of the Nt-PDGLP1 predicted protein sequence identified an N-terminal potential signal peptide consistent with this protein being delivered to the plasma membrane through the secretory pathway (see Supplemental Figure 1B online).
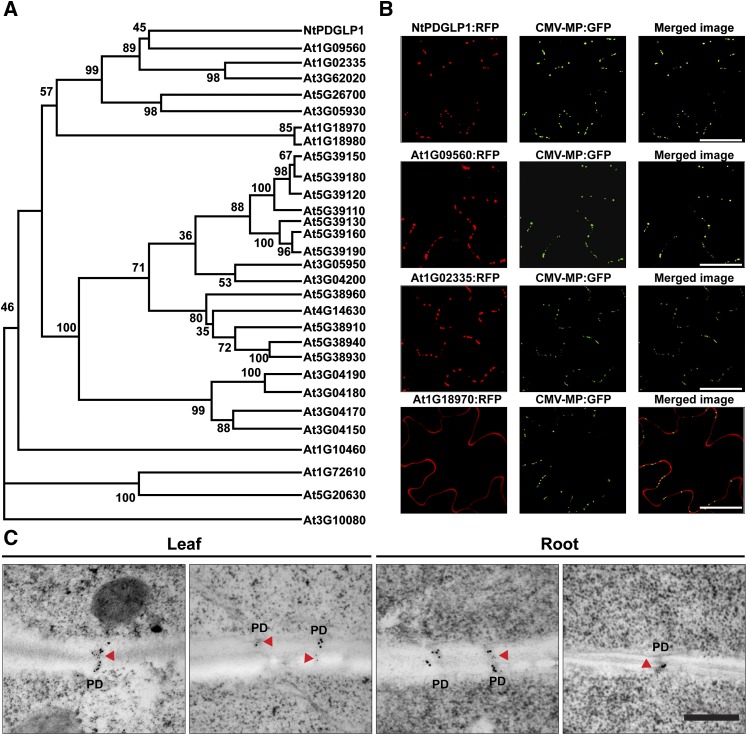
To facilitate our studies on the PDGLP family members, we next performed a molecular phylogenetic analysis to identify potential Arabidopsis orthologs. Here, we found that Nt-PDGLP1 was located in a specific clade of the Arabidopsis GLP family (Figure 1A; see Supplemental Data Set 1 online). Of the five Arabidopsis GLP family members in this clade, only two (At1G09560 and At1G02335) exhibited a punctuate pattern equivalent to that obtained with Nt-PDGLP1 (see Supplemental Figure 1C online) and were colocalized with the PD marker CMV MP-GFP (Figure 1B); consequently, these two GLP members were designated as PDGLP1 and PDGLP2, respectively. Plasmolysis experiments performed on PDGLP1-GFP transgenic plants confirmed that the GFP signal was retained at the cell wall; furthermore, immunogold labeling studies, using antibodies directed against callose and GFP, confirmed that PDGLP1-GFP was located within PD (Figure 1C; see Supplemental Figure 1D online).
Figure 1.
Arabidopsis PDGLP1 and PDGLP2 Are Plasmodesmal Targeted Proteins.
(A) Phylogenetic analysis of the relationship between the Arabidopsis GLP family and the tobacco PDGLP1 identified in our tobacco PECP co-IP studies using the pumpkin phloem Cm-PP16 as bait. The phylogenetic tree was constructed in MEGA 3.1; numbers at each branch point represent the bootstrap values for percentage of 1000 replicate trees.
(B) PDGLP1 (At1G09560) and PDGLP2 (At1G02335) are colocalized with the PD marker CMV MP-GFP. The C-terminal mCherry (RFP) tagged At1G09560, At1G02335, and At1G18970 (GLP4) were agroinfiltrated into leaves of CMV MP-GFP transgenic N. benthamiana plants. Yellow signal in merged images represents colocalization of At1G09560 and At1G02335 with CMV MP-GFP. Images were taken by CLSM. Bars = 10 µm.
(C) PDGLP1-GFP is located within PD. Leaf and root tissues of PDGLP1-GFP transgenic plants were processed for transmission electron microscopy–based double immunogold labeling with anticallose Ab (15-nm gold particles) and anti-GFP Ab (10-nm gold particles). Red arrowheads indicate detection of PDGLP1-GFP within the PD. Bar = 200 nm.
[See online article for color version of this figure.]
PDGLP1 and PDGLP2 Are Expressed Predominantly in Roots
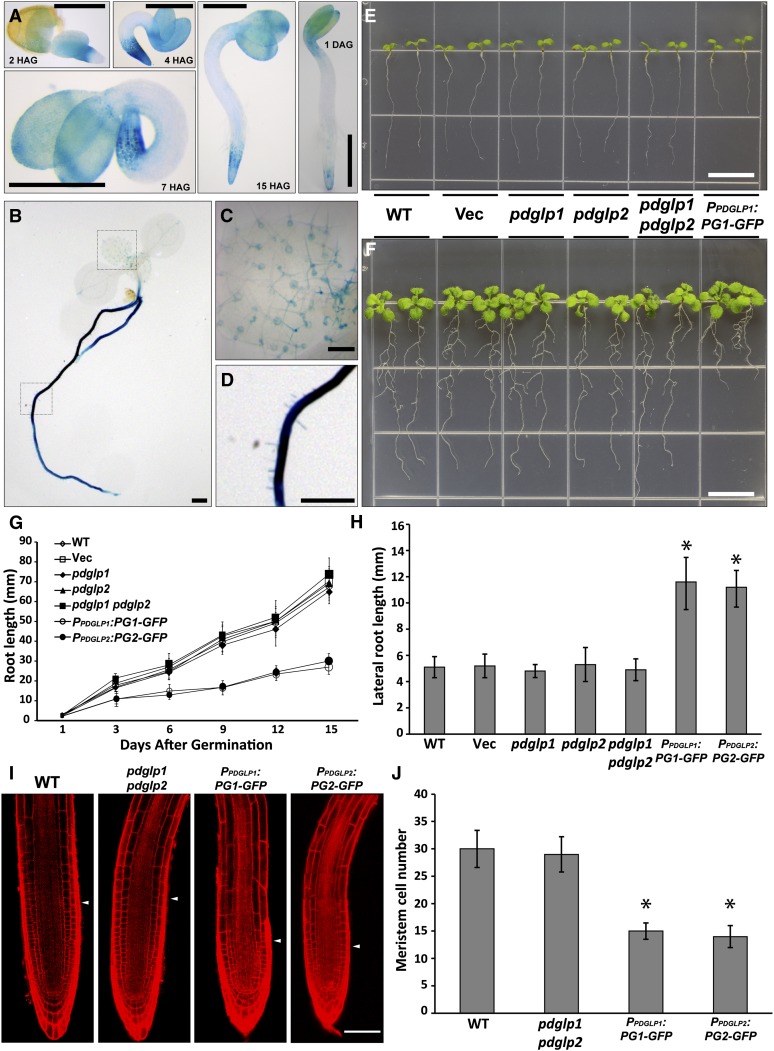
The expression patterns for PDGLP1 and PDGLP2 were characterized using transgenic Arabidopsis plants expressing β-glucuronidase (GUS) under the control of the endogenous PDGLP1 or PDGLP2 promoter. GUS staining revealed that both genes are expressed during germination and early seedling development, with PDGLP1 being strongest in the shoot and root apices (Figure 2A) and PDGLP2 being expressed along the seedling axis (see Supplemental Figure 2 online). Seven days after germination (DAG), PDGLP1 expression was strongest in the root (Figure 2B), and signal in the shoot appeared to be confined to trichome basal cells of new developing leaves (Figure 2C). For PDGLP2 at 3 and 7 DAG, expression was detected in leaf and root vascular tissue; this pattern continued into the mature plant phase (see Supplemental Figure 2 online). These studies established that, past the early germination stage, both PDGLP1 and PDGLP2 are primarily expressed in the Arabidopsis root system.
Figure 2.
PDGLP1 Expression Profile and Phenotypic Analysis of Mutant and Transgenic PDGLP1-GFP Plant Lines.
(A) Histochemical staining of PPDGLP1:GUS transgenic plants reveals PDGLP1 expression pattern in cotyledons and the root tip during early seedling development. HAG, hours after germination. Bar = 1 mm.
(B) and (C) GUS staining performed with 7-d-old transgenic PPDGLP1:GUS plants indicates the presence of strong signal in the root system (B), whereas in the first true leaves, GUS staining was restricted to basal trichome cells (C) (area shown is enlarged from the dashed square indicted in [B]).
(D) Strong GUS staining in the primary root (area shown is enlarged from the dashed square indicted in [B]).
(E) and (F) Phenotype of wild-type (WT), empty vector–transformed transgenic plants (Vec), pdglp1, pdglp2, pdglp1 pdglp2 double mutant, and PPDGLP1:PDGLP1-GFP (PG1-GFP) seedlings at 5 (E) and 10 (F) DAG. Note the short root phenotype for the PPDGLP1:PG1-GFP seedlings. Bars = 10 mm.
(G) Average root length of wild-type (open diamonds), Vec (open squares), pdglp1 (closed diamonds), pdglp2 (triangles), pdglp1 pdglp2 (squares), PPDGLP1:PG1-GFP (open circles), and PPDGLP2:PG2-GFP (closed circles) seedlings measured at the indicated DAG. Values represent mean ± sd; n = 40 seedlings for each plant line tested.
(H) Transgenic PG1-GFP and PG2-GFP plants displayed enhanced lateral root length compared with wild-type, vec, pdglp1, pdglp2, and pdglp1/pdglp2 seedlings. Asterisk indicates significant differences, P < 0.002, based on Student’s t test.
(I) Five-day-old primary roots from wild-type, pdglp1, pdglp2, pdglp1/2, PG1-GFP, and PG2-GFP seedlings. Arrowheads indicate the boundary between the meristem and elongation zones. Images collected by confocal microscopy of propidium iodine–stained roots. Bar = 50 µm.
(J) Analysis of meristem size in the indicated plant lines. Confocal images were analyzed to determine the cortical cell number in the meristem zone of each plant line. Values represent mean ± sd; n = 50 roots. Asterisk indicates significant differences, P < 0.002, based on Student’s t test.
[See online article for color version of this figure.]
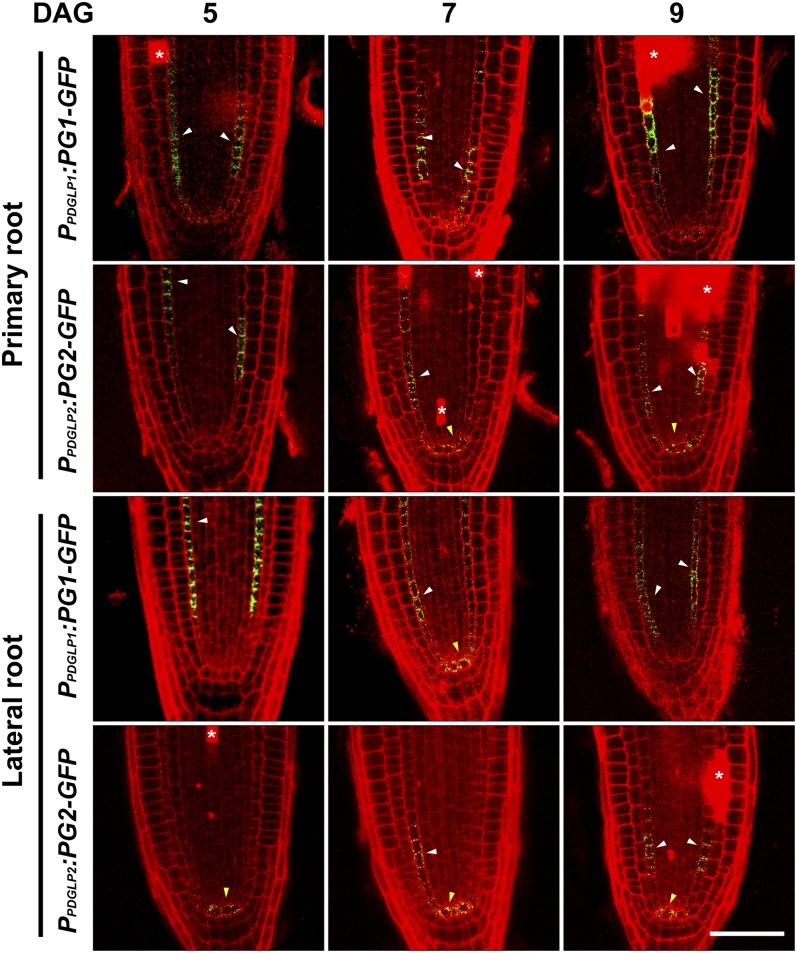
Confocal analysis of the root meristem zones of PPDGLP1:PDGLP1-GFP and PPDGLP2:PDGLP2-GFP transgenic plants was performed to ascertain the cellular location of PDGLP1-GFP and PDGLP2-GFP. In the primary root, GFP signal was detected in the endodermal cells for both proteins (Figure 3). Weak signal was also detected for these two proteins in the quiescent center of the primary root. Analyses performed on lateral roots of the same seedlings used for our primary root studies indicated similar patterns, but here a stronger GFP signal was detected in the quiescent center (Figure 3).
