VIPP1 is a protein conserved among photosynthetic organisms. Although the precise role in chloroplasts remains unclear, VIPP1 has been proposed to play a role in thylakoid membrane biogenesis. This study involves an in-depth analysis of Arabidopsis thaliana vipp1 mutants and demonstrates that VIPP1 is rather required for envelope maintenance in chloroplasts.
Abstract
VESICLE-INDUCING PROTEIN IN PLASTIDS1 (VIPP1), proposed to play a role in thylakoid biogenesis, is conserved in photosynthetic organisms and is closely related to Phage Shock Protein A (PspA), which is involved in plasma membrane integrity in Escherichia coli. This study showed that chloroplasts/plastids in Arabidopsis thaliana vipp1 knockdown and knockout mutants exhibit a unique morphology, forming balloon-like structures. This altered morphology, as well as lethality of vipp1, was complemented by expression of VIPP1 fused to green fluorescent protein (VIPP1-GFP). Several lines of evidence show that the balloon chloroplasts result from chloroplast swelling related to osmotic stress, implicating VIPP1 in the maintenance of plastid envelopes. In support of this, Arabidopsis VIPP1 rescued defective proton leakage in an E. coli pspA mutant. Microscopy observation of VIPP1-GFP in transgenic Arabidopsis revealed that VIPP1 forms large macrostructures that are integrated into various morphologies along the envelopes. Furthermore, live imaging revealed that VIPP1-GFP is highly mobile when chloroplasts are subjected to osmotic stress. VIPP1-GFP showed dynamic movement in the transparent area of spherical chloroplasts, as the fluorescent molecules formed filament-like structures likely derived from disassembly of the large VIPP1 complex. Collectively, our data demonstrate that VIPP1 is a multifunctional protein in chloroplasts that is critically important for envelope maintenance.
INTRODUCTION
Thylakoids, which are specific to organisms performing oxygenic photosynthesis, are the inner membrane systems that orchestrate photosynthetic electron transport and ΔpH-dependent ATP synthesis. In higher plants, they are formed in the chloroplast as a unique membrane network. The biogenesis of thylakoids is apparently a complex process that involves the synthesis and maintenance of pigments, proteins, and lipids (Herrmann, 1999; Vothknecht and Westhoff, 2001). It is also affected by environment (e.g., light, temperature, and nutrients) and by organ development (Tzinas et al., 1987; Monge et al., 1993; Kota et al., 2002). Despite such complexity, several important factors involved in thylakoid formation have been proposed, including VESICLE-INDUCING PROTEIN IN PLASTIDS1 (VIPP1), THYLAKOID FORMATION1 (THF1), CHLOROPLAST SECRETION-ASSOCIATED RAS1 (CPSAR1), and FtsH (Kroll et al., 2001; Sakamoto et al., 2003; Wang et al., 2004; Garcia et al., 2010). Among these factors, FtsH appears to be the only one for which functions as a thylakoid metalloprotease have been well established (Sakamoto et al., 2003; Zhang et al., 2010). How these proteins play roles in thylakoid biogenesis remains unclear, although they are highly conserved in photosynthetic organisms.
A crucial role of VIPP1 in thylakoid biogenesis has been proposed in both chloroplasts and cyanobacteria. Kroll et al. (2001) first reported a vipp1 knockdown Arabidopsis thaliana mutant (vipp1-kd) in which T-DNA was inserted into the promoter region. Aberrant thylakoid membranes along with vesicle membrane structures were detected in vipp1-kd, leading the authors to infer that VIPP1 is involved in vesicle trafficking between the inner envelope and thylakoid membrane. However, evidence supporting this proposition is circumstantial, and the precise function of VIPP1 in vesicle budding, migration, and fusion with thylakoids remains elusive. A recent study of cyanobacteria revealed that the initial detriment induced by VIPP1 depletion is not directly attributable to the loss of thylakoid membranes but rather to the loss of VIPP1 itself (Gao and Xu, 2009).
Consistent with its proposed function in thylakoid biogenesis, VIPP1 was found to be associated with both thylakoid membranes and envelopes in Arabidopsis (Kroll et al., 2001). In Synechocystis, VIPP1 is localized in both thylakoid membranes and plasma membranes (Srivastava et al., 2005). However, contrasting reports have described that VIPP1 was localized exclusively in the inner envelope/plasma membrane but not in thylakoid membranes in the same organisms (Westphal et al., 2001; Aseeva et al., 2004). Such inconsistent results of VIPP1 localization might result from different methods and/or different experimental conditions. Nevertheless, these studies collectively imply that a large portion of VIPP1 is attached to chloroplast envelopes or plasma membranes. VIPP1 was also described as being associated with proteins in the cytosol (Jouhet and Gray, 2009), suggesting that it might be exposed to the outside of chloroplasts. We therefore inferred that the precise localization and role of VIPP1 should be reevaluated.
VIPP1 shows significant sequence homology to Phage Shock Protein A (PspA) in Escherichia coli (Aseeva et al., 2004; Bultema et al., 2010). Both proteins appear to have similar secondary structures, although plant-type VIPP1s contain a C-terminal extension of ∼40 amino acids (see Supplemental Figure 1 online). PspA is induced rapidly in E. coli under stressful conditions that perturb the membrane integrity, such as infection by filamentous phage, heat shock, and ethanol treatment (Brissette et al., 1990). Under such stress conditions, homo-oligomers of PspA are formed. They subsequently bind to the inside surface of damaged plasma membranes to form lattice-like scaffolds, which can subsequently stabilize damaged membranes (Standar et al., 2008). As a consequence of PspA expression, proton leakage through plasma membranes can be mitigated. It is particularly interesting that a large PspA complex in E. coli was shown to move along very rapidly with the membrane surface, suggesting that proton motive force (PMF) maintenance through PspA is important under certain stress conditions (Engl et al., 2009). Similarly to PspA in E. coli, VIPP1s in photosynthetic organisms appear to form a large complex. In Arabidopsis, VIPP1 has been shown to form ring-like homo-oligomers of >1 MD (Aseeva et al., 2004). In the green alga Chlamydomonas reinhardtii, mutual adhesion of the ring particles of VIPP1 to form extremely long rod-like structures in vitro (>1 MD) has been observed (Liu et al., 2007). These reports suggest that an N-terminal region, rather than the plant-specific C-terminal region, is important for VIPP1 to form oligomers (Aseeva et al., 2004). In fact, DeLisa et al. (2004) reported that the expression of Synechocystis VIPP1 in E. coli ΔpspA mutants represses a defect in Tat-dependent protein transport related to the proper function of plasma membranes. A recent in vitro study further indicated that VIPP1 enhances protein transport on the chloroplast Tat pathway in pea (Pisum sativum) (Lo and Theg, 2012). Consequently, the accumulated information related to the similarities between PspA and VIPP1 implies strongly that VIPP1 may function to protect chloroplast envelopes: Envelope localization of VIPP1 also supports this implication.
Given the possible role of VIPP1 in chloroplast biogenesis, we attempted in this study to observe chloroplasts in Arabidopsis vipp1 knockdown and knockout mutants. Somewhat surprisingly, loss of VIPP1 was shown to engender a unique chloroplast morphology that had not been shown or characterized previously. Characterization of this structure along with VIPP1–green fluorescent protein (GFP) fusion protein suggests that VIPP1 forms a large complex at envelopes to maintain the membrane potential. We observed that VIPP1 is highly mobile around the region where envelopes were damaged. We provide evidence that, irrespective of its involvement in thylakoid formation, VIPP1 plays an indispensable role in envelope maintenance.
RESULTS
Unique Chloroplast Morphology Observed in vipp1 Mutants
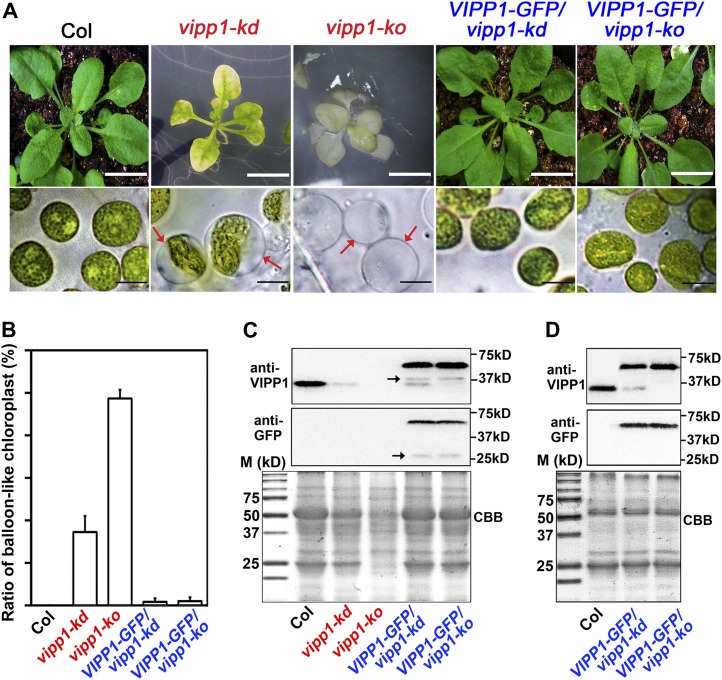
In vipp1-kd, which was previously characterized, a T-DNA insertion in the promoter region reduced VIPP1 accumulation to ∼20% of the wild-type level (Kroll et al., 2001). The mutant plant, which has lost its photoautotrophic growth capability, develops a pale-green phenotype at an early developmental stage when grown on Murashige and Skoog (MS) medium supplemented with Suc (the sublethal phenotype of vipp1-kd is shown in Supplemental Figure 2 online). In this study, we first characterized vipp1-kd carefully to examine whether it had any deficiency during thylakoid development in living mesophyll tissues. Cotyledons or true leaves from vipp1-kd grown on MS medium were observed directly using light microscopy. Unexpectedly, vipp1-kd contained chloroplasts that were distinct from those in the wild type (Figure 1A): Although almost all chloroplasts in the wild-type Columbia (Col) appeared to have green areas (representing thylakoids within chloroplasts) distributed throughout their structures, many if not most of the chloroplasts in vipp1-kd had formed into balloon-like structures in which thylakoids were located in a limited area of the chloroplasts. Transparent regions of different sizes were found inside chloroplasts of the vipp1-kd mutant. Chloroplasts of this type were also found in mesophyll protoplasts prepared from vipp1-kd (see Supplemental Figure 3 online). Our observations showed that ∼30% of the mesophyll chloroplasts in vipp1-kd had such transparent areas, although no abnormal chloroplasts were found in leaves from Col (Figure 1B).
Figure 1.
Balloon-Like Chloroplasts in vipp1 Mutants.
