This study presents detailed genetic, biochemical, functional, and physiological analyses of phospholipid:diacylglycerol acyltransferase (PDAT) in the green microalga Chlamydomonas reinhardtii. Multiple lines of evidence indicate that PDAT is a multifunctional protein and plays an essential role in membrane turnover, degradation, and TAG formation in C. reinhardtii.
Abstract
Many unicellular microalgae produce large amounts (∼20 to 50% of cell dry weight) of triacylglycerols (TAGs) under stress (e.g., nutrient starvation and high light), but the synthesis and physiological role of TAG are poorly understood. We present detailed genetic, biochemical, functional, and physiological analyses of phospholipid:diacylglycerol acyltransferase (PDAT) in the green microalga Chlamydomonas reinhardtii, which catalyzes TAG synthesis via two pathways: transacylation of diacylglycerol (DAG) with acyl groups from phospholipids and galactolipids and DAG:DAG transacylation. We demonstrate that PDAT also possesses acyl hydrolase activities using TAG, phospholipids, galactolipids, and cholesteryl esters as substrates. Artificial microRNA silencing of PDAT in C. reinhardtii alters the membrane lipid composition, reducing the maximum specific growth rate. The data suggest that PDAT-mediated membrane lipid turnover and TAG synthesis is essential for vigorous growth under favorable culture conditions and for membrane lipid degradation with concomitant production of TAG for survival under stress. The strong lipase activity of PDAT with broad substrate specificity suggests that this enzyme could be a potential biocatalyst for industrial lipid hydrolysis and conversion, particularly for biofuel production.
INTRODUCTION
Glycerolipids represent the largest class of lipids, comprising 10 to 90% of eukaryotic cell dry weight (Bell and Coleman, 1980). These compounds play a variety of biological roles, including being major components of membrane matrices and functional complexes (such as photosynthetic reaction centers and electron transport chains), highly concentrated carbon and energy reserves, and signal molecules. Triacylglycerols (TAGs) are the most important carbon and energy storage lipids in eukaryotes and are also a major source of highly reduced carbon molecules for food and fuel.
In eukaryotes, TAGs are known to be synthesized via multiple pathways. The acyl-CoA–dependent Kennedy pathway involves three sequential acylations of glycerol-3-phosphate catalyzed by a suite of enzymes, the last of which is diacylglycerol acyltransferase (DGAT) that catalyzes the acylation of a diacylglycerol (DAG) with an acyl-CoA to form TAG. This pathway has been well studied in yeasts (Lardizabal et al., 2001; Sorger and Daum, 2002), plants (Katavic et al., 1995; Jako et al., 2001; Shockey et al., 2006), and mammals (Cases et al., 1998, 2001). Another TAG synthesis pathway is the acyl-CoA–independent pathway, mediated by phospholipid:DAG acyltransferases (PDATs) that transfer a fatty acyl moiety from a phospholipid (PL) to DAG to form TAG. The PDAT pathway has been reported in yeasts (Dahlqvist et al., 2000; Zhang et al., 2012) and vascular plants (Dahlqvist et al., 2000; Ståhl et al., 2004). Finally, the transacylation pathway uses two DAG molecules as both acyl donors and acceptors to form TAG. This pathway is catalyzed by an acyl-CoA–independent diacylglycerol transacylase (DGTA). DGTA activities have been detected in microsome preparations of intestines of Rattus norvegicus (Lehner and Kuksis, 1993) and developing seeds of safflower (Carthamus tinctorius) (Lehner and Kuksis, 1993; Stobart et al., 1997), but no gene encoding DGTA has been identified in any organism.
Many unicellular microalgae produce large amounts of neutral lipids (20 to 50% of cell dry weight), mainly in the form of TAG, under unfavorable environmental or culture conditions, but the molecular and cellular mechanisms for TAG synthesis and accumulation are less well understood (Hu et al., 2008). The unicellular green microalga Chlamydomonas reinhardtii serves as a model organism for elucidating the pathways and physiological role of TAG synthesis. C. reinhardtii grows rapidly and produces TAG, particularly under adverse abiotic environmental conditions (Weers and Gulati, 1997; Li et al., 2010a, 2010b; Miller et al., 2010), and its entire genome sequence and genetic toolbox are available (Kindle, 1990; Merchant et al., 2007; Molnar et al., 2009). We observed that the accumulation of TAG in C. reinhardtii under stress is accompanied by substantial degradation of chloroplast membranes and thus hypothesized that fatty acids from the chloroplasts and other intracellular membrane systems may be converted into TAG by a PDAT-like protein. Because the predominant lipids in the chloroplast membranes are galactolipids, including monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) (Janero and Barrnett, 1981), we further hypothesized that the PDAT-like protein might also function as a galactolipid:DAG acyltransferase. Recently, the single gene encoding PDAT in C. reinhardtii, referred to as Cr-PDAT in this study, was cloned, and a loss-of-function study indicated that the PDAT pathway contributed to ∼25% of the total TAG accumulated in C. reinhardtii under nitrogen-depleted conditions (Boyle et al., 2012). However, the biochemical and functional properties of Cr-PDAT remain elusive.
In this study, we cloned a full-length cDNA encoding PDAT from C. reinhardtii. The in vitro and in vivo biochemical characteristics and physiological functions of Cr-PDAT were elucidated. Multiple lines of evidence indicate that PDAT is a multifunctional protein and plays an essential role in membrane turnover, degradation, and TAG formation in C. reinhardtii.
RESULTS
Identification and Cloning of a Putative PDAT cDNA from C. reinhardtii
To identify the gene encoding PDAT in C. reinhardtii, we conducted a BLAST search of the Chlamydomonas genome database and GenBank with Sc-PDAT and At-PDAT as query sequences. We obtained a sequence annotated as phosphatidylcholine-sterol O-acyltransferase (LCAT) (accession number XM_001699696). This putative LCAT gene encoded a protein with high similarity to Sc-PDAT (35%) and At-PDAT (34%); therefore, this gene was cloned and designated as Cr-PDAT. The full-length cDNA of Cr-PDAT contained a 3157-bp open reading frame flanked by 5′ and 3′ untranslated regions of 126 and 503 bp, respectively (see Supplemental Figure 1A online). The open reading frame consisted of 15 exons (see Supplemental Figure 1A online) and is predicted to encode a protein of 1041 amino acid residues with a mass of 104,600 D and a pI of 5.96.
Analysis of the amino acid sequence predicted from the Cr-PDAT cDNA indicated that this gene belongs to the membrane-bound O-acyl transferase (MBOAT) family. Like all other MBOAT proteins, a putative N-glycosylation site and a Ser/Thr phosphorylation site were detected in Cr-PDAT, and InterProScan indicated the presence of an LCAT domain between amino acids 267 and 539 of the deduced Cr-PDAT amino acid sequence (see Supplemental Figure 2 online).
Sequence alignment of a number of LCAT-like proteins, including PDAT, LCAT, and phospholipid:sterol acyltransferase, revealed seven characteristic conserved regions (see Supplemental Figure 1B online). A catalytic triad (Ser-Asp-His), which was previously identified in mammalian LCAT (Peelman et al., 1998) and plant and yeast PDAT (Ståhl et al., 2004; Banas et al., 2005), was conserved in all LCAT-like proteins, including Cr-PDAT. This triad is part of the catalytic domain of LCAT enzymes, by which a fatty acid is cleaved from the sn-2 position of phosphatidylcholine (PC) and then transesterified with a free hydroxyl group of cholesterol to generate a cholesterol ester (Peelman et al., 1998). A highly conserved hydrophobic domain (domain I in Supplemental Figure 1B online) was found in all LCAT-like proteins, but its function is unknown. LCAT-like proteins also contain a so-called lid domain (domain II; see Supplemental Figure 1B online), which may be involved in destabilizing the lipid bilayer, facilitating binding of a hydrophobic substrate and diffusing it into the enzyme’s active site cavity (Peelman et al., 1998). A Trp (Trp-285) residue present in this lid domain (see Supplemental Figure 1B online) is thought to bind the cleaved fatty acid in the active site of the At-PDAT (Martinelle et al., 1996; Ståhl et al., 2004). Domain III (corresponding to 360PYDWRL365 in Cr-PDAT) is highly conserved in all sequences (see Supplemental Figure 1B online) and may be involved in PL recognition; several residues, including Glu-173 and the amphipathic helix encompassing residues 175 to 198 in the human LCAT (Hs-LCAT), which determines its substrate specificity and binding (Peelman et al., 1998), reside within this domain.
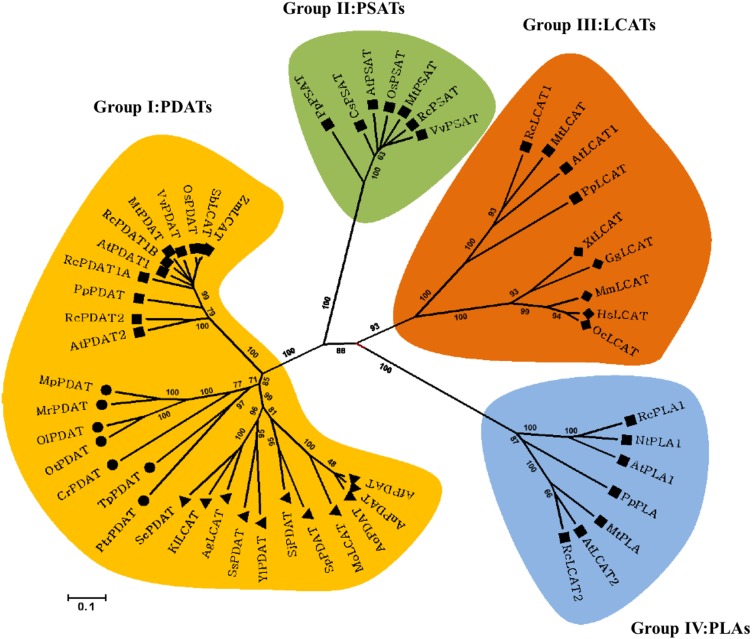
To gain insights into the evolutionary relationship between Cr-PDAT and other MBOAT proteins, we reconstructed a cladogram based on the multiple sequences from plants, animals, fungi, and algae. Based upon our phylogenetic analysis, the MBOAT family proteins were clustered into four major groups, corresponding to LCAT, PDAT, phospholipid:sterol acyltransferase, and phospholipase (PLA) proteins (Figure 1). The PDAT group branched into three subgroups: plant, fungal, and green algal PDAT. The algal PDAT lineage was more closely related to fungal PDAT than plant PDAT. The PLA group was the most divergent group related to PDAT but was closely related to the LCAT group.
Figure 1.
Cladogram of the LCAT-Like Family Proteins from Plants, Fungi, Microalgae, and Animals.
The neighbor-joining method was used to reconstruct the cladogram. The bootstrap value (obtained from 1000 replicates) is shown on each node. The 0.1 scale represents 10% divergence, calculated as the estimated number of replacement. Squares, plants; triangles, fungi; circles, microalgae; diamonds, animals.
[See online article for color version of this figure.]
TMHMM analysis (transmembrane prediction based on a hidden Markov model) and hydropathy profiling indicated that Cr-PDAT is an integral membrane protein with a transmembrane domain of 23 amino acids at its N terminus (see Supplemental Figure 3 online), similar to the structures of At-PDAT and Sc-PDAT (Dahlqvist et al., 2000; Ståhl et al., 2004). PDAT activities were detected in microsome preparations from yeasts and vascular plants (Dahlqvist et al., 2000), suggesting a possible localization of this enzyme in the endoplasmic reticulum (ER); however, these microsome preparations contained other cellular compartments (e.g., Golgi apparatus and plasma membrane); thus, the presence of PDAT in those organelles cannot be ruled out. Three plant PDATs have an aromatic amino acid–rich stretch in the C-terminal end recently shown to act as an ER retrieval motif (McCartney et al., 2004). However, no signal peptide or C-terminal retention sequence for ER targeting was detected in the Cr-PDAT sequence. Instead, a 92–amino acid canonical chloroplast transit peptide was identified at the N terminus of Cr-PDAT, suggesting a possible chloroplast localization.
Expression of the Cr-PDAT Gene in Pichia pastoris
To determine the function and enzymatic activity of Cr-PDAT, full-length PDAT (ORFCrPDAT) and truncated PDAT without the transmembrane domain (ΔTMCrPDAT) were cloned into the yeast expression vector pPICZαB in frame with an α-factor secretion sequence and were expressed in the yeast P. pastoris under the control of a methanol-inducible promoter. The recombinant proteins were purified by affinity chromatography and analyzed by SDS-PAGE. The molecular masses of the recombinant proteins were 95.0 and 120.0 kD, corresponding to the predicted molecular sizes of ΔTMCrPDAT and ORFCrPDAT, respectively. The identities of both recombinant proteins were further confirmed by immunoblotting (see Supplemental Figure 4 online) using an antiserum raised against ΔTMCrPDAT protein expressed in Escherichia coli cells (see Supplemental Figure 4A online). These two recombinant proteins were isolated and used for enzyme activity assays.
