Recycling of a nutrient from old tissues to young tissues is an important process for plant growth and development. This work describes a transporter for the Cu-nicotianamine complex, which is required for delivering Cu to the developing young tissues and seeds through phloem transport.
Abstract
Cu is an essential element for plant growth, but the molecular mechanisms of its distribution and redistribution within the plants are unknown. Here, we report that Yellow stripe-like16 (YSL16) is involved in Cu distribution and redistribution in rice (Oryza sativa). Rice YSL16 was expressed in the roots, leaves, and unelongated nodes at the vegetative growth stage and highly expressed in the upper nodes at the reproductive stage. YSL16 was expressed at the phloem of nodes and vascular tissues of leaves. Knockout of this gene resulted in a higher Cu concentration in the older leaves but a lower concentration in the younger leaves at the vegetative stage. At the reproductive stage, a higher Cu concentration was found in the flag leaf and husk, but less Cu was present in the brown rice, resulting in a significant reduction in fertility in the knockout line. Isotope labeling experiments with 65Cu showed that the mutant lost the ability to transport Cu-nicotianamine from older to younger leaves and from the flag leaf to the panicle. Rice YSL16 transported the Cu-nicotianamine complex in yeast. Taken together, our results indicate that Os-YSL16 is a Cu-nicotianamine transporter that is required for delivering Cu to the developing young tissues and seeds through phloem transport.
INTRODUCTION
Cu, which mostly functions as a cofactor of numerous proteins, is an essential element for plant growth and development (Linder and Goode, 1991). Therefore, Cu plays an important role in photosynthesis, respiration, C and N metabolism, and protection against oxidative stress (Marschner, 2011). The requirement for Cu is lower than that for other micronutrients, such as Fe, Zn, and Mn, but Cu deficiency often occurs in plants growing on low Cu soils, calcareous soils, and on soils with high organic matter content. Visible symptoms of Cu deficiency include stunted growth, distortion of young leaves, and chlorosis/necrosis starting at the apical meristem and extending down the leaf margins (Marschner, 2011). On the other hand, Cu is toxic to plants in excess, which is characterized by root growth inhibition. Redox cycling between Cu(I) and Cu(II) results in production of highly toxic hydroxyl radicals. In addition, Cu is highly reactive to thiols and can possibly displace other essential metals in proteins (Lippard and Berg, 1994). Typically, symptoms of Cu deficiency start when Cu concentration in the vegetative tissues decreases below 5 μg g−1 dry weight, while toxicity levels are observed above 20 μg g−1 dry weight (Marschner, 2011). Therefore, homeostasis of Cu is tightly regulated in plants to keep the proper Cu concentrations in different tissues.
Cu homeostasis is largely regulated by a number of transporters involved in Cu uptake, translocation, and distribution (Pilon, 2011). It is likely that Cu is taken up in Arabidopsis thaliana roots by COPPER TRANSPORTER1 (COPT1), a homolog of the yeast Cu transporter CTR1 (Sancenón et al., 2003). COPT1 is strongly expressed in roots and upregulated by Cu deficiency. Furthermore, knockdown of COPT1 reduces Cu uptake (Sancenón et al., 2003, 2004). In rice (Oryza sativa), there are seven homologs of COPT1, all of which are able to transport Cu either alone or by forming complexes with each other when expressed in the yeast mutant Δctr1 ctr3 (Yuan et al., 2011). The expression of rice COPT1, COPT5, and COPT6 in the roots was induced by Cu deficiency (Yuan et al., 2011). It is not clear which of these is involved in Cu uptake in the roots, although COPT1 showed the strongest expression in the roots. The expression of Arabidopsis ZINC/IRON-REGULATED TRANSPORTER-LIKE PROTEIN2 (ZIP2) was induced by Cu deficiency, and ZIP2 showed transport activity for Cu in yeast (Wintz et al., 2003); however, it is unknown whether this protein is involved in Cu uptake in plants. In contrast with COPT1, which transports Cu(I), ZIP2 transports Cu(II).
A member of the P-type ATPases, Heavy Metal ATPase5 (HMA5), is likely to function as an efflux transporter of Cu in Arabidopsis (Andrés-Colás et al., 2006). HMA5 is mainly expressed in the roots and flowers. Knockout of this gene resulted in higher Cu accumulation in the roots compared with the wild type. This gene is also associated with Cu tolerance in Arabidopsis (Kobayashi et al., 2008). These findings indicate that HMA5 is probably involved in detoxification or xylem loading of Cu by exporting Cu from the roots in Arabidopsis (Andrés-Colás et al., 2006; Kobayashi et al., 2008). In rice, HMA9 was mainly expressed in vascular tissues, including the xylem and phloem, suggesting its involvement in the loading and unloading of heavy metals, including Cu in these tissues (Lee et al., 2007). The expression of HMA9 was increased by excess of Cu (Lee et al., 2007).
Several transporters involved in Cu delivery inside the cell have also been identified. RESPONSIVE-TO-ANTAGONIST1/HMA7 was the first functionally characterized heavy metal ATPase (HMA) in plants and is needed to deliver Cu to the ethylene receptors (Hirayama et al., 1999). P-type ATPase of Arabidopsis1 (PAA1)/HMA6, PAA2/HMA8, and HMA1 are involved in Cu transport into the chloroplasts of Arabidopsis (Shikanai et al., 2003; Abdel-Ghany et al., 2005; Seigneurin-Berny et al., 2006). However, HMA1 was recently reported to be involved in Zn detoxification by decreasing Zn concentration in plastids (Kim et al., 2009). A homolog of the yeast Cox19 protein, At-Cox19 in Arabidopsis, has been implicated in the transport of Cu into mitochondria (Attallah et al., 2007). More recently, it was reported that COPT5 is involved in Cu efflux from vacuoles in Arabidopsis (Garcia-Molina et al., 2011; Klaumann et al., 2011).
All transporters described above are for transport of ionic Cu, either Cu(I) or Cu(II). However, several studies have shown that Cu in the xylem sap is present in the form of a complex with nicotianamine (NA; Stephan and Grun, 1989; Pich et al., 1994; Pich and Scholz, 1996), which has a high affinity for Cu (Rellán-Alvarez et al., 2008; Curie et al., 2009). Cu-NA in the xylem is delivered to different tissues; however, the transporters responsible for the distribution of Cu are unknown. Moreover, Cu is redistributed from older tissues to younger tissues through the phloem, but the transporter involved in this process has not been identified. In this study, we found that YSL16, a member of Yellow stripe-like proteins, is directly involved in distribution and redistribution of Cu in the form of Cu-NA to the developing tissues and seeds of rice, which has a distinct distribution system from dicots.
RESULTS
Expression Pattern of YSL16 at the Vegetative and Reproductive Stages
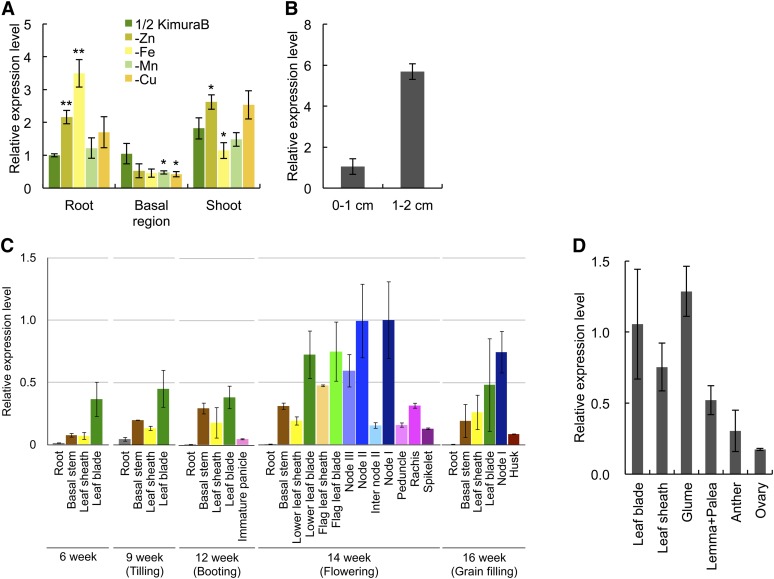
We performed a quantitative real-time RT-PCR to examine the tissue-specific expression of YSL16. At the vegetative stage, YSL16 was expressed in the roots, shoots, and basal regions (2 cm above the connection between the roots and shoots) (Figure 1A). The expression level in these tissues was similar. We also investigated the effect of metal deficiencies on the expression of YSL16. Three marker genes (YSL15, ZIP4, and COPT5) for Fe, Zn, and Cu deficiency, respectively, were used to evaluate the status of the deficiency. YSL15 (Inoue et al., 2009) was induced by more than 35 times in the Fe-deficient roots, and ZIP4 (Ishimaru et al., 2005) and COPT5 (Yuan et al., 2010) were induced by 6.6 and 2.5 times, respectively, in the Zn- and Cu-deficient roots (see Supplemental Figure 1 online). In these roots with different metal deficiencies, the expression of YSL16 was slightly induced by Zn and Fe deficiency, but not by Mn and Cu deficiency (Figure 1A). In the shoots and basal regions, YSL16 expression also fluctuated slightly upon deficiency of either metal, but the change was smaller compared with in the root (Figure 1A).
Figure 1.
Expression Patterns of YSL16 at Different Growth Stages.
(A) Tissue-dependent expression of YSL16 at the vegetative stage. Four-week-old plants were grown in a nutrient solution in the absence of Zn, Fe, Mn, or Cu or presence of these metals (1/2 KimuraB) for 7 d. The roots, basal regions (2 cm above the root-shoot junction), and shoots were sampled for expression analysis. Asterisks above bars indicate significant differences (*P < 0.05; **P < 0.01) compared with the metal-sufficient condition.