(A) Phenotypes of seedlings and chloroplasts from Col (wild type), vipp1 mutants (vipp1-kd and vipp1-ko), and their VIPP1-GFP–complemented lines. Top panels show photographs of different 4-week-old lines of Arabidopsis (bars = 1 cm). Bottom panels show chloroplasts from unfixed leaf tissues examined using bright-field microscopy (bars =10 µm). Red arrows indicate balloon-like chloroplasts/plastids.
(B) Ratio of balloon-like to regular chloroplasts/plastid in leaves from different lines of Arabidopsis shown in (A). Plastids were counted in the leaves of at least three individual plants (n = 50, shown with sd).
(C) Immunoblot analysis of leaf extracts from 6-week-old lines of Arabidopsis shown in (A). Loading was normalized by equal protein amount. Top and middle panels, immunoblots cross-reacted with anti-VIPP1 and anti-GFP, respectively. Arrows indicate degradation products (see the text). Bottom panel, Coomassie blue–stained gel (CBB) as a loading control. Molecular markers (M) are shown on the left.
(D) Immunoblot analysis of Percoll-purified chloroplast proteins from 6-week-old lines of Arabidopsis. Immunoblots were performed as shown in (C). vipp1-kd and vipp1-ko were not included in this immunoblot because no intact chloroplasts/plastids were recovered by Percoll gradient.
To ascertain whether this structure is also detectable in the knockout mutant of vipp1, we isolated and characterized another mutant line in which VIPP1 expression was fully inactivated. This line, termed vipp1-ko, had a T-DNA insertion in the third intron and accumulated no detectable VIPP1 when examined using immunoblot analysis (Figure 1C; see Supplemental Figure 4B online). The mutant grows slowly on MS medium supplemented with 1.5% Suc (Figure 1A; see Supplemental Figure 2 online). Seedlings were smaller than those in Col and vipp1-kd and had pale leaves. vipp1-ko seedlings can survive only on MS medium and perish 1 d after transfer from MS plates to soil. The lethal phenotype of vipp1-ko verified that VIPP1 is essential. Microscopy observations revealed that almost all plastids in the leaves of the vipp1-ko mutant displayed the balloon-like structure (Figures 1A and 1B). Consequently, the balloon-like chloroplasts represent a unique chloroplast morphology that has not been reported for vipp1 mutants.
Expression of VIPP1-GFP Rescues the Aberrant Chloroplast Morphology in vipp1 Mutants
To characterize the balloon-like chloroplast further, we examined whether overexpression of VIPP1-GFP could rescue this chloroplast phenotype as well as nonphotoautotrophic growth of the vipp1 mutants. Results of an earlier study demonstrated that transiently expressed VIPP1-GFP localized to chloroplasts (Aseeva et al., 2004); however, its functionality has not been investigated using a transgenic line. In this study, VIPP1-GFP was introduced into vipp1-kd and vipp1-ko under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The resulting transgenic lines are designated as VIPP1-GFP/vipp1-kd and VIPP1-GFP/vipp1-ko.
Immunoblot analysis of extracts taken from 6-week-old leaves with antibodies against VIPP1 and GFP revealed a band (61 kD) corresponding to VIPP1-GFP, but it was present only in these transgenic lines (Figure 1C, top and middle panels). As expected, vipp1-kd accumulates ∼20% of endogenous VIPP1 levels, while no VIPP1 protein at all is present in vipp1-ko. In addition to these bands, we detected several minor fragments, as shown in Figure 1C (arrows), of which the sizes varied depending on the leaf tissues used. We regarded these fragments as degradation products of VIPP1-GFP that were created during extraction because VIPP1 forms a large complex with various macrostructures during leaf development (see text below) and because it is sensitive to degradation. In fact, no such degradation products were detected when chloroplast proteins were purified by the Percoll gradient and extracted under mild conditions (Figure 1D). It is noteworthy that few or no chloroplasts/plastids were recoverable from Percoll gradient in vipp1-kd and vipp1-ko mutants, probably because of a defect in chloroplast integrity. By contrast, chloroplasts were purified by Percoll from transgenic lines expressing VIPP1-GFP, as they were from Col. Together, these results indicate that full-length VIPP1-GFP was expressed but was not degraded in the chloroplasts of VIPP1-GFP/vipp1-kd and VIPP1-GFP/vipp1-ko. Expression levels of VIPP1-GFP in these transgenic lines appeared to be comparable to those of endogenous VIPP1 in Col.
Under normal light conditions, both VIPP1-GFP/vipp1-kd and VIPP1-GFP/vipp1-ko resembled the wild type and were able to grow photoautotrophically (Figure 1A). These results demonstrate that VIPP1-GFP can substitute for VIPP1 function in vivo. Concomitant with the photoautotrophic growth, both transgenic lines rescued chloroplast morphology: No balloon-like chloroplast was detected in either complemented line (Figures 1A and 1B). Based on these observations, we conclude that the altered chloroplast morphology presented in Figure 1 resulted from the loss of VIPP1. Expression of VIPP1-GFP allowed us to characterize its localization in chloroplasts (see below).
Altered Chloroplast Morphology Probably Results from Stromal Swelling
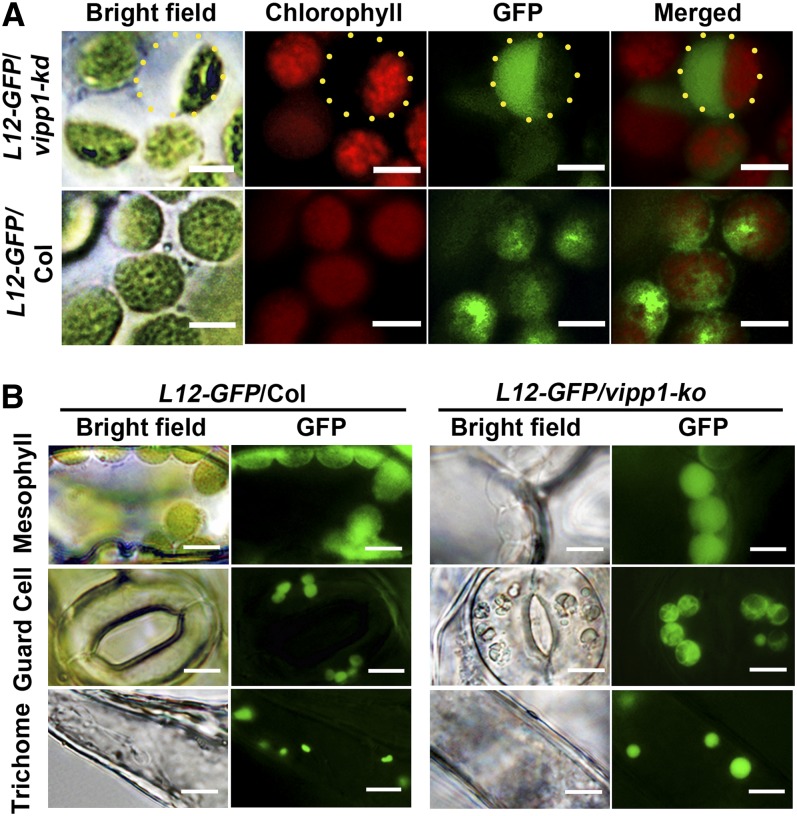
The intriguing chloroplast morphology in vipp1 raised the question of whether the transparent area within chloroplasts represents stroma or intermembrane space between the inner and outer envelopes. To address this question, we generated Col and vipp1-kd in which stroma-localized GFP (L12-GFP) was expressed. L12-GFP contains a transit peptide sequence derived from rice (Oryza sativa) chloroplastic ribosomal protein L12 (Arimura et al., 1999). In Col, L12-GFP overlapped with chlorophyll autofluorescence. Moreover, it was distributed broadly within chloroplasts. By contrast, vipp1-kd had L12-GFP signals that are detected predominantly in the transparent area, as evidenced by merged images (Figure 2A). The result presented above demonstrated that vipp1 mutants have extra stromal space that is not usually observed in Col.
Figure 2.
Swelling of Chloroplasts and Plastids in vipp1 Mutants Visualized Using Stroma-Localized L12-GFP.
(A) Distribution of L12-GFP within chloroplasts. Transgenic lines expressing L12-GFP in Col (L12-GFP/Col) or vipp1-kd (L12-GFP/vipp1-kd) were generated, and their mesophyll chloroplasts were observed by microscopy. Bright-field images and their fluorescent signals corresponding to chlorophyll (red) and GFP (green) are indicated along with merged images. Yellow-dotted circles represent the area of swollen stroma, which is transparent in bright field but detectable by L12-GFP. Bars = 5 µm.
(B) Spherical and enlarged plastids detected in different cell types of the transgenic line expressing L12-GFP in vipp1-ko (L12-GFP/vipp1-ko). Chloroplasts/plastids from mesophyll cells, guard cells, and trichomes were visualized using L12-GFP. Bright-field images (left) and GFP signals (right) are shown. Bars = 10 µm.
Next, we conducted tests to determine if the extra stromal space indeed enlarged the chloroplasts. The varied size of chloroplasts in mesophylls prevented us from confirming this enlargement. By contrast, when we compared L12-GFP signals in guard cells and trichomes, vipp1-ko apparently had more stromal space than Col and consequently had larger chloroplasts/plastids (Figure 2B). Therefore, the swelling of both plastids apparently creates the balloon-like structure in vipp1 mutants.
Stroma Swelling Is Specific to vipp1 and Is Affected by Osmotic Pressure
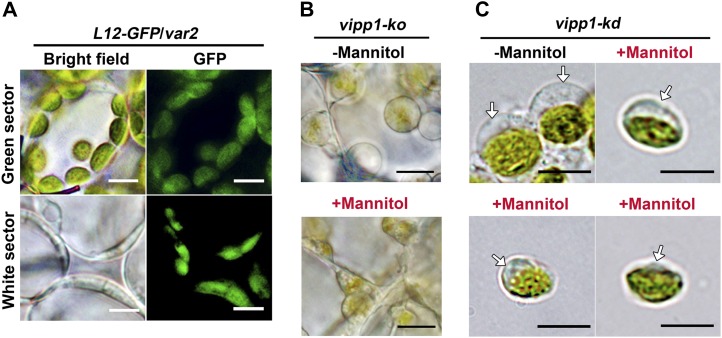
Our careful observation of vipp1 chloroplasts raised the possibility that VIPP1 functions in envelopes in addition to thylakoid formation. To test this possibility, we first observed plastids in another mutant, yellow variegated2 (var2), that is defective in thylakoid development. The VAR2 locus encodes a major isoform of FtsH metalloprotease (FtsH2). Its depletion gives rise to the variegated sectors in mature leaves. As reported previously, plastids visualized by L12-GFP in protoplasts from var2 white sectors were irregular and varied in size (Kato et al., 2007). Importantly, we never observed balloon-like plastids in the var2 white sectors, and improper thylakoid development rendered plastids smaller than those in the green sectors (Figure 3A). Similarly to those of var2, plastids of other mutants (e.g., thf1 and cpsar1) were also shown to have a smaller volume than those of normal chloroplasts (Wang et al., 2004; Garcia et al., 2010). These observations, although circumstantial, suggest that the stromal swelling observed in vipp1 is unique and that it is not caused simply by the defect in thylakoid formation.