Typical PDAT Activity of Cr-PDAT
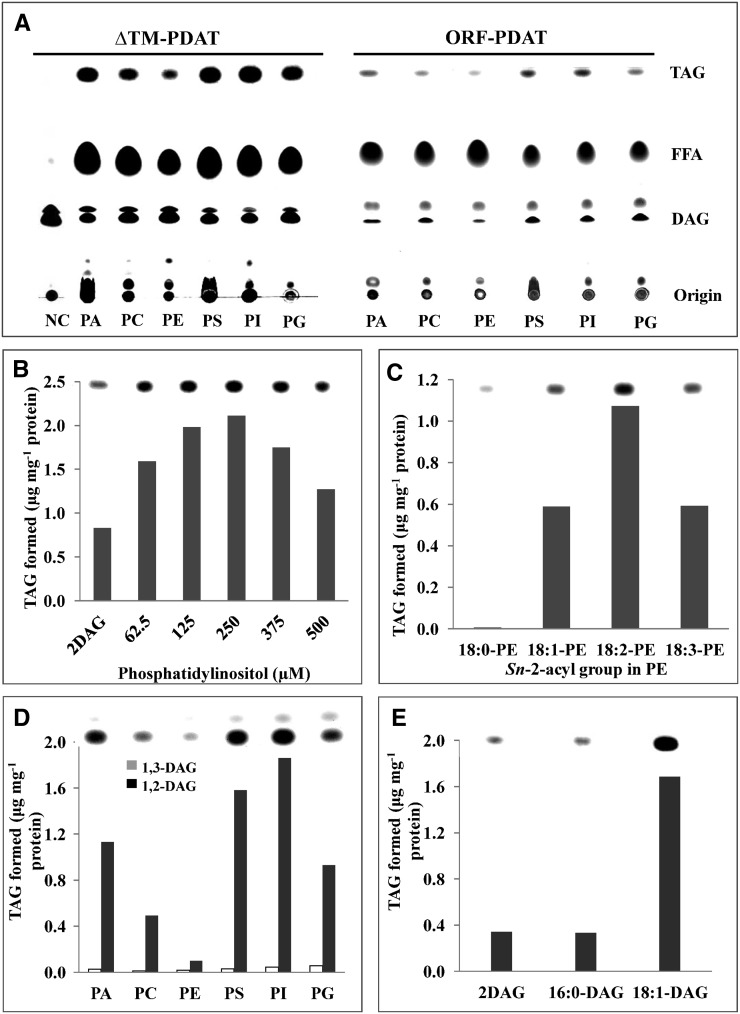
In vitro enzyme activity assays revealed that both ORFCrPDAT and ΔTMCrPDAT mediated the incorporation of DAG into TAG in the presence of different PL substrates (Figure 2A). ΔTMCrPDAT showed slightly higher PDAT activity than ORFCrPDAT under optimized reaction conditions (Figure 2A). The lower activity of the full-length Cr-PDAT was probably due to improper conformational change of the membrane proteins in the aqueous reaction buffer. As shown in Figure 2A, both ORFCrPDAT and ΔTMCrPDAT exhibited a strong preference for anionic PL, including phosphatidic acid (PA), phosphatidylserine (PS), phosphatidylinositol (PI), and phosphatidylglycerol (PG), but showed less activities with the cationic PL (i.e., PC and phosphatidylethanolamine [PE]). Conversely, Sc-PDAT and At-PDAT exhibited strong preferences for PE and PC, respectively (Dahlqvist et al., 2000; Ståhl et al., 2004; Ghosal et al., 2007). The enzymatic activity of ΔTMCrPDAT was determined using PI as an acyl donor and 1,2-DAG as an acyl acceptor. The rate of ΔTMCrPDAT-catalyzed TAG synthesis was proportional to the PI concentration up to 250 μM PI, above which TAG production was inhibited (Figure 2B).
Figure 2.
PDAT Activity and Substrate Specificity of Recombinant Cr-PDAT.
(A) PDAT activity of ORFCrPDAT and ΔTMCrPDAT. The sn-1,2-diolein (18:1 DAG) and different PLs, including PA, PS, PI, PG, PC, and PE, were used as substrates. Heat-inactivated PDAT was used as negative control (NC) for the assay. FFA, free fatty acid.
(B) Effects of different PL concentrations on the biosynthesis of TAG.
(C) Acyl donor side-chain specificity of Cr-PDAT. The substrate specificity was determined with PE containing various sn-2-acyl groups, and sn-1,2-diolein was used as an acyl acceptor.
(D) Acyl acceptor positional specificity of Cr-PDAT. The substrate specificity was determined with 1,2-dioleoyl-sn-glycerol (1,2-DAG) and 1,3-dioleoyl-sn-glycerol (1,3-DAG). Various PLs (PA, PC, PE, PS, PI, and PG) were used as acyl donors.
(E) Acyl acceptor side-chain specificity of Cr-PDAT. The substrate specificity was determined with 1,2-dipalmitoyl-sn-glycerol (16:0-DAG) and 1,2-dioleoyl-sn-glycerol (18:1-DAG). PI (250 µM) was used as an acyl donor. ΔTMCrPDAT was used in the enzymatic assays ([B] to [E]).
The substrate specificity of ΔTMCrPDAT for different acyl groups at the sn-2 position of PL was also investigated using various PE molecules as acyl donors and 1,2-DAG as acyl acceptors (Figure 2C). The highest enzymatic activity was detected when PE had a linoleic fatty-acyl group (18:2) at the sn-2 position of glycerol, whereas the lowest activity occurred when PE had a stearic (18:0) fatty-acyl group at the sn-2 position. The acyl acceptor specificity of Cr-PDAT was examined with 1,3- and 1,2-DAG, respectively. As shown in Figure 1D, Cr-PDAT exhibited a preference for 1,2-DAG over 1,3-DAG for all species of PLs tested as acyl donors (e.g., PA, PC, PE, PS, PI, and PG). ΔTMCrPDAT preferred DAG with unsaturated fatty acid side chains (1,2-18:1) compared with the saturated 1,2-16:0 DAG (Figure 2E).
Cr-PDAT DGTA Activity
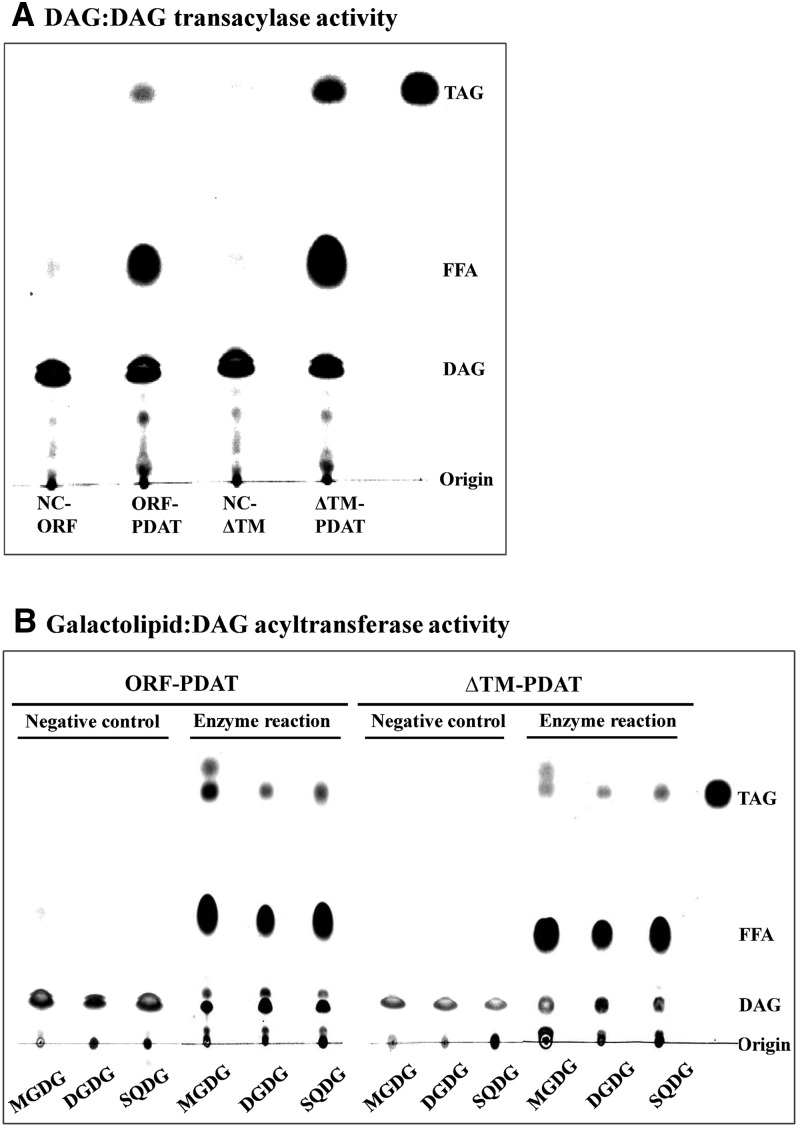
DGTA-catalyzed TAG formation has been observed in some eukaryotic organisms that can use DAG molecules as both acyl donor and acceptor (Lehner and Kuksis, 1993; Stobart et al., 1997). We investigated whether the recombinant ΔTMCrPDAT protein exhibited any DGTA activity by incubating sn-1,2-18:1-DAG with either ORFCrPDAT or ΔTMCrPDAT. TAGs were detected by thin layer chromatography (TLC) analysis, although the conversion efficiency was only one-seventh of its acyltransferase activity (Figure 3A), indicating that Cr-PDAT can indeed use DAG to form TAG.
Figure 3.
DAG:DAG Transacylase and Galactolipid:DAG Acyltransferase Activities of Recombinant Cr-PDAT.
(A) DAG:DAG transacylase activities of Cr-PDAT. FFA, free fatty acid.
(B) Galactolipid:DAG acyltransferase activities of Cr-PDAT.
Sn-1,2-diolein (18:1DAG) was used as an acyl donor and acceptor in the DAG:DAG transacylase assay or as acyl donor in the galactolipid:DAG acyltransferase assay. Heat-inactivated Cr-PDAT was used as negative control (NC).
Cr-PDAT Galactolipid:DAG Acyltransferase Activity
Considering that Cr-PDAT was predicted to be a chloroplast-localized protein and was previously identified in the chloroplast of C. reinhardtii by proteomics analysis (Terashima et al., 2011), we suspected that Cr-PDAT could use galactolipids, the most abundant membrane lipids in chloroplasts, as substrates by a mechanism similar to the PL:DAG acyl transfer reaction. To test this hypothesis, galactolipids (e.g., MGDG and DGDG) and the sulfolipid sulfoquinovosyldiacylglycerol (SQDG) were used for acyltransferase assays with both ORFCrPDAT and ΔTMCrPDAT. As shown in Figure 3B, ORFCrPDAT was able to use MGDG but not DGDG and SQDG as acyl donors to form TAG. However, ΔTMCrPDAT failed to convert MGDG into TAG, indicating that the transmembrane domain of ORFCrPDAT may be critical for its MGDG:DAG acyltransferase activity. An alternative explanation is that ORFCrPDAT is more hydrophobic than ΔTMCrPDAT and might react only with nonpolar lipids, such as MGDG, under our enzyme assay conditions.
Cr-PDAT Lipase Activity with Broad Substrate Specificity
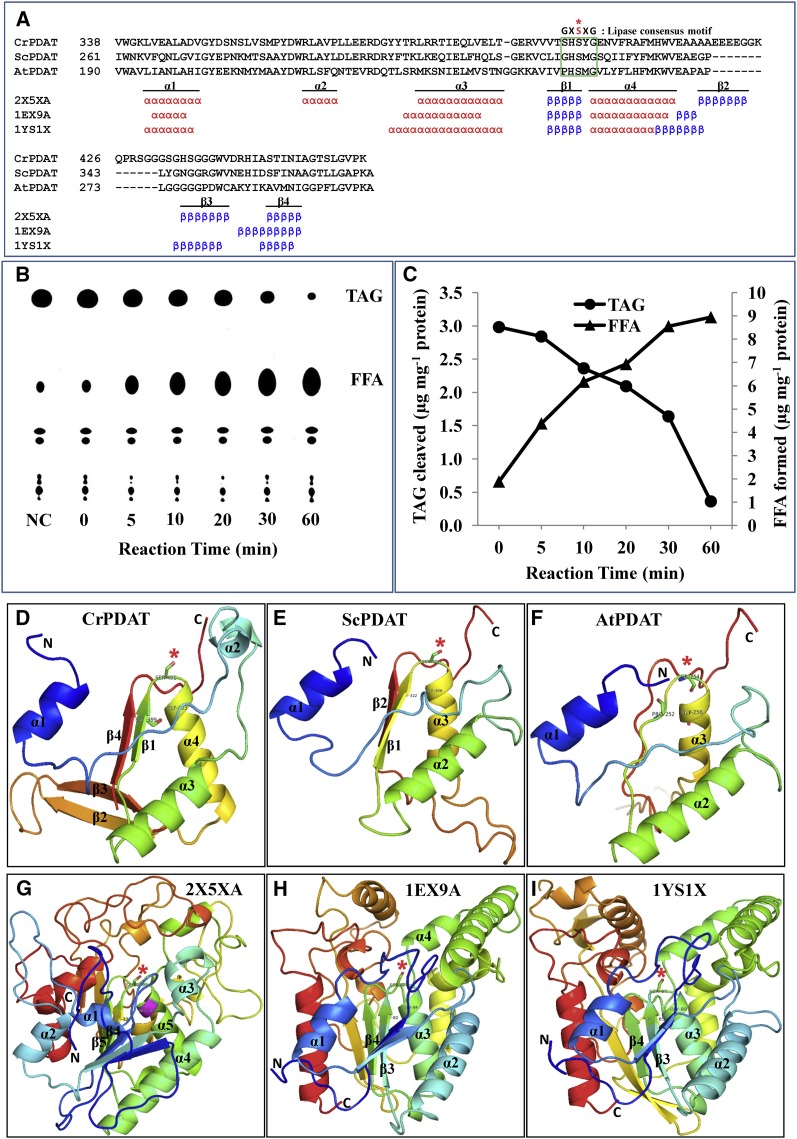
Noticeable amounts of free fatty acids were detected in the acyl transfer reaction catalyzed by ORFCrPDAT or ΔTMCrPDAT but not in the negative control reactions in which heat-inactivated PDAT was introduced (Figures 2 and 3). This points to a possible role of Cr-PDAT as a lipase. To test this, we conducted a structure-based functional analysis. A sequence comparison and secondary structure prediction revealed that the catalytic site motif (G/A/S-X-S-X-G) appears well conserved among these three PDAT and three template proteins belonging to the α/β hydrolase family (Figure 4A). The secondary structure elements of three layers of α/β/α fold were identified in these proteins as well (Figure 4A).
