Capping protein (CP) binds to the fast-growing ends of actin filaments and is negatively regulated by signaling lipids in vitro. This study demonstrates that the end-capping activity of CP is inhibited by membrane signaling phospholipids in living epidermal cells from Arabidopsis thaliana and that CP remodels the actin cytoskeleton in response to changes in these phospholipids.
Abstract
Remodeling of actin filament arrays in response to biotic and abiotic stimuli is thought to require precise control over the generation and availability of filament ends. Heterodimeric capping protein (CP) is an abundant filament capper, and its activity is inhibited by membrane signaling phospholipids in vitro. How exactly CP modulates the properties of filament ends in cells and whether its activity is coordinated by phospholipids in vivo is not well understood. By observing directly the dynamic behavior of individual filament ends in the cortical array of living Arabidopsis thaliana epidermal cells, we dissected the contribution of CP to actin organization and dynamics in response to the signaling phospholipid, phosphatidic acid (PA). Here, we examined three cp knockdown mutants and found that reduced CP levels resulted in more dynamic activity at filament ends, and this significantly enhanced filament-filament annealing and filament elongation from free ends. The cp mutants also exhibited more dense actin filament arrays. Treatment of wild-type cells with exogenous PA phenocopied the actin-based defects in cp mutants, with an increase in the density of filament arrays and enhanced annealing frequency. These cytoskeletal responses to exogenous PA were completely abrogated in cp mutants. Our data provide compelling genetic evidence that the end-capping activity of CP is inhibited by membrane signaling lipids in eukaryotic cells. Specifically, CP acts as a PA biosensor and key transducer of fluxes in membrane signaling phospholipids into changes in actin cytoskeleton dynamics.
INTRODUCTION
A dynamic network of actin filaments is critical for a wide variety of cellular processes, including vesicle trafficking, cell morphogenesis, and rapid cytoplasmic remodeling in response to biotic and abiotic stimuli. Actin filament organization and turnover are governed by dozens of actin binding proteins. However, there remains a knowledge gap between the biochemical properties of actin binding proteins and how they participate in the regulation of actin dynamics and response to stimuli in vivo. Phosphoinositide lipids (PPIs) are thought to play a central role in regulating the actin cytoskeleton during signal transduction and membrane trafficking. Many actin binding proteins directly interact with and are regulated by PPIs in vitro (Saarikangas et al., 2010), but whether this occurs in cells requires further analysis.
The barbed end of an actin filament is the favored site for actin polymerization in vitro and is therefore the presumed end where rapid actin filament elongation occurs in vivo. Capping protein (CP) is a conserved heterodimeric complex composed of α- and β-subunits. At a biochemical level, CP binds to filament barbed ends with high affinity and dissociates slowly, thereby blocking actin assembly and disassembly (Cooper and Sept, 2008). CP also binds to and is negatively regulated by signaling PPIs in vitro (Schafer et al., 1996; Kim et al., 2007; Kuhn and Pollard, 2007). Genetic evidence demonstrates that loss-of-function or null mutants for CP result in defective cell and developmental phenotypes in mammals, flies, and microbes (Amatruda et al., 1992; Hug et al., 1995; Hopmann et al., 1996; Kovar et al., 2005; Kim et al., 2006). Based on its biochemical features and cellular abundance, CP is considered to be the major filament barbed-end capper in vivo (Cooper and Sept, 2008). In support of this, the population of available barbed ends in Dictyostelium and yeast cells is inversely correlated with the amount of cellular CP (Hug et al., 1995; Kim et al., 2004), and loss of CP results in a 10 to 35% increase in filamentous F-actin levels, presumably due to polymerization onto free filament barbed ends (Hug et al., 1995; Hopmann et al., 1996; Kim et al., 2004; Kovar et al., 2005). CP is also essential for generating specific actin-based structures. Yeast null mutants have fewer actin cables and an increased number of actin patches (Amatruda et al., 1992; Kovar et al., 2005). However, cp knockdowns in mammalian cells result in proliferation of bundled actin in filopodia and loss of lamellipodial arrays at the leading edge of crawling cells (Hug et al., 1995; Rogers et al., 2003; Mejillano et al., 2004). Collectively, these data have been used to infer that CP binds filament barbed ends to keep filaments short and to focus polymerization at free filament ends (Pollard and Cooper, 2009). However, the consequence for actin organization caused by loss of CP is variable in different organisms, suggesting that the mechanism of actin turnover differs between cell types. Furthermore, whether and how exactly CP regulates the availability of barbed ends in vivo remains to be further addressed.
CP from Arabidopsis thaliana (At CP) is well characterized in vitro; it binds to filament barbed ends, prevents profilin-actin addition to filaments, and inhibits filament-filament annealing (Huang et al., 2003). In addition, At CP binds to the signaling lipids, phosphatidic acid (PA), and phosphatidylinositol (4,5)-bisphosphate (PIP2) in vitro, and this inhibits its barbed end capping activity and causes filament uncapping (Huang et al., 2006). PA is an abundant membrane phospholipid increasingly recognized as an important signaling intermediate in plant cells, especially in response to extrinsic stresses like cold, wounding, and attack by pathogens (Li et al., 2009; Testerink and Munnik, 2011). It is also important for normal development by regulating the actin cytoskeleton and vesicle trafficking, among other processes (Li et al., 2009; Testerink and Munnik, 2011). For example, organization of the actin cytoskeleton responds to altered PA levels and phospholipase D (PLD) activity (Motes et al., 2005; Pleskot et al., 2010), and applying exogenous PA to suspension-cultured cells and pollen tubes leads to an increase in actin filament levels (Lee et al., 2003; Huang et al., 2006; Pleskot et al., 2010). At CP is the only plant actin binding protein reported to bind PA (Huang et al., 2006) and could therefore act as a direct regulator of phospholipid-mediated changes in cytoarchitecture. However, whether CP is negatively regulated by PPIs or PA in eukaryotic cells is still an open question.
The ability to visualize individual actin filaments in Arabidopsis epidermal cells at high temporal and spatial resolution (Staiger et al., 2009; Smertenko et al., 2010; Henty et al., 2011; Tóth et al., 2012) makes this an ideal system to dissect genetically the role of barbed-end filament regulation. Previous observations demonstrate that most of the newly created filament ends generated by severing activity fail to regrow (Staiger et al., 2009; Smertenko et al., 2010), suggesting that the majority of barbed ends are capped at or soon after their creation. New filaments elongate rapidly at their barbed ends, reaching lengths of several microns before suffering the first severing events or ceasing growth. In addition, new filaments can be constructed by filament-filament annealing in vivo (Smertenko et al., 2010). These data imply that the end-capping activity is important for the control of actin filament assembly in plant cells (Blanchoin et al., 2010; Staiger et al., 2010).
Based on its biochemical features, we hypothesize that CP regulates the properties of filament barbed ends in plant cells (Huang et al., 2006; Blanchoin et al., 2010). Moreover, we predict that CP is a sensor of cellular PA levels and translates fluxes in membrane phospholipids into altered actin dynamics (Huang et al., 2006). Here, we use live-cell imaging to dissect the contribution of CP to actin filament dynamics and response to fluxes in PA. Three cp knockdown mutants have more available filament ends, resulting in up to a sixfold increased filament-filament annealing frequency and a greater proportion of elongating filaments that originate from preexisting plus ends. If CP is negatively regulated by phospholipids in vivo, then we predict that elevated PA should block CP activity and result in more available filament ends, mimicking the cp knockdown mutants. Significantly, treatment of wild-type cells with exogenous PA phenocopies the dynamic behaviors of filament ends observed in cp mutant cells, with annealing frequency increased up to threefold. By contrast, reducing PA levels by inhibition of PLD activity with 1-butanol results in reduced actin filament density in wild-type cells. Finally, the increased filament levels and end dynamics in a cp mutant are completely abrogated by exogenous PA treatment. Thus, we demonstrate that the signaling phospholipid PA negatively regulates CP activity in eukaryotic cells.
RESULTS
Arabidopsis CP Mutants Show Abnormal Cell and Organ Growth
To analyze the function of CP in Arabidopsis, three T-DNA insertion mutants were characterized. The homozygous CP α-subunit (CPA) mutant, cpa-1, contained a T-DNA in the first intron, whereas two CP β-subunit (CPB) mutant alleles had insertions in the seventh intron (cpb-1) and in the last exon (cpb-3), respectively (see Supplemental Figures 1A and 1B online). The levels of CPA and CPB transcripts were evaluated by quantitative real-time PCR (qRT-PCR) on 10-d-old dark-grown cp mutant seedlings (see Supplemental Figure 1C online). The full-length CPA transcript was markedly reduced but not eliminated in the cpa-1 mutant. Considering the T-DNA insertion sites in cpb mutants, we used two sets of primers to detect the existence of CPB full-length transcripts. One pair of primers resides upstream of the T-DNA insertion sites, whereas the other flanks both insertions. The full-length CPB transcripts were decreased approximately twofold in the cpb-1 mutant, whereas in the cpb-3 mutant, no products were detected using flanking primers, suggesting that the remaining mRNA was truncated (see Supplemental Figure 1C online). In yeast, CP α- and β-subunit expression is interdependent; deletion of the gene for one subunit leads to a loss of protein for the other subunit (Amatruda et al., 1992). Arabidopsis CP behaves similarly; reduction of CPA expression caused a decrease in CPB transcript level and vice versa (see Supplemental Figure 1C online).
