This study shows that the bZIP protein HapX, a key regulator of the iron response, is required for rhizosphere competence of the vascular wilt fungus Fusarium oxysporum against soil-inhabiting bacteria and for virulence on tomato (Solanum lycopersicum) plants and immunodepressed mice, establishing a conserved role for HapX-mediated iron homeostasis in fungal infection of plants and mammals.
Abstract
Soilborne fungal pathogens cause devastating yield losses and are highly persistent and difficult to control. During the infection process, these organisms must cope with limited availability of iron. Here we show that the bZIP protein HapX functions as a key regulator of iron homeostasis and virulence in the vascular wilt fungus Fusarium oxysporum. Deletion of hapX does not affect iron uptake but causes derepression of genes involved in iron-consuming pathways, leading to impaired growth under iron-depleted conditions. F. oxysporum strains lacking HapX are reduced in their capacity to invade and kill tomato (Solanum lycopersicum) plants and immunodepressed mice. The virulence defect of ΔhapX on tomato plants is exacerbated by coinoculation of roots with a biocontrol strain of Pseudomonas putida, but not with a siderophore-deficient mutant, indicating that HapX contributes to iron competition of F. oxysporum in the tomato rhizosphere. These results establish a conserved role for HapX-mediated iron homeostasis in fungal infection of plants and mammals.
INTRODUCTION
Soilborne fungal pathogens are ubiquitous, highly persistent, and extremely difficult to control. They cause root rots, wilts, stunting, and seedling damping-off in a wide range of plant species, leading to devastating losses in field and greenhouse crops both in industrialized and developing countries. Agricultural practices, such as crop rotation, resistance breeding, and application of fungicides, are insufficient to prevent root diseases of important crop plants (Haas and Défago, 2005). One of the most important soilborne pathogens is Fusarium oxysporum, the causal agent of vascular wilt disease in more than 100 different plant species (Armstrong and Armstrong, 1981; Dean et al., 2012). Like other root-infecting fungi, F. oxysporum persists in the soil for extended time periods, either in the form of thick-walled chlamydospores or as a saprophyte on dead organic matter. Compounds exudated by the host plant trigger spore germination, followed by directed hyphal growth and penetration of the root, preferentially through natural openings at the junctions of epidermal cells (Lagopodi et al., 2002; Pérez-Nadales and Di Pietro, 2011). Inside the root, the fungus grows inter- and intracellularly until it reaches the vascular tissue, where it colonizes the xylem vessels, provoking wilting and plant death. Some F. oxysporum isolates also cause opportunistic infections in humans, which range from superficial or locally invasive to disseminated, depending on the immune status of the individual (Nucci and Anaissie, 2007). Previous work established that a single isolate of F. oxysporum f sp lycopersici, FGSC 9935, is able to cause disease both in tomato (Solanum lycopersicum) plants and in immunodepressed mice (Ortoneda et al., 2004). The availability of the complete genome sequence (Ma et al., 2010) makes this strain an excellent model for studying the genetic basis of transkingdom pathogenicity in fungi.
During saprophytic and preinfection stages, F. oxysporum competes with other microorganisms in the soil and the plant rhizosphere for limited nutrients and essential elements, such as iron (Simeoni et al., 1987). Ever since the earliest reports on antagonistic disease-suppressing soil microorganisms more than 70 years ago, it has been known that nonpathogenic rhizosphere-colonizing microbes can protect plants against root-infecting pathogens, a mechanism termed biocontrol (Baker, 1968). Fluorescent pseudomonads are effective biocontrol agents against plant pathogenic fungi, bacteria, and nematodes (Mercado-Blanco et al., 2001; Haas and Défago, 2005; Weller, 2007). Pseudomonas spp owe their fluorescence to an extracellular diffusible pigment called pyoverdine (Pvd), which displays a high affinity for Fe3+ ions and functions as a siderophore (Ravel and Cornelis, 2003). In addition to Pvd, secondary siderophores with lower iron affinity, including pyochelin, pseudomonine, quinolobactin, ornicorrugatin (Ocg), and nocardamine, are produced by different Pseudomonas strains (Cornelis and Matthijs, 2002; Matthijs et al., 2008). The battery of siderophores enables fluorescent pseudomonads to efficiently compete for limited iron resources in the soil (Ravel and Cornelis, 2003).
Iron is an essential cofactor for a wide range of cellular processes, but its excess is toxic to the cell (Halliwell and Gutteridge, 1984). Iron homeostasis requires fine-tuned mechanisms to maintain the balance between uptake, storage, and consumption of iron. In the saprophytic model fungus Aspergillus nidulans, maintenance of iron homeostasis is mediated essentially by two transcription factors, SreA and HapX, which are interconnected in a negative feedback loop (Haas et al., 1999; Hortschansky et al., 2007). During iron starvation, the bZIP protein HapX, initially identified as an interactor of the heterotrimeric CCAAT binding core complex (Tanaka et al., 2002; Hortschansky et al., 2007), downregulates the expression of sreA, a repressor of siderophore biosynthesis and of iron-dependent pathways (Mercier et al., 2006; Hortschansky et al., 2007; Schrettl et al., 2010; Hsu et al., 2011). HapX governs iron homeostasis and virulence in the human pathogens Aspergillus fumigatus (Schrettl et al., 2010), Candida albicans (Chen et al., 2011; Hsu et al., 2011), and to a lesser extent in Cryptococcus neoformans (Jung et al., 2010). HapX is conserved throughout the fungal kingdom, but its function during fungal pathogenicity on plants has not been explored so far.