Figure 4.
Cr-PDAT Possesses a Lipase Functional Domain and TAG Lipase Activity.
(A) Sequence alignment and secondary structure elements between the PDAT-like proteins and their referenced templates within α/β hydrolase fold region. The conserved lipase motif is indicated by the green box. The essential Ser (S) residue that constitutes the catalytic triad for the active site is shown as a red asterisk.
(B) TAG lipase activity was detected by monitoring the formation of free fatty acids (18:1) and the degradation of TAG at various time points. Heat-inactivated PDAT was used as negative control (NC); reaction time for the negative control was 60 min. FFA, free fatty acid.
(C) Quantitative analysis of TAG and fatty acids in the enzymatic assay.
(D) to (F) Comparison of the predicted structure of conserved α/β hydrolase fold domains of Cr-PDAT, Sc-PDAT, and At-PDAT, respectively.
(G) to (I) Templates used for construction of structures of Cr-PDAT, Sc-PDAT, and At-PDAT, respectively. The rainbow color code describes the three-dimensional structures from the N (blue) to C termini (red); specific α-helices (α) and β -sheets (β) were identified as well as the active site Ser (asterisk).
The possible lipase activity of ΔTMCrPDAT was tested with triolein as a substrate. As shown in Figures 4B and 4C, triolein was degraded rapidly with concomitant formation of free fatty acids, confirming the TAG lipase function of Cr-PDAT.
In comparative modeling analyses, all three-dimensional structure models exhibited an overall α/β hydrolase fold (α/β/α) with a similar steric position (Figures 4D to 4I), except in the case of the topology of At-PDAT, where the first Gly was replaced with Pro and the first two β-sheets of this common fold region were absent (Figure 4F). Previous studies showed an N-terminal deletion version of Sc-PDAT possessed low phospholipase activity in vitro (∼2% of transacylation activity), which produced 2% free fatty acids during TAG production (Ghosal et al., 2007). At-PDAT has not been shown to have phospholipase activity (Dahlqvist et al., 2000; Ståhl et al., 2004). These data collectively suggest that the first two β-sheets, conserved in Cr-PDAT and Sc-PDAT, may be essential for the lipase activity.
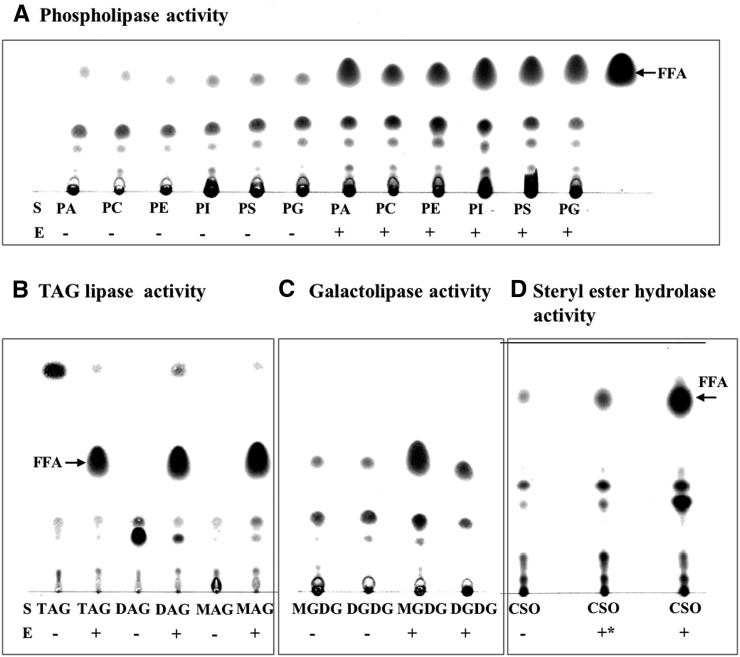
To determine further the substrate specificity of Cr-PDAT as a lipase, we measured lipase activity of recombinant ΔTMCrPDAT with various classes of lipids and confirmed that Cr-PDAT can hydrolyze a variety of PLs (e.g., PA, PC, PE, PI, PS, and PG) (Figure 5A), neutral lipids (e.g., monoacylglycerol, DAG, and TAG) (Figure 5B), galactolipids (e.g., MGDG and DGDG) (Figure 5C), and steryl esters (Figure 5D). As a phospholipase, Cr-PDAT exhibited a preference for anionic PLs (e.g., PA, PS, PI, and PG) over cationic PLs (e.g., PC and PE) (Figure 5B). As a galactolipase, Cr-PDAT showed a preference for MGDG over DGDG (Figure 5C). ΔTMCrPDAT also exhibited strong steryl hydrolase activity toward cholesteryl esters, whereas ORFCrPDAT had detectable but relatively weak steryl hydrolase activity (Figure 5C).
Figure 5.
Broad Substrate Specificity of Cr-PDAT as a Lipase.
(A) Phospholipase activities toward various PLs. FFA, free fatty acid.
(B) Neutral lipid hydrolase activities toward TAG, DAG, and MAG.
(C) Galactolipase activities toward MGDG and DGDG.
(D) Steryl ester hydrolase activities of and toward cholesteryl ester (CSO). ORFCrPDAT (+*) and/or ΔTMCrPDAT (+) were used for enzymatic assays. Reactions without the addition of enzyme (−) were used to show the nonenzymatic hydrolysis of lipid substrates in the aqueous buffer. Free fatty acids (FFA) resulting from the hydrolysis of lipid substrates (S) are marked. Negative control (heat-inactivated PDAT) and positive control (commercial lipase) are shown in Supplemental Figure 12 online.
Overexpression of Recombinant Cr-PDAT in S. cerevisiae
To investigate whether the expression of Cr-PDAT is involved in TAG synthesis in vivo, we introduced the Cr-PDAT gene (pYES:CrPDAT) into S. cerevisiae, with S. cerevisiae transformed with an empty vector as a control (pYES). Both transformants grew in a TAG induction medium for 36 h, and total lipids were extracted for TLC analysis. A basal level of TAG was evident in the pYES cells and increased gradually over 36 h under TAG induction conditions due to endogenous PDAT and DGAT activities. Under the same culture conditions, the pYES:CrPDAT cells produced approximately two- to threefold greater amounts of TAG than the control, confirming the contribution of exogenous Cr-PDAT to overall TAG production in the heterologous organism (Figure 6).
Figure 6.
Enhanced Synthesis of TAG by Heterologous Expression of Cr-PDAT in Yeast Cells.
The neutral lipid fraction was extracted from S. cerevisiae wild-type INVSc1 cells transformed with pYES2:CrPDAT or with the empty vector (pYES2) at the time points indicated. The TAG concentration was determined using Image J software and normalized to 0 h (before induction) of the empty vector control. FFA, free fatty acid.
Transient Upregulation of Cr-PDAT in Response to Stress Associated with Membrane Lipid Degradation and TAG Accumulation
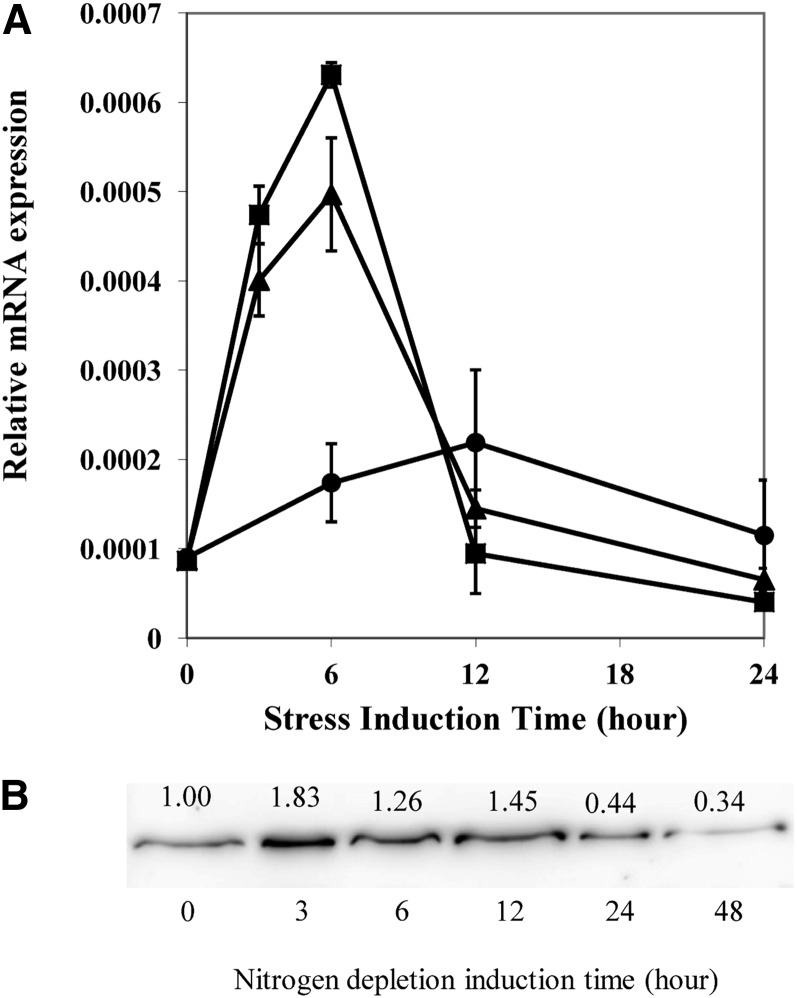
To determine whether Cr-PDAT was involved in stress responses in C. reinhardtii, gene expression was investigated by quantitative real-time PCR (Figure 7A). Whereas little change in the relative mRNA level was observed in cells grown under favorable growth conditions [low light and sufficient nitrogen (LL+N)], the transcription of Cr-PDAT was transiently upregulated to a maximum level at 6 h following nitrogen depletion (LL-N), which was 2.9-fold higher than the control (6 h, LL+N). The maximum mRNA level as 3.6-fold greater at 6 h under high light (HL) and nitrogen-depleted conditions (HL-N) than that of the control (6 h, LL+N). After 6 h, Cr-PDAT transcripts decreased rapidly to the basal level at 12 h and remained more or less constant thereafter, regardless of the growth conditions under which the cells had been maintained.
Figure 7.
Regulation of PDAT in C. reinhardtii (Wild Type).
(A) Expression profile of the Cr-PDAT gene under LL/nitrogen-replete (LL+N, circles), LL/nitrogen-depleted (LL-N, triangles), and HL/nitrogen-depleted (HL-N, squares) conditions. Gene expression was normalized to the expression of the 18 S rRNA gene. Data are presented as means ± se (n = 4).
(B) Immunoblotting analysis of protein level of Cr-PDAT in response to nitrogen deprivation stress. Cells grown in TAP medium under low light for 4 d were transferred to TAP-N medium. Protein concentration was determined using Image J software and normalized to the control (0 h, before induction).
The regulation of Cr-PDAT at the protein level was determined by immunoblotting. Cr-PDAT was transiently upregulated in response to N deprivation (Figure 7B). The protein expression achieved the maximum level at 3 h after the onset of N depletion from the culture medium and then gradually decreased during the following 48 h (Figure 7B).
