An important photoprotective mechanism in plants, algae, and cyanobacteria is the selective replacement of damaged protein subunits within the oxygen-evolving enzyme of photosynthesis. This work reports the purification of the cyanobacterial FtsH protease involved in this process and shows using electron microscopy that it is a hexamer consisting of two types of alternating subunits.
Abstract
FtsH metalloproteases are key components of the photosystem II (PSII) repair cycle, which operates to maintain photosynthetic activity in the light. Despite their physiological importance, the structure and subunit composition of thylakoid FtsH complexes remain uncertain. Mutagenesis has previously revealed that the four FtsH homologs encoded by the cyanobacterium Synechocystis sp PCC 6803 are functionally different: FtsH1 and FtsH3 are required for cell viability, whereas FtsH2 and FtsH4 are dispensable. To gain insights into FtsH2, which is involved in selective D1 protein degradation during PSII repair, we used a strain of Synechocystis 6803 expressing a glutathione S-transferase (GST)–tagged derivative (FtsH2-GST) to isolate FtsH2-containing complexes. Biochemical analysis revealed that FtsH2-GST forms a hetero-oligomeric complex with FtsH3. FtsH2 also interacts with FtsH3 in the wild-type strain, and a mutant depleted in FtsH3, like ftsH2− mutants, displays impaired D1 degradation. FtsH3 also forms a separate heterocomplex with FtsH1, thus explaining why FtsH3 is more important than FtsH2 for cell viability. We investigated the structure of the isolated FtsH2-GST/FtsH3 complex using transmission electron microscopy and single-particle analysis. The three-dimensional structural model obtained at a resolution of 26 Å revealed that the complex is hexameric and consists of alternating FtsH2/FtsH3 subunits.
INTRODUCTION
The ability of oxygenic photosynthetic organisms, such as cyanobacteria and green plants, to convert solar energy into chemical energy via photosynthesis is inhibited at high irradiances in a process termed photoinhibition (Aro et al., 1993; Adir et al., 2003). Such inhibition is considered to be a major factor in limiting crop yields, especially when plants are exposed to other types of abiotic stress (Murata et al., 2007; Takahashi and Badger, 2011). At a molecular level, a major target for photodamage is the photosystem II (PSII) complex of the thylakoid membrane, which functions as the light-driven water:plastoquinone oxidoreductase of the photosynthetic electron transport chain (Wydrzynski and Satoh, 2005). Although PSII contains 20 subunits or more, depending on the species, the D1 reaction center polypeptide, which binds many of the cofactors involved in electron transfer through the complex, is most prone to irreversible oxidative damage in the light (Adir et al., 2003). To maintain PSII activity, irreversibly damaged subunits are replaced by newly synthesized copies in the so-called PSII repair cycle (Adir et al., 2003; Nixon et al., 2005, 2010). This process involves partial disassembly of the damaged PSII complex, degradation and replacement of the damaged subunit by a newly synthesized copy, reattachment of disassembled, undamaged PSII subunits, and light-driven assembly of the Mn4Ca water-splitting catalyst (Nixon et al., 2010).
A crucial step in PSII repair is the recognition and degradation of the damaged D1 subunit. Recent work has identified a critical role for FtsH proteases in both cyanobacteria and chloroplasts (Nixon et al., 2005; Kato and Sakamoto, 2009). This class of protease is embedded in the membrane via one or two transmembrane helices in the N-terminal region of the molecule and contains an AAA+ (ATPase associated with diverse cellular activities) module followed by a protease domain (PD) harboring a Zn2+ binding site (Erzberger and Berger, 2006). FtsH proteases are found in bacteria as well as in chloroplasts and mitochondria and catalyze the degradation of both membrane-bound and soluble proteins (Ito and Akiyama, 2005; Koppen and Langer, 2007). FtsH can also form supercomplexes with members of the Band 7 (or SPFH [for stomatin/prohibitin/flotillin/HflKC]) family of proteins, such as the prohibitins in mitochondria (Steglich et al., 1999).
In the case of the cyanobacterium Synechocystis sp PCC 6803 (hereafter Synechocystis 6803), four FtsH homologs, designated FtsH1-4, encoded by slr1390, slr0228, slr1604, and sll1463, respectively, have been identified from analysis of the genome sequence (Mann et al., 2000). FtsH2 and FtsH4 are dispensable, whereas FtsH1 and FtsH3 are required for cell viability (Mann et al., 2000). Studies on mutants lacking FtsH2 have provided compelling evidence for a physiological role for this FtsH subunit in the degradation of D1 during PSII repair in vivo (Silva et al., 2003; Komenda et al., 2006). FtsH2 is also involved in the removal of D1 following damage by heat (Kamata et al., 2005) and UV-B irradiation (Cheregi et al., 2007) and degrades unassembled and mutant PSII subunits in the thylakoid membrane (Komenda et al., 2006). FtsH2 also regulates, directly or indirectly, the level of the soluble enzyme, glucosyl-glycerol phosphate synthase, involved in osmoprotection (Stirnberg et al., 2007).
Nine FtsH subunits are also targeted to the Arabidopsis thaliana chloroplast (Sakamoto et al., 2003). Mutants lacking At-FtsH2 and At-FtsH5 show impaired rates of D1 degradation and PSII repair (Bailey et al., 2002; Kato et al., 2009). Genetic and coimmunoprecipitation data suggest that FtsH subunits might form both homo-oligomeric (Sakamoto et al., 2003) and hetero-oligomeric complexes in chloroplasts (Sakamoto et al., 2003; Yu et al., 2004; Zaltsman et al., 2005), but as yet no chloroplast FtsH complex has been isolated and characterized in terms of subunit composition and organization.
Information on the structure of intact FtsH complexes, rather than Escherichia coli–expressed soluble fragments, is sparse, and it is only recently that a hexameric structure was confirmed for the heteromeric m-AAA complex of the yeast mitochondrial inner membrane by cryo–electron microscopy (cryo-EM) (Lee et al., 2011). The resolution of the structure, determined at 12 Å, was sufficient to confirm the presence of six FtsH protomers within each complex, obtained by fitting the atomic structures of cytosolic fragments of FtsH into the cryo-EM model (Bieniossek et al., 2006, 2009), but was insufficient to determine the structural organization of the two types of subunit within the ring.
Here, we report the isolation of FtsH2 from Synechocystis 6803 using a glutathione S-transferase (GST) tagging approach (Barker et al., 2008). Our results have revealed that FtsH2 forms a hetero-oligomeric complex with the FtsH3 subunit. Using the GST tag as a marker for the FtsH2 subunit, we have been able to provide direct evidence from negative stain electron microscopy that FtsH2-GST and FtsH3 subunits alternate within a hexameric ring structure.
RESULTS
Phenotype of a GST-Tagged FtsH2 Derivative of Synechocystis 6803
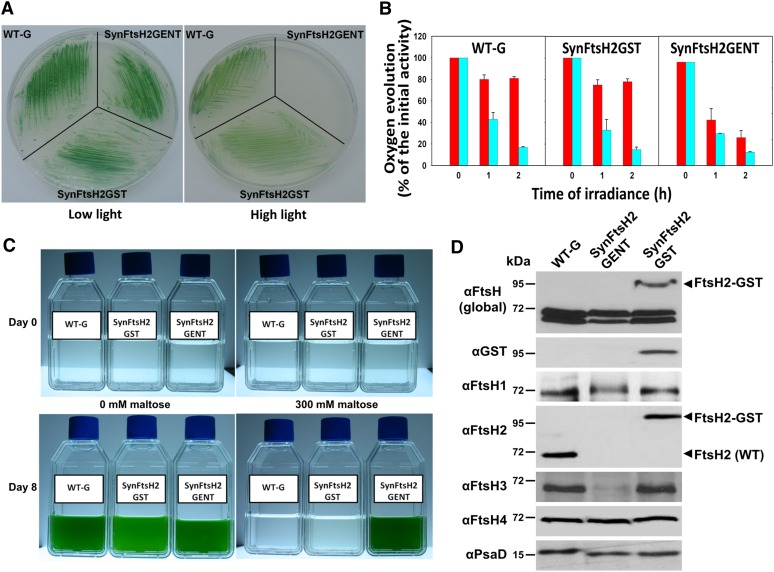
Previous work has shown that E. coli FtsH can tolerate the addition of an affinity tag at the C terminus (Akiyama et al., 1995; Shotland et al., 1997). Consequently, to aid the purification of FtsH2, we constructed a strain of Synechocystis 6803, termed SynFtsH2GST, in which a GST affinity tag that also included a C-terminal Strep II tag, was fused to the C terminus of FtsH2 (see Methods). To probe for potential effects of the GST tag on FtsH2 function, we tested growth of the GST-tagged strain under conditions that are dependent on FtsH2 activity. In contrast with strain SynFtsH2GENT lacking FtsH2, the SynFtsH2GST strain was able to grow under high-light conditions (Figure 1A), with a cellular pigment content indistinguishable from the wild-type strain (WT-G) (see Supplemental Figure 1 online), and to repair PSII as effectively as WT-G as deduced by the ability to maintain PSII activity upon exposure to high irradiances of white light (Figure 1B). The rate of damage to PSII, assessed by determining loss of PSII activity in cells exposed to lincomycin to block protein synthesis, was similar in the WT-G, SynFtsH2GST, and the SynFtsH2GENT strains (Figure 1B). Additionally, the GST-tagged strain, like the WT-G strain, was unable to grow in liquid culture in the presence of 300 mM maltose (Figure 1C). By contrast, SynFtsH2GENT was able to grow because of a perturbation in osmoregulation due to a defect in degrading the soluble enzyme glucosyl-glycerol phosphate synthase involved in the synthesis of the compatible solute glucosyl-glycerol (Stirnberg et al., 2007).
Figure 1.
SynFtsH2GST Expressing FtsH2-GST Behaves Like WT-G.
(A) Growth of WT-G, SynFtsHGENT (lacking FtsH2), and SynFtsH2GST on BG-11 plates under low (5 μE m−2 s−1) and high light (100 μE m−2 s−1).