Figure 3.
Both PDGLP1-GFP and PDGLP2-GFP Are Localized to the Endodermis and Quiescent Center in Both Primary and Lateral Root Meristems.
Confocal microscopy was used to map the cellular domains occupied by PDGLP1-GFP and PDGLP2-GFP in primary and lateral root tips at the indicated DAG. Darts indicate GFP signal in endodermal (white) and quiescent center (yellow) cells. Asterisks indicate propidium iodide–stained dead/dying cells. Bar = 25 µm.
[See online article for color version of this figure.]
The role of PDGLP1 and PDGLP2 in plant growth and development was next analyzed using a combination of pdglp1, pdglp2, and double pdglp1 pdglp2 mutants (see Supplemental Figure 3 online). Neither the pdglp1 or pdglp2 mutants nor the pdglp1 pdglp2 double mutant showed any visible phenotype when compared with wild-type and vector control plants (Figures 2E and 2F). However, transgenic plants expressing PDGLP1-GFP driven by the native promoter, an overexpression (OX) condition, had shortened primary roots, compared with wild-type and vector control plants (Figures 2E to 2G). Interestingly, similar root phenotypes were observed with transgenic plants expressing PDGLP1-4xMyc (Myc) or PDGLP2-Myc driven by the cauliflower mosaic virus 35S promoter (see Supplemental Figure 4A online). This root phenotype was also detected in transgenic plants expressing untagged versions of PDGLP1/2 driven by the 35S promoter (see Supplemental Figure 4B online). In addition, plants expressing either PDGLP2-GFP, PDGLP1-GUS, or PDGLP2-GUS driven by their respective native promoters similarly displayed this shortened primary root phenotype (see Supplemental Figures 5A to 5C online).
The shortened primary roots in all of these various PDGLP1 and PDGLP2 OX plants appeared to be compensated for by an increase in total lateral root length (Figure 2H; see Supplemental Figure 5D online), without any significant effect on the actual number of lateral roots (see Supplemental Figure 5E online). An important additional observation was that the root dry weight for all of these various PDGLP1/2 OX plant lines did not differ significantly from wild-type or vector control plants (see Supplemental Figure 6 online). This finding is of significance, as it suggests that total resource allocation to the root system of these plants was unchanged, but that either higher than normal levels of either PDGLP1 or PDGLP2 or protein dysfunction associated with C-terminally fused GFP, GUS, or Myc can lead to an inhibition in primary over lateral root growth.
An examination of the cellular morphology of the apical regions of primary roots in wild-type and pdglp1/2 seedlings revealed no discernible differences in the size of their root meristematic zones (Figures 2I and 2J). By contrast, primary roots of PDGLP1-GFP and PDGLP2-GFP plants had reduced root meristematic zones (Figures 2I and 2J). In addition, propidium iodide staining indicated the presence of dead cells scattered throughout the meristematic region of these roots (Figure 3; see Supplemental Figure 7 online). Importantly, a similar phenotype was also observed in the primary roots of our transgenic PDGLP1-Myc, PDGLP2-Myc (see Supplemental Figure 4C online), PDGLP1-GUS, and PDGLP2-GUS (see Supplemental Figure 5F online) plants.
Analysis of the primary roots from 1 to 12 DAG indicated that, in these various transgenic plant lines, by 4 DAG cellular organization was perturbed (see Supplemental Figure 7 online) and then this state progressed, giving rise to an increasing number of dead cells. Inspection of the lateral roots in these same transgenic plant lines indicated a milder phenotype, especially in terms of the presence of dead cells (see Supplemental Figures 4C and 5G online). Taken together, these data suggest that PDGLP1 and PDGLP2 are involved in root growth and development and that either elevated protein levels or proteins rendered dysfunctional through C-terminal fusion of a Myc/GFP/GUS tag alter PD function to perturb root cell development and eventual viability.
PDGLP1 Signal Peptide Functions in Protein Delivery to PD
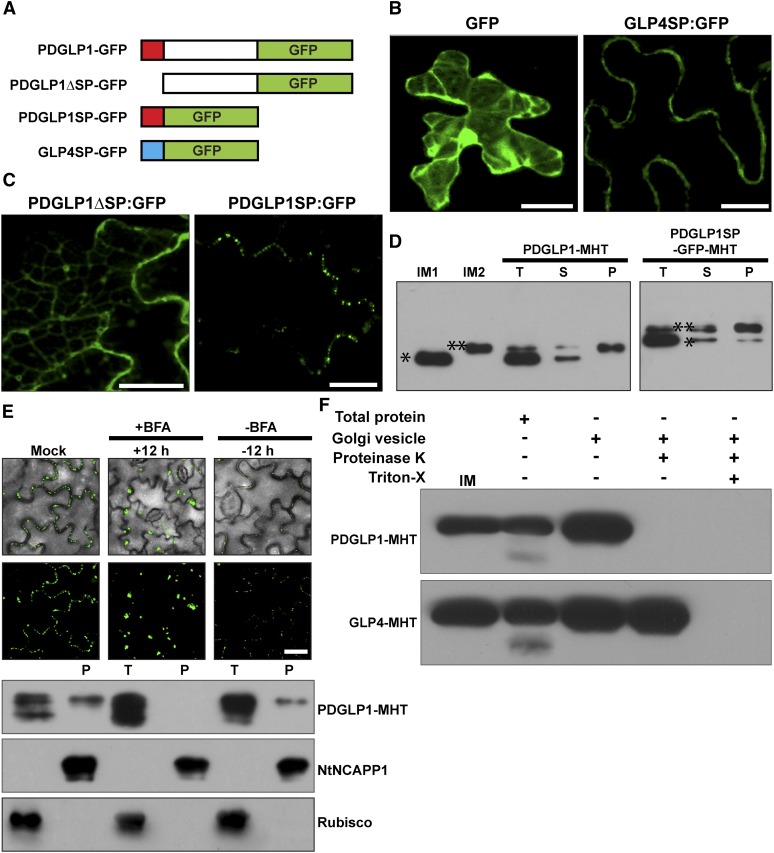
To gain insight into the underlying mechanism responsible for the observed PDGLP1/2 overexpression phenotype, we next performed cell biology studies to examine the role of PDGLP1 and PDGLP2 in NCAP signaling. First, we investigated the targeting mechanism involved in delivery of PDGLP1 to the PD. To this end, a series of mutants was generated in which the PDGLP1 signal peptide (SP) was deleted (PDGLP1ΔSP-GFP) or fused directly to GFP (PDGLP1SP-GFP) (Figure 4A). As controls for these studies, we used GFP alone and GFP fused to the SP of Arabidopsis GLP4 (At1G18970), a non-PD-targeted GLP (Figures 1B and 4A). As expected, GFP fluorescence was detected within the cytoplasm, whereas GLP4SP-GFP was targeted to the cell periphery (Figure 4B). Importantly, PDGLP1ΔSP-GFP failed to accumulate in the PD, but PDGLP1SP-GFP was efficiently targeted to PD (Figure 4C).
Figure 4.
The PDGLP1 SP Is Necessary and Sufficient for Protein Delivery to PD via the ER-Golgi Secretory Pathway.
(A) Arabidopsis PDGLP1 and GLP4 constructs used to test the role of the SP in protein targeting to PD. Putative N-terminal SPs shown as a red (PDGLP1) and blue (GLP4) box. GFP was fused to the C terminus of PDGLP1, a SP-deleted PDGLP1 mutant (PDGLP1ΔSP-GFP), and alone with the PDGLP1 SP (PDGLP1SP-GFP) or the GLP4 SP (GLP4SP-GFP).
(B) The control constructs tested remained either in the cytoplasm and nucleus (GFP alone) or in the cell wall (GLP4SP-GFP).
(C) The PDGLP1ΔSP-GFP mutant is no longer targeted to PD, whereas the PDGLP1SP-GFP construct accumulated in punctate foci along the cell wall, likely representing location to PD. In (B) and (C), the tested constructs were introduced into N. benthamiana leaves by particle bombardment and observed using confocal microscopy after a 24-h incubation period. Bars = 10 µm.
(D) PDGLP1 SP is not cleaved during protein targeting to PD. PDGLP1-MHT and PDGLP1SP-GFP-MHT were first agroinfiltrated in N. benthamiana leaves and then 4 d later leaf tissues were extracted for biochemical experiments. IM1 and IM2, internal markers for PDGLP1 lacking (single asterisk) or containing (double asterisk) the SP; P, PECP fraction; S, soluble protein fraction; T, total protein fraction.
(E) PDGLP1 uses the secretory pathway for its targeting to PD. A PDGLP1-GFP construct was agroinfiltrated into N. benthamiana leaves and, 24 h later, they were imaged by confocal microscopy prior to infiltration with BFA. Following a 12-h BFA treatment, recovery experiments were performed by infiltration of a 0.5% (v/v) DMSO solution. A parallel set of experiments was performed with a PDGLP1-MHT construct, and these leaves were employed to test for the effect of BFA treatment on PDGLP1-GFP subcellular localization. Here, Nt-NCAPP1 and Rubisco antibodies were used to confirm the purity of the PECP fraction. P, PECP fraction; T, total protein fraction. Bar = 10 µm.
(F) PDGLP1 is located on the outside of the Golgi-derived vesicles. PDGLP1-MHT and GLP4-MHT constructs were agroinfiltrated into N. benthamiana leaves and Golgi-derived vesicles were fractionated by velocity sedimentation. Vesicles were either treated with Proteinase K (20 µg/mL) or first pretreated with 0.1% Triton X-100 followed by Proteinase K. Note: PDGLP1-MHT was not detected in Proteinase K–treated vesicles, whereas a combination of Triton X-100 and Proteinase K was necessary to eliminate the GLP4-MHT signal.
[See online article for color version of this figure.]
Our SP experiments suggested that, during passage through the secretory system, the PDGLP1 SP may be retained on PDGLP1 (or GFP) to mediate targeting to the PD. This hypothesis was tested by isolating total protein, soluble protein and PECP fractions from leaves transiently expressing PDGLP1-4xMyc-6xHis (MHT) or PDGLP1SP-GFP-MHT. These biochemical assays confirmed that, whereas the total and soluble protein fractions contained both uncleaved and SP cleaved forms of PDGLP1-MHT, only the uncleaved form was detected in the PECP fraction (Figure 4D). A similar result was obtained for PDGLP1SP-GFP-MHT, except that a small amount of cleavage product was also detected in the PECP fraction.