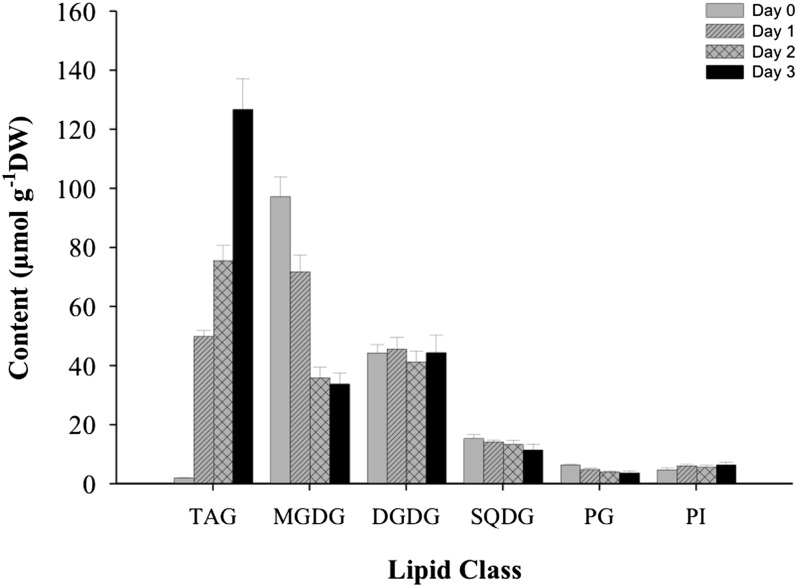
The concentrations of membrane lipids and TAG in C. reinhardtii wild-type cells grown under LL and N-depleted conditions were quantified using electrospray ionization–mass spectrometry (ESI-MS). Membrane lipids, particularly MGDG, SQDG, and PG, were rapidly degraded (Figure 8). For example, MGDG and PG decreased by 63 and 36%, respectively, in the first 2 d of N depletion, while TAG increased ∼40 times during this period (Figure 8).
Figure 8.
Lipid Content of C. reinhardtii (Wild Type) under N-Replete and N-Depleted Conditions.
For TAG induction, cells grown in TAP medium under low light for 4 d were transferred to TAP-N medium. Lipids were extracted and analyzed using ESI-MS. Data are presented as means ± se (n = 4 to 6). DW, dry weight.
PDAT Is Responsible for TAG Biosynthesis in C. reinhardtii
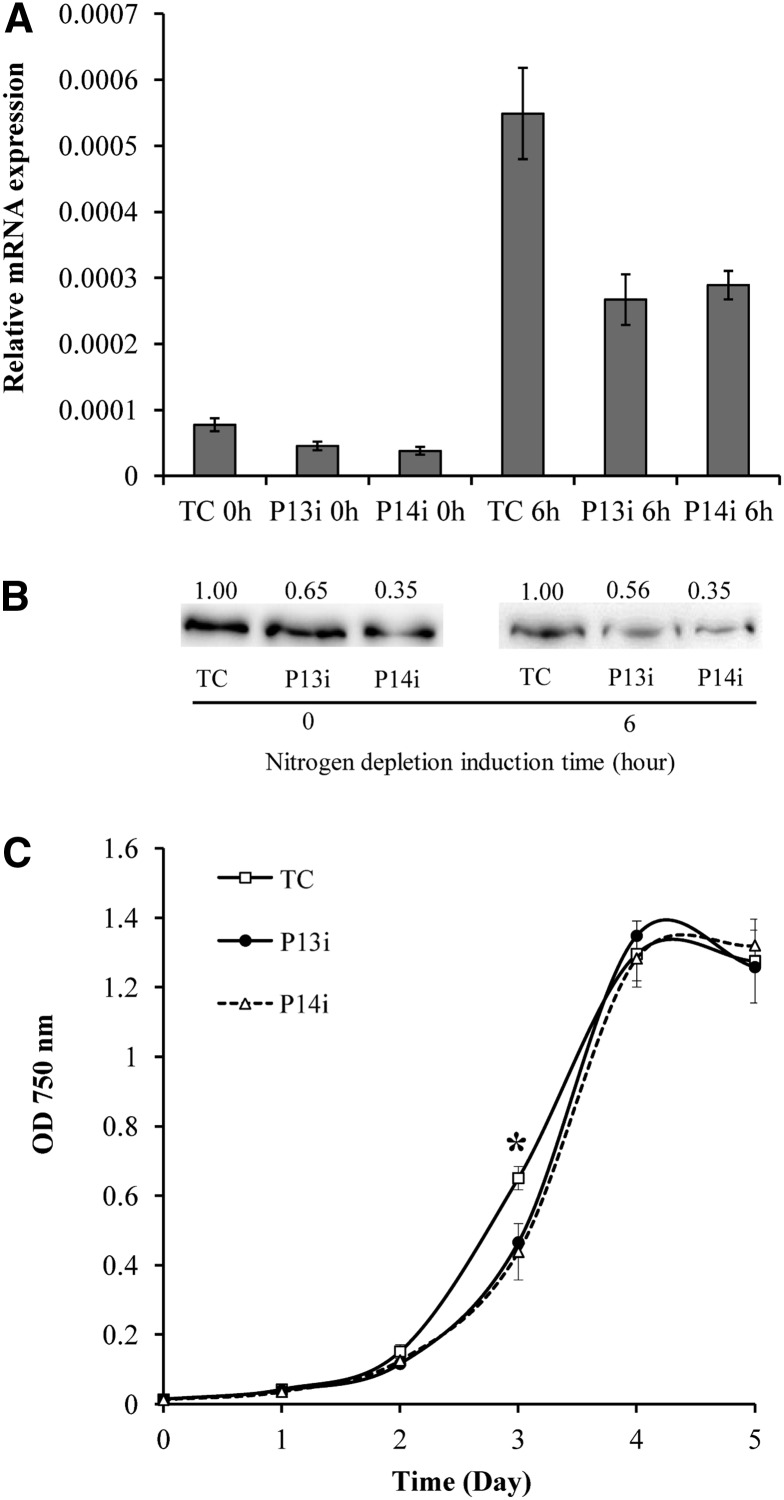
To investigate the function and possible biological role of PDAT in C. reinhardtii, PDAT knockdown mutants were generated with an artificial microRNA (amiRNA)–mediated gene silencing approach (Molnar et al., 2009). A total of 16 PDAT knockdown mutants were selected for gene expression analysis, among which two mutants, P13i and P14i, showed the maximum reduction in PDAT transcript (∼50% lower than the control transformed with an empty vector) under both N-replete and N-depleted conditions (Figure 9A). The knockdown of PDAT in P13i and P14i was confirmed at the protein level by immunoblotting. As shown in Figure 9B, compared with that of the control transformed with an empty vector, PDAT decreased by 35 and 44% in P13i under N-replete and N-depleted conditions, respectively; it decreased by 65% in P14i under both culture conditions. Under N-replete conditions, P13i and P14i exhibited slower growth than the control in the mid-exponential growth phase (day 3), but the difference became insignificant in the stationary growth phase (Figure 9C).
Figure 9.
Generation of amiRNA Knockdown Mutants.
(A) Knockdown of Cr-PDAT in P13i and P14i compared with the control transformed with an empty vector (TC), confirmed with real-time PCR.
(B) Immunoblotting analysis of protein level in the knockdown mutants and the control (TC). Protein concentration was determined using Image J software and normalized to the control.
(C) Growth kinetics of the knockdown mutants (P13i and P14i) and the control (TC) under N-replete conditions. Data are presented as means ± se (n = 4 to 6), with the asterisk indicating P < 0.05.
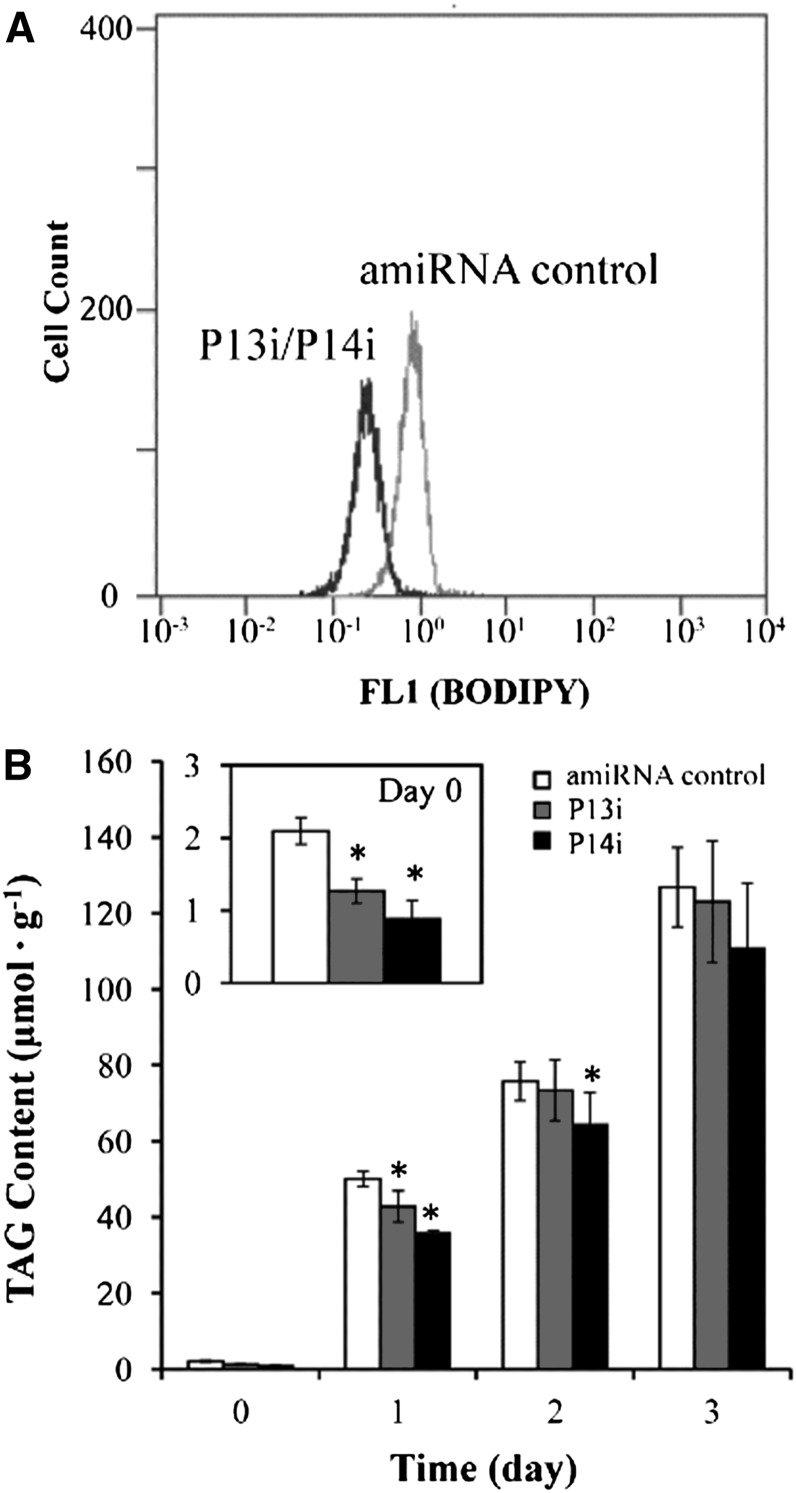
To determine whether the knockdown of PDAT would affect TAG formation in C. reinhardtii, flow cytometry combined with BODIPY staining (a fluorescence dye targeting TAG) was applied to screen for TAG-deficient mutants. As shown in Figure 10A, the fluorescence intensity derived from the PDAT knockdown mutants grown in Tris-acetate phosphate (TAP) for 4 d was significantly lower (P < 0.05) than the control, indicative of a reduction in TAG production in the mutants. A quantitative analysis of TAG by ESI-MS revealed that compared with the control, the TAG content of PDAT knockdown mutants declined by 39.6% in P13i and 57.9% in P14i under N-replete conditions and was reduced by 14.4% in P13i and 28.4% in P14i under N-depleted conditions for 24 h (Figure 10B). These results suggest that PDAT contributed appreciably to the formation of TAG under favorable growth conditions. A 60-fold increase in TAG was evident in both the PDAT knockdown mutants and the control cells under N-depleted conditions after 3 d, and the difference in the maximum TAG concentration in the two types of cells was statistically insignificant (Figure 10B), suggesting that the contribution of the PDAT pathway to TAG synthesis under N deprivation conditions must be minor.
Figure 10.
PDTA Knockdown Depressed TAG Synthesis in P13i and P14i.
(A) Flow cytometry histogram illustrating the TAG content of the cell population measured as BODIPY fluorescence concentration (FL1) in situ.
(B) Total TAG concentration under N-replete and N-depleted conditions in amiRNA knockdown mutants (P13i and P14i) and control cells. Cells grown in TAP medium for 4 d were transferred to TAP-N medium to induce TAG synthesis, which is presented as day 0 (before induction). Data are presented as means ± se (n = 4 to 6), with asterisks indicating P < 0.05.
