Abstract
Histone acetylation and deacetylation play an important role in the modification of chromatin structure and regulation of gene expression in eukaryotes. Chromatin acetylation status is modulated antagonistically by histone acetyltransferases and histone deacetylases (HDACs). In this study, we characterized the function of histone deacetylase701 (HDT701), a member of the plant-specific HD2 subfamily of HDACs, in rice (Oryza sativa) innate immunity. Transcription of HDT701 is increased in the compatible reaction and decreased in the incompatible reaction after infection by the fungal pathogen Magnaporthe oryzae. Overexpression of HDT701 in transgenic rice leads to decreased levels of histone H4 acetylation and enhanced susceptibility to the rice pathogens M. oryzae and Xanthomonas oryzae pv oryzae (Xoo). By contrast, silencing of HDT701 in transgenic rice causes elevated levels of histone H4 acetylation and elevated transcription of pattern recognition receptor (PRR) and defense-related genes, increased generation of reactive oxygen species after pathogen-associated molecular pattern elicitor treatment, as well as enhanced resistance to both M. oryzae and Xoo. We also found that HDT701 can bind to defense-related genes to regulate their expression. Taken together, these results demonstrate that HDT701 negatively regulates innate immunity by modulating the levels of histone H4 acetylation of PRR and defense-related genes in rice.
INTRODUCTION
Histones are subjected to posttranslational modifications, such as acetylation, methylation, phosphorylation, and ubiquitination, which establish a rapid and reversible pattern for gene expression across the genome (Strahl and Allis, 2000; Jenuwein and Allis, 2001). In general, acetylation of histones is associated with transcriptional activation, whereas deacetylation of histones is associated with gene repression. These active and repressive histone marks are generated by the combined action of specific histone acetyltransferases and histone deacetylases (HDACs), respectively (Kurdistani and Grunstein, 2003). The structures of HDACs involved in histone modifications are highly conserved in yeast, animals, and plants, suggesting that they act through a similar mechanism in transcriptional regulation. Plant HDACs can be divided into four major groups or families. In addition to a plant-specific HD2 family, three other major families are designated as RPD3, HDA1, and SIR2 based on their primary homology to yeast counterparts. The plant-specific HD2 family may reflect the divergence of developmental processes and environmental responses of plant systems from those of yeast and mammals (Pandey et al., 2002).
The function of Arabidopsis thaliana HD2 family members have been well studied in recent years. For example, At-histone deacetylase1 (HDT1), At-HDT2, and At-HDT3 share similar expression patterns in ovules, embryos, shoot apical meristems, and leaves (Wu et al., 2000). Knockdown of HDT1 in Arabidopsis results in seed abortion (Wu et al., 2003) and causes reduced silencing of Arabidopsis rDNA (Lawrence et al., 2004). In addition, knockdown of HDT1 and HDT2 in the asymmetric leaf1 (as1) or as2 mutant background causes the formation of abaxialized and filamentous leaves (Ueno et al., 2007). At-HDT2C is involved in abscisic acid–stress response; its overexpression leads to insensitivity to abscisic acid and increased tolerance to salt and drought stresses (Sridha and Wu, 2006).
Phylogenic analysis of Arabidopsis, rice (Oryza sativa), maize (Zea mays), and other plant HD2 genes has grouped the dicot and monocot sequences into two distinct clades, suggesting that monocots and dicots may have different HD2 ancestors and that the monocot HD2 members may have been evolved and diversified from the dicot homologs (Pandey et al., 2002). Although there are at least 17 HDAC genes in the rice genome (www.chromdb.org), the function of most family members is unclear.
In this study, we investigated the role of the HDAC gene HDT701 in resistance to the rice pathogens Magnaporthe oryzae and Xanthomonas oryzae pv oryzae (Xoo), which are the causal agents of blast and bacterial blight diseases, respectively. We found that overexpression of HDT701 in transgenic rice leads to enhanced susceptibility to both blast and bacterial blight. By contrast, silencing of HDT701 causes enhanced resistance to both pathogens and the increased generation of reactive oxygen species (ROS) triggered by the pathogen-associated molecular pattern (PAMP) elicitors flg22 and chitin. We also found that HDT701 modulates the levels of histone H4 acetylation in rice plants and negatively regulates the transcription and histone H4 acetylation levels of defense-related genes. These results reveal that, by modulating the histone H4 acetylation levels of defense-related genes in rice, HDT701 functions as a negative regulator of innate immunity to rice pathogens.
RESULTS
Transcription of HDT701 Is Altered after Rice Blast Infection and PAMP Treatment
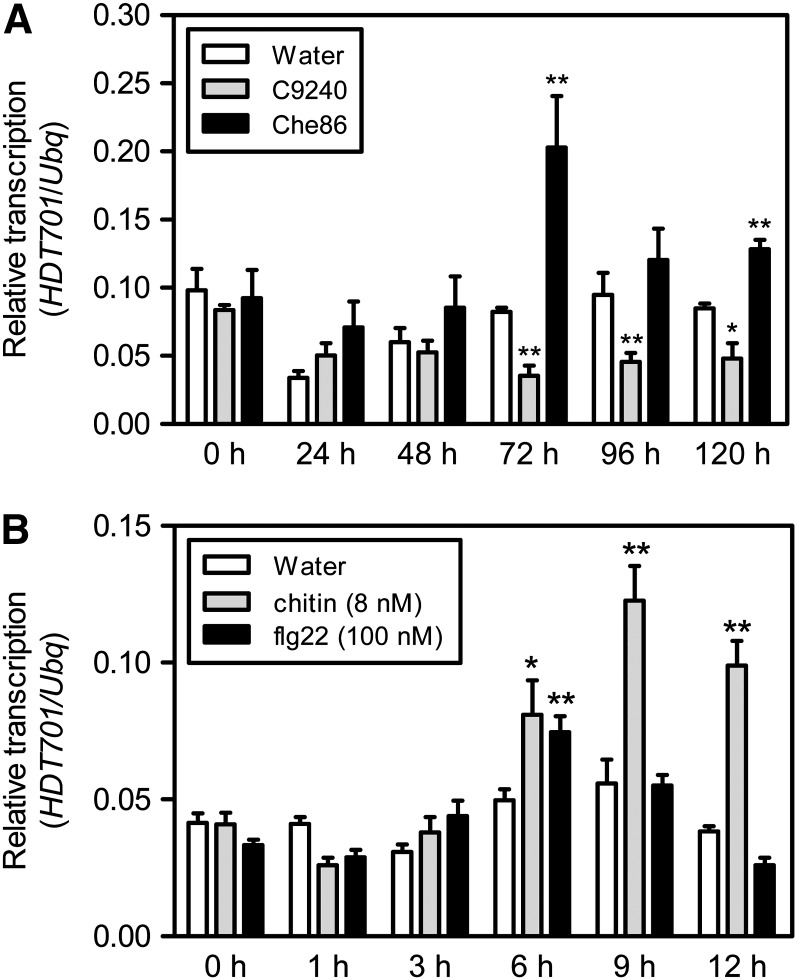
To identify chromatin-related genes involved in the defense response to M. oryzae and Xoo, we searched the transcription profiles of all chromatin-related genes in the rice massively parallel signature sequencing (MPSS) database (http://mpss.udel.edu/rice/). Among the genes that were induced or repressed by one or both pathogens, HDT701 showed an opposite expression during compatible versus incompatible reactions (i.e., the transcription level of HDT701 was increased in the compatible reaction but decreased in the incompatible reaction). To confirm the data from the MPSS libraries, we performed quantitative RT-PCR using the RNA samples isolated from the leaves of wild-type rice cv Nipponbare plants inoculated with the compatible M. oryzae isolate Che86, the incompatible isolate C9240, or water as the control at 0, 24, 48, 72, 96, and 120 h. Consistent with the MPSS data, the transcription level of HDT701 started to increase between 48 and 72 h in the compatible reaction, whereas it decreased after 72 h during the incompatible response and remained unchanged in the water control after inoculation (Figure 1A). Next, we investigated whether the transcription of HDT701 was affected by treatment with the PAMP elicitors chitin or flg22. Quantitative RT-PCR analysis was performed using the RNA isolated from rice leaf disks treated with chitin or flg22 at different time points (0, 1, 3, 6, 9, and 12 h). The analysis showed the transcription level of HDT701 was increased at 6 h and reached a peak at 9 h after chitin treatment (Figure 1B). By contrast, the transcription of HDT701 was increased at 3 h and reached a peak at 6 h after flg22 treatment (Figure 1B). These results suggested that HDT701 might play a role in rice PAMP-triggered immunity (PTI).
Figure 1.
Expression of HDT701 in Nipponbare after M. oryzae Inoculation and PAMP Elicitor Treatments.
(A) Time-course transcription analysis of HDT701 in the compatible interaction with M. oryzae isolate Che86, in the incompatible interaction with M. oryzae isolate C9240, and in the water treatment by quantitative RT-PCR. Error bars indicate the sd from three biological replicates (n = 3), and asterisks indicate statistically significant differences compared with water treatment (t test, *P < 0.05, **P < 0.01).
(B) Time-course transcription analysis of HDT701 after chitin, flg22, and water treatments by quantitative RT-PCR analysis. Error bars indicate the sd from three biological replicates (n = 3), and asterisks indicate statistically significant differences compared with water treatment (t test, *P < 0.05, **P < 0.01).