We next used brefeldin A (BFA), an inhibitor of protein transport from the ER to the Golgi (Nebenführ et al., 2002), to further confirm the involvement of the secretory pathway in PDGLP1 delivery to the plasma membrane/PD. In contrast with the mock treatment, a 12-h application of BFA to Nicotiana benthamiana leaves transiently expressing PDGLP1-GFP abolished accumulation of the GFP signal within PD, and a 12-h wash/recovery period resulted in restoration of the control punctuate pattern of GFP fluorescence (Figure 4E). Parallel experiments were performed with PDGLP1-MHT and, here, leaves were used to isolate total protein and PECP fractions. Protein gel blotting assays showed that PDGLP1-MHT was not detected in the PECP fraction isolated from BFA treated leaves but that signal could again be detected in this fraction following a 12-h recovery period (Figure 4E, bottom panel). For these assays, anti-NtNCAPP1 (Lee et al., 2003) was used as a PECP control, and anti-ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) served as a control against contamination of the PECP fraction. Finally, expression of a PDGLP1-GFP construct that contained an ER retention signal failed to be targeted to PD (see Supplemental Figure 8 online). Taken together, these findings support the hypothesis that PDGLP1 delivery to the PD involves the secretory pathway.
Proteinase protection assays were next performed on purified Golgi vesicles to ascertain the physical orientation of the PDGLP1. Golgi vesicles isolated from leaves transiently expressing PDGLP1-MHT or GLP4-MHT constructs were treated with ± proteinase K and ± Triton X-100 and then proteins were extracted for immunoblot analysis. These assays established that the PDGLP1-MHT signal was abolished by proteinase K treatment alone, whereas proteolytic digestion of GLP4-MHT required both proteinase K and Triton X-100 treatment (Figure 4F). Based on these results, the SPs for PDGLP1 and GLP4 appear to anchor PDGLP1 and GLP4 to the outside and inside surfaces of the secretory vesicle, respectively. These orientations would result in GLP4 being secreted into the cell wall, whereas following vesicle fusion, PDGLP1 would be anchored to the plasma membrane by its uncleaved SP, leaving the protein within the cytoplasmic phase of the PD microchannels.
PDGLP1 Interacts with a Subset of Proteins in the PECP Fraction
PDGLP1 and PDGLP2 function(s) may involve an interaction with other proteins located in the PD. This possibility was explored using recombinant PDGLP1 in combination with a PECP preparation. Here, we first performed glutathione S-transferase (GST) pull-down assays to establish whether our recombinant purified PDGLP1-MHT could directly interact with Cm-PP16. Interestingly, we found that both PDGLP1-MHT and PDGLP1ΔSP-MHT directly interacted with Cm-PP16 (Figures 5A to 5C), indicating that the SP was not essential for PDGLP1-CmPP16 interaction. Next, PDGLP1 protein overlay assays were conducted using PECP fractions isolated from Bright Yellow 2 (BY-2) suspension cells (Lee et al., 2003); a number of PDGLP1interacting proteins were detected (Figure 6A). In view of this finding, co-IP experiments were next employed to isolate potential PDGLP1 interacting proteins (Figure 6B), and liquid chromatography–tandem mass spectroscopy was used for their identification (Table 1). It is noteworthy that, of the seven proteins identified, PDGLP1, actin, NCAPP1, and β-1,3-glucanase were identified in both PDGLP1 and Cm-PP16 co-IP experiments (see Supplemental Table 1 online), consistent with these proteins forming PD components of a PDGLP1 complex involved with NCAP cell-to-cell trafficking. Naturally, PDGLP1 was one of the seven isolated proteins, but the presence of PDGLP2 was of note as this suggested it could interact with either PDGLP1 or a protein that is a component of a PDGLP1 complex.
Figure 5.
PDGLP1 Interacts Directly with the NCAP Cm-PP16.
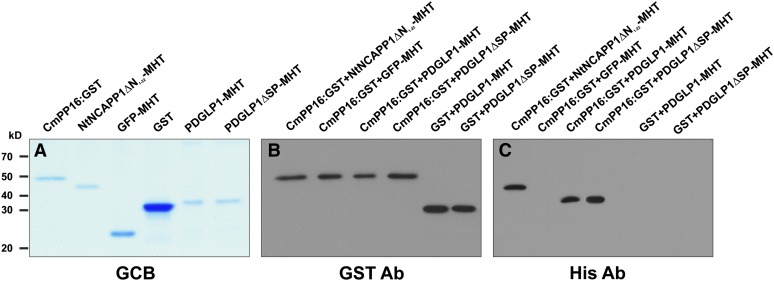
(A) Recombinant CmPP16-GST, NtNCAPP1ΔN1-22-MHT, GFP-MHT, GST, PDGLP1-MHT, and PDGLP1ΔSP-MHT were expressed in and purified from plants using a ZYMV viral vector system. Proteins were separated on a 12% SDS-PAGE gel and visualized by staining with Gelcode Blue (GCB).
(B) and (C) An in vitro GST pull-down assay demonstrates that PDGLP1 directly interacts with Cm-PP16. Pull-downed proteins were separated on a 12% SDS-PAGE gel and then subjected to immunoblot analysis using a GST antibody (B) or a His antibody (C). GFP and GST were used as negative controls, and NtNCAPP1ΔN1-22-MHT served as a positive control.
[See online article for color version of this figure.]
Figure 6.
PDGLP1 Interacts with a Subset of Proteins in the PECP Fraction to Form a PDGLP1-Specific Complex.
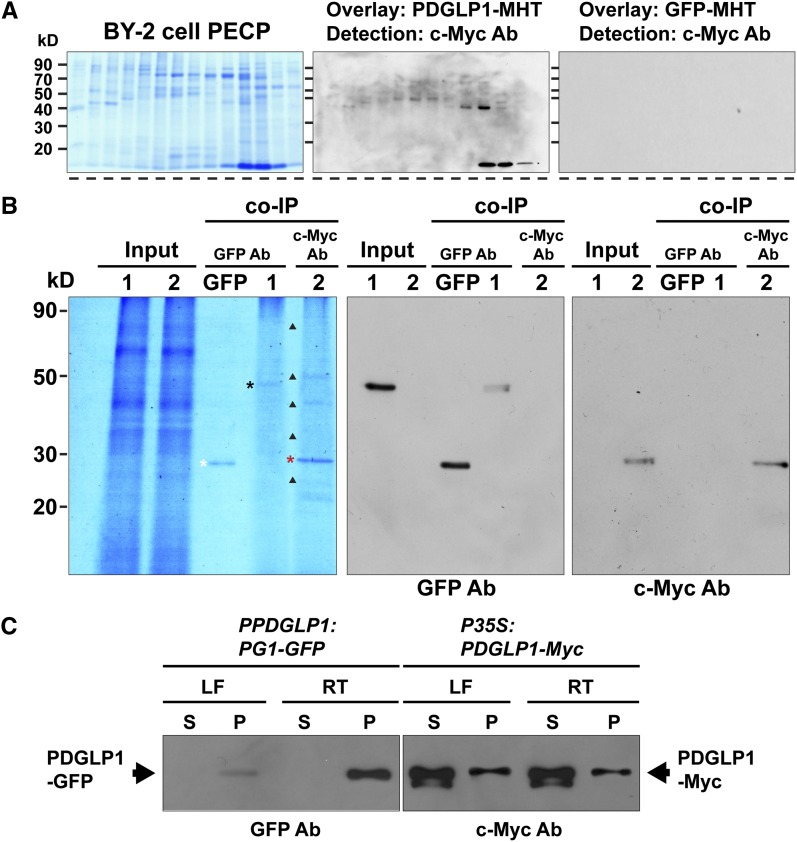
(A) Protein overlay assays establish that PDGLP1-MHT interacts with a range of proteins contained in the BY-2 cell PECP preparation. Proteins in the PECP were separated by fast protein liquid chromatography, and fractions were then separated by SDS-PAGE, followed by transfer to nitrocellulose membrane. Membrane was overlaid with purified PDGLP1-MHT and then subjected to immunoblot analysis, using an anti-c-Myc monoclonal antibody, to identify interacting proteins. GFP-MHT was used as a negative control.
(B) PDGLP1-interacting proteins identified by co-IP experiments. Purified recombinant GFP, PDGLP1-GFP (lane 1), or PDGLP1-Myc (lane 2) was incubated with an Arabidopsis PECP fraction (Input) followed by co-IP using anti-GFP or anti-c-Myc monoclonal antibodies (mAb). White, black, and red asterisks indicate immunoprecipitated GFP, PDGLP1-GFP, and PDGLP1-Myc, respectively. GFP served as the negative control. Immunoprecipitated bait proteins were confirmed by immunoblot analysis using anti-GFP (middle panel) or anti-c-Myc mAb (right panel).
(C) Both PDGLP1-GFP and PDGLP1-Myc are highly enriched in the Arabidopsis PECP fraction. Transgenic PPDGLP1:PDGLP1-GFP or P35S:PDGLP1-Myc plants were used to prepare soluble and PECP fractions, which were then separated by SDS-PAGE and tested by protein gel blot analysis using GFP or anti-c-Myc monoclonal antibodies. PECP fractions were prepared from leaf (LF) and root (RT) tissues. S, soluble fraction; P, PECP fraction.
[See online article for color version of this figure.]
Table 1. PDGLP1-Myc Interaction Partners Identified from an Arabidopsis PECP Preparation by Co-IP Assays.
| Protein Identity | Molecular Mass (kD) | No. of Unique Peptides | Coverage (%) | Arabidopsis Accession No. |
|---|---|---|---|---|
| Putative ABC transporter | 80 | 5 | 21 | AT5G58270 |
| Actin | 48 | 2 | 20 | AT3G53750 |
| NCAPP1 | 40 | 3 | 24 | AT5G15140 |
| β-1,3-glucanase | 40 | 6 | 38 | AT3G57270.1 |
| Phosphate-responsive1 | 33 | 5 | 29 | AT4G08950.1 |
| PDGLP1 | 23 | 13 | 83 | AT1G09560 |
| PDGLP2 | 23 | 2 | 12 | AT1G02335 |
PDGLP1-GFP Is Defective in Interacting with PECP Proteins
Parallel PECP co-IP experiments, performed using purified recombinant PDGLP1-GFP, yielded significantly different results (Figure 6B, lane 1), compared with PDGLP1-MHT (Figure 6B, lane 2). The C-terminally located GFP impeded the ability of PDGLP1 to bind to its IPs, as it bound weakly only to itself. This might reflect a steric impediment to protein–protein interaction caused by the increase in size of the GFP attached to the PDGLP1. This finding provided a possible explanation for the abnormal root phenotype of transgenic Arabidopsis seedlings expressing either PDGLP1-GFP (Figures 2E and 2F) or PDGLP1-GUS (see Supplemental Figure 5A online). Given that the interaction between PDGLP1 and the identified IPs is essential for the cell-to-cell trafficking of a subset of NCAPs, the dysfunctional PDGLP1-GFP and PDGLP1-GUS would block this NCAP pathway, causing a perturbation to the developmental process(es) mediated by these specific NCAPs.
Purified recombinant PDGLP1-Myc/MHT, with its smaller tag, could bind to proteins in the PECP preparation (Figures 6A and 6B), which raised the question as to why the transgenic Arabidopsis plants carrying P35S:PDGLP1-Myc or P35S:PDGLP2-Myc also exhibited the abnormal root phenotype (see Supplemental Figure 4 online). A potential answer to this question was provided by our analysis of the levels of these proteins in the soluble and PECP fractions. When expressed under its endogenous promoter, PDGLP1-GFP was detected only in the PECP fraction, whereas under the strong cauliflower mosaic virus 35S promoter, PDGLP1-Myc was present in both the soluble and PECP fractions (Figure 6C). Thus, although the smaller Myc tag did not prevent PDGLP1 from binding to potential interacting proteins (Figure 6B), the dramatic increase in the levels of PDGLP1-Myc, within the cytoplasmic phase, could well serve to sequester essential PD components required for normal NCAP delivery to and/or movement through the PD microchannels.