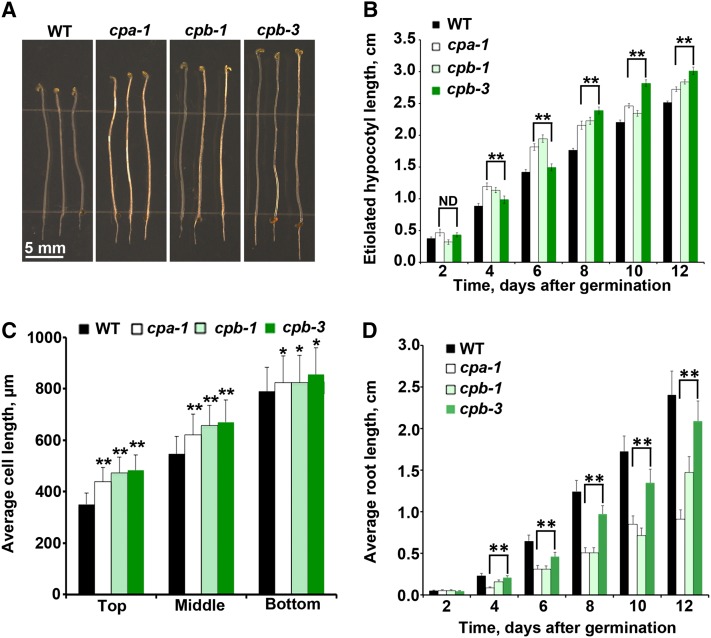
To investigate whether the reduction of CP results in cell and organ growth phenotypes, cp seedlings were evaluated under dark- or light-grown conditions. In the dark, hypocotyls from cp seedlings were noticeably longer than the wild type (Figure 1A), and these differences were significant over a developmental time series (Figure 1B). The growth of hypocotyl epidermal cells occurs along an acropetal gradient in Arabidopsis (Gendreau et al., 1997), with cells at the base finishing expansion prior to those at the apex. To examine the correlation between organ elongation phenotype and axial cell expansion, epidermal cell lengths from 5-d-old hypocotyls were measured and binned into thirds: top (located directly below the apical hook), middle, and bottom (basal cells adjacent to roots). When compared with the wild type, cp mutants have significantly longer cells in all three regions of the hypocotyl (Figure 1C). However, cell widths in cp mutants were not significantly altered. Under light-grown conditions, cp seedlings had significantly shorter root lengths compared with wild-type roots (Figure 1D). At later developmental stages, however, cp mutants did not show any clearly visible cell or organ growth phenotypes. Collectively, these data demonstrate that reduced CP levels affect cell and organ growth early in development. In addition, CP plays different roles or has varying levels of importance depending on the tissue type or developmental stage.
Figure 1.
Loss of CP Alters Organ and Epidermal Cell Length.
(A) Representative images of 11-d-old cp (cpa-1, cpb-1, and cpb-3) homozygous mutants and wild-type (WT) seedlings grown in continuous darkness. Bar = 5 mm.
(B) Etiolated hypocotyls from 4- to 12-d-old cpa-1, cpb-1, and cpb-3 mutant seedlings are significantly longer than the wild type (**P < 0.005; ND, no significant difference; t test). Measurements were taken from at least 50 seedlings per genotype on every other day. Values represent means ± se.
(C) Epidermal cell lengths were measured on 5-d-old dark-grown hypocotyls. Cell lengths from cp mutants are significantly longer compared with the wild type in the top (**P < 0.001, t test), middle (**P < 0.001, t test), and bottom (*P < 0.05, t test) thirds of hypocotyls. Values are means ± se (n > 50 cells from at least 10 hypocotyls per genotype).
(D) Light-grown roots from cpa-1, cpb-1, and cpb-3 mutant seedlings are significantly shorter than wild-type roots (**P < 0.005, t test). Values are means ± se from at least 50 seedlings per genotype taken on alternating days between 2 and 12 d postgermination.
In other species, CP is an obligate heterodimer, and neither subunit alone is sufficient to bind actin (Amatruda et al., 1992; Sizonenko et al., 1996; Wear et al., 2003; Kim et al., 2004). To disrupt both subunits and further reduce functional CP protein levels, two homozygous double mutants (cpa-1 cpb-1 and cpa-1 cpb-3) were generated. The levels of CPA and CPB transcripts in double mutants were not much further reduced compared with the single mutants (see Supplemental Figure 1C online). Moreover, the double mutants did not have a stronger etiolated hypocotyl length phenotype than the single mutants (see Supplemental Figure 2 online). For example, single cpa and cpb mutants had 10 to 20% longer hypocotyls than the wild type on day 12 (Figure 1B), and the double mutants were also ∼10 to 20% longer compared with wild-type hypocotyls (see Supplemental Figure 2 online). The similar transcription levels for CP genes and hypocotyl growth phenotypes in single and double mutants suggest that knocking down a single subunit is sufficient to reduce functional CP protein levels in Arabidopsis. In this case, studying the three single cp mutant alleles should provide strong genetic evidence for CP function in Arabidopsis.
Cortical Actin Arrays in Epidermal Cells of cp Mutants Have More Actin Filaments
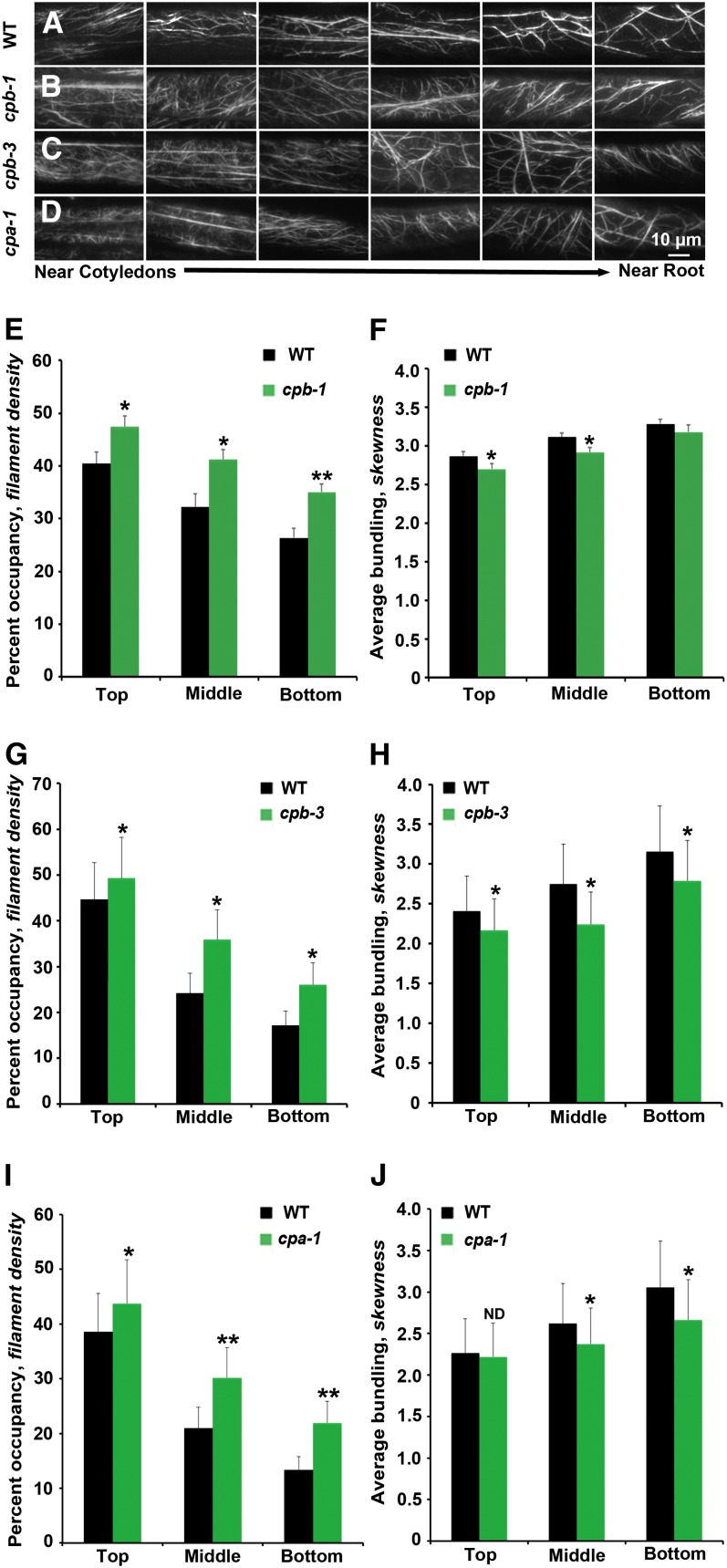
In hypocotyl epidermal cells, the abundance of actin filaments in the cortical array and the extent of actin filament bundling are correlated with the gradient of axial cell expansion (Henty et al., 2011). As one moves from cells undergoing fast axial expansion (near the cotyledons) toward the root where cell growth has ceased, actin arrays in the cortical cytoplasm of 5-d-old hypocotyl epidermal cells display decreased filament density and increased bundling (Henty et al., 2011). To examine whether reduced CP levels cause changes in overall cytoskeletal architecture, we observed the cortical actin array in hypocotyl epidermal cells from cp mutants and wild-type cells expressing the reporter gene GFP-fABD2 (for second actin binding domain of Arabidopsis FIMBRIN1) (Sheahan et al., 2004; Staiger et al., 2009). Representative epidermal cell images along individual hypocotyls from cp mutants showed a similar trend of actin architecture along the gradient compared with wild-type cells. However, the overall actin arrays appeared to be somewhat more dense than in comparable wild-type cells (Figures 2A to 2D).