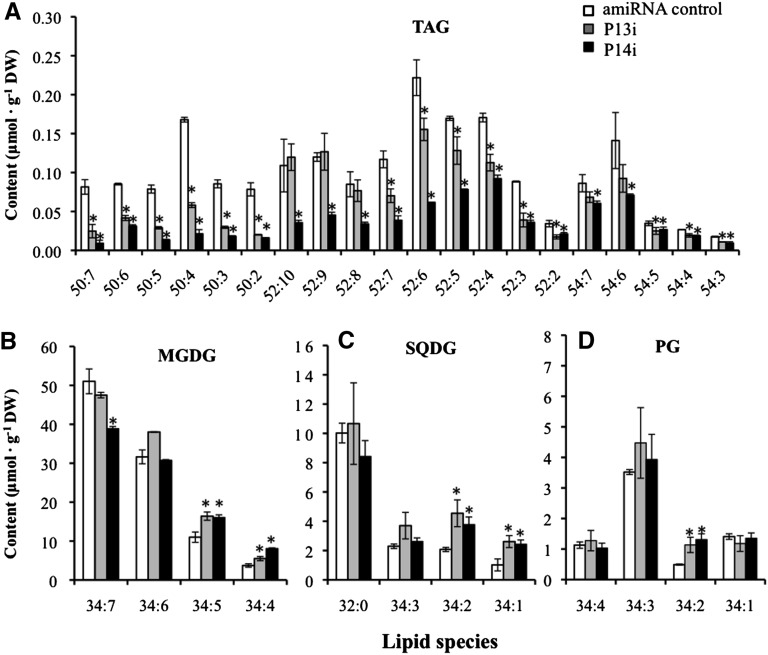
TAG in C. reinhardtii comprised three subclasses based on total number of carbon atoms in its fatty acyl groups, namely, TAG C50, C52, and C54 (see Supplemental Figure 5 online). Under N-replete conditions, the TAG C50 molecular species showed a more drastic decline in content than the C52 and C54 molecular species in P13i and P14i compared with the control (Figure 11A). Tandem mass spectrometry analysis indicated that TAG C50 contained two C16 and one C18 fatty acyl group, and C16 was predominant at the sn-2 position of glycerol (see Supplemental Figure 6 online). This suggests that Cr-PDAT prefers DAG molecules with C16 at the sn-2 position in vivo. Because the DAG molecules with C16 at the sn-2 position are generally believed to be synthesized via a prokaryotic pathway in chloroplasts (Frentzen, 1998), the large decrease in TAG C50 indicates that PDAT is involved in TAG biosynthesis in C. reinhardtii chloroplasts. After being subjected to nitrogen starvation, the decrease in TAG molecular species was not significant (P > 0.05) between the PDAT knockdown mutants and the control at day 3 (see Supplemental Figure 7 online).
Figure 11.
The Concentrations of Lipid Molecular Species in amiRNA Knockdown Mutants (P13i and P14i) and Controls Transformed with an amiRNA Empty Vector under N-Replete Conditions.
Concentrations of TAG (A), MGDG (B), SQDG (C), and PG (D) molecular species. Algal cells grown in TAP medium for 4 d were used for ESI-MS analysis. The lipid molecular species are presented as the number of carbon atoms:total double bonds in the fatty acyl groups. Data are presented as means ± se (n = 4 to 6), with asterisks indicating P < 0.05. DW, dry weight.
PDAT-Mediated TAG Synthesis Substrates Derived from Chloroplast Membrane Turnover and Degradation
To determine if PDAT uses membrane lipids (e.g., PLs and glycolipids) as the substrates for the acyl transfer reaction or hydrolysis in vivo, major membrane lipids of the wild type and PDAT knockdown mutants were analyzed by ESI-MS. Although changes in total membrane lipid concentrations (i.e., MGDG, DGDG, etc.) in PDAT knockdowns were almost undetectable under N-replete conditions (see Supplemental Figure 8 online), presumably due to lipid homeostasis that maintained major membrane lipid species at a constant level, knockdown of PDAT dramatically changed the concentrations of several molecular species belonging to the chloroplast membrane lipids, including MGDG, SQDG, and PG (Figures 11B to 11D). The molecular lipid species are denoted according to the number of carbon atoms: total double bonds in the fatty acyl groups, which was annotated based on the mass-to-charge ratio of lipid ions. The higher concentrations of MGDG 34:5 and 34:4, SQDG 34:2 and 34:1, and PG 34:2 in the mutants indicate that Cr-PDAT may participate in chloroplast membrane remodeling or turnover using chloroplast membrane–derived PLs and glycolipids as substrates to form TAG under N-replete conditions. The difference in the membrane lipids, particularly the concentrations of MGDG and PG, between PDAT knockdown mutants and the control strain was insignificant after 2 d of N depletion (see Supplemental Figure 9 online). This suggests that chloroplast membrane remodeling or turnover becomes less active under stress for an extended period of time (>24 h), resulting in fewer membrane lipids available for PDAT-mediated TAG synthesis. It is worth noting that DGDG molecular species varied little in the PDAT knockdown mutants (see Supplemental Figure 10 online), consistent with the in vitro enzymatic assays in which Cr-PDAT did not use DGDG as a substrate for transacylation (Figure 3B) and DGDG was a less favorable substrate than MGDG for hydrolysis (Figure 5).
Although Cr-PDAT showed the highest selectivity on PI among tested membrane lipids in vitro, there was no increase in PI concentration detected in Cr-PDAT mutants compared with the control under both N-replete (see Supplemental Figure 8 online) and N-depleted (see Supplemental Figure 11 online) conditions. This indicates that PDAT and PI may reside in different subcellular compartments. Cr-PDAT harboring a chloroplast transit peptide is localized in chloroplasts (Terashima et al., 2011), whereas PI is absent from the thylakoid membranes (Janero and Barrnett, 1981) and is likely enriched in extraplastidic membranes of C. reinhardtii.
DISCUSSION
Cr-PDAT Is a Multifunctional Enzyme
The biosynthesis of TAG is a common metabolic pathway that occurs in almost all plants, animals, fungi, and some bacteria. Much research has focused on the Kennedy pathway, in which DGAT catalyzes the acylation of DAG to form TAG. Recently, an acyl-CoA–independent pathway catalyzed by PDAT was discovered in algae, yeasts, and plants (Banaś et al., 2000; Dahlqvist et al., 2000; Boyle et al., 2012). However, the biochemical characteristics and physiological importance of PDAT in algae have not been investigated. In this study, in vitro enzyme assays of recombinant full-length and ΔTM Cr-PDAT provided evidence that Cr-PDAT is a multifunctional enzyme with acyltransferase and lipase activities with broad substrate specificity. For its transacylation function, Cr-PDAT can use PLs, galactolipids, and DAG as acyl donors for TAG synthesis. For its lipase function, Cr-PDAT can release free fatty acids from a variety of lipid substrates, including PLs, galactolipids, and neutral lipids. Cr-PDAT may contribute to membrane remodeling and degradation, with TAG being a by-product of this process.
Galactolipids are the major lipid building blocks of chloroplast membranes. In vascular plants, drastic decreases in MGDG content occur during freezing, desiccation, or ozone treatment (Sakaki et al., 1990a, 1990b; Gigon et al., 2004; Li et al., 2008a). In this study, we observed that the MGDG content of C. reinhardtii decreased continuously over 48 h under N-depleted conditions (Figure 8). It is well documented that, under stress, a decrease in MGDG is usually accompanied by TAG accumulation (Sakaki et al., 1990a, 1990b; Gigon et al., 2004), but the pathways responsible for this conversion are poorly understood. A pathway mediated by a galactolipid:galactolipid galactosyltransferase (GGGT) located on the chloroplast envelope membrane (van Besouw and Wintermans, 1978; Heemskerk et al., 1983) may play a role in the lipid remodeling process (Sakaki et al., 1990b; Benning and Ohta, 2005; Moellering and Benning, 2011). GGGT transfers a galactosyl moiety from an MGDG molecule to a second galactolipid molecule (MGDG or DGDG), resulting in the formation of DAG, the precursor for TAG synthesis, and oligogalactoglycerolipids. Recently, SENSITIVE TO FREEZING2 (SFR2), a gene essential for freezing tolerance, has been identified as the GGGT-encoding gene in Arabidopsis thaliana. The sfr2 mutant showed ∼50% reduction in TAG content compared with the wild type in response to freezing treatment, indicating a role for GGGT in TAG synthesis in vivo (Moellering et al., 2010).
In this study, we discovered that Cr-PDAT acted as a galactolipid:DAG acyltransferase, transferring a fatty acyl group from MGDG to DAG to form TAG. As such, knockdown of Cr-PDAT led to the accumulation of certain MGDG species and reduced TAG accumulation in the mutants. Therefore, the PDAT pathway represents a previously unrecognized mechanism for galactolipid remodeling, turnover, and degradation. From this perspective, TAG can be regarded as a coproduct of Cr-PDAT–mediated chloroplast membrane remodeling or degradation. From a broader perspective, the formation of this coproduct is part of lipid homeostasis within the chloroplast or the whole cell. Cr-PDAT showed high substrate specificity for galactolipid:DAG transacylation in that only MGDG could be efficiently utilized as an acyl donor. The MGDG-dependent galactolipid:DAG acyltransferase function of Cr-PDAT is consistent with the fact that this protein harbors a chloroplast transit peptide and was detected in the chloroplast (Terashima et al., 2011), where MGDG exclusively resides. Galactolipid:DAG acyltransferase activity has not been reported for any previously known PDAT. It is unknown if such a function is restricted to Cr-PDAT or is common to most, if not all, photosynthetic organisms.
When various PLs were used as acyl donors and diolein was used as an acyl acceptor, Cr-PDAT showed a preference for anionic PLs (e.g., PG, PI, PA, and PS) over cationic PLs (e.g., PC and PE) (Figure 2A). This is distinct from Sc-PDAT and At-PDAT, which exhibited higher substrate specificities toward PC and PE than the other PLs (Banaś et al., 2000; Dahlqvist et al., 2000; Ghosal et al., 2007). This difference can be explained in part by the distinct cellular PL compositions among yeasts, plants, and green algae. In yeast and vascular plants, PC and PE represent the most abundant PLs, whereas PG and PI are the major PLs, and PC is absent in C. reinhardtii (Giroud et al., 1988). Based on these observations, we speculate that cellular membranes undergo highly dynamic and efficient remodeling and turnover to maintain proper functions under various environmental conditions; thus, the substrate specificity of PDAT has coevolved with the lipid composition of various organisms.
In addition to PL/galactolipid:DGAT activity, Cr-PDAT also exhibited moderate DGTA activity. An important role for DGTA in TAG synthesis has been proposed in developing seeds of sunflower (Helianthus annuus) (Fraser et al., 2000), endosperm of castor bean (Ricinus communis) (Mancha and Stymne, 1997), and developing safflower seeds (Stobart et al., 1997). Although one DGTA has been purified and characterized from rat intestinal microsomal preparations (Lehner and Kuksis, 1993), understanding of this pathway has been hindered by the lack of the genomic information on this enzyme. Recently, Sc-PDAT was found to have DGTA activity, which was ∼11 or 2% of its PDAT activity, when PC and PE were used as acyl donors, respectively (Ghosal et al., 2007). At-PDAT also displayed DGTA activity of about one-tenth of its PDAT activity (Ståhl et al., 2004). Under our experimental conditions, the DGTA activity of Cr-PDAT was approximately one-seventh of its PDAT activity. These results suggest that the DGTA function is a common feature of PDAT in plants, yeasts, and green algae. Considering that DAG can be produced via multiple routes in cells, such as by dephosphorylation of PA catalyzed by phosphatidic acid phosphatase in the Kennedy pathway or GGGT-mediated remodeling of galactolipids, PDAT may work in concert with these enzymes to further convert DAG into TAG via PDAT-mediated DAG:DAG or PL/galactolipid:DAG transacylation routes.
A number of enzymes involved in lipid metabolism have been reported to be multifunctional proteins, catalyzing both transacylation reactions and lipid hydrolysis. For example, oleosin, the structural protein of plant lipid bodies, is a bifunctional enzyme that exhibits both monoacylglycerol acyltransferase and phospholipase activities (Parthibane et al., 2012). The yeast TAG lipase Tgl4p exerts multiple functions as a TAG lipase, steryl ester hydrolase, phospholipase, and acyltransferase (Rajakumari and Daum, 2010a). The multiple functions of the above chimeric enzymes are due to the fusion of two distinct functional domains (acyltransferase and lipase domains) into a single protein.
By contrast, Cr-PDAT contains a single functional LCAT domain, which shares the Ser/Asp-Glu/His triad with lipases, esterases, and proteases (Peelman et al., 1998). Both LCAT and lipase belong to the α/β hydrolase fold family; like lipase, LCAT contains a potential lid domain involved in substrate binding (Peelman et al., 1998). The ability of an enzyme to catalyze an adventitious secondary activity at the active site responsible for the primary activity has been termed “catalytic promiscuity” (Copley, 2003; Khersonsky et al., 2006). Despite the potential catalytic promiscuity in the LCAT domain, the functions of most LCAT-like proteins appear to be highly divergent. For instance, out of the six LCAT-like proteins identified in Arabidopsis, LCAT3 (At3g03310) and LCAT4 (At4g19860) function as phospholipase A1 (Noiriel et al., 2004; Chen et al., 2012), whereas PDAT1 (At13640) shows PDAT activity but no phospholipase activity (Ståhl et al., 2004).