Figure 3.
Chloroplast Swelling Is Characteristic of vipp1 Mutants and Is Affected by Osmolytes.
(A) Microscopy observation of chloroplasts/plastids in var2 leaves expressing L12-GFP (L12-GFP/var2). Cells from green and white sectors. Bright-field images (left) and GFP fluorescence (right) are shown. Bars = 10 µm.
(B) In situ observations of vipp1-ko plastids in leaves infiltrated with (+) or without (−) 1 M mannitol. Bars = 10 µm.
(C) Observation of intact chloroplasts purified from vipp1-kd protoplasts. Purified chloroplasts were incubated with isolation medium supplemented with (+) or without (−) 1 M mannitol in vitro. Arrows indicate the transparent stromal area. Bars = 10 µm.
To confirm that the balloon-like chloroplasts were associated with an alteration in envelopes, we examined the effects, if any, of osmotic pressure outside chloroplasts on chloroplast morphologies in Col and vipp1. Similarly to the balloon-like structure revealed in this study, swollen spherical chloroplasts were created not only by treating the isolated Col chloroplast with lower osmotic concentration solutions; they could also be shrunk back to their original state when replaced in a solution with hypertonic Suc or mannitol medium (Hongladarom and Honda, 1966). Reversible behavior of this kind is related directly to changes in osmotic pressure inside the chloroplast, which is normally controlled by the inner envelope. To ascertain whether the spherical shape of vipp1 chloroplasts was induced by an increased osmotic potential inside the chloroplast/plastid, hypertonic mannitol solution was used to treat the chloroplasts/plastids of vipp1 mutants. Small discs of vipp1-ko leaves were vacuum infiltrated with 1 M mannitol solution for 2 min and checked by light microscopy. Compared with the control Col plant, plastids in vipp1 cells were small and irregular in size (Figure 3B). Furthermore, we isolated chloroplasts from vipp1-kd, resuspended in 1 M mannitol media and kept them on ice for 5 min. Similarly to the result observed for leaf discs, the chloroplasts were considerably smaller than chloroplasts in isotonic media (0.33 M mannitol). The transparent region of chloroplasts, representing the stromal fraction, shrank into smaller structures (Figure 3C). We concluded that the chloroplast/plastid swelling was induced by increased osmotic pressure inside the organelle of vipp1 mutants.
Ultrastructure of Chloroplasts in vipp1
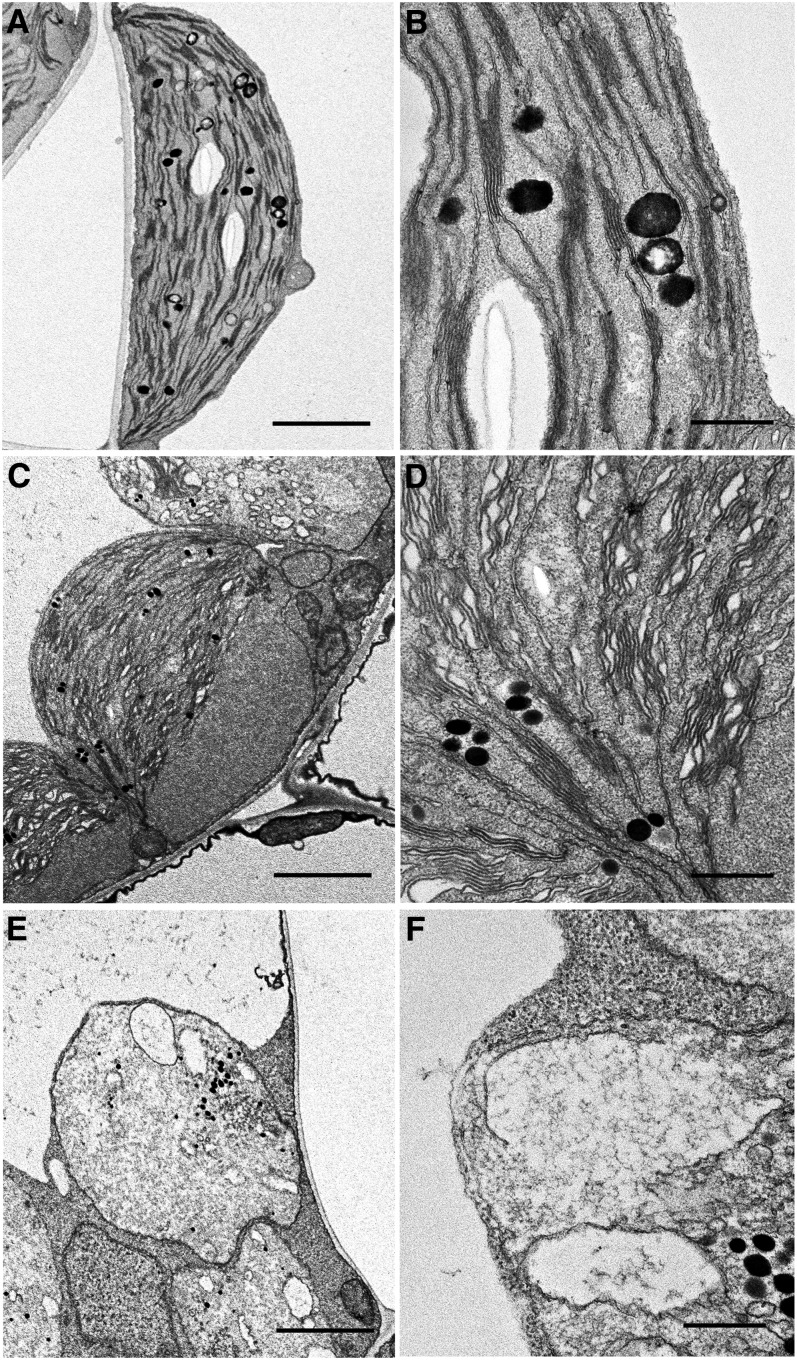
The ultrastructure of the swollen chloroplasts observed in vipp1-kd and vipp1-ko was further characterized using transmission electron microscopy (TEM). Col leaves grown in MS medium supplemented with Suc developed typical chloroplasts with normal stacking of thylakoid membranes and accumulation of starch granules (Figures 4A and 4B). By contrast, vipp1-kd chloroplasts exhibited a varied appearance (Figures 4C and 4D). Importantly, TEM analysis showed that despite fixation of the samples, chloroplasts were observed as those in living tissues with extra stromal area, again confirming stromal swelling in vipp1 mutants. Furthermore, these swollen chloroplasts contained granal stacks that differed slightly from those in Col: more lumenal area existed between the thylakoid membranes, which appeared ruffled and less stacked. It is particularly interesting that such types of swollen thylakoids have also been observed in a VIPP1 knockdown line of C. reinhardtii carrying VIPP1 artificial microRNA under high light conditions (Nordhues et al., 2012). TEM analysis of vipp1-ko revealed a much more extreme plastid structure that was clearly distinguishable from chloroplasts in Col. All plastids lacked normal granal stacks and had vacuolated membrane structures of various sizes (Figures 4E and 4F). Those irregular inner membrane structures, along with increased quantities of plastoglobules, are frequently observed in var2 white sectors and other mutants with defective chloroplast development. Consequently, our TEM analysis further demonstrated extraordinary chloroplast architecture in vipp1 mutants. VIPP1 is an important protein for proper chloroplast development.
Figure 4.
Chloroplasts and Plastids in vipp1 Mutants Examined Using TEM.
Chloroplast ultrastructure of Col ([A] and [B]), vipp1-kd ([C] and [D]), and vipp1-ko ([E] and [F]) seedlings was observed using TEM. Spherical chloroplasts with extra stromal space, detected in unfixed tissues, are also detected using electron microscopy in vipp1 mutants ([C] and [E]). Magnified electron micrographs corresponding to grana thylakoids show irregular granal stacks in vipp1-kd (D) and vacuolated membrane structures in vipp1-ko (F). Bars = 2.5 µm in (A), (C), and (E) and 500 nm in (B), (D), and (F).
Functional Complementation of the E. coli ΔpspA Mutant with VIPP1-GFP
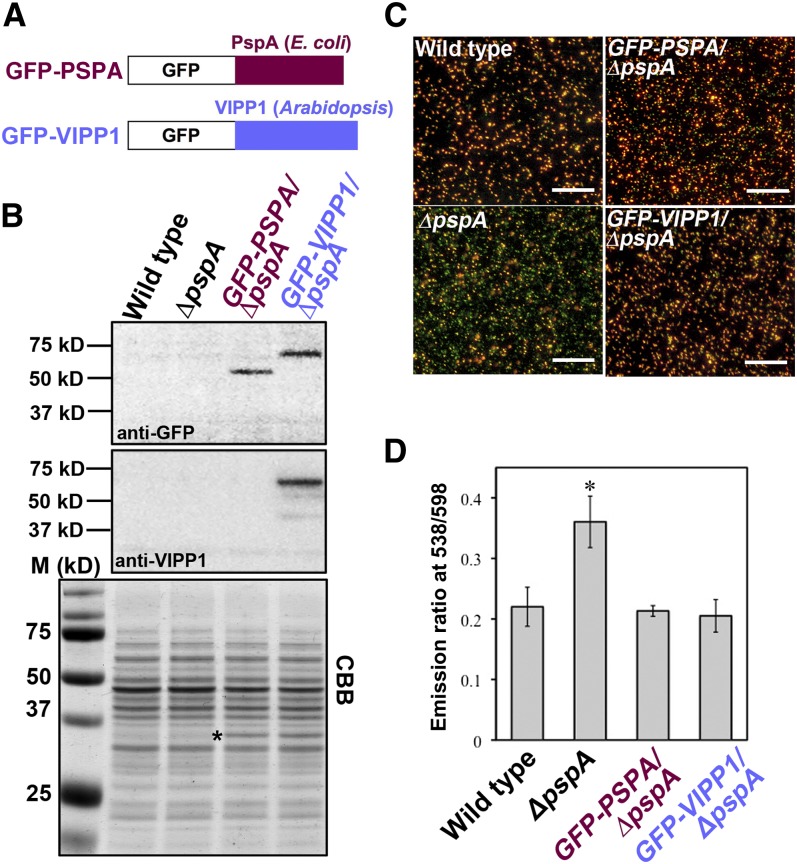
We demonstrated that VIPP1 plays an important role in the chloroplast envelope, perhaps similarly to PspA. Results of numerous studies suggest that PspA is involved in the maintenance of PMF, possibly by mediating a generalized reduction in proton permeability in response to localized disturbances in membrane integrity (Jovanovic et al., 2006; Kobayashi et al., 2007; Standar et al., 2008). We therefore examined the expression of VIPP1-GFP to ascertain whether it rescues membrane functionality in a ΔpspA mutant of MG1655, in which PMF has been shown to be perturbed. Two constructs, one containing E. coli pspA fused to GFP and the other containing Arabidopsis VIPP1 cDNA (lacking sequences corresponding to the transit peptide) fused to GFP (Figure 5A), were prepared to express these genes under the Trc promoter. MG1655 cells transformed with these constructs were first subjected to immunoblot analysis. GFP-PspA (∼53 kD) and GFP-VIPP1 (∼61 kD) accumulated to comparable levels based on equal cell numbers (Figure 5B).
