The UV-B photoreceptor UVR8 uses specific Trp amino acids in UV-B perception. This study examines the functional importance of all 14 UVR8 tryptophans in responses in plants.
Abstract
Arabidopsis thaliana UV RESISTANCE LOCUS8 (UVR8) is a photoreceptor specifically for UV-B light that initiates photomorphogenic responses in plants. UV-B exposure causes rapid conversion of UVR8 from dimer to monomer, accumulation in the nucleus, and interaction with CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1), which functions with UVR8 in UV-B responses. Studies in yeast and with purified UVR8 implicate several tryptophan amino acids in UV-B photoreception. However, their roles in UV-B responses in plants, and the functional significance of all 14 UVR8 tryptophans, are not known. Here we report the functions of the UVR8 tryptophans in vivo. Three tryptophans in the β-propeller core are important in maintaining structural stability and function of UVR8. However, mutation of three other core tryptophans and four at the dimeric interface has no apparent effect on function in vivo. Mutation of three tryptophans implicated in UV-B photoreception, W233, W285, and W337, impairs photomorphogenic responses to different extents. W285 is essential for UVR8 function in plants, whereas W233 is important but not essential for function, and W337 has a lesser role. Ala mutants of these tryptophans appear monomeric and constitutively bind COP1 in plants, but their responses indicate that monomer formation and COP1 binding are not sufficient for UVR8 function.
INTRODUCTION
Light is a key regulator of plant development, acting throughout the lifecycle. Different parameters of the light environment are detected by specific photoreceptors coupled to networks of signal transduction pathways (Jiao et al., 2007). It is well established that photomorphogenic responses are mediated by the phytochrome photoreceptors, which detect principally red and far-red light, and the cryptochromes, phototropins, and zeitlupe family proteins, which detect UV-A and blue light (Christie, 2007; Li and Yang, 2007; Franklin and Quail, 2010). In addition, relatively low, nondamaging levels of UV-B light (280 to 315 nm) elicit photomorphogenic responses in plants (Frohnmeyer and Staiger, 2003; Ulm and Nagy, 2005; Jenkins, 2009; Heijde and Ulm, 2012), including the expression of hundreds of genes and the suppression of hypocotyl elongation. A subset of genes induced via the UV-B photomorphogenic pathway encode proteins that help to prevent or repair damage by UV-B and therefore promote plant growth in sunlight (Ulm et al., 2004; Brown et al., 2005; Favory et al., 2009). Recent research has shown that the Arabidopsis thaliana UV RESISTANCE LOCUS8 (UVR8) protein specifically mediates photomorphogenic UV-B responses (Brown et al., 2005; Jenkins, 2009; Heijde and Ulm, 2012) by acting as a UV-B photoreceptor (Rizzini et al., 2011; Christie et al., 2012; Wu et al., 2012). It is therefore important to characterize the UVR8 protein and to understand how it functions to mediate UV-B responses.
UVR8 was first identified in a screen for Arabidopsis mutants hypersensitive to UV-B light (Kliebenstein et al., 2002). The uvr8 mutant had reduced levels of flavonoid sunscreen pigments and impaired expression of the CHALCONE SYNTHASE (CHS) gene in response to UV-B light. Further uvr8 alleles were isolated in transgene expression screens using the CHS and ELONGATED HYPOCOTYL5 (HY5) gene promoters to drive luciferase expression (Brown et al., 2005; Favory et al., 2009). Brown et al. (2005) demonstrated that UVR8 acts in a UV-B–specific manner and orchestrates expression of a set of genes that protect against potential damage by UV-B exposure, including genes encoding flavonoid biosynthesis enzymes, DNA repair enzymes, proteins involved in maintaining chloroplast function, and proteins concerned with amelioration of oxidative stress. Further genes regulated by UVR8 encode the HY5 and related HY5 HOMOLOG (HYH) transcription factors (Brown et al., 2005; Brown and Jenkins, 2008; Favory et al., 2009). HY5 has a key role in mediating the UV-B response of many UVR8 regulated genes and in conferring protection against UV-B light (Ulm et al., 2004; Brown et al., 2005; Brown and Jenkins, 2008; Stracke et al., 2010), whereas HYH has a secondary role, acting redundantly with HY5 in the regulation of some genes (Brown and Jenkins, 2008). Some of the genes regulated by UVR8 seem to be involved in morphogenesis, because the uvr8 mutant is altered in the UV-B–induced suppression of hypocotyl extension (Favory et al., 2009) and regulation of leaf expansion (Wargent et al., 2009). Hence, UVR8 regulates a range of photomorphogenic UV-B responses.
UVR8 is constitutively expressed (Kaiserli and Jenkins, 2007) and conserved among plant species, including in nonvascular plants, such as mosses and algae (Rizzini et al., 2011), suggesting that the protein appeared early in the evolution of photosynthetic plants to help them survive exposure to UV-B in sunlight. An action spectrum of UVR8 function, derived from dose–response studies of the induction of HY5 transcripts indicates that UVR8 is most effective at 280 nm with significant action at 290 and 300 nm (Brown et al., 2009). Because 280 nm wavelengths do not reach the surface of the earth, the longer wavelength action of UVR8 is the most physiologically relevant.
UVR8 was found to encode a seven-bladed β-propeller protein (Kliebenstein et al., 2002). Elucidation of the crystal structure of UVR8 (Christie et al., 2012; Wu et al., 2012) shows that it exists as a homodimer that is maintained by salt-bridge interactions between charged amino acids at the dimeric interaction surface. Mutational analysis shows that specific Arg, Asp, and Glu amino acids are key to salt-bridge formation (Christie et al., 2012). Exposure of UVR8 to UV-B light causes dissociation of the dimer into monomers; monomerization is observed with the purified protein (Christie et al., 2012; Wu et al., 2012), in plants, and in heterologous systems (Rizzini et al., 2011). Four tryptophans of UVR8 (W94, W233, W285, and W337) located adjacent to salt-bridge amino acids at the dimeric interface are sufficiently close that their electronic orbitals overlap. These excitonically coupled tryptophans are arranged to form two pyramids across the dimeric interface (Christie et al., 2012). Absorption of UV-B light by one or more pyramid tryptophans results in a loss of exciton coupling and leads to disruption of the salt-bridges and hence monomerization. W285 and W233 are most strongly implicated in UV-B absorption by UVR8 (Rizzini et al., 2011; Christie et al., 2012; Wu et al., 2012).
UV-B exposure stimulates the rapid accumulation of UVR8 in the nucleus (Kaiserli and Jenkins, 2007). Nuclear localization of UVR8 is required but is not sufficient for its function in regulating target gene expression. UVR8 binds to chromatin containing the HY5 gene and additional target genes via histones (Brown et al., 2005; Cloix and Jenkins, 2008), suggesting the basis of a mechanism for UVR8 function in regulating transcription. It seems likely that UVR8 interacts with other proteins associated with chromatin to promote remodeling and the recruitment or activation of transcription factors that stimulate transcription of its target genes.
Oravecz et al. (2006) reported that the CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) protein acts as a positive regulator of photomorphogenic UV-B responses. The cop1-4 mutant is impaired in the induction of flavonoid biosynthesis and the expression of numerous genes in response to UV-B, including HY5. This function is in contrast with the role of COP1 as a repressor of photomorphogenesis in dark-grown seedlings, where it acts as an E3 ubiquitin ligase to target positive regulators of photomorphogenesis, such as HY5 for proteolytic destruction by the proteasome (Yi and Deng, 2005). In UV-B responses, COP1 is required for regulation of many of the genes regulated by UVR8, suggesting that the two proteins function together. Consistent with this hypothesis, it was found that UVR8 interacts directly with COP1 in plants in response to UV-B exposure (Favory et al., 2009). This interaction is reproduced in heterologous systems (Rizzini et al., 2011). UVR8 interacts with two further proteins, REPRESSOR OF UV-B PHOTOMORPHOGENESIS1 (RUP1) and RUP2, which act as negative regulators of UVR8-mediated responses in vivo (Gruber et al., 2010).
Experiments with purified UVR8 and with the protein expressed in yeast have implicated a few specific tryptophans in photoreception. However, UVR8 has 14 conserved tryptophans, and the contribution of most of these to UVR8 structure and function has not been investigated systematically. Furthermore, it is essential to extend the functional analysis of the tryptophans from experiments in vitro and in yeast to studies with plants. Thus, in this article we use mutational analysis to study the function of all 14 tryptophans of UVR8 in transgenic uvr8-1 mutant plants. The findings extend our knowledge of the role of tryptophans in UVR8 function, establish the roles of specific tryptophans in UVR8-mediated responses in plants, and show that monomerization and interaction with COP1 are not sufficient to initiate a UVR8-mediated response in vivo.