(B) PSII repair activity of WT-G, SynFtsH2GST, and SynFtsH2GENT assessed by measuring PSII oxygen-evolving activity in cells exposed to 300 μE m−2 s−1 white light at 29°C with or without lincomycin (cyan and red columns, respectively), a protein synthesis inhibitor; initial rates of oxygen evolution were 731 ± 46, 802 ± 60, and 545 ± 43 μmol O2 mg chlorophyll−1 h−1 for WT-G, SynFtsH2GST, and SynFtsH2GENT, respectively (means of two to three measurements for each of the two biological replicates ± se).
(C) Growth of WT-G, SynFtsHGENT, and SynFtsH2GST in BG-11 in either the presence or absence of 300 mM maltose.
(D) Immunoblotting analysis of the different strains using antibodies specific for all FtsH subunits (global FtsH), GST, and each of the FtsH subunits. PsaD was used a loading control. Protein samples were separated by SDS-PAGE using 8% (w/v) polyacrylamide gels lacking urea.
Control immunoblots confirmed the presence of the FtsH2-GST fusion in the membrane and, importantly, that there was no detectable accumulation of FtsH2 liberated by cleavage of the larger fusion protein (Figure 1D). Accumulation of FtsH1 and FtsH4, determined using FtsH-specific antibodies, was not significantly affected by inactivation of FtsH2 (Figure 1D), whereas a dramatic reduction in the amount of FtsH3 was observed in SynFtsH2GENT, which was restored to wild-type levels in the SynFtsH2GST strain (Figure 1D). Together, these data suggested that (1) the FtsH2-GST fusion protein was functional in vivo and still retained the ability to degrade both membrane proteins and soluble targets and (2) accumulation of FtsH3 was heavily dependent on FtsH2, possibly through formation of a common complex.
Purification of FtsH2-GST
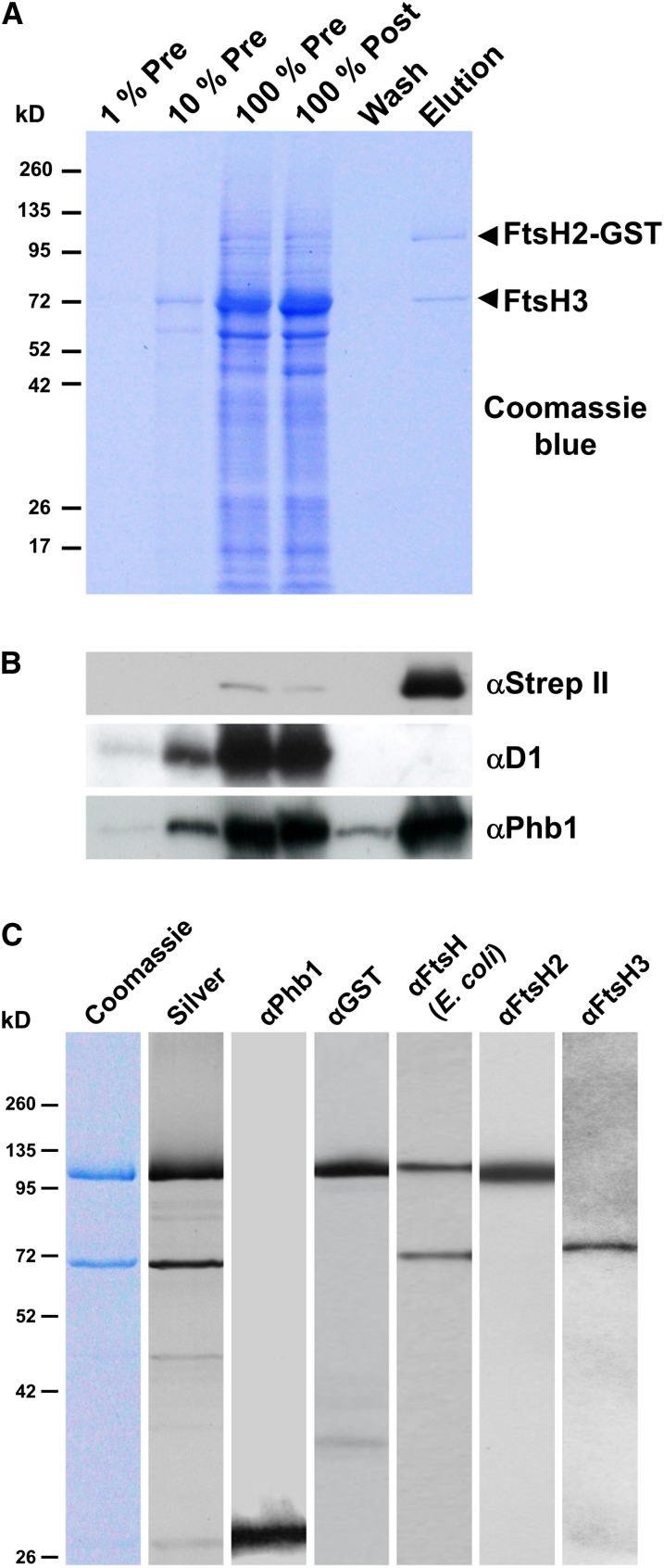
GST-tagged FtsH2 was isolated from detergent-solubilized membrane extracts by binding to glutathione-agarose resin and eluting with reduced glutathione (Figure 2A). SDS-PAGE followed by Coomassie blue staining revealed the presence of two major protein bands in the eluate (Figure 2A). Some minor copurifying proteins could be visualized by silver staining (Figure 2C), but the D1 subunit was not detected (Figure 2B). The upper band migrating at ∼100 kD was detected by antibodies specific for FtsH2, GST, and the Strep II tag (Figures 2B and 2C) and was assigned to the full-length FtsH2-GST fusion protein. This was confirmed by microsequencing that yielded the sequence MKFSXXXALL (where X is an unidentified amino acid), which matched the predicted N-terminal sequence of FtsH2, encoded by slr0228 (MKFSWRTALL). The lower band cross-reacted with antibodies to FtsH but not with antibodies to FtsH2, indicating the presence of a different FtsH subunit(s) (Figure 2C). N-terminal sequencing for this band yielded the sequence SKNNKKXXNA (where X is an unidentified amino acid), which corresponds to the first 10 predicted amino acid residues of FtsH3, encoded by slr1604, after removal of the N-terminal Met residue (MSKNNKKWRNA). This assignment was confirmed in immunoblots using specific antibodies (Figure 2C). No other FtsH subunit was found in this band by mass spectrometry (data not shown). Overall, these data indicated that FtsH2-GST and FtsH3 form hetero-oligomeric complexes in vivo. Interestingly, low levels of the prohibitin homolog, Phb1, encoded by slr1106, were also detected immunochemically (Figures 2B and 2C).
Figure 2.
Affinity Purification of FtsH2-GST.
(A) Detergent-solubilized thylakoid membranes from SynFtsH2GST before chromatography (Pre), the extract after binding to the glutathione resin (Post), the wash fraction just before elution (Wash), and the fraction eluted by glutathione (Elution) were separated by SDS-PAGE. Arrowheads indicate the positions of the FtsH2-GST and FtsH3 proteins.
(B) Analysis of column fractions by immunoblotting with antibodies specific for the Strep II tag (αStrep II), D1 (αD1), and Prohibitin (αPhb1). 100% Pre corresponds to 1 μg chlorophyll a.
(C) Analysis of purified sample by Coomassie blue and silver staining and by immunoblotting with antibodies specific for Prohibitin (Phb1), GST, E. coli FtsH, FtsH2, and FtsH3.
[See online article for color version of this figure.]
FtsH2 also Interacts with FtsH3 in the Wild Type
To confirm that FtsH2 and FtsH3 formed a complex in the parental wild-type strain (WT-G), immunoprecipitation experiments using FtsH2- and FtsH3-specific antibodies were performed. To improve the sensitivity of detection, [35S] radiolabeled samples of WT-G and SynFtsH2GST were used. As expected, GST-tagged FtsH2 and FtsH3 were immunoprecipitated from SynFtsH2GST extracts using both sets of antibodies (Figure 3A). In the case of WT-G, both FtsH2 and FtsH3 were again coimmunoprecipitated (Figure 3A). It must be noted that the relative intensity of the bands detected by autoradiography is not a direct measure of the stoichiometry of FtsH2 and FtsH3 in the complex as it is dependent on a number of factors, including the number of sulfur-containing amino acid residues in each subunit (19 in FtsH2 and 12 in FtsH3).
Figure 3.
FtsH2 Interacts with FtsH3 in WT-G.
(A) Immunoprecipitation analysis performed on radiolabeled crude thylakoid membranes (TM) isolated from SynFtsH2GST and the wild type (WT-G) using the indicated antibodies were analyzed by immunoblotting (Blot) and autoradiography (Autorad). The positions of the FtsH2-GST, FtsH2, and FtsH3 proteins are indicated, * and ** indicate non-FtsH related cross-reactions.
(B) Crude thylakoid membrane proteins isolated from WT-G and mutant slr0228:cmR lacking FtsH2 (FtsH2−) were separated by SDS-PAGE (amounts corresponding to 2.0 µg [100%], 1.0 μg [50%], and 0.5 μg [25%] of chlorophyll a were loaded) and immunoblotted using antibodies specific for FtsH2 and FtsH3. Correct sample loadings were confirmed by detection of cytochrome f.
(C) Membrane proteins isolated from WT-G were separated by 2D-Clear Native (CN)/SDS-PAGE and blotted onto PVDF membrane. The blot was stained by Sypro Orange (Stained blot) and then sequentially probed with antibodies specific for FtsH2, FtsH3, and Phb1 (Blots). Arrows indicate possible FtsH/Phb1 supercomplexes. Positions of unassembled proteins (U.P.), PSII reaction center core lacking CP43 (RC47), monomeric [RCC(1)], dimeric [RCC(2)], and supercomplexes (RCCS1) of PSII are indicated.