Figure 2.
The Architecture of Cortical Actin Arrays in Epidermal Cells of cp Mutants Is Altered.
(A) to (D) Montages of VAEM micrographs of epidermal cells taken from representative hypocotyls of wild-type (WT) (A), cpb-1 (B), cpb-3 (C), and cpa-1 (D) seedlings expressing GFP-fABD2. Seedlings were grown in continuous darkness for 5 d and images collected to display cells near the cotyledon at left and cells near the root at right. Bar = 10 µm.
(E), (G), and (I) Average filament density or percentage of occupancy was measured along the axial gradient of cell expansion in hypocotyls and binned into three regions. The hypocotyls of 5-d-old dark-grown seedlings for all three mutants, cpb-1 (E), cpb-3 (G), and cpa-1 (I), have significantly increased filament density in the top, middle, and bottom thirds of 5-d-old dark-grown seedlings when compared with their respective wild-type siblings. Values given are means ± se (n ≥ 300 images from ≥ 150 cells per region, 30 hypocotyls were measured; t test: *P < 0.05 and **P < 0.01).
(F), (H), and (J) The extent of filament bundling (skewness) was measured and binned for the same regions as in density measurements. Epidermal cells from cpb-1 (F), cpb-3 (H), and cpa-1 (J) mutant seedlings have less filament bundling in different regions of the 5-d-old dark-grown seedlings when compared with their respective wild-type siblings. The same images used for (E), (G), and (I) were analyzed for skewness (*P < 0.05; ND, no significant difference; t test).
[See online article for color version of this figure.]
Two parameters, skewness and density, were previously established to measure the extent of actin filament bundling and the percentage of occupancy of actin filaments in epidermal cells, respectively (Higaki et al., 2010; Henty et al., 2011). As shown in Figure 2, cp mutants have a significantly increased percentage of occupancy of actin filaments in all three regions of the hypocotyl compared with the wild type (Figures 2E, 2G, and 2I). Additionally, cp cells were somewhat less bundled than comparable wild-type cells (Figures 2F, 2H, and 2J).
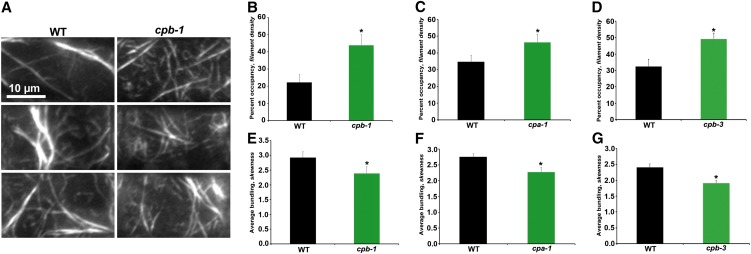
To determine whether actin architecture is altered in other organs, we examined epidermal cells from the root elongation zone of light-grown seedlings (Figure 3A). The percentage of occupancy of actin filaments was significantly increased in cp mutant cells (Figures 3B to 3D) and nearly doubled in cpb-1 cells (Figure 3B), whereas bundling was slightly decreased compared with wild-type cells (Figures 3E to 3G). These results confirm that reduced CP levels in plants caused an increase in the density of actin filament arrays, consistent with higher F-actin levels.
Figure 3.
Actin Architecture in Epidermal Cells from the Root Elongation Zone of cp Mutants Is Altered.
(A) Montages of VAEM micrographs from representative epidermal cells taken from the elongation zone of roots from light-grown wild-type (WT) and cpb-1 seedlings. Bar = 10 µm.
(B) to (D) Average filament density or percentage of occupancy was measured for epidermal cells within the root elongation zone. cpb-1 (B), cpa-1 (C), and cpb-3 (D) seedlings had significantly increased filament density in 7-d-old light-grown seedlings when compared with wild-type sibling seedlings.
(E) to (G) Filament bundling (skewness) was measured on the same images as for density measurements. Filaments in cpb-1 (E), cpa-1 (F), and cpb-3 (G) seedlings had less bundling when compared with wild-type sibling seedlings.
Values given are means ± se (n > 60 images per seedling from 25 seedlings per genotype; *P < 0.05, t test).
[See online article for color version of this figure.]
CP Contributes to Actin Stochastic Dynamics by Modulating the Properties of Filament Ends
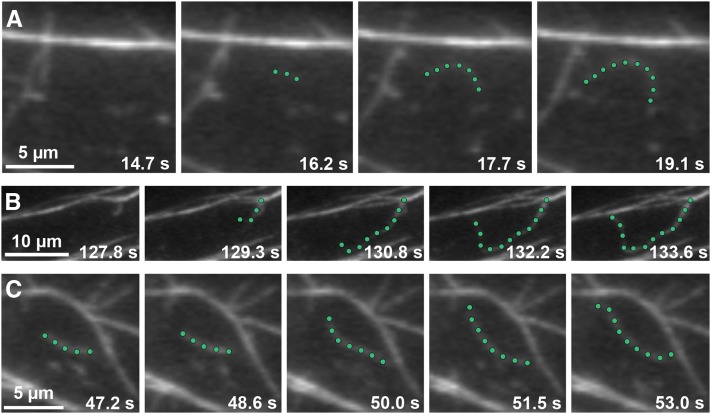
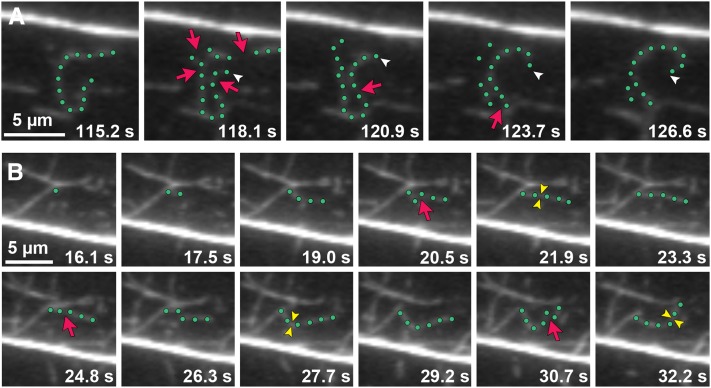
Actin filament turnover in Arabidopsis epidermal cells has been described as “stochastic dynamics,” which has two major components: rapid filament elongation at barbed ends and filament disassembly by severing activity (Staiger et al., 2009; Smertenko et al., 2010; Henty et al., 2011). New filament growth originates from three locations: de novo in the cytoplasm, the side of existing filaments or bundles, and the ends of preexisting fragments (Figures 4A to 4C; see also Supplemental Movies 1 to 3 and Supplemental Movie Legends 1 online). Numerous small fragments are generated by the severing of existing filaments; however, only occasionally do the newly created barbed ends regrow (Figure 5A; see Supplemental Movie 4 online). This suggests that the majority of barbed ends are capped at or soon after their creation (Staiger et al., 2009). Smertenko et al. (2010) recently reported new filament construction by the annealing of severed ends, and we have observed similar events in hypocotyl epidermal cells (Figure 5B; see Supplemental Movie 5 online). Given that CP blocks filament-filament annealing in vitro (Huang et al., 2003), this and other properties of filament ends could be regulated by CP in live cells.
Figure 4.
Growing Actin Filaments Originate from Three Locations in Wild-Type Epidermal Cells.