(B) Spatial expression of YSL16 in the roots. Different root segments (0 to 1 cm and 1 to 2 cm, with 0 being defined as the root tip) were sampled for expression analysis.
(C) The expression level of YSL16 in different tissues at different growth stages. Different tissues of rice grown in a field were sampled for expression analysis at different growth stages.
(D) Expression of YSL16 in reproductive organs. The expression level was determined by quantitative real-time RT-PCR. Actin and HistoneH3 were used as internal standards.
The expression level relative to the expression in control root (A), root segment (0 to 1 cm) (B), node I at flowering stage (C), or leaf blade (D) is shown. Data are given as the means ± sd of three biological replicates.
[See online article for color version of this figure.]
Spatial expression analysis showed that YSL16 was mainly expressed in the mature root zone (1 to 2 cm from the root tip) but not in the root tip (0 to 1 cm from the root tip, with 0 being defined as the very tip) (Figure 1B).
The expression of YSL16 was further investigated in different tissues at different growth stages of rice grown in a field. YSL16 was expressed at a higher level in the leaf blade during the vegetative growth stages (Figure 1C). During the reproductive stage, YSL16 was expressed in all tissues examined, but at different levels (Figure 1C). Stronger expression was found in nodes I, II, and III and the basal stem, which includes unelongated nodes (Figure 1C). Furthermore, YSL16 was expressed in all reproductive organs, but the expression level is relatively higher in the glume than in other organs, including the lemma, palea, anther, and ovary (Figure 1D).
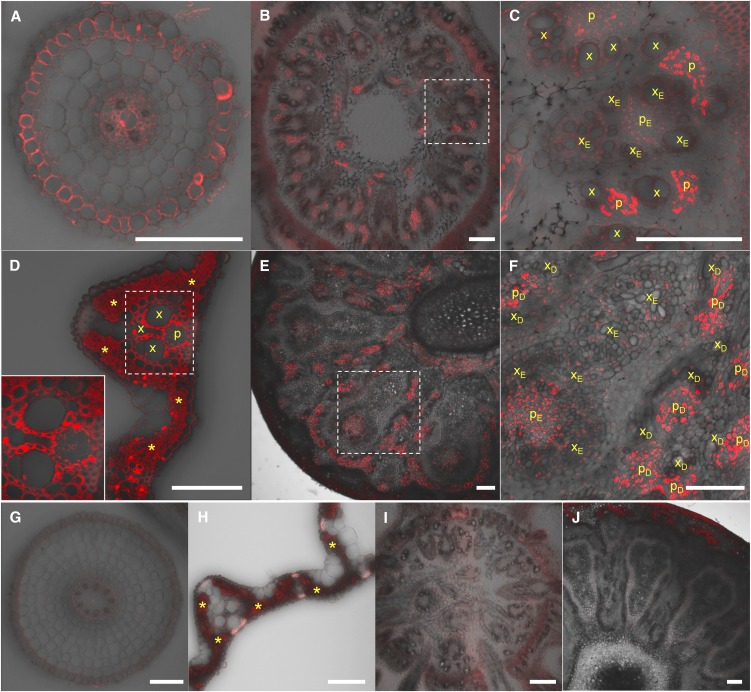
Cell-Specific Expression of YSL16
To examine the cell specificity of YSL16 expression, we generated transgenic lines carrying the YSL16 promoter (2.3 kb) fused with GFP (for green fluorescent protein). Immunostaining with an antibody against GFP showed that YSL16 was expressed at the exodermis and stele of the roots (Figure 2A). At the basal node, YSL16 was found in the phloem region of vascular bundles with a weaker expression in the enlarged vascular bundle (Figures 2B and 2C). In the leaf blade, YSL16 was mainly expressed in vascular tissues (Figure 2D). At the reproductive stage, the signal was observed in the phloem region of the diffuse and enlarged vascular bundles of node I with a weaker expression in the enlarged vascular bundle (Figures 2E and 2F). No signal was detected in tissues of wild-type rice (Figures 2G to 2J), indicating the specificity of the antibody.
Figure 2.
Cell Specificity of YSL16 Expression.
Immunostaining with an anti-GFP antibody was performed in different tissues of pOsYSL16-GFP transgenic rice ([A] to [F]) and wild-type rice ([G] to [J]). Root ([A] and [G]), basal node ([B], [C], and [I]), leaf blade ([D] and [H]), and node I ([E], [F], and [J]). Areas boxed with broken lines in (B), (D), and (E) are magnified in (C) and insets in (D) and (F), respectively. Fluorescence from the secondary antibody (red) and the transmitted light image (gray scale) are shown. The xylem and phloem regions of regular vascular bundles (x and p), in enlarged vascular bundles (xE and pE), and in diffuse vascular bundles (xD and pD) are shown. Bars = 100 µm.
Subcellular Localization of YSL16
The subcellular localization of YSL16 was determined by transient assay in onion (Allium cepa) epidermal cells and rice leaf protoplasts. A construct of the YSL16 open reading frame fused with GFP was cointroduced into the onion epidermal cells with 35S-DsRed as a marker of the cytosol and nucleus. The GFP signal was not colocalized with DsRed signal in the cells expressing YSL16-GFP (see Supplemental Figures 2A to 2D online). Furthermore, YSL16-GFP was localized to the outer region of the cell rather than the nuclei and cytosol marked by DsRed (see Supplemental Figure 2E online). By contrast, GFP alone was colocalized with DsRed signal (see Supplemental Figure 2F online). When YSL16-GFP was expressed in rice leaf protoplasts, the GFP signal was also observed in the plasma membrane (see Supplemental Figure 2G online). These results indicate that YSL16 is a plasma membrane–localized transporter.
Physiological Role of YSL16 at the Vegetative Stage
To investigate the physiological role of YSL16, we obtained a T-DNA insertion line of YSL16 (RMD_05Z11CP34) from the Rice Mutant Database (Zhang et al., 2006). In this line, a T-DNA was inserted into the third intron of YSL16 (see Supplemental Figure 3A online). This T-DNA insertion resulted in no mRNA transcription of YSL16 in this line, indicating that this is a knockout line of YSL16 (see Supplemental Figure 3B online).
When this knockout line (ysl16) was hydroponically cultivated in a normal nutrient solution with wild-type rice, the root dry weight of the knockout line was slightly less than that of wild-type rice (Figure 3A). The root length of the knockout line was significantly shorter than that of wild-type rice (Figure 3B). The growth of the shoot was similar between the knockout line and the wild-type rice (Figure 3C).
Figure 3.
Biomass and Metal Concentrations of Wild-Type Rice and the ysl16 Knockout Line.
(A) to (C) Growth of wild-type (WT) rice and the ysl16 knockout line. Root dry weight (DW) (A), root length (B), and shoot dry weight (C).
(D) and (E) Concentration of Fe, Zn, Cu, and Mn in the roots (D) and shoots (E).
(F) Cu concentration in the cell sap of root tips (0 to 5 mm).
Wild-type rice and the ysl16 knockout line were cultivated in one-half-strength Kimura B nutrient solution for 3 weeks before sampling. The metal concentration was determined by ICP-MS. Data are given as means ± sd of three biological replicates. Asterisks above bars indicate significant differences (*P < 0.05) between wild-type rice and the mutant.
Mineral analysis showed that the concentration of Cu was 22 and 20% lower in the roots and shoots of the knockout line, respectively, compared with the wild-type rice (Figures 3D and 3E). However, there was no difference in the concentration of other micronutrients, including Fe, Zn, and Mn, in the roots and shoots between the two lines (Figures 3D and 3E). In the root tips (0 to 5 mm from the root tip), the concentration of Cu in the cell sap was higher in the wild-type rice than in the knockout line (Figure 3F).
We also determined the leaf position–dependent concentration of four metals. Since the first leaf was too small and not expanded, we sampled the leaves from leaf 2 to 6 (Figure 4). The knockout line showed a higher Cu concentration in the older leaves but lower concentration in the younger leaves and leaf sheath compared with the wild-type rice (Figure 4A). By contrast, there was no difference in the concentration of Fe, Zn, and Mn in the different leaves between the knockout line and wild-type rice (Figures 4B to 4D). These results indicate that Cu distribution in the knockout line is altered. The NA concentration was also determined in the old and young leaves. The mutant line contained slightly higher NA levels than wild-type rice in the old leaves (i.e., leaves 2 and 3; see Supplemental Figure 4 online), whereas no significantly difference was observed in the youngest leaf (i.e., leaf 8).
Figure 4.
Metal Concentration in Different Leaves.
Wild-type rice, the ysl16 mutant, and two complementation lines were cultivated in one-half-strength Kimura B nutrient solution until the six-leaf stage. Individual leaf blades (leaf blades 2 to 6) and leaf sheath were sampled for metal analysis of Cu (A), Fe (B), Zn (C), and Mn (D) with ICP-MS. Data are given as means ± sd of three biological replicates. Asterisks above bars indicate significant differences (**P < 0.01) compared with wild-type rice. DW, dry weight; WT, the wild type.
To confirm that this altered Cu distribution is caused by the knockout of YSL16, we performed a complementation test by introducing a construct with the genomic sequence of YSL16 containing its own promoter and the 3′ untranslated region to the knockout line. Two independent complementation lines showed a similar expression level of YSL16 to wild-type rice (see Supplemental Figure 3B online). Analysis of these lines showed that the transgene restored wild-type Cu distribution to the different leaves (Figure 4A). This result confirms that YSL16 is involved in Cu distribution.