Figure 5.
Functional Complementation of the E. coli ΔpspA Mutant with VIPP1-GFP and PMF Recovery.
(A) Schematic illustrations of GFP-PspA and GFP-VIPP1 fusion proteins.
(B) Immunoblot analysis of E. coli parental strain (wild type), the ΔpspA mutant (ΔpspA), and ΔpspA mutants complemented with E. coli pspA (GFP-PSPA/ΔpspA) and Arabidopsis GFP-VIPP1 (GFP-VIPP1/ΔpspA). Total cell extracts from the same amount of overnight culture were denatured and loaded on each lane. Antibodies against GFP (anti-GFP, top) and VIPP1 (anti-VIPP1, middle) were used to detect corresponding signals on the blots. A Coomassie blue–stained gel image (CBB; bottom) is shown as a loading control. A band denoted by an asterisk was regarded as the β-lactamase encoded by the ampicillin-resistant marker gene in pEC1.
(C) Fluorescence images of E. coli strains shown in (B) after staining with JC-1. Images were obtained using fluorescence microscopy (filter set U-MWIB2; Olympus) after JC-1 staining for 20 min. Bars = 50 µm.
(D) Ratio of fluorescence emission at 538/598 nm calibrated from the JC-1-stained E. coli strains shown in (C). At least five independent cultures for each E. coli strain were subjected to fluorescence measurement (excitation at 485 nm).
Fluorescence ratio imaging was used to measure the membrane potential (ΔΨ) component of PMF in individual cells after loading with JC-1, a cationic dye that is sensitive to membrane potential. At low membrane potential, JC-1 exists as a green fluorescent monomer, although the dye forms red fluorescent aggregates at higher potentials. First, we measured the difference in fluorescence under microscopy. When overnight-cultured cells from the wild type and ΔpspA were stained with JC-1, we found that the lack of PspA results in a lower ΔΨ, as represented by the shift of fluorescence from red-orange to green (Figure 5C). Both ΔpspA lines carrying either GFP-PspA or GFP-VIPP1 showed red-orange fluorescence similar to that of the wild type. To confirm this recovery in the transformants, we next measured the different emission signals using a fluorescence spectrophotometer. With excitation at 485 nm, two emission peaks appeared at 538 and 598 nm, which corresponded to the monomer and oligomeric states of JC-1, respectively. Figure 5D shows the emission ratio of 538 nm/598 nm, reflecting the membrane potential. Consistent with the microscopy observation, the fluorescence ratio in the ΔpspA mutant was significantly higher than that of the wild type, although the expression of GFP-VIPP1 and that of GFP-PspA recovered the defective membrane potential. These observations collectively indicated that VIPP1 indeed contributes to the maintenance of the ΔΨ component of PMF, just as PspA does.
Lattice-Like and Connected Structure of VIPP1 in Envelopes
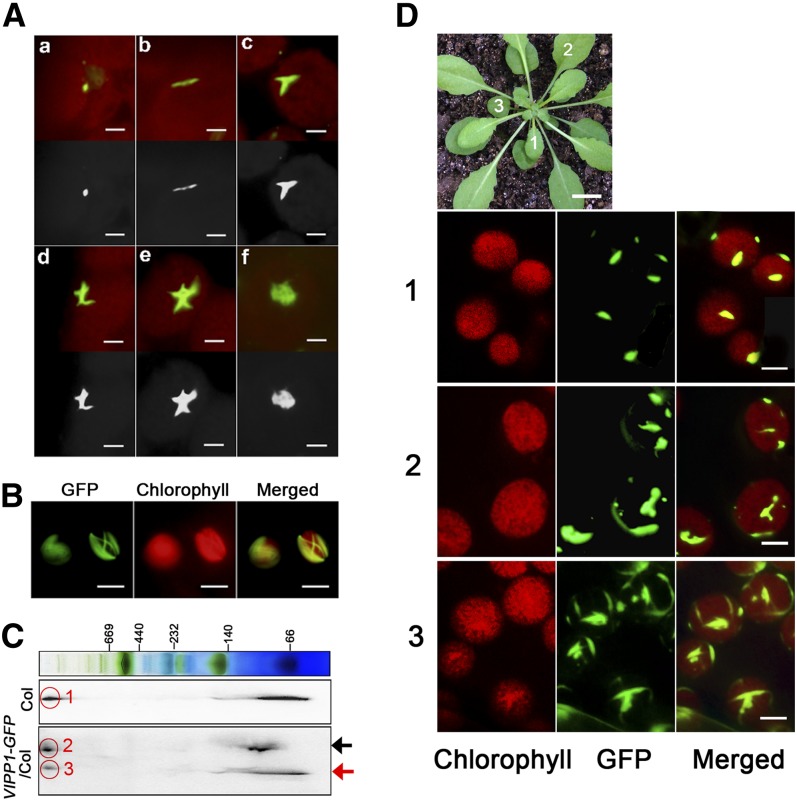
VIPP1 was inferred to play a role in envelope maintenance. Therefore, we attempted to conduct detailed characterization of its association with the envelope. Consistent with previous studies (Li et al., 1994; Kroll et al., 2001), most VIPP1 was localized on the envelope in our experiment (see Supplemental Figure 5 online). We further investigated the subchloroplast localization of VIPP1 in living leaf tissues. Microscopy observation of GFP signals in VIPP1-GFP/vipp1-kd protoplasts revealed that GFP signals appeared to be localized along the envelope and that it was assembled into various structures (Figure 6A). In addition, VIPP1-GFP occasionally formed highly connected networks. The filaments composed by VIPP1-GFP were mutually crossed to form a lattice-like structure (Figure 6B). We also assessed the supercomplex formation of VIPP1-GFP based on two-dimensional blue-native SDS-PAGE gel analysis (Figure 6C). Signals corresponding to VIPP1-GFP and native VIPP1 formed a supercomplex at a similar position, indicating that the GFP signals that were observed by fluorescence microscopy represented functional complexes. It is noteworthy that our initial observation of VIPP1-GFP signals revealed only one or a few clustered signals per chloroplast, which raised the question of whether the VIPP1 complex shows any changes during leaf development. Indeed, GFP signals from different leaves in an identical plant exhibited markedly altered morphologies (Figure 6D). In a younger leaf (marked 1), only one or two VIPP1-GFP aggregates per chloroplast were detected as spot-like structures of similar size. By contrast, chloroplasts in more matured leaves (marked 2 and 3) tended to have highly complex morphologies: mutually connected rod-like super structures and lattice-like structures that resembled those observed in mesophyll protoplasts (Figure 6B).
Figure 6.
Microscopy Observation of VIPP1-GFP Reveals Various Supercomplexes along the Chloroplast Envelope.
(A) Images of VIPP1-GFP supercomplexes in chloroplasts of VIPP1-GFP/vipp-kd protoplasts photographed using fluorescence microscopy: Various morphologies, such as a dot (a), line (b), fork (c), cross (d), five-point star (e), and web (f) were detected. Top panels show the original images obtained from microscopy observation, and bottom panels show their black and white images converted by Photoshop software. Bars = 2.5 µm.
(B) Lattice-like structures of VIPP1-GFP occasionally formed along the chloroplast envelope. Bars = 10 µm.
(C) Supercomplex of VIPP1 and VIPP1-GFP in Col and VIPP1-GFP/Col, respectively, monitored using Blue Native-SDS-PAGE. Total chloroplast proteins of Col and VIPP1-GFP/Col were separated by Blue Native-SDS-PAGE and probed with antibodies against VIPP1. Black and red arrows indicate positions of VIPP1-GFP and VIPP1, respectively. The signals within the red circles respectively correspond to the supercomplex of VIPP1 in Col (1), of VIPP1-GFP in VIPP1-GFP/Col (2), and of VIPP1 in VIPP1-GFP/Col (3).
(D) Morphologies of VIPP1-GFP at different stages of leaf development. Top, photograph of a VIPP1-GFP/vipp1-kd plant (bar = 1.0 cm). Leaves marked with numbers (1, 2, and 3) in this plant were subjected to microscopy observation. Chloroplasts from each leaf were observed to detect chlorophyll autofluorescence (left) and GFP (middle). Merged images of both signals are shown on the right panels. Bars = 5.0 µm.
VIPP1 Is Disassembled and Is Highly Mobile along the Region of the Damaged Envelope
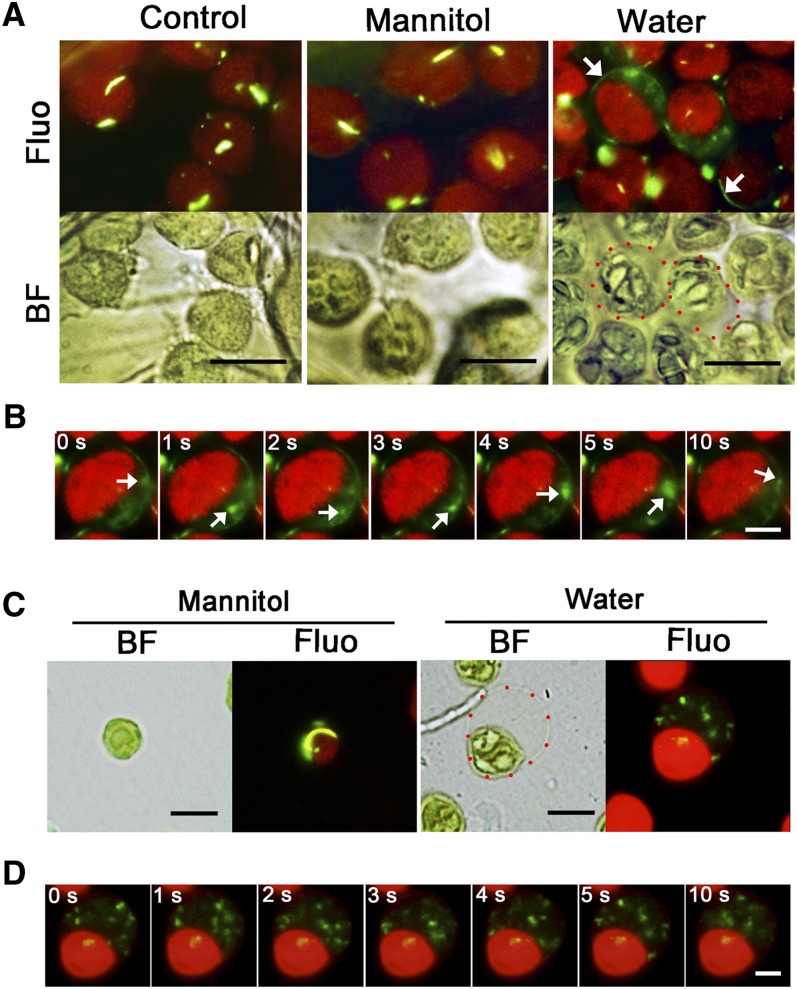
Microscopy observations revealed that some faint green signals moved occasionally in the background. PspA in E. coli has been shown to move during a process related to restoring membrane integrity (Engl et al., 2009). Therefore, we reasoned that this observation actually reflected VIPP1 movement. We therefore decided to visualize VIPP1 movement by live imaging of chloroplast envelopes that had been damaged by osmotic stress. Detached leaves from 4-week-old VIPP1-GFP/vipp1-kd plants were vacuum infiltrated with hypotonic solution (water) or isotonic solution (0.33 M mannitol) and chloroplasts were observed directly under bright-field and fluorescence microscopy (Figure 7A). In hypotonic solution, some of the chloroplasts in mesophyll cells showed stromal swelling and resembled the balloon-like structures detected in vipp1 mutants, indicating that the envelope had been subjected to osmotic stress (Figure 7A, red dotted circles). In such chloroplasts, VIPP1-GFP was found to move rapidly at the transparent area of the balloon structures (Figure 7B; see Supplemental Movie 1 and Supplemental Movie Legends 1 online). Careful observation of the imaging revealed that a preexisting rod-like supercomplex appeared to be disassembled; instead, a newly formed VIPP1-GFP complex was formed around the envelope (Figure 7A, white arrows). By contrast, we neither observed swollen chloroplasts in the control leaves (no treatment or in isotonic solution; Figure 7A, left and middle panels) nor detected VIPP1 movement (see Supplemental Movies 2 and 3 online). These results strongly suggest that VIPP1 is highly mobile around the envelope when membrane integrity was compromised.
