This study used high-throughput screening to identify five immune priming chemicals that potentiate but do not directly induce defense responses. These compounds inhibit salicylic acid (SA) glucosyltransferases and increase SA during pathogen infection. Thus, SA glucosylation can be a target for developing novel crop protectants.
Abstract
Plant activators are compounds, such as analogs of the defense hormone salicylic acid (SA), that protect plants from pathogens by activating the plant immune system. Although some plant activators have been widely used in agriculture, the molecular mechanisms of immune induction are largely unknown. Using a newly established high-throughput screening procedure that screens for compounds that specifically potentiate pathogen-activated cell death in Arabidopsis thaliana cultured suspension cells, we identified five compounds that prime the immune response. These compounds enhanced disease resistance against pathogenic Pseudomonas bacteria in Arabidopsis plants. Pretreatments increased the accumulation of endogenous SA, but reduced its metabolite, SA-O-β-d-glucoside. Inducing compounds inhibited two SA glucosyltransferases (SAGTs) in vitro. Double knockout plants that lack both SAGTs consistently exhibited enhanced disease resistance. Our results demonstrate that manipulation of the active free SA pool via SA-inactivating enzymes can be a useful strategy for fortifying plant disease resistance and may identify useful crop protectants.
INTRODUCTION
Like animals, plants activate their innate immune system upon recognition of pathogens. Pathogen-associated molecular patterns or pathogen effectors are recognized by either transmembrane-type pattern recognition receptors or cytosolic nucleotide binding and Leu-rich receptors and trigger a range of defense responses (Dangl and Jones, 2001). During the recognition process, salicylic acid (SA) accumulates at high levels (Vlot et al., 2009). Despite its importance as a phytohormone that is essential for resistance against biotrophic pathogens, its biosynthesis, metabolic pathways, and signal transduction are largely unknown. Once understood, the SA regulatory mechanisms may be exploited for developing crop protection technologies through molecular breeding or chemical applications.
Exogenously applied SA stimulates defense responses and confers disease resistance in plants. A number of chemicals that mimic SA action have been developed. These compounds are commonly called plant activators, and various types of compounds have been examined (Kessmann et al., 1994; Schreiber and Desveaux, 2008). For example, benzothiadiazole (commercial product Actigard) is a synthetic analog of SA (Görlach et al., 1996; Lawton et al., 1996). However, strong induction of a defense response is often accompanied by growth inhibition (Shirano et al., 2002; Zhang et al., 2003; Noutoshi et al., 2005), limiting the usefulness of these compounds in the field. Similarly, probenazole (Oryzemate) suppresses growth when sprayed on leaves, but not when taken up by the roots. Soil amendments of probenazole have been shown to provide durable immune-priming effects (Yoshioka et al., 2001) and thus have been used to protect paddy field rice (Oryza sativa) from blast fungus and bacterial leaf blight (Watanabe et al., 1977). Similar plant activators, such as tiadinil (V-get) and isotianil (Routine or Stout), were developed based on earlier versions of these agrochemicals. Compared with commonly used pesticides that directly target pathogens, plant activators are not pathogen specific and have not been overcome by microbes and thus have proven to be durable in the field (Watanabe et al., 1977).
A wide variety of compounds have been screened to identify plant activators that are more effective and may be applicable over a broad range of crops. For instance, several chemical screening procedures were reported using Arabidopsis thaliana seedlings in combination with a promoter reporter system for defense genes as markers of activity (Serrano et al., 2007; Knoth et al., 2009). However, the compounds identified in these screenings constitutively activate defense responses and are often associated with arrested growth and yield reduction. Therefore, there has been a need to develop screening schemes for chemicals that prime immunity but do not directly activate defense genes.
In this study, we report the establishment of a high-throughput chemical screening procedure to identify plant immune-priming compounds that potentiate but do not directly induce immune responses in Arabidopsis cell suspension cultures. A commercial library of 10,000 structurally unbiased small organic molecules was screened using the bacterial pathogen, Pseudomonas syringae pv tomato DC3000 avrRpm1 (Pst-avrRpm1). We found that active compounds inhibit both a known and a previously unknown SA glucosyltransferase (SAGT). Our results demonstrate that manipulation of the active free SA pool via SA-inactivating enzymes can be a useful strategy for fortifying plant disease resistance.
RESULTS
Screening for Plant Immune-Priming Compounds
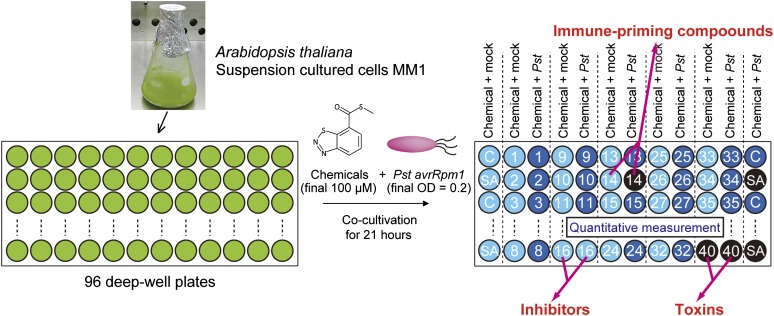
For high-throughput quantitative screening for plant immune-priming compounds, we used the Arabidopsis MM1 cell suspension cultures (Menges and Murray, 2002) challenged with Pst-avrRpm1 (Mackey et al., 2002). In this system, Arabidopsis cells induced immune-related cell death as a defense response 20 h after inoculation, as has been observed in planta (Mackey et al., 2002). Using Evans blue dye, which stains dead cells, we established a high-throughput screening system with 96 deep-well plates to measure cell death (Figure 1; see Supplemental Figures 1 to 4 online and Methods). We expected that immune-priming chemicals would increase Pst-avrRpm1–induced cell death, but cells that were not exposed to the pathogen should not show any effects. Confirming this expectation, some known plant activators, such as SA or tiadinil, enhanced Pst-avrRpm1–induced cell death (see Supplemental Figure 5 online). Chemicals that induced cell death in the absence of Pst-avrRpm1 were eliminated as toxic to the cells. We also expected that inhibitors of immune responses would be isolated, since known inhibitors, such as staurosporin, K252a, okadaic acid, and diphenyleneiodonium (Lamb and Dixon, 1997), all inhibit cell death in a concentration-dependent manner (see Supplemental Figure 6 online). The potential cell death inhibitors will be described elsewhere.
Figure 1.
Schematic Representation of the High-Throughput Screening Protocol for Plant Immune-Priming Compounds.
Arabidopsis MM1 suspension cells were dispensed into each well of 96-deep well plates, and each chemical compound (final 25 µg/mL) was applied to columns. The bacteria Pst-avrRpm1 was added to only one column for each chemical. The final volume of each well is 100 µL. After 21 h of cocultivation, plant cells were incubated with Evans blue dye (0.05%) and washed with 1 mL of water. The dye was extracted and the absorbance at 595 nm was measured. DMSO (0.5%) and SA (100 µM) were used as negative and positive controls, respectively. The data were represented as relative to the mean of negative controls with Pst-avrRpm1. The detailed protocol is described in Methods and Supplemental Figure 4 online.