The significant reduction in the steady state level of FtsH3, to ∼10% of wild-type levels in the absence of FtsH2 as assessed in immunoblotting experiments (Figure 3B), is largely due to destabilization of FtsH3 in the membrane as determined in radioactive pulse-chase experiments (see Supplemental Figure 2 online), consistent with loss of a stabilizing binding partner for FtsH3. FtsH2 also comigrated with FtsH3 when solubilized membrane extracts were analyzed by two-dimensional Clear Native/SDS-PAGE (Figure 3C). The most abundant complex migrated with an apparent mass of ∼650 kD, larger than the size of 410 kD predicted for a hexamer, which might reflect the presence of additional subunits in the complex in vivo. Smaller amounts of even larger FtsH complexes were also detected as well as a 550-kD complex that comigrated with dimeric PSII [RCC(2)]. The larger FtsH complexes comigrated with Phb1 and might represent FtsH/Phb1 supercomplexes (arrowed in Figure 3C). Overall, these data indicated that untagged FtsH2 also forms a complex with FtsH3 in the wild type.
Analysis of a Mutant Deficient in FtsH3
The ftsH3 gene is vital for cell viability in Synechocystis 6803 (Mann et al., 2000). To gain insights into the physiological role of FtsH3, we constructed a strain, SynFtsH3reg, lacking the chromosomal copy of ftsH3 but containing a plasmid-borne copy of ftsH3 whose expression was driven by the nirA promoter, which can be downregulated by the inclusion of NH4+ in the culture medium (Qi et al., 2005). Addition of 13 mM NH4+ to the medium led to the parallel loss of both FtsH2 and FtsH3 within 3 d, whereas no such disappearance was observed in WT-G cultivated under identical conditions (Figure 4A). Cells of SynFtsH3reg depleted of FtsH3 nearly stopped growing after 4 d of cultivation in the presence of 13 mM NH4+ (see Supplemental Figure 3A online) and showed a decrease in the cellular content of chlorophyll a (see Supplemental Figure 3B online) and in PSII oxygen-evolving activity (see Supplemental Figure 3C online). They were also not able to efficiently degrade the D1 protein during 120 min of exposure to high irradiances of visible light in the presence of chloramphenicol (Figure 4B). Instead, the electrophoretically resolved D1 band became smeared, indicating its oxidation (Lupínková and Komenda, 2004). A similar impairment of growth upon addition of NH4+ was also displayed by ftsH2− mutants (data not shown; Drath et al., 2008). By contrast, under the same conditions, WT-G cells grew normally (see Supplemental Figure 3A online) and their pigment content did not change substantially (see Supplemental Figure 3B online), despite decreased oxygen evolution (see Supplemental Figure 3C online), and cells showed an approximate 50% decrease in the amount of D1 after 30 min of high-light treatment (Figure 4B). Overall, these data provide further support for a close relationship between FtsH2 and FtsH3 and evidence for a role for FtsH3 in degrading damaged D1 during PSII repair.
Figure 4.
Analysis of the SynFtsH3reg Strain.
(A) Immunoblot analysis of crude thylakoid membranes isolated from WT-G and SynFtsH3reg at various times following addition of 13 mM NH4Cl to the growth medium using antibodies specific for FtsH3 and FtsH2. Detection of cytochrome f was used to confirm equal sample loadings. Mutant slr0228:cmR lacking FtsH2 (FtsH2−) was used as a control. All strains were grown in BG11 medium in the presence of 13 mM NH4Cl and 5 mM Glc at an irradiance of 10 μE m−2 s−1. A total of 2 μg of chlorophyll a was loaded per lane.
(B) High light–induced D1 degradation in WT-G and SynFtsH3reg. Cells of WT-G and SynFtsH3reg grown for 7 d in BG11 medium containing 13 mM NH4Cl at an irradiance of 10 μE m−2 s−1 were exposed to high light (500 μE m−2 s−1) in the presence of chloramphenicol (50 μg mL−1), and cells were harvested after various times of illumination for protein analysis using antibody specific for the D1 protein. A total of 2 μg of chlorophyll a was loaded per lane. Detection of cytochrome f was used to confirm equal sample loadings.
(C) Two-dimensional analysis of PSII complexes in cells of WT-G and SynFtsH3reg mutants. WT-G and FtsH3reg cells were grown in BG11 medium without ammonium ions (WT-G 0d and FtsH3reg 0d) and then transferred to the BG11 medium containing 13 mM NH4Cl and cultivated for 5 d (WT-G 5d and FtsH3reg 5d) at an irradiance of 40 μmol photons m−2 s−1. Complexes are designated as in Figure 3C. A total of 5 μg of chlorophyll a was loaded per lane. The arrows designate the RC47 complex, which accumulates more in the mutant than the wild type (arrow 1), the SbtA spot, that accumulates more in the wild type than the mutant (arrow 2), and the NdhH spots of two NDH complexes that accumulate less in the mutant than the wild type (arrows 3 and 4). U.P., unassembled proteins.
To assess the global impact of FtsH3 depletion, membrane proteins isolated from SynFtsH3reg were compared with WT-G. While the two-dimensional pattern of membrane proteins in WT-G cultivated for 5 d in the presence of 13 mM NH4+ was similar to that of the control SynFtsH3reg cells cultivated without NH4+, the protein profile of SynFtsH3reg grown in the presence of 13 mM NH4+ (Figure 4C) showed marked similarities to that of a previously characterized ftsH2− strain (Komenda et al., 2006): The level of the PSII subcomplex lacking CP43, termed RC47, accumulated in both strains due to inhibition of PSII repair-related D1 degradation, and expression of the SbtA protein, which is involved in bicarbonate uptake and whose expression is dependent on FtsH2 (Zhang et al., 2007), was noticeably less than in WT-G and in SynFtsH3reg grown without NH4+. These data show that downregulation of ftsH3 expression caused a similar phenotype to ftsH2− consistent with participation of a hetero-oligomeric FtsH2/FtsH3 in PSII repair and induction of CO2-concentrating mechanisms.
One notable feature seen just in the NH4+-treated SynFtsH3reg cells was a decrease in the level of NDH-1 complexes (Figure 4C), which also participate in the delivery of CO2/bicarbonate into the cells (Zhang et al., 2004). No additional changes to the two-dimensional protein profile were detected when SynFtsH3reg was incubated for up to 7 d in NH4+ (data not shown).
Isolation of a FtsH1/FtsH3 Heterocomplex
Previous mutagenesis experiments have shown that ftsH2 and ftsH4 are dispensable for cell viability, whereas ftsH1 and ftsH3 are crucial (Mann et al., 2000). Given that our data showed the formation of FtsH2/FtsH3 complexes in vivo and that FtsH2, and hence the FtsH2/FtsH3 complex, is not required for cell viability (Mann et al., 2000), we investigated the possibility that FtsH3 might participate in more than one type of FtsH complex. To test this, a fully segregated GST-tagged derivative of FtsH3, as well as GST-tagged versions of FtsH1 and FtsH4, and a FtsH2 control, were isolated by affinity chromatography and analyzed by SDS-PAGE and immunoblotting using FtsH-specific antibodies (Figure 5). The results indicated that FtsH3 forms an FtsH1/FtsH3 heterocomplex as well as the previously described FtsH2/FtsH3 heterocomplex, and that FtsH4 forms a homo-oligomeric complex. N-terminal sequencing of the FtsH4-GST band yielded the sequence AIKPQP that matched the predicted N-terminal sequence of FtsH4, encoded by sll1463 (MAIKPQP). No N-terminal sequence data could be obtained from FtsH1-GST, possibly because the N terminus was blocked.
Figure 5.
Affinity Purification of GST-Tagged FtsH Complexes from Each of the SynFtsHGSTery Mutants.
(A) Purified FtsH-GST proteins were separated by SDS-PAGE and analyzed by silver staining.
(B) Immunoblot analysis using antibodies specific for GST, global FtsH, and each FtsH homolog.
(C) Table summarizing the interactions between the FtsH homologs.
Electron Microscopy and Single-Particle Analysis of the FtsH2-GST/FtsH3 Complex
We used transmission electron microscopy of negatively stained samples to determine the overall dimensions of the isolated FtsH2-GST/FtsH3 complex, the number of protomers present in the complex, and their arrangement within the complex using the GST tag as a means of differentiating the FtsH2 subunits from FtsH3. Although there was some aggregation of the sample on the grid, possibly due to interactions between GST tags on different complexes (Natalello et al., 2008), a sufficient number of negatively stained protein complexes could be visualized in the 18 micrographs used to allow detailed analysis. An asymmetric structural map was obtained by analyzing all discrete two-dimensional projection views observed in the electron micrographs of negatively stained protein complexes (Figure 6A). These views covered a broad range of orientation, including top and side elevations, as well as views that could be attributed to tilting particles (see the Euler angle distribution in Supplemental Figure 4A online). We took advantage of this to calculate a three-dimensional map, or reconstruction, by angular reconstitution (Van Heel, 1987). Figure 6B shows five typical exemplar class averages, taken from a total of 263 class averages, representing 2964 single two-dimensional particles used to construct the final three-dimensional map, progressing from a view attributed to the top elevation through to a view assigned to the side. This three-dimensional map is shown in Figure 6C as a surface-rendered molecular envelope, each at the same orientation as the class averages given in Figure 6B. According to Fourier shell correlation analysis (see Supplemental Figure 4B online), the three-dimensional map has a resolution of ∼26 Å assuming asymmetry and Fourier-shell correlation at 3σ with a conservative correlation coefficient of 0.5. It is apparent from the comparison of the two-dimensional class averages (Figure 6B) with the post-three-dimensional reconstruction, two-dimensional reprojection images (Figure 6D) that a majority of orientations were incorporated into the three-dimensional map.
Figure 6.
Single-Particle Analysis of the GST-Tagged FtsH2/FtsH3 Complex.
(A) Typical region in a micrograph of negatively stained FtsH2-GST complexes. FtsH particles are circled. Additional images showing this complex can be viewed in Barker et al. (2008). Bar = 50 nm.
(B) A selection of five characteristic two-dimensional views of the FtsH2-GST complex, taken from a total of 263 different class averages.
(C) Surface-rendered views of the final three-dimensional map calculated by angular reconstitution viewed from the same angles as presented in (B). Positions of density attributed to three GST tags are indicated by arrows and colored in pink for one of the views.