In this study, we addressed the role of HapX and iron homeostasis in the infection process of F. oxysporum. We show that HapX is a major regulator of the transcriptional response to iron limitation and establish its relevance during fungal infection of plants. Moreover, we provide evidence for its function during iron competition of F. oxysporum against siderophore-producing pseudomonads. These results reveal a key role for HapX in iron homeostasis, virulence, and rhizosphere competence of this important fungal pathogen.
RESULTS
Loss of HapX Impairs Fungal Growth under Iron-Limiting Conditions without Affecting Iron Acquisition
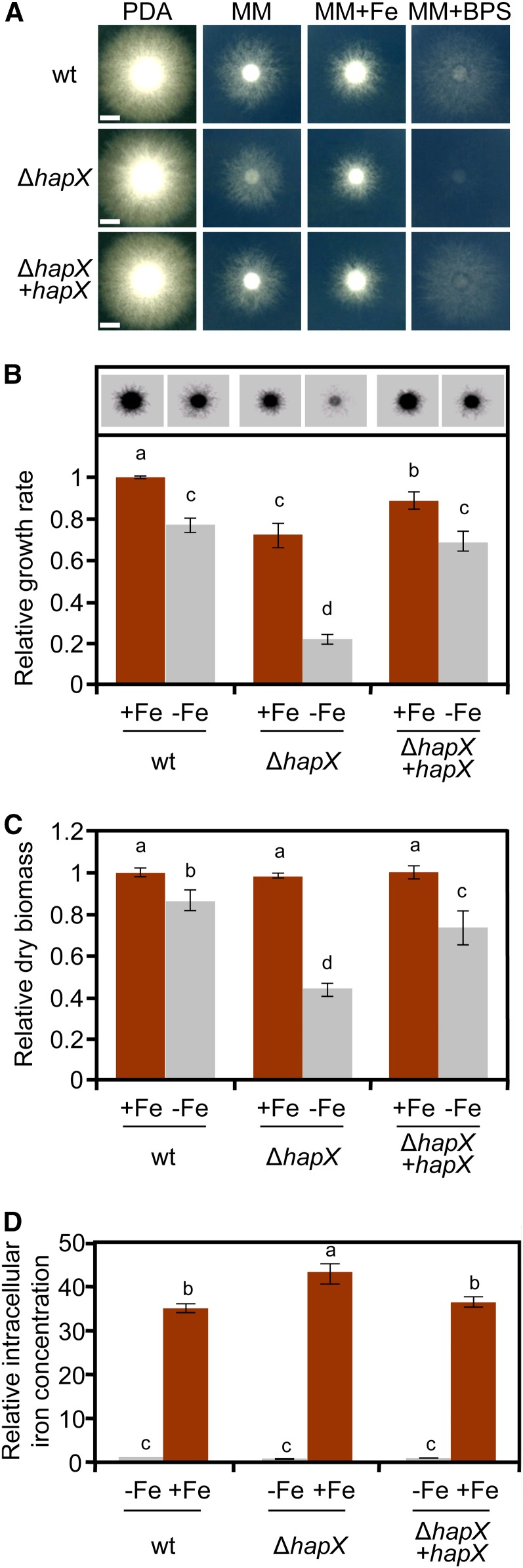
A BLASTP search of the F. oxysporum genome database identified a single predicted HapX ortholog, FOXG_07577, which displays 32% overall identity with HapX from A. fumigatus. Alignment of the amino acid sequence revealed the presence of all the characteristic domains of this class of transcription factors (see Supplemental Figure 1 online), including an N-terminal region essential for the interaction with the CCAAT binding complex (Hortschansky et al., 2007), a bZIP domain, and several Cys-rich motifs putatively involved in iron sensing (Hortschansky et al., 2007; Schrettl et al., 2010). To study the role of HapX in F. oxysporum, we replaced the entire FOXG_07577 coding sequence with the hygB resistance cassette to generate several ΔhapX deletion mutants (see Supplemental Figure 2 online). The ΔhapX strains showed no growth defects on rich media, but mycelial growth was markedly reduced under iron-limiting conditions and was almost undetectable in the presence of the iron chelator bathophenanthrolinedisulfonic acid disodium salt (BPS) (Figures 1A and 1B). Likewise, biomass production of the ΔhapX mutant in liquid culture was similar to the wild-type strain under iron-replete conditions but was reduced by more than 50% in iron-depleted medium (Figure 1C). Reintroduction of the intact hapX allele into the ΔhapX mutant, yielding the complemented strain ΔhapX+hapX (see Supplemental Figure 2 online), fully restored wild-type growth (Figures 1A to 1C).