Figure 7.
Dynamic Movement of VIPP1 in the Balloon-Like Chloroplasts.
(A) In situ observation of chloroplasts in VIPP1-GFP/vipp1-kd leaves after vacuum filtration with 0.33 M mannitol or with distilled water or with no treatment (Control). Chloroplast micrographs were taken in situ under bright-field (BF) or fluorescence (Fluo) conditions. A real-time image of the same chloroplasts was recorded simultaneously as described in Methods. Red-dotted circles in the image of the water-filtrated leaf indicate representative balloon-like chloroplasts. White arrows indicate newly formed filament structures. Bars = 10 µm.
(B) Time-lapse fluorescence images of swollen chloroplast in VIPP1-GFP/vipp1-kd leaves. The movement of one mobile complex is indicated in each image by a white arrow. Bar = 5 µm. For original images, see Supplemental Movies 1 to 3 online.
(C) Observation of VIPP in purified chloroplasts. Intact chloroplasts purified from VIPP1-GFP/vipp1-kd were resuspended in 0.33 M mannitol or distilled water and were observed using microscopy under bright-field (BF) or fluorescence (Fluo) conditions. A real-time image of the same chloroplasts was recorded under the fluorescence field. A red-dotted circle in the image of the water-treated chloroplast indicates the area corresponding to swollen stroma. Bars = 10 µm.
(D) Time-lapse fluorescence images of the swollen chloroplast in (C). Bar = 5 µm. For original images, see Supplemental Movies 4 and 5 online.
To visualize the mobile property of VIPP1 carefully, osmotic stress was also induced in Percoll-purified chloroplasts from VIPP1-GFP/vipp1-kd leaves. Supporting our observation in intact leaves, we observed highly mobile VIPP1-GFP in the transparent area of balloon-like chloroplasts, in which small dot-like signals move rapidly, as though being mutually connected and dissociated (Figures 7C and 7D; see Supplemental Movie 4 online). These balloon-like chloroplasts were detected only under hypotonic conditions. Normal-appearing chloroplasts under isotonic conditions had static VIPP1-GFP signals forming a large rod or lattice structure (Figure 7C, left panel; see Supplemental Movie 5 online). Collectively, our imaging analysis revealed that VIPP1 complex formation is a dynamic process that is associated with damage to the chloroplast envelope.
DISCUSSION
Chloroplasts maintain their integrity through the envelope membrane, which controls the transfer of ions, organic compounds, and peptides. The outer envelope has often been regarded as a passive compartment in plant cells, whereas the inner envelope, which contains many specific transporters, acts as a barrier against permeation between the stroma and cytoplasm (Douce and Joyard, 1990; Fuks and Homblé, 1999; Inoue, 2007; Fischer, 2011). Protein transport across the chloroplast envelope is an energy-requiring process that depends on the membrane integrity (Flügge and Hinz, 1986; Scott and Theg, 1996). Envelopes appear to be damaged by environmental stress, such as heat (McCain et al., 1989), drought (Dekov et al., 2000), UV-B (Peng and Zhou, 2009), and element toxicity (Vassilev et al., 1995). In fact, little is known about the maintenance of plastid envelope integrity. In this study, we highlighted VIPP1 as a candidate for this process, based on the balloon-like structure of chloroplasts that is unique to vipp1 mutants. We presented a line of evidence that (1) this structure actually results from chloroplast/plastid swelling, (2) VIPP1 is functionally orthologous to E. coli PspA, and (3) VIPP1 is present as a large, lattice-like complex that is dynamically mobile in the chloroplast envelope when damaged by osmotic stress. Together, our observations provide insight into a mechanism of plastid envelope maintenance in which VIPP1 plays a pivotal role.
Swelling Chloroplasts and Loss of VIPP1
Mature chloroplasts in mesophyll cells are generally lens shaped and in species such as Arabidopsis contain numerous granal stacks (Figures 4A and 4B) (Sakamoto et al., 2008). A defect in thylakoid formation often results in nonphotoautotrophic growth and aberrant plastid morphology. For example, the microscopy observation of plastids in var2 white sectors indicated that the loss of thylakoids affects the plastid shape, making it irregular and smaller than chloroplasts in green sectors (Kato et al., 2007). Additionally, plastids in thf1 (Wang et al., 2004) and cpsar1 (Garcia et al., 2010) showed various morphologies. These observations implicate chloroplast architecture as being closely related to the structure of thylakoids; the spatial distribution of thylakoids might serve partly as a foundation of normal chloroplast morphology, and the loss of thylakoids results in wrinkled plastid morphologies. Notably, the spherical plastids observed in vipp1-ko differ from the plastids described above. Spherical chloroplasts in vipp1-kd appear to contain normal thylakoids but still show a balloon-like structure. Such circumstances led us to infer that the primary function of VIPP1 is not restricted to thylakoid biogenesis and that it is related to the plastid architecture through maintenance of envelopes. Because vipp1-kd was initially characterized as showing “high chlorophyll fluorescence,” the initial work on VIPP1 specifically emphasized thylakoids rather than the envelope (Kroll et al., 2001). In this seminal work, chloroplasts were examined only by TEM after fixation, which might have prevented the detection of their balloon structures. Close examination of these data reinforced our finding in this work because a chloroplast in 3-week-old leaves appears to contain normal-appearing granal stacks, but it has a swollen stromal area (Figure 1C of Kroll et al., 2001).
A series of experiments was conducted in this study to verify our assumption that the balloon-like chloroplast results from stromal swelling. Chloroplast morphology of this type was shown to correlate with altered osmotic pressure (Figures 3B and 3C). For example, MscS-LIKE PROTEIN2 (MSL2) and MSL3 are necessary for releasing ions from plastids to the cytosol. A mutant lacking both proteins exhibits spherical plastids, which appear to result from increased osmotic pressure within the plastids (Haswell and Meyerowitz, 2006). Along with the spherical morphology, increased volume is another feature related to the high osmotic pressure inside plastids. Similarly to the msl2 msl3 double mutant, vipp1-ko contains large plastids in guard cells and trichomes (Figure 2B). Relevant to these observations, we had difficulty purifying chloroplasts from vipp1 mutants by Percoll gradient. Finally, the fact that the balloon-like chloroplasts in vipp1-kd were converted to a normal morphology in hypertonic solution (1 M mannitol) demonstrates that the osmotic potential is high inside chloroplasts, which causes water influx from the cytosol. Collectively, our finding of stromal swelling in vipp1 mutants strongly suggests that VIPP1 is important for maintaining the envelope integrity. It is noteworthy that a recent report on C. reinhardtii VIPP1 knockdown (Nordhues et al., 2012) did not refer to such swollen chloroplasts, probably because C. reinhardtii is unicellular and has a large single chloroplast that occupies a substantial portion of the cell, making observations of swelling difficult.
Orthologous Function of VIPP1 with PspA
Both VIPP1 and bacterial PspA have similar secondary structures, forming large homo-oligomeric rings with high molecular mass (Bultema et al., 2010). Recent observations have indicated that PspA is involved in maintaining the integrity of the plasma membrane in E. coli (Kobayashi et al., 2007). DeLisa et al. (2004) found that VIPP1 from Synechocystis spp PCC6803 can enhance Tat-dependent protein export that depends on PMF and that it can thereby complement the export defect of an E. coli ΔpspA null mutant. In this study, a fluorescence molecule (JC-1) was used to assess PMF. The red shift of GFP-VIPP1/ΔpspA fluorescence compared with that of the ΔpspA mutant showed directly that Arabidopsis VIPP1 can restore PMF effectively (Figures 5C and 5D). Our result indicates that VIPP1 is functionally orthologous to PspA. It is noteworthy that both PspA and VIPP1 are hydrophilic proteins that contain no sequence characteristics of integral membrane proteins (Cserzö et al., 1997). Nevertheless, Kobayashi et al. (2007) demonstrated that PspA can bind directly to membrane phospholipids and that it repairs proton leakage of the damaged membrane. It is therefore possible that VIPP1 can also attach directly to the membrane lipids that are damaged transiently by various stresses (see below). Overall, these observations led us to propose that VIPP1 is involved in sustaining/repairing chloroplast envelope membranes.