(D) Two-dimensional reprojections of the views shown in (C).
Bar in (D) = 10 nm for (B) to (D).
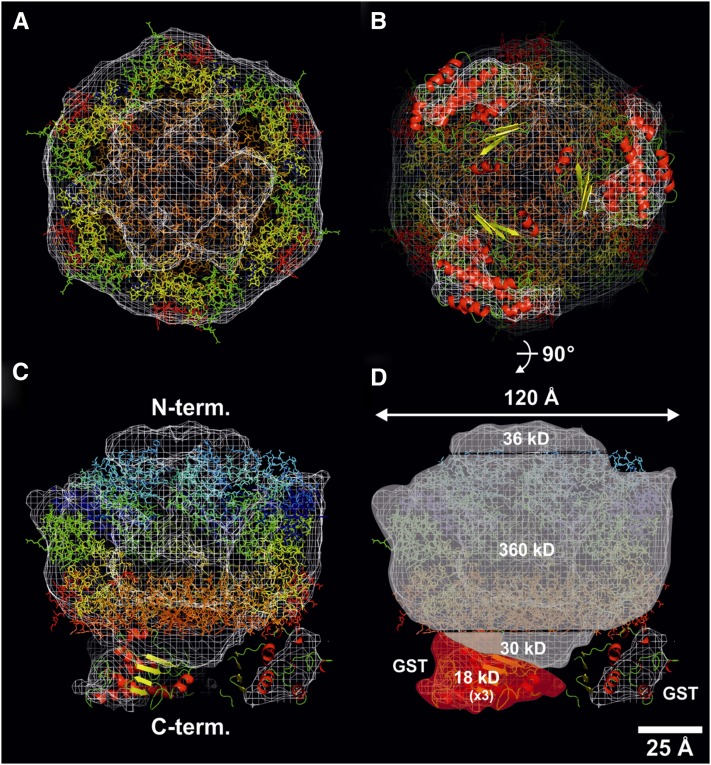
To aid interpretation of the three-dimensional reconstruction, the crystallographically derived hexameric structure of a soluble fragment of FtsH, containing residues 146 to 603 of the 610–amino acid sequence of Thermotoga maritima FtsH (Bieniossek et al., 2009), encompassing the conserved AAA domain and PD, was modeled by visual inspection into the molecular envelope, contoured at a threshold of 2.5 σ (Figure 7A). The crystal structure fitted remarkably well into the central core, providing clear evidence to support the presence of six FtsH subunits in the GST-tagged FtsH complex. The model shows a central ball-shaped complex of ∼120 Å in diameter, with three minor densities extending out from one side, which could be attributed to the presence of three GST tags, which were fitted using an atomic structure (Figures 7B and 7C). The location of the GST tags with respect to the central body of the complex suggests an alternating arrangement of three FtsH2-GST and three FtsH3 subunits in a hexameric complex (Figure 7B).
Figure 7.
Three-Dimensional Modeling of the GST-Tagged FtsH2/FtsH3 Complex.
An isomesh rendered molecular envelope, contoured with a threshold of 2.5 σ for the final calculated asymmetric three-dimensional map, is shown in white. The modeling in of the crystallography-derived hexameric soluble fragment of apoFtsH from T. maritima (3KDS.pdb) was done by visual inspection with atoms colored as a spectral rainbow from the N terminus (blue) to the C terminus (red).
(A) A 5-nm-thick cross section viewed from the C-terminal PDs of 3KDS.pdb downwards, revealing its hexameric nature.
(B) A 4-nm-thick cross section, viewed from the GST tags upwards, toward the C-terminal domains of the hexameric biological assembly of 3KDS.pdb.
(C) A 5-nm-thick cross section, viewed from the side; the best fit for the six FtsH protomers is shown with the N-terminal AAA+ domain uppermost.
(D) Four regions shown include an N-terminal cap, whose volume is calculated to be ∼36 kD (assuming 0.844 Å3 per D at 2.5 σ), a central core of 360 kD, a C-terminal bottom of 30 kD, and three smaller outlying domains of 18 kD. In (B) to (D), these outlying domains, closest to the C-terminally orientated central core 3KDS.pdb file, are assigned to the GST tag (1GTA.pdb). Three such domains are observed, even though no threefold symmetry operators were applied during angular reconstitution. The maximum diameter of the complex was observed to be 120 Å. Bar = 25 Å.
The fusion of the GST tag to the C terminus of FtsH2 also allowed us to determine the orientation of the complex with respect to the N and C termini (Figure 7C). The central core of the complex has a maximum diameter of 120 Å, close to the value of 130 Å determined by cryo-EM for the m-AAA protease (Lee et al., 2011). By assuming a protein density of 0.844 Å3 per Dalton (van Heel et al., 1996), masses of 360 kD could be assigned to the central portion containing the AAA module and PD, 36 kD to the N-terminal cap consisting of the transmembrane helices and lumenal loops, 18 kD to the three outlying domains assigned to the 27-kD GST tag, leaving a C-terminal bottom region of 30 kD (Figure 7D) likely to contain the C-terminal region of FtsH, which is relatively disordered and not present in the T. maritima crystal structure used in the fitting, plus possibly some of the GST tag. Thus, the overall mass of the isolated FtsH2-GST/FtsH3 complex using the above assumptions was estimated to be 480 kD, close to the theoretical mass of 489 kD.
DISCUSSION
Previous work has identified an important role for FtsH2 in PSII repair and in other quality control processes in the thylakoid membrane of Synechocystis 6803 (reviewed in Nixon et al., 2010). We show here that FtsH2 forms a hetero-oligomeric complex with FtsH3. This conclusion is based on a number of independent lines of evidence, including copurification of FtsH2 and FtsH3 by affinity chromatography (Figure 2), coimmunoprecipitation of FtsH2 and FtsH3 (Figure 3A), comigration of FtsH2 and FtsH3 by native gel electrophoresis (Figure 3C), and an enhanced rate of degradation of FtsH3 in the absence of FtsH2 (see Supplemental Figure 2 online), leading to reduced accumulation of FtsH3 (Figure 3B). Repression of FtsH3 synthesis also led to decreased FtsH2 accumulation in the membrane (Figure 4A) and a similar impairment in D1 degradation as observed earlier in ftsH2− mutants (Figure 4B). Overall, these data indicate that the major FtsH complex involved in degrading D1 during PSII repair in Synechocystis 6803 is an FtsH2/FtsH3 heterocomplex. Other physiological processes currently assigned to FtsH2, such as a role in osmoregulation and carbon concentrating mechanisms, are also likely to be dependent on the activity of the FtsH2/FtsH3 hetero-oligomer.
In addition, our structural analysis of the FtsH2-GST/FtsH3 heterocomplex (Figure 7) has provided new insights into the subunit organization of this class of FtsH complex. The combination of structural data obtained by single-particle electron microscopy and the available high-resolution crystal structures for the conserved soluble domains of FtsH allowed us to conclude that the number of FtsH subunits within the complex was six. The additional GST tag of ∼27 kD was of sufficient size for it to be detected in the transmission electron microscopy reconstructions of the complex, thereby providing information on the location of the FtsH2 subunits within the complex. A similar experimental strategy involving the use of yellow fluorescent protein fusions has also been exploited to study the location of subunits within the cyanobacterial NDH-1 complex (Birungi et al., 2010). Antibodies can also be used to identify subunits during single-particle analysis, but there can be significant experimental drawbacks, including partial decoration of the complex (Perálvarez-Marín et al., 2011). Our structural model suggests that the FtsH2-GST and FtsH3 subunits alternate within the complex. Given the averaging procedure used to generate the final structure, a random positioning of the subunits within the hexamer would not have given rise to the three additional densities that we assign to the GST tag but instead would have been averaged out over the ring. The equimolar ratio of the FtsH2-GST and FtsH3 subunits in the FtsH2/FtsH3 complex is also broadly in line with the relative intensities of the Coomassie blue–stained bands detected by SDS-PAGE (Figure 2), given the limitations of using protein stains to estimate protein levels, especially when one protein is fused with GST (Tal et al., 1985).
We considered the possibility that the alternating subunits observed in the FtsH2-GST/FtsH3 complex might reflect steric constraints imposed by the presence of the GST tag, which would prevent the location of two tagged FtsH2 subunits next to each other. However, there are important arguments speaking against this possibility. First, the strain expressing FtsH2-GST/FtsH3 behaves like the wild type (Figure 1). This shows that the protease fulfills its physiological function well, and it is difficult to imagine that the protease with incorrectly arranged subunits would do so. Second, structural models of the GST-tagged complex allow accommodation of a GST tag even on each subunit without significant steric problems.
Sequencing of the isolated FtsH2 and FtsH3 subunits by Edman degradation indicated that there is no removal of a signal peptide during integration of the subunits into the thylakoid membrane. Consequently, FtsH2 and FtsH3 are likely to contain two transmembrane helices in the N-terminal region connected by short lumenally exposed loops of ∼80 to 100 residues, which might be involved in recognizing substrates, such as the D1 subunit (Bailey et al., 2002). The recent structural model for the m-AAA protease obtained by cryo-EM has highlighted the fact that the short loops lying on the opposite side of the membrane to the AAA+ and PD are actually splayed out. This feature is absent in our structural model most likely due to a combination of limited resolution and the fact that the loop regions seem to be more flexible than the rest of the complex (Lee et al., 2011).