Unlike the vascular plants, C. reinhardtii contains a single LCAT-like protein, Cr-PDAT, that displays significant catalytic promiscuity. We speculate that a multifunctional Cr-PDAT may be particularly relevant to C. reinhardtii and other organisms with small and compact genomes; synthesis of a single PDAT with multiple functions allows better resource utilization and management for maintaining lipid homeostasis necessary for growth and reproduction, as well as for adapting to a changing environment.
Physiological Role of Cr-PDAT in Vivo
The multiple enzyme activities of Cr-PDAT suggest possible physiological roles of this enzyme in C. reinhardtii. Heterologous overexpression and gene knockdown experiments, along with its regulation at the transcript and protein level, suggest that Cr-PDAT is involved in TAG synthesis in vivo. The mRNA level of Cr-PDAT was upregulated under N-depleted or HL conditions, similar to the gene expression patterns of other acyltransferases related to TAG synthesis, suggesting a role for Cr-PDAT in TAG synthesis in vivo.
Knockdown of Cr-PDAT led to increased concentration of molecular species of MGDG, SQDG, and PG in vivo, in addition to the impaired TAG synthesis (Figure 11). The ESI-MS results indicated that in the PDAT knockdown mutants, TAG species with the most drastic reduction contained two C16 and one C18 fatty-acyl groups (see Supplemental Figure 7 online), consistent with observations of increased chloroplast membrane lipid species enriched in C16 fatty-acyl groups at the sn-2 position of glycerol (Giroud et al., 1988). We conclude that Cr-PDAT uses chloroplast membrane lipids, particularly MDGG, SQDG, and PG, as substrates to synthesize TAG in vivo. Lipidomics analyses also indicate that Cr-PDAT preferentially uses DAG synthesized via a prokaryotic pathway as substrates in vivo. Considering that Cr-PDAT harbors a chloroplast transit peptide and is present in the chloroplast proteome (Terashima et al., 2011), we suggest that Cr-PDAT is involved in TAG synthesis in the chloroplast. The precise localization (e.g., thylakoid membrane or chloroplast envelope) of Cr-PDAT within the chloroplast, however, remains unknown.
TAG in eukaryotic cells is thought to be a deposit for fatty acids, which in turn serve as a carbon and energy reserve. However, recent studies have revealed that TAG homeostasis is linked to membrane lipid turnover and remodeling, processes that are vital to maintaining proper structure and fluidity of cellular membranes (Rajakumari and Daum, 2010a, 2010b; Gaspar et al., 2011). Membrane remodeling and turnover involve a deacylation-reacylation cycle in which fatty acids are removed from PLs and are then reacylated to yield lysophospholipids (Lands, 1960; Kennedy, 1961; Lands and Merkl, 1963; Merkl and Lands, 1963; Bates et al., 2009). Fatty acids at the sn-2 position of PLs can be removed by (1) phospholipase A2, which produces free fatty acids and 1-acyl lysophospholipids (Winstead et al., 2000; Kudo and Murakami, 2002); and (2) PDAT, which can bypass the production of free fatty acids by incorporating the cleaved fatty acids directly into TAG. In the latter case, TAG synthesis is regarded as a buffer system for detoxification of lipotoxic free fatty acids (Listenberger et al., 2003; Petschnigg et al., 2009).
Our results indicate that the contribution of PDAT to TAG synthesis in C. reinhardtii is prominent in the logarithmic growth phase under favorable growth conditions but less crucial under stress conditions. A similar physiological role of PDAT has been observed in the yeast S. cerevisiae, where it is a major contributor to TAG accumulation in the exponential growth phase but not in the stationary phase (Oelkers et al., 2000, 2002). We also observed that Cr-PDAT knockdown resulted in a small but noticeable decrease in the specific growth rate in logarithmically growing cells. Collectively, we concluded that PDAT-mediated membrane remodeling and TAG synthesis is pivotal in vigorously growing cells and indispensable for normal growth under favorable culture conditions. It also plays an important role in chloroplast degradation and TAG synthesis at the early stages (first 1 to 2 d) of N depletion. As stress persists, however, Cr-PDAT may become less critical. Massive accumulation of TAG in C. reinhardtii under N depletion or other stress conditions may be attributable to other TAG synthesis pathways such as the Kennedy pathway.
Although our results suggest the Cr-PDAT pathway may play a minor role in TAG synthesis under N-depleted conditions over extended period of time (Figure 10; see Supplemental Figure 7 online), we cannot rule out the possibility that PDAT-mediated TAG production may overlap with other TAG synthesis enzymes in C. reinhardtii, compensating for the knockdown of PDAT under stress. In a previous study, an Arabidopsis knockout mutant with a T-DNA insertion in the PDAT1 locus did not display altered seed fatty acid content or composition (Mhaske et al., 2005), but RNA interference silencing of PDAT1 in a dagt1-1 background plant resulted in a 70 to 80% decrease in seed oil content (Zhang et al., 2012), suggesting that DGAT can compensate for PDAT knockout in a wild-type background. This may be also true for C. reinhardtii, which contains at least one DGAT1 and five DGAT2 genes. The unaltered TAG content in knockdown mutants of C. reinhardtii under stress conditions can also be explained by the TAG lipase activity of Cr-PDAT, which was likely activated when cells were grown under N-depleted conditions for 3 d. To address these questions, further research into the precise regulation of Cr-PDAT functions in vivo is needed.
Biotechnological Implications
In this study, we demonstrated that in an aqueous reaction system, recombinant Cr-PDAT can efficiently hydrolyze neutral lipids (TAG, DAG, and MAG), PLs, and galactolipids to produce free fatty acids (Figures 4 and 5). The multifunction of Cr-PDAT as an acyltransferase and a lipase with broad substrate specificity offers great potential as a biocatalyst for the conversion of crude algal oil into biofuels and non-fuel oils. Large-scale production of Cr-PDAT and evaluation of PDAT-based direct conversion of crude algal oils into free fatty acids and free fatty acids methyl esters is in progress.
METHODS
Strains, Media, and Growth Conditions
The wild-type Chlamydomonas reinhardtii strain cc-1690 was used for the cDNA library construction and gene expression analysis, and the cell wall–deficient wild-type strain cw-15 was used to generate the mutants P13i and P14i using an amiRNA knockdown method. All strains were grown in TAP medium containing 7.5 mM NH4Cl (Harris, 1989) at 23°C under continuous illumination of 40 μmol photons m−2 s−1. For HL (400 μmol photons m−2 s−1), cells in the exponential growth phase raised under LL conditions (40 μmol photons m−2 s−1) were harvested by centrifugation, washed once with TAP medium and then resuspended in fresh TAP medium at a starting cell density of 106 cells mL−1. Cultures were then exposed to HL for 3 d. To impose nitrogen deprivation (−N), cells in the stationary growth phase (corresponding to a cell density of 2 × 107 cells mL−1) were collected by centrifugation and washed with nitrogen-deficient TAP medium (TAP-N), and equal amounts of cells were resuspended in TAP-N medium. Cell aliquots were collected at defined time intervals after being transferred to TAP or TAP-N medium. All cultures were grown on a shaking table with continuous shaking at 130 rpm.
Yeast, Bacterial Strains, and Culture Conditions
The Pichia pastoris wild-type X-33 strain (Invitrogen) was used to express the secretory recombinant Cr-PDAT proteins. The wild type Saccharomyces cerevisiae strain INVSc1 (MATα his3D1 leu2 trp1-289 ura3-52 MAT his3D1 leu2 trp1-289 ura3-52; Invitrogen) was used as a heterologous host for intracellular overexpression of Cr-PDAT. Escherichia coli strain TOP10 (Invitrogen) was used for recombinant DNA work. All strains and transformants generated were grown according to manufacturer’s instructions.
RNA Isolation and cDNA Library Construction
Harvested C. reinhardtii cells (∼7 g wet weight) were pulverized in a mortar with liquid nitrogen, and extracts were transferred to a tube containing Trizol reagent (Invitrogen). Total RNA isolation was conducted following the procedure described by Li et al. (2008b). To construct a high-quality cDNA library, RNA was isolated from cells subjected to the various treatments described above (e.g., LL, HL, and −N). Poly(A)+ RNA was purified from pooled RNA using the Oligotex mRNA purification kit (Qiagen). About 5 µg of mRNA was used to synthesize double-stranded cDNA, and the cDNA was ligated into the λ-ZAPII arms to construct a cDNA library using the GigapackIII Gold packaging kit (Stratagene). An aliquot of the cDNA library was mass excised by ExAssist helper phage (Stratagene) to recover pBluescript plasmids.
Quantitative Real-Time PCR
For real-time RT-PCR analysis, the first strand of the cDNA was synthesized using the Taqman reverse transcription system (Applied Biosystems) per the manufacturer’s instructions. Real-time RT-PCR was performed on an ABI Prism 7500 sequence detection system (Applied Biosystems) following the protocol previously described by Li et al. (2008b). Primer sequences used for expression studies are listed in the Supplemental Table 1 online. RNA levels were normalized using the 18S rRNA gene as the internal standard.
Identification and Cloning of cDNA Encoding PDAT in C. reinhardtii
The S. cerevisiae PDAT (Sc-PDAT) and Arabidopsis thaliana PDAT (At-PDAT) protein coding sequences were used to identify the C. reinhardtii genes with the highest similarity using the BLAST server (http://blast.ncbi.nlm.nih.gov/). One hit revealed a significant similarity to these known PDAT sequences. Several primers (see Supplemental Table 1 online) were designed based on the sequences conserved with other PDAT orthologs to clone the coding region of cDNA from the C. reinhardtii cDNA library. All PCR reactions used in this study were performed in the presence of 5% DMSO. PCR products of the expected size were cloned into the pCR2.1 cloning vector (Invitrogen) and verified by sequencing. Full-length cDNA was obtained following screening the cDNA library (see Supplemental Methods 1 online).
Bioinformatics Analysis
Protein sequence alignments and similarity analyses were conducted with the CLC Main Workbench program (CLC Bio). Primary structure analyses, topological organization predictions, and posttranslational modification predictions were performed with ExPASy proteomic tools available at http://www.expasy.ch/tools/. Protein motifs were identified using InterProScan at http://www.ebi.ac.uk/Tools/pfa/iprscan/. For analyses of putative transit peptide and signal sequences, ChloroP 1.1 (Emanuelsson et al., 1999) and SignalP 4.0 (Petersen et al., 2011) were used.
Phylogenetic analyses were conducted using neighbor-joining methods. The neighbor-joining tree was constructed with MEGA 5.0. Molecular distances within the aligned sequence were calculated according to the position correction model. Branch points were tested for significance by bootstrapping with 1000 replications (Tamura et al., 2007). The alignment used for the analysis is available as Supplemental Data Set 1 online.
To select potential templates for building three-dimensional structures of the PDAT-like protein (Cr-PDAT, Sc-PDAT, and At-PDAT), the template identification tool in SWISS-MODEL (http://swissmodel.expasy.org/workspace/) was used as a comparative modeling method. Secondary structure elements were constructed based on sequence alignments generated by the above methods. The three-dimensional models of the PDAT-like proteins were built with the SWISS-MODEL workspace program using selected structures of α/β hydrolases of Paucimonas lemoignei (Protein Data Bank accession ID: 2X5XA, chain A), Pseudomonas aeruginosa (Protein Data Bank accession ID: 1EX9A, chain A), and Burkholderia cepacia (Protein Data Bank accession ID: 1YS1) as templates. All figures illustrating molecular modeling results were prepared in PyMOL (Molecular Graphics System).
Heterologous Expression of Cr-PDAT in P. pastoris and S. cerevisiae
For each host system, two types of plasmids containing full-length Cr-PDAT (designated as ORFCrPDAT) and the Cr-PDAT coding sequence without the putative membrane-spanning region (ΔTMCrPDAT), respectively, were constructed. Details on the heterologous expression of ORFCrPDAT and ΔTMCrPDAT in E. coli, P. pastoris, and S. cerevisiae are given in Supplemental Methods 1 online.
Expression of Cr-PDAT in E. coli and Production of Anti-PDAT Antibodies
To produce antibodies against Cr-PDAT, ΔTMCrPDAT was amplified using ΔTM-PDATEF/ΔTM-PDATER (see Supplemental Table 1 online) and cloned into the expression vector pQE30 (Qiagen). The resulting pQE30 constructs were transformed into the E. coli strain M15 containing pREP4 (Qiagen). For expression, positive transformants were expressed by adding isopropyl-β-d-thiogalactopyranoside (0.1 mM) after 4 h of growth. Cells were grown for additional 6 h before being harvested (see Supplemental Figure 4 online). The mouse anti-ΔTMCrPDAT polyclonal antibodies were produced by Precision Antibody.