Remobilization of Cu from Old Leaves
To examine whether YSL16 is involved in the redistribution of Cu from old leaves to young leaves, we determined Cu mobility from old leaves in wild-type rice, the knockout line, and two independent complementation lines. Plants grown hydroponically in the presence of Cu were exposed to a Cu-free nutrient solution for 8 d, and the Cu concentration in different leaves was compared before and after Cu deficiency treatment. In the wild-type rice and the complementation lines, the Cu concentration was decreased in the old leaves (leaves 2 to 5) (Figures 5A, 5C, and 5D). By contrast, the Cu concentration was hardly changed in the old leaves of the knockout line (Figure 5B). The mobility of Cu in the old leaves (leaf blades 2 to 5) was between 10 and 33% in the wild-type rice and two independent complementation lines, depending on leaf position (Figure 5E). By contrast, the mobilization hardly occurred in the knockout line. This result indicates that YSL16 is involved in the export of Cu from old leaves.
Figure 5.
Remobilization of Cu from Older Leaves.
(A) to (D) Concentration of Cu in wild-type rice (A), YSL16 knockout line (B), and two independent complementation lines ([C] and [D]) before and after Cu deficiency treatment. Rice seedlings were grown in one-half-strength Kimura B nutrient solution until the six-leaf stage and then in nutrient solution without Cu for 8 d. Cu concentration in the different mature leaves before and after Cu deficiency treatment was determined with ICP-MS.
(E) Mobility of Cu from older leaves. Mobility of Cu (%) was calculated as decrease in Cu content/total Cu content before treatment × 100.
Asterisks above bars indicate significant differences (**P < 0.01) between treatments before and after Cu deficiency ([A] to [D]) or compared with wild-type rice (E). DW, dry weight; WT, the wild type.
65Cu Mobility between Different Leaves
To confirm that Cu is redistributed directly from a leaf to an older or younger leaf, we performed a label tracer experiment with a stable isotope, 65Cu. Two different forms of Cu (65CuCl2 or 65Cu-NA) were fed to the cut end of a leaf (leaf 4) for 30 h, and then the distribution to lower leaves (old) and upper leaves (young) was determined. When fed with 65Cu-NA, the distribution to the lower leaves (leaves 2 and 3) was very low in all lines and there was no difference between wild-type rice and the mutant (Figure 6A). Distribution to upper expanded leaves (leaves 5 and 6) was also very low, and no difference in the Cu distribution was found between different lines (Figure 6A). However, a higher distribution (4.7 to 5.8% of total 65Cu fed) to the newest developing leaf (leaf 7) in the wild-type rice and two complementation lines was found compared with the mutant (Figure 6A). No detected 65Cu was moved in the wild-type rice, knockout line, and two complementation lines when fed with 65CuCl2 (Figure 6B). These results further indicate that YSL16 is directly involved in remobilization of Cu from old to young leaves in the form of Cu-NA.
Figure 6.
65Cu Mobility between Different Leaves.
65Cu mobility between different leaves. Leaf 4 of seven-leaf stage plants (wild-type rice, mutant, and two complementation lines) was cut at 2 cm from the tip and exposed to a solution containing 50 µM 65Cu-NA (A) or 65CuCl2 (B). After 30 h, different leaf blades were sampled. The redistribution percentage of total 65Cu fed is shown. Data are given as means ± sd of three biological replicates. Asterisks above bars indicate significant differences (**P < 0.01) compared with wild-type (WT) rice.
Role of YSL16 at the Reproductive Stage
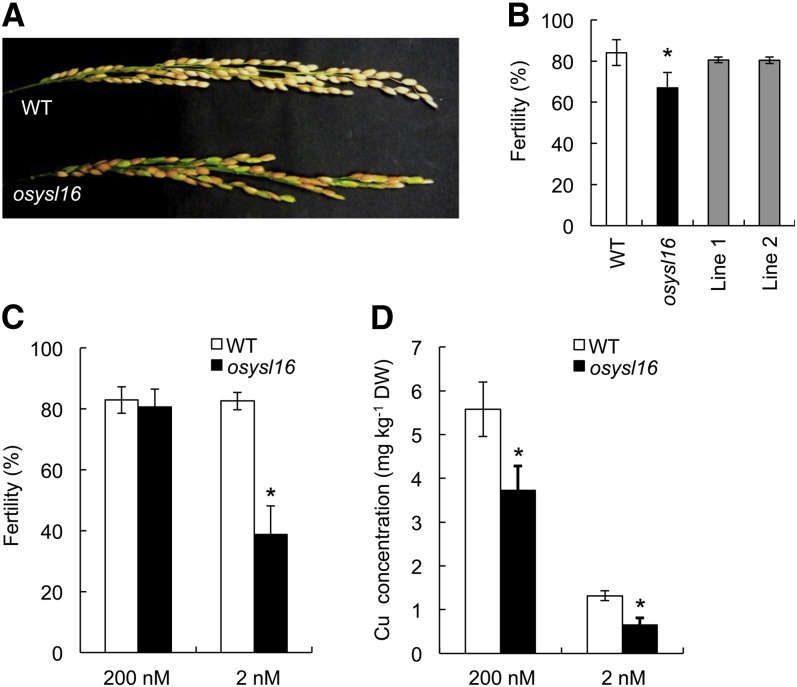
When the knockout line was grown in soil until ripening, we found that the fertility was significantly reduced compared with that of wild-type rice (Figures 7A and 7B). The percentage of filled grain was 84.0% in the wild-type rice, in contrast with 67.0% in the knockout line when grown in soil (Figure 7B). Two independent complementation lines had similar fertility to the wild type, further confirming that the low fertility was caused by mutation of YSL16 (Figure 7B).
Figure 7.
Knockout of YSL16 Resulted in Decreased Fertility.
(A) and (B) Fertility of rice grown in soil. Wild-type rice (WT), the ysl16 knockout line, and two independent complementation lines were grown in soil until ripening.
(A) Panicles of the wild type and ysl16 knockout line at harvest.
(B) Fertility of the wild type, ysl16 knockout line, and two independent complementation lines. The fertility was calculated as follows: filled seed number/total seed number × 100.
(C) and (D) The fertility of rice grown hydroponically at different Cu levels. Both wild-type rice and the ysl16 knockout line were grown in a nutrient solution containing 200 or 2 nM Cu until ripening. Fertility (C) and Cu concentration (D) in brown rice.
Data are given as means ± sd of three biological replicates. Asterisks above bars indicate significant differences (*P < 0.05) between wild-type rice and the mutant. DW, dry weight.
To examine whether the reduced fertility of the knockout line was caused by a Cu deficiency, we grew the knockout line and wild-type rice in nutrient solution containing high (200 nM) and low (2 nM) concentrations of Cu. In the presence of 200 nM Cu, the fertility did not differ between wild-type rice and the knockout line (Figure 7C). However, at 2 nM Cu, the fertility was significantly reduced in the knockout line, but not in the wild-type rice. To examine whether the reduced fertility is due to pollen viability, we compared pollen viability using KI-I2 staining. At 200 nM Cu, there was no difference in pollen viability between the wild-type rice and knockout line (see Supplemental Figures 5A and 5C online). However, at 2 nM Cu, the pollen viability was significantly reduced in the knockout line (see Supplemental Figures 5B and 5C online). The Cu concentration in the brown rice was significantly decreased in the knockout line at either Cu level (Figure 7D). These results indicate that the decreased fertility in the knockout line is caused by a low concentration of Cu.
Mineral analysis in different tissues showed that the Cu concentration was higher in the flag leaf blade and husk but lower in the culm, rachis, and brown rice in the knockout line than in the wild-type rice and two complementation lines (Figure 8A). However, there was no difference in the concentration of Fe, Zn, and Mn in tissues among wild-type rice, the knockout mutant, and two complementation lines (Figure 8B; see Supplemental Figure 6 online).
Figure 8.
Metal Concentration in Different Rice Tissues at Harvest.
(A) and (B) Wild-type rice (WT), the ysl16 knockout line, and two independent complementation lines were grown in soil until ripening. Different tissues above the node I were separated and subjected to analysis of Cu (A) and Fe (B). Metal concentration was determined by ICP-MS.
(C) Mobility of 65Cu from the flag leaf to panicle. The flag leaf of wild-type rice, the knockout line, and two complementation lines grown in soil was cut at 2 cm from the tip and then exposed to 50 µM 65Cu-NA or 65CuCl2 for 2 weeks. At ripening, the panicles were sampled for 65Cu determination.
Data are given as means ± sd of three ([A] and [B]) or six (C) biological replicates. Asterisks above bars indicate significant differences (*P < 0.05; **P < 0.01) compared with wild-type rice. DW, dry weight.
To examine the remobilization of Cu from the leaf to the panicle, we fed the cut end of the flag leaf with 65CuCl2 or 65Cu-NA for 2 weeks. When fed with 65Cu2+, there was almost no 65Cu remobilization from the flag leaf to the panicle in all lines (Figure 8C). However, when fed with 65Cu-NA, 29.1, 28.1, and 28.8% of the 65Cu was moved to the panicle in wild-type rice and two complementation lines, respectively (Figure 8C), in contrast with 2.7% in the knockout line. These results indicate that YSL16 also plays an important role in moving Cu from the flag leaf to the grain and that Cu-NA is the form of transport.
Expression of Genes Related to Fe and Cu Homeostasis
To investigate the effect of YSL16 on the homeostasis of Fe and Cu, we compared the expression of some genes involved in Fe or Cu homeostasis in different tissues in wild-type rice and the knockout line subjected to an Fe or Cu deficiency. Among three Cu-responsive marker genes, the expression of COPT1 (Yuan et al., 2010) in root tips was significantly stronger in the mutant (see Supplemental Figure 7A online). The expression of COPT5 (Yuan et al., 2010) in the younger leaves (leaf 5 and leaf 6), basal nodes, and root tip was significantly stronger in the mutant (see Supplemental Figure 7A online). By contrast, the expression of HMA9 (Lee et al., 2007) was significantly upregulated in the oldest leaf tested (leaf 2) and downregulated in the basal node in the mutant (see Supplemental Figure 7A online). The altered expression in the young leaves and root tips of the mutant is consistent with a decreased Cu concentration in these tissues (Figures 3F and 4A).