Dysfunction in Root PD Manifested by an Increase in Symplasmic Permeability
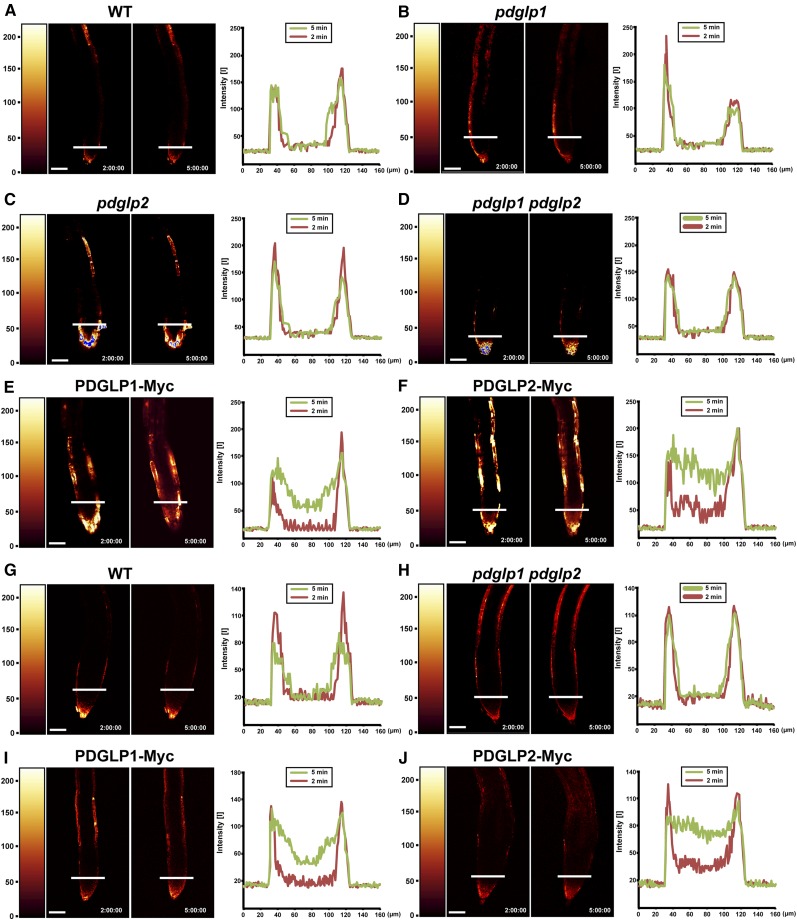
To further explore the basis for the perturbation in root development observed in our tagged PDGLP1/2 transgenic plants, we next probed the PD status in the roots of these lines using carboxyfluorescein diacetate (CFDA) (Zhu et al., 1998). Ester loading of CFDA at the root tip provides an effective means to probe the symplasmic connectivity of the various tissues. Root tips of 5- to 7-d-old seedlings were briefly dipped into CFDA and then the time course for carboxyfluorescein (CF) diffusion, within the root tissue, was analyzed using the two-dimensional process function in the Leica confocal laser scanning microscopy (CLSM) software. Although CF could be detected in the outer layers of wild-type, pdglp1, pdglp2, and pdglp1 pdglp2 roots 5 min after CFDA treatment (Figures 7A to 7D), no signal was detected within the inner regions of these root tips. In marked contrast, parallel experiments performed on root tips of PDGLP1-Myc or PDGLP2-Myc plants revealed that, within a similar 5-min CFDA treatment period, CF signal could be detected in the inner region of the root tip (Figures 7E and 7F). Similar results were obtained for lateral roots, in that CF was not detected within the inner region of wild-type and pdglp1 pdglp2 lateral roots, but a clear signal was detected for both PDGLP1-Myc and PDGLP2-Myc (Figures 7G to 7J). These experiments were performed more than 30 times, using primary and lateral roots for each transgenic plant line, and consistent patterns of CF movement were observed for each of the plant lines tested. Taken together, these studies support the hypothesis that overexpression of PDGLP1-Myc and PDGLP2-Myc causes an increase in PD permeability above that present in wild-type, pdglp1, pdglp2, and pdglp1 pdglp2 root tips.
Figure 7.
Symplasmic Permeability Is Enhanced in the Root Tips of Transgenic PDGLP1-Myc and PDGLP2-Myc Plants.
A brief (10-s) application of CFDA (60 µg/mL) to the primary ([A] to [F]) or lateral ([G] to [J]) root tips was used to load CF into the cytoplasm of Arabidopsis root cap and epidermal cells. CF distribution within the root tip was analyzed by confocal microscopy at 2 and 5 min after CFDA application. In wild-type (WT) (A), pdglp1 (B), pdglp2 (C), and pdglp1 pdglp2 (D) primary root tips, CF was restricted to the outer layer of cells. For the PDGLP1-Myc (E) and PDGLP2-Myc (F) seedlings, CF was detected in the central region of the primary root tip within 5 min of CFDA application. For wild-type (G) and pdglp1 pdglp2 (H) lateral root tips, CF was restricted to the outer layer of cells. By contrast, in the PDGLP1-Myc (I) and PDGLP2-Myc (J) seedlings, CF was detected in the central region of the lateral root tip within 5 min of CFDA application. Right panels present quantification of the fluorescence intensity measurements made at the locations indicated by the white bars on the individual roots. Red and green traces represent fluorescence intensity profiles collected at 2 and 5 min after CFDA application, respectively. Color bars on the left side of the confocal image indicate relative fluorescence intensity. Bars = 60 µm.
[See online article for color version of this figure.]
PDGLP1/2 Function in Regulation of Phloem Delivery to the Root System
Transgenic plants with increased levels of PDGLP1 or PDGLP2, with either a Myc/MHT or GFP/GUS fusion at the C terminus, exhibit a change in root architecture, reflected by reduced primary root and enhanced lateral root growth. Interestingly, total root dry weight remained unchanged between the wild type and all the various forms of transgenic PDGLP1/2 plants tested (see Supplemental Figure 6 online). This finding suggested that a PDGLP1/2-induced dysfunction in root PD might have caused a shift in relative sink strength between the primary and lateral root apices.
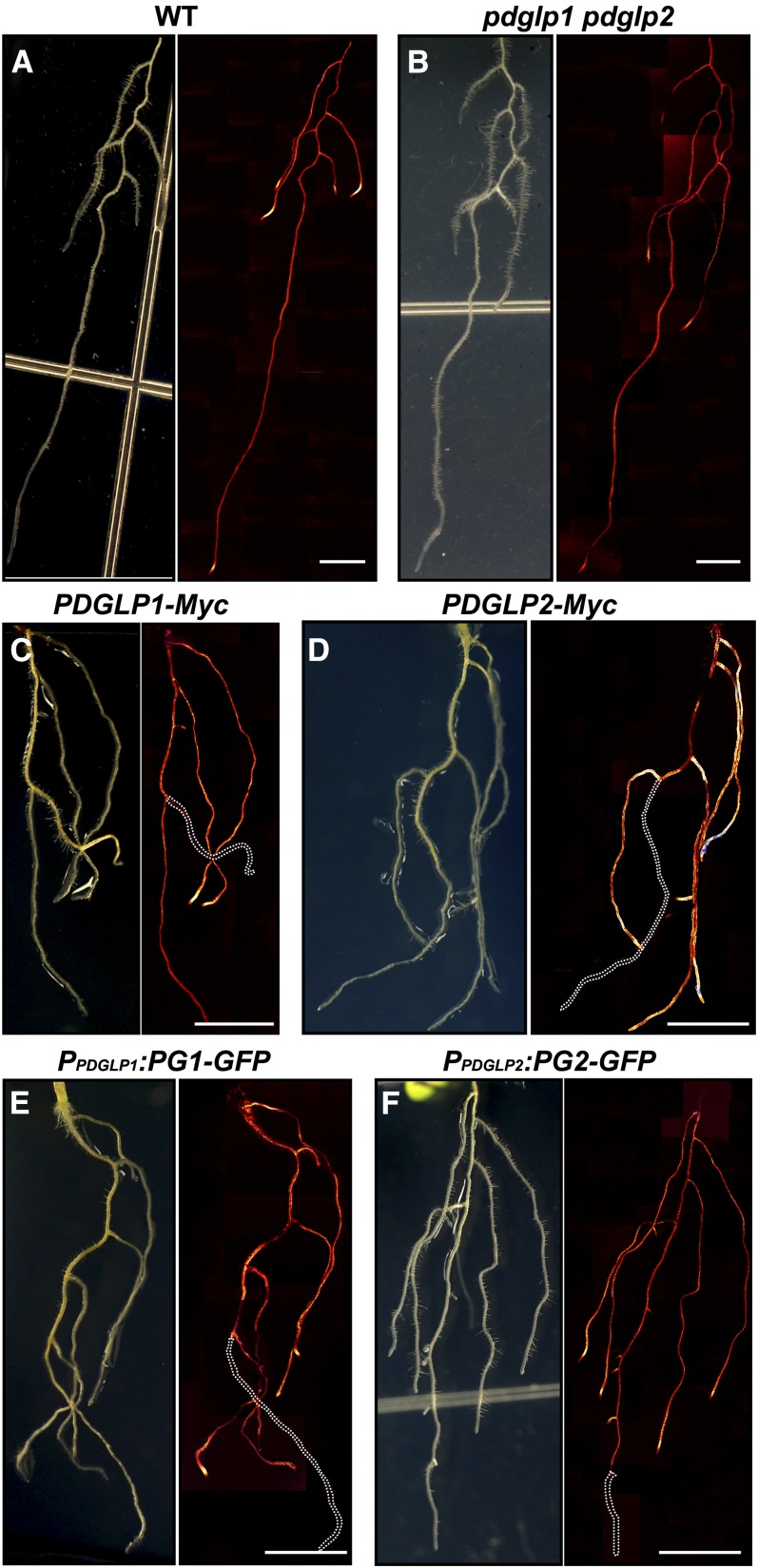
To test this hypothesis, for each Arabidopsis genotype, CFDA was loaded onto single cotyledons of 6-d-old seedlings to observe the efficacy of phloem unloading at the primary and lateral root tips (Oparka et al., 1994). CLSM images of the entire root structure were collected 90 min after CF loading into the phloem of the treated cotyledons. Both the wild type and the pdglp1 pdglp2 double mutant had equivalent patterns of CF delivery and unloading into the primary and lateral root systems (Figures 8A and 8B). By contrast, both PDGLP1-Myc and PDGLP2-Myc transgenic plants exhibited a strong CF signal in their lateral roots, but the CF signal was absent from the primary root (Figures 8C and 8D); note the absence of dye in the primary root indicated by the white dotted lines. Equivalent results were obtained with PDGLP1-GFP and PDGLP2-GFP transgenic plants (Figures 8E and 8F).
Figure 8.
Expressing Myc/GFP-Tagged Forms of PDGLP1 or PDGLP2 in Transgenic Arabidopsis Seedlings Alters the Pattern of Phloem Delivery into the Root System.
Phloem translocation into the Arabidopsis root system was examined by applying CFDA (60 µg/mL) onto a single cotyledon of 8-d-old seedlings. Confocal analysis of phloem delivery of CF into the roots was performed 90 min after CFDA application.
(A) and (B) In wild-type (WT) (A) and pdglp1 pdglp2 (B) seedlings, CF was unloaded in the root tip regions of both primary and lateral roots.
(C) to (F) In PDGLP1-Myc (C), PDGLP2-Myc (D), PDGLP1-GFP (E), and PDGLP2-GFP (F) transgenic seedlings, CF unloading was detected only in the lateral roots. White dotted lines indicate the position of the primary root into which phloem delivery of CF did not take place. Bars = 5 mm.