Figure 1.
Loss of hapX Impairs Growth of F. oxysporum under Iron-Limiting Conditions but Not Iron Uptake.
(A) Growth of the indicated fungal strains on solid media differing in availability of iron. Cultures were grown for 3 d at 28°C. wt, wild type.
(B) Relative mycelial growth of the indicated strains on MM with or without Fe was estimated by measuring the area corresponding to fungal colonies on the inverted contrast images in (A), and normalized to the growth of the wild type strain on MM+Fe.
(C) Relative dry biomass of the indicated strains grown in liquid MM with or without Fe for 5 d at 28°C. Bars represent se from three independent experiments with three technical replicates each. Values with the same letter are not significantly different according to Mann-Whitney test (P = 0.05).
(D) The indicated strains were grown in iron-depleted MM for 24 h and transferred for 1 h to iron-depleted MM (−Fe) or iron-replete MM (+Fe). Intracellular iron concentration was determined colorimetrically and expressed relative to the wild-type strain under iron-depleted conditions. Bars represent se from three independent experiments with three technical replicates each. Values with the same letter are not significantly different according to Mann-Whitney test (P = 0.05).
Bars in (A) = 5 mm.
[See online article for color version of this figure.]
To test whether impaired growth of ΔhapX under iron-depleted conditions is caused by the inability of the mutant to obtain iron from the environment, we measured intracellular iron content 1 h after a shift from iron-depleted to iron-replete conditions (Tamarit et al., 2006). Levels of iron in the ΔhapX mutant were slightly higher than those detected in the wild type and the complemented strain (Figure 1D), suggesting that HapX is not essential for iron uptake in F. oxysporum.
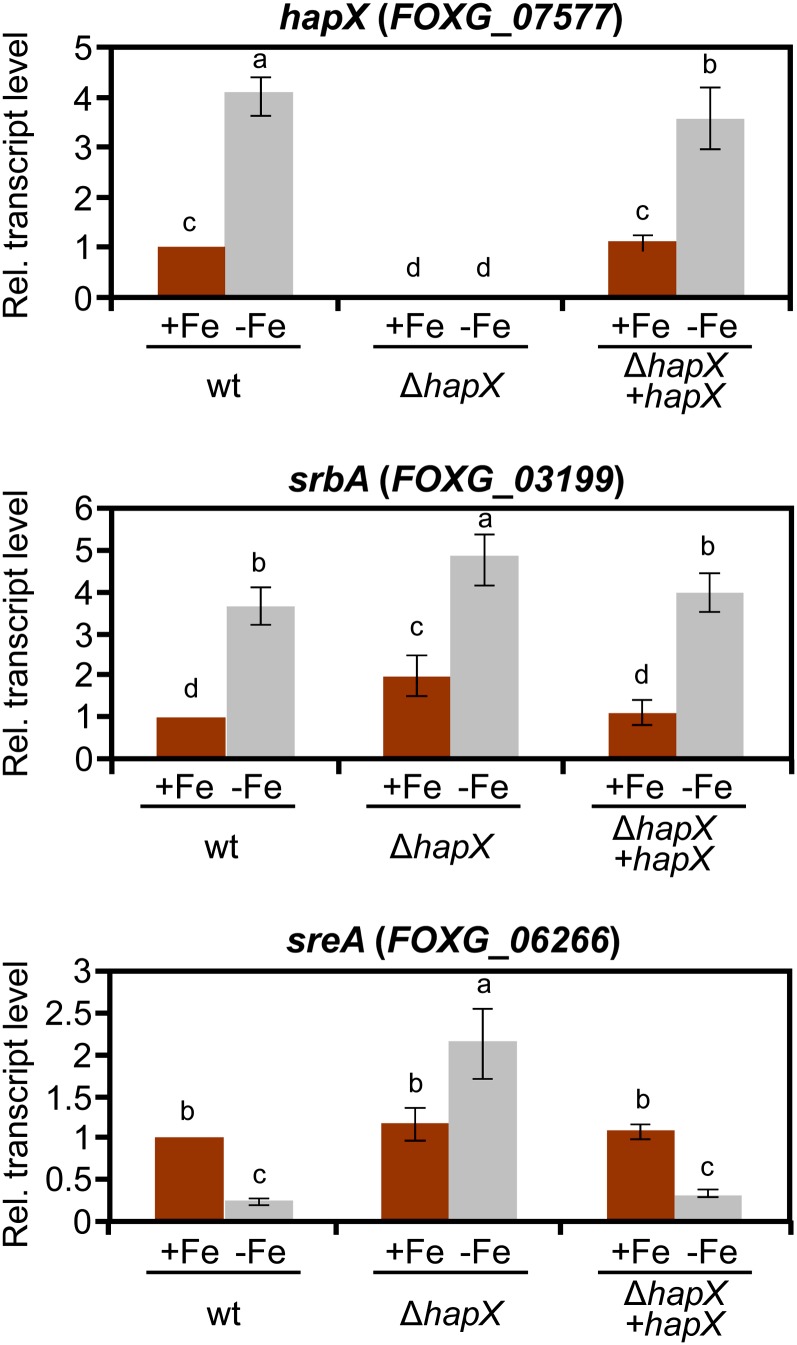
We next examined the role of iron and HapX in the transcription of known iron regulatory genes. Transcript levels of hapX were significantly upregulated in the wild type during iron starvation, indicating that hapX is an iron-repressed gene (Figure 2). Expression of the srbA gene encoding a sterol regulatory element binding protein that regulates iron acquisition in response to hypoxia (Blatzer et al., 2011a) was also induced by iron depletion, and this process was independent of HapX. However, transcript levels of sreA encoding a negative regulator of siderophore biosynthesis decreased under iron starvation conditions, and this decrease was strictly dependent on HapX (Figure 2).