The effect of Cu or Fe deficiency on the expression of COPT1, COPT5, and HMA9 was also compared in the roots and shoots of wild-type rice and the mutant. Cu deficiency resulted in the upregulation of COPT1 and COPT5 and the downregulation of HMA9 in the roots of both wild-type rice and ysl16 (see Supplemental Figure 7B online). However, there was no difference in the expression of these genes between the wild-type rice and the mutant except that the expression of HMA9 was slightly lower in ysl16 mutant roots (see Supplemental Figure 7B online). In the shoots, the expression of COPT1 and COPT5 was higher in the mutant than in wild-type rice under Cu deficiency (see Supplemental Figure 7C online). Fe deficiency hardly affected the expression of these three genes in both the roots and shoots (see Supplemental Figures 7B and 7C online).
The expression of four marker genes related to Fe homeostasis, including IRT1, IRO2, Ferritin, and YSL2 (Zheng et al., 2009) in different tissues, was similar between the wild-type rice and the mutant (see Supplemental Figure 8A online). The expression of all these genes responded to Fe deficiency, with upregulation of IRT1, IRO2, and YSL2 and downregulation of Ferritin in both the roots and shoots (see Supplemental Figures 8B and 8C online). However, there was no difference in the expression between wild-type rice and the mutant. Cu deficiency hardly affected the expression of these genes (see Supplemental Figures 8B and 8C online). These results indicate that knockout of YSL16 does not affect Fe homeostasis in rice.
Transport Substrate of OsYSL16
The transport substrate of YSL16 was investigated using the yeast expression system. Since only the distribution of Cu was altered in the knockout line (Figures 3D to 3F, 4, and 8) and Cu is present in the form of the Cu-NA complex in the xylem (Pich and Scholz, 1996; Curie et al., 2009), we first determined the transport activity of the Cu-NA complex. YSL16 was expressed under the control of the Gal-inducible promoter in a yeast mutant (Δctr1 ctr3) defective in Cu uptake (Sancenón et al., 2003). A dose-dependent experiment showed that the uptake of Cu-NA was significantly higher in yeast expressing YSL16 than in yeast expressing the empty vector at a concentration higher than 5 µM in the presence of Gal (Figure 9A). This concentration is similar to that of Cu found in the cell sap of root tips (Figure 3F). YSL16-GFP described above also showed similar transport activity for Cu-NA to YSL16 (see Supplemental Figure 9A online), indicating that GFP fusion did not affect the function of YSL16. In a time-course uptake experiment, a significant increase in Cu uptake was observed at and after 20 min of exposure to the Cu-NA complex in the presence of galactose (Figure 9B).
Figure 9.
Transport Activity of YSL16 for the Cu-NA Metal Complex in Yeast.
(A) Dose-dependent transport activity of YSL16 for 65Cu-NA in yeast. Yeast expressing YSL16 or not (empty vector) was exposed to a solution containing various concentrations of 65Cu-NA for 2 h. DW, dry weight.
(B) Time-course experiment of Cu-NA uptake. Yeast expressing YSL16 or not (empty vector) was exposed to a solution containing 5 μM Cu-NA and Gal and sampled at different time points. Cu concentration was determined by ICP-MS or AAS. Data are given as means ± sd of three biological replicates. Asterisks indicate significant differences (*P < 0.05; **P < 0.01) between yeast expressing YSL16 and the empty vector control.
(C) and (D) Yeast complementation test of Fe-NA (C) and Zn-NA (D). YSL16 was introduced into yeast strain Δfet3 fet4, which is defective in Fe uptake (C), or yeast strain Δzrt1 zrt2, which is defective in Zn uptake (D). Zm-YS1 was used as a positive control for Fe-NA uptake. The yeast was cultured on plates containing 5 µM Fe-NA at different pH levels or 5 µM Zn-NA at 30°C for 5 and 3 d for Δfet3 fet4 and Δzrt1 zrt2, respectively.
We also investigated the transport activity of YSL16 for ionic Cu and the Cu-deoxymugineic acid (DMA) complex. However, YSL16 did not show transport activity for these two forms (see Supplemental Figure 9B online). Furthermore, we tested whether YSL16 is able to transport other metal-NA complex, including Zn-NA and Fe-NA in yeast. Expression of YSL16 did not restore the growth of a yeast mutant (Δfet3 fet4), which is defective in Fe uptake in the presence of Fe-NA at pH 6.5 (Figure 9C), whereas expression of maize (Zea mays) YS1, which was used as a positive control, complemented the growth. Expression of YSL16 was also not able to rescue the growth of ZHY3 (Δzrt1 zrt2), a mutant defective in zinc uptake, in the presence of Zn-NA (Figure 9D). These results suggest that YSL16 does not have transport activity for Fe-NA and Zn-NA.
DISCUSSION
YSL16 Is a Transporter of the Cu-NA Complex in Rice
YSL16 is one of the 18 members of YSL transporters in the rice genome (Curie et al., 2009). In Arabidopsis, there are eight members. According to their amino acid sequence similarity, the YSL family can be divided into four groups (Zheng et al., 2011). Os-YSL16 belongs to group I, and all members in this group except Os-YSL9 and 16 have been functionally characterized. Studies have shown that these YSL group I members are involved in the transport of metals (Fe, Zn, Ni, Mn, and Cu) complexed with phytosiderophores and/or NA (Curie et al., 2009). For example, Zm-YS1, barley (Hordeum vulgare) YS1, and Os-YSL15 and Os-YSL18 transport the Fe(III)-phytosiderophore complex (Curie et al., 2001; Murata et al., 2006; Inoue et al., 2009; Aoyama et al., 2009). Recently, Arabidopsis YSL3 was also shown to have transport activity for the Fe(III)-phytosiderophore complex in yeast, although Arabidopsis does not produce phytosiderophores (Chu et al., 2010). By contrast, Os-YSL2, At-YSL1, 2, and 3, and Tc-YSL3 from the metal hyperaccumulator Alpine pennycress (Thlaspi caerulescens), transport ferrous NA complex [Fe(II)-NA] (DiDonato et al., 2004; Koike et al., 2004; Le Jean et al., 2005; Waters et al., 2006; Gendre et al., 2007; Chu et al., 2010). In addition, Os-YSL2 also showed transport activity for the manganese-NA complex (Koike et al., 2004), At-YSL2 for Cu-NA (DiDonato et al., 2004), although this activity was not detected in another study (Schaaf et al., 2005), and TcYSL3 for Ni-NA (Gendre et al., 2007). Our results indicate that Os-YSL16 transports the Cu-NA complex, but not Cu2+, Cu-DMA, Fe(II)-NA, or Zn-NA, based on a yeast expression assay (Figure 9; see Supplemental Figure 9 online). Knockout of Os-YSL16 only affected the distribution of Cu but not of other metals, including Fe, Zn, and Mn (Figures 4 and 8; see Supplemental Figure 6 online). These results indicate that Os-YSL16 is a transporter of the Cu-NA complex. This transport substrate fits with the physiological role of YSL16 as discussed below.
YSL16 Is Required for the Distribution of Cu to Developing Leaves
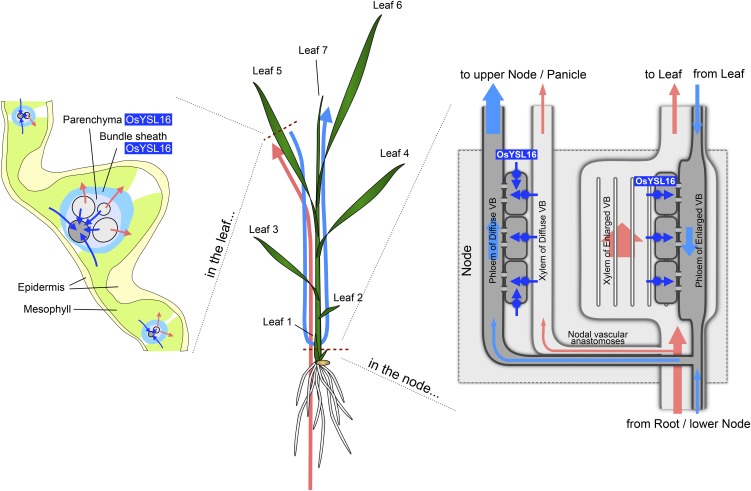
Cu taken up by the roots will be loaded to the xylem and then delivered to different tissues. To deliver Cu to developing young tissues with lower transpiration compared with fully expanded older leaves, phloem-mediated transport of Cu is required. Vascular bundles in the young unexpanded leaves are connected to the ones originating from a node located at the upper part of the unelongated stem (Kawahara et al., 1975; Hoshikawa, 1989). Therefore, the Cu-NA complex in the xylem must be transported out of the xylem and then transferred to the phloem of another vascular bundle connected to the young leaf. GFP reporter expression controlled by the YSL16 promoter is localized to the phloem region of vascular bundles in the unelongated stem (Figures 2B and 2C). Recently, it was reported that YSL16 promoter activity was also detected in the xylem in promoter-β-glucuronidase transgenic plants (Kakei et al., 2012). This inconsistency might be attributed to the length of promoter used. We used 2.3 kb of the promoter region, whereas a short promoter (1.5 kb) was used in the study by Kakei et al. (2012). Localization of YSL16 only in the phloem of unelongated stems is consistent with the phenotype of the knockout line; knockout of YSL16 resulted in a decreased Cu concentration in the young leaves but increased Cu concentration in the older leaves (Figure 4A). These results indicate that YSL16 is involved in the phloem transport of Cu-NA, which is required for delivering Cu to the developing tissues at the unelongated nodes (Figure 10).