Screening and Isolation of Novel Plant Immune-Priming Compounds
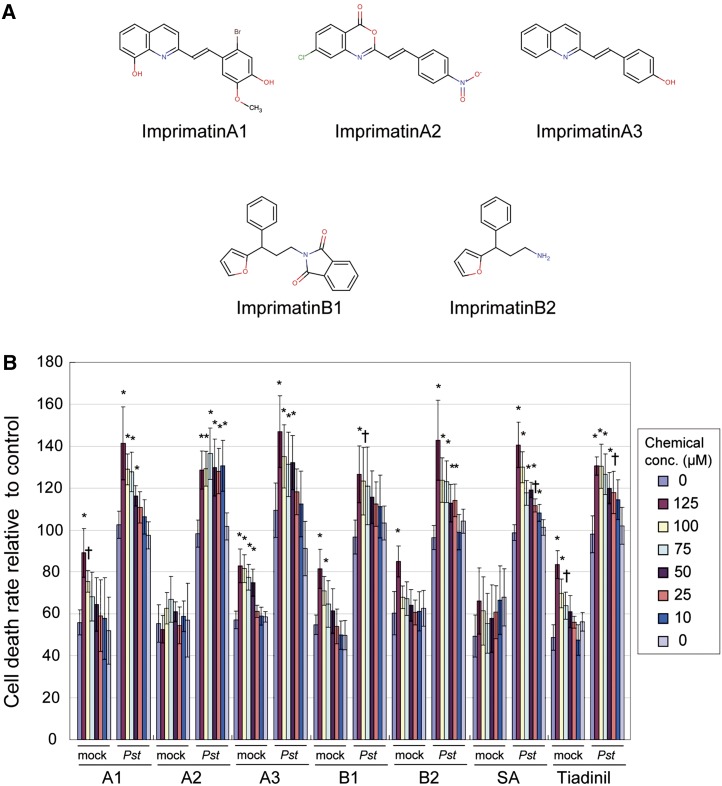
We screened a commercially available synthetic library composed of 10,000 organic chemicals (ChemBridge). After three replications of the screening procedure and dose–response confirmation experiments, we identified several candidate compounds that reproducibly enhanced Pst-avrRpm1–induced cell death at 100 µM without toxic effects. We designated these candidates Imprimatins for immune-priming chemicals. Two distinct molecular structural backbones of the compounds were found, and we named these groups Imprimatins A and B (Figure 2A). By searching online databases (NAMIKI, http://www.namiki-s.co.jp/chemcupid/, and the ChemMine; Girke et al., 2005), we found two structurally similar molecules to the Imprimatins. These were designated as -A3 and -B2 (Figure 2A). Imprimatin A3 has a ring structure like Imprimatins A1 and -A2, whereas Imprimatin B2 has the same ring structure as Imprimatin B1. Imprimatins A1, -A3, -B1, and -B2 potentiated Pst-avrRpm1–induced cell death in a concentration-dependent manner (Figure 2B). These compounds exhibited weak toxic effects at higher concentration ranges. Imprimatin A2 showed a similar impact on cell death at all concentrations tested (Figure 2B). Like tiadinil, all these compounds showed clear cell death priming effects at lower concentrations; thus, we further characterized these compounds.
Figure 2.
Compounds Isolated from the Chemical Screening.
(A) Chemical structures of Imprimatins A1, A2, A3, B1, and B2.
(B) The effects of Imprimatins on Arabidopsis suspension cell death upon inoculation with Pst-avrRpm1. For each compound, chemicals and bacteria were applied as indicated in the right panel. The cell death was measured as the concentration of Evans blue dye extracted from samples after staining. As a mock treatment, MgCl2 was added instead of pathogen. Cell death intensity is represented as relative value to the mean of the control (Pst + DMSO) of each experimental group (as 100). Error bars represent se (n = 4). *P < 0.01 and †P < 0.05; two-tailed Student’s t test with post-hoc Bonferroni’s correction.
Imprimatin-Induced Disease Resistance in Plants
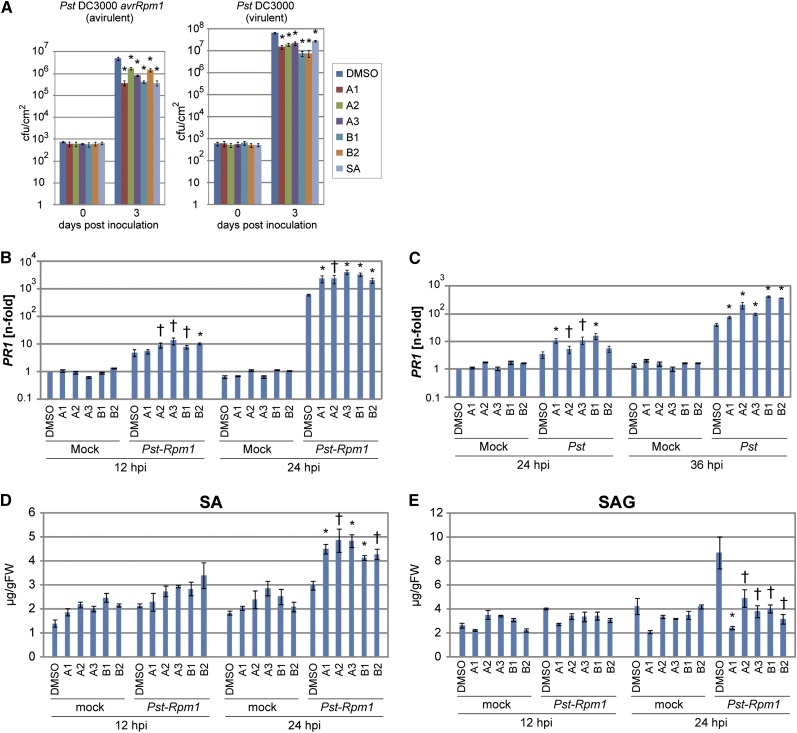
We next tested whether the imprimatins could induce disease resistance in whole plants. To test this, the roots of hydroponically grown Arabidopsis seedlings were immersed in solutions containing each of the Imprimatins for 3 d, at which time Pst-avrRpm1 was inoculated into leaves. Each of these chemical treatments suppressed bacterial growth (Figure 3A). The Imprimatins also reduced growth of both avirulent and virulent strains of P. syringae pv tomato DC3000. These results indicate that a cell culture–based screening system can be used to find novel plant protectants that function in planta.
Figure 3.
Characterization of Disease Resistance Responses in Arabidopsis Plants Treated with Imprimatins.