There are reports of mitochondrial FtsH subunits participating in both homo- and hetero-oligomeric complexes (Koppen et al., 2007; Piechota et al., 2010) with the relative proportions in some cases dependent on cell type (Koppen et al., 2007). Consequently, we do not exclude the possibility that under certain growth conditions FtsH2 might form significant amounts of other types of FtsH2 complex, such as homocomplexes, in addition to the FtsH2/FtsH3 complex described here. The fact that FtsH3 is required for cell viability whereas FtsH2 is dispensable (Mann et al., 2000) suggests that FtsH3 is not restricted to forming complexes with FtsH2. The data in Figure 5 show that FtsH3 also forms a complex with FtsH1. As accumulation of FtsH3 is reduced to much <25% of wild-type levels in the absence of FtsH2 (Figure 3B), the amount of the FtsH1/FtsH3 complex appears to be substantially less than that of the FtsH2/FtsH3 complex. However, given that both FtsH1 and FtsH3 are vital for cell viability (Mann et al., 2000), the FtsH1/FtsH3 complex is likely to play a crucial role in the cell, although we cannot exclude the possibility that FtsH1 and FtsH3 also form some homocomplexes in addition to the heterocomplex. By contrast the FtsH4 subunit does not copurify with any of the other FtsH subunits and seems to form a homo-oligomer. Future work will clarify the location and substrate specificity of these different types of FtsH complex.
The formation of FtsH2/FtsH3 and FtsH1/FtsH3 hetero-oligomers in the cyanobacterium Synechocystis 6803 has close parallels with current views on the structure of FtsH complexes in chloroplasts. In the case of Arabidopsis, genetic and biochemical data have been interpreted in terms of the formation of hetero-oligomeric FtsH complexes with the most abundant complexes composed of FtsH subunits found within two distinct branches of the phylogenetic tree: type B subunits comprising At-FtsH2 and At-FtsH8 and type A subunits comprising At-FtsH1 and At-FtsH5 (Sakamoto et al., 2003; Yu et al., 2004; Zaltsman et al., 2005). In this regard, it is important to note the FtsH1 and FtsH2 subunits found in Synechocystis 6803 are most closely related to the chloroplast type B subunits, whereas FtsH3 is more related to the type A subunits (Sakamoto et al., 2003). As yet, no FtsH complex has been isolated from chloroplasts and characterized, which has led to much discussion about their possible subunit composition and organization (e.g., Zhang et al., 2010). Given the close evolutionary relationship between cyanobacteria and chloroplasts, it seems reasonable that the hetero-oligomeric complexes found in chloroplasts also contain three of each type of subunit arranged in an alternating fashion around the ring. However, recent work has concluded that chloroplast FtsH heterocomplexes might be composed of two type A and four type B subunits, but deviations from a 1:1 stoichiometry because of contamination by FtsH monomers or partially assembled FtsH complexes could not be excluded (Moldavski et al., 2012). The single-particle approach described here helps overcome this potential problem.
By contrast, the FtsH4 subunit of Synechocystis 6803 appears to form homocomplexes (Figure 5). Phylogenetic analyses suggest that FtsH4 is most closely related to the At-FtsH7 and At-FtsH9 nucleus-encoded chloroplast-targeted FtsH subunits (Sakamoto et al., 2003; Yu et al., 2004), about which little is currently known. The advantage of expressing heterocomplexes rather than homocomplexes remains to be clarified, although a role in determining substrate specificity has been suggested (Koppen et al., 2007).
FtsH hexameric complexes can form larger supercomplexes, of unknown function, with members of the Band 7 family of proteins, including HflK/C in E. coli and prohibitins in mitochondria (Ito and Akiyama, 2005). Preliminary data presented here (Figures 2B and 3C) suggest that FtsH/Phb1 supercomplexes might also accumulate in Synechocystis 6803. The relatively low levels of Phb1 found in the affinity-purified FtsH2-GST/FtsH3 preparation could reflect the transient nature of the complex or sensitivity of the complex to detergent treatment (Boehm et al., 2009).
How FtsH complexes function is still unclear. The recent cryo-EM data obtained for the yeast m-AAA complex (Lee et al., 2011) hint toward the possibility that the hexameric complexes might contain a 13-Å gap between the N-terminal transmembrane segment and the AAA+ domain through which unfolded, but not folded, polypeptides could be guided to the central pore loops of the AAA ring. The distance between a proposed initial substrate binding site and the central pore region (Lee et al., 2011) is compatible with a requirement for target substrates to possess an unfolded segment of 20–amino acid residues or more (Chiba et al., 2000). Once engaged with the FtsH complex, ATP-driven conformational changes within the AAA ring would drive the movement of the unfolded target protein into the proteolytic chamber for subsequent highly processive degradation into small peptides, which then exit through ∼25-Å lateral openings within the body of the complex (Lee et al., 2011). We have shown that degradation of D1 during PSII repair in Synechocystis 6803 is also dependent on the length of the N-terminal tail of D1 (Komenda et al., 2007). Consequently, our current view is that D1 degradation in Synechocystis 6803 is mainly initiated from the N terminus and proceeds in a highly processive manner without formation of distinct degradation products (Nixon et al., 2010).
In summary, we isolated a hetero-oligomeric FtsH complex of the thylakoid membrane involved in PSII repair. Our data confirm that the isolated complex is hexameric and provide direct evidence for an alternating arrangement of the two subunits within the complex. Ultimately, a detailed understanding of the complex will require a high-resolution structure probably determined by x-ray crystallography. Given the low amounts of the natural complex that can be purified, this will most likely require high-level coexpression of FtsH2 and FtsH3 in a heterologous system.
METHODS
Cyanobacterial Strains and Growth Conditions
All strains used in this study are derived from the Glc-tolerant Synechocystis sp PCC 6803 strain (WT-G) (Williams, 1988). Strain SynFtsH2GENT is equivalent to the strain Syn0228GENT that contains a deletion of the 3′ end of the ftsH2 (slr0228) gene and replacement by a gentamycin resistance cassette (Komenda et al., 2006). Strain slr0228:cmR contains the insertion of a chloramphenicol resistance cassette into the ftsH2 (slr0228) gene (Komenda et al., 2006). Unless stated otherwise, strains were grown in liquid BG-11 mineral medium or on solid BG-11 plates containing 1.5% (w/v) agar, both containing 5 mM TES-KOH, pH 8.2, and supplemented with 5 mM Glc, at a light intensity of 20 µE m−2 s−1 white fluorescent light and at 29°C. Light sensitivity growth assays were conducted on plates at low (5 µE m−2 s−1) and high (100 µE m−2 s−1) light intensities, and osmoregulation was assessed in liquid cultures by the addition of 300 mM maltose to BG-11 mineral medium.
Construction of the FtsH2-GST–Tagged Strain (SynFtsH2GST)
To tag FtsH2 with GST, strain SynFtsH2GENT was transformed with plasmid p0228GSTSTREP that contains an in-frame fusion of the coding sequence for GST and a STREP II tag to the 3′-end of ftsH2 (slr0228), plus flanking DNA to facilitate homologous recombination into the genome. Plasmid p0228GSTSTREP was constructed in three steps (see Supplemental Figure 5 online). First, the last third of the slr0228 gene plus some downstream region (genome coordinates: 2530235-2531612) were amplified using the 0228fd (5′-GGGGGATCCGGACCGGGTGGTAGCTGGTAT-3′) and the 0228rv (5′-GGGGGTACCATGGCATCCTCCGTTGCAATT-3′) primers. The resulting DNA fragment was cloned into the pGEM-T Easy vector (Promega) to yield p0228. To generate the p0228GSTSTREP plasmid, the intermediate p0228TEVHISSTREP plasmid was produced from p0228 as follows. The coding sequence for a tobacco etch virus protease site (TEV; encoding ENLYFQG) followed by a nona-His tag (His9) and a STREP-II affinity tag (encoding WSHPQFEK) were introduced after the penultimate slr0228 codon by overlap extension PCR (Ho et al., 1989). The primers in the first two separate reactions were 0228fd with 0228tevhisstprv (5′-CACGTGATGGTGATGGTGATGGTGATGGTGGCCTTGAAAATAGAGATTTTCAGCGCTTAGTTGGGGAATTAACTGTTC-3′) and 0228tevhisstpfd (5′-TATTTTCAAGGCCACCATCACCATCACCATCACCATCACGTGGTATACGGATCCAATTGGTCCCATCCC-3′) with 0228rv using the p0228 plasmid as template DNA in both reactions. The second PCR reaction used the 0228fd and 0228rv primers with the previous two PCR reaction products as DNA template yielding p0228TEVHISSTREP after ligation into the pGEM-T Easy vector. In the last step, the p0228TEVHISSTREP plasmid was digested in a two-step process to replace the sequence between the Eco47III and BamHI restriction sites with the gst gene. The gst gene was amplified from the pGEX-6P-3 vector (GE Healthcare) using PCR with the GSTfd (5′-GGGGTATACATGTCCCCTATACTAGGTTAT-3′; introduces blunt cutter Bst1107I at the 5′-end) and GSTrv (5′-GGGGGATCCATCCGATTTTGGAGGATGGTC-3′; introduces BamHI at the 3′-end) primer pair. The resulting PCR fragment was digested with Bst1107I and BamHI restriction enzymes. Also, p0228TEVHISSTREP was digested using BamHI and AlwNI (cuts once in the pGEMTeasy vector) to yield three fragments of 2146, 1638 (fragment a), and 739 bp. In a second reaction, AlwNI and Eco47III (blunt cutter) were used to digest p0228TEVHISSTREP into two fragments of 2824 (fragment b) and 1699 bp. After these digestions, the fragments a and b and the gst gene were ligated together, yielding the p0228GSTSTREP vector. This process was necessary due to the presence of two BamHI sites in the p0228TEVHISSTREP plasmid. After transformation of the p0228GSTSTREP vector into Syn0228GENT, transformants were selected for restored ability to grow under high-light conditions (100 µE m−2 s−1). A resulting high-light resistant strain, termed SynFtsH2GST, was confirmed by PCR and restriction digestion to contain the correct slr0228:gst fusion (see Supplemental Figure 6 online).
Construction of the Erythromycin-Resistant GST-Tagged Strains of FtsH
A universal tagging cassette (see Supplemental Figure 7B online) consisting of the coding sequences for the thrombin cleavage site, GST, and strep II tag and a selectable marker conferring erythromycin resistance was made to enable a simple, one-step tagging strategy. The gst gene was amplified from pGEX-6P-3 (GE Healthcare) using the GST-F/R primer pair (see Supplemental Table 1 online) and cloned into pGEM-T Easy (Promega) to yield the intermediate vector pGST (see Supplemental Figure 7A online). An erythromycin resistance marker (Elhai and Wolk, 1988) was then ligated into pGST via the HpaI site to generate pGST-ErmA (see Supplemental Figure 7C online).