Figure 10.
Schematic Presentation of the Role of YSL16 in Cu Distribution.
YSL16 is a transporter of the Cu-NA complex, which is mainly expressed in vascular tissues of leaves and the phloem region of nodes. Its major role is to load the Cu-NA complex into the phloem, which is required for the proper distribution and redistribution of Cu from older leaves to the developing tissues, including young leaves and seeds. Pink and blue arrows indicate Cu movement through the xylem and phloem, respectively. Dashed red line shows the position of observation.
YSL16 is also expressed in the roots (Figure 1), at the exodermis, and stele (Figure 2A). Knockout of YSL16 resulted in a slight reduction in the Cu concentration of the roots and shoots (Figures 3D and 3E). Considering that YSL16 is a transporter of Cu-NA, it is unlikely that YSL16 is directly involved in the uptake of Cu. YSL16 was highly expressed in the mature root zone, and knockout of YSL16 resulted in a decrease in Cu concentration in the root tips and in root growth (Figures 1B, 3A, 3B, and 3F). These results suggest that YSL16 plays an important role in delivering Cu to the root tips, which is necessary for cell division and elongation.
YSL16 Is Involved in the Redistribution of Cu from Old Leaves to Developing Leaves
YSL16 is expressed in the vascular tissues of the leaf blade (Figure 2D). When YSL16 was knocked out, the mobility of Cu from old leaves was impaired (Figures 5 and 6). Cu is thought not to be efficiently redistributed from older leaves to younger leaves and meristems (Marschner, 2011). However, Cu mobilization from older tissues to younger tissues has been observed in plants such as Ricinus communis (Stephan et al., 1994) and wheat (Triticum aestivum; Hocking, 1994). In this study, 10 to 33% of total Cu in the older leaves was moved out from the older leaves in the wild-type rice during 8 d of Cu deficiency (Figure 5E). A labeling experiment with stable isotope 65Cu showed that 5.8% of total 65Cu fed from leaf 4 as 65Cu-NA was mobilized to the youngest leaf (leaf 7) in wild-type rice after 30 h of labeling (Figure 6A); however, this redistribution was greatly reduced in the knockout line (Figure 6A). Moreover, when fed with ionic 65Cu, the mobilization from old to young leaves was almost undetectable in both the wild-type rice and knockout line (Figure 6B). These results indicate that, in the leaves, YSL16 is required for transporting Cu-NA from the mesophyll cells to the phloem (Figure 10). This contributes to the recycling of Cu in the older leaves of wild-type rice (Figures 4A and 10).
YSL16 Plays an Important Role in the Distribution of Cu to the Seeds at the Nodes
Flowering and efficient seed set require Cu (Marschner, 2011). Gramineous plants, including rice, have a distinct system of delivering minerals to the seeds. A recent study showed that transporters at the upper nodes beneath the panicles play an important role in distributing minerals to the panicles in rice (Yamaji and Ma, 2009). YSL16 showed the highest expression at node I and node II (Figure 1C) and was mainly expressed in the phloem region (Figures 2E and 2F). Knockout of YSL16 resulted in increased Cu concentration in the leaf blade and husk, but decreased Cu concentration in the culm, rachis, and brown rice (Figure 8A). Feeding 65Cu-NA but not 65CuCl2 to the cut end of flag leaf resulted in high mobilization to the panicle in the wild-type rice and complementation lines but not in the knockout line (Figure 8C). These results indicate that YSL16 plays an important role in delivering Cu-NA into the phloem connected to the panicles, a role similar to that in the unelongated nodes at the vegetative stage as discussed above (Figure 10). Therefore, the reduced fertility in the knockout line is the result of impaired Cu distribution (Figures 7 and 8A). This is supported by the observation that increasing the Cu supply improved the fertility of the knockout line (Figures 7C and 7D; see Supplemental Figure 5 online).
Other YSL transporters have also been reported to be involved in long-distance transport of metals. Arabidopsis YSL2 is expressed in many cell types in both roots and shoots and is suggested to be involved in the lateral movement of metals in the vasculature and in Fe and Zn homeostasis (DiDonato et al., 2004; Schaaf et al., 2005). YSL1 and 3 are strongly expressed in cauline leaves and moderately expressed in inflorescence stems, which have been suggested to be responsible for metal export for retranslocation into developing seeds (Le Jean et al., 2005; Chu et al., 2010). At-YSL1 and 3 mainly function as Fe transporters, but the double mutant also exhibited lower levels of Cu in the flowers and seeds (Waters et al., 2006), suggesting that YSL1 and 3 are also involved in Cu distribution. However, these two proteins did not exhibit transport activity for Cu-NA in yeast, and their exact role in Cu distribution has not been elucidated. In rice, YSL2 is mainly expressed in the phloem cells of the vascular bundles, especially in the companion cells of Fe-deficient leaves (Koike et al., 2004) and has been suggested to be involved in the transport of Fe and Mn through the phloem, including the translocation of Fe and Mn into rice grains (Koike et al., 2004). YSL18 is strongly expressed in the flowers and is probably responsible for the translocation of iron in reproductive organs and phloem in joints (Aoyama et al., 2009). However, the expression of these genes in the node was not examined; therefore, it is not clear whether they have similar roles as YSL16 in metal distribution.
YSL16 Plays a Major Role in Cu Distribution but Not in Fe Distribution
Recently, two studies reported on the function of YSL16 at the seedling stage of rice. Using a different knockout line, it was reported that knockout of YSL16 did not cause any changes in phenotype and mineral accumulation (Lee et al., 2012) but that enhanced expression of YSL16 slightly increased translocation of Fe from the roots to shoots, although the exact mechanism is unknown. On the other hand, using knockdown approaches, YSL16 was reported to be involved in the allocation of Fe in rice by unloading Fe(III)-DMA from the xylem (Kakei et al., 2012). However, the authors of this study found that Fe allocation into new leaves was significantly higher in the knockdown lines than in wild-type rice, raising the question of the exact role of YSL16 in Fe distribution. In this study, we found that unlike Cu, there was no difference in the Fe concentration of old and young leaves between wild-type rice and the mutant (Figures 4B). Knockout of YSL16 also did not affect the Fe concentration of different tissues, including brown rice, husk, culm, rachis, and leaf blade and sheath at harvest (Figure 8B). These results show that YSL16 is not involved in Fe distribution. This is also supported by the expression results of four genes related to Fe homeostasis; all these genes showed similar expression between wild-type rice and the mutant (see Supplemental Figure 8 online) in contrast with genes related to Cu homeostasis, which showed higher expression in the young tissues of the mutant (see Supplemental Figure 7 online). Therefore, it is unlikely that knockout of YSL16 directly affects Fe homeostasis. Since Fe in the xylem is mainly present in the form of the Fe-citrate complex in rice (Yokosho et al., 2009), the role of YSL16 in xylem unloading of Fe would be minor. In fact, in the presence of Fe2+, which is rich in paddy soil, there was no difference in the growth between wild-type rice and knockdown lines (Kakei et al., 2012). The inconsistencies between these previous studies and this study are due to the following two reasons; first, the previous studies did not analyze leaf position-dependent mineral concentration as we did (Figure 4) and therefore could not reveal differences in Cu mobility, as we did. Second, the phenotype was not observed under Cu-sufficient conditions at any point in the growth period in their studies. Cu is an essential element for plant growth, but its requirement is low (Marschner, 2011). As shown in Figures 7C and 7D, the clear difference in phenotype between the wild-type rice and the knockout line was only observed when the Cu concentration in the nutrient solution was 2 nM. Any contamination from water and chemicals will result in failure to observe the difference in phenotype. We used distilled water for preparing low Cu nutrient solution.
In conclusion, YSL16 is mainly a phloem-localized transporter of the Cu-NA complex. Its major role is to load the Cu-NA complex into the phloem, which is required for remobilization of Cu from older leaves to developing tissues, including young leaves and seeds.
METHODS
Plant Materials and Growth Conditions
Wild-type rice (Oryza sativa cv Nipponbare and cv Zhonghua 11) and a T-DNA insertion line (RMD_05Z11CP34; http://rmd.ncpgr.cn/) (Zhang et al., 2006) were used. The homozygous lines of the T-DNA insertion were screened by PCR using Os-YSL16–specific primers (5′-GACAGCGACTGCTGTTCTCATAAAC-3′ and 5′-TTGCGGCTTTACCACTGAAG-3′) and a T-DNA left border primer (5′-ATGTGTTATTAAGTTGTCTAAG-3′).
For hydroponic experiments, seeds of wild-type rice and the knockout line were soaked in water overnight at 25°C in the dark and then transferred to a net floating on 0.5 mM CaCl2 solution. On day 7, seedlings were transferred to a 3.5-liter plastic pot containing one-half-strength Kimura B solution and grown in a greenhouse at 25 to 30°C. After 10 d, the seedlings (four plants per pot) were transferred to a 1.2-liter pot containing freshly prepared nutrient solution. The nutrient solution contained the macronutrients (mM): (NH4)2SO4 (0.18), MgSO4·7H2O (0.27), KNO3 (0.09), Ca(NO3)2·4H2O (0.18), and KH2PO4 (0.09); and the micronutrients (μM): MnCl2·4H2O (0.5), H3BO3 (3), (NH4)6Mo7O24·4H2O (1), ZnSO4·7H2O (0.4), Fe-EDTA (20), and 0.2 μM CuSO4·5H2O. The pH of this solution was adjusted to 5.5, and the nutrient solution was renewed every 2 d. All experiments were repeated at least three times with three replicates each, and representative results of one experiment are shown.