(A) Disease resistance against avirulent and virulent Pst strains induced by the treatment of Arabidopsis plants with Imprimatins. Seedlings hydroponically grown on rockwool for 3 weeks under short-day conditions were drenched in water containing each chemical (100 µM) for 3 d, followed by Pst strain inoculation into leaves. Bacterial growth inside leaves was measured 3 d after inoculation. Error bars represent se (n = 4). *P < 0.01; two-tailed Student’s t test. cfu, colony-forming units.
(B) and (C) PR1 expression analysis in Imprimatin-treated Arabidopsis plants infected with avirulent (B) and virulent (C) Pst strains. The chemical-treated seedlings were spray inoculated with each strain of bacteria (OD600 = 0.05). PR1 transcript levels were determined by quantitative RT-PCR with cDNA prepared from samples at the indicated time points. The expression values of the individual genes were normalized using Actin2 as an internal standard. Error bars represent se (n = 3). *P < 0.01 and †P < 0.05; two-tailed Student’s t test. These results are representative of three independent replicates.
(D) and (E) Measurement of endogenous SA (D) and SAG (E) during pathogen infection in the Imprimatin-treated Arabidopsis plants. The levels of SA and SAG were measured by HPLC using samples extracted from Arabidopsis seedlings inoculated with Pst-avrRpm1 as above. Error bars represent se (n = 3). *P < 0.01 and †P < 0.05; two-tailed Student’s t test. FW, fresh weight.
Defense Responses in Imprimatin-Treated Plants
To investigate the mode of action of the Imprimatins, we examined transcript levels of PATHOGENESIS-RELATED1 (PR1), a marker gene for SA-dependent immunity signaling (Ward et al., 1991), after treatment. Exposure of Arabidopsis seedlings to Imprimatins for 24 h did not induce transcription of PR1 (Figures 3B and 3C). These data indicate that Imprimatins are not functional analogs of SA. However, PR1 gene expression in treated plants was notably higher 12 and 24 h after Pst-avrRpm1 inoculation (Figure 3B). Similarly, PR1 expression was higher in plants infected with the virulent strain (Pst) (Figure 3C).
These results suggest that Imprimatin signaling functions in a pathogen-dependent manner, but free SA levels are also higher in plants treated with the Imprimatins, which may account for the generally higher expression of PR1 in these plants (Figure 3D). High free-SA concentrations are usually correlated with higher levels of its major inactive metabolite SA-2-O-β-d-glucoside (SAG) upon pathogen infection (Lee and Raskin, 1998). However, Imprimatin-treated plants accumulated much less SAG than DMSO-treated control plants, suggesting that Imprimatins block glucosylation of SA (Figure 3E).
Inhibition of SAGT
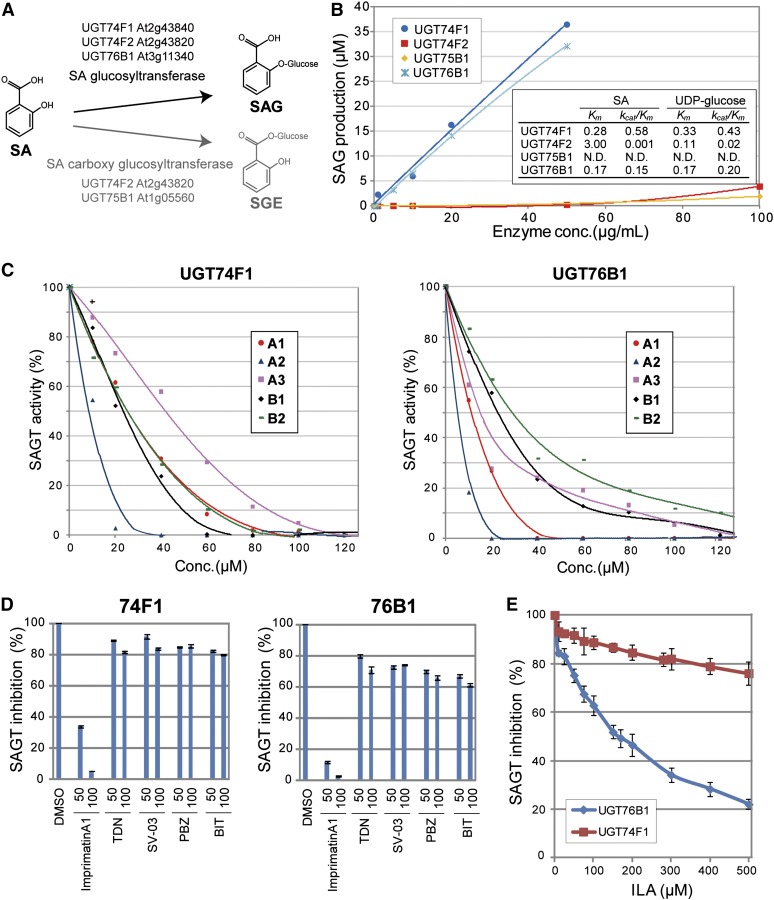
Plant hormones, such as auxins, abscisic acid, and gibberellins, can be inactivated by conjugation with a Glc moiety (Kleczkowski and Schell, 1995). Glucosylation of SA results in either SAG or a SA Glc ester (SGE) (Figure 4A). After SA infiltration into leaves, SGE is formed transiently at early stages, while SAG levels continuously increase over time (Dempsey et al., 2011). During pathogen infection, the vast majority of free SA is converted to SAG (Lee and Raskin, 1998), which is transported to the vacuole for degradation (Dean et al., 2005). In Arabidopsis, UGT74F1, UGT74F2, and UGT75B1 recognize SA as a substrate (Lim et al., 2002). Our enzyme kinetic analysis of these proteins demonstrates that UGT74F1 had higher activity than UGT74F2 and UGT75B1, an observation that is consistent with a previous report (Lim et al., 2002) (Figure 4B). We found that other UGT enzymes have SAGT activity, including UGT76B1 (Figure 4B). Michaelis-Menten kinetics indicate that the SAGT activity of UGT76B1 was similar to that of UGT74F1 (Figure 4B; see Supplemental Figure 7 online).
Figure 4.
Inhibition of SAGT Activity.
(A) Schematic representation of SA glucosylation patterns in Arabidopsis and the enzymes involved in each process.
(B) Michaelis-Menten enzyme kinetics of SAGT. Concentration-dependent SAG production of UGT74F1, UGT74F2, and UGT76B1 in vitro were measured by HPLC using affinity-purified His-tagged recombinant proteins expressed in Escherichia coli. The kinetic parameter Km (mM) and the specificity constant kcat/Km (mM−1 s−1) were determined from the Lineweaver-Burk plots (see Supplemental Figure 7 online). These results are the means of three independent replicates.
(C) Inhibition of SAGT activities of UGT74F1 (left panel) and UGT76B1 (right panel) by Imprimatins. The SAG produced by each enzyme was analyzed by HPLC in a reaction mixture containing SA at each Km concentration (Conc.). Values were plotted relative to the control of four independent experiments.