To construct the transformation vectors, the full-length ftsH open reading frame and 655 bp of downstream DNA sequence were used as the flanking sequence to facilitate homologous recombination into the genome. To enable insertion of the gst-tagging cassette, overlap-extension PCR was used to introduce an EcoRV and an XbaI site immediately before the STOP codon (see Supplemental Figure 8A online). In the first step of overlap extension PCR, two separate reactions were performed using primer sets FtsH-F/FtsH-OE-R and FtsH-OE-F/FtsH-R for each ftsH gene (see Supplemental Table 1 online), with genomic DNA from WT-G Synechocystis 6803 as template. In the second step, PCR fragments from the previous reactions were used as DNA template, together with primer set FtsH-F/R, with the resulting PCR products then cloned into pGEM-T Easy vector to create pGEMFtsHx vectors where FtsHx is the particular FtsH subunit (see Supplemental Figures 8B and 8C online). Consequently, the gst-tagging cassette, released from pGST-ErmA using EcoRV and SpeI, was ligated into each of the pGEMFtsHx vectors via EcoRV and XbaI sites to yield the four final transformation vectors pFtsHxGSTery, where FtsHx represents the particular FtsH subunit (see Supplemental Figures 8D and 8E online). After transformation of WT-G, transformants were selected for erythromycin resistance (15 µg mL−1) and complete segregation was confirmed by PCR (see Supplemental Figure 9 online). The resulting mutants are termed SynFtsHxGSTery, where FtsHx represents the particular FtsH subunit.
Construction of the SynFtsH3reg Strain
For regulatable expression of FtsH3 under control of the nirA promoter from Synechococcus sp PCC 7942, the ftsH3 (slr1604) gene was cloned in frame into the pCER20 plasmid using NdeI and XbaI restriction sites (Qi et al., 2005). The resulting plasmid (pFtsH3reg), able to replicate autonomously in Synechocystis, was transformed into the WT-G strain of Synechocystis 6803 via triparental mating (Elhai and Wolk, 1988), and transformants were selected on medium supplemented with gentamycin at 10 μg mL−1.
To delete the wild-type copy of the ftsH3 (slr1604) gene, a linear deletion construct was prepared replacing most of the ftsH3 gene (nucleotides 161 to 1691) with an erythromycin resistance cassette using a megaprimer PCR method as described by Dobáková et al. (2009). This construct containing 600 bp upstream and downstream regions of the ftsH3 gene with the erythromycin resistance cassette (obtained from plasmid pPV142 of Staphylococcus simulans) in the middle was constructed in two steps using long fusion primers complementary to the ftsH3 gene in one direction and the erythromycin resistance cassette in the other. This linear deletion construct was used for transformation of Synechocystis 6803 WT-G containing plasmid pFtsH3reg. Transformants were selected and segregated on erythromycin-containing agar plates, and full segregation was confirmed by PCR. Unlike in the wild type, deletion of ftsH3 was possible in the strain containing plasmid pFtsH3reg, showing functional expression of FtsH3 from this plasmid. The obtained mutant was designated SynFtsH3reg. To suppress expression of FtsH3, the strain was initially grown in BG-11 medium containing no ammonium ions (ferric ammonium citrate replaced by ferric citrate) and after reaching an OD750 of 0.5 (exponential phase of growth), the culture was diluted with BG-11 medium containing 17 mM NH4Cl instead of NaNO3 to give a final concentration of ∼13 mM NH4Cl. The culture was diluted every day with this medium to OD750 0.2 in order to maintain sufficient ammonium in the medium.
Preparation of Crude Thylakoid Membranes
Crude Synechocystis thylakoid membranes for gel electrophoresis (SDS-PAGE) analyses were prepared by glass bead (212 to 300 µm in diameter) breakage at 4°C followed by differential centrifugation (Boehm et al., 2009). Chlorophyll a content was determined by extraction into methanol and absorption measurements at 666 and 750 nm (Komenda and Barber, 1995).
Spectroscopic and Polarography Methods
Measurement of chlorophyll concentrations after methanol extraction and absorption spectra of cells in vivo were measured using a Shimadzu UV3000 spectrophotometer, and the light-saturated steady state rate of oxygen evolution in cell suspensions was assessed polarographically in a thermostated chamber at 29°C in the presence of artificial electron acceptors p-benzoquinone (0.5 mM final concentration) and potassium ferricyanide (1 mM final concentration), all as described by Komenda et al. (2007).
Assessment of PSII Repair Efficiency
Cultures (50 mL) diluted to 2 µg chlorophyll/mL were shaken in 250-mL Erlenmeyer flasks and exposed to photoinhibitory light (300 µE m−2 s−1) over a period of 120 min either in the presence or absence of the protein synthesis inhibitor lincomycin (100 µg/mL). The PSII oxygen-evolving activity of samples taken during the time course was measured in the presence of artificial electron acceptors (see above).
SDS-PAGE and Immunoblotting
Unless stated otherwise, membrane protein samples and selected fractions of the GST affinity purification procedure were separated on 10% (v/v) denaturing one-dimensional SDS-PAGE gels containing 6 M urea as described previously (Boehm et al., 2009). Gels were either stained with Coomassie Brilliant Blue R 250 or by silver staining (Blum et al., 1987) or electroblotted onto polyvinylidene difluoride (PVDF) membrane using the iBlot system (Invitrogen). Immunoblotting analyses were performed using specific primary antibodies and anti-rabbit or anti-mouse horseradish peroxidase–conjugated secondary antibodies respectively (both GE Healthcare). Signals were visualized using a chemiluminescent kit (SuperSignal West Pico; Pierce). Primary antibodies used in this study were as follows: (1) a polyclonal antibody specific for Escherichia coli–expressed recombinant GST from Schistosoma japonicum generated in rabbit (Sigma-Aldrich), at 1 in 2000 dilution; (2) a monoclonal antibody specific for Strep-tag II generated in mouse (Qiagen), at 1 in 2000 dilution; (3) a polyclonal antipeptide antibody raised against residues 297 to 312 of E. coli FtsH (Tomoyasu et al., 1993), which is potentially cross-reactive with all Synechocystis sp PCC 6803 FtsH homologs, kindly provided by Teru Ogura (University of Kumamoto, Japan); (4) polyclonal antipeptide antibodies raised against residues 578 to 592 of FtsH1, residues 98 to 115 of Synechocystis FtsH2, residues 59 to 75 of Synechocystis FtsH3, all at 1 in 1000 dilution, and residues 556 to 574 of Synechocystis FtsH4, at 1 in 10,000 dilution; (5) a rabbit polyclonal antiserum (#304-F) raised against residues 325 to 353 of precursor D1 from pea (Pisum sativum; Nixon et al., 1990) at a dilution of 1 in 5000; (6) a rabbit polyclonal antiserum (global FtsH) raised against residues 286 to 304 of FtsH2, which are conserved in all four FtsH subunits, at a dilution of 1 in 10,000; (7) a rabbit antiserum raised against the prohibitin homolog, Phb1, encoded by slr1106 (Boehm et al., 2009); (8) a rabbit antiserum raised against Chlamydomonas reinhardtii PsaD, kindly provided by Jean-David Rochaix, used at a dilution of 1 in 5000. The cytochrome f subunit of the cytochrome b6f complex was detected by directly incubating unblocked blots with chemiluminescent reagents.
Two-Dimensional Gel Electrophoresis
Isolated membranes were solubilized with 1% n-dodecyl-β-d-maltoside (β-DM) and analyzed by two-dimensional electrophoresis. The first step was performed at 4°C through a 5 to 14% polyacrylamide gel using a clear variant of the blue-native PAGE described by Schägger and von Jagow (1991) in which Coomassie Brilliant Blue was omitted from all solutions and the upper electrophoresis buffer contained instead 0.05% sodium deoxycholate and 0.02% β-DM. Samples containing 5 μg of chlorophyll were loaded in each lane. The whole lane was excised from the native gel, incubated for 30 min in 25 mM Tris-HCl, pH 7.5, containing 1% SDS, and placed on the top of a denaturing linear 12 to 20% gradient polyacrylamide gel containing 7 M urea (Komenda et al., 2005). Proteins separated in the gel were either stained by Coomassie Brilliant Blue or transferred onto a PVDF membrane for immunoblotting.
Affinity Purification of FtsH2-GST
The cells of a 10-liter culture of SynFtsH2GST, grown at ∼100 µE m−2 s−1 and bubbled with air, were first concentrated to ∼1 liters using a tangential flow cell concentrator and then harvested by centrifugation (Sorvall RC6+; F10; 9000 rpm; 15 min; 4°C). The cell pellet was washed in 500 mL KPN buffer (40 mM K-phosphate, pH 8.0, 100 mM NaCl, and 1 mM DTT) and finally resuspended in 50 mL KPN buffer (chlorophyll a concentration = 1 mg/mL) supplemented with one tablet of complete EDTA-free protease inhibitor (Roche Diagnostics). Cells were broken by being passed twice through a French Press at 1250 p.s.i., and intact cells were removed by centrifugation (Sorvall RC6+; F21; 5000 rpm; 5 min; 4°C). Crude thylakoid membranes were then harvested by ultracentrifugation (Beckmann L8-70M; Ti45; 38,500 rpm; 60 min; 4°C) and resuspended in buffer A (50 mM HEPES, pH 7.2, 1.2 M betaine, 5% [v/v] glycerol, 100 mM NaCl, 5 mM MgCl2, 10 µM ZnCl2, and 1 mM DTT; chlorophyll a concentration = 1 mg/mL). A 10% (w/v) β-DM stock solution in buffer A was added to solubilize the membrane protein complexes at a final concentration of 1% (w/v) β-DM for 30 min on ice with occasional gentle agitation. Insolubilized material was pelleted (Beckmann L8-70M; Ti70; 31,700 rpm; 30 min; 4°C) and the chlorophyll a concentration determined. Swollen glutathione resin (5 mL; Sigma-Aldrich), equilibrated with 60 mL buffer A, was then added to the sample. After an incubation period of 3 h on a rotating wheel, the sample was transferred to a Proteus one-step batch midi spin column (Generon) and washed with GST washing buffer (50 mM HEPES, pH 7.2, 5% [v/v] glycerol, 100 mM NaCl, 5 mM MgCl2, 10 µM ZnCl2, 1 mM DTT, and 0.03% [w/v] β-DM) until the washes were clear. A last wash was performed with 10 mL GST washing buffer for one-dimensional SDS-PAGE analysis. FtsH2-GST protein complexes were eluted twice with 5 mL GST elution buffer (50 mM Tris/HCl, pH 8.0, 80 mM NaCl, 5 mM MgCl2, 12.5 µM ZnCl2, 0.005% [w/v] β-DM, 25 mM reduced glutathione, and 1.4 mM β-mercaptoethanol, added freshly). The eluted fractions were then pooled and concentrated to 1 mL using a Vivaspin 20 protein concentrator (Sartorius Stedim Biotech) with a 100 kD molecular mass cutoff.