Expression Pattern Analysis
For the tissue-specific expression analysis of YSL16 at the vegetative stage, the seedlings (cv Nipponbare) prepared above were exposed to a solution without Cu, Fe, Zn, or Mn. After 1 week, roots, shoots, and basal regions (2 cm above the roots) were sampled and subjected to RNA extraction. For spatial expression analysis, different root segments (0 to 1 cm and 1 to 2 cm) were sampled for RNA extraction. To investigate the expression pattern of YSL16 at different growth stages, different tissues from plants (cv Nipponbare) grown in a paddy field were taken.
For expression analysis of genes related to Fe homeostasis or Cu homeostasis, wild-type rice and the osysl16 mutant were grown in a nutrient solution as described above to the six-leaf stage, and leaf blades 2 to 6, the basal nodes, and root tips (0 to 1 cm) were separately sampled for RNA extraction. Samples of the roots and shoots were also taken from the wild-type rice and the mutant grown in Fe-free, Cu-free, or normal solution for 10 d.
Total RNA was extracted using an RNeasy plant mini kit (Qiagen) and then converted to cDNA after DNase I treatment using a SuperScript II kit (Invitrogen) following the manufacturer’s instructions. Specific cDNAs were amplified by Sso Fast EvaGreen Supermix (Bio-Rad), and quantitative real-time PCR was performed on CFX384 (Bio-Rad). The primer efficiency was 103.5%. Both HistoneH3 and Actin were used as internal standards, and normalized relative expression was calculated by the ΔΔCt (cycle threshold) method using CFX Manager software (Bio-Rad). The primers used for quantitative real-time PCR were listed in Supplemental Table 1 online.
Transient Expression in Onion Epidermal Cells and Rice Protoplasts
For observation of subcellular localization of YSL16, YSL16 cDNA containing the SalI restriction site, but not the stop codon, was amplified by RT-PCR using the primers 5′-ACGAGTCGACATGGACCGCCACGCGCTGG-3′ and 5′-TACCTCATGACGTTTCCCGGTATGAACTTC-3′. The amplified cDNA fragment was then subcloned in frame in front of the GFP coding region in the pBluescript vector, generating the OsYSL16-sGFP construct under the control of the 35S promoter. The construct or the 35S-sGFP control was then bombarded into onion epidermal cells together with 35S-DsRed (Clontech). After incubation in the dark at 25°C for 20 h, the fluorescence was observed with confocal laser scanning microscopy (LSM700; Carl Zeiss).
For transient expression of YSL16 in rice cells, YSL16-GFP was introduced into rice leaf protoplasts. The rice leaf protoplasts were prepared from seedlings (2 weeks old, cv Nipponbare) grown hydroponically. Preparation of the protoplasts and transformation by the plasmid was performed by a polyethylene glycol–mediated method according to Chen et al. (2006). The GFP signal and autofluorescence of the chloroplasts were observed by laser scanning microscope (LSM700; Carl Zeiss).
Cell-Specific Expression Profile of Os-YSL16
To investigate the cell specificity of the YSL16 expression profile, we amplified the 2.3-kb promoter region of YSL16 using primers 5′-ATCTGGTACCGACGATCTCTAGCCTATGC-3′ and 5′-TACCAAGCTTGCTGCCGGCCGCACCCGACG-3′. The fragment was then subcloned into the binary vector pPZP2H-sGFP (Fuse et al., 2001) to generate ProYSL16-GFP using the cloning sites KpnI and HindIII, and this construct was subsequently introduced into Agrobacterium tumefaciens (strain EHA101). Callus was induced from mature embryos of rice cultivar Nipponbare for Agrobacterium-mediated transformation. Transformation was performed according to the protocol of Chen et al. (2003).
To observe the expression profile, immunostaining with an antibody against GFP was performed according to Yamaji and Ma (2007). Tissues including node I, leaf blade, basal node, and root of ProYSL16-GFP transgenic rice and wild-type rice grown in nutrient solution were sampled. The signal was observed with a confocal laser scanning microscope (LSM700; Carl Zeiss).
Complementation Test
For complementation test of the YSL16 T-DNA insertion line (RMD_05Z11CP34), a genomic DNA fragment of YSL16, including an ∼2 kb promoter region, coding region, and ∼1 kb downstream region was amplified using primers 5′-ACAGTCGACGGCTGTACTCCACCAATATTC-3′ and 5′-ACAGGATCCTGTGCTAGCACTTGTCCTCGA-3′. The fragment was then subcloned into the binary vector pTF101.1 (Frame et al., 2002) to generate pYSL16-com using the cloning sites SalI and BamHI and subsequently introduced into Agrobacterium (strain EHA101). Transformation of the knockout line was performed as described above. Phenotypic analysis of two independent lines was performed as described below.
Phenotypic Analysis of the YSL16 Knockout Line
Both wild-type rice (cv Zhonghua 11) and the knockout line were grown in one-half-strength Kimura B solution. After 3 weeks, the root length was measured and then plants were separated into roots and shoots. Dry weights were recorded after drying in an oven at 70°C for 3 d.
In a separate experiment, plants were separated into different leaf blades (leaves 2 to 6), leaf sheaths, and roots for investigation of metal distribution. The first leaf blade was too small to be sampled. A similar experiment was also performed with two independent complementation lines.
For the soil culture experiment, 3-week-old seedlings of the wild type (cv Zhonghua 11), ysl16 mutant, and two independent complementation lines prepared as above were transplanted to a 3.5-liter plastic pot filled with rice paddy soils previously treated with coating fertilizer (14-12-14 for nitrogen-phosphorus-potassium) at a rate of 1 g/kg soil. They were grown in a closed greenhouse with natural light. Tap water was supplied daily. After seed filling, the leaf blade and leaf sheath of the flag leaf, culm, rachis, husk, and brown rice were sampled for metal analysis. Fertility (percentage of filled grains) was determined by placing seeds in an 8.5% NaCl solution and calculated as follows: the number of sinking seeds/the total number of seeds × 100.
To investigate the effect of different Cu levels on fertility, we grew the wild-type rice and ysl16 knockout line hydroponically in the presence of 200 or 2 nM Cu until ripening. Distilled and deionized water was used to prepare the nutrient solution with 2 nM Cu. The fertility of the wild type and ysl16 knockout line was determined as described above. The pollen viability was evaluated as described below.
Pollen Grain Viability Staining
To estimate the viability of pollen from the wild type (cv Zhonghua 11) and ysl16 mutant under different Cu levels, spikelets were collected from each plant just before anthesis and anthers were randomly sampled for staining with 1% I2-KI solution. Photographs were taken under light microscopy observation. The pollen viability ratio was calculated as follows: black-stained pollen/total pollen in the screen × 100. Four biological replicates were performed.
Cu Mobility Analysis of Old Leaves
To determine Cu mobility in the wild type (cv Zhonghua 11), ysl16 mutant, and two complementation lines, four seedlings were grown in one-half-strength Kimura B solution. At the stage of full expansion of the sixth leaf, half of the plants were harvested for metal analysis of different leaf blades (leaves 2 to 5). The roots of the remaining half of plants were washed in 5 mM CaCl2 solution for 12 h to remove apoplastic Cu and were then subjected to a nutrient solution without Cu. After 8 d of further growth, different leaf blades (leaves 2 to 5) were sampled for metal analysis as described below. The mobility of Cu was calculated as follows: decrease in Cu content/total Cu content × 100.
Cu in Root Tip Cell Sap
Root tips (0 to 5 mm) were excised from 3-week-old plants grown hydroponically as described above and placed on a filter set in a tube. The samples were immediately frozen at −80°C and stored until use. After brief thawing at room temperature, the cell sap was collected by centrifugation at 20,600g for 10 min (Xia et al., 2010) and subjected to Cu determination after digestion as described below.
Isotope Labeling Experiment with 65Cu
To determine Cu redistribution between the different leaves, plants at the seven-leaf stage, which had been cultivated in Cu-free solution for 2 weeks, were used for feeding experiments with the stable isotope 65Cu. Leaf 4 was cut 2 cm from the tip with a razor and subsequently exposed to 1 mL of solution containing 50 µM of 65CuCl2 or 65Cu-NA according to Carey et al. (2011). 65CuCl2 (99.7% 65Cu) was purchased from Taiyo Nippon Sanso. The 65Cu-NA complex was prepared by mixing 65CuCl2 with NA at 20% excess of NA, followed by incubating at 60°C for 10 min in MES buffer at pH 6.4 (Schaaf et al., 2004). After feeding for 30 h, leaf blades 2 to 7 were sampled and 65Cu concentration was determined by inductively coupled plasma mass spectrometry (ICP-MS) in the isotope mode. For each treatment, three to six biological replicates were performed. The experiment was repeated three times.
To determine the mobilization of Cu from the flag leaf to panicles, the flag leaf of wild-type rice, two complementation lines, and the mutant grown in soil was cut at 2 cm from the tip with a razor and fed with 50 µM of 65CuCl2 or 65Cu-NA for 2 weeks after flowering. The feeding solution was renewed three times. Six biological replicates were performed. At harvest, the flag leaf and panicles were sampled for determination of 65Cu. The mobilization was calculated as the net 65Cu increase amount (i.e., the total 65Cu amount minus natural amount of 65Cu) divided by the 65Cu uptake amount from the feeding solution.