As exemplified by the study of E. coli, severe damage caused by the lack of VIPP1 can be associated with maintenance of PMF across plastid envelopes. The chloroplast inner envelope is energized by ATP-driven proton pumps (Keegstra et al., 1989; van den Wijngaard and Vredenberg, 1997). The pumping of protons from the stroma to the cytoplasm sets up a trans-envelope PMF (Heber and Heldt, 1981; Berkowitz and Peters, 1993). The alkalization of the stroma activates several pH-sensitive enzymes of the Calvin cycle, such as fructose-bisphosphatase (Racker and Schroeder, 1958), thereby permitting photosynthesis to occur. Given the proposed role of VIPP1 in envelope integrity, proton leakage from the cytosol to stroma in vipp1 engenders a lowered pH environment in the stroma, which in turn negatively regulates the Calvin cycle and concomitant photosynthetic activity. The decreased stromal pH might concomitantly disturb the proton gradient across thylakoid membranes, which in turn affects lumenal protein transport via the Tat pathway (Mould and Robinson, 1991; Robinson et al., 2000). In this scenario, the fundamental role of VIPP1 in envelopes can affect thylakoid biogenesis indirectly. Recently, Lo and Theg (2012) demonstrated in vitro that VIPP1’s primary role in the chloroplast Tat pathway is independent of energy potential, but it acts to stimulate substrate binding to thylakoid membranes. Although implying an additive role for VIPP1 in reorganizing thylakoid membrane structure in vitro, this observation is consistent with our presumption that VIPP1 functions to repair damaged membranes to maintain its integrity, as does PspA in E. coli.
In this study, we observed the ultrastructure of plastids not only in vipp1-kd but also in vipp1-ko using TEM. A previous study of vipp1-kd showed an invaginated vesicle-like structure derived from the inner envelope, which led the authors to propose that VIPP1 is involved in vesicle-mediated thylakoid formation. In contrast with these observations, no such vesicles were found; instead, we found vacuolated membrane bodies that consist of single membranes (Sakamoto et al., 2009). Although vacuolated membrane bodies are related to defects in thylakoid biogenesis, they appear to derive from thylakoids. Together with the functions of VIPP1 in the envelope and PMF described previously, we suggest that VIPP1 is unlikely to be involved in forming vesicles by itself. A recent study of C. reinhardtii also questioned the putative role of VIPP1 in vesicle-mediated thylakoid membrane formation and instead suggested that VIPP1 was involved in the biogenesis/assembly of thylakoid protein complexes (Nordhues et al., 2012). A report of an in vitro study of chloroplast Tat transport proposed that VIPP1 regulates chloroplast Tat activity via structural modification of thylakoid membranes (Lo and Theg, 2012). It is therefore plausible that VIPP1 functions in thylakoid membranes in addition to envelopes, as evidenced by the fact that a residual portion of VIPP1 (∼20% of total VIPP1 proteins) has been detected in thylakoid membranes (Li et al., 1994; Kroll et al., 2001). VIPP1 is a multifunctional protein in chloroplasts.
The VIPP1 Complex and Its Dynamic Behavior Associated with Envelope Damage
In VIPP1-GFP/vipp1-kd, VIPP1-GFP was found to aggregate into various structures (Figures 6A, 6B, and 6D). The basic unit of these structures might be the annular particle. In E. coli, PspA has been reported to form a large and regular scaffold structure, which presumably stabilizes stressed membranes physically through multiple interactions over large membrane surface areas (Standar et al., 2008). Similarly, a lattice-like structure of VIPP1 was detected in Arabidopsis leaves in a development-dependent manner (Figure 6D). In chloroplasts derived from older leaves, the lattice-like structure of VIPP1 perhaps promotes the closure of transient holes or leaks. Taking advantage of visualized VIPP1-GFP aggregates in chloroplasts, further analysis will reveal its relation to membrane stresses.
Our finding that VIPP1 becomes highly mobile in response to osmotic stress further supports an important role of VIPP1 in envelopes. As our observations showed, PspA has been shown to display a range of curved and linear motions along the plasma membrane, which is MreB dependent and important for PMF maintenance under pIV-secretin stress (Engl et al., 2009). A notable observation in our study is that although the lattice structures found in mature leaves were somewhat static, VIPP1-GFP showed dynamic movement in the area corresponding to swelled stroma. This result strongly suggests that VIPP1 responds to envelope stress very rapidly by supplying less aggregated VIPP1 proteins to the swelling area. We assume that a filament-like structure represents a newly formed VIPP1 complex that is reassembled around the damaged inner envelope. New VIPP1 can be supplied either by de novo synthesis of VIPP1 or by disassembly of preexisting complexes. Although both cases are possible, our live imaging implies that the latter case of the complex disassembly participates in this filament formation. How this movement is regulated in response to membrane stress remains as a subject for additional study. In C. reinhardtii, the large VIPP1 complex has been shown to interact with CDJ2/HSP70B (Liu et al., 2005). Moreover, it is disassembled by HSP70B(Dnak)-CKJ2-CGE1 chaperones in an ATP-dependent manner (Liu et al., 2007). Given that ATP synthesis in chloroplasts depends on a proton gradient across thylakoid membranes, VIPP1 aggregation and disassembly can be regulated by internal PMF, as proton leakage may allow VIPP1 to form large complexes.
In conclusion, we propose that VIPP1 plays a critical role in envelope maintenance, which has not been completely elucidated. The emergence of PspA-like proteins during evolution of photosynthetic organisms suggests that the proper control of PMF is necessary for the maintenance of envelope and thylakoid membranes and thereby affects thylakoid formation in chloroplasts.
METHODS
Plant Materials
Arabidopsis thaliana ecotype Col was used as the wild type in this study. The vipp1 knockdown line vipp1-kd was reported as hcf155 previously by Kroll et al. (2001). The vipp1 knockout line vipp1-ko was created through T-DNA insertion (SAIL_5_F07, obtained from the Nottingham Arabidopsis Stock Centre). Both vipp1-kd and vipp1-ko mutants are semilethal. Therefore, plants were grown on MS agar plates supplemented with Suc and maintained as a heterozygote.
The chimeric construct VIPP1-GFP was prepared as described by Aseeva et al. (2004) and was under the control of the CaMV 35S promoter. The VIPP1-GFP fusion construct was cloned into the plant expression vector pGreen0029 (Hellens et al., 2000). The resulting plasmid was named pGreen0029-VIPP1 and was transformed into Col by Agrobacterium tumefaciens through the floral dip method (Clough and Bent, 1998). Transgenic plants that expressed VIPP1-GFP were selected and crossed with heterozygous vipp1-kd and vipp1-ko to yield VIPP1-GFP/vipp1-kd and VIPP1-GFP/vipp1-ko plants that expressed VIPP1-GFP and that were homozygous for the corresponding vipp1 mutation. Another transgenic plant, L12-GFP/Col expresses L12-GFP (containing the transit peptide from rice [Oryza sativa] ribosomal protein L12) under the control of the CaMV 35S promoter and was kindly provided by Shin-ichi Arimura (University of Tokyo). This plant was described in an earlier report by Arimura et al. (1999). L12-GFP/vipp1-kd and L12-GFP/vipp1-ko were generated by crossing L12-GFP/Col with a vipp1-kd heterozygote and vipp1-ko heterozygote, respectively. L12-GFP/var2 was generated in our previous work (Kato et al., 2007). Surface-sterilized seeds were sown onto 0.7% MS agar plates supplemented with 1.5% (w/v) Suc. Plants were maintained under light (∼100 µmol m−2 s−1) with 12-h/12-h light/dark cycles at a constant temperature of 22°C. After a 2-week growth period on MS medium, the wild type and complement lines were transferred to soil, while two homozygous vipp1 mutants were grown only on MS. Phenotypes of the mutants are shown in Supplemental Figure 2 online. For all microscopy observations, leaves grown on MS agar plates were used.
Protoplast and Chloroplast Isolation
Fully expanded leaves of Arabidopsis were excised and suspended gently in enzyme solution (0.1% [w/v] cellulase [Onozuka R10; Yakult], 0.05% [w/v] Pectolyase Y-23 [Kyowa Chemical], 400 mM mannitol, 10 mM CaCl2, 20 mM KCl, 5 mM EGTA, and 20 mM MES, pH 5.7). After incubation at room temperature for 1.5 h, protoplasts were collected by centrifugation at 60g for 1 min and washed with ice-cold wash buffer (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES, pH 5.7). After centrifugation at 60g for 1 min, the isolated protoplasts were resuspended with 500 μL wash buffer and used for examination or for the subsequent step.
Intact chloroplasts were separated from protoplasts by filtration through a 20-µm nylon mesh and collected by centrifugation. The pellet was resuspended gently in the buffer (0.33 M mannitol, 1 mM MgCl2, and 50 mM HEPES-KOH, pH 7.7) and loaded on top of Percoll step gradients (40 and 70% [v/v] in the same buffer) and spun for 5 min at 2000g at 4°C. Chloroplasts between 40 and 70% Percoll were diluted and spun for 2 min at 1300g and washed once with the same buffer for additional analysis.
RT-PCR
mRNA isolation from Arabidopsis was performed using the NucleoSpin RNA II isolation kit (Macherey-Nagel) following the manufacturer’s instructions. For synthesis of cDNA, the M-MLV RT (H-)-Kit (Promega) was used. PCR analysis from cDNA was performed using the primers Vipp1-RT-fw (5′-CTAGCAAAAGTCGTTAGCTTTCCTTCGCAG-3′) and Vipp1-RT-rv (5′-CACCATGGCTCTCAAAGCTTCACCTGT-3′).
Protein Extraction, SDS-PAGE, and Immunoblot Analysis
Total proteins from Arabidopsis leaves were extracted using buffer (125 mM Tris-Cl, pH 6.8, 2% [w/v] SDS, 5% [v/v] glycerol, 5% [v/v] 2-mercaptoethanol, and 0.05% [w/v] bromophenol blue) supplemented with 1 mM phenylmethanesulfonyl fluoride. For Col and two complemented lines (VIPP1-GFP/vipp1-kd and VIPP1-GFP/vipp1-ko), proteins from Percoll-purified chloroplasts were extracted with the same buffer and subjected to immunoblot analysis. An equal amount of proteins (30 µg) was loaded onto an SDS-PAGE gel. The resolved gels were stained with Coomassie Brilliant Blue R 250 as loading control. For immunoblot analysis, total proteins were electroblotted onto polyvinylidene difluoride membrane (ATTO) after SDS-PAGE, and the membranes were blocked with 5% (w/v) milk in PBST buffer (50 mM sodium phosphate buffer, pH 7.5, 155 mM NaCl, and 0.05% [v/v] Tween 20) for 1 h. After three washes with PBST buffer, the membranes were incubated with anti-VIPP1 (Aseeva et al., 2007) or anti-GFP (sc-8334; Santa Cruz) for 2 h. After three more washes with PBST buffer, the membranes were incubated with second antibodies for 2 h. Proteins were detected using an ECL chemiluminescence detection system (Amersham Biosciences) as described previously (Sakamoto et al., 2003).