HDT701 Is Expressed during the Entire Rice Life Cycle, and the Protein Is Localized in the Nucleus
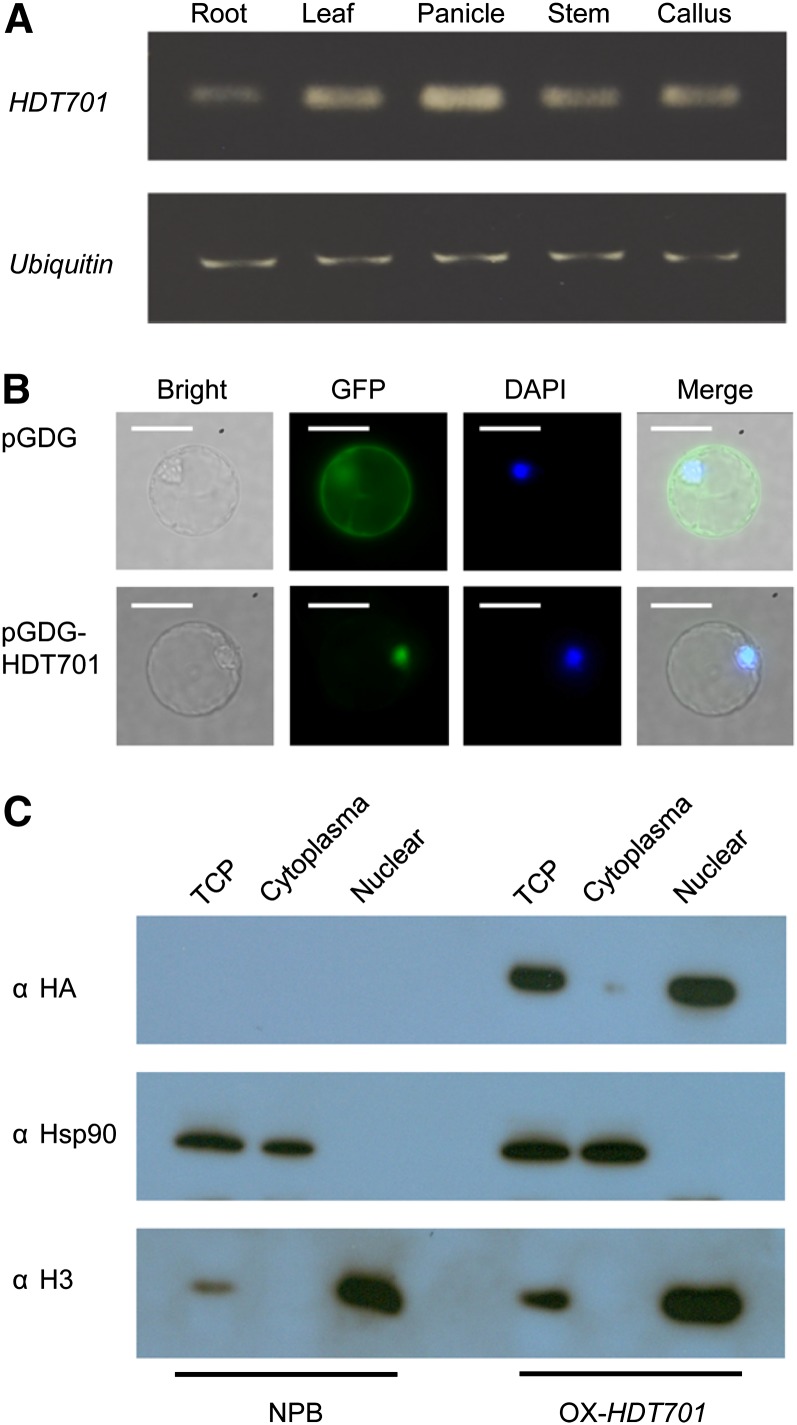
To explore the biological role of the HDT701 gene in rice, we next examined the temporal and spatial transcription of HDT701 in different rice tissues using the rice ubiquitin gene as the internal control. We found that HDT701 was expressed throughout the lifecycle of the rice plants and in almost all rice tissues examined, with a slightly higher expression in the flower organs (Figure 2A). To determine the subcellular localization of HDT701 in rice cells, we fused the coding region of HDT701 with the green fluorescent protein (GFP) fragment under the control of the cauliflower mosaic virus 35S promoter and expressed it transiently in rice protoplasts via the polyethylene glycol–mediated transfection method (Chen et al., 2006). Fluorescence microscopy indicated that the GFP signal was localized in the nucleus of the transfected rice protoplasts, whereas the green fluorescent signal in the GFP control vector was universally distributed in the rice protoplast cells (Figure 2B). We also detected a hemagglutinin (HA)-tagged HDT701 protein in different subcellular fractions isolated from overexpression (OX) plants (OX-HDT701) (see below) using heat shock protein90 (HSP90) as a cytoplasmic protein marker and histone H3 as a nuclear protein marker. The immunoblot analysis demonstrated that the HA-tagged HDT701 protein predominantly accumulated in the nucleus (Figure 2C).
Figure 2.
Expression of HDT701 in Rice Plants and Subcellular Localization of the Protein.
(A) Quantitative RT-PCR analysis of HDT701 transcription in different tissues of Nipponbare plants.
(B) Epifluorescence microscopy images deriving from GFP or 4′, 6-diamidino-2-phenylindole (DAPI) of rice protoplasts transiently expressing GFP or GFP-HDT701 fusion protein.
(C) Immunoblot detection of HA-tagged HDT701 protein in different subcellular fractions isolated from the HDT701 OX plants. NPB, Nipponbare; TCP, total cell protein.
Bars in (B) = 10 μm.
HDT701 Regulates the Levels of Histone H4 Acetylation in Rice Plants
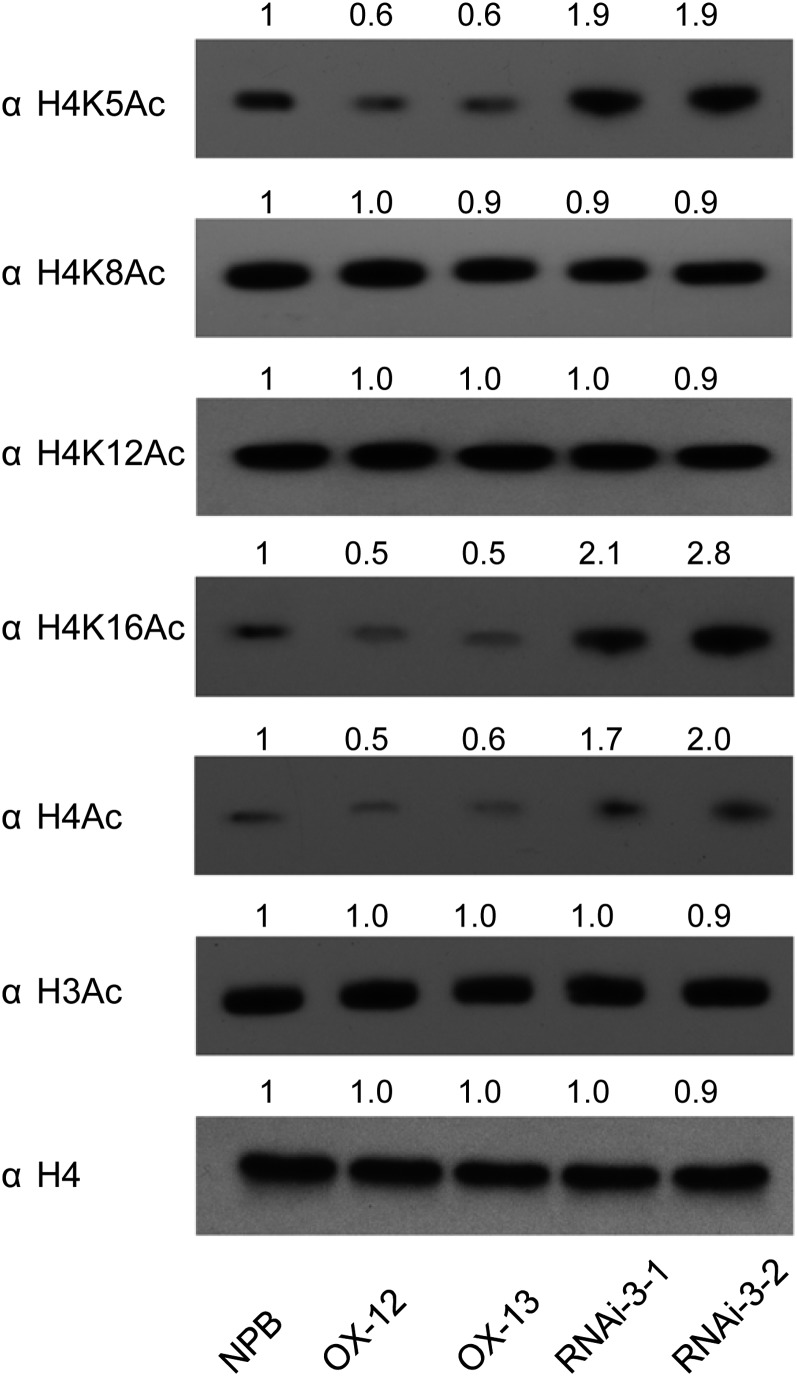
Although several plant HDACs, such as Os-SIRT1 and At-HDA19, have been reported to regulate histone H3 acetylation levels in planta (Zhou et al., 2005; Huang et al., 2007), little information is available regarding whether the HD2 family of HDACs can affect histone acetylation levels. To examine the putative histone deacetylase function of HDT701, we isolated the nuclear-rich protein from OX and RNA interference (RNAi) transgenic plants of HDT701 (see below) and performed immunoblot analysis using antibodies against bulk H3 or H4 acetylation and an antibody against unmodified histone H3 as the loading control. The immunoblot analysis clearly showed that the histone H3 acetylation level was not changed in the OX and RNAi plants but that the histone H4 acetylation level was decreased in the OX plants and increased in the RNAi plants compared with wild-type Nipponbare plants (Figure 3). K5, K8, K12, and K16 are four Lys residues on the histone H4 N terminus that can be acetylated. To determine which of these Lys residues can be deacetylated by HDT701, we performed immunoblot analysis using antibodies against different acetylated Lys residues of histone H4. The acetylation levels of histone H4K5 and H4K16 were increased in the RNAi plants but were decreased in the OX plants (Figure 3). However, we could not detect obvious changes in the levels of H4K8 and H4K12 acetylation with the antibodies tested (Figure 3). Thus, we concluded that HDT701 is an active histone H4 deacetylase that can regulate histone H4K5 and H4K16 acetylation levels in planta. Considering the altered transcription of HDT701 after M. oryzae infection (Figure 1A), we determined the global acetylation levels of H4K5 and H4K16 in infected Nipponbare plants. The assays showed that there were no obvious changes at the global acetylation levels of H4K5 and H4K16 in both compatible and incompatible reactions (see Supplemental Figure 1 online), possibly because of the limited sensitivity of immunoblotting and/or less transcriptional changes of HDT701 in infected Nipponbare plants compared with the HDT701 OX and RNAi plants.