RESULTS
UVR8 Structure Identifies Distinct Sets of Tryptophans
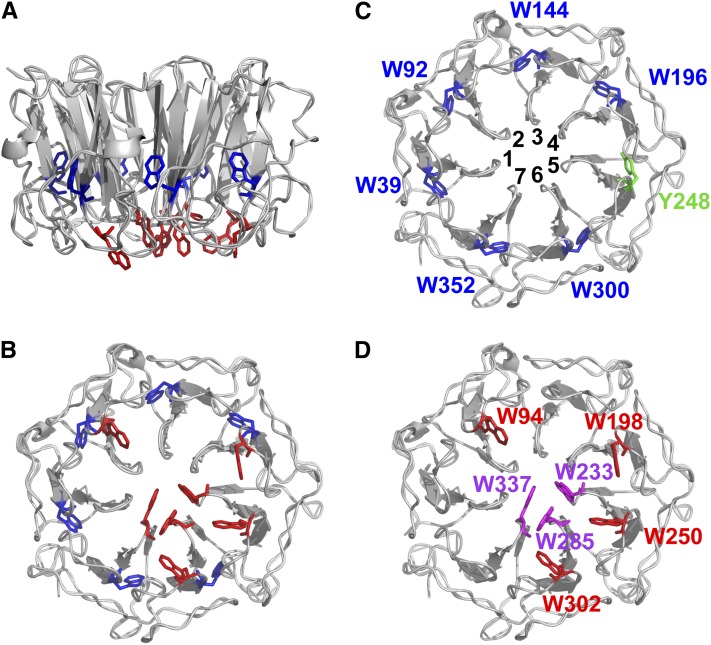
UVR8 has 14 tryptophans that are highly conserved in plant species (Rizzini et al., 2011; Wu et al., 2011; Christie et al., 2012). The crystal structure of UVR8 (Figures 1A and 1B) (Christie et al., 2012; Wu et al., 2012) shows the positions of all the tryptophans, except one (W400), which is in the C terminus. The remaining 13 tryptophans are organized into two groups. First, within the protein core is a ring of six tryptophans and one Tyr (Y248), each contributed by a different blade of the β-propeller (Figure 1C). Two of these tryptophans (W144 and W352) are conserved in the structurally related proteins RCC1 (Renault et al., 1998) and HERC2 (Bekker-Jensen et al., 2010; Wu et al., 2011), and three (W39, W196, and W300) are conserved between UVR8 and HERC2; the remaining Trp is W92. These aromatic residues form hydrogen bonds and hydrophobic interactions between adjacent blades that help to maintain the β-propeller structure. Second, seven tryptophans are located in the dimeric interface (Figure 1D); these include the triad tryptophans W233, W285, and W337 and also W94, which forms the apex of the excitonically coupled, cross-dimer Trp pyramid implicated in photoreception (Christie et al., 2012). W198, W250, and W302 are at the periphery of the dimeric interaction surface at the opposite side of the triad to W94.
Figure 1.
UVR8 Has Distinct Sets of Tryptophans.
(A) The arrangement of all UVR8 tryptophans (except W400) in the monomer viewed from the side. The structure is shown for amino acids 14 to 380. Tryptophans in the protein core and at the interaction surface are shown in blue and red, respectively.
(B) As in (A), but viewed from the dimeric interaction surface.
(C) The tryptophans in the core viewed from the dimeric interaction surface. Each Trp is associated with a different propeller blade (numbered). Y248 from blade 5 completes the ring of aromatic residues.
(D) The tryptophans at the dimeric interaction surface. The triad tryptophans are shown in magenta.
The images were produced using PyMOL.
Most of the Conserved Tryptophans of UVR8 Are Not Required for Function in Plants
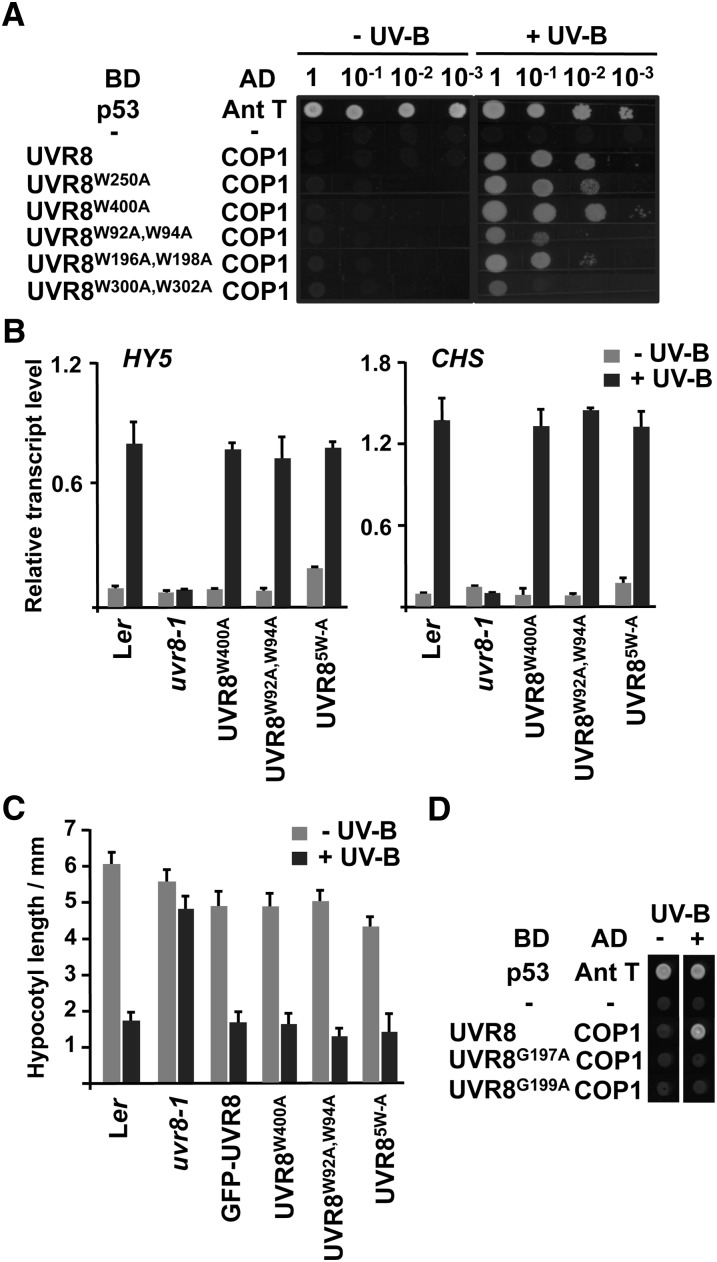
To examine the roles of tryptophans in UVR8 function, we mutated all of them singly and in several combinations and examined the activity of the mutant proteins in yeast and transgenic plants. The data obtained for all the tryptophans in this study are summarized in Table 1. In most cases, Trp was mutated to Ala, because of its nonpolar, nonaromatic nature. In yeast, a very low fluence rate of narrowband UV-B light stimulates interaction between UVR8 and COP1 (Rizzini et al., 2011). Mutation of eight UVR8 tryptophans, in some cases as double mutants (UVR8W250A; UVR8W400A; UVR8W92A,W94A; UVR8W196A,W198A; UVR8W300A,W302A), did not prevent the UV-B–stimulated interaction with COP1 (Figure 2A).
Table 1. Summary of Responses of UVR8 Trp Mutants.
| Ring Tryptophans |
Dimeric Interface Tryptophans |
C Terminus |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Response | Wild Type | W39 | W92 W196 W300 | W144 W352 | W94 W198 W250 W302 | W233 | W285 | W337 | W400 |
| UVR8-COP1 interaction in yeast | UV-B light induces interaction | No interaction as Ala mutant; wild-type interaction as Phe or Tyr mutant | As the wild type | As W39 | As the wild type | Interaction in darkness and UV-B light | As W233 | As W233 | As the wild type |
| Functional assays in plants | UV-B light induces gene expressiona, hypocotyl growth suppression; survival in high UV-B light | No wild-type responses | As the wild type | No protein expression in plants | As the wild type | Detectable but much reduced responses; no survival in high UV-B light | No wild-type responses as Ala or Phe mutant | Slightly reduced responses; survival in high UV-B light | As the wild type |
| Dimer/monomer in plantsb | Dimer in absence of UV-B light; UV-B light induces monomerization | Constitutive monomer | As the wild type | – | As the wild type | Dimer and monomer in absence of UV-B light; UV-B light induces monomerization | Ala mutant constitutive monomer; Phe mutant constitutive dimer | As W233 | As the wild type |
| UVR8-COP1 interaction in plants | UV-B light induces interaction | No interaction | As the wild type | – | As the wild type | Constitutive interaction | Constitutive interaction as Ala mutant; no interaction as Phe mutant | As W233 | As the wild type |
All UVR8 Trp mutants are Ala mutants unless otherwise stated.