(D) Effect of known plant activators on the SAGT activities of UGT74F1 (left panel) and UGT76B1 (right panel). SAG levels were measured with tiadinil (TDN), probenazole (PBZ), SV-03 (SV), or 1,1-dioxo-1,2-benzothiazol-3-one (BIT) at the indicated concentrations (µM). Imprimatin A1 was used as a positive control. Data shown are relative to the DMSO control. Error bars represent se (n = 3).
(E) Effects of isoleucic acid (ILA) on the SAGT activities of UGT74F1 and UGT76B1. SAG was measured in the presence of different concentrations of ILA. Data represent relative values to each control. Error bars represent se (n = 3).
Each of the Imprimatins effectively blocked the SAGT activity of UGT74F1 and UGT76B1 (Figure 4C). Half-maximal inhibitory concentrations (IC50) show that the range of effective SAGT inhibition was similar to that for enhancement of cell death induced by Pst-avrRpm1 (Table 1, Figure 2B). For example, Imprimatin A2 exhibited higher enhancement effects even at a lower concentration range. The inhibition constants (Ki) of these compounds basically align with the IC50 values (Table 1; see Supplemental Figure 8 online). Interestingly, these enzymatic kinetic studies also indicate that the Imprimatins inhibited SAGT activity in a competitive manner with SA as substrate (see Supplemental Figures 8A and 8C and Supplemental Table 1 online), but not all the Imprimatins competed with UDP-Glc as donor (Wang, 2009) (see Supplemental Figures 8B and 8D and Supplemental Table 1 online). Our data demonstrate that the increase in Pst-avrRpm1–induced cell death is due to the interruption of SA inactivation by Imprimatins.
Table 1. IC50 Values and Ki of Imprimatins for SAGTs in Arabidopsis.
|
Ki (µM) |
||||||
|---|---|---|---|---|---|---|
|
IC50 (µM) |
UGT74F1 |
UGT76B1 |
||||
| Imprimatin | UGT74F1 | UGT76B1 |
SA | UDP-Glc | SA | UDP-Glc |
| A1 | 25.3 ± 5.5 | 11.3 ± 1.8 | 34.3 | 68.4 | 2.2 | 6.9 |
| A2 | 8.7 ± 1.4 | 5.7 ± 0.8 | 50.7 | n.a. | 4.2 | 6.8 |
| A3 | 42.5 ± 4.1 | 15.7 ± 5.1 | 90.8 | n.a. | 12.6 | n.a. |
| B1 | 23.5 ± 9.5 | 22.2 ± 4.4 | 47.6 | 47.7 | 3.8 | 30.6 |
| B2 | 21.6 ± 4.4 | 32.0 ± 8.4 | 40.3 | 57.2 | 54.6 | 35.8 |
Inhibitory effects of Imprimatins on UGT74F1 and UGT76B1 were determined from four experiments. Data are expressed as mean ± sd. The Ki values of Imprimatins were determined from Dixon plots shown in Supplemental Figure 8 online. n.a., not applicable.
The modes of action of commercially available plant activators are unknown, but probenazole and SV-03, the active core of the tiadinil metabolite, are thought to act upstream and downstream of SA, respectively (Nakashita et al., 2002; Yasuda et al., 2006). Thus, we tested if probenazole, tiadinil, and their functional metabolites also inhibit SAGT activity. These plant activators did not effectively inhibit SAGT activity of either UGT74F1 or UGT76B1 even at the highest concentration (100 µM) (Figure 4D). These results indicate that the Imprimatin mode of action is different from those of the commercially available plant activators.
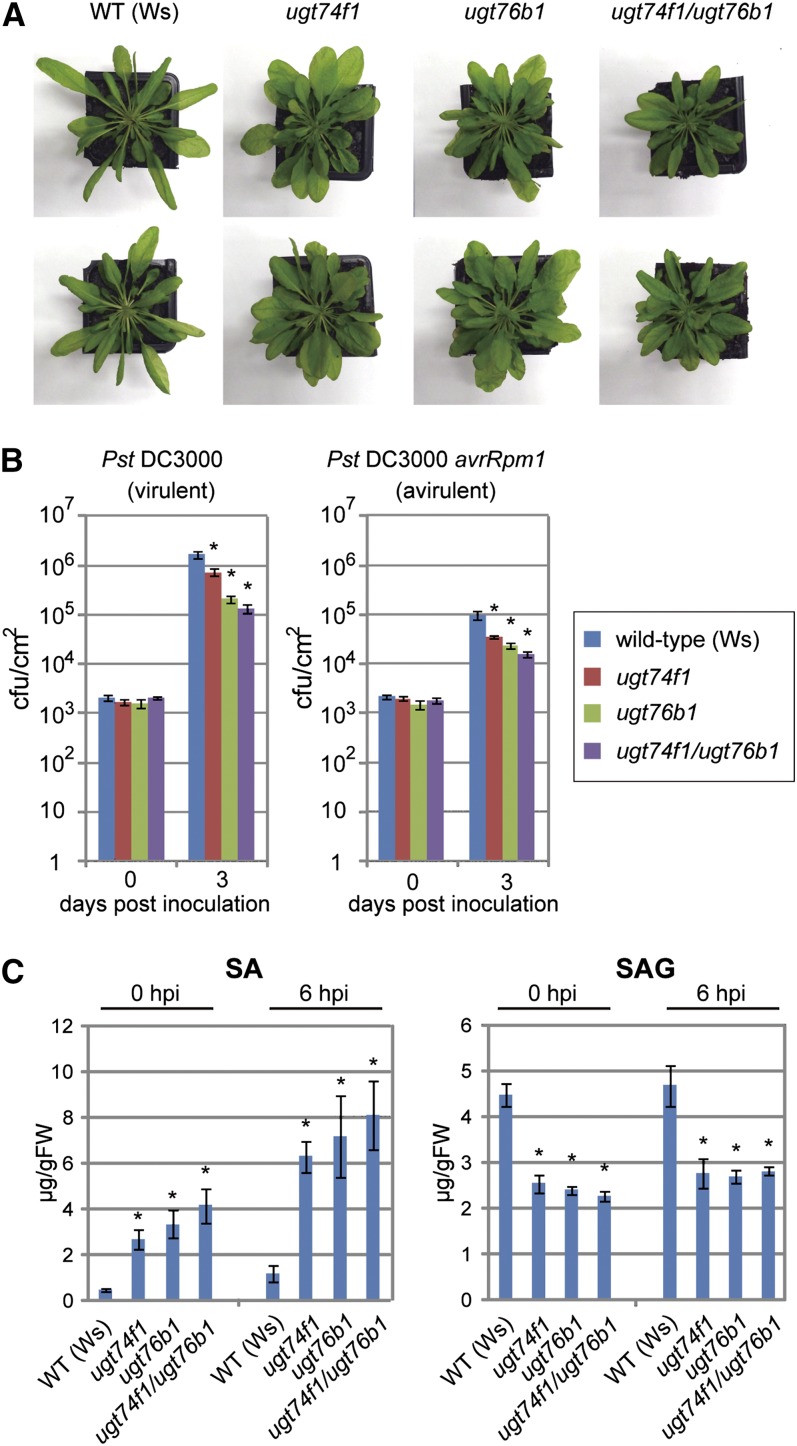
Loss of UGT74F1 and UGT76B1 Induces Disease Resistance in Arabidopsis Plants
Arabidopsis T-DNA insertion mutant lines for UGT74F1 and UGT76B1 were obtained from FLAGdb-FST (see Supplemental Figures 9A and 9C online). We crossed ugt74f1 and ugt76b1 to generate ugt74f1 ugt76b1 double knockout mutants. Rosettes of the double mutant were smaller than either of the single mutants (Figure 5A). This might be due to the accumulation of free SA as reported earlier (von Saint Paul et al., 2011). The rosette leaves of all three mutants were wider and rounder compared with the wild type. ugt76b1, and ugt74f1 to a lesser extent, were more resistant to both virulent and avirulent Pst strains than the wild type (Figure 5B). Increased resistance was consistently observed in the ugt74f1 ugt76b1 double mutants compared with each single mutant. The endogenous levels of free SA in all three mutants were higher and SAG levels were lower than in the wild type (Figure 5C). After inoculation of Pst-avrRpm1, all mutants also accumulated higher SA and lower SAG than in the wild type. The amounts of free SA in these mutants were correlated with their disease resistance levels. These data indicate that UGT74F1 and UGT76B1 convert SA to SAG in Arabidopsis.