Time-lapse VAEM series show examples of actin filaments originating and growing rapidly from three different cellular locations: de novo (A), the side of a bundle (B), and the end of a preexisting fragment (C) (see Supplemental Movies 1 to 3 online). Representative actin filaments are highlighted with green dots. Images were recorded from epidermal cells in the actively growing region (top third) of 5-d-old dark-grown wild-type hypocotyls. Bars = 5 µm in (A) and (C) and 10 µm in (B).
Figure 5.
The Dynamic Behavior of Actin Filament Ends in Epidermal Cells Can Be Tracked.
(A) Time-lapse VAEM series shows an example of an actin filament elongating from a newly created barbed end. The highlighted filament (green dots) stops growing and is severed (red arrows) into several fragments. One filament end (white arrowheads) resumes growth within 5 s after severing (see Supplemental Movie 4 online).
(B) New filaments can be constructed by filament-filament annealing of severed fragments. The highlighted growing filament gets severed (red arrows). Newly created ends join together within ∼1.5 s to form a new filament (yellow arrowheads). Three annealing events occur in the sequential time-lapse frames (see Supplemental Movie 5 online).
Images were taken by time-lapse VAEM from cpb-1 epidermal cells. Bars = 5 µm.
To better understand and quantify the behavior of filament ends, we developed several parameters to analyze this further by time-lapse imaging of axially expanding epidermal cells. We tracked >50 filaments from their first appearance to complete disappearance and examined the behavior of every severed end from each of these filaments. The regrowth and annealing frequencies were defined as the percentage of presumed barbed ends undergoing regrowth or annealing from the total number of severed ends per filament, respectively. As shown in Table 1 and Supplemental Table 1 online, regrowth frequency in wild-type cells was ∼3%, which is close to previously reported values (4.4%; Staiger et al., 2009). Values for regrowth frequency in cp mutants were not significantly different from the wild type. Annealing events also occurred infrequently; from ∼200 newly created barbed ends, only ∼2% were observed to anneal in wild-type epidermal cells (Table 1; see Supplemental Table 1 online). Significantly, the annealing frequency in cp mutants increased up to fourfold compared with the wild type (Table 1; see Supplemental Table 1 online). Time-lapse images from a cpb-1 cell show an extreme example of annealing, where three events occurred within one filament (Figure 5B; see Supplemental Movie 5 online). Finally, we quantified the percentage of filament origins from three different locations in wild-type epidermal cells and cp mutants. All three cp mutants have a significantly increased proportion of filaments that originate from ends (∼30%) when compared with the wild type (∼20%; Table 1; see Supplemental Table 1 online).
Table 1. Actin Dynamics Parameters for cpb-1 and Wild-type Epidermal Cells.
| Stochastic Dynamics Parameters | Wild Type | cpb-1 |
|---|---|---|
| Elongation rate; µm/s | 1.64 ± 0.45 | 1.67 ± 0.54ND |
| Severing frequency; breaks/µm/s | 0.012 ± 0.006 | 0.009 ± 0.005* |
| Max. filament length; µm | 11.9 ± 3.5 | 15.0 ± 5.0* |
| Max. filament lifetime; s | 21.4 ± 7.1 | 26.5 ± 9.8* |
| Regrowth of severed ends; % | 2.9 ± 1.4 | 3.5 ± 1.4ND |
| Annealing of severed ends; % | 2.1 ± 1.0 | 9.3 ± 3.0* |
| Filament origin; % per cell | ||
| de novo | 27.9 ± 12.7 | 27.6 ± 15.9 |
| ends | 21.8 ± 8.7 | 28.2 ± 11.3† |
| side | 50.3 ± 14.7 | 44.2 ± 15.4 |
Measurements taken from epidermal cells in the top third of 5-d-old hypocotyls of the cpb-1 mutant and corresponding wild-type sibling plants. Values given are means ± se, with n > 50 filaments from n > 10 epidermal cells and at least 10 hypocotyls per line. For filament origin: n > 30 cells from at least 10 hypocotyls per line. ND, not significantly different from the wild-type control value by Student’s t test; P value > 0.05. *Significantly different from the wild-type control value by Student’s t test; P value ≤ 0.05. †Significantly different from wild-type control group value by analysis of variance; P value < 0.0001.
We also quantified several other stochastic dynamics parameters to see whether CP contributes to actin turnover in addition to regulating filament-end dynamics (Table 1; see Supplemental Table 1 online). The maximum actin filament length and lifetime were significantly increased in cp mutants, and the severing frequency was modestly reduced compared with the wild type. However, the filament elongation rate in cp mutants showed no substantial difference compared with the wild type.
When nonelongating cells from the base of 5-d-old hypocotyls were examined, the trends for most parameters were consistent with data for the fast-growing cells at the top of hypocotyls. It is noteworthy that the annealing frequency in the cpb-1 mutant was even higher (13.3%), with an increase of sixfold compared with the wild type (see Supplemental Table 2 online).
These data demonstrate that CP modulates the properties of filament ends in plant cells; its loss increases annealing frequency, as well as the proportion of filaments that originate from ends. In addition, CP contributes to actin filament turnover by decreasing maximum filament lengths and lifetimes.
cpb-1 Is Insensitive to the Variation of Cellular PA Levels
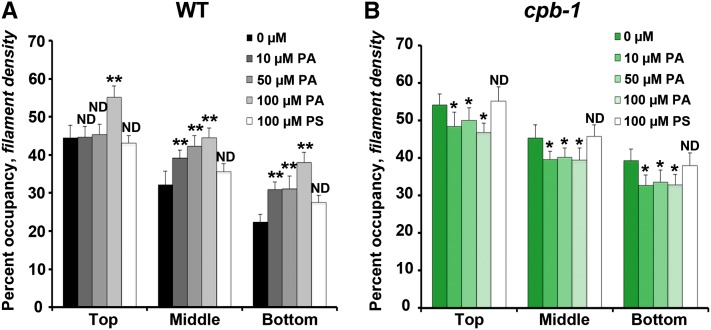
Previous studies show that applying exogenous PA to suspension-cultured cells and pollen tubes causes an increase in actin filament levels (Lee et al., 2003; Huang et al., 2006; Pleskot et al., 2010), and this was inferred to be due to inhibition of CP activity (Huang et al., 2006). We extended these findings by applying PA for 30 min at various concentrations to dark-grown hypocotyls and observed the overall organization of the cortical actin array in epidermal cells. Typically, the actin cytoskeleton appeared somewhat more dense in wild-type cells treated with PA (see Supplemental Figure 3 online). We confirmed these changes by measuring density in wild-type epidermal cells treated with PA. As shown in Figure 6A, treatment with PA (10 to 100 µM) resulted in a 10 to 20% increase in percentage of occupancy, especially in the middle and bottom regions of the hypocotyl. These differences are specific because treatment with another acidic phospholipid, phosphatidlyserine (PS), had no significant effect on percentage of occupancy (Figure 6A). If PA treatments prevent CP interaction with filament ends in wild-type cells, then cp mutants should be insensitive to exogenous lipid. To test this hypothesis, we performed the same treatments on cpb-1, as shown in Figure 6B. There was no significant increase in the percentage of occupancy of actin arrays in cpb-1 cells at any concentration tested and; in fact, filament density actually decreased modestly at all PA concentrations (Figure 6B).
Figure 6.
Density of the Actin Array in cpb-1 Hypocotyl Epidermal Cells Is Insensitive to Exogenous PA Treatment.
Percentage of occupancy or density was measured and binned into three regions along the axial gradient of cell expansion in 5-d-old etiolated wild-type (WT) (A) or cpb-1 (B) hypocotyls after incubation with 0, 10, 50, or 100 µM PA or with 100 µM PS for 30 min. Wild-type seedlings treated with PA have significantly increased filament density in the middle and bottom thirds of hypocotyls in a dose-dependent manner (A). By contrast, PA treatments of cpb-1 seedlings did not increase the density, but rather led to a modest decline in density at all concentrations tested (B). Treatments with another acidic phospholipid, PS, had no measurable effect on actin filament levels in either wild-type or cpb-1 seedlings. Values given are means ± se (n = 300 images from 150 cells per region; 10 hypocotyls were measured for each treatment per genotype; *P < 0.05 and **P < 0.001, analysis of variance).