HY5 and CHS transcripts.
As determined by the SDS-PAGE assay with nonboiled samples.
, Not tested because proteins were not expressed.
Figure 2.
Mutation of Eight UVR8 Tryptophans Has Little Effect on Function in Vivo.
(A) Plasmids containing a GAL4 binding domain (BD) or activation domain (AD), fused as indicated to UVR8 Trp mutants or COP1, were cotransformed into yeasts, which were then spotted as a dilution series on full selective media plates. As controls, yeasts were cotransformed with plasmids containing mammalian p53 and antigen T (Ant T; positive control) or no inserts (−; negative control). Yeasts were grown under 0.1 μmol m−2 s−1 of UV-B light (+UV-B) or in darkness (−UV-B) for 72 h.
(B) Quantitative assays of HY5 and CHS transcripts, normalized to control ACTIN2 transcript levels, in wild-type Ler, uvr8-1, and uvr8-1 transformed with GFP-UVR8 mutants; UVR85W-A is GFP-UVR8W196A,W198A,W250A,W300A,W302A. Plants were exposed (+UV-B) or not (−UV-B) to 3 μmol m−2 s−1 of UV-B light for 3 h. Data are means of three experiments ±se. The values for +UV-B are significantly greater than those for −UV-B (P < 0.01) for all genotypes except uvr8-1, which shows no significant difference (P > 0.05).
(C) Hypocotyl lengths of wild-type Ler, uvr8-1, and uvr8-1 transformed with GFP-UVR8 mutants. Seedlings were grown for 4 d in 2 μmol m−2 s−1 of white light with (+UV-B) or without (−UV-B) 1.5 μmol m−2 s−1 of UV-B light. Mean is shown ±se, n = 30. The values for +UV-B are significantly lower than those for −UV-B (P < 0.01) for all genotypes except uvr8-1, which shows no significant difference (P > 0.05).
(D) Yeast two-hybrid assay, performed as in (A), with UVR8 mutations from within the deleted region of uvr8-1.
To examine function in plants, UVR8 proteins with mutations in one or more of the previously mentioned eight tryptophans were expressed as green fluorescent protein (GFP) fusions in the uvr8-1 mutant. Several lines were selected for each mutant protein in which the level of expression of the GFP fusion was very similar to that of wild-type GFP-UVR8 (see Supplemental Figure 1 online), which fully complements the UV-B induction of HY5 gene expression in uvr8-1 (Kaiserli and Jenkins, 2007). As shown in Figure 2B, each of the mutant UVR8 proteins, in one case with mutations in five of the tryptophans (GFP-UVR8W400A; GFP-UVR8W92A,W94A; GFP-UVR8W196A,W198A,W250A,W300A,W302A [referred to as GFP- UVR85W-A]) restored wild-type levels of UV-B induction of HY5 and CHS expression in uvr8-1 plants. Similarly, the suppression of hypocotyl extension by UV-B light, which is impaired in uvr8-1, was fully complemented by the UVR8 mutants (Figure 2C). Moreover, each GFP-UVR8 mutant line survived exposure to elevated levels of UV-B light, as did the wild type, in contrast with the highly sensitive uvr8-1 (see Supplemental Figure 2 online).
The uvr8-1 mutant has a five amino acid deletion (W196GWGR200; Kliebenstein et al., 2002). Mutation of W196 and W198 to Ala did not confer a loss of function; therefore, we tested the importance of the other deleted amino acids. UVR8G197A and UVR8G199A showed no interaction with COP1 in yeast, indicating that mutation of these residues may account for the loss of UVR8 function in uvr8-1 (Figure 2D); immunoblot analysis demonstrated that the mutant proteins were expressed in the yeast cells (see Supplemental Figure 3A online).
Some Mutations of Putative Structural Tryptophans Impair UVR8 Interaction with COP1 and Stability and Function in Vivo
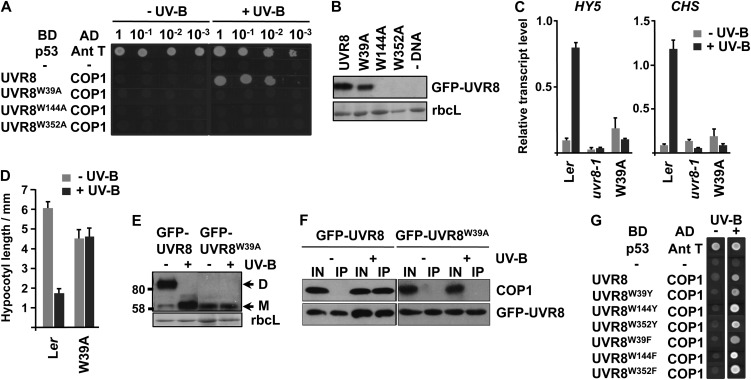
Mutation of three of the UVR8 ring tryptophans (W39, W144, and W352) (Figure 1C) to Ala prevented interaction of UVR8 with COP1 in yeast (Figure 3A); however, the mutant proteins were expressed in the cells (see Supplemental Figure 3B online). Despite several attempts, we failed to make transgenic Arabidopsis expressing GFP-UVR8W144A and GFP-UVR8W352A, and these proteins did not express transiently in Nicotiana benthamiana leaves (Figure 3B). GFP-UVR8W39A did express in N. benthamiana (Figure 3B) and in transgenic plants (see Supplemental Figure 1 online), but the fusion did not complement either the impaired gene expression (Figure 3C) or hypocotyl growth suppression phenotypes of uvr8-1 (Figure 3D), and the plants were sensitive to UV-B light, similar to uvr8-1 (see Supplemental Figure 2 online).
Figure 3.
Effect of Mutation of Specific UVR8 Ring Tryptophans on Function in Vivo.
(A) Plasmids containing a GAL4 binding domain (BD) or activation domain (AD), fused as indicated to UVR8 Trp mutants or COP1, were cotransformed into yeasts, which were then spotted as a dilution series on full selective media plates. As controls, yeasts were cotransformed with plasmids containing mammalian p53 and antigen T (Ant T; positive control) or no inserts (−; negative control). Yeasts were grown under 0.1 μmol m−2 s−1 of UV-B light (+UV-B) or in darkness (−UV-B) for 72 h.
(B) Transient expression of wild-type UVR8 and UVR8 Trp mutant proteins fused to GFP in N. benthamiana leaves compared with a nontransfected control (−DNA). Leaf proteins were fractionated by SDS-PAGE, and an immunoblot was incubated with anti-GFP antibody. Ponceau S–stained ribulose-1,5-bis-phosphate carboxylase/oxygenase large subunit (rbcL) protein is shown as a loading control.
(C) Quantitative assays of HY5 and CHS transcripts, normalized to control ACTIN2 transcript levels, in wild-type Ler, uvr8-1, and uvr8-1 transformed with GFP-UVR8W39A. Plants were exposed (+UV-B) or not (−UV-B) to 3 μmol m−2 s−1 of UV-B light for 3 h. Data are means of three experiments ±se. The values for +UV-B are significantly greater than those for −UV-B (P < 0.01) for the wild type but not for uvr8-1 and GFP-UVR8W39A (P > 0.05).
(D) Hypocotyl lengths of wild-type Ler and uvr8-1 transformed with GFP-UVR8W39A. Seedlings were grown for 4 d in 2 μmol m−2 s−1 of white light with (+UV-B) or without (−UV-B) 1.5 μmol m−2 s−1 of UV-B light. Mean is shown ±se, n = 30. The value for +UV-B is significantly lower than that for −UV-B (P < 0.01) for the wild type but not for GFP-UVR8W39A (P > 0.05).
(E) Dimer/monomer status of UVR8 in whole cell extracts obtained from uvr8-1 plants transformed with GFP-UVR8 or GFP-UVR8W39A. Extracts were exposed (+) or not (−) to 3 μmol m−2 s−1 of UV-B light for 30 min. SDS-loading buffer was added to the extracts, and unboiled samples were run on a 7.5% SDS-PAGE gel. An immunoblot was probed with anti-UVR8 antibody. UVR8 dimer (D) and monomer (M) bands are indicated. Ponceau staining of rbcL is shown as a loading control.