Figure 3.
Histone Modifications of the Wild-Type Nipponbare (NPB), HDT701 OX, and HDT701 RNAi Plants.
Histone-enriched protein isolated from 4-week-old rice leaves and the covalent modification status of histones were analyzed by immunoblotting using the antibodies against different histone-modification modules as indicated. The band intensities of various histone modifications from OX and RNAi plants are shown as numbers normalized to Nipponbare (NPB) levels above the figure; the intensity of Nipponbare was set to 1.
OX of HDT701 in Transgenic Rice Increases Susceptibility to Rice Pathogens
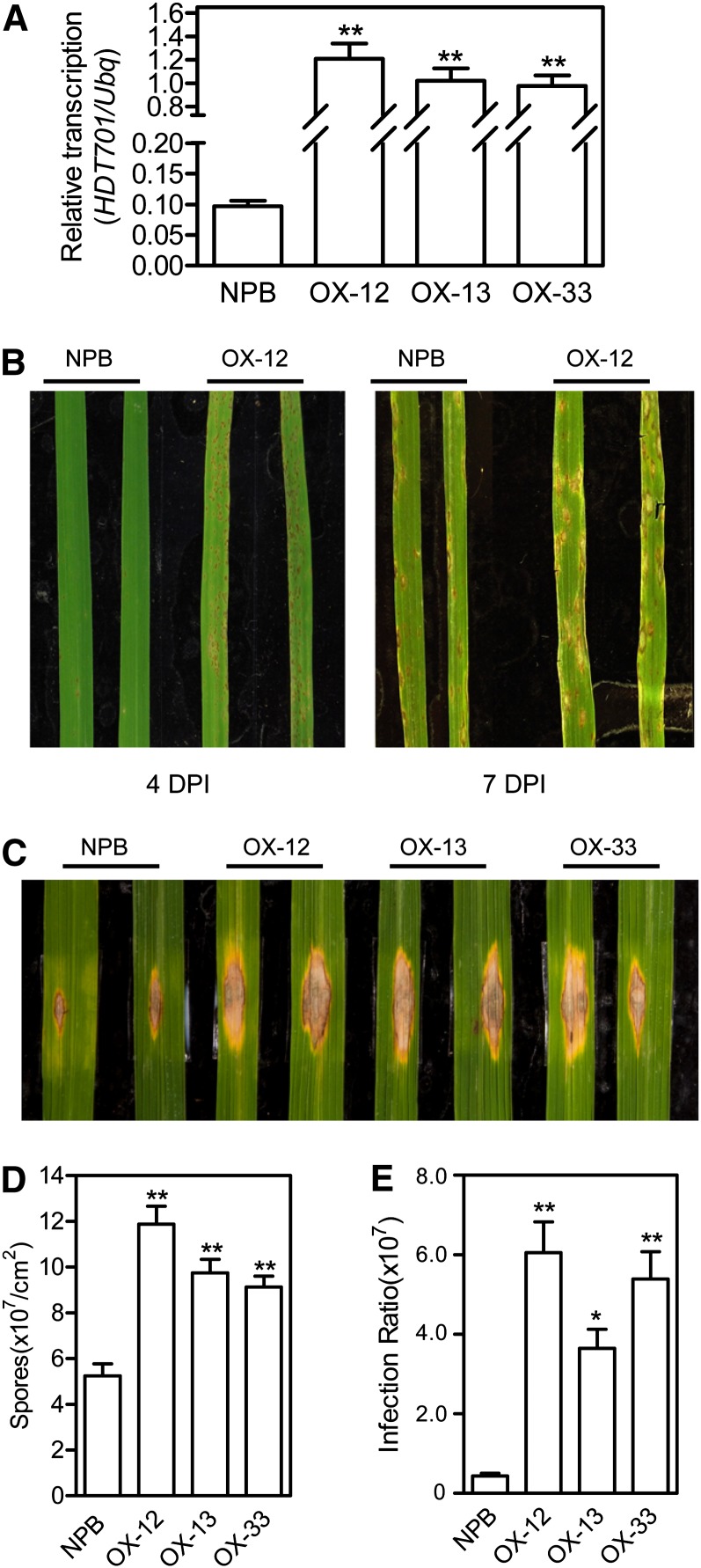
To elucidate the function of HDT701 in defense responses to rice pathogens, we made an OX construct in which the HDT701 full-length cDNA was fused with the HA epitope tag at its 5′ terminus under the control of the maize ubiquitin promoter. The construct was transformed into Nipponbare via the Agrobacterium tumefaciens-mediated transformation method. The responses of three of 20 OX-HDT701 independent homozygous lines to both M. oryzae and Xoo were studied. Quantitative RT-PCR analysis confirmed the increased transcription of HDT701 in the transgenic lines (Figure 4A). Consistent with previous findings (Hu et al., 2009), the morphology of the OX-HDT701 plants did not differ from that of Nipponbare plants.
Figure 4.
Disease Phenotypes of the HDT701 OX Plants.
(A) Transcription analysis of the HDT701 OX plants by quantitative RT-PCR. Error bars indicate the sd from three biological replicates (n = 3), and asterisks indicate statistically significant differences compared with Nipponbare (NPB) (t test, **P < 0.01).
(B) Disease phenotypes of the HDT701 OX plants and wild-type Nipponbare plants after spray inoculation with M. oryzae isolate CHNOS. DPI, days postinoculation.
(C) Disease phenotypes of the HDT701 OX plants and wild-type Nipponbare plants after punch inoculation with M. oryzae isolate Che86.
(D) Numbers of spores produced by the HDT701 OX plants and wild-type Nipponbare plants after punch inoculation. Error bars indicate the sd from eight biological replicates (n = 8), and asterisks indicate statistically significant differences compared with Nipponbare (t test, **P < 0.01).
(E) Relative fungal biomass in the HDT701 OX plants and wild-type Nipponbare plants after punch inoculation. Error bars indicate the sd from eight biological replicates (n = 8), and asterisks indicate statistically significant differences compared with Nipponbare (t test, *P< 0.05, **P<0.01).
First, we inoculated 3-week-old OX-HDT701 plants with the compatible M. oryzae isolate CHNOS, which showed weak virulence on Nipponbare plants, by the spray-inoculation method. Disease symptoms, such as cell death lesions, developed faster on the OX-HDT701 plants than on the wild-type Nipponbare plants at 4 d after inoculation (DAI) (Figure 4B, left). At 7 DAI, disease symptoms were more severe on the OX-HDT701 plants (Figure 4B, right). To confirm the result from the spray inoculation, we evaluated the disease symptoms of the OX-HDT701 plants after punch inoculation (Ono et al., 2001) with the compatible M. oryzae isolates Che86 and RO1-1 (these isolates were used because punch inoculation with isolate CHNOS did not result in large lesions). Among 6-week-old plants that had been punch inoculated, lesions induced by both isolates were larger on the OX-HDT701 plants than on the Nipponbare plants (Figure 4C; see Supplemental Figure 2A online). The number of blast spores on the inoculated leaves was also significantly greater on the OX-HDT701 leaves than on Nipponbare leaves (Figure 4D). To measure fungal biomass in the inoculated leaves accurately, we isolated the total DNA from the infected rice leaves and quantified fungal biomass using a DNA-based quantitative PCR method with two sets of specific primers against M. oryzae Pot2 and rice ubiquitin (Kawano et al., 2010). The analysis showed that fungal biomass was greater in the infected OX-HDT701 leaves than in the wild-type Nipponbare leaves (Figure 4E). In addition to examining resistance to compatible M. oryzae isolates, we also inoculated the OX-HDT701 plants with the incompatible M. oryzae isolate C9240 and found that there was no lesion development on the OX-HDT701 plants or on the Nipponbare or RNAi knockdown plants (see Supplemental Figures 2B and 2C online) and no difference in the relative fungal biomass among Nipponbare, OX, and RNAi plants (see Supplemental Figure 2B online), which suggested that HDT701 is not involved in resistance gene–mediated immunity or effector-triggered immunity (ETI) in rice plants.