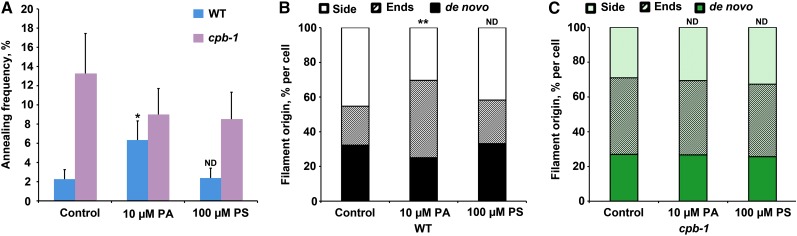
To further evaluate whether PA inhibits CP binding to filament ends, we measured single filament parameters after a modest PA treatment (10 µM for 30 min; see Supplemental Table 2 online). Single filaments, especially the filament ends, in wild-type epidermal cells treated with PA had dynamic behaviors similar to those in cpb-1 cells. The most convincing evidence was that the annealing frequency in PA-treated wild-type cells increased threefold compared with untreated cells (Figure 7A; see Supplemental Table 2 online). In addition, the proportion of filaments that originated from ends increased almost twofold after PA treatment of wild-type cells (Figure 7B; see Supplemental Table 2 online). On the other hand, annealing frequency and filament origins in cpb-1 mutant cells were insensitive to PA treatment (Figures 7A and 7C; see Supplemental Table 2 online). Single filament dynamics in cells treated with 100 µM PS showed no significant difference from untreated cells (Figure 7; see Supplemental Table 2 online). These data confirm that PA prevents CP binding to filament ends and, as a result, causes the increased percentage of occupancy of actin and enhanced filament-filament annealing in plant cells. Importantly, cp mutant cells are insensitive to PA treatments, providing genetic evidence that the signaling lipid regulates actin dynamics by negatively regulating CP activities.
Figure 7.
Annealing Frequency and Filament Origin Are Insensitive to Exogenous PA Treatment in cpb-1 Epidermal Cells.
Annealing frequency and filament origins were measured in cpb-1 and wild-type (WT) epidermal cells from the bottom third of hypocotyls following treatments with PA and PS.
(A) Annealing frequency from wild-type (WT) cells treated with PA was approximately threefold higher than untreated cells (blue; *P < 0.05, t test); cpb-1 cells after PA treatment had no obvious increase in annealing frequency compared with control cells (purple). Treatments with PS had no significant effect on annealing frequency in either cpb-1 or wild-type cells. Values given are means ± se (n > 50 filaments from > 10 epidermal cells and at least 10 hypocotyls for each treatment per genotype).
(B) The percentage of filaments that originate from ends was increased in wild-type cells after PA treatment.
(C) Filament origins in cpb-1 cells treated with phospholipids were not significantly different from untreated cells. Measurements were taken from n = 30 cells from at least 10 hypocotyls for each treatment per genotype; **P < 0.0001, analysis of variance.
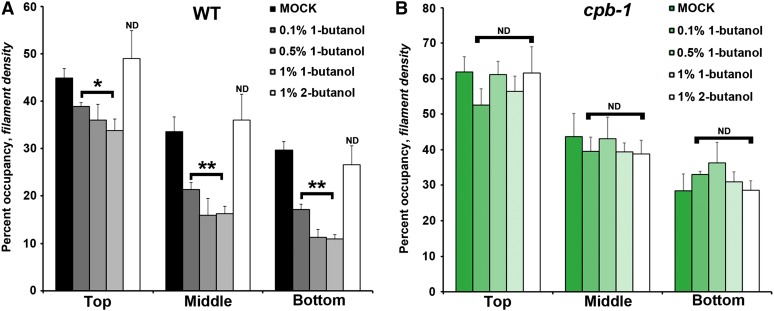
An alcohol isomer that inhibits PLD-dependent PA production (Munnik et al., 1995), 1-butanol, can effectively reduce cellular PA levels in plant cells (Motes et al., 2005; Pleskot et al., 2012). Applying 1-butanol to Arabidopsis seedlings causes actin cytoskeleton defects in different cell types, including hypocotyl epidermal cells (Motes et al., 2005). To quantitatively describe these defects, we treated dark-grown hypocotyls with 1-butanol at concentrations of between 0.1 and 1% (v/v) for 15 min and measured density in wild-type epidermal cells. As shown in Figure 8A, wild-type cells treated with 1-butanol had a significantly decreased percentage of occupancy of actin filaments. By contrast, treatment with the inactive secondary alcohol, 2-butanol at 1% (v/v), had no significant effect on percentage of occupancy (Figure 8A). Not surprisingly, reduction of CP abrogated the effect of 1-butanol treatments compared with wild-type cells (Figure 8B).
Figure 8.
Density of the Actin Array in cpb-1 Hypocotyl Epidermal Cells Is Insensitive to 1-Butanol Treatment.
Percentage of occupancy or density was measured and binned into three regions along the axial gradient of cell expansion in 5-d-old etiolated wild-type (WT) (A) or cpb-1 (B) hypocotyls after incubation with 0, 0.1, 0.5, or 1% 1-butanol or with 1% 2-butanol for 15 min. Wild-type seedlings treated with 1-butanol have significantly decreased filament density in the top, middle, and bottom regions of hypocotyls, and this reduction is dose dependent (A). By contrast, 1-butanol treatments of cpb-1 seedlings did not decrease the density (B). Treatments with 2-butanol had no measurable effect on actin filament levels in either wild-type or cpb-1 seedlings. Values given are means ± se (n = 300 images from 150 cells per region; 10 hypocotyls were measured for each treatment per genotype; *P < 0.05 and **P < 0.001, analysis of variance).
Collectively, the variation of cellular PA levels causes actin cytoskeleton defects in wild-type cells. Increasing PA levels stimulates actin polymerization (Figure 6A), whereas reducing PA results in a decreased percentage of occupancy of actin filaments (Figure 8A). Actin architecture in the cpb-1 mutant is insensitive to both the elevation and reduction of PA (Figures 6B and 8B). These data indicate that CP can sense cellular PA levels and functions as a downstream target of PA that transduces the PA signal into changes in actin cytoskeleton organization and dynamics.
DISCUSSION
By observing directly the dynamic behaviors of individual actin filaments with live-cell imaging techniques, we dissected the contribution of CP to actin organization and stochastic dynamics. Furthermore, we tested the hypothesis that the filament end-capping activity of CP is negatively regulated by membrane phospholipids in vivo. Reduced CP levels in Arabidopsis caused significantly enhanced hypocotyl and epidermal cell lengths, whereas roots were markedly shorter. All three cp mutants had more dense actin filament arrays than the wild type. CP was partially responsible for the availability of filament ends in live cells. For example, reduced CP levels resulted in more free ends, with an up to sixfold increase in annealing frequency and doubling of the number of growing filaments that originated from ends. In addition, CP contributes to actin turnover by keeping filaments short and short-lived, rather than regulating filament elongation rates. Increased actin filament levels and filament end dynamics following exogenous PA treatments mimicked those in cp mutants. These specific actin responses to exogenous PA were completely abrogated in cp mutants. Moreover, reducing PA levels in wild-type epidermal cells had the opposite effect to elevated PA, it reduced actin filament density. The cpb-1 mutant was also insensitive to reduced PA levels. Thus, we provide genetic evidence that the end-capping activity of CP is inhibited by a membrane signaling lipid in eukaryotic cells.
Previous data from diverse eukaryotic cells demonstrate that loss of CP increases the population of free barbed ends and elevates filamentous actin levels (Hug et al., 1995; Hopmann et al., 1996; Kim et al., 2004; Kovar et al., 2005). Here, we confirmed these results in plant cells and tracked the dynamic behaviors of individual filament ends. Three independent cp knockdown alleles had a greater number of available filament ends in cells, allowing more newly growing filaments to originate from barbed ends. As a result, reduced CP levels caused a significantly increased abundance of actin filaments in the cortical array of epidermal cells.
Filament-filament annealing has long been recognized in vitro and is inhibited by CP binding to barbed ends (Andrianantoandro et al., 2001; Huang et al., 2003). Recent data indicate that annealing also occurs in vivo (Okreglak and Drubin, 2007; Smertenko et al., 2010); this could be a favorable mechanism to generate a high density of actin filaments rapidly in response to biotic and abiotic stimuli (Abraham et al., 1999; Miyoshi et al., 2006). Considering that actin filaments elongate at ∼1.7 µm/s, this means a plant cell must add 629 ATP-loaded monomers on to each plus end per second (Staiger et al., 2009). The ATP on each subunit is hydrolyzed rapidly after assembly, resulting in the consumption of millions of ATPs per second in each cell for actin polymerization. Compared with this huge energy expenditure from monomer addition, filament-filament annealing could provide a way to build a dense actin network with much greater efficiency. In this study, we demonstrated that CP prevents annealing events in vivo. Reduced CP levels resulted in an up to sixfold increase in filament-filament annealing frequency compared with wild-type cells.
However, CP is not responsible for all behaviors of filament ends in plant cells. The regrowth frequency from recently severed ends was not affected in cp mutants, suggesting that there are multiple filament end regulators. Among ∼200 newly created ends tracked, only ∼3% resumed growth (Staiger et al., 2009). Both villin/gelsolin, which caps barbed ends after severing (Khurana et al., 2010; Bao et al., 2012), and ACTIN-INTERACTING PROTEIN1 (AIP1) are likely to be partially redundant with CP in Arabidopsis. A recent study demonstrates that loss of AIP1 in moss cells results in fewer severing events, suggesting that ADF/cofilin activity is reduced in the absence of AIP1 (Augustine et al., 2011).