(F) Coimmunoprecipitation of GFP-UVR8 and COP1 in whole cell extracts obtained from uvr8-1 plants transformed with GFP-UVR8 or GFP-UVR8W39A exposed (+) or not (−) to 3 μmol m−2 s−1 of UV-B light for 3 h. Coimmunoprecipitation assays were performed under the same conditions. Input samples (15 μg, IN) and eluates (IP) were loaded on a SDS-PAGE gel, and the immunoblot was probed with anti-COP1 and anti-GFP antibodies.
(G) Yeast two-hybrid assay, performed as in (A), with Tyr and Phe mutants of specific ring tryptophans.
UVR8 forms a dimer in plant tissue that is converted to the monomer after UV-B exposure (Rizzini et al., 2011). The dimer is observed after SDS-PAGE as long as the protein is not boiled in SDS sample buffer (Rizzini et al., 2011; Christie et al., 2012; Wu et al., 2012). In contrast with GFP-UVR8, the GFP-UVR8W39A mutant appeared as a monomer in the gel assay both before and after UV-B treatment (Figure 3E). Interaction of UVR8 with COP1 in plants can be monitored using a coimmunoprecipitation assay (Favory et al., 2009). Wild-type GFP-UVR8 interacted with COP1 after UV-B exposure, but, by contrast, the GFP-UVR8W39A mutant failed to interact with COP1 (Figure 3F).
Because Ala mutations of the tryptophans mentioned above impaired expression and function of UVR8, we tested the effect of mutations to the aromatic amino acids Phe and Tyr. Each Trp-Phe and Trp-Tyr mutant of W39, W144, and W352 showed UV-B light stimulated interaction with COP1 in yeast, similar to the wild type (Figure 3G).
W233, W285, and W337 Are Key to UVR8 Function in Yeast and Plants
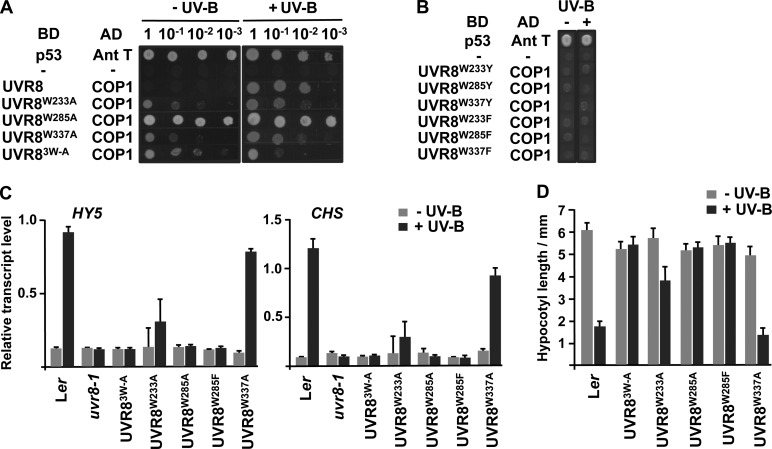
The triad tryptophans (Figure 1D) were mutated to examine their role in UVR8 function in vivo. Mutation of W233, W285, and W337 to Ala, both singly and in combination, caused interaction with COP1 in yeast in darkness as well as in UV-B light (Figure 4A). By contrast, mutations to Phe or Tyr prevented interaction in both darkness and UV-B light (Figure 4B), although the proteins were expressed in the cells (see Supplemental Figure 3C online). A similar observation was reported previously for W285A and W285F (Rizzini et al., 2011).
Figure 4.
Mutation of Triad Tryptophans Has Differential Effects on UVR8 Function in Vivo.
(A) Plasmids containing a GAL4 binding domain (BD) or activation domain (AD), fused as indicated to UVR8 Trp mutants or COP1, were cotransformed into yeasts, which were then spotted as a dilution series on full selective media plates. As controls, yeasts were cotransformed with plasmids containing mammalian p53 and antigen T (Ant T; positive control) or no inserts (−; negative control). Yeasts were grown under 0.1 μmol m−2 s−1 of UV-B light (+UV-B) or in darkness (−UV-B) for 72 h. UVR83W-A is GFP-UVR8W233A,W285A,W337A.
(B) Yeast two-hybrid assay, performed as in (A), with Tyr and Phe mutants of triad tryptophans.
(C) Quantitative assays of HY5 and CHS transcripts, normalized to control ACTIN2 transcript levels, in wild-type Ler, uvr8-1, and uvr8-1 transformed with GFP-UVR8 mutants. Plants were exposed (+UV-B) or not (−UV-B) to 3 μmol m−2 s−1 of UV-B light for 3 h. Data are means of three experiments ±se. The values for +UV-B are significantly greater than those for −UV-B (P < 0.01) for the wild type and GFP-UVR8W337A but not for the other genotypes (P > 0.05).
(D) Hypocotyl lengths of wild-type Ler and uvr8-1 transformed with GFP-UVR8 mutants. Seedlings were grown for 4 d in 2 μmol m−2 s−1 of white light with (+UV-B) or without (−UV-B) 1.5 μmol m−2 s−1 of UV-B light. Mean is shown ±se, n = 30. The values for +UV-B are significantly lower than those for −UV-B for the wild type (P < 0.01), GFP-UVR8W337A (P < 0.01), and GFP-UVR8W233A (P < 0.05) but not for the other genotypes (P > 0.05).
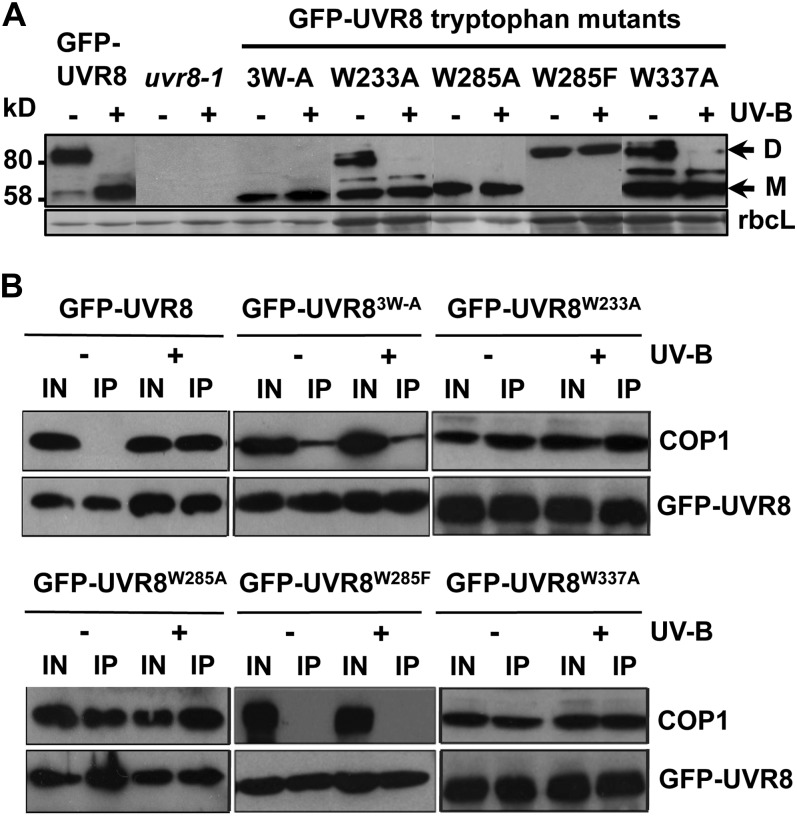
The function of the triad tryptophans in plants was examined by expression of GFP fusions in uvr8-1. For each mutation, transgenic lines were selected that had very similar levels of expression to GFP-UVR8 (see Supplemental Figure 1 online), which fully complements uvr8-1 (Kaiserli and Jenkins, 2007). We reported recently (Christie et al., 2012) that GFP-UVR8W285A fails to restore the loss of UV-B–induced HY5 gene expression in uvr8-1, and a similar result was observed for CHS expression (Figure 4C). Furthermore, GFP-UVR8W285A failed to complement the impaired hypocotyl growth-suppression phenotype (Figure 4D), and the transgenic plants were as sensitive to UV-B light as uvr8-1 (see Supplemental Figure 2 online). Similarly, GFP-UVR8W285F was unable to complement uvr8-1 in each of these functional assays.