Figure 5.
Arabidopsis SAGT Knockout Mutants.
(A) Phenotypes of SAGT knockout mutants. Wild-type (WT; Wassilewskija [Ws]), ugt74f1, ugt76b1, and ugt74f1 ugt76b1 Arabidopsis plants were grown on soil for 4 weeks under short-day conditions.
(B) Disease resistance of the ugt74f1 and ugt76b1 single and double mutants to both avirulent and virulent Pst strains. Bacteria were infiltrated into rosette leaves of 4-week-old plants grown under short-day conditions. Pathogen growth in leaves was monitored at 0 and 3 d after inoculation. Error bars represent se (n = 12). *P < 0.01; two-tailed Student’s t test. cfu, colony-forming units.
(C) Endogenous SA and SAG levels in the ugt74f1 and ugt76b1 single and double mutants. Plants were grown on soil for 4 weeks under short-day conditions, and Pst-avrRpm1 was inoculated by spraying. Rosette leaves were sampled for measurement using HPLC. Error bars represent se (n = 4). *P < 0.01; two-tailed Student’s t test. FW, fresh weight; hpi, hours post inoculation.
DISCUSSION
We screened 10,000 compounds in suspension-cultured cells and identified several small molecules that increase disease resistance in planta. Conventional in planta chemical screening requires large quantities of chemicals and a strictly controlled growth space. Our high-throughput method only requires 60 μL of suspension cells and 0.25 µg of compound. More importantly, this screening system differentiates chemicals that prime the immune system from direct cell death activators or toxins by evaluating cell viability in the absence of pathogen challenge.
We identified five novel compounds that enhanced disease resistance in plants. They are classified into two groups by structural similarity. The Imprimatin A group is characterized by a conjugated imine moiety as backbone. These compounds contain a trans-olefin–bonded bicyclic heterocyclic benzene derivative containing a nitrogen and a benzene derivative on each side. Imprimatins B1 and -B2 commonly have a 3-(2-furyl)-3-phenylpropylamino group. Interestingly, a chemical structure search for Imprimatin A matched with avenalumin, a phytoalexin in oat (Avena sativa; Mayama et al., 1981). However, the avenalumin structure had been corrected as avenanthramide (Miyagawa et al., 1995), and Imprimatin A is not similar to avenanthramide. Just as quercetin serves as an artificial substrate for UGT74F1 (Cartwright et al., 2008), Imprimatins also might be glucosylated by SAGTs, resulting in perturbation of SA metabolism by competition. Further structure-activity relationship analyses or drug metabolism studies in planta will reveal the core motif required for activity.
In this study, we identified SA metabolism as a target of several structurally related compounds. In fact, we expected to isolate SA-related compounds that modify synthesis, signaling, or metabolism of SA in this screen because SA can potentiate defense responses, including immunity-related cell death in suspension cells (Shirasu et al., 1997). Although the structural backbones of these compounds fall into two classes, each of the Imprimatins in this study perturbed two distinct SAGTs, UGT74F1 and UGT76B1. As these compounds are able to compete with SA for the inhibition of SAGTs (see Supplemental Figure 8 and Supplemental Table 1 online), we envision that the active sites of the two SAGTs may be structurally similar. However, the ability to compete with UDP-Glc differs among the Imprimatins, reflecting their structural differences. At this moment, the inhibitory effects of these five compounds on other glucosyltransferases are not known.
UGT76B1 was identified as a SAGT in this study, which is rather surprising because it has glucosyltransferase activity for isoleucic acid (ILA) but very weak activity for SA in vitro, based on a mass spectrometric analysis (von Saint Paul et al., 2011). Our enzyme kinetic study clearly demonstrated that the activity of UGT76B1 is comparable to that of UGT74F1, a known SAGT (Figure 4B). This is consistent with the high levels of SA in ugt76b1 mutants and SAG in plants overexpressing UGT76B1 (von Saint Paul et al., 2011). We also demonstrated that the SAGT activity of UGT76B1 was inhibited by ILA (Figure 4E). This is specific to UGT76B1, as ILA did not affect the SAGT activity of UGT74F1 (Figure 4E). As the affinities of SA and ILA to UGT76B1 are similar, it is possible that this enzyme is able to use both SA and ILA as substrates. These data also suggest that application of high concentrations of ILA to Arabidopsis plants would likely result in inhibition of UGT76B1 SAGT activity. This might also explain the weak increase in disease resistance of plants sprayed with 1 mM ILA (von Saint Paul et al., 2011).
One of the differences between UGT74F1 and UGT76B1 is their mRNA expression patterns. UGT74F1 expresses constitutively at low levels and modestly increases in response to environmental conditions, whereas transcription of UGT76B1 is greater than UGT74F1 and highly induced by both abiotic and biotic stresses (Dempsey et al., 2011; von Saint Paul et al., 2011) (see Supplemental Figure 10 online). Free SA levels are usually low, but drastically increase after infection (Lee and Raskin, 1998; Wildermuth et al., 2001). Importantly, most of the free SA is immediately converted to SAG (Lee and Raskin, 1998). The expression patterns of the two SAGT enzymes suggest that UGT74F1 functions at a basic level, and UGT76B1 becomes important as the plant removes free SA under stress conditions. The disease resistance of ugt76b1 was consistently greater than that of ugt74f1. Alternatively, it is possible that UGT74F1 might also have an important role during disease resistance in highly localized areas. Remarkable upregulation of UGT74F1 mRNA was detected in ugt74f1 (see Supplemental Figure 9E online). These could be nonfunctional transcripts possibly produced through feedback compensation mechanisms to maintain SA homeostasis. SAG was still detectable in the ugt76b1 ugt74f1 double mutants (Figure 5C), suggesting that there are other glucosyltransferases that can accept SA as a substrate. In fact, UGT74F2 has weak SAGT activity (Lim et al., 2002) (Figure 4B), and its transcription was upregulated by SA treatment (see Supplemental Figure 10C online). The primary intrinsic substrate of UGT74F2 is likely to be anthranilic acid (Quiel and Bender, 2003), but it may still be able to compensate for the loss of UGT74F1 or UGT76B1.