Next, we examined the resistance of the OX-HDT701 plants to the bacterial blight pathogen Xoo. The OX-HDT701 plants were inoculated with Xoo race 6 using the scissor-clipping method (Wang et al., 1996). Disease symptoms were evaluated by measuring lesion length at 12 DAI (see Supplemental Figure 2D online). The lesions were ∼30% longer on the OX-HDT701 plants than on the Nipponbare plants (see Supplemental Figure 2E online).
Taken together, the results from both M. oryzae and Xoo inoculations demonstrate that OX of HDT701 in rice plants enhances the susceptibility to both pathogens.
Knockdown of HDT701 in Transgenic Rice Enhances Resistance to Rice Pathogens
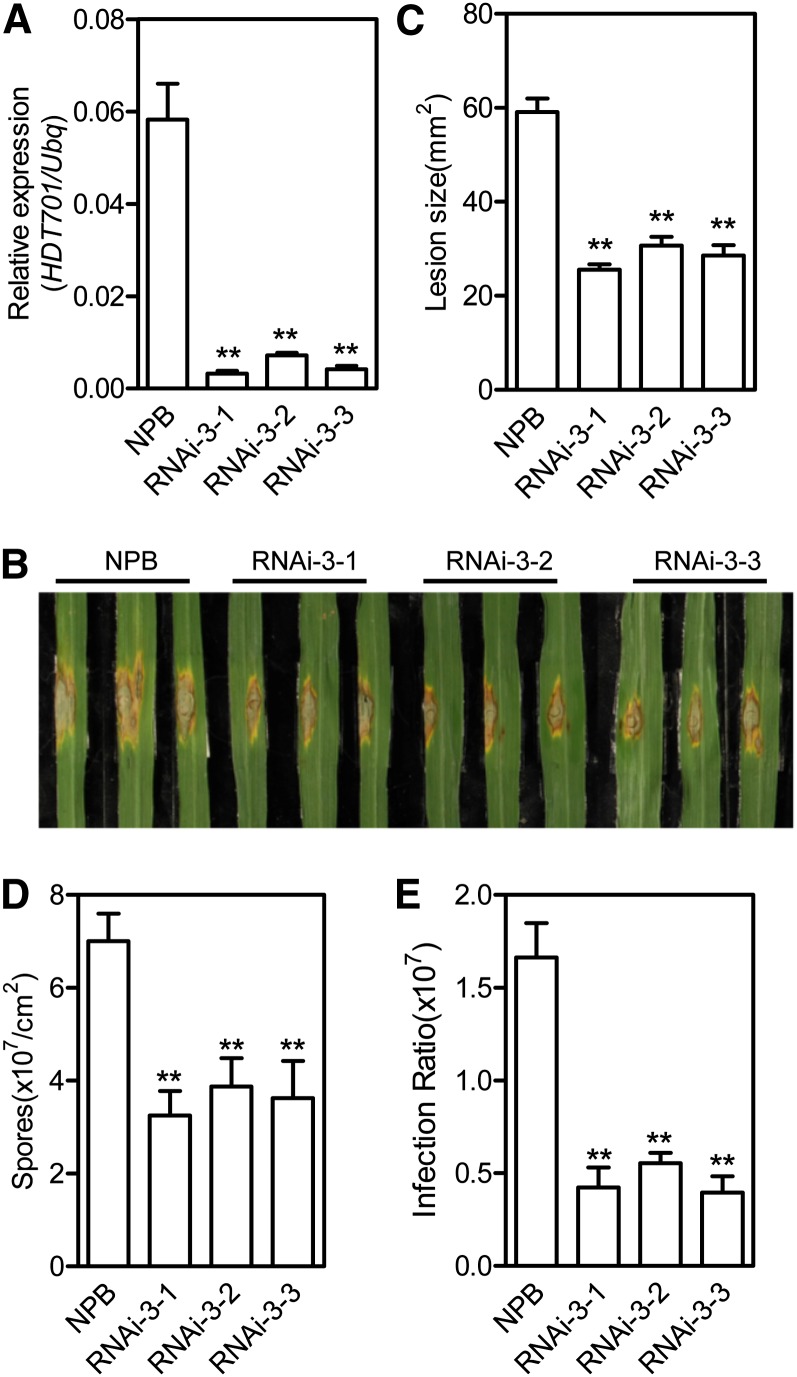
To elucidate further the function of HDT701 in response to rice pathogens, we generated HDT701 RNAi transgenic plants using two RNAi constructs, RNAi-3 and RNAi-5, which were designed to target the specific 3′ and 5′ untranslated regions (UTRs) of HDT701, respectively. We first examined the silencing effect of RNAi-3 using quantitative RT-PCR (Figure 5A). HDT701 RNAi plants did not show any observable morphological changes compared with wild-type Nipponbare plants. After punch inoculation, lesions were smaller on RNAi-3 RNAi plants than on Nipponbare plants (Figure 5B). Development of disease symptoms was quantified by measuring lesion size at 12 DAI. The lesion size on HDT701 RNAi-3 plants was approximately one-half of that on Nipponbare plants (Figure 5C). In addition, fungal sporulation and relative fungal biomass assays showed that fewer spores and less fungal biomass were associated with the HDT701 RNAi-3 plants compared with wild-type Nipponbare plants, indicating that fungal growth was suppressed in the HDT701 RNAi plants (Figures 5D and 5E). Similarly, an enhanced resistance phenotype to M. oryzae was observed in RNAi-5 transgenic rice plants (see Supplemental Figures 3A and 3B online). To measure the resistance level of the HDT701 RNAi plants to Xoo, we inoculated the RNAi-3 plants by the scissor-clipping method. The lesions were ∼20% shorter on HDT701 RNAi plants than on Nipponbare plants (see Supplemental Figures 3C and 3D online), indicating that knockdown of HDT701 in rice plants increases resistance to Xoo. After considering all of the inoculation results obtained from both the HDT701 OX and RNAi plants, we conclude that HDT701 is a negative regulator of disease resistance in rice.
Figure 5.
Disease Phenotypes of the HDT701 RNAi Plants.
(A) Transcription analysis of the HDT701 gene in the RNAi-3 transgenic plants by quantitative RT-PCR. Error bars indicate the sd from three biological replicates (n = 3), and asterisks indicate statistically significant differences compared with Nipponbare (NPB) (t test, **P < 0.01).
(B) Disease phenotypes of the HDT701 RNAi-3 plants and wild-type Nipponbare plants after punch inoculation with M. oryzae isolate RO1-1.
(C) Lesion sizes on the HDT701 RNAi-3 plants and wild-type Nipponbare plants after punch inoculation. Error bars indicate the sd from eight biological replicates (n = 8), and asterisks indicate statistically significant differences compared with Nipponbare (t test, **P < 0.01).
(D) Numbers of spores produced on the HDT701 RNAi-3 plants and wild-type Nipponbare plants after punch inoculation. Error bars indicate the sd from eight biological replicates (n = 8), and asterisks indicate statistically significant differences compared with Nipponbare (t test, **P < 0.01).
(E) Relative fungal biomass of the HDT701 RNAi-3 plants and wild-type Nipponbare plants after punch inoculation. Error bars indicate the sd from eight biological replicates (n = 8), and asterisks indicate statistically significant differences compared with Nipponbare (t test, **P < 0.01).