[See online article for color version of this figure.]
The observed pattern of CF delivery in these various transgenic plant lines could reflect a cessation or retardation of growth at the primary root tip, a change in the unloading priority of the phloem translocation system, or a combination of both. To test between these possibilities, CFDA loading experiments were performed on plants on which we had previously excised the lateral roots. Here, CF delivery into the primary root system of PDGLP1-Myc, PDGLP2-Myc, PDGLP1-GFP, and PDGLP2-GFP transgenic plant lines was restored, as was unloading at the primary root tips (see Supplemental Figure 9 online). In addition, auxin treatment, which induces lateral root formation (Okushima et al., 2007), resulted in a similar phenotype, as in the root system of these auxin-treated plants CF was not delivered into the primary root tip (see Supplemental Figures 10A and 10B online). Finally, excision of the wild-type primary root tip (meristem region) was also found to block CF delivery into this region of the root system (see Supplemental Figure 10C online). Taken together, these studies offered support for the hypothesis that expression of tagged forms of PDGLP1/2 causes a perturbation both to root meristem function and growth, resulting from the reduction in meristem size and/or a change in the priority of nutrient delivery through the phloem.
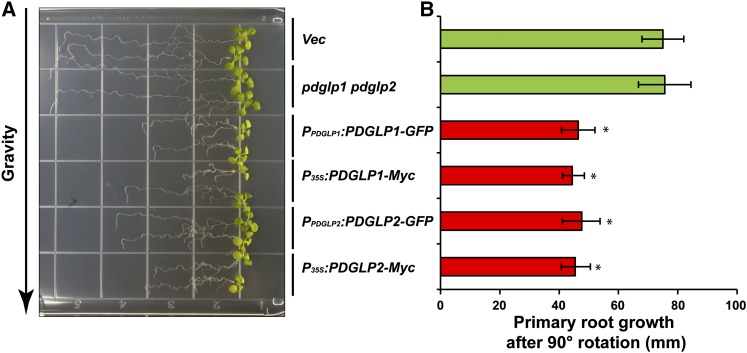
Primary Roots of Transgenic PDGLP1/2-Myc Plants Display a Normal Gravitropic Response
Experiments in which transgenic PDGLP1/2-Myc and PDGLP1/2-GFP were given a change in the gravity vector were conducted to further test the capacity of the primary root tip to undergo a differential growth response. Although the primary root length of 9-d-old PDGLP1/2-Myc/GFP seedlings was ∼50% of that for the vector control seedlings, they all displayed a normal gravitropic response (Figure 9A). Measurements made of primary root extension after imposition of a 90° rotation indicated that all PDGLP1/2-Myc/GFP seedlings had similar growth rates (Figure 9B). These results support the hypothesis that the smaller meristematic region of these transgenic roots has retained the capacity for complex signal transduction, involving an abiotic stimulus and localized growth mediated by auxin signaling.
Figure 9.
Primary Roots of Tagged PDGLP1 and PDGLP2 Transgenic Plants Display a Normal Gravitropic Response.
(A) Control (Vec), pdglp1 pdglp2 double mutant, PPDGLP1:PG1-GFP, PPDGLP2:PG2-GFP, P35S:PDGLP1-Myc, and P35S:PDGLP2-Myc plants were grown vertically on Murashige and Skoog plates for 8 d and then plates were rotated 90° to test the capacity of these lines to undergo differential growth in response to the altered gravitropic signal. Note that the primary roots of all tagged plant lines displayed a normal gravitropic response.
(B) Extent of primary root growth following plate rotation. Values represent mean ± sd; n = 50 roots. Asterisk indicates significant differences, P < 0.002, based on Student’s t test.
[See online article for color version of this figure.]
DISCUSSION
PDGLP1 and PDGLP2 Are Components in the NCAP Pathway
In this study, we identified and characterized Arabidopsis PDGLP1 and PDGLP2. In Arabidopsis, there are 29 members in the GLP family, and PDGLP1 and PGGLP2 were colocated with Nt-PDGLP1 in a clade containing five family members (Figure 1A). Of these five GLPs, only PDGLP1 and PDGLP2 were found to be colocalized with PD markers (Figure 1B; see Supplemental Figure 1A online). This is noteworthy, as all members of this clade exhibited at least 87% sequence similarity, with the regions of diversity being located within the N-terminal SP which, in itself, reflects a common component in the GLP family (Bernier and Berna, 2001). Our immunogold labeling studies (Figures 1C), in conjunction with our PDGLP1-RFP, CMV MP-GFP/TMV MP-GFP colocalization experiments (Figure 1B; see Supplemental Figure 1 online), provided strong support for our hypothesis that PDGLP1 is localized to PD. Thus, the previously established structural variants, within the N-terminal region of the GLP family (Woo et al., 2000), likely reflect the evolution of a novel SP on At PDGLP1 and At PDGLP2 that now allows for their targeting to PD.
PDGLP1 and PDGLP2 Function as Regulatory Components of Root Growth
Our GUS reporter analyses established that both PDGLP1 and PDGLP2 expression begins early during germination (Figure 2A; see Supplemental Figure 2 online), consistent with an earlier GLP study (Grzelczak et al., 1985). At later stages of development, these two genes become predominantly coexpressed in the root system. Interestingly, GUS staining indicated PDGLP1/2 expression throughout the root (Figures 2B and 2D), whereas the signal associated with PDGLP1-GFP and PDGLP2-GFP was confined to the endodermal layer (Figure 3). This discrepancy could reflect that these tagged proteins are dysfunctional (i.e., acting as dominant-negative proteins) and therefore are being targeted for turnover. Although the phenotype of the pdglp1 pdglp2 double mutant was equivalent to the wild type, PPDGLP1:PDGLP1-GFP transgenic lines displayed reduced primary root growth (Figures 2E and 2F; see Supplemental Figures 5A and 5B online), which is likely due to the overall reduction in meristem size (Figures 2I and 2J) and the enhancement of lateral root growth (see Supplemental Figure 5D online). This finding is similar to a previous report concerning the nonresponding to oxylipins2 mutant, in which regulation of lateral root development was shown to be dependent on the expression level of GLPs in the root (Vellosillo et al., 2007). Importantly, this reduced primary root growth phenotype was also observed for all tagged forms of PDGLP1 and PDGLP2 plants (see Supplemental Figures 4A, 4B, 5A, 5B, 5G, and 5H online), in which enhanced lateral root growth was detected (Figure 2H; see Supplemental Figure 5D online), but the number of lateral roots (see Supplemental Figure 5E online) as well as total root dry weight (see Supplemental Figure 6 online) remained the same as in wild-type plants.
This change in root phenotype, reflecting a reduction in root meristem size, can be replicated by imposing specific physiological conditions, such as phosphate deficiency or application of an exogenous auxin treatment (Li et al., 2006; Okushima et al., 2007). However, as neither PDGLP1 nor PDGLP2 expression was influenced by exogenous auxin treatment, the observed phenotype is more likely caused by the action of tagged dysfunctional/dominant-negative forms of PDGLP1/2. The possibility cannot be discounted that these effects are due to an abnormal toxic effect of these tagged PDGLP1/2 proteins. Although the radial pattern of cell layers in the primary root tip was not different between the wild type and our tagged PDGLP1/2 plant lines, the observed decrease in the root meristem size could well be the result of a decrease in the rate of cell division in these primary roots (Dello Ioio et al., 2008). Thus, we propose the hypothesis that the dysfunctional forms of PDGLP1/2 elicit the observed effect on root growth through their inability to support the cell-to-cell trafficking of agents involved in regulating the cell cycle (Vatén et al., 2011). Our PDGLP1/2-tagged plant lines and the SHORT-ROOT mutant (shr) display similar root phenotypes (Helariutta et al., 2000). Thus, it is possible that in the roots of our transgenic plants, cell-to-cell movement of SHR could be compromised (Nakajima et al., 2001). However, this is unlikely, as the PDGLP1/2-tagged roots displayed normal root architecture, in contrast with that of shr, in which the root ground tissue (endodermis and cortex) is perturbed (Helariutta et al., 2000).
PDGLP1 Uses the Secretory Pathway for Delivery to PD
All GLP family proteins have an N-terminally located SP (Bernier and Berna, 2001; Dunwell et al., 2008), which has been proposed to function in the secretion of GLPs into the cell wall, suggesting various functions for these proteins in the apoplasm (Bernier and Berna, 2001; Zimmermann et al., 2006; Davidson et al., 2010). Nevertheless, the mechanism by which the GLP SP functions in protein targeting to the cell wall remains to be elucidated. In this study, we established that the PDGLP1 SP itself was necessary and sufficient to target PDGLP1 to the PD (Figures 4A to 4C). Furthermore, biochemical studies revealed that SP-containing PDGLP1 was highly enriched in our PECP preparations; a similar result was obtained based on parallel experiments conducted with PDGLP1SP-GFP-MHT (Figure 4D). By contrast, both SP-cleaved and uncleaved forms of PDGLP1 and PDGLP1SP-GFP-MHT were present in the soluble fractions (Figure 4D). Thus, it is clear that the PDGLP1 SP functions in an unexpected manner, in that it is not cleaved during its delivery to the plasma membrane and, subsequently, it appears to function in sequestrating PDGLP1 to the PD. Indeed, cleavage of this SP may well occur during the PDGLP1 recycling process.
It is well established that for most proteins being processed on the conventional secretory pathway, the N-terminal, or internal SP plays a pivotal role in directing protein sorting to the ER. Subsequently, such sorted proteins are delivered either to the cellular exterior or the plasma membrane via secretory vesicles. However, there are situations in which proteins containing or lacking a SP can be transported to the cell surface on an unconventional non-ER-Golgi secretory pathway (Nickel and Rabouille, 2009). In this regard, BFA is a well-characterized inhibitor of the secretory pathway; thus, it is often used to block protein sorting through the Golgi (Mellman and Warren, 2000; Nebenführ et al., 2002). In our studies, BFA treatment inhibited PDGLP1-GFP targeting to PD (Figure 4E). In addition, our biochemical experiments confirmed that, in the presence of BFA, PDGLP1-MHT was not detected in the PECP preparation, but that upon removal of BFA, PDGLP1-MHT was once again detected in the PECP fraction (Figure 4D). These findings support the hypothesis that PDGLP1 is delivered to the cell periphery by the conventional ER-Golgi secretory pathway.
Several PD proteins, such as PLASMODESMATA-LOCATED PROTEIN1 and a plant-specific class 1 REVERSIBLY GLYCOSYLATED POLYPEPTIDE, have also been reported to use this same secretory pathway for delivery to PD (Sagi et al., 2005; Thomas et al., 2008). However, unlike these PD proteins, PDGLP1/2 does not possess any membrane-spanning or conventional membrane-anchoring motifs. Thus, the SP is the only means to retain PDGLP1 attached to the membrane vesicle. In this regard, our proteinase protection assays provided clear evidence that GLP4-MHT is located inside the secretory vesicle, whereas PDGLP1-MHT is attached to the outer surface of the secretory vesicle. This location of GLP4-MHT is consistent with the conventional model for the operation of the ER-Golgi secretory pathway. Presently, we have no information as to the mechanism involved in positioning of PDGLP1 to the outer vesicle surface. However, on a speculative note, we suggest that the PDGLP1 SP could well reflect a novel mechanism for targeting proteins, via the secretory pathway, to the PD.