Figure 2.
Deletion of hapX Affects Expression of Iron Regulatory Genes.
Quantitative real-time RT-PCR analysis was performed in the indicated strains germinated for 16 h in iron-depleted MM and transferred for 10 h to iron-depleted (−Fe) or iron-replete MM (+Fe). Transcript levels of hapX, sreA, and srbA genes are expressed relative to those of the wild-type (wt) strain grown under iron-replete conditions. Bars represent se from three independent experiments with three technical replicates each. Values with the same letter are not significantly different according to the Mann-Whitney test (P = 0.05).
[See online article for color version of this figure.]
Deletion of hapX Leads to Deregulation of Siderophore Biosynthesis
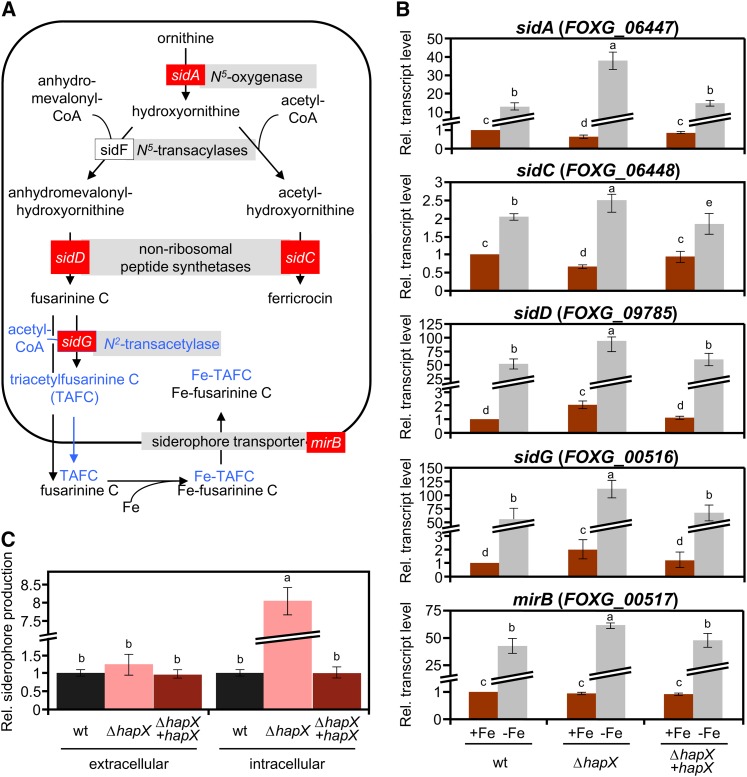
With the aim of further characterizing the iron uptake system in F. oxysporum, we interrogated the genome database using BLASTP and found putative structural orthologs of all the key siderophore biosynthetic genes characterized in Aspergillus (Figure 3A; see Supplemental Table 1 online). In addition, we identified a second ortholog of sidC, FOXG_17422, which is specific for the genus Fusarium (see Supplemental Figure 3 online). To explore the role of HapX in the regulation of siderophores, we monitored the expression of key genes involved in iron uptake: sidA (encoding Orn monooxygenase), sidC and sidD (encoding two siderophore nonribosomal peptide synthetases [NRPSs]), sidG (encoding fusarinine C [FsC]–acetyl CoA–N2-transacetylase), and mirB (encoding a putative siderophore transporter). Transcript levels of these genes were sharply increased in mycelia grown under iron starvation as compared with iron-sufficient conditions (Figure 3B). In line with these data, a chrome azurol S (CAS) assay detected significant amounts of extracellular siderophores in culture supernatants of fungal strains grown under iron starvation conditions (Figure 3C) but not under iron-sufficient conditions. Transcriptional induction of siderophore genes by iron starvation was higher in the ΔhapX mutant (200, 25, 140, 180, and 65%, respectively, compared with the wild type), although total levels of extracellular siderophores did not differ significantly between the two strains. Strikingly, however, intracellular siderophore content in mycelia of the ΔhapX mutant was eight times higher than in the wild type and the ΔhapX+hapX strain (Figure 3C).
Figure 3.