We showed that SAGT genes negatively regulate disease resistance in Arabidopsis, which is consistent with the model established by observation of the chemical effects of the five Imprimatins. Because the null mutations in UGT74F1 and UGT76B1 result in much smaller seedlings, the application of inhibitors at a later stage in development would seem to have a clear advantage over genetic manipulation for increasing disease defense readiness. In addition, the protection conferred by these compounds is likely to be as durable as other plant activators. Thus, we propose that using chemicals for controlled modulation of SA metabolism can be an effective strategy for crop protection.
METHODS
Chemicals
A chemical library of 10,000 small compounds (DIVERSet NovaCore NQ612, 5 mg/mL DMSO) was purchased from Chembridge. Additional candidate compounds were obtained from other chemical vendors: Imprimatin A1 (Synthon-Lab; SL066106, 2-[(E)-2-(2-bromo-4-hydroxy-5-methoxyphenyl)ethenyl] quinolin-8-ol), Imprimatin A2 (Princeton; OSSK_706278, 7-chloro-2-[(E)-2- (4-nitrophenyl)ethenyl]-4H-3,1-benzoxazin-4-one), Imprimatin A3 (ENAMINE; T0516-9496, 4-[(E)-2-(quinolin-2-yl)ethenyl]phenol), Imprimatin B1 (Pharmeks; P2001S-228941, 2-(3-(2-furyl)-3-phenylpropyl)benzo[c]azoline-1,3-dione), and Imprimatin B2 (Pharmeks; P2000S-41848, 3-(2-furyl)-3-phenylpropylamine). ILA was synthesized from l-Ile as described by Poterała and Plenkiewicz (2011). SAG for establishing the standard curve in HPLC analyses was kindly provided by Shigeo Tanaka (Tanaka et al., 1990).
High-Throughput Screening for Plant Immune Activators
Suspension cultures of Arabidopsis thaliana MM1 were basically maintained as described (Maor et al., 2007). Arabidopsis MM1 cells were grown in flasks in 100 mL of liquid Murashige and Skoog (MS) medium containing 3% Suc supplemented with 0.5 mg/L MES, pH 5.7, 0.5 mg/L naphthaleneacetic acid, and 0.05 mg/L 6-benzylaminopurine under long-day conditions at 22°C on a shaker (120 rpm). Bacteria were grown overnight at 28°C in liquid Luria-Bertani media supplemented with 50 µg/mL of kanamycin and rifampicin. The following day, cells were collected by centrifugation and resuspended in 10 mM MgCl2 (OD600 = 2.0). Cells (58.5 µL) were dispensed into each well of 96 deep well plates (nunc 260252; Nalge Nunc International) using a multichannel pipette with truncated tips. A total of 0.5 μL of each chemical compound (final 25 µg/mL) was added to wells (see Supplemental Figure 4 online). DMSO (1%) and sodium salicylate (SA) (100 µM) were applied to lanes 1 and 12 of the plate in alternate shifts as negative and positive controls, respectively. After 1 h with shaking, 41 μL of the pathogen suspension (10 mL of Pst-avrRpm1 [OD600 = 2.0], 2.86 mL of MES, and 27.5 mL of hormone-free MS medium) was added to one of the duplicate wells. Final concentrations were Pst OD600 = 0.2 and 14.3 mM MES. To evaluate chemical toxicity, medium without bacteria (10 mM MgCl2) was added to the other wells. The 96-well plates (FALCON 353912; Beckton Dickinson) were used as a lid. The plates were incubated with shaking for 21 h under 16-h/8-h (light/dark) conditions at 22°C. Then, 5 μL of 1% Evans blue was added into each well and mixed. After 30 min incubation, cells were washed four times with 1 mL of water. The dye was eluted with 400 μL of 50% methanol and 1% SDS, and absorbance at 595 nm was measured with a microplate reader. Cell death rates were calculated relative to pathogen-induced cell death in 1% DMSO as 100%. All experiments were repeated three times, and samples that recorded >120% absorbance without a significant increase in absorbance in the absence of the pathogen were selected as positives.
Chemical Treatment of Plants and RNA Experiments
Arabidopsis wild-type seedlings (Columbia) grown on half-MS agar plates (1% Suc) for 1 week under short-day conditions (8 h/16 h light/dark) were transferred onto rockwool and hydroponically cultivated at 22°C. After 3 weeks, plants were transferred into small pots supplemented with or without 100 µM solution of each chemical for 3 d before spray inoculation with bacteria (OD600 = 0.05 with 0.02% Silwet L-77). Rosetta leaves were collected in 2-mL tubes and frozen in liquid nitrogen. Samples were crushed with four zirconia balls (diameter of 2 mm) using a Shake Master Neo (Bio Medical Science). Total RNAs were extracted with PureLink Micro-to-Midi Total RNA purification system with the on-column DNase treatment procedure (Invitrogen), and RNA concentrations and purity were measured with a spectrometer at 260 and 260/280 nm (BioPhotometer plus; Eppendorf). cDNAs were synthesized with PrimeScript RT reagent kit with gDNA Eraser (Perfect Real Time; Takara). Quantitative RT-PCR amplifications were performed in 96-well plates with a LightCycler 480 real-time thermocycler (Roche Diagnostics) using a KAPA SYBR Fast quantitative PCR kit (Kapa Biosystems). Quantification of the target transcript was performed using the LightCycler 480 internal Absolute Quantification 2nd Derivative Max software and normalized by Actin2. The primers used in this study are listed in Supplemental Table 2 online.
Measurement of Endogenous SA and SAG in Arabidopsis Plants
Rosette leaves of spray-inoculated Arabidopsis seedlings with or without chemical treatment described above were harvested into 2-mL tubes and pulverized in liquid nitrogen. As for SAGT mutants, Arabidopsis plants (Wassilewskija) were grown on soil for 4 weeks and rosette leaves were harvested. Samples were extracted twice with 1 mL of 95% methanol supplemented with 5% formic acid at 80°C. The extracts were pooled, dried at 40°C in a vacuum rotary evaporator, extracted with 1 mL of water containing 5% formic acid at 80°C for 10 min, and passed through an Oasis 1cc HLB cartridge (Waters) that had been prewashed with 5% formic acid. SAG was eluted from the column with 1 mL of 25% methanol-5% formic acid. SA was extracted from the eluent with 1 mL of 87% methanol-5% formic acid. SAG was quantified by reverse-phase HPLC (Hitachi LaChrom Elite with L2485 fluorescence detector) on a 5-µm C18 column (Phenomenex) with 10% acetonitrile containing 0.1% trifluoroacetic acid at 1 mL/min over 20 min. For SA, HPLC separation used a linear gradient with 18 to 55% acetonitrile containing 0.1% trifluoroacetic acid at 1 mL/min over 20 min. SA and SAG were detected fluorometrically (excitation 295 nm; emission 370 nm), and each retention time was 8.3 and 16.4 min, respectively. Standard curves for SA and SAG were prepared from similarly diluted samples containing 0.1 to 5000 µM of each compound. Typical recovery rates of SA and SAG from spiked samples were 94.6 and 83.8%, respectively.