Formation of Protein Complexes in PD Is a Key Requirement for PDGLP1 Function
Our co-IP experiments provided strong evidence that PDGLP1-MHT is a component of a PD-located protein complex (Figure 6). These studies revealed that PDGLP1-MHT interacts with NCAPP1 as well as with actin, a β-1,3-glucanase, phosphate responsive 1, a putative ABC transporter, and PDGLP2 (Table 1). Members of the GLP family have been shown to form both homo- and heterodimers (Woo et al., 2000; Rodríguez-López et al., 2001). Thus, detection of both PDGLP1 and PDGLP2 in our co-IP experiments is fully consistent with the presence of a PDGLP1-PDGLP2 complex within the PECP fraction. Identification of NCAPP1, in this PDGLP1 complex, is also of importance as it was earlier identified in a PECP experiment in which Cm-PP16 was used as the bait (Lee et al., 2003), and in this study Cm-PP16 was used as bait to identify PDGLP1. Thus, a PDGLP1-NCAPP1-CmPP16 interaction might be involved in transferring the Cm-PP16 NCAP from the cytoplasmic phase into the orifice of the PD microchannel.
Our studies showed that PDGLP1-GFP is processed through the secretory pathway and becomes localized to the PD (Figure 1B; see Supplemental Figure 1C online). However, in contrast with PDGLP1-MHT, the GFP tag on the C terminus of PDGLP1 abolished its ability to bind with the putative interaction proteins present in the Arabidopsis PECP preparation (Figure 6B, lane 1). This finding suggests that the more bulky GFP tag can sterically block PDGLP1 motifs required for protein–protein interaction. Consistent with these co-IP results, PPDGLP1:PDGLP1-GFP, PPDGLP2:PDGLP2-GFP, PPDGLP1:PDGLP1-GUS, and PPDGLP2:PDGLP2-GUS transgenic plant lines all exhibited the shortened primary root and enhanced lateral root growth phenotype (see Supplemental Figure 5 online). Taken together, these results provide additional support for the hypothesis that PDGLP1 functions, at the level of the PD, in the form of a protein complex.
Perhaps the most interesting finding was that, although the PDGLP1-MHT could interact with the identified slate of PECP interacting proteins (Figures 6A and 6B), our P35S:PDGLP1-Myc and P35S:PDGLP2-Myc transgenic plant lines all displayed the shortened primary root and enhanced lateral root growth phenotype (see Supplemental Figure 4 online). Biochemical assays performed on these transgenic lines indicated that PDGLP1-Myc was present in both the PECP and soluble fractions, whereas parallel assays conducted using PPDGLP1:PDGLP1-GFP plants only detected PDGLP1-GFP in the PECP fraction (Figure 6C). These results suggest that overexpression of PDGLP1-Myc results in saturation of a PDGLP1/2-specific PD pathway and/or PD target sites that then cause it to accumulated in a cytoplasmic compartment. Accumulation of high levels of PDGLP1-Myc in the soluble fraction may well establish a dominant-negative condition, through the formation of dysfunctional PDGLP1-IP complexes that then interrupt NCAP trafficking, in a similar manner to that earlier reported for NCAPP1 (Lee et al., 2003).
Perturbation to PDGLP1 and PDGLP2 Function Alters Phloem Partitioning in the Root System
CFDA has been used to probe PD-mediated symplasmic connectivity of the various cell types within the root tip (Zhu et al., 1998). Our studies indicated that, compared with the wild type, the primary and lateral roots of both PDGLP1 and PDGLP2 OX seedlings exhibited enhanced CF movement from the periphery into the inner tissues (Figure 7). This change in symplasmic coupling may reflect an effect of PDGLP1/2 OX on PD density. Alternatively, elevated levels of PDGLP1 or PDGLP2 could cause an increase in PD SEL, which could then have an effect on solute movement within the root (Rutschow et al., 2011). Irrespective of the mechanism underlying this increase in symplasmic coupling, such a change would be expected to increase the rate of phloem unloading in these PDGLP1/2 OX roots. As this increase in symplasmic coupling was observed in both primary and lateral root tips, this heightens the mystery as to why phloem delivery to the primary root unloading zone was reduced relative to the lateral roots (Figure 8). One possibility is that expression of tagged PDGLP1/2, within the vascular tissues of the primary root, might well cause a perturbation to symplasmic unloading in this tissue. Alternatively, the decrease in the size of the primary root meristem would result in it being a less competitive sink for phloem delivery of photosynthate.
Photoassimilate supply from source to sink tissues is regulated, in part, by relative sink strength and local sink strength can be affected by phytohormones, such as auxin (Mravec et al., 2009). Excision of the primary root meristem from wild-type seedlings prevented delivery of CF into such roots (see Supplemental Figure 10C online); this provides a direct example of the effect of reduced sink strength on photosynthate allocation within the root system. A similar situation was achieved by treating wild-type roots with auxin; here, increased sink strength in the lateral roots, over the primary root tip, resulted in cessation of phloem translocation into this weaker sink (see Supplemental Figure 10B online). Our removal of lateral roots from PDGLP1-Myc, PDGLP2-Myc, PPDGLP1:PDGLP1-GFP, or PPDGLP2:PDGLP2-GFP seedlings could restore phloem delivery and unloading to their primary roots (see Supplemental Figure 9 online). Here, removal of the lateral roots, acting as stronger sinks, allowed the primary root tip, a weaker sink region, to compete for photosynthate.
It is well known that non-cell-autonomous signaling between the endodermis and the stele plays a pivotal role in regulating root development (Helariutta et al., 2000; Carlsbecker et al., 2010; Lehesranta et al., 2010; Tsukagoshi et al., 2010; Wu and Gallagher, 2011). In our PDGLP1-Myc, PDGLP2-Myc, PDGLP1-GFP, and PDGLP2-GFP plant lines, OX of these various forms of PDGLP1/2 likely gives rise to a situation in which normal NCAP trafficking through the root PD is compromised. This perturbation to local signaling, in the primary root tip, could cause a reduction in cell division. Interestingly, OX of these proteins in the lateral roots does not appear to cause such a serious impediment to cell proliferation. Thus, the lateral roots outcompete the impacted primary root for photosynthate, resulting in a reduction in primary and an enhancement in lateral root growth. As the total root dry weight remained unchanged between wild-type and PDGLP1/2 OX plant lines (see Supplemental Figure 6 online), the possibility exists that, in wild-type roots, the PDGLP1/2 complex, located in the phloem PD, regulates phloem partitioning between the various sink organs in the root in a manner that might be complementary to the role of Lateral Root Development3 (Ingram et al., 2011).
METHODS
Plant Materials
Arabidopsis thaliana plants were grown under long-day conditions (16 h light/8 h dark) at a constant temperature of 22°C. For phloem dye unloading experiments, Arabidopsis seedlings were grown in continuous light conditions, as described previously (Oparka et al., 1994). The pdglp1 and pdglp2 mutants were obtained from the ABRC (CS863003 and CS856095, respectively), and the double mutant pdglp1 pdglp2 was generated by crosses between pdglp1 and pdglp2. Genotypes of these mutants were confirmed by PCR analysis using the appropriate primer sets (see Supplemental Table 2 online). Pumpkin (Cucurbita maxima cv Big Max) and Nicotiana benthamiana plants were grown as described previously (Taoka et al., 2007; Ma et al., 2010) for use in recombinant protein expression and purification and BFA experiments, respectively. For all comparisons and statistical analyses, at least three independent replicate experiments were conducted and evaluated using JMP 7 (SAS Institute).
Phylogenetic Analysis
The Arabidopsis Information Resource database was searched using GLP genes as queries, and ClustalW (http://www.Arabidopsis.org/cgi-bin/bulk/sequences/seqtoclustalw.pl) was used for alignment. Their accession numbers and sequence information can be found in Supplemental Data Set 1 online. MEGA 3.1 software constructed the GLP neighbor-joining tree (Ham et al., 2009), and a consensus tree was built from 1000 bootstrap replicates.
Plasmid Construction
For generating transgenic Arabidopsis plants, PDGLP1 and PDGLP2 were subcloned into pGWB17 for gene overexpression and pGWB3 for GUS staining (Nakagawa et al., 2007); these constructs were transformed into Agrobacterium tumefaciens strain C58C1. For transient expression of GFP and RFP fusion proteins in N. benthamiana, Nt-PDGLP1, PDGLP1, PDGLP2, GLP4, GLP4SP, PDGLP1ΔSP, and PDGLPSP-GFP were first amplified for subcloning into pENTR/D-TOPO (Invitrogen). To construct RFP and GFP fusion proteins, pGWB453/pGWB454 and pGWB4 (Nakagawa et al., 2007), respectively, were used with Gateway LR Clonase II enzyme mix (Invitrogen). PDGLPSP-GFP was digested with BamHI and XbaI and replaced with the original GFP in pGSH (Taoka et al., 2007). The pMLBART/TMV or CMV MP-GFP was used as a PD marker (Taoka et al., 2007). To engineer a PDGLP1 construct containing an ER retention signal, the PDGLP1 clone was digested with BamHI and EcoRI and then inserted into pBIN19-ER-mGFP5 (Hwang and Gelvin, 2004). The ER marker WAK2-RFP was obtained from the ABRC (CD3-960). PDGLP1SP-GFP or GLP4SP-GFP was amplified from pGWB4/PDGLP1SP-GFP or GLP4SP-GFP, respectively, and then inserted into pGWB17. Transient protein expression was performed by infiltrating Agrobacterium carrying pGWB17/PDGLP1, pGWB17/PDGLP2, pGWB4/PDGLP1SP-GFP, or pGWB4/GLP4SP-GFP into N. benthamiana leaves (Taoka et al., 2007).
For pull-down assays, GFP, Nt-NCAPP1ΔN1-22, PDGLP1, and PDGLP1ΔSP were subcloned into a Zucchini yellow mosaic virus (ZYMV)–based viral vector as described previously (Ma et al., 2010; Li et al., 2011). GST and Cm-PP16-GST were amplified from pGWB24/CmPP16 (Li et al., 2011) and inserted into the modified ZYMV viral vector without a c-MycX4-His6 tag (Lin et al., 2002).
RT-PCR and Mutant Screening
Total RNA of pdglp1, pdglp2, and pdglp1/2 mutant plants was extracted from 7-d-old seedlings. cDNA was synthesized using the SuperScript III first-strand synthesis system (Invitrogen). PDGLP1, PDGLP2, and ACTIN transcripts were amplified for 25, 33, and 20 cycles (96°C for 30 s, 57°C for 30 s, and 72°C for 30 s, respectively) using appropriate primer sets (see Supplemental Table 2 online).
For mutant screening, three mutant lines for pdglp1 (CS863003, CS879560, and salk_099426C) and two mutant lines for pdglp2 (CS856095 and salk_047180) were obtained from the ABRC. For the CS863003 and CS856095 mutant lines, PDGLP1 and PDGLP2 transcripts were not detected; thus, they were used for generating the double mutant pdglp1 pdglp2. Mutant genotyping was performed by PCR using appropriate primer sets (see Supplemental Table 2 online) (25 cycles each at 96°C for 30 s, 57°C for 30 s, and 72°C for 30 s).
Propidium Iodide and GUS Staining
Propidium iodide (at 10 µg/mL) was used to stain root cell walls to facilitate CLSM analysis of root architecture as described previously (Helariutta et al., 2000). GUS staining of PPDGLP1:GUS and PPDGLP2:GUS seedlings was performed as described previously (Foster et al., 2002). Images for GUS staining were obtained with a Zeiss Stereo Discovery V12 stereomicroscope or a Zeiss Axioskop 2 Plus microscope.
Electron Microscopy and Immunogold Labeling
Root tips and leaves of 5-d-old PPDGLP1:PG1-GFP plants were dissected and frozen rapidly with a HPM100 high-pressure freezer (Leica Microsystems). Specimens were freeze-substituted in anhydrous acetone with 0.1% uranyl acetate and 0.25% glutaraldehyde at −80°C for 72 h, and the temperature of the freeze substitution medium was then raised from −80 to −45°C over a 48-h period. After being rinsed with anhydrous acetone, Lowicryl HM20 resin (Electron Microscopy Sciences) was used for embedding the samples by a stepwise increase in resin concentration (0, 33, 66, and 100%) over a 48-h period at −45°C and then polymerized under UV light for 24 h. An AFS2 automatic freeze substitution machine (Leica Microsystems) was used in the processing of these specimens. Sample blocks were sectioned (80- to 120-nm thickness) and mounted on nickel slot grids coated with formvar (Electron Microscopy Sciences).