Siderophore Biosynthesis Is Upregulated in the ΔhapX Mutant during Iron Starvation.
(A) Siderophore biosynthetic pathways based on the information available in Aspergillus. Genes marked in red were used for transcriptional analysis in F. oxysporum (B). Putative conversion of FsC to TAFC suggested by genome inspection, but not detected in the siderophore analysis, is shown in blue.
(B) Quantitative real-time RT-PCR analysis of the indicted genes was performed in fungal strains grown as described in Figure 2. Transcript levels of sidA, sidC, sidD, sidG, and mirB genes are expressed relative to those of the wild-type (wt) strain grown under iron-replete conditions. Bars represent se from three independent experiments with three technical replicates each.
(C) CAS assay-mediated quantification of extra- and intracellular siderophore production during iron starvation, normalized to the wild-type strain. Bars represent se from three independent experiments with three technical replicates each. Values with the same letter are not significantly different according to the Mann-Whitney test (P = 0.05).
[See online article for color version of this figure.]
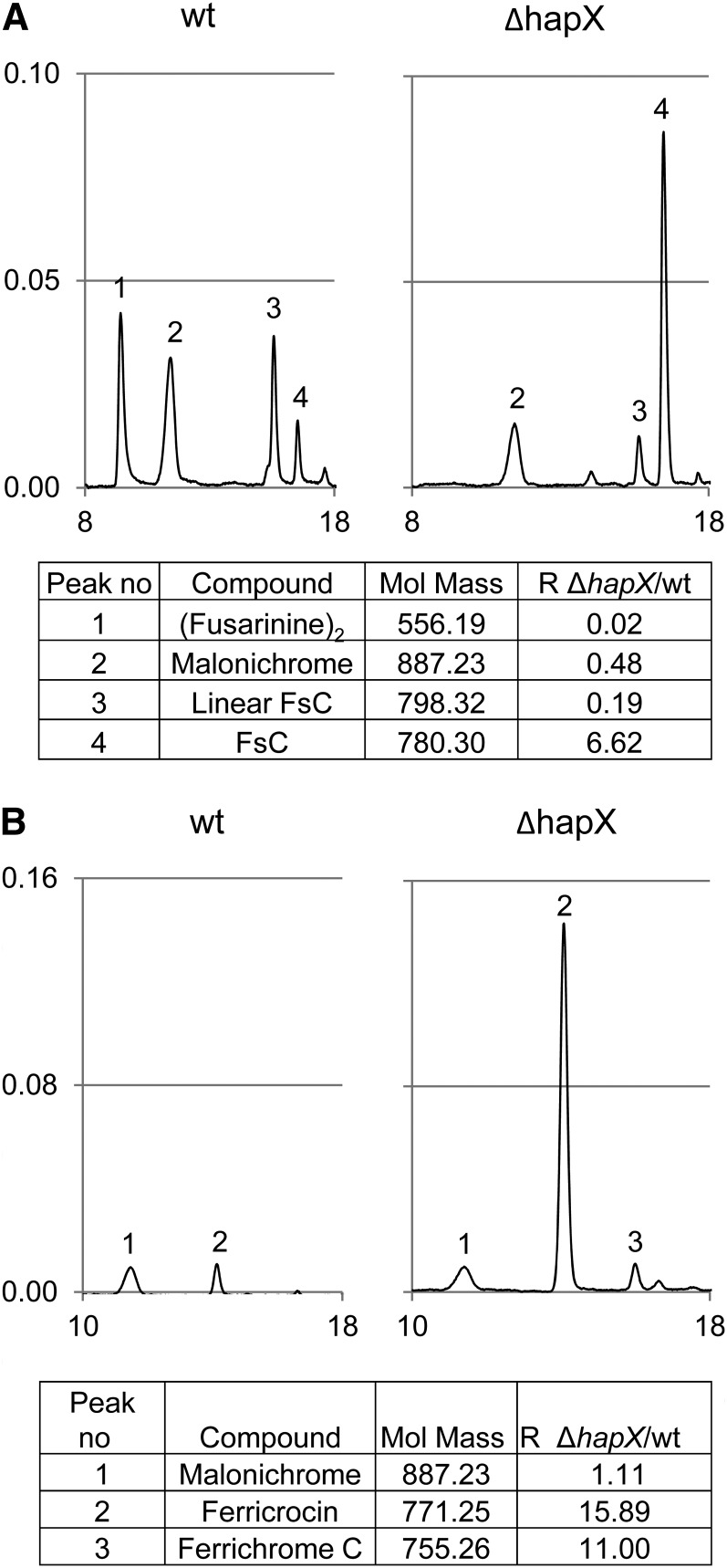
Analysis of culture supernatants by a combination of reversed-phase high-performance liquid chromatography (HPLC) and high-resolution electrospray ionization mass spectroscopy (MS) detected two different siderophores, FsC and malonichrome (Figure 4A). Malonichrome (peak 2) is a ferrichrome-type compound consisting of a cyclic hexapeptide with the structure Gly-Ala-Gly-(N5-malonyl-N5-hydroxyornithine)3 and was previously described as an extracellular siderophore of Fusarium roseum (Emery, 1980). FsC (peak 4) is a cyclic tripeptide consisting of three N5-anhydromevalonyl-N5-hydroxyornithine (termed fusarinine) residues linked by ester bonds and was reported from a variety of fungi, including Aspergillus spp and Fusarium spp (Haas et al., 2008). In addition to cyclic FsC, F. oxysporum supernatants contained linear FsC and dimeric fusarinine (fusarinine)2 (peaks 3 and 1, respectively), two degradation products derived from FsC by hydrolysis of one or two ester bonds, respectively (Moore and Emery, 1976). The rate of FsC degradation detected in the wild type was higher than in the ΔhapX mutant (Figure 4A, compare peaks 1, 3, and 4), possibly because of differences in the biochemical composition of the wild-type and mutant supernatants or the fungal biomass produced (Figure 1C). It is also worth noting that the pH of the ΔhapX culture supernatant (4.7) was much lower than that of the wild type (7.2).