GFP-UVR8W233A gave partial complementation of UVR8 function. In each experiment undertaken with uvr8-1/GFP-UVR8W233A plants, HY5 and CHS transcript levels were greater in UV-B light than in its absence, with quantification suggesting that the response to UV-B light was ∼20 to 25% of that in wild-type plants. However, the transcript levels in the different experiments were quite variable, and the mean values in UV-B light were not significantly greater than those in the absence of UV-B light when analyzed by Student’s t test (Figure 4C). Hypocotyl growth suppression by UV-B light was partially restored in uvr8-1/GFP-UVR8W233A transgenic plants, and the reduction of hypocotyl length was statistically significant (Figure 4D), but the plants showed little difference in UV-B sensitivity compared with uvr8-1 (see Supplemental Figure 2 online).
GFP-UVR8W337A substantially, but not fully, complemented uvr8-1 in HY5 and CHS gene expression, giving ∼80% of the wild-type response (Figure 4D). The plants survived elevated UV-B light but did not seem as healthy as the GFP-UVR8 control (see Supplemental Figure 2 online). However, hypocotyl growth suppression by UV-B light in uvr8-1/GFP-UVR8W337A plants was very similar to the wild-type response (Figure 4C).
Mutations of W233, W285, and W337 Affect UVR8 Monomerization and Interaction with COP1
The ability of W233, W285, and W337 mutants of UVR8 to form dimers and to monomerize after UV-B exposure was examined in plants using SDS-PAGE with nonboiled samples. As shown in Figure 5A, GFP-UVR8W285A and GFP-UVR8W233A,W285A,W337A appeared as constitutive monomers in the above assay, whereas GFP-UVR8W233A and GFP-UVR8W337A appeared as a mixture of dimer and monomer minus UV-B light and monomer after UV-B exposure. By contrast, GFP-UVR8W285F appeared as a constitutive dimer in plants.
Figure 5.
Mutation of Triad Tryptophans Affects UVR8 Dimer/Monomer Status and Interaction with COP1 in Vivo.
(A) Immunoblot of whole cell extracts obtained from uvr8-1 plants transformed with GFP-UVR8 or GFP-UVR8 triad Trp mutants. UVR83W-A is GFP-UVR8W233A,W285A,W337A. Extracts were exposed (+) or not (−) to 3 μmol m−2 s−1 of UV-B light for 30 min. SDS-loading buffer was added to the extracts, and unboiled samples were run on a 7.5% SDS-PAGE gel. An immunoblot was probed with anti-UVR8 antibody. UVR8 dimer (D) and monomer (M) bands are indicated. Ponceau staining of ribulose-1,5-bis-phosphate carboxylase/oxygenase large subunit (rbcL) is shown as a loading control.
(B) Coimmunoprecipitation assays with whole cell extracts obtained from GFP-UVR8 or GFP-UVR8 triad Trp mutant plants exposed (+) or not (−) to 3 μmol m−2 s−1 of UV-B light for 3 h. Coimmunoprecipitation assays were performed under the same conditions. Input samples (15 μg, IN) and eluates (IP) were loaded on a SDS-PAGE gel and the immunoblot was probed with anti-COP1 and anti-GFP antibodies.
The coimmunoprecipitation assay revealed a constitutive interaction between COP1 and each of the Ala mutants GFP-UVR8W233A, GFP-UVR8W285A, and GFP-UVR8W337A, with a weaker interaction in the triple mutant GFP-UVR8W233A,W285A,W337A (Figure 5B). Again, by contrast, GFP-UVR8W285F did not interact with COP1. Because the Ala mutants bound COP1 even when plants were not exposed to UV-B, we examined whether there was evidence for a cop1 mutant phenotype. Growth of the Trp-Ala mutants seemed normal under non–UV-B conditions (Figure 4D; see Supplemental Figure 2 online), and hypocotyl extension and cotyledon expansion in darkness resembled that of control GFP-UVR8 seedlings rather than cop1-4 (see Supplemental Figure 4 online).
DISCUSSION
Some Tryptophans Are Important in Maintaining UVR8 Structure
To date, the study of UVR8 tryptophans has focused on the role of the triad tryptophans at the dimeric interface in UV-B photoreception. However, the data presented in Figure 3 show that the ring tryptophans within the UVR8 core are important in maintaining structure, which underpins photoreceptor function. The aromatic residues contributed by the seven blades of the propeller (Figure 1C) have a very similar molecular environment. Each residue forms a hydrogen bond with a backbone carbonyl group of the preceding blade as well as hydrophobic interactions with adjacent residues that together will strengthen linkages between blades and help to maintain the β-propeller structure.
Mutation of the ring tryptophans W144 and W352 to Ala most likely disrupts the hydrogen bond and hydrophobic interactions in the UVR8 core sufficiently to cause misfolding and instability of the protein, leading to the failure to express in plants. Although UVR8W144A and UVR8W352A are expressed in yeast, they fail to interact with COP1, probably because the protein is unable to adopt the correct conformation for COP1 binding. UVR8W39A does express in plants but appears as a monomer in the gel assay (Figure 3E), indicating that the dimer is unable to form or is substantially weakened. W39 is involved in linking blades 1 and 7 of the protein, which is particularly important in preserving the overall structure. Moreover, UVR8W39A fails to interact with COP1 in plants and to functionally complement uvr8-1. It is likely that impaired interactions in the core of UVR8W39A affect the monomer structure sufficiently to prevent COP1 interaction. The Tyr and Phe mutants of W144, W352, and W39 do interact with COP1 in a UV-B–dependent manner in yeast, indicating that these aromatic residues permit formation of the hydrophobic interactions that stabilize the UVR8 core. It is interesting that mutation of the remaining three ring tryptophans (W92, W196, and W300) does not prevent function in vivo. In fact, two of these tryptophans are mutated to Ala in the UVR85W-A mutant, which complements uvr8-1. A possible explanation of the more benign effect of mutations to W92, W196, and W300 is that, in contrast with W39, W144, and W352, these residues have adjacent Tyr residues (Y90, Y194, and Y298) that could move into the space caused by mutation of the tryptophans to Ala and form a water-mediated hydrogen bond with the backbone carbonyl group of the preceding blade as well as hydrophobic interactions.
Specific Tryptophans at the Dimeric Interface Are Concerned with Photoreception in Vivo
The dimeric interface has a specific spatial arrangement of tryptophans. The triad tryptophans, W233, W285, and W337, are close together and excitonically coupled. W94, which is to one side of the triad in the interacting surface (Figure 1D), is excitonically coupled with the triad on the opposite monomer and forms the apex of a pyramid with the triad as the base (Christie et al., 2012). The three tryptophans (W198, W250, W302) at the opposite side of the triad to W94 form an outer ring of aromatic residues together with Y201, Y253, and F305, which is suggested to shield the triad from solvent (Christie et al., 2012). However, mutation of these three outer tryptophans to Ala, in the context of the UVR85W-A mutant, does not prevent UVR8 function in plants. The UVR85W-A mutant fully complements the impaired gene expression, hypocotyl growth suppression, and UV-B tolerance phenotypes of uvr8-1. Similarly, Ala mutations of W198, W250, and W302 do not prevent interaction with COP1 in yeast. We conclude that these tryptophans are not required for UVR8 function in vivo. However, we cannot exclude the possibility that mutations in these amino acids cause subtle changes in UVR8 function under particular conditions that have not been examined in this study. For instance, it will be interesting to examine the dose–response relationships of these mutants to see whether there are differences in response at low, nonsaturating doses.
Previous studies in yeast and in vitro have implicated the triad tryptophans in UV-B photoreception. The UVR8W285F mutant expressed in yeast does not monomerize in response to UV-B light (Rizzini et al., 2011), and the purified protein is a constitutive dimer that fails to respond to UV-B light based on circular dichroism (CD) spectroscopy (Christie et al., 2012) and fluorescence spectroscopy (Wu et al., 2012). Here we show that UVR8W285F is a constitutive dimer that is nonfunctional in plants; it fails to restore UV-B induction of gene expression or hypocotyl growth suppression in response to UV-B light in uvr8-1, and the plants are highly sensitive to UV-B light. In contrast with UVR8W285F, UVR8W285A seems to be a constitutive monomer in yeast (Rizzini et al., 2011) and in plants (Figure 5A) when assayed by SDS-PAGE with nonboiled samples. Although this electrophoretic technique effectively identifies a weaker dimer and is a very convenient assay for cell extracts, it does not rigorously show whether a protein is monomeric or dimeric. Thus, in contrast with the gel assay, size exclusion chromatography shows that purified UVR8W285A is a dimer that does not monomerize in response to UV-B light (Christie et al., 2012). Purified UVR8W233F is also a constitutive dimer when examined by size exclusion chromatography but appears as a monomer when examined by the gel assay in yeast extracts (Rizzini et al., 2011). Nonetheless, it is clear that Ala mutation of W285 weakens the dimer, and it is quite likely that the protein exists as a monomer under the conditions found within the plant cell. Regardless of its dimer/monomer status, UVR8W285A, like UVR8W285F, is nonfunctional in plants, as shown by the lack of induction of HY5 (Christie et al., 2012) and CHS transcripts (Figure 4C) in response to UV-B light, the lack of hypocotyl growth suppression by UV-B light (Figure 4D), and the high sensitivity of the transgenic uvr8-1/UVR8W285A plants to UV-B light (see Supplemental Figure 2 online). The pivotal importance of W285 in UVR8 photoreception is evident, because the other triad tryptophans, including W233, are totally unable to compensate when it is mutated.