For immunogold labeling, sections were blocked for 30 min using PBS with 0.2% Tween 20 (PBST) supplemented with 2% nonfat milk, and then double-labeled with an anti-GFP antibody (Rockland; 1:50 dilution) and an anticallose antibody (Biosupplies; 1:200 dilution) in PBST with 1% nonfat milk for 3 h at room temperature. After a thorough wash with PBST, the anti-GFP and anticallose antibodies were labeled with an anti-rabbit IgG secondary antibody conjugated to 10-nm gold particles (1:10 dilution in PBST) and with an anti-mouse IgG secondary antibody conjugated to 15-nm gold particles (1:10 dilution in PBST), respectively. Immunolabeled sections were poststained with 2% uranylacetate and Reynolds lead and observed with a Hitachi H-7000 (Pleasant) transmission electron microscope operated at 100 kV.
Recombinant Protein Purification and PECP Preparation
Purification of recombinant proteins from ZYMV viral vector-infected pumpkin leaves was performed as described previously (Ma et al., 2010; Li et al., 2011). Briefly, ZYMV-based constructs were coated onto gold particles (1 µm) and bombarded onto pumpkin cotyledons using a Helios gene gun system (Bio-Rad). Recombinant proteins were purified using a two-step method. A HisTrap FF column (GE Healthcare) was employed for the first purification step, and a c-Myc tagged protein mild purification kit (MBL International) was used for the second step, according to the manufacturer’s instructions.
PECP preparation from either BY-2 suspension cultured cells or Arabidopsis was performed as described previously (Lee et al., 2003). Briefly, 300 g of BY-2 cells or 100 g of Arabidopsis seedlings was homogenized using a precooled Bead-Beater homogenizer (BioSpeck Products) containing 0.5-mm glass beads, homogenization buffer, and proteinase inhibitor. After four rounds of homogenization, homogenate was centrifuged at 300g for 5 min; the pellet was resuspended and subjected to additional rounds (approximately five) of centrifugation. The PECP fraction was extracted by incubation with PECP extraction buffer, overnight at 4°C. All steps were performed at 4°C and the PECP fraction was stored at −80°C.
Protein Overlay Assays
Protein overlay assays were performed as described previously (Taoka et al., 2007; Li et al., 2011). Briefly, a cation-exchange fractionated BY-2 cell PECP preparation was separated on a 13% SDS-PAGE gel, and proteins were then transferred to nitrocellulose membrane. Membranes were subsequently incubated with recombinant PDGLP1-Myc or GFP-Myc diluted in BSA buffer. After washing, the membrane blots were processed for immunoblot analysis, as described below.
Immunoblot Analysis
Immunoblot analyses were performed as follows. Membranes were immunoblotted with the appropriate primary antibody preparations (anti-c-Myc monoclonal antibody [mAb] at 1:5000 dilution, anti-Rubisco Ab at 1:1000 dilution, anti-NCAPP1 Ab at 1:1000 dilution, anti-GFP mAb at 1:5000 dilution, anti-His mAb at 1:1000 dilution, and anti-GST Ab at 1:5000) in the blocking agent (5% nonfat milk in 1× tris-buffered saline). After washing, membranes were incubated with horseradish peroxidase–conjugated anti-rabbit or mouse (1:20,000 or 1:50,000 dilution, respectively; Sigma-Aldrich) in the blocking agent for 1 h and then developed with chemiluminescence reagent (Perkin-Elmer Life Sciences).
BFA Treatment
Five-week-old N. benthamiana plants were agroinfiltrated with PPDGLP1:PDGLP1-GFP or PPDGLP1:PDGLP1-Myc constructs, and 24 h later leaves were infiltrated with BFA (50 µg/mL in 0.5% [v/v] DMSO). GFP fluorescent signal was analyzed 12 h after BFA treatment. BFA was subsequently removed from the treated area by infiltration of a 0.5% (v/v) DMSO solution; recovery of GFP signal at the PD was observed after a 12-h recovery period.
Co-IP Experiments
Co-IP was performed as described previously (Taoka et al., 2007; Ham et al., 2009). Briefly, aliquots of an Arabidopsis PECP preparation (500 µg protein/mL) were dialyzed, overnight at 4°C, against binding/wash buffer (0.14 M NaCl, 8.0 mM sodium phosphate, 2.0 mM potassium phosphate, and 10 mM KCl, pH 7.4). Recombinant PDGLP1-MHT and GFP were purified as described previously (Ma et al., 2010). Immobilized anti-c-Myc or anti-GFP IgG (Pierce Biotechnology) was incubated with purified recombinant PDGLP1-MHT, or GFP, and Arabidopsis PECP preparation for 2 h at 4°C, and then immunoprecipitation was performed using the ProFound c-Myc tag immunoprecipitation/co-IP application set (Pierce Biotechnology), following the manufacturer’s instructions. Elution fractions were separated on a 13% SDS-PAGE gel, and protein was visualized with Gelcode Blue Stain reagent (Pierce Biotechnology). Protein bands were excised from the gel and processed for liquid chromatography–tandem mass spectroscopy analysis as described previously (Ham et al., 2009).
For pull-down assays, purified recombinant CmPP16-GST or GST was mixed with each purified His6-fused recombinant protein (700 ng) and pulled down with affinity glutathione-Sepharose 4B beads (GE Healthcare) as described previously (Li et al., 2011).
Golgi-Derived Vesicle Preparation and Proteinase Protection Assays
Golgi-derived vesicles were fractionated, as described previously (Munoz et al., 1996). Briefly, 50 g of agroinfiltrated N. benthamiana leaves were homogenized using a Bead-Beater homogenizer (BioSpeck Products) containing 0.5-mm glass beads in homogenizing buffer (0.5 M Suc, 0.1 M KH2PO4, pH 6.65, 5 mM MgCl2, and 1 mM DTT). The homogenate was filtered through Miracloth (Calbiochem) and then centrifuged at 1000g for 5 min, followed by Suc gradient sedimentation, as described previously (Munoz et al., 1996). Activity of IDPase, a marker enzyme for plant-derived Golgi bodies, was tested in each fraction. Proteinase protection assays were performed as described previously (Matern et al., 2000).
Fluorescent Dye Coupling Analyzed by CSLM
CFDA (Sigma-Aldrich) (60 µg/mL) was ester loaded into Arabidopsis cotyledons or root apices as described previously (Oparka et al., 1994; Zhu et al., 1998), and spatiotemporal fluorescence was detected by CLSM (model DM RXE 6 TCS-SP2 AOBS; Leica Microsystems) as described previously (Zhu et al., 1998).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: PDGLP1, At1g09560; PDGLP2, At1g02335; and GLP4, At1g18970.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transient Expression of Nt PDGLP1-RFP, PDGLP1-GFP, and PDGLP2-GFP Results in Protein Accumulation in a Punctate Pattern at the Cell Periphery.
Supplemental Figure 2. PDGLP2 Expression Pattern Determined Using Transgenic PPDGLP2:GUS Plants.
Supplemental Figure 3. Molecular Characterization of the pdglp1 and pdglp2 Mutants.
Supplemental Figure 4. Ectopic Expression of PDGLP1-Myc, PDGLP2-Myc, and Nontagged PDGLP1 Yielded Short Root Phenotypes.
Supplemental Figure 5. Transgenic Plants Expressing C-Terminal Fusions of GUS or GFP on PDGLP1 and PDGLP2 Also Displayed a Short Root Phenotype.
Supplemental Figure 6. No Significant Difference in Root Dry Weight Was Detected Between Seedlings of the Indicated Plant Lines.
Supplemental Figure 7. Abnormal Cellular Architecture in Primary Roots of Transgenic P35S:PDGLP1-Myc and PPDGLP1:PG1-GFP Develops Shortly After Germination.
Supplemental Figure 8. A KDEL-Tagged Form of PDGLP1 Is Retained Within the ER.
Supplemental Figure 9. Excision of Lateral Roots Restores Phloem Delivery of CF into the Primary Roots of Transgenic PDGLP1-Myc, PG1-GFP, and PG2-GFP Plants.
Supplemental Figure 10. Auxin Application or Excision of the Primary Root Tip of Wild-Type Arabidopsis Seedlings Phenotypically Mimics CF Delivery into the Root Systems of Tagged PDGLP1/2 Transgenic Seedlings.
Supplemental Table 1. Identification of Cm-PP16 Interacting Proteins Purified from a Tobacco BY-2 Cell PECP Fraction.
Supplemental Table 2. PCR Primers Employed in This Study.
Supplemental Data Set 1. Sequences Used to Generate the Phylogeny Presented in Figure 1A.
Acknowledgments
This work was supported by National Science Foundation Grant IOS-0918433 (to W.J.L.) and USDA Grant 2010-34446-21694 (to B.-H.K).
AUTHOR CONTRIBUTIONS
B.-K.H., G.L., and W.J.L. designed the research project. F.Z. cloned Nt-PDGLP1. B.-K.H. and G.L. performed all molecular, cellular, and biochemical experiments. B.-H.K. performed the immunogold studies. B.-K.H., G.L., B.-H.K., and W.J.L. analyzed the data and wrote the article.