PPIs are versatile regulatory molecules in eukaryotic cells and play diverse roles in various cellular events, including intracellular signaling responses, membrane trafficking, and modulating cytoskeletal organization (Saarikangas et al., 2010). Disruption of cellular PPI levels produces actin cytoskeletal defects (Tolias et al., 2000; Yamamoto et al., 2001; El Sayegh et al., 2007). For example, increasing cellular PIP2 in mammalian cells induces actin stress fiber formation and inhibits membrane ruffling, which is induced by actin polymerization under the plasma membrane (Yamamoto et al., 2001). Overexpression of type I phosphatidylinositol phosphate 5-kinase α (PIP5KI), which synthesizes PIP2, stimulates actin comet tail formation during vesicle trafficking (Rozelle et al., 2000). These and other data implicate that the actin cytoskeleton responds to various signaling cascades triggered by PPI fluxes in vivo.
Although numerous actin binding proteins are regulated by PPIs in vitro (Saarikangas et al., 2010), few studies provide evidence for the interaction of actin binding proteins and PPIs in vivo. Elevation of PIP2 in monkey kidney cells results in the inhibition of several conserved actin binding proteins (Yamamoto et al., 2001). Reduction of PIP2 levels in rat cells switches the subcellular localization of cofilin from plasma membrane to F-actin and activates its severing activity (van Rheenen et al., 2007). In addition, yeast cells expressing a cofilin with mutated PIP2 binding site have longer cytoplasmic actin cables than wild-type cells (Ojala et al., 2001). Disrupting the interaction of gelsolin and PIP2 in vivo results in aberrant actin organization, including increased filament lengths and fewer filament branches (El Sayegh et al., 2007). However, the exact mechanisms by which the activities of these actin binding proteins are regulated by PPIs and what the consequences for actin dynamics are remain unclear.
The interaction of CP and PPIs has been well studied in vitro. PIP2 can bind to CP, and this interaction inhibits capping activity (Schafer et al., 1996; Huang et al., 2003, 2006). By directly observing capping and uncapping of actin filament ends with total internal reflection fluorescence microscopy, one study reported that PIP2 can efficiently remove CP from filament barbed ends (Kim et al., 2007). On the contrary, another study provides evidence that although PIP2 prevents CP binding to filament barbed ends, it does not efficiently dissociate CP from ends (Kuhn and Pollard, 2007), indicating that the exact mechanism by which PIP2 regulates CP is still controversial (Cooper and Sept, 2008).
Despite the uncapping and anticapping activities of PIP2 in vitro, little is known about the physiological significance of PIP2–CP interactions. Increasing PIP2 concentration at the plasma membrane in CV1 cells through PIP5KI overexpression results in loss of CP from detergent-resistant cytoskeleton fractions (Yamamoto et al., 2001). Several older studies report that the activation of actin assembly in permeabilized human platelets results from uncapping of actin filament barbed ends by PPIs (Hartwig et al., 1995; Barkalow et al., 1996). These data imply that PIP2 inhibits end-capping activity of CP in vivo, which is important for the robust actin assembly required in diverse cellular processes.
CP from Arabidopsis is regulated by both PA and PIP2, with a modest preference for PIP2 (Huang et al., 2003, 2006). Because PA is markedly more abundant in plant membranes than is PIP2, the authors speculate that PA binding is physiologically relevant for CP activity. PA is considered a major signaling lipid that mediates many normal cell and developmental functions in plants (Li et al., 2009; Testerink and Munnik, 2011). Cellular levels of PA also change rapidly in response to biotic and abiotic stresses, such as water deficit, wounding, and microbial attack. Evidence for connections between PA signaling, PLD activity, and the cytoskeleton is emerging rapidly (Li et al., 2009; Testerink and Munnik, 2011). Recombinant plant PLDβ isoforms bind to actin filaments directly, and their activity is modulated by actin in a polymerization-dependent manner; specifically, monomeric actin inhibits PLDβ activity, whereas filamentous actin enhances it (Kusner et al., 2002; Pleskot et al., 2010). Hindering PLD activity and PA production with 1-butanol treatments results in an increase in bundled actin filaments in multiple plant cells and organs (Motes et al., 2005; Pleskot et al., 2010). Moreover, exogenous PA treatments cause an increase in actin filament levels (Lee et al., 2003; Huang et al., 2006; Pleskot et al., 2010).
In this study, we observed an increased density of actin filaments in the cortical array of epidermal cells following exogenous PA treatments. Lowering PA levels by inhibiting its production from PLD by 1-butanol treatments had the opposite effect. Furthermore, careful examination of actin dynamics after exogenous PA treatments revealed significant changes in the properties of filament ends. An increase in available ends was observed in wild-type cells treated with PA, with up to a threefold enhancement of filament-filament annealing and more growing filaments that originated from ends. Importantly, the increased actin filament levels and changes in end dynamics in cpb-1 mutant cells were completely insensitive to PA treatment. These results demonstrate that CP activities are negatively regulated by PA, resulting in the uncapping of filament ends, and thereby allowing assembly of new F-actin from a large pool of profilin-actin. Considering its concentration in cells (∼1 µM), CP is abundant enough to respond to signaling cascades triggered by PA/PLD. Additionally, the activation of actin assembly via the inhibition of CP through the PA/PLD pathway may further enhance PLD activity (Kusner et al., 2002; Pleskot et al., 2010). This positive feedback mechanism would facilitate signal amplification during responses to developmental cues or biotic and abiotic stresses. Collectively, we provide in vivo evidence that the end-capping activity of CP is inhibited by membrane lipids in eukaryotic cells. Thus, CP acts as a PA biosensor and key transducer of fluxes in membrane signaling lipids into changes in actin cytoskeleton dynamics. To date, At CP is the only plant actin binding protein known to directly bind and be regulated by PA in vitro (Huang et al., 2006); however, we cannot exclude the possibility that PA regulates other actin binding proteins in plant cells. In fact, the slight reduction of actin filament density after exogenous PA treatments in cpb-1 mutant cells suggests that there may be other proteins involved in PA-induced actin cytoskeleton changes.
The actin cytoskeleton plays a vital role in the regulation of anisotropic cell expansion. Investigations of actin and actin binding proteins in plant cell growth have provided insights into the coordination between F-actin array formation and cell expansion (Smith and Oppenheimer, 2005; Hussey et al., 2006). Numerous pharmacological and genetic data demonstrate that disrupting actin cytoskeleton arrays results in cell expansion defects (Smith and Oppenheimer, 2005; Hussey et al., 2006). For example, loss of ADF4 causes excessively bundled actin arrays and enhances the expansion of hypocotyl epidermal cells, which suggests that increased actin bundling is required for cell expansion (Henty et al., 2011). It has been hypothesized that actin bundles contribute to the delivery of cell wall–containing vesicles to plasma membrane sites where growth occurs (Hussey et al., 2006). It has also been argued that actin filament cables are responsible for distributing Golgi around the cell, which then dictates where cellulose synthase–containing vesicles are delivered to cortical microtubules for insertion into the plasma membrane (Gutierrez et al., 2009). Our data show that reduced CP levels cause a significantly more dense but less bundled actin array; however, these changes in actin array architecture still correlate with excessive elongation of hypocotyl epidermal cells. These findings imply that the extent of filament density and/or bundling do not directly correlate with cell elongation. In addition, epidermal cells from petioles of the adf4 mutant are shorter than wild-type petiole cells, which also exhibit enhanced bundling (Henty et al., 2011). Moreover, cp mutants have significantly reduced root length, but cells from the root elongation zone have actin arrays with an increased density of actin filaments and decreased extent of bundling. These data further suggest that there is no direct correlation between gross actin array architecture and the expansion of cells from different tissues or developmental stages. Previous studies demonstrate that actin filaments undergo rapid turnover and incessant rearrangements (Staiger et al., 2009; Smertenko et al., 2010; Henty et al., 2011). This raises the question of why plant cells need the actin network to be extremely dynamic. We hypothesize that there is a close interplay between actin dynamics and delivery of material for cell growth. One potential mechanism whereby cells might control their expansion is via precise regulation of actin dynamics. Based on the detailed analysis of stochastic dynamic parameters, the actively expanding hypocotyl epidermal cells from cp mutants have increased filament lengths and lifetimes as well as reduced severing frequency compared with wild-type cells. Consequently, the mutants have disrupted turnover of actin filaments; adf4 mutants also have the similar dynamic properties (Henty et al., 2011). Therefore, we predict that perturbation of actin dynamics and the increase in filament lengths and lifetimes leads to aberrant subcellular trafficking. As a consequence, cp and adf4 mutants with altered actin dynamicity show increased axial expansion of hypocotyl epidermal cells.