HDT701 Is Involved in PTI
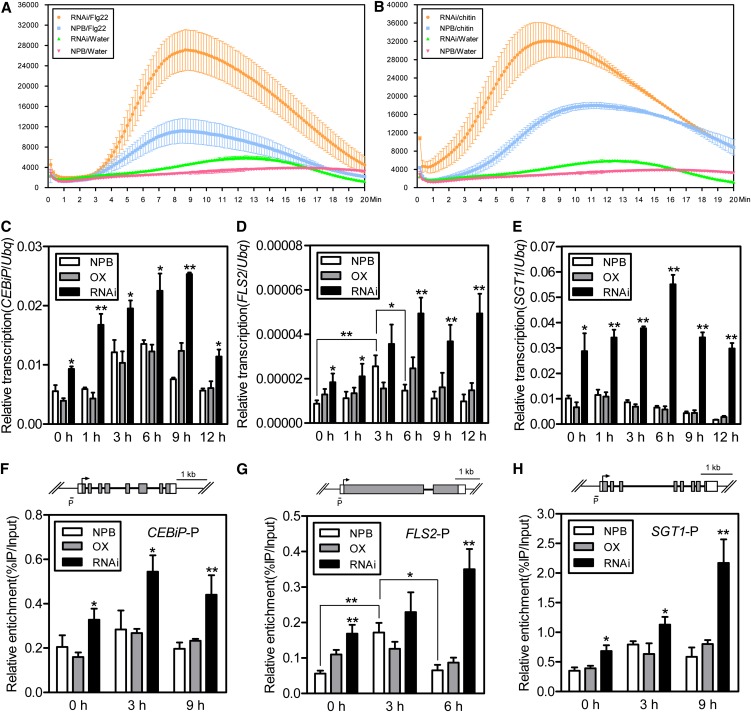
The mildly resistant phenotype of the HDT701 RNAi plants, the induced transcription of HDT701 after PAMP elicitor treatment, and the lack of disease symptoms in the incompatible reaction in both the HDT701 OX and knockdown plants suggested that HDT701 might be involved in modulating PTI rather than ETI, because the latter would have caused more robust and stronger defense responses. To test this hypothesis, we examined the responses of the Nipponbare, HDT701 OX, and RNAi plants to the PAMP elicitors flg22 and chitin. Leaf disks from 6-week-old plants were separately immersed in chitin or flg22 solution, and the level of ROS was measured using the luminol chemiluminescence assay (Schwacke and Hager, 1992). In flg22- or chitin-treated tissues, knockdown of HDT701 resulted in a twofold increase in ROS production compared with wild-type Nipponbare (Figures 6A and 6B). Unexpectedly, we could not detect the significant ROS changes in the OX plants compared with that in the Nipponbare plants in both flg22 and chitin treatments (see Supplemental Figures 4A and 4B online). To elucidate the molecular mechanism underlying the enhanced ROS generation in the HDT701 RNAi plants, we monitored the dynamic expression changes of several pattern recognition receptor (PRR) and PTI-related genes using quantitative RT-PCR at different time points (0, 1, 3, 6, 9, and 12 h). Interestingly, we found that the transcription levels of FLS2, CEBiP, and SGT1 were increased in the HDT701 RNAi plants, even in leaves that were not treated with the PAMP elicitors (Figures 6C to 6E). It is noteworthy that the transcription level of FLS2 in the wild-type Nipponbare plants was also significantly increased at 3 h and reduced to the basal level 6 h after flg22 treatment (Figure 6D).
Figure 6.
ROS Generation, Defense Gene Expression, and Histone Acetylation Levels in the HDT701 OX and RNAi Plants.
(A) ROS level in HDT701 RNAi plants after flg22 treatment. Error bars indicate the sd (n = 3). NPB, Nipponbare.
(B) ROS level in HDT701 RNAi plants after chitin treatment. Error bars indicate the sd (n = 3).
(C) Time-course transcription analysis of CEBiP in Nipponbare (NPB), HDT701 OX, and RNAi plants after chitin treatment. Error bars indicate the sd from three biological replicates (n = 3), and asterisks indicate statistically significant differences compared with Nipponbare (t test, *P < 0.05, **P < 0.01).
(D) Time-course transcription analysis of FLS2 in Nipponbare, HDT701 OX, and RNAi plants after flg22 treatment. Error bars indicate the sd from three biological replicates (n = 3), and asterisks indicate statistically significant differences compared with Nipponbare or between the two comparisons marked with brackets (t test, *P < 0.05, **P < 0.01).
(E) Time-course transcription analysis of SGT1 in Nipponbare, HDT701 OX, and RNAi plants after chitin treatment. Error bars indicate the sd from three biological replicates (n = 3), and asterisks indicate statistically significant differences compared with Nipponbare (t test, *P < 0.05, **P < 0.01).
(F) to (H) Time-course histone H4 acetylation level of CEBiP, FLS2, and SGT1 in Nipponbare, HDT701 OX, and RNAi plants after chitin ([F] and [H]) and flg22 (G) treatments. Diagrams of the CEBiP, FLS2, and SGT1 gene structures are shown above their respective ChIP analysis panels, in which the gray boxes indicate exons, white boxes indicate UTRs, the arrow shows translation start point, the thick black lines between gray boxes indicate introns, and the letter P, together with the short black lines, indicate PCR fragments that correspond to the promoter regions. Error bars indicate the sd from three biological replicates (n = 3), and asterisks indicate statistically significant differences compared with Nipponbare or between the two comparisons marked with brackets (t test, *P < 0.05, **P < 0.01).
Histone acetylation is closely associated with gene transcription; therefore, we next examined the histone H4 acetylation level in the promoter regions of the PRR and PTI-related genes. Consistent with the transcription result, the chromatin immunoprecipitation (ChIP) analysis against the H4Ac antibody coupled with quantitative PCR showed that the histone H4 acetylation level in the promoter regions of FLS2, CEBiP, and SGT1 was higher in the HDT701 RNAi plants than in the wild-type Nipponbare and HDT701 OX plants after chitin or flg22 treatment (Figures 6F to 6H). Interestingly, the histone H4 acetylation level in Nipponbare plants was also significantly increased 3 h after flg22 treatment and reduced to the basal level at 6 h (Figure 6G). By contrast, the transcription and H4 acetylation levels of Spin1, a K homology domain gene involved in rice flowering time control (Vega-Sánchez et al., 2008), remained unchanged in the wild-type and HDT701 OX and RNAi plants after flg22 treatment (see Supplemental Figures 4C and 4E online) and chitin treatment (see Supplemental Figures 4D and 4F online). Considering that the transcription of HDT701 was induced after PAMP treatment and that HDT701 can regulate histone H4K5 and H4K16 acetylation levels in planta, we next examined the H4K5 and H4K16 acetylation levels of Nipponbare leaf tissues after PAMP treatment. However, we did not detect any obvious global acetylation level changes between different time points in both chitin- and flg22-treated Nipponbare samples using immunoblot analysis (see Supplemental Figure 5 online). These results indicate that HDT701 plays a negative role in rice PTI by suppressing the levels of histone acetylation and expression of the PTI-associated genes.
HDT701 Directly Binds to Defense-Related Genes to Regulate Their Expression
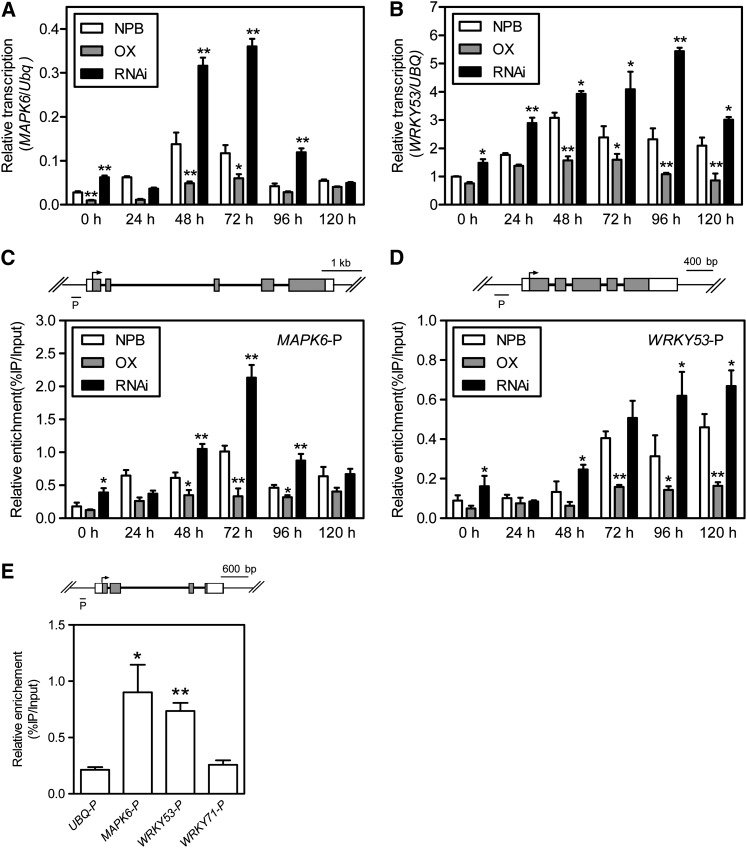
To explore further the mechanism of HDT701-mediated defense in M. oryzae-infected plants, we assayed the transcription level of several downstream defense-related genes in the HDT701 OX and RNAi plants after inoculation with the compatible M. oryzae isolate Che86 using quantitative RT-PCR. The analysis revealed that one mitogen-activated protein (MAP) kinase gene, MAPK6, and one transcription factor, WRKY53, were upregulated in the RNAi plants but downregulated in the OX plants (Figures 7A and 7B). In agreement with the transcription levels, the histone H4 acetylation levels were increased in the promoter regions of both MAPK6 and WRKY53 in the RNAi plants but were decreased in the OX plants (Figures 7C and 7D).
Figure 7.
Expression and Histone H4 Acetylation Levels of MAPK6 and WRKY53 in the HDT701 OX and RNAi Plants after M. oryzae Inoculation. NPB, Nipponbare.
(A) and (B) Time-course transcription analysis of MAPK6 and WRKY53, respectively, after M. oryzae inoculation. Error bars indicate the sd from three biological replicates (n = 3), and asterisks indicate statistically significant differences compared with Nipponbare (NPB) (t test, *P < 0.05, **P < 0.01).
(C) and (D) Time-course relative histone H4 acetylation level analysis on promoter region of MAPK6 and WRKY53, respectively, after M. oryzae inoculation. Diagrams of the MAPK6 and WRKY53 gene structures are shown above the ChIP analysis panel. Error bars indicate the sd from three biological replicates (n = 3), and asterisks indicate statistically significant differences compared with Nipponbare (t test, *P < 0.05, **P < 0.01).