Ala and Phe mutants of W233 show very little responsiveness to UV-B light in biophysical assays of the purified proteins, similar to the equivalent mutants of W285 (Christie et al., 2012; Wu et al., 2012). Moreover, W233 is very important in maintaining exciton coupling in the Trp cluster responsible for photoreception. In plants, UVR8W233A seems to have a weakened dimer but, unlike UVR8W285A, it is not constitutively monomeric and shows a mixture of dimer and monomer under non–UV-B conditions. In addition, UVR8W233A shows dimer-to-monomer conversion in uvr8-1 plants and has detectable activity in gene expression and hypocotyl growth suppression in response to UV-B light. These data indicate that W233 is important but not essential for UVR8 activity in vivo. They also show that W285 cannot fully compensate for mutation of W233, despite its key role in UVR8 photoreception.
Purified UVR8W337F and UVR8W337A proteins have a partially reduced response to UV-B light, assayed by CD spectroscopy, but show UV-B–induced dimer-to-monomer conversion (Christie et al., 2012). In plants, UVR8W337A has a weaker dimer than wild-type UVR8, similar to UVR8W233A, and it monomerizes in response to UV-B light. Expression of UVR8W337A in uvr8-1 substantially, but not completely, restores UV-B–induced gene expression, and this may explain why the plants are not as completely tolerant of elevated UV-B light as the wild type. However, the plants expressing UVR8W337A are very similar to the wild type in their hypocotyl growth suppression response. Thus, W337 has a significant role in UVR8 function, but it is not as important in photoreception as W285 and W233.
Although W94 is part of the excitonically coupled Trp pyramid, it is not essential for UVR8 function. In vitro, UVR8W94A shows only a small reduction in response to UV-B light compared with wild-type UVR8. In plants, mutation of W94, in the context of the double UVR8W92A,W94A mutant, has no apparent effect on UVR8 function. However, as mentioned above, it is possible that the functional significance of some tryptophans, such as W94, may be revealed under particular conditions, and hence it will be valuable to examine the role of W94 under varying fluence rates and wavelengths of UV-B light.
Thus, our in vivo analysis of the functions of the pyramid tryptophans substantially extends the observations reported for purified UVR8 mutant proteins and shows how each Trp contributes to UVR8-mediated photomorphogenic responses in plants. The observed order of importance of the tryptophans in responses in vivo, W285>W233>W337>W94, is essentially consistent with the in vitro assay of photoreception by CD spectroscopy (Christie et al., 2012). The in vivo data support the conclusion from in vitro experiments that W285 is the principal chromophore of UVR8; mutation of W285 to Phe in the purified photoreceptor retunes its spectral sensitivity so that it is able to sense UV-C light (Christie et al., 2012). W233 also has a key role in UV-B photoreception. Apart from its ability to absorb UV-B light, W233 is important in maintaining excitonic coupling within the cross-dimer Trp pyramid (Christie et al., 2012). However, it is interesting that mutation of W233 to Ala does not eliminate UVR8 activity in vivo, whereas very little photochemical activity was detectable in vitro (Christie et al., 2012; Wu et al., 2012). Evidently other tryptophans, most likely W285, can partly compensate for mutation of W233, although this is not seen with mutation of W285. Thus, it is not simply the case that W285 and W233 act as functionally redundant UV-B chromophores for UVR8. Further research is required to determine the precise roles and functional relationships of W285, W233, and the other tryptophans within the pyramid.
Interaction with COP1 Is Not Sufficient for UVR8 Function
Genetic studies indicate that COP1 is required for UVR8-mediated responses, and UV-B light stimulates the direct interaction of UVR8 with COP1. This interaction is believed to initiate signaling, although the steps leading to transcriptional regulation are not understood. The constitutively dimeric UVR8W285F mutant does not interact with COP1 either in yeast (Rizzini et al., 2011) or in plants (Figure 5B), consistent with its inability to mediate responses. In fact, the Phe and Tyr mutants of all the triad tryptophans fail to interact with COP1 in yeast (Figure 4B). By contrast, UVR8W285A interacts with COP1 in yeast both in darkness and in UV-B light, and a similar constitutive interaction is seen in plants. Similarly, UVR8W233A and UVR8W337A interact constitutively with COP1 in plants and yeast, although in yeast, the interaction in darkness is less strong than for UVR8W285A. It has been reported that overexpression of UVR8 leads to a cop1-like phenotype under UV-B light, probably because COP1 is sequestered, restricting its involvement with other light signaling pathways and its availability to act as an E3 ubiquitin ligase (Favory et al., 2009). However, the constitutive binding of COP1 to the Ala mutants of UVR8 triad tryptophans did not cause a cop1-like phenotype either in light-grown (see Supplemental Figure 2 online) or dark-grown (see Supplemental Figure 4 online) plants. This is most likely because transgenic lines were selected in which the level of expression of the mutant UVR8 proteins was close to that of wild-type UVR8, so no overexpression phenotypes were observed.
It is not entirely clear why the Ala mutants of the triad tryptophans interact constitutively with COP1, whereas the Tyr and Phe mutants do not. Undoubtedly, introduction of the small, nonaromatic Ala residue is more disruptive to the structural relationships of the triad tryptophans and adjacent amino acids than the aromatic mutations. Wu et al. (2012) reported that mutation of W285 to Ala substantially alters the orientations of W337 and W233 and the adjacent Asp D129. However, it is not known how these changes lead to constitutive interaction with COP1. It is possible that they affect the conformation of the protein and, in consequence, both weaken the salt-bridges that hold the dimer together and expose the region that interacts with COP1 even under non–UV-B conditions. By contrast, mutation of W285 to Phe does not alter its structural relationships with adjacent amino acids (Wu et al., 2012), and hence UVR8W285F does not bind to COP1 in darkness.
The current model of UVR8 function is that UV-B light induces monomerization and that consequent binding to COP1 initiates responses. However, the results presented here for the Ala mutants of the triad tryptophans indicate that COP1 binding to UVR8 is not sufficient to induce a response. Each of these mutants interacts with COP1 constitutively, but there is no response with any of them in the absence of UV-B light, and the extent of response after UV-B exposure is not correlated with COP1 binding. Moreover, if the monomeric form of these mutants is present in the absence of UV-B light, which is suggested by both the gel assay (Figure 5A) and the interaction with COP1 (Figure 5B), it can be concluded that monomer formation per se is also not sufficient for UVR8 function. Rather, the extent of response of the triad mutants to UV-B light in vivo is closely correlated with the effect of the mutation on photoreception. Thus, for example, UVR8W285A and UVR8W337A both interact with COP1 constitutively, but the former is nonresponsive to UV-B light both in vitro and in vivo, whereas the latter has a strong response to UV-B light in vitro, as judged by CD spectroscopy (Christie et al., 2012), which is correlated with a near–wild-type level of response in vivo. It therefore seems that UV-B photoreception generates some form of signal that is required for UVR8 activity apart from the induction of monomerization and COP1 binding. For instance, photoreception may initiate a conformational change in the protein that is required for function. Thus, a lack of photochemical activity may impair the production of a functional UVR8 regardless of whether it is monomeric and can bind COP1. Further research is therefore needed to understand the mechanism of UVR8 photoreception and how photoreception affects the protein to generate a functional monomer that can interact with COP1 to initiate photomorphogenic responses.
METHODS
Plant Materials
Seeds of wild-type Arabidopsis thaliana ecotype Landsberg erecta (Ler) were obtained from the Nottingham Arabidopsis Stock Center. The uvr8-1 mutant allele in the Ler background (Kliebenstein et al., 2002) was obtained from Dan Kliebenstein (University of California, Davis). The UVR8pro:GFP-UVR8 transgenic line in the uvr8-1 background (line 6-2) was described by Kaiserli and Jenkins (2007). All the GFP-UVR8 mutant transgenic lines produced in this study are in the uvr8-1 background.