METHODS
Plant Material
Arabidopsis thaliana T-DNA insertion lines cpa-1 (SALK_080009), cpb-1 (SALK_014783), and cpb-3 (SALK_101017) were obtained from the ABRC (Ohio State University). Homozygous mutant plants were identified by PCR using primers described in Supplemental Table 3 online. These lines were crossed to wild-type Columbia-0 expressing the GFP-fABD2 reporter (Sheahan et al., 2004; Staiger et al., 2009), and homozygous mutants, as well as wild-type siblings, were recovered from F2 populations.
RNA Extraction and qRT-PCR
Dark-grown Arabidopsis seedlings were harvested at 10 d after germination, flash frozen, and ground to powder under liquid nitrogen. RNA was prepared with TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Two-step qRT-PCR was performed using 2× SYBR Green master mix (Applied Biosystems). Gene-specific primers for CPA and CPB (see Supplemental Figure 1 and Supplemental Table 3 online) were used, and relative expression levels were compared after normalization to GAPD transcript levels. Three biological and technical replicates were performed per genotype. Results were analyzed using comparative ΔCT (for cycle threshold) with GAPD controls (Khurana et al., 2010; Henty et al., 2011).
Plant Growth
Surface-sterilized seeds were plated onto 0.5× Murashige and Skoog medium supplemented with 1% Suc and stratified at 4°C for 3 d. After exposure to white light for 4 h, seedlings were grown in the dark for 5 d at 21°C prior to imaging of hypocotyl epidermal cells. A single-blind experimental design was used for all phenotypic analyses. To analyze root growth, seedlings were grown vertically on 0.5× Murashige and Skoog medium without Suc, solidified with 0.6% agar, under long-day conditions (16 h light/8 h dark) at 21°C (Dyachok et al., 2011). All image measurements were performed with Image J (http://rsb.info.nih.gov/ij/).
Phospholipid Preparation and Plant Treatment
The 1,2-diacyl-sn-glycero-3-phosphate from egg yolk (P9511; PA) and PS (P7769) were purchased from Sigma-Aldrich. Phospholipids were dissolved in chloroform, and the solvent was evaporated under a stream of N2. Before use, phospholipids were mixed with water and sonicated in a water bath for 5 min. For phospholipid or butanol treatments, 5-d-old dark-grown seedlings were soaked in PA or PS for 30 min or in butanol for 15 min prior to imaging. Water was used for control experiments.
Time-Lapse Imaging of Actin Filament Dynamics
The cortical actin cytoskeleton in epidermal cells from dark-grown hypocotyls expressing GFP-fABD2 was examined by time-lapse variable-angle epifluorescence microscopy (VAEM) as described previously (Staiger et al., 2009; Henty et al., 2011). A single-blind experimental design was used to compare single actin filament parameters between homozygous cp mutants and wild-type sibling lines. Maximum filament length and lifetime, filament severing frequency, and elongation rates were determined as previously (Staiger et al., 2009; Henty et al., 2011). Filament origin was defined as a percentage of the first 10 to 15 filaments observed per cell that originated either de novo, from the side of a bundle or filament, or from the end of a preexisting fragment. For this parameter, >30 cells from at least 10 hypocotyls per genotype were measured. The behavior of all newly created ends following filament severing events was also tracked. Regrowth or annealing frequency of the presumed plus ends was determined as a percentage; the number of events observed was divided by the total number of ends for each growing filament and multiplied by 100.
Quantitative Analyses of Cytoskeleton Architecture
Two parameters, skewness and density, were employed to quantitatively analyze actin cytoskeleton architecture (Higaki et al., 2010; Khurana et al., 2010; Henty et al., 2011); these correlate with the extent of actin filament bundling and percentage of occupancy of the GFP signal in an image, respectively. Every epidermal cell in a file along the long axis of 5-d-old etiolated hypocotyls, or the elongation zone of 7-d-old light-grown root tips, was documented with a series of overlapping VAEM micrographs. A fixed exposure time and gain setting were selected for cp mutants and their respective controls. Micrographs were analyzed in Image J using the methods described previously (Higaki et al., 2010; Henty et al., 2011). For statistical analysis, raw skewness and density values were binned into thirds based on position along the hypocotyl. For these measurements, at least 500 images of hypocotyl epidermal cells per binned section or >60 images of root epidermal cells were collected from 25 individual seedlings per genotype.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: At CPA, At3g05520; and At CPB, At1g71790.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Physical Maps and Molecular Characterization of the Arabidopsis Capping Protein (At CP) T-DNA Insertion Alleles.
Supplemental Figure 2. Two cp Double Mutants Have a Modest Hypocotyl Length Phenotype.
Supplemental Figure 3. Actin Arrays in Wild-type Epidermal Cells Are More Dense after PA Treatments.
Supplemental Movie 1. An Elongating Actin Filament Originates de Novo.
Supplemental Movie 2. An Elongating Actin Filament Originates from the Side of a Bundle.
Supplemental Movie 3. An Elongating Actin Filament Originates from a Preexisting End.
Supplemental Movie 4. An Actin Filament Elongates Rapidly from a Newly Created Barbed End.
Supplemental Movie 5. New Filaments Can Be Constructed by Filament-Filament Annealing of Severed Fragments.
Supplemental Movie Legends 1. Legends for Supplemental Movies 1 to 5.
Acknowledgments
We thank our colleagues in the Staiger lab for valuable feedback during these studies as well as David Kovar, Viktor Žárský, Martin Potocký, and Roman Pleskot for helpful comments on the article. This research was funded by a grant from the Physical Biosciences Program of the Office of Basic Energy Sciences at the U.S. Department of Energy (DE-FG02-09ER15526) to C.J.S. The total internal reflection fluorescence microscopy facility was supported in part by funds from the Bindley Bioscience Center.
AUTHOR CONTRIBUTIONS
J.L., S.H., L.B., and C.J.S designed the research. J.L., J.L.H.-R., S.H., and X.W performed experiments. J.L., J.L.H.-R., S.H., and C.J.S. analyzed data. J.L., L.B., and C.J.S. wrote the article.
Glossary
- PPI
phosphoinositide lipid
- CP
capping protein
- PIP2
phosphatidylinositol (4,5)-bisphosphate
- PA
phosphatidic acid
- PLD
phospholipase D
- qRT-PCR
quantitative real-time PCR
- PS
phosphatidlyserine
- VAEM
variable-angle epifluorescence microscopy
References
- Abraham V.C., Krishnamurthi V., Taylor D.L., Lanni F. (1999). The actin-based nanomachine at the leading edge of migrating cells. Biophys. J. 77: 1721–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda J.F., Gattermeir D.J., Karpova T.S., Cooper J.A. (1992). Effects of null mutations and overexpression of capping protein on morphogenesis, actin distribution and polarized secretion in yeast. J. Cell Biol. 119: 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E., Blanchoin L., Sept D., McCammon J.A., Pollard T.D. (2001). Kinetic mechanism of end-to-end annealing of actin filaments. J. Mol. Biol. 312: 721–730 [DOI] [PubMed] [Google Scholar]
- Augustine R.C., Pattavina K.A., Tüzel E., Vidali L., Bezanilla M. (2011). Actin interacting protein1 and actin depolymerizing factor drive rapid actin dynamics in Physcomitrella patens. Plant Cell 23: 3696–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao C., Wang J., Zhang R., Zhang B., Zhang H., Zhou Y., Huang S. (May 5, 2012). Arabidopsis VILLIN2 and VILLIN3 act redundantly in sclerenchyma development via bundling of actin filaments. Plant J. http://dx.doi.org/.10.1111/j.1365-1313X.2012.05044.x [DOI] [PubMed] [Google Scholar]
- Barkalow K., Witke W., Kwiatkowski D.J., Hartwig J.H. (1996). Coordinated regulation of platelet actin filament barbed ends by gelsolin and capping protein. J. Cell Biol. 134: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L., Boujemaa-Paterski R., Henty J.L., Khurana P., Staiger C.J. (2010). Actin dynamics in plant cells: A team effort from multiple proteins orchestrates this very fast-paced game. Curr. Opin. Plant Biol. 13: 714–723 [DOI] [PubMed] [Google Scholar]
- Cooper J.A., Sept D. (2008). New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell Mol. Biol. 267: 183–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyachok J., Zhu L., Liao F., He J., Huq E., Blancaflor E.B. (2011). SCAR mediates light-induced root elongation in Arabidopsis through photoreceptors and proteasomes. Plant Cell 23: 3610–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sayegh T.Y., Arora P.D., Ling K., Laschinger C., Janmey P.A., Anderson R.A., McCulloch C.A. (2007). Phosphatidylinositol-4,5 bisphosphate produced by PIP5KIgamma regulates gelsolin, actin assembly, and adhesion strength of N-cadherin junctions. Mol. Biol. Cell 18: 3026–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E., Traas J., Desnos T., Grandjean O., Caboche M., Höfte H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M.C., Ehrhardt D.W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Hartwig J.H., Bokoch G.M., Carpenter C.L., Janmey P.A., Taylor L.A., Toker A., Stossel T.P. (1995). Thrombin receptor ligation and activated Rac uncap actin filament barbed ends through phosphoinositide synthesis in permeabilized human platelets. Cell 82: 643–653 [DOI] [PubMed] [Google Scholar]