Fractionation of Chloroplasts
Fractionation of chloroplasts was performed as described (Li et al., 1994) with some modifications. Chloroplasts were lysed hypertonically by freezing and thawing in 0.1 M Suc solution. The lysate was diluted with one volume of TE buffer (10 mM Tris, pH 7.5, and 2 mM EDTA). The lysate was loaded onto an 8.0-mL Suc step gradient with 2.0 mL of 1.2 M Suc, 3.0 mL of 1.0 M Suc, and 3.0 mL of 0.46 M Suc. The gradient was centrifuged at 200,000g for 1 h. The stroma, envelope membrane, and thylakoid fractions were collected from the supernatant, 0.46 M/1.0 M Suc interface, and the pellet, respectively. SDS-PAGE and immunoblotting were conducted as mentioned above. The primary antibodies anti-LHCb1 (AS01004; Agrisera), anti-FBPase, and anti-Tic110 (Aseeva et al., 2007) were chosen to detect the representative proteins in the different fractions of chloroplast.
Blue Native Gel and Two-Dimensional SDS-PAGE
Blue Native-PAGE was performed as described (Schägger et al., 1994) with some modifications. Isolated chloroplasts from Col or VIPP1-GFP/Col were incubated with 1% n-dodecyl-β-maltoside for 1.0 h on ice. Samples were loaded onto 0.75-mm-thick 5 to 13.5% acrylamide gradient gels. Then, electrophoresis was performed at 4°C. For two-dimensional analysis, excised Blue Native-PAGE lanes were soaked in SDS sample buffer (125 mM Tris-Cl, pH 6.8, 2% [w/v] SDS, 5% [v/v] glycerol, 5% [v/v] 2-mercaptoethanol, and 0.05% [w/v] bromophenol blue) for 1.0 h and layered onto 1-mm-thick 15% SDS polyacrylamide gels containing 8 M urea. SDS-PAGE and immunoblotting were conducted as described above.
Microscopy Observation
For observation of chloroplasts/plastids in living leaf tissues, a piece of Arabidopsis leaf (1 × 1 mm) was excised and examined using a fluorescence microscope (DSU-BX51; Olympus) equipped with a disk scanning unit. For detecting signals from GFP and chlorophyll autofluorescence, different filter sets (U-MNIBA2 for green/GFP, U-MWIG2 for red/chlorophyll, and U-MWIB2 for both) were selected.
For TEM, Arabidopsis rosette leaves from 3-week-old plants grown on MS plates were cut into 1 × 1-mm pieces and fixed in 2% (w/v) paraformaldehyde and 2% (v/v) glutaraldehyde in 0.05 M cacodylic acid buffer, pH 7.4, and postfixed in 2% (v/v) osmium tetroxide in the same buffer at 4°C for 3 h. Samples were further dehydrated with a graded ethanol series (50, 70, 90, and 100%). Ethanol was subsequently replaced using a series of epoxy resin (Quetol 651; Nissin EM) dilutions (50, 70, 90, and 100%). Then, the resin was hardened for 2 d at 60°C. The chloroplast structure was evaluated on a transverse ultrathin section cut (Ultratome V; LKB Produkter). Sections were stained with 2% lead citrate and examined using a transmission electron microscope (JEM-1200EX; JEOL) at 80 kV, which was performed at Tokai Electron Microscopy.
Osmotic Pressure Test
Leaves from the vipp1-ko mutant were cut into small pieces (1 × 1 mm). Then, they were incubated with osmotic buffer (1 M mannitol, 1 mM MgCl2, 50 mM HEPES-KOH, pH 8.0, and 2 mM EDTA). The osmotic buffer was vacuum infiltrated by placing the incubated leaf pieces into a syringe and by pumping gently. The infiltrated leaf pieces were examined directly using microscopy. Isolated chloroplasts, collected from a brief spin-down at 1300g for 2 min, were suspended gently in osmotic buffer and incubated for 5 min on ice before examination using microscopy.
Motility Assay of VIPP1-GFP
To monitor the movement of VIPP1-GFP in chloroplasts of VIPP1-GFP/vipp1-kd, a piece of Arabidopsis leaf (1 × 1 mm) was excised and vacuum filtrated with 0.33 M mannitol or distilled water. Samples were mounted on a slide for observation on an Olympus BX51 upright microscope with a ×100 oil objective. A filter set (U-MWIB2 [excitation filter at 460 to 490 nm, emission long-pass filter at 510 nm]; Olympus) was selected for detecting signals from GFP and chlorophyll autofluorescence. Photographs were taken using a microscope-mounted video imaging system that consisted of a cooled charge-coupled device camera (DP70; Olympus) operated by the DP controller (Olympus) software package. Live images were captured at 5 frames/s. For observation of VIPP1-GFP movement in isolated chloroplasts, intact chloroplasts isolated from VIPP1-GFP/vipp1-kd through the Percoll gradient method were used. The pellets of intact chloroplasts were resuspended with 0.33 M mannitol solution or distilled water. Shortly thereafter, the samples were loaded on a slide for observation with the same device and method as used for the leaf pieces.
Expression of GFP-PspA and GFP-VIPP1 in the E. coli ΔpspA Mutant
Bacterial strains (MG1655, ΔpspA, and GFP-PSPA/ΔpspA) and plasmid (pEC1) were gifts from Martin Buck (Lloyd et al., 2004; Engl et al., 2009). A part of Arabidopsis VIPP1 cDNA (without the sequence corresponding to the transit peptide, N-terminal 71 amino acids) was PCR amplified using pGreen0029-VIPP1 as a template and a pair of primers, forward, 5′-CCGGAATTCACTATGAATCTTTTTGAACGAT-3′, and reverse, 5′-CCCAAGCTTCTAAAAGTCGTTAGCTTTCCTT-3′, thereby introducing EcoRI and HindIII restriction sites (underlined) into the 5′ and 3′ end of the cDNA, respectively. The PCR product was cleaved using the restriction enzymes and ligated into the EcoRI-HindIII sites of the GFP reporter plasmid pEC1 (Engl et al., 2009). The resulting plasmid, termed pEC1-VIPP1, was transformed into the ΔpspA mutant. Transformants were cultured in liquid medium overnight at 37°C. Then, they were subcultured into fresh Luria-Bertani and grown to an OD600 of 0.8. For immunoblot analysis, the same amount of bacteria was taken based on absorbance at 600 nm; it was denatured with SDS loading buffer and analyzed using SDS-PAGE and immunoblotting.
To determine the membrane potential of the E. coli strains used for this study, a membrane potential sensor cationic dye was used: JC-1 (Molecular Probes). An original solution of JC-1 was diluted to 5 mg/mL in DMSO and kept frozen at −20°C. Cells from an overnight Luria-Bertani culture were subcultured into fresh Luria-Bertani and grown to an A600 of 0.8. One milliliter of culture was spun down and resuspended in 1 mL of permeabilization buffer (10 mM Tris, pH 7.5, 1 mM EDTA, and 10 mM Glc). Then, 2 μL of 5 mg/mL JC-1 was added to the bacterial solution and incubated for 20 min at room temperature. The bacterial suspension was applied onto a microscope slide and checked using a microscope. Fluorescent bacteria were visualized using a fluorescence microscope with a filter set (excitation 460 to 490 nm, emission >510 nm, U-MWIB2; Olympus). The fluorescence emission of JC-1 shifts from red (598 nm) to green (538 nm) depending on the membrane potential (Becker et al., 2005). In addition to microscopy observation, a fluorescence spectrophotometer (F-7000; Hitachi) was used to scan the emission profile of bacteria. The fluorescence intensities of the cell suspensions were measured using excitation wavelengths of 485 nm and scanning emission wavelengths of 500 to 650 nm.
Accession Numbers
Sequence data of VIPP1 can be found in the Arabidopsis Genome Initiative database under accession number At1g65260.1. Sequence information of PspA can be found in GenBank under accession number U00096.2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Alignment of VIPP1/PspA from Arabidopsis and E. coli.
Supplemental Figure 2. Phenotypes of vipp1-kd and vipp1-ko.
Supplemental Figure 3. Phenotype of the Balloon-Like Chloroplast in Protoplasts Isolated from vipp1-kd.
Supplemental Figure 4. Characterization of a Homozygous vipp1-ko Mutant Line.
Supplemental Figure 5. Suborganelle Localization of VIPP1 in Arabidopsis Chloroplasts.
Supplemental Movie 1. Real-Time Movie of VIPP1-GFP/vipp1-kd Chloroplasts of an Arabidopsis Leaf Observed in Situ, Vacuum-Filtrated with Distilled Water.
Supplemental Movie 2. Real-Time Movie of VIPP1-GFP/vipp1-kd Chloroplasts of an Arabidopsis Leaf Observed in Situ with No Treatment.
Supplemental Movie 3. Real-Time Movie of VIPP1-GFP/vipp1-kd Chloroplasts of an Arabidopsis Leaf Observed in Situ, Vacuum-Filtrated with 0.33M Mannitol.
Supplemental Movie 4. Real-Time Movie of a Percoll-Purified Chloroplast from VIPP1-GFP/vipp1-kd Treated with Distilled Water.
Supplemental Movie 5. Real-Time Movie of a Percoll-Purified Chloroplast from VIPP1-GFP/vipp1-kd Treated with 0.33M Mannitol.
Supplemental Movie Legends 1. Legends for the Supplemental Movies.
Acknowledgments
We thank Martin Buck for providing bacterial strains (MG1655, ΔpspA, and GFP-PSPA/ΔpspA) and plasmid (pEC1). We also thank Shinichi Arimura for the L12-GFP/Col transgenic Arabidopsis and Jürgen Soll for providing antibodies against Tic110 and FBPase. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (No. 21-09310 to W.S.), the Oohara Foundation (to W.S.), and the Deutsche Forschungsgemeinschaft (SFB-TR1 Project A6 to U.C.V.). L.Z. was partly supported by a postdoctoral fellowship from the Japan Society for the Promotion of Science.
AUTHOR CONTRIBUTIONS
W.S. and L.Z. designed the research, performed experiments, analyzed the data, and wrote the article. S.O. performed experiments. Y.K. and U.C.V. analyzed the data.