Figure 4.
Characterization of Siderophores Produced by F. oxysporum.
Siderophores were analyzed by MS/HPLC of culture supernatants (A) and cellular extracts (B) after iron saturation. In the representative HPLC chromatograms, the y and x axis denote absorption at 430 nm and retention time in minutes, respectively. Note that deletion of hapX leads to a decrease in degradation of FsC to linear FsC and (fusarinine)2 as well as to higher production of ferricrocin and ferrichrome C. Mol. Mass, molecular mass; R, ΔhapX/wild-type (wt) ratio of siderophore production.
Three intracellular ferrichrome-type siderophores were detected in mycelial extracts: malonichrome, ferricrocin, and ferrichrome C (Figure 4B). Similar to malonichrome, ferricrocin and ferrichrome C are cyclic hexapeptides whose structures are Gly-Ser-Gly-(N5-acetyl-N5-hydroxyornithine)3 and Gly-Ala-Gly-(N5-acetyl-N5-hydroxyornithine)3, respectively (Haas et al., 2008). Mycelia of the ΔhapX mutant showed a dramatic increase of ferricrocin and ferrichrome C levels (16- and 11-fold, respectively), confirming the results obtained in the CAS assay.
Deletion of hapX Causes Derepression of Iron Regulated Genes and Accumulation of Protoporphyrin IX under Iron Starvation Conditions
Because hapX deletion had no apparent effect on iron uptake (Figure 1D), we asked whether the growth defects of the ΔhapX mutant under iron-limiting conditions could result from iron misuse. To test this hypothesis, global RNA expression profiles of the wild type and the ΔhapX mutant were compared under iron-sufficient and iron-limiting conditions. To search for genes that are repressed under iron starvation in a HapX-dependent manner, we established two selection criteria: (1) a minimum twofold downregulation under steady state iron starvation versus iron sufficiency in the wild type (genes repressed by iron starvation); (2) a minimum twofold upregulation during steady state iron-starved growth in the ΔhapX mutant relative to the wild type (genes derepressed in ΔhapX under iron starvation). Among the 114 genes identified in the screen, 23 (20%) can be directly assigned to iron-dependent processes, such as respiration, the tricarboxylic acid (TCA) cycle, amino acid metabolism, iron-sulfur-cluster biosynthesis, heme biosynthesis, oxidative stress detoxification, vacuolar iron storage, and iron regulation (see Supplemental Data Set 1 online). A comparison with previous genome-wide transcriptional profiling studies in A. fumigatus (Schrettl et al., 2010), C. albicans (Chen et al., 2011), and Schizosaccharomyces pombe (Mercier et al., 2008) defines a list of gene orthologs that share HapX-dependent repression under iron starvation conditions (Table 1). Notably, in Saccharomyces cerevisiae, which lacks a HapX ortholog, a large proportion of this gene set is posttranscriptionally repressed during iron starvation by Cth1 and Cth2 (Puig et al., 2008).
Table 1. Iron Metabolism-Related Genes Repressed by HapX Orthologs under Iron Starvation.