Immunoblotting
To isolate total proteins, 5 mL cell cultures were centrifuged at 1000g for 5 min and washed with 50 mM Tris-HCl, pH 6.8. Cell pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 6.8, containing 2% SDS and 10 mM EDTA and a protease inhibitor cocktail [Sigma-Aldrich]). Cells were incubated for 1 h at room temperature, and insoluble cell debris was removed by centrifugation at 13,000g for 30 min at 4°C prior to loading samples onto the SDS-PAGE. Total protein concentration of supernatants (whole cell extract) was determined using the SPN protein assay kit (G Biosciences). Protein samples dissolved in SDS-PAGE sample buffer were separated on SDS-PAGE and transferred to nitrocellulose membranes. The membranes were incubated for 12 h with recombinant anti-ΔTMCrPDAT antibody diluted 1:100 with PBS containing 5% (w/v) nonfat milk and 0.1% (v/v) Tween 20, followed by incubation with anti-mouse IgG conjugated with horseradish peroxidase (Bio-Rad) diluted 1:2000 in the same buffer. Antigen-antibody complexes were visualized using an enhanced chemiluminescence detection kit (Pierce).
Extraction of Lipids from Yeast Cells and TLC Analysis
For total lipid extraction, yeast cells were collected by centrifugation and washed twice with distilled water. Cells were broken by vigorous shaking with 200 μL glass beads (diameter 0.5 mm; Sigma-Aldrich) by a mini-beadbeater (Biospec Products) for 1 min. Total lipids were then extracted according to the Bligh and Dyer method (Bligh and Dyer, 1959) and separated on a silica TLC using petroleum ether/diethyl ether/acetic acid (70:30:1, v/v/v) as the solvent system (Ghosal et al., 2007). For the visualization of separated lipids, the developed TLC plates were air dried, sprayed uniformly with 8% (w/v) H3PO4 containing 10% (w/v) copper (II) sulfate pentahydrate, and charred at 180°C for 10 min. The lipids were quantified using densitometry and image analysis scaled to a dilution series of the corresponding lipid standard (e.g., 1 to 20 µg triolein).
Enzyme Assays
For the acyltransferase activity assay, the reaction mixture contained 1 μg recombinant PDAT in 50 mM potassium phosphate buffer, pH 7.2, with 250 μM lipid donors (e.g., PA, PC, PE, PS, PI, PG, MGDG, DGDG, and SQDG) and 250 μM lipid acceptor (1,2- or 1,3-DAG) in a final reaction volume of 200 μL. The suspension was thoroughly mixed and incubated at 30°C for 30 min. Lipids were extracted and analyzed as described above. Lipolytic acyl hydrolase activities of recombinant PDAT was performed under the same conditions used for acyltransferase activity assay with various lipid substrates. Novozyme lipase B (Sigma-Aldrich) was used as positive control for the lipase activity assays. Heat-inactivated PDAT was used as negative control for all of the enzyme activity assays.
amiRNA-Mediated Cr-PDAT Gene Silencing
The amiRNA was designed to target C. reinhardtii PDAT following instructions provided by Molnar et al. (2009). The resulting forward oligonucleotides 5′-ctagtTCGACGCTTATTCAAGCTTAAtctcgctgatcggcaccatgggggtggtggtgatcagcgctaTTAACCTTGAATAAGCGTCGAg-3′ and reverse oligonucleotides 5′-ctagcTCGACGCTTATTCAAGGTTAAtagcgctgatcaccaccacccccatggtgccgatcagcgagaTTAAGCTTGAATAAGCGTCGAa-3′ were annealed and cloned into the SpeI site of a pChlamyRNA3 vector (available from the Chlamydomonas Center at the University of Minnesota, St. Paul, MN) to produce the pChlamyRNA3:CrPDAT vector. This vector was linearized by KpnI and transformed into the C. reinhardtii cell wall mutant cw15 by the glass bead method (Kindle, 1990). Transformants were selected on TAP plates with paromomycin (10 μg mL−1). Of the several mutants obtained, P13i and P14i showed stable suppression of Cr-PDAT over the course of half a year and were selected for further study.
TAG Analysis with Flow Cytometry
Flow cytometry was used to screen for C. reinhardtii amiRNA interference mutants with a TAG-less phenotype. Thirty microliters of cell culture was diluted with 270 μL of PBS buffer prior to flow cytometric analysis. Cells were stained with a 1:100 dilution of a stock solution (100 µM) of BODIPY (Sigma-Aldrich) and incubated in the dark for 5 min at room temperature. Stained cells were analyzed using a benchtop flow cytometer (Beckman Coulter) equipped with an argon laser (excitation wavelength 488 nm). Green fluorescence intensity (FL1) from BODIPY was measured at 525 nm. Red fluorescence intensity (FL3) from chlorophyll was measured at 670 nm and set as a trigger.
Extraction of Lipids from C. reinhardtii Cells
Five milliliters of C. reinhardtii cells were harvested by centrifugation at 1000g for 5 min. Cell pellets were washed with 5 mL PBS buffer and centrifuged at 1000g for 5 min. Cells were resuspended in 3 mL of chloroform:methanol (2:1, v/v) and agitated vigorously at room temperature for 1 h. Extracts were mixed with 0.75 mL potassium chloride (0.7%, w/v). After further centrifugation at 1000g for 5 min, extracts were split into two phases, with total lipids in the lower organic phase. The lower phase was transferred into another vial using a glass pipette. The organic solvent was evaporated under a stream of nitrogen. Total lipids were recovered in 0.5 mL chloroform/methanol (1:1) for ESI-MS (see below). To quantify the lipid content, another aliquot of the original cell culture (5 mL) was used to measure cell dry weight as previously described (Li et al., 2010a).
Lipid Analysis and Quantification with ESI-MS
Mass spectrometry analysis was performed with an Agilent 6460 triple quadrupole liquid chromatography/mass spectrometer equipped with an electrospray ion source (Agilent). Single-stage mass spectrometry was used for the detection of total positive and negative lipid ions. Precursor ions and neutral loss scanning were employed for membrane glycerolipid identification according to previously described methods (Welti et al., 2002, 2003; Hsu and Turk, 2009); sequential neutral loss scanning was applied for TAG identification (Han and Gross, 2001, 2003). Nitrogen was used as a nebulizing gas (at 0.3 Bar) and a dry gas (4 liters min−1 at 200°C). The spray capillary voltage was 3700 V for the negative ion mode and 4200 V for the positive ion mode analysis. Prior to direct infusion into the ESI source, 10 μL lipid extract was mixed with internal standards (see Supplemental Table 2 online), the ionization reagent (200 µM NaI for positive ion mode and 500 µM NH4OH for negative ion mode), and chloroform:methanol (1:1) for a final volume of 100 µL. A five-microliter sample mixture was directly infused into ESI by loop injections with methanol as a mobile phase at a flow rate at 0.1 mL min−1.
For analysis of TAG in cells grown under normal culture conditions, 3-μL tripalmitin-supplemented samples were injected into a 5 cm × 4.6-mm Eclipse C-18 column at 40°C with liquid chromatography solvent at 0.3 mL min−1. The liquid chromatography gradient was as follows: 40% methanol, 50% acetonitrile, and 10% isopropanol, 1 min, followed by 10% methanol, 50% acetonitrile, and 40% isopropanol, 30 min. NaI (10 mM) in acetone was added by Hampton syringe pump at a flow rate of 6 μL min−1 via a tee connection into the eluent stream of HPLC prior to mass spectrometry. For fatty acyl group identification, TAG lipid ions selected as precursor ions were subjected to collision energy (50 V). The fatty acyl groups were identified based on the fragments deriving from neutral loss (NL) of individual fatty acyl groups. Fragments corresponding to the NL of sn-2 fatty acyl groups are known to yield lower abundances than those from the NL of sn-1/3 fatty acyl groups (Guella et al., 2003); thus, the abundances of tandem mass spectrometry product fragments were used for regiochemical determination.
For quantification of lipid content, calibration standards of each lipid class (see Supplemental Table 2 online) were titrated relative to a constant amount of internal standards. The peak intensity ratios of calibration standards relative to internal standards were plotted against their molar concentration ratios to establish the standard curve.
Accession Numbers
Accession numbers of protein sequences used in this study are given in Supplemental Table 3 online.
Supplemental Data
The following materials are available in the online version of this article:
Supplemental Figure 1. Gene Structure of Cr-PDAT and Amino Acid Sequence Alignment with Other LCAT-Like Proteins.
Supplemental Figure 2. Full-Length Nucleotide Sequence and Deduced Amino Acid Sequence of the Chlamydomonas PDAT Gene.
Supplemental Figure 3. Cr-PDAT Is a Membrane Protein with One Predicted Transmembrane Domain.
Supplemental Figure 4. Recombinant Cr-PDAT Protein Expression in E. coli and P. pastoris.
Supplemental Figure 5. TAG Composition of Wild-Type C. reinhardtii Grown under N-Replete Conditions.
Supplemental Figure 6. Fatty Acyl Groups of TAG C50 Determined by MS/MS Analysis.
Supplemental Figure 7. Concentration of TAG Molecular Species in Cr-PDAT amiRNA Knockdown Mutants Grown under N-Depleted Conditions.
Supplemental Figure 8. Total Membrane Lipid Concentrations in the Cr-PDAT Knockdown Mutants under N-Replete Conditions.
Supplemental Figure 9. MGDG, SQDG, and PG Molecular Species Concentrations in the Cr-PDAT Knockdown Mutants under N-Depleted Conditions.
Supplemental Figure 10. DGDG Species Concentrations in the Cr-PDAT Knockdown Mutants under N-Replete and -Depleted Conditions.
Supplemental Figure 11. PI Concentrations in the Cr-PDAT Knockdown Mutants under N-Depleted Conditions.
Supplemental Figure 12. Positive and Negative Controls for Lipase Activity Assays.
Supplemental Table 1. Primers Used for PDAT Gene Cloning and PCR Analysis.
Supplemental Table 2. Lipid Standards for ESI-MS Analysis.
Supplemental Table 3. GenBank Accession Numbers of Protein Sequences Used in This Study.
Supplemental Methods 1. cDNA Library Screening and Heterologous Expression of Cr-PDAT in E. coli, P. pastoris, and S. cerevisiae.
Supplemental Data Set 1. Text File of the Alignment Used to Generate the Phylogenetic Tree in Figure 1.
Acknowledgments
This work was supported by the U.S. Department of Energy (Grant DE-EE-0003372) and by the Science Foundation of Arizona (Grant SRG 0438-09).
AUTHOR CONTRIBUTIONS
K.Y. performed molecular cloning, recombinant protein expression, enzymatic assays, and bioinformatics analyses. D.H. developed the mass spectrometry method and performed in vivo functional characterization. Y.L. performed gene expression analysis and generated knockdown mutants. Q.H. designed the research. All of the authors contributed to analyzing the data and writing the article.