Radioactive Labeling
For coimmunoprecipitation, cells were radioactively labeled according to Komenda et al. (2005). An amount of cells containing 75 μg of chlorophyll a was resuspended in 250 all of BG-11, incubated under shaking at 60 μE m–2 s–1 for 30 min and then mixed with l-[35S]Met and l-[35S]Cys (trans-label, MP Biochemicals, B; >1000 Ci mmol–1; final activity 400 μCi mL–1). The suspension was then exposed to 500 μE m–2 s–1 at 29°C for 30 min, and after the incubation period, cells were frozen in liquid nitrogen to be used for crude thylakoid isolation. Pulse-chase experiments were performed as described by Komenda et al. (2006).
Coimmunoprecipitation of FtsH2 and FtsH3 from Crude Thylakoid Membranes
The coimmunoprecipitation experiment was performed using a modified method described by Komenda et al. (2005). Isolated membranes (10 μg of chlorophyll) were resuspended in 25 mM MES-NaOH, pH 6.5, containing 10 mM CaCl2, 10 mM MgCl2, and 25% glycerol (B buffer) and solubilized by the addition of 10% β-DM (β-DM to chlorophyll a ratio 20:1 [w/w], 0.4% β-DM final concentration). After pelleting insolubilized material, the supernatant was incubated overnight with antibody raised against FtsH2 or FtsH3 (dilution 15×) at 4°C. Samples were then incubated with protein A–Sepharose 4B (Sigma-Aldrich) for 1 h at 4°C and protein A–Sepharose bound protein immunoglobulin complexes were pelleted. The resin was washed twice with 1 mL of B buffer containing 0.1% (w/v) β-DM and twice with B buffer. Protein complexes were eluted from the resin at 50°C with 1× SDS sample buffer (25 mM Tris-HCl, pH 7.5, 1 M Suc, 2% [w/v] SDS, and 2% [w/v] DTT) and analyzed by SDS-PAGE. Separated proteins were transferred onto a PVDF membrane, which was then dried and exposed to a phosphor imager plate (GE Healthcare) for 3 d. The membrane was then used for immunodetection of FtsH2 and FtsH3.
Protein Sequencing
N-terminal protein sequencing was performed by the University of Leeds Protein Sequencing Facility, the Medical Research Council Laboratory of Molecular Biology, and the University of Cambridge Protein and Nucleic Acid Chemistry Facility, according to their instructions.
Single-Particle Analysis
Samples were applied to glow-discharged copper grids and negatively stained with 2% (w/v) uranyl acetate. Images were recorded at room temperature using a Philips CM100 TEM, operating at 80 kV and ×50,850 magnification. Micrographs were chosen for minimal astigmatism/drift and scanned using a Nikon LS9000 densitometer. Fourier power spectra for each micrograph displayed first minima in the range of 19 to 21 Å. A data set of ∼3400 particles was compiled using “boxer” of the EMAN software package (Ludtke et al., 2004). Further processing was performed using Imagic-5 (Image Science) at a sampling frequency of 2.5 Å/pixel on the specimen scale. Reference-free alignment, multivariate statistical analysis, and iterative refinement resulted in two-dimensional class averages (Ruprecht and Nield, 2001). Eulerian angles were then assigned a priori by angular reconstitution (Van Heel, 1987) (see Supplemental Figure 4 online) and iterative refinements implemented. The resolution of the final three-dimensional map, which comprised 263 class averages, merged from a broad range of relative orientation subpopulations representing 2964 particles, was estimated conservatively (0.5 correlation coefficient) by Fourier shell correlation (van Heel and Schatz, 2005). Reprojections were taken from the final three-dimensional model and used to identify atypical views and further refine averages.
Coordinate data sets were obtained from the Research Collaboratory Structural Bioinformatics Data Bank (www.rcsb.org) for 3KDS.pdb (structure of the cytosolic region of the Thermotoga maritima FtsH protease at 2.60 Å; Bieniossek et al., 2009) and 1GTA.pdb (structure of GST at 2.4 Å; McTigue et al., 1995). Structures were modeled into the final calculated three-dimensional map using PyMol (DeLano, 2008). Surface-rendered views are shown with a threshold of 2.5 σ.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: FtsH1 (Slr1390), NC_000911.1; FtsH2 (Slr0228), Q55700.1; FtsH3 (Slr1604), NP_440330.1; and FtsH4 (Sll1463), NC_000911.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Absorption Spectra of Photoautotrophically Grown Cells.
Supplemental Figure 2. Enhanced Degradation of FtsH3 in Strains Lacking FtsH2.
Supplemental Figure 3. Phenotype of SynFtsH3reg.
Supplemental Figure 4. Image Processing Euler Map.
Supplemental Figure 5. Plasmids Used to Construct SynFtsH2GST.
Supplemental Figure 6. Genotype Analysis of the FtsH2 Mutants.
Supplemental Figure 7. Construction of gst Tagging Cassette.
Supplemental Figure 8. Construction of Transformation Vectors to Make SynFtsHxGSTery Mutants.
Supplemental Figure 9. Genotype Analysis of the Four SynFtsHxGSTery Mutants.
Supplemental Table 1. Oligonucleotide Primers Used to Generate and Analyze Mutants.
Acknowledgments
We thank the UK Biotechnology and Biological Sciences Research Council (Grant BB/F020554/1) and Engineering and Physical Sciences Research Council (Grant EP/F002070X/1) for financial support. J.K. and V.K. were supported by projects Algatech (CZ.1.05/2.1.00/03.0110), RVO61388971, and P501/12/G055 of the Grant Agency of the Czech Republic. J.N. currently holds a Royal Society University Research Fellowship. We thank Teru Ogura for providing antibodies to E. coli FtsH and Jean-David Rochaix for sending the PsaD-specific antibodies.
AUTHOR CONTRIBUTIONS
J.K., P.J.N., and J.N. designed research. M. Boehm, M. Barker, J.Y., V.K., M.T., J.K., and J.N. performed research. M. Boehm, J.Y., M. Barker, M.T., J.K., J.N., and P.J.N. analyzed the data. M. Boehm, J.K., P.J.N., and J.N. wrote the article. P.J.N. and J.N. contributed equally to this work.
Glossary
- PSII
photosystem II
- PD
protease domain
- cryo-EM
cryo–electron microscopy
- GST
glutathione S-transferase
- PVDF
to be defined
- β-DM
n-dodecyl-β-d-maltoside
References
- Adir N., Zer H., Shochat S., Ohad I. (2003). Photoinhibition - A historical perspective. Photosynth. Res. 76: 343–370 [DOI] [PubMed] [Google Scholar]
- Akiyama Y., Yoshihisa T., Ito K. (1995). FtsH, a membrane-bound ATPase, forms a complex in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 270: 23485–23490 [DOI] [PubMed] [Google Scholar]
- Aro E.M., Virgin I., Andersson B. (1993). Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Bailey S., Thompson E., Nixon P.J., Horton P., Mullineaux C.W., Robinson C., Mann N.H. (2002). A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277: 2006–2011 [DOI] [PubMed] [Google Scholar]
- Barker M., Boehm M., Nixon P.J., Nield J. (2008). Structural analysis of an FtsH2/FtsH3 complex isolated from Synechocystis sp. PCC 6803. In Photosynthesis. Energy from the Sun, J.F. Allen, E. Gannt, J. Golbeck, and B. Osmond, eds (Dordrecht, The Netherlands: Springer), pp. 738–740.
- Bieniossek C., Niederhauser B., Baumann U.M. (2009). The crystal structure of apo-FtsH reveals domain movements necessary for substrate unfolding and translocation. Proc. Natl. Acad. Sci. USA 106: 21579–21584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C., Schalch T., Bumann M., Meister M., Meier R., Baumann U. (2006). The molecular architecture of the metalloprotease FtsH. Proc. Natl. Acad. Sci. USA 103: 3066–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birungi M., Folea M., Battchikova N., Xu M., Mi H., Ogawa T., Aro E.-M., Boekema E.J. (2010). Possibilities of subunit localization with fluorescent protein tags and electron microscopy examplified by a cyanobacterial NDH-1 study. Biochim. Biophys. Acta 1797: 1681–1686 [DOI] [PubMed] [Google Scholar]
- Blum H., Beier H., Gross H.J. (1987). Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8: 93–99 [Google Scholar]
- Boehm M., Nield J., Zhang P., Aro E.M., Komenda J., Nixon P.J. (2009). Structural and mutational analysis of band 7 proteins in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 191: 6425–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheregi O., Sicora C., Kós P.B., Barker M., Nixon P.J., Vass I. (2007). The role of the FtsH and Deg proteases in the repair of UV-B radiation-damaged photosystem II in the cyanobacterium Synechocystis PCC 6803. Biochim. Biophys. Acta 1767: 820–828 [DOI] [PubMed] [Google Scholar]
- Chiba S., Akiyama Y., Mori H., Matsuo E., Ito K. (2000). Length recognition at the N-terminal tail for the initiation of FtsH-mediated proteolysis. EMBO Rep. 1: 47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. (2008). The PyMOL Molecular Graphics System. (Palo Alto, CA: DeLano Scientific).