(E) Relative binding of HA-HDT701 to the promoters of UBQ, MAPK6, WRKY53, and WRKY71. Diagram of the Ubiquitin gene structure is shown above the ChIP analysis panel. The diagrams of the MAPK6, WRKY53, and WRKY71 gene structure are shown in (C), (D), and Supplemental Figure 6D online, respectively. Error bars indicate the sd from three biological replicates (n = 3), and asterisks indicate statistically significant differences compared with ubiquitin promoter region (UBQ-P) (t test, *P < 0.05, **P < 0.01).
H4 acetylation acts as a general regulatory mechanism associated with gene transcription and global H4 acetylation changes in the HDT701 OX and RNAi plants; therefore, it is possible that HDT701-mediated histone deacetylation would affect both defense-related and non–defense-related genes. Analysis of the transcriptional and H4 acetylation levels of non–defense-related gene Spin1 revealed that its expression and H4 acetylation levels did not show differences among the Nipponbare, HDT701 OX, and RNAi plants (see Supplemental Figures 6A and 6C online). In addition, we found that the transcription and histone H4 acetylation levels of WRKY71 were induced at early inoculation time points (24 and 48 h) in the RNAi plants but also were induced at late time points (96 and 120 h) in the Nipponbare and OX plants (see Supplemental Figures 6B and 6D online), indicating that the transcription and H4Ac levels of WRKY71 were affected upon silencing of HDT701. However, considering that the late peak of the transcription and H4Ac levels of WRKY71 compared with that of MAPK6 and WRKY53 in M. oryzae–infected Nipponbare plants as well as the similar peak transcription and H4Ac levels of WRKY71 among the Nipponbare, OX, and RNAi plants after M. oryzae inoculation (see Supplemental Figures 6B and 6D online), we speculate that MAPK6 and WRKY53 but not WRKY71 might be direct targets of HDT701 in the defense pathway. To test this assumption, we next checked the possible binding of HDT701 to MAPK6, MAPK5, and WRKY71 using ChIP against the HA antibody in the OX plants. Quantitative PCR analysis showed that DNA ChIP immunoprecipitated with the HA antibody was enriched in the promoter region of MAPK6 and WRKY53 compared with the rice reference gene ubiquitin and WRKY71 (Figure 7E). Taken together, these results demonstrate that HDT701 can directly bind to a subset of defense-related genes and transcription factors to regulate their transcription during the interaction with M. oryzae.
DISCUSSION
Negative Regulation of HDT701 in Rice Innate Immunity
Epigenetic regulation, such as by histone modification and chromatin remodeling, plays important roles in the host defense against pathogens in animal systems (Hamon and Cossart, 2008). HDACs can positively or negatively regulate the Toll-like receptor pathway and interferon signaling pathways in animal innate immunity (Shakespear et al., 2011). However, only a few HDAC genes have been reported to regulate the defense pathways in plants. Arabidopsis HDA19, one of the RPD3-type histone deacetylase genes, plays a positive role in basal resistance to pathogens (Zhou et al., 2005). The transcription of HDA19 is induced by jasmonic acid and ethylene. Arabidopsis plants overexpressing HDA19 display enhanced resistance to the fungal pathogen Alternaria brassicicola and increased expression of ERF1, suggesting that HDA19 may be involved in the jasmonic acid–mediated defense pathway against necrotrophic pathogens (Zhou et al., 2005). HDA19 also physically interacts with and represses the transcriptional activity of two WRKY family transcription factors, WRKY38 and WRYK62, both of which act as negative regulators in plant basal defense (Kim et al., 2008). A recent report showed that HDA19 was involved in the repression of salicylic acid (SA)–mediated defense responses in Arabidopsis (Choi et al., 2012). Another Arabidopsis HDAC, SRT2, which is a homolog of yeast Sir2, negatively regulates plant basal defense against the pathogen Pseudomonas syringae pv tomato DC3000 by suppressing the transcription of the SA biosynthesis-related genes PAD4, EDS5, and SID2 (Wang et al., 2010). Whether the repression of these SA-related genes is regulated directly by SRT2, however, remains unclear.
In this study, we performed an in-depth functional analysis of HDT701 in rice innate immunity. Initially, we showed that the transcription of HDT701 is reduced in the incompatible reaction but induced in the compatible reaction, suggesting that rice pathogens might target HDT701 so as to suppress the defense response during infection. We also demonstrated that the HDT701 OX plants are more susceptible to rice pathogens, whereas the HDT701 RNAi plants are more resistant, which indicates that HDT701 has a negative role in the rice defense response. Finally, we showed that HDT701 is involved in the PTI-mediated pathway by modulating the histone acetylation and expression levels of defense-related genes.
HDT701 Function on Key PTI-Related Components
Plants have evolved two layers of defense responses against pathogen invasion: PTI and ETI (Jones and Dangl, 2006). PTI is activated when PAMP elicitors are recognized by PRRs in plants, which stimulates basal defense to pathogens. On the other hand, ETI is activated when the actions and/or structures of avirulence gene products are recognized by the cognate resistance proteins in plants, leading to a robust and rapid defense response, such as the hypersensitive reaction. Considering that HDT701 RNAi plants exhibit a relatively mild resistance to rice pathogens, we speculate that the HDT701 gene is involved in rice PTI. This speculation is supported by the results of the expression profiles and histone H4 acetylation levels of several PRR and PTI-related genes and also by the generation of ROS after flg22 and chitin treatment in the HDT701 RNAi transgenic plants.
Chitin is a major component of the fungal cell wall and acts as a PAMP elicitor that triggers PTI in several plant species (Wan et al., 2008). CEBiP, the plasma membrane glycoprotein with two extracellular LysM motifs, is important for chitin perception and signal transduction in rice (Kishimoto et al., 2010; Shimizu et al., 2010). CEBiP can form homo-oligomers or hetero-oligomers with a receptor kinase CERK1 (Shimizu et al., 2010). Knockdown of CEBiP in cultured rice cells decreases defense responses to chitin, and transgenic rice plants with knockdown CEBiP are more susceptible than the wild type to rice blast infection (Kishimoto et al., 2010). Another PAMP elicitor, flagellin, is derived from flagellated bacteria and triggers immune responses in animals and plants. In Arabidopsis, the flagellin signal is perceived by the leucine-rich repeat transmembrane receptor kinase flagellin sensitive2 (FLS2) (Gómez-Gómez and Boller, 2000). Os-FLS2, the rice ortholog of the Arabidopsis flagellin receptor FLS2, is essential for flagellin signal perception in rice. Overexpression of Os-FLS2 in cultured rice cells generated stronger immune reactions (Takai et al., 2008). In this study, we found that both the transcription and histone H4 acetylation levels of CEBiP and FLS2 are higher in the HDT701 RNAi plants than in wild-type plants (Figures 6C, 6D, 6F, and 6G), indicating a negative role of HDT701 in PAMP signal perception. Considering that transcription of HDT701 is induced under chitin or flagellin treatment, it is possible that pathogens suppress both CEBiP- and FLS2-mediated pathways by modulating the transcription of HDT701 after infection. The elevated expression of HDT701 might cause negative feedback regulation to weaken PTI in infected plants, as we observed in the OX transgenic lines. As shown in Figure 6G, the FLS2 H4Ac level in the wild-type Nipponbare plant was significantly increased 3 h after flg22 treatment and was reduced to the basal level 6 h after the treatment. By contrast, the FLS2 H4Ac level in the OX transgenic rice was not changed after the treatment. We speculate that the overexpression of HDT701 might activate an unknown feedback mechanism that prevents the increase of the FLS2 H4Ac level in the transgenic rice after flg22 treatment. Another possibility is that the negative feedback role of HDT701 is downstream of FLS2, which is a membrane protein and acts as the receptor of flg22.
PAMP elicitors trigger a series of defense responses, including ROS generation. Os-Rac1, a member of the Pho-type small GTPase family, is a key modulator of innate immunity and positively regulates disease resistance. Rac1 functions through the RAR1-SGT1-HSP cytosolic defensome complex and mediates accumulation of the ROS generated by NADPH oxidases (Nakashima et al., 2008). We found that the transcription levels and the histone H4 acetylation levels of SGT1 were higher in the HDT701 RNAi plants than in the wild-type plants (Figure 6E). In addition, ROS generation was higher in the HDT701 RNAi plants than in wild-type plants after flg22 and chitin treatment. These results demonstrate that SGT1 is regulated at least partially by HDT701 via its HDAC activity (Figure 6H). Intriguingly, we could not detect significant changes in ROS generation after PAMP treatment in the OX-HDT701 plants relative to Nipponbare plants (see Supplemental Figures 4A and 4B online), which is consistent with the unchanged transcription and histone H4 acetylation levels of SGT1 in these plants, suggesting that other feedback regulation mechanisms might exist in the OX-HDT701 plants.