- Henty J.L., Bledsoe S.W., Khurana P., Meagher R.B., Day B., Blanchoin L., Staiger C.J. (2011). Arabidopsis actin depolymerizing factor4 modulates the stochastic dynamic behavior of actin filaments in the cortical array of epidermal cells. Plant Cell 23: 3711–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T., Kutsuna N., Sano T., Kondo N., Hasezawa S. (2010). Quantification and cluster analysis of actin cytoskeletal structures in plant cells: Role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J. 61: 156–165 [DOI] [PubMed] [Google Scholar]
- Hopmann R., Cooper J.A., Miller K.G. (1996). Actin organization, bristle morphology, and viability are affected by actin capping protein mutations in Drosophila. J. Cell Biol. 133: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Kovar D.R., Staiger C.J. (2003). Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 278: 44832–44842 [DOI] [PubMed] [Google Scholar]
- Huang S., Gao L., Blanchoin L., Staiger C.J. (2006). Heterodimeric capping protein from Arabidopsis is regulated by phosphatidic acid. Mol. Biol. Cell 17: 1946–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug C., Jay P.Y., Reddy I., McNally J.G., Bridgman P.C., Elson E.L., Cooper J.A. (1995). Capping protein levels influence actin assembly and cell motility in Dictyostelium. Cell 81: 591–600 [DOI] [PubMed] [Google Scholar]
- Hussey P.J., Ketelaar T., Deeks M.J. (2006). Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57: 109–125 [DOI] [PubMed] [Google Scholar]
- Khurana P., Henty J.L., Huang S., Staiger A.M., Blanchoin L., Staiger C.J. (2010). Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 22: 2727–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Galletta B.J., Schmidt K.O., Chang F.S., Blumer K.J., Cooper J.A. (2006). Actin-based motility during endocytosis in budding yeast. Mol. Biol. Cell 17: 1354–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., McCully M.E., Bhattacharya N., Butler B., Sept D., Cooper J.A. (2007). Structure/function analysis of the interaction of phosphatidylinositol 4,5-bisphosphate with actin-capping protein: Implications for how capping protein binds the actin filament. J. Biol. Chem. 282: 5871–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Yamashita A., Wear M.A., Maéda Y., Cooper J.A. (2004). Capping protein binding to actin in yeast: Biochemical mechanism and physiological relevance. J. Cell Biol. 164: 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R., Wu J.-Q., Pollard T.D. (2005). Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol. Biol. Cell 16: 2313–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.R., Pollard T.D. (2007). Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J. Biol. Chem. 282: 28014–28024 [DOI] [PubMed] [Google Scholar]
- Kusner D.J., Barton J.A., Wen K.-K., Wang X., Rubenstein P.A., Iyer S.S. (2002). Regulation of phospholipase D activity by actin. Actin exerts bidirectional modulation of mammalian phospholipase D activity in a polymerization-dependent, isoform-specific manner. J. Biol. Chem. 277: 50683–50692 [DOI] [PubMed] [Google Scholar]
- Lee S., Park J., Lee Y. (2003). Phosphatidic acid induces actin polymerization by activating protein kinases in soybean cells. Mol. Cells 15: 313–319 [PubMed] [Google Scholar]
- Li M., Hong Y., Wang X. (2009). Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim. Biophys. Acta 1791: 927–935 [DOI] [PubMed] [Google Scholar]
- Mejillano M.R., Kojima S.-i., Applewhite D.A., Gertler F.B., Svitkina T.M., Borisy G.G. (2004). Lamellipodial versus filopodial mode of the actin nanomachinery: Pivotal role of the filament barbed end. Cell 118: 363–373 [DOI] [PubMed] [Google Scholar]
- Miyoshi T., Tsuji T., Higashida C., Hertzog M., Fujita A., Narumiya S., Scita G., Watanabe N. (2006). Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: Evidence of frequent filament severing. J. Cell Biol. 175: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motes C.M., Pechter P., Yoo C.M., Wang Y.S., Chapman K.D., Blancaflor E.B. (2005). Differential effects of two phospholipase D inhibitors, 1-butanol and N-acylethanolamine, on in vivo cytoskeletal organization and Arabidopsis seedling growth. Protoplasma 226: 109–123 [DOI] [PubMed] [Google Scholar]
- Munnik T., Arisz S.A., de Vrije T., Musgrave A. (1995). G protein activation stimulates phospholipase D signaling in plants. Plant Cell 7: 2197–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala P.J., Paavilainen V.O., Lappalainen P. (2001). Identification of yeast cofilin residues specific for actin monomer and PIP2 binding. Biochemistry 40: 15562–15569 [DOI] [PubMed] [Google Scholar]
- Okreglak V., Drubin D.G. (2007). Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 178: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleskot R., Potocký M., Pejchar P., Linek J., Bezvoda R., Martinec J., Valentová O., Novotná Z., Zárský V. (2010). Mutual regulation of plant phospholipase D and the actin cytoskeleton. Plant J. 62: 494–507 [DOI] [PubMed] [Google Scholar]
- Pleskot R., Pejchar P., Bezvoda R., Lichtscheidl I.K., Wolters-arts M., Marc J., Žárský V., Potocký M. (March 19, 2012). Turnover of phosphatidic acid through distinct signalling pathways affects multiple aspects of tobacco pollen tube tip growth. Front. Physio. 3: 54 http://dx.doi.org/10.3389/fpls.2012.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Cooper J.A. (2009). Actin, a central player in cell shape and movement. Science 326: 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.L., Wiedemann U., Stuurman N., Vale R.D. (2003). Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozelle A.L., Machesky L.M., Yamamoto M., Driessens M.H.E., Insall R.H., Roth M.G., Luby-Phelps K., Marriott G., Hall A., Yin H.L. (2000). Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr. Biol. 10: 311–320 [DOI] [PubMed] [Google Scholar]
- Saarikangas J., Zhao H., Lappalainen P. (2010). Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 90: 259–289 [DOI] [PubMed] [Google Scholar]
- Schafer D.A., Jennings P.B., Cooper J.A. (1996). Dynamics of capping protein and actin assembly in vitro: Uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135: 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan M.B., Staiger C.J., Rose R.J., McCurdy D.W. (2004). A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136: 3968–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizonenko G.I., Karpova T.S., Gattermerir D.J., Cooper J.A. (1996). Mutational analysis of capping protein function in Saccharomyces cerevisiae. Mol. Biol. Cell 7: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A.P., Deeks M.J., Hussey P.J. (2010). Strategies of actin reorganisation in plant cells. J. Cell Sci. 123: 3019–3028 [DOI] [PubMed] [Google Scholar]
- Smith L.G., Oppenheimer D.G. (2005). Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 21: 271–295 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Poulter N.S., Henty J.L., Franklin-Tong V.E., Blanchoin L. (2010). Regulation of actin dynamics by actin-binding proteins in pollen. J. Exp. Bot. 61: 1969–1986 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Sheahan M.B., Khurana P., Wang X., McCurdy D.W., Blanchoin L. (2009). Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J. Cell Biol. 184: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerink C., Munnik T. (2011). Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 62: 2349–2361 [DOI] [PubMed] [Google Scholar]
- Tolias K.F., Hartwig J.H., Ishihara H., Shibasaki Y., Cantley L.C., Carpenter C.L. (2000). Type Ialpha phosphatidylinositol-4-phosphate 5-kinase mediates Rac-dependent actin assembly. Curr. Biol. 10: 153–156 [DOI] [PubMed] [Google Scholar]
- Tóth R., Gerding-Reimers C., Deeks M.J., Menninger S., Gallegos R.M., Tonaco I.A.N., Hübel K., Hussey P.J., Waldmann H., Coupland G. (2012). Prieurianin/endosidin 1 is an actin-stabilizing small molecule identified from a chemical genetic screen for circadian clock effectors in Arabidopsis thaliana. Plant J. 71: 338–352 [DOI] [PubMed] [Google Scholar]
- van Rheenen J., Song X., van Roosmalen W., Cammer M., Chen X., Desmarais V., Yip S.-C., Backer J.M., Eddy R.J., Condeelis J.S. (2007). EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 179: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wear M.A., Yamashita A., Kim K., Maéda Y., Cooper J.A. (2003). How capping protein binds the barbed end of the actin filament. Curr. Biol. 13: 1531–1537 [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Hilgemann D.H., Feng S.Y., Bito H., Ishihara H., Shibasaki Y., Yin H.L. (2001). Phosphatidylinositol 4,5-bisphosphate induces actin stress-fiber formation and inhibits membrane ruffling in CV1 cells. J. Cell Biol. 152: 867–876 [DOI] [PMC free article] [PubMed] [Google Scholar]