Transport Activity Assay in Yeast
The cDNA of YSL16 or the YSL16-GFP fusion, generated as described above, was subcloned into the pYES2 vector (Invitrogen). The pYES2-YSL16, pYES2-YSL16-GFP, or the vector control pYES2 were introduced into a yeast mutant Δctr1ctr3 defective in Cu uptake (Sancenón et al., 2003) according to the manufacturer’s protocol (S.C. EasyComp transformation kit; Invitrogen). Transformants were selected on uracil-deficient medium and grown in synthetic complete (SC-uracil) yeast medium containing 2% Glc, 0.67% yeast nitrogen base without amino acids (Difco), 0.2% appropriate amino acids, and 2% agar at pH 6.0. One colony was selected from each transformation strain and grown in the liquid SC-uracil medium. For the Cu uptake assay, an appropriate amount evaluated by OD600 of cells precultured in Cu-sufficient SC-uracil medium were transferred to a freshly prepared Cu-deficient SC-uracil medium using yeast nitrogen base without Cu (US Biological) until OD600 of 0.2. When the OD600 reached 2.0 to 3.0 after 12 to 24 h in culture, yeast cells were harvested and transferred to freshly prepared Cu-deficient SC-uracil medium containing 2% Gal for induction of the GAL promoter and 25 mM MES adjusted to pH 5.6. The precultured yeast was adjusted to an OD600 value of 3.0 by reducing the amount of liquid. Cells were cultured for 2 h and then 65Cu-NA was added to the solution at a final concentration of 0, 2, 5, 10, or 20 µM. For experiments involving different forms of Cu uptake, a final concentration of 5 µM CuCl2, Cu-DMA, or Cu-NA was added to the cell culture medium after 2 h of Gal induction. The uptake experiment was also similarly conducted with yeast expressing Os-YSL16, OsYSL16-GFP, or empty vector using 10 µM Cu-NA. After 2 h of incubation with gentle shacking, yeast cells were harvested by centrifugation at 8000g for 3 min and then washed three times with milli-Q water. In a time-course experiment, cells were collected at 0, 5, 20, 40, 60, 120, and 240 min after exposure to 5 µM Cu-NA, and the yeast cells were collected by centrifugation at 8000g for 3 min and then washed three times with milli-Q water. The samples were digested with 0.5 mL 2 n HCl after drying overnight. The Cu concentration was determined by inductively coupled plasma-mass spectrometer (ICP-MS) or atomic absorption spectrometry (AAS) as described below. Cu-NA and Cu-DMA were prepared by mixing CuCl2 with NA (20% excess) or DMA (double amount), respectively, at 60°C for 10 min (Schaaf et al., 2004). DMA was purified from root exudates of Fe-deficient wheat (Triticum aestivum). NA was purchased from Hasegawa.
For the Fe-NA and Zn-NA uptake experiment, yeast strain DDY4 (Δfet3fet4) (Dix et al. 1997) and ZHY3 (Δzrt1zrt2) (Zhao and Eide, 1996) were used, respectively. The pYES2-OsYSL16 and the vector control pYES2 were introduced into the corresponding yeast mutant. For the case of DDY4, pYES2-ZmYS1 was also introduced and used as a positive control. A similar method was used for the liquid uptake assay as for Cu-NA mentioned above. For the plate experiment, SC-Ura medium at pH 6.5 was used for the assay. Fe(II)-NA or Zn-NA was freshly prepared as described by Schaaf et al. (2004) and added to the medium just before solidification of the medium. For spotting sample, freshly prepared DDY4 or ZHY3 mutant culture was centrifuged and resuspended in different amounts of sterile water, resulting in different dilutions at OD (600 nm) of 0.2, 0.02, 0.002, and 0.0002. After 10 μL of each dilution was spotted on the plates, the plates were incubated at 30°C for 5 and 3 d for DDY4 and ZHY3, respectively.
Metal Determination
After drying in an oven at 70°C for 2 d, the samples of different tissues were digested with 5 mL of 11 n HNO3 on a heater at up to 150°C. The metal concentration in the digest solution was determined by AAS (Z-2000; Hitachi) or ICP-MS (Agilent 7700).
NA Determination
NA was extracted from older leaves (leaves 2 and 3) and the youngest leaf (leaf 8) of wild-type rice and ysl16 mutant seedlings (six weeks old) and quantified using HPLC according to Cheng et al. (2007). Briefly, ∼100 mg of fresh leaf samples was ground in liquid nitrogen and extracted with 500 μL of water at 80°C for 30 min, followed by 10 min centrifugation (18,000g). The supernatant solution was transferred to centrifuge tubes with filters (Amicon Ultra) and further centrifuged for 10 to 30 min at 18,000g. The supernatant solution was determined with HPLC using a cation exchange column (Shim-pack; Amino-Li, Shimadzu) with an amino acid analysis kit and o-phthalaldehyde reagent (Shimadzu). Chemically synthesized NA (T-Hasegawa) was used as a standard. Three biological replicates were made for each leaf.
Statistical Analysis of Data
The statistical analysis was performed using Student’s t test (for comparison between two samples) or Tukey’s test (for more than three samples), assuming unequal variances. Significance was defined as P < 0.05 (*) or P < 0.01 (**).
Accession Number
Sequence data from this article can be found in the GenBank/EMBL databases under accession number AB673450 for YSL16.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of Marker Genes Related to Fe, Zn, or Cu Deficiency.
Supplemental Figure 2. Subcellular Localization of Rice YSL16.
Supplemental Figure 3. Gene Structure and T-DNA Insertion Line of YSL16.
Supplemental Figure 4. The Concentration of Nicotianamine in Older and Younger Leaves of Wild-Type Rice and the Mutant.
Supplemental Figure 5. Pollen Viability of Wild-Type Rice and the ylsl16 Mutant.
Supplemental Figure 6. Concentration of Zn and Mn in Different Tissues.
Supplemental Figure 7. Expression of Genes Related to Cu Homeostasis: COPT1, COPT5, and HMA9.
Supplemental Figure 8. Expression of Genes Related to Fe Homeostasis: IRT1, IRO2, Ferritin, and YSL2.
Supplemental Figure 9. Yeast Cu Uptake Assay.
Supplemental Table 1. Primer Sequences Used for Real-Time RT-PCR Analysis.
Acknowledgments
The research was supported by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 22119002 to J.F.M. and No. 23688009 to N.Y). L.Z. is a postdoctoral fellow of Japan Society for the Promotion of Science. We thank Changyin Wu at National Center of Plant Gene Research (Wuhan, China), Huazhong Agricultural University for providing T-DNA insertion line RMD_05Z11CP34. We also thank Dennis J. Thiele for providing yeast mutant Δctr1 ctr3.
AUTHOR CONTRIBUTIONS
J.F.M. designed the research. L.Z., N.Y., K.Y., and J.F.M. performed the experiments. L.Z. and J.F.M. analyzed the data, and L.Z., N.Y., and J.F.M. wrote the article.
Glossary
- GFP
green fluorescent protein
- NA
nicotianamine
- DMA
deoxymugineic acid
- ICP-MS
inductively coupled plasma mass spectrometry
- SC
synthetic complete
References
- Abdel-Ghany S.E., Müller-Moulé P., Niyogi K.K., Pilon M., Shikanai T. (2005). Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17: 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-Colás N., Sancenón V., Rodríguez-Navarro S., Mayo S., Thiele D.J., Ecker J.R., Puig S., Peñarrubia L. (2006). The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J. 45: 225–236 [DOI] [PubMed] [Google Scholar]
- Aoyama T., Kobayashi T., Takahashi M., Nagasaka S., Usuda K., Kakei Y., Ishimaru Y., Nakanishi H., Mori S., Nishizawa N.K. (2009). OsYSL18 is a rice iron(III)-deoxymugineic acid transporter specifically expressed in reproductive organs and phloem of lamina joints. Plant Mol. Biol. 70: 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attallah C.V., Welchen E., Pujol C., Bonnard G., Gonzalez D.H. (2007). Characterization of Arabidopsis thaliana genes encoding functional homologues of the yeast metal chaperone Cox19p, involved in cytochrome c oxidase biogenesis. Plant Mol. Biol. 65: 343–355 [DOI] [PubMed] [Google Scholar]
- Carey A.M., Norton G.J., Deacon C., Scheckel K.G., Lombi E., Punshon T., Guerinot M.L., Lanzirotti A., Newville M., Choi Y., Price A.H., Meharg A.A. (2011). Phloem transport of arsenic species from flag leaf to grain during grain filling. New Phytol. 192: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Jin W., Wang M., Zhang F., Zhou J., Jia Q., Wu Y., Liu F., Wu P. (2003). Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J. 36: 105–113 [DOI] [PubMed] [Google Scholar]
- Chen S., Tao L., Zeng L., Vega-Sanchez M.E., Umemura K., Wang G.L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Cheng L., Wang F., Shou H., Huang F., Zheng L., He F., Li J., Zhao F.J., Ueno D., Ma J.F., Wu P. (2007). Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiol. 145: 1647–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.H., Chiecko J., Punshon T., Lanzirotti A., Lahner B., Salt D.E., Walker E.L. (2010). Successful reproduction requires the function of Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol. 154: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C., Cassin G., Couch D., Divol F., Higuchi K., Le Jean M., Misson J., Schikora A., Czernic P., Mari S. (2009). Metal movement within the plant: Contribution of nicotianamine and yellow stripe 1-like transporters. Ann. Bot. (Lond.) 103: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C., Panaviene Z., Loulergue C., Dellaporta S.L., Briat J.F., Walker E.L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409: 346–349 [DOI] [PubMed] [Google Scholar]
- DiDonato R.J., Jr, Roberts L.A., Sanderson T., Eisley R.B., Walker E.L. (2004). Arabidopsis Yellow Stripe-Like2 (YSL2): A metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J. 39: 403–414 [DOI] [PubMed] [Google Scholar]