HDT701 Regulation of the Expression of Defense-Related Genes
In the plant defense signaling pathway, MAP kinase (MAPK) pathway components and WRKY transcription factors are important players that modulate rice innate immunity (Chen and Ronald, 2011). MAPK cascades transduce defense signals to downstream proteins via protein phosphorylation (Zhang and Klessig, 2001). Several MAPKs have been documented to regulate plant defense response as well as other signaling pathways. For example, the abscisic acid–induced Os-MAPK5 can positively regulate tolerance to drought, salt, and cold. Conversely, Os-MAPK5 can negatively modulate defense gene expression and disease resistance to M. oryzae (Xiong and Yang, 2003). Os-MAPK6 is posttranslationally activated by the fungal PAMP elicitor chitin and is required for chitin elicitor-induced biosynthesis of diterpenoid phytoalexin, which acts as a toxin that suppresses M. oryzae growth (Kishi-Kaboshi et al., 2010). In addition, the MAPK6-mediated cascade is activated by the RAC1 defensome complex to transduce PIT-mediated immunity (Lieberherr et al., 2005; Kawano et al., 2010). Similarly, the WRKY transcription factor family has been extensively studied for its role in the rice defense response (Liu et al., 2005; Liu et al., 2007; Qiu et al., 2007; Shimono et al., 2007; Chujo et al., 2008; Peng et al., 2008; Tao et al., 2009). Overexpression of WRKY53 induced after chitin treatment in cultured rice cells (Chujo et al., 2007) and in rice plants leads to enhanced resistance to M. oryzae, indicating its positive role in basal defense (Chujo et al., 2009). Expression of WRKY71 is induced by pathogen infection, and overexpressing WRKY71 in rice cells leads to upregulation of several elicitor-induced defense-related genes (Chujo et al., 2008). In our transcriptional analysis and ChIP analysis, we found that the transcription and histone acetylation levels of MAPK6 and WRKY53 were increased in the RNAi plants but decreased in the OX plants. Moreover, we observed that HDT701 could bind to the promoter region of MAPK6 and WRKY53 but not WRKY71 (Figure 7E), suggesting that HDT701 may regulate basal resistance through a specific group of defense-related genes. Although we demonstrated that several components in the defense pathways are negatively modulated by HDT701, additional defense-related genes modulated by HDT701 might be detected by a whole-genome transcriptome analysis and ChIP sequencing assay of the RNAi plants.
METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa ssp japonica cv Nipponbare) was used in this study. Rice seeds were surface-sterilized and transferred to one-half–strength Murashige and Skoog medium. After germination, rice seedlings were transplanted into soil and kept in a growth chamber at 26/20°C under a 14-h light/10-h dark cycle or in a greenhouse at 25°C.
Rice Blast Inoculation
Isolates C9240, Che86, CHNOS, and RO1-1 of Magnaporthe oryzae were used for inoculations. For spray inoculation, 3-week-old rice seedlings were sprayed with a spore suspension (1.2 × 105 spores/mL) containing 0.05% Tween-20. Six-week-old plants were punch inoculated as described previously (Ono et al., 2001). Briefly, 10 μL of a spore suspension (5.0 × 105 spores/mL) containing 0.05% Tween-20 was added to the press-injured spots on fully expanded rice leaves; the inoculated spots were wrapped with transparent scotch tape, inoculated leaves were photographed 12 DAI, and lesion size was measured using ImageJ (http://rsbweb.nih.gov/ij/). For determination of in planta sporulation after punch inoculation, leaf strips containing a lesion spot were excised and submerged in 200 μL of distilled water containing 0.05% Tween-20 in a 1.5-mL microcentrifuge tube. After the suspension was vigorously mixed, spores were counted with a microscope. Relative infection ratio was determined using DNA-based quantitative PCR as described previously (Kawano et al., 2010).
Gene Transcription Analysis
Total RNA was extracted from rice tissues using TRIzol reagent (Invitrogen) and was treated with DNaseI (Invitrogen) to remove genomic DNA contamination. The first strand of cDNA was synthesized from 1 μg of total RNA using the Reverse Transcription System (Promega) according to the manufacturer’s instructions. Quantitative PCR were performed using SYBR Green Supermix (Bio-Rad) on an ImyiQ2 real-time PCR detection system (Bio-Rad), and data were analyzed using IQ5 software (Bio-Rad). Gene-specific primers that were used are listed in Supplemental Table 1 online.
Subcellular Localization in Rice Protoplasts
Rice protoplast isolation and transfection were performed as described previously (Chen et al., 2006). To generate the pGDG-HDT701 construct, we obtained the full-length of HDT701 open reading frame from Nipponbare leaf cDNA by PCR using primers HDT701-OG-F/R and subcloned the fragment into the pGDG vector after digestion with BamHI (Goodin et al., 2002). A 1-μg quantity of pGDG-HDT701 plasmid was transfected into rice protoplasts using the polyethylene glycol 400–mediated transfection method, and transfected rice protoplasts were observed with a Nikon Eclipse E600 fluorescence microscope (Nikon). Images were captured with a SPOT 2 Slider charge-coupled camera.
Nuclear-Cytoplasmic Protein Fractionation
A total of 1 g of rice leaves from 4-week-old plants was ground in liquid nitrogen into fine powder and resuspended in 5 mL of buffer A (0.4 M of Suc, 10 mM of Tris, pH 8.0, 10 mM of MgCl2, 0.1 mM of phenylmethylsulfonyl fluoride [PMSF], 5 mM of β-mercaptoethanol, 1× protease inhibitor cocktail). After it was passed through two layers of Miracloth, the filtrate was centrifuged at 1500g for 20 min to separate the crude cytoplasmic and nuclear fraction. The supernatant, which contained the cytoplasmic fraction, was centrifuged at 16,000g for 15 min to remove the cellular debris. The pellet, which contained the nuclear fraction, was purified by washing in buffer B (0.25 M of Suc, 10 mM of Tris, pH 8.0, 10 mM of MgCl2, 1% Triton X-100, 0.1 mM of PMSF, 5 mM of β-mercaptoethanol, 1× protease inhibitor cocktail) and buffer C (1.7 M of Suc, 10 mM of Tris, pH 8.0, 2 mM of MgCl2, 0.15% Triton X-100, 0.1 mM of PMSF, 5 mM of β-mercaptoethanol, 1× protease inhibitor cocktail) followed by centrifuging at 16,000g at 4°C. The final pellet was resuspended in 200 μL of nuclear lysis buffer (20 mM of Tris-HCl, pH 7.5, 20 mM of KCl, 2 mM of EDTA, 2.5 mM of MgCl2, 20% glycerol, 250 mM of Suc, and 5 mM of DTT).
Plasmid Construction and Rice Transformation
To generate the overexpression vector of HDT701, we obtained the full-length coding region of the gene from Nipponbare leaf cDNA by PCR using primers HDT701-OG-F/R and cloned the fragment into the pCXUN-HA vector between the maize (Zea mays) ubiquitin promoter and the nopaline synthase terminator (Chen et al., 2009). To generate the RNAi vectors of HDT701, we PCR-amplified the specific regions of 5′ UTR or 3′ UTR of the gene and cloned the fragment into the pENTR entry vector (Invitrogen). With the gateway system, the target fragment was cloned into the pANDA vector (Miki et al., 2005). These plasmids were transferred into the calli induced from the mature seeds of Nipponbare using the Agrobacterium tumefaciens–mediated rice transformation method as previously described (Qu et al., 2006).
Histone Extraction
Histone-enriched proteins were extracted from rice leaves using the sulfuric acid–extraction method as described previously (Ding et al., 2010). Briefly, nuclei isolated from 2 g of rice leaf tissue were incubated in 0.4 M of H2SO4 for 45 min and precipitated in 20% trichloroacetic acid. The pellet was washed in acetone and resuspended in 1× SDS loading buffer. The covalent modification status of histones was analyzed by immunoblotting.
Immunoblot Analysis
Extracted protein was loaded on 10 or 15% SDS-polyacrylamide gel for protein separation. After it was transferred to polyvinylidene fluoride fluoropolymer membranes, the protein was detected using antibodies against HA (Roche), histone H3 (ab1791, Abcam), acetyl-histone H3 (06-599, Millipore), acetyl-histone H4 (06-866, Millipore), acetyl-histone H4K5 (07-327, Millipore), or acetyl-histone H4K16 (07-329, Millipore). Quantification of the band intensities on the immunoblots was performed using the ImageJ software according to the instructions (http://rsb.info.nih.gov/ij/docs/menus/analyze.html#gels).
Detection of ROS Accumulation
Leaf disks (4 mm in diameter) were excised from 6-week-old rice plants and incubated in distilled water for 12 h. Three leaf disks were treated with either 1 μL of 10 μM flg22 peptide (100 nM final concentration) or 1 μL of 0.8 μM chitin (8 nM final concentration) in 100 μL of luminol solution, immun-star hp substrate (Bio-Rad), and 1 μL of horseradish peroxidase-streptavidin (Jackson Immunoresearch). Luminescence was recorded every 20 s for 20 min on a Glomax 20/20 luminometer (Promega).
ChIP Assay
The ChIP assay was performed as described previously (Johnson et al., 2002; Ding et al., 2010). Briefly, 2 g of leaf tissue from 4-week-old rice plants grown in a growth chamber was used for the ChIP assay. Antibody against acetyl-histone H4 (Millipore) or HA (Roche) was used for the immunoprecipitation. The immunocomplex was harvested with Dyanbeads (Invitrogen), and the precipitated DNA was dissolved in TE (10 mM of Tris, 1 mM of EDTA, pH 8.0) for quantitative PCR using the rice ubiquitin gene as the control.