Enzyme Assay for SA Glucosyltransferase
The cDNAs of UGT75B1 (At1g05560), UGT74F1 (At2g43840), UGT74F2 (At2g43820), and UGT76B1 (At3g11340) were amplified by PCR using Phusion DNA polymerase (Finnzymes) with primers listed in Supplemental Table 2 online. The PCR fragments of UGT75B1, UGT74F1, and UGT74F2 were cloned into pGEM-T Easy vector (Promega) after incubation with Ex-Taq DNA polymerase (Takara) for 3′-adenine attachment. After sequence validation, they were recloned into the NheI-EcoRI site of pET28a vector (Novagen) and transformed into Escherichia coli Rosetta (DE3; Novagen). The PCR fragment of UGT76B1 was cloned using a ZeroBlunt TOPO PCR cloning kit (Invitrogen). After sequence validation, the PCR-reamplified cDNA fragment was cloned using a pENTR/D-TOPO cloning kit to transfer the insert into pDEST17 using Gateway LR clonase (Invitrogen). The plasmid was transformed into E. coli Arctic Express (DE3; Agilent Technologies). Transformants were grown using the Overnight Express Autoinduction System (Novagen), and cells were lysed with BugBuster (Novagen). Expressed proteins were purified on His-tag resin (Novagen) according to the manufacturer’s protocol. Enzyme assays were basically performed as described by Lee and Raskin (1998). Each assay (40 µL) for the measurement of Km for SA was performed with 2.5 µg of UGT74F1 or 5 µg of UGT76B1, 0.05 to 3 mM SA, 5 mM UDP-Glc, and 14 mM 2-mercaptoethanol in 50 mM MES, pH 8.0, buffer. Reaction mixes were incubated at 30°C for 30 min before stopping by addition of 8 μL of 50% (v/v) trichloroacetic acid and analyzed by reverse-phase HPLC with a linear 10 to 65% acetonitrile gradient containing 0.1% trifluoroacetic acid at 1 mL/min over 24 min. SA and SAG were detected by absorbance (296 nm) or fluorescence (excitation 295 nm; emission 370 nm). Specific enzyme activity was expressed as nmol of substrate converted to its Glc conjugate/nanokatal by 1 mg of protein in 30 min. The Km values of each enzyme were calculated from Lineweaver-Burk plots (see Supplemental Figure 7 online). For the analysis of IC50 and inhibition assays using agrochemicals and ILA, SA and UDP-Glc were added to the reaction at the concentration of Km values for each enzyme with concentrations of compounds ranging from 0 to 200 µM. For the determination of Ki, three different concentrations of the substrate around the Km value at a fixed concentration of the other substrate were used. Each Ki value was analyzed with a Dixon plot (see Supplemental Figure 8 online).
Pathology Tests
In planta pathogen growth assays were performed as described with modifications to allow for treatment with chemical compounds (Weigel and Glazebrook, 2002). Arabidopsis seedlings grown on MS agar plates for 1 week under short-day conditions (8 h/16 h light/dark) were transferred onto rockwool and hydroponically cultivated at 22°C. After 3 weeks, plants were transferred into small pots. Plants were watered with either a 100 or 200 µM solution of each chemical for 3 d before inoculation with bacteria by needleless syringe (OD600 = 0.002). Soil-grown plants were used for testing SAGT mutants.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: UGT75B1 (At1g05560), UGT74F1 (At2g43840), UGT74F2 (At2g43820), UGT76B1 (At3g11340), Actin2 (At3g18780), and BSMT1 (At3g11480).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Defense Responses of Arabidopsis Suspension-Cultured Cells Infected with Virulent and Avirulent Pathogenic Pst.
Supplemental Figure 2. Comparison of Cell Death of Arabidopsis MM1 Suspension-Cultured Cells in Response to Infection with Pseudomonas syringae pv tomato DC3000.
Supplemental Figure 3. Effect of Chemical Solvent DMSO on Cell Death of MM1 Cells.
Supplemental Figure 4. Screening Protocol.
Supplemental Figure 5. Effect of the Commercial Plant Disease Resistance Inducer Tiadinil and a Phytohormone Salicylic Acid on the Interaction between MM1 Cells and Pst-avrRpm1.
Supplemental Figure 6. Effect of the Known HR Cell Death Inhibitors on the Interaction between MM1 Cells and Pst-avrRpm1.
Supplemental Figure 7. Lineweaver-Burk Plots of UGT74F1 and UGT76B1 for SA and UDP-Glc.
Supplemental Figure 8. Dixon Plots for the Inhibition Constants (Ki) of Imprimatins.
Supplemental Figure 9. Arabidopsis T-DNA Insertion Mutants for SAGT.
Supplemental Figure 10. Expression of Glucosyltransferase Genes in Arabidopsis thaliana.
Supplemental Table 1. Inhibitory Mode of Action of Imprimatins for SAGTs in Arabidopsis.
Supplemental Table 2. Primers Using for Genotyping of SAGT Mutants and qRT-PCR Analysis.
Acknowledgments
We thank Shigeo Tanaka for the gracious gifts of SAG and SGE. We thank Nihon Nohyaku Co. for kindly providing tiadinil and SV-03. We also thank Kazuo Takahashi for advice on statistical analysis. This work was supported in part by a grant from the Gatsby Foundation to K.S., by a Grant-in-Aid for Scientific Research (KAKENHI, No. 19039034 to K.S. and Y.N., 24228008 and 19678001 to K.S, and 22780036 to Y.N.), by the Special Coordination Fund for Promoting Sciences and Technology of the Ministry of Education, Culture, Sports, Science and Technology of Japan to Y.N., and grants from The Sumitomo Foundation, The Kurata Memorial Hitachi Science and Technology Foundation to Y.N.
AUTHOR CONTRIBUTIONS
Y. Noutoshi designed the research, performed most of the experiments, and wrote the article. M.O. carried out enzyme kinetics studies. Y.M., T.O., H.S., D.S., Y.J., A.H., and Y.K. contributed to the measurement of endogenous SA and SAG. T.K. and Y. Nishina synthesized l-Ile acid. K.S. designed the research and wrote the article.