Glossary
- PMF
proton motive force
- MS
Murashige and Skoog
- Col
Columbia
- CaMV
cauliflower mosaic virus
- TEM
transmission electron microscopy
- GFP
green fluorescent protein
References
- Arimura S., Takusagawa S., Hatano S., Nakazono M., Hirai A., Tsutsumi N. (1999). A novel plant nuclear gene encoding chloroplast ribosomal protein S9 has a transit peptide related to that of rice chloroplast ribosomal protein L12. FEBS Lett. 450: 231–234 [DOI] [PubMed] [Google Scholar]
- Aseeva E., Ossenbühl F., Eichacker L.A., Wanner G., Soll J., Vothknecht U.C. (2004). Complex formation of Vipp1 depends on its alpha-helical PspA-like domain. J. Biol. Chem. 279: 35535–35541 [DOI] [PubMed] [Google Scholar]
- Aseeva E., Ossenbühl F., Sippel C., Cho W.K., Stein B., Eichacker L.A., Meurer J., Wanner G., Westhoff P., Soll J., Vothknecht U.C. (2007). Vipp1 is required for basic thylakoid membrane formation but not for the assembly of thylakoid protein complexes. Plant Physiol. Biochem. 45: 119–128 [DOI] [PubMed] [Google Scholar]
- Becker L.A., Bang I.S., Crouch M.L., Fang F.C. (2005). Compensatory role of PspA, a member of the phage shock protein operon, in rpoE mutant Salmonella enterica serovar Typhimurium. Mol. Microbiol. 56: 1004–1016 [DOI] [PubMed] [Google Scholar]
- Berkowitz G.A., Peters J.S. (1993). Chloroplast inner envelope ATPase acts as a primary H+ pump. Plant Physiol. 102: 261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissette J.L., Russel M., Weiner L., Model P. (1990). Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87: 862–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultema J.B., Fuhrmann E., Boekema E.J., Schneider D. (2010). Vipp1 and PspA: Related but not twins. Commun. Integr. Biol. 3: 162–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cserzö M., Wallin E., Simon I., von Heijne G., Elofsson A. (1997). Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10: 673–676 [DOI] [PubMed] [Google Scholar]
- Dekov I., Tsonev T., Yordanov I. (2000). Effects of water stress and high-temperature stress on the structure and activity of photosynthetic apparatus of Zea mays and Helianthus annuus. Photosynthetica 38: 361–366 [Google Scholar]
- DeLisa M.P., Lee P., Palmer T., Georgiou G. (2004). Phage shock protein PspA of Escherichia coli relieves saturation of protein export via the Tat pathway. J. Bacteriol. 186: 366–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R., Joyard J. (1990). Biochemistry and function of the plastid envelope. Annu. Rev. Cell Biol. 6: 173–216 [DOI] [PubMed] [Google Scholar]
- Engl C., Jovanovic G., Lloyd L.J., Murray H., Spitaler M., Ying L., Errington J., Buck M. (2009). In vivo localizations of membrane stress controllers PspA and PspG in Escherichia coli. Mol. Microbiol. 73: 382–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K. (2011). The import and export business in plastids: Transport processes across the inner envelope membrane. Plant Physiol. 155: 1511–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge U.I., Hinz G. (1986). Energy dependence of protein translocation into chloroplasts. Eur. J. Biochem. 160: 563–570 [DOI] [PubMed] [Google Scholar]
- Fuks B., Homblé F. (1999). Passive anion transport through the chloroplast inner envelope membrane measured by osmotic swelling of intact chloroplasts. Biochim. Biophys. Acta 1416: 361–369 [DOI] [PubMed] [Google Scholar]
- Gao H., Xu X.D. (2009). Depletion of Vipp1 in Synechocystis sp. PCC 6803 affects photosynthetic activity before the loss of thylakoid membranes. FEMS Microbiol. Lett. 292: 63–70 [DOI] [PubMed] [Google Scholar]
- Garcia C., Khan N.Z., Nannmark U., Aronsson H. (2010). The chloroplast protein CPSAR1, dually localized in the stroma and the inner envelope membrane, is involved in thylakoid biogenesis. Plant J. 63: 73–85 [DOI] [PubMed] [Google Scholar]
- Haswell E.S., Meyerowitz E.M. (2006). MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr. Biol. 16: 1–11 [DOI] [PubMed] [Google Scholar]
- Heber U., Heldt H.W. (1981). The chloroplast envelope: Structure, function, and role in leaf metabolism. Annu. Rev. Plant Physiol. 32: 139–168 [Google Scholar]
- Hellens R., Mullineaux P., Klee H. (2000). Technical Focus: A guide to Agrobacterium binary Ti vectors. Trends Plant Sci. 5: 446–451 [DOI] [PubMed] [Google Scholar]
- Herrmann R.G. (1999). Biogenesis and evolution of photosynthetic (thylakoid) membranes. Biosci. Rep. 19: 355–365 [DOI] [PubMed] [Google Scholar]
- Hongladarom T., Honda S.I. (1966). Reversible swelling and contraction of isolated spinach chloroplasts. Plant Physiol. 41: 1686–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K. (2007). The chloroplast outer envelope membrane: The edge of light and excitement. J. Integr. Plant Biol. 49: 1100–1111 [Google Scholar]
- Jouhet J., Gray J.C. (2009). Interaction of actin and the chloroplast protein import apparatus. J. Biol. Chem. 284: 19132–19141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic G., Lloyd L.J., Stumpf M.P., Mayhew A.J., Buck M. (2006). Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J. Biol. Chem. 281: 21147–21161 [DOI] [PubMed] [Google Scholar]
- Kato Y., Miura E., Matsushima R., Sakamoto W. (2007). White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiol. 144: 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K., Olsen L.J., Theg S.M. (1989). Chloroplastic precursors and their transport across the envelope membranes. Annu. Rev. Plant Physiol. 40: 471–501 [Google Scholar]
- Kobayashi R., Suzuki T., Yoshida M. (2007). Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol. Microbiol. 66: 100–109 [DOI] [PubMed] [Google Scholar]
- Kota Z., Horvath L.I., Droppa M., Horvath G., Farkas T., Pali T. (2002). Protein assembly and heat stability in developing thylakoid membranes during greening. Proc. Natl. Acad. Sci. USA 99: 12149–12154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll D., Meierhoff K., Bechtold N., Kinoshita M., Westphal S., Vothknecht U.C., Soll J., Westhoff P. (2001). VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. USA 98: 4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.M., Kaneko Y., Keegstra K. (1994). Molecular cloning of a chloroplastic protein associated with both the envelope and thylakoid membranes. Plant Mol. Biol. 25: 619–632 [DOI] [PubMed] [Google Scholar]
- Liu C., Willmund F., Golecki J.R., Cacace S., Hess B., Markert C., Schroda M. (2007). The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas. Plant J. 50: 265–277 [DOI] [PubMed] [Google Scholar]
- Liu C.M., Willmund F., Whitelegge J.P., Hawat S., Knapp B., Lodha M., Schroda M. (2005). J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol. Biol. Cell 16: 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd L.J., Jones S.E., Jovanovic G., Gyaneshwar P., Rolfe M.D., Thompson A., Hinton J.C., Buck M. (2004). Identification of a new member of the phage shock protein response in Escherichia coli, the phage shock protein G (PspG). J. Biol. Chem. 279: 55707–55714 [DOI] [PubMed] [Google Scholar]
- Lo S.M., Theg S.M. (2012). Role of vesicle-inducing protein in plastids1 in cpTat transport at the thylakoid. Plant J. 71: 656–668
- McCain D.C., Croxdale J., Markley J.L. (1989). Thermal damage to chloroplast envelope membranes. Plant Physiol. 90: 606–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monge E., Perez C., Pequerul A., Madero P., Val J. (1993). Effect of iron chlorosis on mineral-nutrition and lipid-composition of thylakoid biomembrane in Prunus persica (L.) bastch. Plant Soil 154: 97–102 [Google Scholar]
- Mould R.M., Robinson C. (1991). A proton gradient is required for the transport of two lumenal oxygen-evolving proteins across the thylakoid membrane. J. Biol. Chem. 266: 12189–12193 [PubMed] [Google Scholar]
- Nordhues A., et al. (2012). Evidence for a role of VIPP1 in the structural organization of the photosynthetic apparatus in Chlamydomonas. Plant Cell 24: 637–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q., Zhou Q. (2009). Influence of lanthanum on chloroplast ultrastructure of soybean leaves under ultraviolet-B stress. J. Rare Earths 27: 304–307 [Google Scholar]
- Racker E., Schroeder E.A. (1958). The reductive pentose phosphate cycle. II. Specific C-1 phosphatases for fructose 1,6-diphosphate and sedoheptulose 1,7-diphosphate. Arch. Biochem. Biophys. 74: 326–344 [DOI] [PubMed] [Google Scholar]
- Robinson C., Woolhead C., Edwards W. (2000). Transport of proteins into and across the thylakoid membrane. J. Exp. Bot. 51(Spec No): 369–374 [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Miyagishima S.-y., Jarvis P. (2008). Chloroplast biogenesis: Control of plastid development, protein import, division and inheritance. In The Arabidopsis Book 6: e0110, doi/10.1199/tab.0110 [DOI] [PMC free article] [PubMed]
- Sakamoto W., Uno Y., Zhang Q., Miura E., Kato Y., Sodmergen (2009). Arrested differentiation of proplastids into chloroplasts in variegated leaves characterized by plastid ultrastructure and nucleoid morphology. Plant Cell Physiol. 50: 2069–2083 [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Zaltsman A., Adam Z., Takahashi Y. (2003). Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15: 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., Cramer W.A., von Jagow G. (1994). Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217: 220–230 [DOI] [PubMed] [Google Scholar]
- Scott S.V., Theg S.M. (1996). A new chloroplast protein import intermediate reveals distinct translocation machineries in the two envelope membranes: Energetics and mechanistic implications. J. Cell Biol. 132: 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R., Pisareva T., Norling B. (2005). Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 5: 4905–4916 [DOI] [PubMed] [Google Scholar]
- Standar K., Mehner D., Osadnik H., Berthelmann F., Hause G., Lünsdorf H., Brüser T. (2008). PspA can form large scaffolds in Escherichia coli. FEBS Lett. 582: 3585–3589 [DOI] [PubMed] [Google Scholar]
- Tzinas G., Argyroudiakoyunoglou J.H., Akoyunoglou G. (1987). The effect of the dark interval in intermittent light on thylakoid development - Photosynthetic unit formation and light harvesting protein accumulation. Photosynth. Res. 14: 241–258 [DOI] [PubMed] [Google Scholar]
- van den Wijngaard P.W., Vredenberg W.J. (1997). A 50-picosiemens anion channel of the chloroplast envelope is involved in chloroplast protein import. J. Biol. Chem. 272: 29430–29433 [DOI] [PubMed] [Google Scholar]
- Vassilev A., Iordanov I., Chakalova E., Kerin V. (1995). Effect of cadmium stress on growth and photosynthesis of young barley (Hordeum vulgare) plants. 2. Structural and functional changes in the photosynthetic apparatus. Bulg. J. Plant Physiol. 21: 12–21 [Google Scholar]
- Vothknecht U.C., Westhoff P. (2001). Biogenesis and origin of thylakoid membranes. Biochim. Biophys. Acta 1541: 91–101 [DOI] [PubMed] [Google Scholar]
- Wang Q., Sullivan R.W., Kight A., Henry R.L., Huang J., Jones A.M., Korth K.L. (2004). Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 136: 3594–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal S., Heins L., Soll J., Vothknecht U.C. (2001). Vipp1 deletion mutant of Synechocystis: A connection between bacterial phage shock and thylakoid biogenesis? Proc. Natl. Acad. Sci. USA 98: 4243–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Kato Y., Zhang L., Fujimoto M., Tsutsumi N., Sodmergen, Sakamoto W. (2010). The FtsH protease heterocomplex in Arabidopsis: Dispensability of type-B protease activity for proper chloroplast development. Plant Cell 22: 3710–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]