- Dix D., Bridgham J., Broderius M., Eide D. (1997). Characterization of the FET4 protein of yeast. Evidence for a direct role in the transport of iron. J. Biol. Chem. 272: 11770–11777 [DOI] [PubMed] [Google Scholar]
- Frame B.R., Shou H., Chikwamba R.K., Zhang Z., Xiang C., Fonger T.M., Pegg S.E., Li B., Nettleton D.S., Pei D., Wang K. (2002). Agrobacterium-mediated transformation of maize embryos using a simple binary vector system. Plant Physiol. 129: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse T., Sasaki T., Yano M. (2001). Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 18: 219–222 [Google Scholar]
- Garcia-Molina A., Andrés-Colás N., Perea-García A., Del Valle-Tascón S., Peñarrubia L., Puig S. (2011). The intracellular Arabidopsis COPT5 transport protein is required for photosynthetic electron transport under severe copper deficiency. Plant J. 65: 848–860 [DOI] [PubMed] [Google Scholar]
- Gendre D., Czernic P., Conéjéro G., Pianelli K., Briat J.F., Lebrun M., Mari S. (2007). TcYSL3, a member of the YSL gene family from the hyper-accumulator Thlaspi caerulescens, encodes a nicotianamine-Ni/Fe transporter. Plant J. 49: 1–15 [DOI] [PubMed] [Google Scholar]
- Hirayama T., Kieber J.J., Hirayama N., Kogan M., Guzman P., Nourizadeh S., Alonso J.M., Dailey W.P., Dancis A., Ecker J.R. (1999). RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97: 383–393 [DOI] [PubMed] [Google Scholar]
- Hocking P.J. (1994). Dry-matter production, mineral nutrient concentrations, and nutrient distribution and redistribution in irrigated spring wheat. J. Plant Nutr. 17: 1289–1308 [Google Scholar]
- Hoshikawa K. (1989). The Growing Rice Plant. (Tokyo: Nobunkyo; ). [Google Scholar]
- Inoue H., Kobayashi T., Nozoye T., Takahashi M., Kakei Y., Suzuki K., Nakazono M., Nakanishi H., Mori S., Nishizawa N.K. (2009). Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 284: 3470–3479 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y., Suzuki M., Kobayashi T., Takahashi M., Nakanishi H., Mori S., Nishizawa N.K. (2005). OsZIP4, a novel zinc-regulated zinc transporter in rice. J. Exp. Bot. 56: 3207–3214 [DOI] [PubMed] [Google Scholar]
- Le Jean M., Schikora A., Mari S., Briat J.F., Curie C. (2005). A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J. 44: 769–782 [DOI] [PubMed] [Google Scholar]
- Kakei Y., Ishimaru Y., Kobayashi T., Yamakawa T., Nakanishi H., Nishizawa N.K. (2012). OsYSL16 plays a role in the allocation of iron. Plant Mol. Biol. 79: 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara K., Chonan N., Matsuda T. (1975). Studies on morphogenesis in rice plants 8. The morphology of vascular bundles in the dwarf part of stem. Jpn. J. Crop. Sci. 44: 61–67 [Google Scholar]
- Kim Y.Y., Choi H., Segami S., Cho H.T., Martinoia E., Maeshima M., Lee Y. (2009). AtHMA1 contributes to the detoxification of excess Zn(II) in Arabidopsis. Plant J. 58: 737–753 [DOI] [PubMed] [Google Scholar]
- Klaumann S., Nickolaus S.D., Fürst S.H., Starck S., Schneider S., Ekkehard Neuhaus H., Trentmann O. (2011). The tonoplast copper transporter COPT5 acts as an exporter and is required for interorgan allocation of copper in Arabidopsis thaliana. New Phytol. 192: 393–404 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kuroda K., Kimura K., Southron-Francis J.L., Furuzawa A., Kimura K., Iuchi S., Kobayashi M., Taylor G.J., Koyama H. (2008). Amino acid polymorphisms in strictly conserved domains of a P-type ATPase HMA5 are involved in the mechanism of copper tolerance variation in Arabidopsis. Plant Physiol. 148: 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S., Inoue H., Mizuno D., Takahashi M., Nakanishi H., Mori S., Nishizawa N.K. (2004). OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 39: 415–424 [DOI] [PubMed] [Google Scholar]
- Lee S., Kim Y.Y., Lee Y., An G. (2007). Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 145: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Ryoo N., Jeon J.S., Guerinot M.L., An G. (2012). Activation of rice Yellow Stripe1-Like 16 (OsYSL16) enhances iron efficiency. Mol. Cells 33: 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder M.C., Goode C.A. (1991). Biochemistry of Copper. (New York: Plenum Press; ). [Google Scholar]
- Lippard S.J., Berg J.M. (1994). Principles of Bioinorganic Chemistry. (Herndon, VA: University Science Books; ). [Google Scholar]
- Marschner P. (2011). Mineral Nutrition of Higher Plants, 3rd ed. (London: Academic Press; ). [Google Scholar]
- Murata Y., Ma J.F., Yamaji N., Ueno D., Nomoto K., Iwashita T. (2006). A specific transporter for iron(III)-phytosiderophore in barley roots. Plant J. 46: 563–572 [DOI] [PubMed] [Google Scholar]
- Pilon M. (2011). Moving copper in plants. New Phytol. 192: 305–307 [DOI] [PubMed] [Google Scholar]
- Pich A., Scholz G. (1996). Translocation of copper and other micronutrients in tomato plants (Lycopersicon esculentum Mill.): Nicotianamine-stimulated copper transport in the xylem. J. Exp. Bot. 47: 41–47 [Google Scholar]
- Pich A., Scholz G., Stephan U.W. (1994). Iron-dependent changes of heavy metals, nicotianamine, and citrate in different plant organs and in the xylem exudate of two tomato genotypes: nicotianamine as possible copper translocator. Plant Soil 165: 189–196 [Google Scholar]
- Rellán-Alvarez R., Abadía J., Alvarez-Fernández A. (2008). Formation of metal-nicotianamine complexes as affected by pH, ligand exchange with citrate and metal exchange. A study by electrospray ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 22: 1553–1562 [DOI] [PubMed] [Google Scholar]
- Sancenón V., Puig S., Mateu-Andrés I., Dorcey E., Thiele D.J., Peñarrubia L. (2004). The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J. Biol. Chem. 279: 15348–15355 [DOI] [PubMed] [Google Scholar]
- Sancenón V., Puig S., Mira H., Thiele D.J., Peñarrubia L. (2003). Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Biol. 51: 577–587 [DOI] [PubMed] [Google Scholar]
- Schaaf G., Ludewig U., Erenoglu B.E., Mori S., Kitahara T., von Wirén N. (2004). ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J. Biol. Chem. 279: 9091–9096 [DOI] [PubMed] [Google Scholar]
- Schaaf G., Schikora A., Häberle J., Vert G.A., Ludewig U., Briat J.F., Curie C., von Wirén N. (2005). A putative function for the arabidopsis Fe-Phytosiderophore transporter homolog AtYSL2 in Fe and Zn homeostasis. Plant Cell Physiol. 46: 762–774 [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D., Gravot A., Auroy P., Mazard C., Kraut A., Finazzi G., Grunwald D., Rappaport F., Vavasseur A., Joyard J., Richaud P., Rolland N. (2006). HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J. Biol. Chem. 281: 2882–2892 [DOI] [PubMed] [Google Scholar]
- Shikanai T., Müller-Moulé P., Munekage Y., Niyogi K.K., Pilon M. (2003). PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan U.W., Grun M. (1989). Physiological disorders of the nicotianamine-auxotroph tomato mutant chloronerva at different levels of iron nutrition. II. Iron deficiency response and heavy metal metabolism. Biochem. Physiol. Pflanz. 185: 189–200 [Google Scholar]
- Stephan U.W., Schmidke I., Pich A. (1994). Phloem translocation of Fe, Cu, Mn, and Zn in Ricinus seedlings in relation to the concentrations of nicotianamine, an endogenous chelator of divalent metal-ions, in different seedling parts. Plant Soil 165: 181–188 [Google Scholar]
- Waters B.M., Chu H.H., Didonato R.J., Roberts L.A., Eisley R.B., Lahner B., Salt D.E., Walker E.L. (2006). Mutations in Arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 141: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintz H., Fox T., Wu Y.Y., Feng V., Chen W., Chang H.S., Zhu T., Vulpe C. (2003). Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J. Biol. Chem. 278: 47644–47653 [DOI] [PubMed] [Google Scholar]
- Xia J., Yamaji N., Kasai T., Ma J.F. (2010). Plasma membrane-localized transporter for aluminum in rice. Proc. Natl. Acad. Sci. USA 107: 18381–18385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N., Ma J.F. (2007). Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol. 143: 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N., Ma J.F. (2009). A transporter at the node responsible for intervascular transfer of silicon in rice. Plant Cell 21: 2878–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosho K., Yamaji N., Ueno D., Mitani N., Ma J.F. (2009). OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol. 149: 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Chu Z., Li X., Xu C., Wang S. (2010). The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell 22: 3164–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Li X., Xiao J., Wang S. (2011). Molecular and functional analyses of COPT/Ctr-type copper transporter-like gene family in rice. BMC Plant Biol. 11: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li C., Wu C., Xiong L., Chen G., Zhang Q., Wang S. (2006). RMD: A rice mutant database for functional analysis of the rice genome. Nucleic Acids Res. 34: 745–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Eide D. (1996). The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271: 23203–23210 [DOI] [PubMed] [Google Scholar]
- Zheng L., Fujii M., Yamaji N., Sasaki A., Yamane M., Sakurai I., Sato K., Ma J.F. (2011). Isolation and characterization of a barley yellow stripe-like gene, HvYSL5. Plant Cell Physiol. 52: 765–774 [DOI] [PubMed] [Google Scholar]
- Zheng L., Huang F., Narsai R., Wu J., Giraud E., He F., Cheng L., Wang F., Wu P., Whelan J., Shou H. (2009). Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol. 151: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]