Glossary
- SA
salicylic acid
- SAGT
SA glucosyltransferase
- SAG
SA-2-O-β-d-glucoside
- SGE
salicylic acid Glc ester
- IC50
half-maximal inhibitory concentrations
- Ki
inhibition constants
- ILA
isoleucic acid
- MS
Murashige and Skoog
References
- Cartwright A.M., Lim E.K., Kleanthous C., Bowles D.J. (2008). A kinetic analysis of regiospecific glucosylation by two glycosyltransferases of Arabidopsis thaliana: Domain swapping to introduce new activities. J. Biol. Chem. 283: 15724–15731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Dean J.V., Mohammed L.A., Fitzpatrick T. (2005). The formation, vacuolar localization, and tonoplast transport of salicylic acid glucose conjugates in tobacco cell suspension cultures. Planta 221: 287–296 [DOI] [PubMed] [Google Scholar]
- Dempsey D.A., Vlot A.C., Wildermuth M.C., Klessig D.F. (2011). Salicylic acid biosynthesis and metabolism. The Arabidopsis Book 9: e0156, doi/10.1199/tab.0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T., Cheng L.C., Raikhel N. (2005). ChemMine. A compound mining database for chemical genomics. Plant Physiol. 138: 573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlach J., Volrath S., Knauf-Beiter G., Hengy G., Beckhove U., Kogel K.H., Oostendorp M., Staub T., Ward E., Kessmann H., Ryals J. (1996). Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8: 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessmann H., Staub T., Hofmann C., Maetzke T., Herzog J., Ward E., Uknes S., Ryals J. (1994). Induction of systemic acquired disease resistance in plants by chemicals. Annu. Rev. Phytopathol. 32: 439–459 [DOI] [PubMed] [Google Scholar]
- Kleczkowski K., Schell J. (1995). Phytohormone conjugates: Nature and function. Crit. Rev. Plant Sci. 14: 283–298 [Google Scholar]
- Knoth C., Salus M.S., Girke T., Eulgem T. (2009). The synthetic elicitor 3,5-dichloroanthranilic acid induces NPR1-dependent and NPR1-independent mechanisms of disease resistance in Arabidopsis. Plant Physiol. 150: 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C., Dixon R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Lawton K.A., Friedrich L., Hunt M., Weymann K., Delaney T., Kessmann H., Staub T., Ryals J. (1996). Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10: 71–82 [DOI] [PubMed] [Google Scholar]
- Lee H.I., Raskin I. (1998). Glucosylation of salicylic acid in Nicotiana tabacum cv. Xanthi-nc. Phytopathology 88: 692–697 [DOI] [PubMed] [Google Scholar]
- Lim E.K., Doucet C.J., Li Y., Elias L., Worrall D., Spencer S.P., Ross J., Bowles D.J. (2002). The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J. Biol. Chem. 277: 586–592 [DOI] [PubMed] [Google Scholar]
- Mackey D., Holt B.F., III, Wiig A., Dangl J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Maor R., Jones A., Nühse T.S., Studholme D.J., Peck S.C., Shirasu K. (2007). Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol. Cell. Proteomics 6: 601–610 [DOI] [PubMed] [Google Scholar]
- Mayama S., Tani T., Ueno T., Hirabayashi K., Nakashima T., Fukami H., Mizuno Y., Irie H. (1981). Isolation and structure elucidation of genuine oat phytoalexin, avenalumin I. Tetrahedron Lett. 22: 2103–2106 [Google Scholar]
- Menges M., Murray J.A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Miyagawa H., Ishihara A., Nishimoto T., Ueno T., Mayama S. (1995). Induction of avenanthramides in oat leaves inoculated with crown rust fungus, Puccinia coronata f. sp. aenae. Biosci. Biotechnol. Biochem. 59: 2305–2306 [Google Scholar]
- Nakashita H., Yoshioka K., Yasuda M., Nitta T., Arai Y., Yoshida S., Yamaguchi I. (2002). Probenazole induces systemic acquired resistance in tobacco through salicylic acid accumulation. Physiol. Mol. Plant Pathol. 61: 197–203 [Google Scholar]
- Noutoshi Y., Ito T., Seki M., Nakashita H., Yoshida S., Marco Y., Shirasu K., Shinozaki K. (2005). A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J. 43: 873–888 [DOI] [PubMed] [Google Scholar]
- Poterała M., Plenkiewicz J. (2011). Synthesis of new chiral ionic liquids from a-hydroxycarboxylic acids. Tetrahedron Asymmetry 22: 294–299 [Google Scholar]
- Quiel J.A., Bender J. (2003). Glucose conjugation of anthranilate by the Arabidopsis UGT74F2 glucosyltransferase is required for tryptophan mutant blue fluorescence. J. Biol. Chem. 278: 6275–6281 [DOI] [PubMed] [Google Scholar]
- Schreiber K., Desveaux D. (2008). Message in a bottle: Chemical biology of induced disease resistance in plants. Plant Pathol. J. 24: 245–268 [Google Scholar]
- Serrano M., Robatzek S., Torres M., Kombrink E., Somssich I.E., Robinson M., Schulze-Lefert P. (2007). Chemical interference of pathogen-associated molecular pattern-triggered immune responses in Arabidopsis reveals a potential role for fatty-acid synthase type II complex-derived lipid signals. J. Biol. Chem. 282: 6803–6811 [DOI] [PubMed] [Google Scholar]
- Shirano Y., Kachroo P., Shah J., Klessig D.F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14: 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K., Nakajima H., Rajasekhar V.K., Dixon R.A., Lamb C. (1997). Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9: 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Hayakawa K., Umetani Y., Tabata M. (1990). Glucosylation of isomeric hydroxybenzoic acids by cell suspension cultures of Mallotus japonicus. Phytochemistry 29: 1555–1558 [Google Scholar]
- Vlot A.C., Dempsey D.A., Klessig D.F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47: 177–206 [DOI] [PubMed] [Google Scholar]
- von Saint Paul V., Zhang W., Kanawati B., Geist B., Faus-Kessler T., Schmitt-Kopplin P., Schäffner A.R. (2011). The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell 23: 4124–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2009). Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett. 583: 3303–3309 [DOI] [PubMed] [Google Scholar]
- Ward E.R., Uknes S.J., Williams S.C., Dincher S.S., Wiederhold D.L., Alexander D.C., Ahl-Goy P., Metraux J.P., Ryals J.A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Igarashi H., Matsumoto K., Seki S., Mase S., Sekizawa Y. (1977). The characteristics of probenazole (Oryzemate®) for the control of rice blast. J. Pestic. Sci. 2: 291–296 [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Yasuda M., Kusajima M., Nakajima M., Akutsu K., Kudo T., Yoshida S., Nakashita H. (2006). Thiadiazole carboxylic acid moiety of tiadinil, SV-03, induces systemic acquired resistance in tobacco without salicylic acid accumulation. J. Pestic. Sci. 31: 329–334 [Google Scholar]
- Yoshioka K., Nakashita H., Klessig D.F., Yamaguchi I. (2001). Probenazole induces systemic acquired resistance in Arabidopsis with a novel type of action. Plant J. 25: 149–157 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Goritschnig S., Dong X., Li X. (2003). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]