- Dobáková M., Sobotka R., Tichý M., Komenda J. (2009). Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 149: 1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drath M., Kloft N., Batschauer A., Marin K., Novak J., Forchhammer K. (2008). Ammonia triggers photodamage of photosystem II in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 147: 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhai J., Wolk C.P. (1988). Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167: 747–754 [DOI] [PubMed] [Google Scholar]
- Erzberger J.P., Berger J.M. (2006). Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35: 93–114 [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Ito K., Akiyama Y. (2005). Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59: 211–231 [DOI] [PubMed] [Google Scholar]
- Kamata T., Hiramoto H., Morita N., Shen J.R., Mann N.H., Yamamoto Y. (2005). Quality control of Photosystem II: An FtsH protease plays an essential role in the turnover of the reaction center D1 protein in Synechocystis PCC 6803 under heat stress as well as light stress conditions. Photochem. Photobiol. Sci. 4: 983–990 [DOI] [PubMed] [Google Scholar]
- Kato Y., Miura E., Ido K., Ifuku K., Sakamoto W. (2009). The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol. 151: 1790–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Sakamoto W. (2009). Protein quality control in chloroplasts: A current model of D1 protein degradation in the photosystem II repair cycle. J. Biochem. 146: 463–469 [DOI] [PubMed] [Google Scholar]
- Komenda J., Barber J. (1995). Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QB site and dependent on protein synthesis. Biochemistry 34: 9625–9631 [DOI] [PubMed] [Google Scholar]
- Komenda J., Barker M., Kuviková S., de Vries R., Mullineaux C.W., Tichy M., Nixon P.J. (2006). The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 281: 1145–1151 [DOI] [PubMed] [Google Scholar]
- Komenda J., Tichý M., Eichacker L.A. (2005). The PsbH protein is associated with the inner antenna CP47 and facilitates D1 processing and incorporation into PSII in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 46: 1477–1483 [DOI] [PubMed] [Google Scholar]
- Komenda J., Tichy M., Prásil O., Knoppová J., Kuviková S., de Vries R., Nixon P.J. (2007). The exposed N-terminal tail of the D1 subunit is required for rapid D1 degradation during photosystem II repair in Synechocystis sp PCC 6803. Plant Cell 19: 2839–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen M., Langer T. (2007). Protein degradation within mitochondria: Versatile activities of AAA proteases and other peptidases. Crit. Rev. Biochem. Mol. Biol. 42: 221–242 [DOI] [PubMed] [Google Scholar]
- Koppen M., Metodiev M.D., Casari G., Rugarli E.I., Langer T. (2007). Variable and tissue-specific subunit composition of mitochondrial m-AAA protease complexes linked to hereditary spastic paraplegia. Mol. Cell. Biol. 27: 758–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Augustin S., Tatsuta T., Gerdes F., Langer T., Tsai F.T. (2011). Electron cryomicroscopy structure of a membrane-anchored mitochondrial AAA protease. J. Biol. Chem. 286: 4404–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke S.J., Chen D.-H., Song J.-L., Chuang D.T., Chiu W. (2004). Seeing GroEL at 6 A resolution by single particle electron cryomicroscopy. Structure 12: 1129–1136 [DOI] [PubMed] [Google Scholar]
- Lupínková L., Komenda J. (2004). Oxidative modifications of the Photosystem II D1 protein by reactive oxygen species: from isolated protein to cyanobacterial cells. Photochem. Photobiol. 79: 152–162 [DOI] [PubMed] [Google Scholar]
- Mann N.H., Novac N., Mullineaux C.W., Newman J., Bailey S., Robinson C. (2000). Involvement of an FtsH homologue in the assembly of functional photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 479: 72–77 [DOI] [PubMed] [Google Scholar]
- McTigue M.A., Williams D.R., Tainer J.A. (1995). Crystal structures of a schistosomal drug and vaccine target: glutathione S-transferase from Schistosoma japonica and its complex with the leading antischistosomal drug praziquantel. J. Mol. Biol. 246: 21–27 [DOI] [PubMed] [Google Scholar]
- Moldavski O., Levin-Kravets O., Ziv T., Adam Z., Prag G. (2012). The hetero-hexameric nature of a chloroplast AAA+ FtsH protease contributes to its thermodynamic stability. PLoS ONE 7: e36008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N., Takahashi S., Nishiyama Y., Allakhverdiev S.I. (2007). Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta 1767: 414–421 [DOI] [PubMed] [Google Scholar]
- Natalello A., Santarella R., Doglia S.M., de Marco A. (2008). Physical and chemical perturbations induce the formation of protein aggregates with different structural features. Protein Expr. Purif. 58: 356–361 [DOI] [PubMed] [Google Scholar]
- Nixon P.J., Barker M., Boehm M., de Vries R., Komenda J. (2005). FtsH-mediated repair of the photosystem II complex in response to light stress. J. Exp. Bot. 56: 357–363 [DOI] [PubMed] [Google Scholar]
- Nixon P.J., Metz J.G., Roegner M., Diner B.A. (1990). A Synechocystis PCC 6803 psbA deletion strain and its transformation with a psbA gene from a higher plant. In Current Research in Photosynthesis, Vol. I, M. Baltscheffsky, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 471–474.
- Nixon P.J., Michoux F., Yu J., Boehm M., Komenda J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. (Lond.) 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perálvarez-Marín A., Tae H., Board P.G., Casarotto M.G., Dulhunty A.F., Samsó M. (2011). 3D mapping of the SPRY2 domain of ryanodine receptor 1 by single-particle cryo-EM. PLoS ONE 6: e25813. [DOI] [PMC free article] [PubMed]
- Piechota J., Kolodziejczak M., Juszczak I., Sakamoto W., Janska H. (2010). Identification and characterization of high molecular weight complexes formed by matrix AAA proteases and prohibitins in mitochondria of Arabidopsis thaliana. J. Biol. Chem. 285: 12512–12521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q., Hao M., Ng W.O., Slater S.C., Baszis S.R., Weiss J.D., Valentin H.E. (2005). Application of the Synechococcus nirA promoter to establish an inducible expression system for engineering the Synechocystis tocopherol pathway. Appl. Environ. Microbiol. 71: 5678–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht J., Nield J. (2001). Determining the structure of biological macromolecules by transmission electron microscopy, single particle analysis and 3D reconstruction. Prog. Biophys. Mol. Biol. 75: 121–164 [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Zaltsman A., Adam Z., Takahashi Y. (2003). Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15: 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. (1991). Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199: 223–231 [DOI] [PubMed] [Google Scholar]
- Shotland Y., Koby S., Teff D., Mansur N., Oren D.A., Tatematsu K., Tomoyasu T., Kessel M., Bukau B., Ogura T., Oppenheim A.B. (1997). Proteolysis of the phage lambda CII regulatory protein by FtsH (HflB) of Escherichia coli. Mol. Microbiol. 24: 1303–1310 [DOI] [PubMed] [Google Scholar]
- Silva P., Thompson E., Bailey S., Kruse O., Mullineaux C.W., Robinson C., Mann N.H., Nixon P.J. (2003). FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp PCC 6803. Plant Cell 15: 2152–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich G., Neupert W., Langer T. (1999). Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 19: 3435–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg M., Fulda S., Huckauf J., Hagemann M., Krämer R., Marin K. (2007). A membrane-bound FtsH protease is involved in osmoregulation in Synechocystis sp. PCC 6803: The compatible solute synthesizing enzyme GgpS is one of the targets for proteolysis. Mol. Microbiol. 63: 86–102 [DOI] [PubMed] [Google Scholar]
- Takahashi S., Badger M.R. (2011). Photoprotection in plants: A new light on photosystem II damage. Trends Plant Sci. 16: 53–60 [DOI] [PubMed] [Google Scholar]
- Tal M., Silberstein A., Nusser E. (1985). Why does Coomassie Brilliant Blue R interact differently with different proteins? A partial answer. J. Biol. Chem. 260: 9976–9980 [PubMed] [Google Scholar]
- Tomoyasu T., Yamanaka K., Murata K., Suzaki T., Bouloc P., Kato A., Niki H., Hiraga S., Ogura T. (1993). Topology and subcellular localization of FtsH protein in Escherichia coli. J. Bacteriol. 175: 1352–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heel M. (1987). Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy 21: 111–123 [DOI] [PubMed] [Google Scholar]
- van Heel M., Harauz G., Orlova E.V., Schmidt R., Schatz M. (1996). A new generation of the IMAGIC image processing system. J. Struct. Biol. 116: 17–24 [DOI] [PubMed] [Google Scholar]
- van Heel M., Schatz M. (2005). Fourier shell correlation threshold criteria. J. Struct. Biol. 151: 250–262 [DOI] [PubMed] [Google Scholar]
- Williams J.G.K. (1988). Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 167: 766–778 [Google Scholar]
- Wydrzynski T., Satoh K. (2005). Photosystem II: The Light-Driven Water:Plastoquinone Oxidoreductase. (Dordrecht, The Netherlands: Kluwer Academic Publishers). [Google Scholar]
- Yu F., Park S., Rodermel S.R. (2004). The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 37: 864–876 [DOI] [PubMed] [Google Scholar]
- Zaltsman A., Ori N., Adam Z. (2005). Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopsis. Plant Cell 17: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Kato Y., Zhang L., Fujimoto M., Tsutsumi N., Sodmergen, Sakamoto W. (2010). The FtsH protease heterocomplex in Arabidopsis: Dispensability of type-B protease activity for proper chloroplast development. Plant Cell 22: 3710–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Battchikova N., Jansen T., Appel J., Ogawa T., Aro E.-M. (2004). Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16: 3326–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Sicora C.I., Vorontsova N., Allahverdiyeva Y., Battchikova N., Nixon P.J., Aro E.-M. (2007). FtsH protease is required for induction of inorganic carbon acquisition complexes in Synechocystis sp. PCC 6803. Mol. Microbiol. 65: 728–740 [DOI] [PubMed] [Google Scholar]