Plasmid Constructs and Transformation
Site-directed mutagenesis was performed using the QuikChange Site-Directed Mutagenesis and Multi QuikChange Site-Directed Mutagenesis kits (Stratagene) following the manufacturer’s instructions. The primers used for site-directed mutagenesis are listed in Supplemental Table 1 online. For plant transformation, modified UVR8 sequences were subcloned into the pEZR(K)L-C vector downstream of enhanced GFP and the CaMV 35S promoter using EcoR1 and Sal1 restriction sites, as in the original GFP-UVR8 construct (Brown et al., 2005). DNA sequencing confirmed that the fusions were made correctly. The fusions were introduced into uvr8-1 mutant plants by Agrobacterium tumefaciens–mediated transformation, and at least three independent homozygous T3 lines (T2 for GFP-UVR8W337A) were tested for complementation and protein expression (see Supplemental Figure 1 online).
Data presented for hypocotyl length, UV-B sensitivity, monomer/dimer status, and coimmunoprecipitation experiments used the following lines (see Supplemental Figure 1 online): GFP-UVR8W39A, line 1-2; GFP-UVR8W400A, line 9-2; GFP-UVR8W92A,W94A, line 8-1; GFP-UVR8W196A,W198A,W250A,W300A,W302A, line 4-1; GFP-UVR8W233A,W337A,W285A, line 11-5; GFP-UVR8W233A, line 5-2; GFP-UVR8W337A, line 8; GFP-UVR8W285A, line 7-1; GFP-UVR8W285F, line 6-6. For the quantitative transcript measurements, data presented are means of at least three independent experiments undertaken with all the lines shown in Supplemental Figure 1 online. Similar results were obtained for all lines expressing a particular fusion.
Yeast Two-Hybrid Assays
Yeast two-hybrid vectors were introduced into the yeast strain AH109. Competent cells were grown on yeast peptone dextrose plates for 2 d. A single colony was resuspended in 300 μL of 40% polyethylene glycol, 1× TE, and 0.1 M of LiAc, and 1 μg of each plasmid, pGADT7 and pGBKT7, was added followed by incubation at 42°C for 15 min. The samples were centrifuged at 1000g for 5 min, and the pellet was resuspended in yeast peptone dextrose medium for at least 1 h at room temperature. Samples were centrifuged at 1000g for 5 min, and the pellets were resuspended in 0.8% NaCl and left at room temperature for 3 h. The cells were then spread on plates containing synthetic dropout (SD) medium (Clontech) with minus Leu, minus Trp dropout supplement (SD−Leu−Trp) (Clontech) to select for transformation of both plasmids. Plates were left for 3 d at 30°C in darkness.
A single colony was picked and resuspended in 100 μL of 0.8% NaCl, and 5 μL was used for spotting on plates containing SD−Leu−Trp or minus Leu, minus Trp, minus His, minus adenine dropout supplement (SD−Leu−Trp−His−Ade) (Clontech) to select for colonies with interacting proteins. Plates were left for 3 d at 30°C either in darkness or under 0.1 μmol m−2 s−1 of narrowband UV-B light (Philips TL20W/01RS tube; spectrum presented in Christie et al., 2012). The data presented are representative of at least three independent experiments. Only results with selective medium are shown, but for all transformations presented, colonies grew on nonselective medium. In addition, control experiments were performed to show that growth on selective medium was not caused by autoactivation.
To confirm expression of bait and prey proteins in each experiment, protein was extracted using a protocol modified from Grefen et al. (2009). A single colony of yeast containing the plasmids of interest was grown overnight in 10 mL of liquid SD−Leu−Trp at 30°C with constant shaking. When the culture had reached an OD at 550 nm between 1.0 and 2.0, 2 mL was pelleted, the supernatant was discarded, and the cells were resuspended in the appropriate volume of LL buffer (50 mM of Tris-HCL, pH 6.8, 4% SDS, 8 M of urea, 30% glycerol, 0.1 M of DTT, 0.005% [w/v] bromophenol blue) calculated from the OD. The mixture was then vortexed for 1 min and incubated at 65°C for 30 min. After centrifugation, proteins were separated on a 10% SDS-PAGE gel, and an immunoblot was incubated with anti-hemagglutinin (Cell Signaling Technology) or anti-MYC (Cell Signaling Technology) antibodies.
Quantitative Transcript Measurements
Plants were grown for 3 weeks in 20 μmol m−2 s−1 of constant white light at 21°C (warm light fluorescent tubes; Osram) and then exposed to 3 μmol m−2 s−1 of narrowband UV-B light for 3 h. RNA extraction was performed using the RNeasy Plant Mini Kit (Qiagen). cDNA synthesis was performed as described by Brown et al. (2005). Quantitative PCR was performed using the MX3000 Stratagene real-time PCR system and a Brilliant III SYBR Green qPCR kit (Stratagene) following the manufacturer’s instructions. The target amplicon was quantified by comparing the amplification in cDNA samples with standards of known amount. The PCR conditions were as follows: 3 min at 95°C, 40 cycles of 10 s at 95°C, 20 s at 60°C, followed by a 60 to 95°C dissociation protocol. The primers used for amplification of HY5, CHS, and ACTIN2 are shown in Supplemental Table 2 online. Stratagene MX software was used to automatically calculate the cycle threshold (Ct) value for each reaction. The Ct values for each standard dilution were plotted against the log of the initial template quantity, and a standard curve was produced. The initial template quantity in each cDNA sample was determined by comparing the Ct values to the standard curve. Each reaction was performed in duplicate. As a control for variation in RNA quantification, reverse transcription efficiency, and template preparation, the expression of CHS and HY5 transcripts was normalized against the amount of ACTIN2 control transcripts in each sample. The data shown are the means of at least three independent experiments using different transgenic lines. Levels of HY5 and CHS transcripts measured for uvr8-1 and the UVR8 mutants in each experiment were determined relative to the level for wild-type plants exposed to UV-B light to facilitate comparison between experiments. Statistical differences in mean transcript levels were determined using Student’s t test.
Measurements of Hypocotyl Length
Hypocotyl length was measured for seedlings grown on 0.8% agar plates containing one-half–strength Murashige and Skoog salts. Stratified seeds were exposed to 120 μmol m−2 s−1 of white light for 4 h in a growth chamber. Seedlings were grown in 2 μmol m−2 s−1 of white light supplemented (or not supplemented in controls) with 1.5 μmol m−2 s−1 of narrowband UV-B light for 4 d. The hypocotyl lengths were measured using ImageJ software. The data presented are from three independent experiments. Statistical differences in hypocotyl lengths were determined using Student’s t test.
Sensitivity to UV-B Light
UV-B sensitivity assays were performed as described by Brown et al. (2005) using a broadband UV-B source (UV-B-313 fluorescent tubes [Q-Panel]; spectrum presented in Christie et al., 2012).
UVR8 Dimer/Monomer Status
Plants were grown under 100 μmol m−2 s−1 of white light for 12 d. Whole cell extracts from leaf tissue were prepared as described by Kaiserli and Jenkins (2007). UVR8 dimer/monomer status was examined essentially as described by Rizzini et al. (2011). Whole cell extracts were kept on ice and exposed (or not exposed in controls) to 3 μmol m−2 s−1 of narrowband UV-B light for 30 min. Then, 4× loading buffer containing 250 mM of Tris-HCl, pH 6.8, 2% SDS, 20% β-mercaptoethanol, 40% glycerol, and 0.5% bromophenol blue was added to the samples, but they were not boiled. The proteins were loaded on a 7.5% SDS-PAGE gel and an immunoblot incubated with anti-UVR8 antibody (Kaiserli and Jenkins, 2007). The data shown are representative of at least three independent experiments.