Accession Numbers
Sequence data from this article can be found in the Rice Genome Annotation Project website (http://rice.plantbiology.msu.edu/) under the following accession numbers: HDT701, Os05g51830; ubiquitin, Os03g13170; FLS2, Os04g52780; CEBiP, Os09g33630; SGT1, Os01g43540; MAPK6, Os06g06090; SPIN1, Os03g60110; WRKY53, Os05g27730; and WRKY71, Os02g08440.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of Histone Modifications of the Nipponbare Plants after M. oryzae Spray Inoculation.
Supplemental Figure 2. Disease Phenotypes of HDT701 OX Plants after Pathogen Inoculation.
Supplemental Figure 3. Disease Phenotypes of HDT701 RNAi Plants after Pathogen Infection.
Supplemental Figure 4. ROS Generation, Defense Gene Expression, and Histone Acetylation Levels in the HDT701 OX and RNAi Plants.
Supplemental Figure 5. Comparison of Histone Modifications in the Nipponbare Plants after PAMP Treatment.
Supplemental Figure 6. Expression and Histone H4 Acetylation Levels of Spin1 and WRKY71 in Nipponbare, HDT701 OX, and RNAi Plants after M. oryzae Inoculation.
Supplemental Table 1. Primers Used in This Study.
Acknowledgments
We thank Ko Shimamoto for providing the pANDA plasmid and Michael Goodin for providing the pGDD vector. We thank Bruce Jaffee for critical reading of this article. This study was supported by the Plant Genome Program of the U.S. National Science Foundation (0701745) to B.C.M. and G.L.W and the 973 Project (2012CB114005) from the Ministry of Sciences and Technology, China to G.L.W and Y.L.P.
AUTHOR CONTRIBUTIONS
B.D. and G.L.W. designed the research. B.D., M.B., and Y.N. performed the research. B.D., B.C.M., and G.L.W. wrote the article.
Glossary
- HDAC
histone deacetylase
- PRR
pattern recognition receptor
- ROS
reactive oxygen species
- PAMP
pathogen-associated molecular pattern
- PTI
pathogen-associated molecular pattern–triggered immunity
- GFP
green fluorescent protein
- HA
hemagglutinin
- OX
overexpression
- RNAi
RNA interference
- DAI
days after inoculation
- ETI
effector-triggered immunity
- UTR
untranslated region
- ChIP
chromatin immunoprecipitation
- SA
salicylic acid
- PMSF
phenylmethylsulfonyl fluoride
- Xoo
Xanthomonas oryzae pv oryzae
- MPSS
massively parallel signature sequencing
References
- Chen S., Songkumarn P., Liu J., Wang G.L. (2009). A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Tao L., Zeng L., Vega-Sanchez M.E., Umemura K., Wang G.L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Chen X., Ronald P.C. (2011). Innate immunity in rice. Trends Plant Sci. 16: 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.M., Song H.R., Han S.K., Han M., Kim M., Park J., Lee Y.H., Jeon J.S., Noh Y.S., Noh B. (2012). HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 71: 135–146
- Chujo T., Sugioka N., Masuda Y., Shibuya N., Takemura T., Okada K., Nojiri H., Yamane H. (2009). Promoter analysis of the elicitor-induced WRKY gene OsWRKY53, which is involved in defense responses in rice. Biosci. Biotechnol. Biochem. 73: 1901–1904 [DOI] [PubMed] [Google Scholar]
- Chujo T., et al. (2007). Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim. Biophys. Acta 1769: 497–505 [DOI] [PubMed] [Google Scholar]
- Chujo T., et al. (2008). Characterization of an elicitor-induced rice WRKY gene, OsWRKY71. Biosci. Biotechnol. Biochem. 72: 240–245 [DOI] [PubMed] [Google Scholar]
- Ding B., Zhu Y., Bu Z.Y., Shen W.H., Yu Y., Dong A.W. (2010). SDG714 regulates specific gene expression and consequently affects plant growth via H3K9 dimethylation. J. Integr. Plant Biol. 52: 420–430 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Goodin M.M., Dietzgen R.G., Schichnes D., Ruzin S., Jackson A.O. (2002). pGD vectors: Versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31: 375–383 [DOI] [PubMed]
- Hamon M.A., Cossart P. (2008). Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe 4: 100–109 [DOI] [PubMed] [Google Scholar]
- Hu Y., Qin F., Huang L., Sun Q., Li C., Zhao Y., Zhou D.X. (2009). Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 388: 266–271 [DOI] [PubMed] [Google Scholar]
- Huang L., Sun Q., Qin F., Li C., Zhao Y., Zhou D.X. (2007). Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 144: 1508–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. (2001). Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Johnson L., Cao X., Jacobsen S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12: 1360–1367 [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kawano Y., Akamatsu A., Hayashi K., Housen Y., Okuda J., Yao A., Nakashima A., Takahashi H., Yoshida H., Wong H.L., Kawasaki T., Shimamoto K. (2010). Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 7: 362–375 [DOI] [PubMed] [Google Scholar]
- Kim K.C., Lai Z., Fan B., Chen Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Kaboshi M., Okada K., Kurimoto L., Murakami S., Umezawa T., Shibuya N., Yamane H., Miyao A., Takatsuji H., Takahashi A., Hirochika H. (2010). A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 63: 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K., Kouzai Y., Kaku H., Shibuya N., Minami E., Nishizawa Y. (2010). Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant J. 64: 343–354 [DOI] [PubMed] [Google Scholar]
- Kurdistani S.K., Grunstein M. (2003). Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4: 276–284 [DOI] [PubMed] [Google Scholar]
- Lawrence R.J., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S. (2004). A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13: 599–609 [DOI] [PubMed] [Google Scholar]
- Lieberherr D., Thao N.P., Nakashima A., Umemura K., Kawasaki T., Shimamoto K. (2005). A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-protein in rice. Plant Physiol. 138: 1644–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Bai X., Wang X., Chu C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164: 969–979 [DOI] [PubMed] [Google Scholar]
- Liu X.Q., Bai X.Q., Qian Q., Wang X.J., Chen M.S., Chu C.C. (2005). OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1. Cell Res. 15: 593–603 [DOI] [PubMed] [Google Scholar]
- Miki D., Itoh R., Shimamoto K. (2005). RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 138: 1903–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A., Chen L., Thao N.P., Fujiwara M., Wong H.L., Kuwano M., Umemura K., Shirasu K., Kawasaki T., Shimamoto K. (2008). RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 20: 2265–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E., Wong H.L., Kawasaki T., Hasegawa M., Kodama O., Shimamoto K. (2001). Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 98: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R., Müller A., Napoli C.A., Selinger D.A., Pikaard C.S., Richards E.J., Bender J., Mount D.W., Jorgensen R.A. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Bartley L.E., Chen X., Dardick C., Chern M., Ruan R., Canlas P.E., Ronald P.C. (2008). OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant 1: 446–458 [DOI] [PubMed] [Google Scholar]
- Qiu D., Xiao J., Ding X., Xiong M., Cai M., Cao Y., Li X., Xu C., Wang S. (2007). OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 20: 492–499 [DOI] [PubMed] [Google Scholar]
- Qu S., Liu G., Zhou B., Bellizzi M., Zeng L., Dai L., Han B., Wang G.L. (2006). The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172: 1901–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R., Hager A. (1992). Fungal elicitors induce a transient release of active oxygen species from cultured spruce cells that is dependent on Ca2+ and protein-kinase activity. Planta 187: 136–141 [DOI] [PubMed] [Google Scholar]
- Shakespear M.R., Halili M.A., Irvine K.M., Fairlie D.P., Sweet M.J. (2011). Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 32: 335–343 [DOI] [PubMed] [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., Shibuya N. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M., Sugano S., Nakayama A., Jiang C.J., Ono K., Toki S., Takatsuji H. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19: 2064–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridha S., Wu K. (2006). Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46: 124–133 [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Allis C.D. (2000). The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Takai R., Isogai A., Takayama S., Che F.S. (2008). Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol. Plant Microbe Interact. 21: 1635–1642 [DOI] [PubMed] [Google Scholar]
- Tao Z., Liu H., Qiu D., Zhou Y., Li X., Xu C., Wang S. (2009). A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 151: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y., Ishikawa T., Watanabe K., Terakura S., Iwakawa H., Okada K., Machida C., Machida Y. (2007). Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19: 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Sánchez M.E., Zeng L., Chen S., Leung H., Wang G.L. (2008). SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice. Plant Cell 20: 1456–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Zhang X.C., Neece D., Ramonell K.M., Clough S., Kim S.Y., Stacey M.G., Stacey G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Gao F., Wu J., Dai J., Wei C., Li Y. (2010). Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol. 51: 1291–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.L., Song W.Y., Ruan D.L., Sideris S., Ronald P.C. (1996). The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Mol. Plant Microbe Interact. 9: 850–855 [DOI] [PubMed] [Google Scholar]
- Wu K., Tian L., Malik K., Brown D., Miki B. (2000). Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 22: 19–27 [DOI] [PubMed] [Google Scholar]
- Wu K., Tian L., Zhou C., Brown D., Miki B. (2003). Repression of gene expression by Arabidopsis HD2 histone deacetylases. Plant J. 34: 241–247 [DOI] [PubMed] [Google Scholar]
- Xiong L., Yang Y. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6: 520–527 [DOI] [PubMed] [Google Scholar]
- Zhou C., Zhang L., Duan J., Miki B., Wu K. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]