Glossary
- TAG
triacylglycerol
- DGAT
diacylglycerol acyltransferase
- DAG
diacylglycerol
- PDAT
phospholipid:DAG acyltransferase
- DGTA
diacylglycerol transacylase
- MGDG
monogalactosyldiacylglycerol
- DGDG
digalactosyldiacylglycerol
- LCAT
phosphatidylcholine-sterol O-acyltransferase
- MBOAT
membrane-bound O-acyl transferase
- ER
endoplasmic reticulum
- PL
phospholipid
- PA
phosphatidic acid
- PS
phosphatidylserine
- PI
phosphatidylinositol
- PG
phosphatidylglycerol
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- TLC
thin layer chromatography
- SQDG
sulfoquinovosyldiacylglycerol
- ESI-MS
electrospray ionization–mass spectrometry
- amiRNA
artificial microRNA
- GGGT
galactolipid:galactolipid galactosyltransferase
- TAP
Tris-acetate phosphate
- HL
high light
- LL
low light
- NL
neutral loss
References
- Banaś A., Dahlqvist A., Ståhl U., Lenman M., Stymne S. (2000). The involvement of phospholipid:diacylglycerol acyltransferases in triacylglycerol production. Biochem. Soc. Trans. 28: 703–705 [PubMed] [Google Scholar]
- Banas A., Carlsson A.S., Huang B., Lenman M., Banas W., Lee M., Noiriel A., Benveniste P., Schaller H., Bouvier-Navé P., Stymne S. (2005). Cellular sterol ester synthesis in plants is performed by an enzyme (phospholipid:sterol acyltransferase) different from the yeast and mammalian acyl-CoA:sterol acyltransferases. J. Biol. Chem. 280: 34626–34634 [DOI] [PubMed] [Google Scholar]
- Bates P.D., Durrett T.P., Ohlrogge J.B., Pollard M. (2009). Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 150: 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell R.M., Coleman R.A. (1980). Enzymes of glycerolipid synthesis in eukaryotes. Annu. Rev. Biochem. 49: 459–487 [DOI] [PubMed] [Google Scholar]
- Benning C., Ohta H. (2005). Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J. Biol. Chem. 280: 2397–2400 [DOI] [PubMed] [Google Scholar]
- Bligh E.G., Dyer W.J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Boyle N.R., et al. (2012). Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287: 15811–15825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S., Smith S.J., Zheng Y.W., Myers H.M., Lear S.R., Sande E., Novak S., Collins C., Welch C.B., Lusis A.J., Erickson S.K., Farese R.V., Jr (1998). Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 95: 13018–13023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S., Stone S.J., Zhou P., Yen E., Tow B., Lardizabal K.D., Voelker T., Farese R.V., Jr (2001). Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J. Biol. Chem. 276: 38870–38876 [DOI] [PubMed] [Google Scholar]
- Chen G., Greer M.S., Lager I., Yilmaz J.L., Mietkiewska E., Carlsson A.S., Stymne S., Weselake R.J. (2012). Identification and characterization of an LCAT-like Arabidopsis thaliana gene encoding a novel phospholipase A. FEBS Lett. 586: 373–377 [DOI] [PubMed] [Google Scholar]
- Copley S.D. (2003). Enzymes with extra talents: Moonlighting functions and catalytic promiscuity. Curr. Opin. Chem. Biol. 7: 265–272 [DOI] [PubMed] [Google Scholar]
- Dahlqvist A., Stahl U., Lenman M., Banas A., Lee M., Sandager L., Ronne H., Stymne S. (2000). Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 97: 6487–6492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., von Heijne G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser T., Waters A., Chatrattanakunchai S., Stobart K. (2000). Does triacylglycerol biosynthesis require diacylglycerol acyltransferase (DAGAT)? Biochem. Soc. Trans. 28: 698–700 [PubMed] [Google Scholar]
- Frentzen M. (1998). Acyltransferases from basic science to modified seed oils. Eur. J. Lipid Sci. Technol. 100: 161–166 [Google Scholar]
- Gaspar M.L., Hofbauer H.F., Kohlwein S.D., Henry S.A. (2011). Coordination of storage lipid synthesis and membrane biogenesis: Evidence for cross-talk between triacylglycerol metabolism and phosphatidylinositol synthesis. J. Biol. Chem. 286: 1696–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal A., Banas A., Ståhl U., Dahlqvist A., Lindqvist Y., Stymne S. (2007). Saccharomyces cerevisiae phospholipid:diacylglycerol acyl transferase (PDAT) devoid of its membrane anchor region is a soluble and active enzyme retaining its substrate specificities. Biochim. Biophys. Acta 1771: 1457–1463 [DOI] [PubMed] [Google Scholar]
- Gigon A., Matos A.R., Laffray D., Zuily-Fodil Y., Pham-Thi A.T. (2004). Effect of drought stress on lipid metabolism in the leaves of Arabidopsis thaliana (ecotype Columbia). Ann. Bot. (Lond.) 94: 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroud C., Gerber A., Eichenberger W. (1988). Lipids of Chlamydomonas reinhardtii. Analysis of molecular species and intracellular site(s) of biosynthesis. Plant Cell Physiol. 29: 587–595 [Google Scholar]
- Guella G., Frassanito R., Mancini I. (2003). A new solution for an old problem: The regiochemical distribution of the acyl chains in galactolipids can be established by electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17: 1982–1994 [DOI] [PubMed] [Google Scholar]
- Han X., Gross R.W. (2001). Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 295: 88–100 [DOI] [PubMed] [Google Scholar]
- Han X., Gross R.W. (2003). Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: A bridge to lipidomics. J. Lipid Res. 44: 1071–1079 [DOI] [PubMed] [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press). [DOI] [PubMed]
- Heemskerk J.W., Bogemann G., Wintermans J.F.G.M. (1983). Turnover of galactolipids incorporated into chloroplast envelopes an assay for galactolipid:galactolipid galactosyltransferase. Biochim. Biophys. Acta 754: 181–189 [Google Scholar]
- Hsu F.F., Turk J. (2009). Electrospray ionization with low-energy collisionally activated dissociation tandem mass spectrometry of glycerophospholipids: Mechanisms of fragmentation and structural characterization. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2673–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., Darzins A. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J. 54: 621–639 [DOI] [PubMed] [Google Scholar]
- Jako C., Kumar A., Wei Y., Zou J., Barton D.L., Giblin E.M., Covello P.S., Taylor D.C. (2001). Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 126: 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janero D.R., Barrnett R. (1981). Cellular and thylakoid-membrane glycolipids of Chlamydomonas reinhardtii 137+. J. Lipid Res. 22: 1119–1125 [PubMed] [Google Scholar]
- Katavic V., Reed D.W., Taylor D.C., Giblin E.M., Barton D.L., Zou J., Mackenzie S.L., Covello P.S., Kunst L. (1995). Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol. 108: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E.P. (1961). Biosynthesis of complex lipids. Fed. Proc. 20: 934–940 [PubMed] [Google Scholar]
- Khersonsky O., Roodveldt C., Tawfik D.S. (2006). Enzyme promiscuity: Evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 10: 498–508 [DOI] [PubMed] [Google Scholar]
- Kindle K.L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87: 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo I., Murakami M. (2002). Phospholipase A2 enzymes. Prostaglandins Other Lipid Mediat. 68-69: 3–58 [DOI] [PubMed] [Google Scholar]
- Lands W.E. (1960). Metabolism of glycerolipids. 2. The enzymatic acylation of lysolecithin. J. Biol. Chem. 235: 2233–2237 [PubMed] [Google Scholar]
- Lands W.E., Merkl I. (1963). Metabolism of glycerolipids. III. Reactivity of various acyl esters of coenzyme A with alpha’-acylglycerophosphorylcholine, and positional specificities in lecithin synthesis. J. Biol. Chem. 238: 898–904 [PubMed] [Google Scholar]
- Lardizabal K.D., Mai J.T., Wagner N.W., Wyrick A., Voelker T., Hawkins D.J. (2001). DGAT2 is a new diacylglycerol acyltransferase gene family: Purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyltransferase activity. J. Biol. Chem. 276: 38862–38869 [DOI] [PubMed] [Google Scholar]
- Lehner R., Kuksis A. (1993). Triacylglycerol synthesis by an sn-1,2(2,3)-diacylglycerol transacylase from rat intestinal microsomes. J. Biol. Chem. 268: 8781–8786 [PubMed] [Google Scholar]
- Li W., Wang R., Li M., Li L., Wang C., Welti R., Wang X. (2008a). Differential degradation of extraplastidic and plastidic lipids during freezing and post-freezing recovery in Arabidopsis thaliana. J. Biol. Chem. 283: 461–468 [DOI] [PubMed] [Google Scholar]
- Li Y., Sommerfeld M., Chen F., Hu Q. (2008b). Consumption of oxygen by astaxanthin biosynthesis: A protective mechanism against oxidative stress in Haematococcus pluvialis (Chlorophceae). J. Plant. Physiol. 165: 1783–1797 [DOI] [PubMed] [Google Scholar]
- Li Y.T., Han D.X., Hu G.R., Dauvillee D., Sommerfeld M., Ball S., Hu Q. (2010a). Chlamydomonas starchless mutant defective in ADP-glucose pyrophosphorylase hyper-accumulates triacylglycerol. Metab. Eng. 12: 387–391 [DOI] [PubMed] [Google Scholar]
- Li Y.T., Han D.X., Hu G.R., Sommerfeld M., Hu Q. (2010b). Inhibition of starch synthesis results in overproduction of lipids in Chlamydomonas reinhardtii. Biotechnol. Bioeng. 107: 258–268 [DOI] [PubMed] [Google Scholar]
- Listenberger L.L., Han X., Lewis S.E., Cases S., Farese R.V., Jr, Ory D.S., Schaffer J.E. (2003). Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA 100: 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancha M., Stymne S. (1997). Remodelling of triacylglycerols in microsomal preparations from developing castor bean (Ricinus communis L) endosperm. Planta 203: 51–57 [Google Scholar]
- Martinelle M., Holmquist M., Clausen I.G., Patkar S., Svendsen A., Hult K. (1996). The role of Glu87 and Trp89 in the lid of Humicola lanuginosa lipase. Protein Eng. 9: 519–524 [DOI] [PubMed] [Google Scholar]
- McCartney A.W., Dyer J.M., Dhanoa P.K., Kim P.K., Andrews D.W., McNew J.A., Mullen R.T. (2004). Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J. 37: 156–173 [DOI] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkl I., Lands W.E. (1963). Metabolism of glycerolipids. IV. Synthesis of phosphatidylethanolamine. J. Biol. Chem. 238: 905–906 [PubMed] [Google Scholar]
- Mhaske V., Beldjilali K., Ohlrogge J., Pollard M. (2005). Isolation and characterization of an Arabidopsis thaliana knockout line for phospholipid: diacylglycerol transacylase gene (At5g13640). Plant Physiol. Biochem. 43: 413–417 [DOI] [PubMed] [Google Scholar]
- Miller R., et al. (2010). Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering E.R., Benning C. (2011). Galactoglycerolipid metabolism under stress: A time for remodeling. Trends Plant Sci. 16: 98–107 [DOI] [PubMed] [Google Scholar]
- Moellering E.R., Muthan B., Benning C. (2010). Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 330: 226–228 [DOI] [PubMed] [Google Scholar]
- Molnar A., Bassett A., Thuenemann E., Schwach F., Karkare S., Ossowski S., Weigel D., Baulcombe D. (2009). Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 58: 165–174 [DOI] [PubMed] [Google Scholar]
- Noiriel A., Benveniste P., Banas A., Stymne S., Bouvier-Navé P. (2004). Expression in yeast of a novel phospholipase A1 cDNA from Arabidopsis thaliana. Eur. J. Biochem. 271: 3752–3764 [DOI] [PubMed] [Google Scholar]
- Oelkers P., Cromley D., Padamsee M., Billheimer J.T., Sturley S.L. (2002). The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 277: 8877–8881 [DOI] [PubMed] [Google Scholar]
- Oelkers P., Tinkelenberg A., Erdeniz N., Cromley D., Billheimer J.T., Sturley S.L. (2000). A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J. Biol. Chem. 275: 15609–15612 [DOI] [PubMed] [Google Scholar]
- Parthibane V., Rajakumari S., Venkateshwari V., Iyappan R., Rajasekharan R. (2012). Oleosin is bifunctional enzyme that has both monoacylglycerol acyltransferase and phospholipase activities. J. Biol. Chem. 287: 1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peelman F., Vinaimont N., Verhee A., Vanloo B., Verschelde J.L., Labeur C., Seguret-Mace S., Duverger N., Hutchinson G., Vandekerckhove J., Tavernier J., Rosseneu M. (1998). A proposed architecture for lecithin cholesterol acyl transferase (LCAT): Identification of the catalytic triad and molecular modeling. Protein Sci. 7: 587–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T.N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C.F., Natter K., Kohlwein S.D. (2009). Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 284: 30981–30993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S., Daum G. (2010a). Multiple functions as lipase, steryl ester hydrolase, phospholipase, and acyltransferase of Tgl4p from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 285: 15769–15776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S., Daum G. (2010b). Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol. Biol. Cell 21: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki T., Kondo N., Yamada M. (1990a). Pathway for the synthesis of triacylglycerols from monogalactosyldiacylglycerols in ozone-fumigated spinach leaves. Plant Physiol. 94: 773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki T., Saito K., Kawaguchi A., Kondo N., Yamada M. (1990b). Conversion of monogalactosyldiacylglycerols to triacylglycerols in ozone-fumigated spinach leaves. Plant Physiol. 94: 766–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey J.M., Gidda S.K., Chapital D.C., Kuan J.C., Dhanoa P.K., Bland J.M., Rothstein S.J., Mullen R.T., Dyer J.M. (2006). Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18: 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger D., Daum G. (2002). Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl-glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J. Bacteriol. 184: 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl U., Carlsson A.S., Lenman M., Dahlqvist A., Huang B.Q., Banas W., Banas A., Stymne S. (2004). Cloning and functional characterization of a phospholipid:diacylglycerol acyltransferase from Arabidopsis. Plant Physiol. 135: 1324–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobart K., Mancha M., Lenman M., Dahlqvist A., Stymne S. (1997). Triacylglycerols are synthesised and utilized by transacylation reactions in microsomal preparations of developing safflower (Carthamus tinctorius L) seeds. Planta 203: 58–66 [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Terashima M., Specht M., Hippler M. (2011). The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr. Genet. 57: 151–168 [DOI] [PubMed] [Google Scholar]
- van Besouw A., Wintermans J.F. (1978). Galactolipid formation in chloroplast envelopes. I. Evidence for two mechanisms in galactosylation. Biochim. Biophys. Acta 529: 44–53 [DOI] [PubMed] [Google Scholar]
- Weers P.M.M., Gulati R.D. (1997). Growth and reproduction of Daphnia galeata in response to changes in fatty acids, phosphorus, and nitrogen in Chlamydomonas reinhardtii. Limnol. Oceanogr. 42: 1584–1589 [Google Scholar]
- Welti R., Li W., Li M., Sang Y., Biesiada H., Zhou H.E., Rajashekar C.B., Williams T.D., Wang X. (2002). Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277: 31994–32002 [DOI] [PubMed] [Google Scholar]
- Welti R., Wang X., Williams T.D. (2003). Electrospray ionization tandem mass spectrometry scan modes for plant chloroplast lipids. Anal. Biochem. 314: 149–152 [DOI] [PubMed] [Google Scholar]
- Winstead M.V., Balsinde J., Dennis E.A. (2000). Calcium-independent phospholipase A(2): Structure and function. Biochim. Biophys. Acta 1488: 28–39 [DOI] [PubMed] [Google Scholar]
- Zhang H.X., Damude H.G., Yadav N.S. (2012). Three diacylglycerol acyltransferases contribute to oil biosynthesis and normal growth in Yarrowia lipolytica. Yeast 29: 25–38 [DOI] [PubMed] [Google Scholar]