Coimmunoprecipitation Assay
Plants were grown under 100 μmol m−2 s−1 of constant white light at 21°C for 12 d and then were put in darkness for 16 h. The plants were exposed (or not exposed in controls) to 3 μmol m−2 s−1 of narrowband UV-B light for 3 h. Whole cell extracts were prepared as described by Kaiserli and Jenkins (2007) in the absence or presence of 3 μmol m−2 s−1 of narrowband UV-B light, and the coimmunoprecipitation assays were performed under the same conditions. Extract samples were incubated for 30 min on ice with 50 μL of anti-GFP microbeads (μMacs; Miltenyi Biotec). The microcolumn was equilibrated using 200 μL of high salt lysis buffer (450 mM of NaCl, 1% Triton X-100, 50 mM of Tris-HCl, pH 8, 5 mM of phenylmethylsulfonyl fluoride, protease inhibitors [Complete Mini; Roche]). The lysate containing the microbeads was applied onto the column and non–GFP-tagged proteins left to run through. The column was washed five times with 200 μL of high salt lysis buffer and once with 300 mM of NaCl, Tris-HCl, pH 7.5. To elute proteins, 20 μL of elution buffer (0.1 M triethylamine, pH 11.8, 0.1% Triton X-100) was applied to the column and left for 5 min at room temperature. A further 50 μL of elution buffer was added, and the eluate was collected in a tube containing 3 μL of 1 M of 2-(N-morpholino)-ethane-sulfonic acid, pH 3.0, to neutralize the sample. The eluates were analyzed by SDS-PAGE and an immunoblot incubated with anti-GFP (Cell Signaling Technology) and anti-COP1 (Jang et al., 2010) antibodies. The data shown are representative of at least three experiments.
Transient Expression in Nicotiana benthamiana
Transient expression of GFP-UVR8 fusions was performed in leaves of 4-week-old N. benthamiana plants. Agrobacterium with the desired mutant GFP-UVR8 plasmid DNA was inoculated in 10 mL of Luria-Bertani broth containing 30 μg mL−1 of gentamycin and 50 μg mL−1 of kanamycin. The culture was left at 30°C, constantly shaking (220 rpm) until it reached an OD at 550 nm of at least 1.0. The culture was pelleted by centrifugation at 2000g for 10 min and washed in 10 mL of sterile 10 mM MgCl2. The cell suspension was diluted to OD550 of 0.2 with 10 mM MgCl2 solution to a final volume of 20 mL. A total of 200 μM of acetosyringone was added, and the solution was left at room temperature for 3 h. Using a syringe, the Agrobacterium medium was infiltrated into the leaves of N. benthamiana. The N. benthamiana plants were put in a growth chamber at 30°C in white light for ∼60 h. To prepare a whole cell extract, plants were ground in liquid N2 with a mortar and pestle, and total protein was extracted in 500 μL of extraction buffer (25 mM of Tris-HCL, pH 7.5, 1 mM of EDTA, 10% glycerol, 5 mM of DTT, 0.1% Triton X-100, and protease inhibitor mix [1 tablet of protease inhibitor mix] [Complete Mini, Roche] per 10 mL of extraction buffer). The mixture was centrifuged at 16,000g for 10 min at 4°C, and the supernatant, containing total protein extract, was collected. Proteins were fractionated by SDS-PAGE. An immunoblot was incubated with anti-GFP antibody.
Light Measurements
Fluence rates of white light were measured using a Skye RS232 meter fitted with a quantum sensor (Skye Instruments). Fluence rates of UV-B light (280 to 315 nm) were measured using a RS232 meter with a SKU 430 sensor.
Accession Numbers
The Arabidopsis Genome Initiative locus identifier for UVR8 is At5g63860 and for COP1 is At2g32950.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Levels of GFP-UVR8 Expression in the Transgenic Lines.
Supplemental Figure 2. Sensitivity to UV-B of Transgenic Lines Expressing GFP-UVR8 Mutants.
Supplemental Figure 3. Expression of UVR8 Mutants and COP1 in Yeast Two-Hybrid Vectors.
Supplemental Figure 4. Phenotype of Dark-Grown Seedlings Expressing Ala Mutants of Triad Tryptophans.
Supplemental Table 1. Primers Used for Site-Directed Mutagenesis of pSK and pGBK Vectors Containing UVR8.
Supplemental Table 2. Primers Used for Quantitative PCR.
Acknowledgments
We thank John Christie and all members of the Jenkins and Christie laboratories for discussion of the research and guidance with the methods, Brian Smith for help in interpreting UVR8 structure, Emanuela Sani for assistance with quantitative PCR, Jason Wargent for advice on statistics, and In-Cheol Jang and Nam-Hai Chua for the COP1 antibody. G.I.J. received research support from the UK Biotechnology and Biological Sciences Research Council and the Leverhulme Trust. A.O. was supported by a Biotechnology and Biological Science Research Council PhD Studentship.
AUTHOR CONTRIBUTIONS
A.O. performed the research and analyzed data. G.I.J. designed the research, analyzed data, and wrote the article.
Glossary
- GFP
green fluorescent protein
- CD
circular dichroism
- Ler
ecotype Landsberg erecta
- SD
synthetic dropout
- Ct
cycle threshold
References
- Bekker-Jensen S., Rendtlew Danielsen J., Fugger K., Gromova I., Nerstedt A., Lukas C., Bartek J., Lukas J., Mailand N. (2010). HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 12: 80–86 [DOI] [PubMed] [Google Scholar]
- Brown B.A., Cloix C., Jiang G.H., Kaiserli E., Herzyk P., Kliebenstein D.J., Jenkins G.I. (2005). A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. USA 102: 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B.A., Headland L.R., Jenkins G.I. (2009). UV-B action spectrum for UVR8-mediated HY5 transcript accumulation in Arabidopsis. Photochem. Photobiol. 85: 1147–1155 [DOI] [PubMed] [Google Scholar]
- Brown B.A., Jenkins G.I. (2008). UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 146: 576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie J.M. (2007). Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58: 21–45 [DOI] [PubMed] [Google Scholar]
- Christie J.M., Arvai A.S., Baxter K.J., Heilmann M., Pratt A.J., O’Hara A., Kelly S.M., Hothorn M., Smith B.O., Hitomi K., Jenkins G.I., Getzoff E.D. (2012). Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloix C., Jenkins G.I. (2008). Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Mol Plant 1: 118–128 [DOI] [PubMed] [Google Scholar]
- Favory J.J., et al. (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K.A., Quail P.H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohnmeyer H., Staiger D. (2003). Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol. 133: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Obrdlik P., Harter K. (2009). The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS). Methods Mol. Biol. 479: 217–233 [DOI] [PubMed] [Google Scholar]
- Gruber H., Heijde M., Heller W., Albert A., Seidlitz H.K., Ulm R. (2010). Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 20132–20137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M., Ulm R. (2012). UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 17: 230–237 [DOI] [PubMed] [Google Scholar]
- Jang I.-C., Henriques R., Seo H.S., Nagatani A., Chua N.-H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G.I. (2009). Signal transduction in responses to UV-B radiation. Annu. Rev. Plant Biol. 60: 407–431 [DOI] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Kaiserli E., Jenkins G.I. (2007). UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J., Lim J.E., Landry L.G., Last R.L. (2002). Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 130: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.H., Yang H.Q. (2007). Cryptochrome signaling in plants. Photochem. Photobiol. 83: 94–101 [DOI] [PubMed] [Google Scholar]
- Oravecz A., Baumann A., Máté Z., Brzezinska A., Molinier J., Oakeley E.J., Adám É., Schäfer E., Nagy F., Ulm R. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L., Nassar N., Vetter I., Becker J., Klebe C., Roth M., Wittinghofer A. (1998). The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 392: 97–101 [DOI] [PubMed] [Google Scholar]
- Rizzini L., Favory J.-J., Cloix C., Faggionato D., O’Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G.I., Ulm R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Stracke R., Favory J.-J., Gruber H., Bartelniewoehner L., Bartels S., Binkert M., Funk M., Weisshaar B., Ulm R. (2010). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33: 88–103 [DOI] [PubMed] [Google Scholar]
- Ulm R., Baumann A., Oravecz A., Máté Z., Adám E., Oakeley E.J., Schäfer E., Nagy F. (2004). Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R., Nagy F. (2005). Signalling and gene regulation in response to ultraviolet light. Curr. Opin. Plant Biol. 8: 477–482 [DOI] [PubMed] [Google Scholar]
- Wargent J.J., Gegas V.C., Jenkins G.I., Doonan J.H., Paul N.D. (2009). UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation. New Phytol. 183: 315–326 [DOI] [PubMed] [Google Scholar]
- Wu D., Hu Q., Yan Z., Chen W., Yan C., Huang X., Zhang J., Yang P., Deng H., Wang J., Deng X., Shi Y. (2012). Structural basis of ultraviolet-B perception by UVR8. Nature 484: 214–219 [DOI] [PubMed] [Google Scholar]
- Wu M., Grahn E., Eriksson L.A., Strid A. (2011). Computational evidence for the role of Arabidopsis thaliana UVR8 as UV-B photoreceptor and identification of its chromophore amino acids. J. Chem. Inf. Model. 51: 1287–1295 [DOI] [PubMed] [Google Scholar]
- Yi C., Deng X.W. (2005). COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15: 618–625 [DOI] [PubMed] [Google Scholar]