Glossary
- PD
plasmodesmata
- MP
movement protein
- NCAP
non-cell-autonomous protein
- SEL
size exclusion limit
- TMV
Tobacco mosaic virus
- co-IP
coimmunoprecipitation
- PECP
plasmodesmal-enriched cell wall protein
- CMV
Cucumber mosaic virus
- GFP
green fluorescent protein
- RFP
red fluorescent protein
- GUS
β-glucuronidase
- DAG
days after germination
- OX
overexpression
- SP
signal peptide
- ER
endoplasmic reticulum
- Rubisco
ribulose-1,5-bisphosphate carboxylase/oxygenase
- GST
glutathione S-transferase
- BY-2
Bright Yellow 2
- CFDA
carboxyfluorescein diacetate
- CLSM
confocal laser scanning microscopy
- ZYMV
Zucchini yellow mosaic virus
- PBST
PBS with 0.2% Tween 20
- CF
carboxyfluorescein
References
- Aoki K., Kragler F., Xoconostle-Cazares B., Lucas W.J. (2002). A subclass of plant heat shock cognate 70 chaperones carries a motif that facilitates trafficking through plasmodesmata. Proc. Natl. Acad. Sci. USA 99: 16342–16347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.K., Chatterjee M., Yu Y.Y., Suh S.G., Miller W.A., Hannapel D.J. (2006). Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18: 3443–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier F., Berna A. (2001). Germins and germin-like proteins: Plant do-all proteins. But what do they do exactly? Plant Physiol. Biochem. 39: 545–554 [Google Scholar]
- Blackman L.M., Boevink P., Cruz S.S., Palukaitis P., Oparka K.J. (1998). The movement protein of cucumber mosaic virus traffics into sieve elements in minor veins of nicotiana clevelandii. Plant Cell 10: 525–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Timmermans M.C.P. (2010). Small RNAs are on the move. Nature 467: 415–419 [DOI] [PubMed] [Google Scholar]
- Crawford K.M., Zambryski P.C. (2000). Subcellular localization determines the availability of non-targeted proteins to plasmodesmatal transport. Curr. Biol. 10: 1032–1040 [DOI] [PubMed] [Google Scholar]
- Davidson R.M., Manosalva P.M., Snelling J., Bruce M., Leung H., Leach J.E. (2010). Rice Germin-Like Proteins: Allelic diversity and relationships to early stress responses. Rice 3: 43–55 [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M.T., Aoyama T., Costantino P., Sabatini S. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Himber C., Voinnet O. (2005). DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 37: 1356–1360 [DOI] [PubMed] [Google Scholar]
- Dunwell J.M., Gibbings J.G., Mahmood T., Naqvi S.M.S. (2008). Germin and germin-like proteins: Evolution, structure, and function. Crit. Rev. Plant Sci. 27: 342–375 [Google Scholar]
- Fernandez-Calvino L., Faulkner C., Walshaw J., Saalbach G., Bayer E., Benitez-Alfonso Y., Maule A. (2011). Arabidopsis plasmodesmal proteome. PLoS ONE 6: e18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T.M., Lough T.J., Emerson S.J., Lee R.H., Bowman J.L., Forster R.L., Lucas W.J. (2002). A surveillance system regulates selective entry of RNA into the shoot apex. Plant Cell 14: 1497–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzelczak Z.F., Rahman S., Kennedy T.D., Lane B.G. (1985). Germin - Compartmentation of the protein, its translatable mRNA, and its biosynthesis among roots, stems, and leaves of wheat seedlings. Can. J. Biochem. Cell Biol. 63: 1003–1013 [Google Scholar]
- Ham B.K., Brandom J.L., Xoconostle-Cázares B., Ringgold V., Lough T.J., Lucas W.J. (2009). A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21: 197–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood V., Yu T.-S., Huang N.-C., Lucas W.J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42: 49–68 [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Hwang H.H., Gelvin S.B. (2004). Plant proteins that interact with VirB2, the Agrobacterium tumefaciens pilin protein, mediate plant transformation. Plant Cell 16: 3148–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram P., Dettmer J., Helariutta Y., Malamy J.E. (2011). Arabidopsis Lateral Root Development 3 is essential for early phloem development and function, and hence for normal root system development. Plant J. 68: 455–467 [DOI] [PubMed] [Google Scholar]
- Ishiwatari Y., Fujiwara T., McFarland K.C., Nemoto K., Hayashi H., Chino M., Lucas W.J. (1998). Rice phloem thioredoxin h has the capacity to mediate its own cell-to-cell transport through plasmodesmata. Planta 205: 12–22 [DOI] [PubMed] [Google Scholar]
- Jo Y., Cho W.K., Rim Y., Moon J., Chen X.Y., Chu H., Kim C.Y., Park Z.Y., Lucas W.J., Kim J.Y. (2011). Plasmodesmal receptor-like kinases identified through analysis of rice cell wall extracted proteins. Protoplasma 248: 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R.A., Atkinson R.G., Forster R.L.S., Lucas W.J. (1998). An RNA-based information superhighway in plants. Science 279: 1486–1487 [DOI] [PubMed] [Google Scholar]
- Kehr J., Buhtz A. (2008). Long distance transport and movement of RNA through the phloem. J. Exp. Bot. 59: 85–92 [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Rim Y., Wang J., Jackson D. (2005). A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 19: 788–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Canio W., Kessler S., Sinha N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293: 287–289 [DOI] [PubMed] [Google Scholar]
- Kragler F., Monzer J., Shash K., Xoconostle-Cazares B., Lucas W.J. (1998). Cell-to-cell transport of proteins: Requirement for unfolding and characterization of binding to a putative plasmodesmal receptor. Plant J. 15: 367–381 [Google Scholar]
- Kurata T., et al. (2005a). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398 [DOI] [PubMed] [Google Scholar]
- Kurata T., Okada K., Wada T. (2005b). Intercellular movement of transcription factors. Curr. Opin. Plant Biol. 8: 600–605 [DOI] [PubMed] [Google Scholar]
- Lee J.-Y., Taoka K., Yoo B.-C., Ben-Nissan G., Kim D.-J., Lucas W.J. (2005). Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17: 2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Yoo B.C., Rojas M.R., Gomez-Ospina N., Staehelin L.A., Lucas W.J. (2003). Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science 299: 392–396 [DOI] [PubMed] [Google Scholar]
- Lehesranta S.J., Lichtenberger R., Helariutta Y. (2010). Cell-to-cell communication in vascular morphogenesis. Curr. Opin. Plant Biol. 13: 59–65 [DOI] [PubMed] [Google Scholar]
- Li M.-Y., Qin C.-B., Welti R., Wang X.-M. (2006). Double knockouts of phospholipases Dzeta1 and Dzeta2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol. 140: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Ham B.K., Lucas W.J. (2011). CmRBP50 protein phosphorylation is essential for assembly of a stable phloem-mobile high-affinity ribonucleoprotein complex. J. Biol. Chem. 286: 23142–23149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.S., Hou R.F., Yeh S.D. (2002). Construction of in vitro and in vivo infectious transcripts of a Taiwan strain of Zucchini yellow mosaic virus. Bot. Bull. Acad. Sinica (Taiwan) 43: 261–269 [Google Scholar]
- Lucas W.J. (2006). Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 344: 169–184 [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Bouche-Pillon S., Jackson D.P., Nguyen L., Baker L., Ding B., Hake S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980–1983 [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Ham B.K., Kim J.Y. (2009). Plasmodesmata - Bridging the gap between neighboring plant cells. Trends Cell Biol. 19: 495–503 [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Lee J.Y. (2004). Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell Biol. 5: 712–726 [DOI] [PubMed] [Google Scholar]
- Ma Y., Miura E., Ham B.K., Cheng H.W., Lee Y.J., Lucas W.J. (2010). Pumpkin eIF5A isoforms interact with components of the translational machinery in the cucurbit sieve tube system. Plant J. 64: 536–550 [DOI] [PubMed] [Google Scholar]
- Martin A., Adam H., Díaz-Mendoza M., Zurczak M., González-Schain N.D., Suárez-López P. (2009). Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 136: 2873–2881 [DOI] [PubMed] [Google Scholar]
- Matern H., Yang X., Andrulis E., Sternglanz R., Trepte H.H., Gallwitz D. (2000). A novel Golgi membrane protein is part of a GTPase-binding protein complex involved in vesicle targeting. EMBO J. 19: 4485–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule A.J. (2008). Plasmodesmata: Structure, function and biogenesis. Curr. Opin. Plant Biol. 11: 680–686 [DOI] [PubMed] [Google Scholar]
- Mellman I., Warren G. (2000). The road taken: Past and future foundations of membrane traffic. Cell 100: 99–112 [DOI] [PubMed] [Google Scholar]
- Molnar A., Melnyk C.W., Bassett A., Hardcastle T.J., Dunn R., Baulcombe D.C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875 [DOI] [PubMed] [Google Scholar]
- Mravec J., et al. (2009). Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140 [DOI] [PubMed] [Google Scholar]
- Munoz P., Norambuena L., Orellana A. (1996). Evidence for a UDP-glucose transporter in Golgi apparatus-derived vesicles from pea and its possible role in polysaccharide biosynthesis. Plant Physiol. 112: 1585–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Nebenführ A., Ritzenthaler C., Robinson D.G. (2002). Brefeldin A: Deciphering an enigmatic inhibitor of secretion. Plant Physiol. 130: 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel W., Rabouille C. (2009). Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol. 10: 148–155 [DOI] [PubMed] [Google Scholar]
- Noueiry A.O., Lucas W.J., Gilbertson R.L. (1994). Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 76: 925–932 [DOI] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka K.J., Duckett C.M., Prior D.A.M., Fisher D.B. (1994). Real-time imaging of phloem unloading in the root tip of Arabidopsis. Plant J. 6: 759–766 [Google Scholar]
- Oparka K.J., Roberts A.G., Boevink P., Santa Cruz S., Roberts I., Pradel K.S., Imlau A., Kotlizky G., Sauer N., Epel B. (1999). Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97: 743–754 [DOI] [PubMed] [Google Scholar]
- Robards A.W., Lucas W.J. (1990). Plasmodesmata. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41: 369–419 [Google Scholar]
- Rodríguez-López M., Baroja-Fernández E., Zandueta-Criado A., Moreno-Bruna B., Muñoz F.J., Akazawa T., Pozueta-Romero J. (2001). Two isoforms of a nucleotide-sugar pyrophosphatase/phosphodiesterase from barley leaves (Hordeum vulgare L.) are distinct oligomers of HvGLP1, a germin-like protein. FEBS Lett. 490: 44–48 [DOI] [PubMed] [Google Scholar]
- Rutschow H.L., Baskin T.I., Kramer E.M. (2011). Regulation of solute flux through plasmodesmata in the root meristem. Plant Physiol. 155: 1817–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi G., Katz A., Guenoune-Gelbart D., Epel B.L. (2005). Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the Golgi apparatus. Plant Cell 17: 1788–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., Yanofsky M.F., Weigel D. (2000). Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289: 779–782 [DOI] [PubMed] [Google Scholar]
- Simpson C., Thomas C., Findlay K., Bayer E., Maule A.J. (2009). An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K., Ham B.K., Xoconostle-Cázares B., Rojas M.R., Lucas W.J. (2007). Reciprocal phosphorylation and glycosylation recognition motifs control NCAPP1 interaction with pumpkin phloem proteins and their cell-to-cell movement. Plant Cell 19: 1866–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C.L., Bayer E.M., Ritzenthaler C., Fernandez-Calvino L., Maule A.J. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H., Busch W., Benfey P.N. (2010). Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Vatén A., et al. (2011). Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 21: 1144–1155 [DOI] [PubMed] [Google Scholar]
- Vellosillo T., Martínez M., López M.A., Vicente J., Cascón T., Dolan L., Hamberg M., Castresana C. (2007). Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 19: 831–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E., Veit B., Sinha N., Hake S. (1991). The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350: 241–243 [DOI] [PubMed] [Google Scholar]
- Waigmann E., Chen M.H., Bachmaier R., Ghoshroy S., Citovsky V. (2000). Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J. 19: 4875–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E., Ueki S., Trutnyeva K., Citovsky V. (2004). The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. CRC Crit. Rev. Plant Sci. 23: 195–250 [Google Scholar]
- Wolf S., Deom C.M., Beachy R.N., Lucas W.J. (1989). Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246: 377–379 [DOI] [PubMed] [Google Scholar]
- Woo E.J., Dunwell J.M., Goodenough P.W., Marvier A.C., Pickersgill R.W. (2000). Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 7: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wright K.M., Wood N.T., Roberts A.G., Chapman S., Boevink P., Mackenzie K.M., Oparka K.J. (2007). Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: Evidence from FRAP. Traffic 8: 21–31 [DOI] [PubMed] [Google Scholar]
- Wu S., Gallagher K.L. (2011). Mobile protein signals in plant development. Curr. Opin. Plant Biol. 14: 563–570 [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cázares B., Xiang Y., Ruiz-Medrano R., Wang H.L., Monzer J., Yoo B.C., McFarland K.C., Franceschi V.R., Lucas W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283: 94–98 [DOI] [PubMed] [Google Scholar]
- Xu X.M., Jackson D. (2010). Lights at the end of the tunnel: New views of plasmodesmal structure and function. Curr. Opin. Plant Biol. 13: 684–692 [DOI] [PubMed] [Google Scholar]
- Xu X.M., Wang J., Xuan Z.Y., Goldshmidt A., Borrill P.G.M., Hariharan N., Kim J.Y., Jackson D. (2011). Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science 333: 1141–1144 [DOI] [PubMed] [Google Scholar]
- Yoo B.-C., Kragler F., Varkonyi-Gasic E., Haywood V., Archer-Evans S., Lee Y.M., Lough T.J., Lucas W.J. (2004). A systemic small RNA signaling system in plants. Plant Cell 16: 1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Lucas W.J., Rost T.L. (1998). Directional cell-to-cell communication in the Arabidopsis root apical meristem - I. An ultrastructural and functional analysis. Protoplasma 203: 35–47 [Google Scholar]
- Zimmermann G., Bäumlein H., Mock H.P., Himmelbach A., Schweizer P. (2006). The multigene family encoding germin-like proteins of barley. Regulation and function in basal host resistance. Plant Physiol. 142: 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]