| Gene | Gene Product | Wild Type ± Fe, log2 | Wild Type/hapx −Fe, log2 | Af | Ca | Sp | Sc |
|---|---|---|---|---|---|---|---|
| FOXG_02862 | Copper-resistance protein Crd2 | 3.93 | −2.44 | x | |||
| FOXG_06292 | 3-isopropylmalate dehydratase (Ile/Leu/Val biosynthesis) | 3.17 | −4.09 | x | x | x | x |
| FOXG_15294 | Mycelial catalase Cat2 (heme) | 3.10 | −1.84 | ||||
| FOXG_04395 | Dihydroxy acid dehydratase (Ile/Val biosynthesis) | 3.07 | −1.56 | x | x | x | x |
| FOXG_04047 | Vacuolar iron transporter Ccc1 | 2.74 | −2.79 | x | x | x | |
| FOXG_10653 | Pyridine nucleotide-disulphide oxidoreductase | 2.71 | −1.62 | x | x | ||
| FOXG_05207 | Aconitate hydratase, mitochondrial | 2.54 | −2.06 | ||||
| FOXG_10442 | Nitrite/sulfite reductase ferredoxin-like half domain | 2.49 | −2.37 | ||||
| FOXG_04849 | Cytochrome P450 monooxygenase | 2.39 | −1.61 | ||||
| FOXG_09278 | Succinate dehydrogenase iron-sulfur protein; TCA cycle | 2.37 | −1.21 | x | x | x | |
| FOXG_11281 | NADH-dependent glutamate synthase (GLT1) | 2.32 | −2.22 | x | x | x | x |
| FOXG_12892 | lysF, Aconitase family (aconitate hydratase, Lys biosynthesis)a | 2.30 | −1.94 | x | x | x | |
| FOXG_12260 | Peroxidase; catalase/peroxidase HPI | 2.27 | −1.48 | ||||
| FOXG_01544 | Succinate dehydrogenase, flavoprotein subunit; TCA cycle | 2.24 | −1.25 | x | x | x | |
| FOXG_01159 | Homocitrate synthase (Lys biosynthesis) | 2.21 | −1.00 | x | |||
| FOXG_09472 | Cytochrome P450 | 2.20 | −1.08 | ||||
| FOXG_05118 | Acyl-CoA dehydrogenase | 1.84 | −1.05 | ||||
| FOXG_03713 | acoA, Aconitase family (aconitate hydratase), TCA cyclea | 1.73 | −1.41 | x | x | x | x |
| FOXG_08910 | Cytochrome P450 monooxygenase | 1.63 | −1.97 | ||||
| FOXG_00142 | Cytochrome c peroxidase | 1.54 | −1.33 | x | x | ||
| FOXG_06266 | Siderophore transcription factor SreAa | 1.23 | −1.60 | x | x | x | |
| FOXG_06399 | FMN-dependent dehydrogenase | 1.17 | −2.22 | ||||
| FOXG_12815 | cycA, Cytochrome ca | 1.12 | −1.49 | x | x | x | x |
| FOXG_01103 | Hemerythrin HHE cation binding domain-containing protein | 1.06 | −1.22 |
F. oxysporum genes that fulfill the following criteria in microarray-based transcriptional profiling: (1) More than twofold downregulated in the wild type under steady state iron starvation versus iron sufficiency (Wild Type ± Fe) and (2) more than twofold upregulated during steady state iron-starved growth in ΔhapX compared with the wild type (Wild Type/hapx − Fe). Genes are ranked based on the level of upregulation in the Wild Type ± Fe condition. The x indicates genes previously identified to be similarly regulated by the HapX orthologs in A. fumigatus (Af; Schrettl et al., 2010), C. albicans (Ca; Chen et al., 2011), and S. pombe (Sp; Mercier et al., 2008) or by Cth1 and Cth2 in S. cerevisiae (Sc; Puig et al., 2008).
Genes whose differential expression was confirmed separately by real-time RT-PCR.
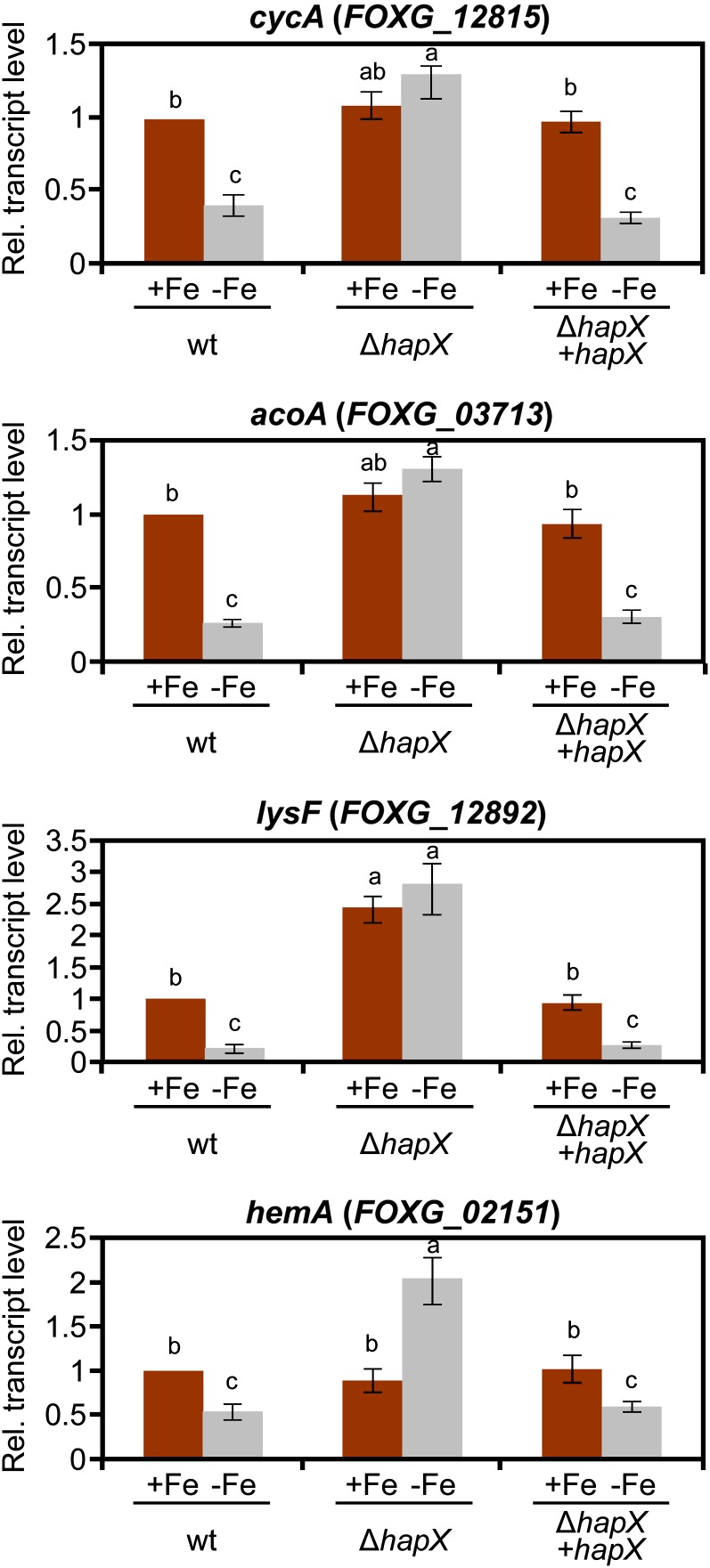
Real-time RT-PCR was performed to measure transcript levels of representative genes from different iron-consuming pathways: cycA encoding the heme protein cytochrome C (respiration), acoA and lysF encoding the iron-sulfur proteins aconitase and homoaconitase (TCA cycle and Lys biosynthesis, respectively), and hemA encoding the α-amino-levulinic acid synthase (heme biosynthesis), (Hortschansky et al., 2007; Schrettl et al., 2010). All of these were confirmed to exhibit strong repression under iron-depleted conditions in the wild type but not in the ΔhapX mutant (Figure 5). Inspection of the regulatory regions revealed the presence of multiple CCAAT motifs, representing potential binding sites of the Hap protein complex (see Supplemental Figure 4 online).
Figure 5.
Deletion of hapX Causes Deregulation of Genes Involved in Iron Use.
Quantitative real-time RT-PCR analysis was performed in the indicated strains grown as described in Figure 2. Transcript levels of cycA, acoA, lysF, and hemA genes are expressed relative to those of the wild-type (wt) strain grown under iron-replete conditions. Bars represent se from three independent experiments with three technical replicates each. Values with the same letter are not significantly different according to the Mann-Whitney test (P = 0.05).
[See online article for color version of this figure.]
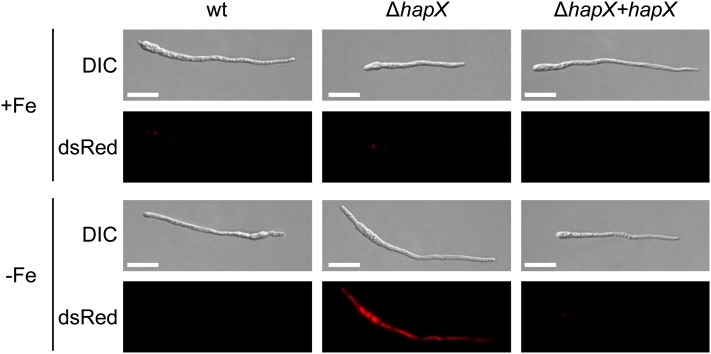
Under iron-depleted conditions, hyphae of the ΔhapX mutant exhibited a characteristic red autofluorescence (Figure 6), which indicates the accumulation of protoporphyrin IX (PpIX). HPLC analysis detected high levels of PpIX in mycelial extracts of the ΔhapX strain (141.67 ± 11.35 pmol ⋅ mg protein−1) but not in those of the wild type and the ΔhapX+hapX strains. PpIX accumulation was previously shown in Aspergillus to result from derepression of heme biosynthesis (Hortschansky et al., 2007; Schrettl et al., 2010). Collectively, these results demonstrate that HapX is important for iron starvation–induced repression of iron-using pathways in F. oxysporum.
Figure 6.
PpIX Accumulates in the ΔhapX Mutant during Iron Starvation.
Fungal strains were grown for 24 h in MM with or without iron and were observed microscopically using the Nomarski technique (DIC) to visualize germlings or a dsRed-fluorescence filter to detect PpIX autofluorescence. wt, wild type.
Bars = 20 μm.
[See online article for color version of this figure.]
HapX Is Required for Transcriptional Activation of a Subset of Genes under Iron Starvation Conditions
We next asked whether HapX could function as an activator of gene expression under iron starvation. To this aim, the transcriptional profiling data were analyzed according to the following criteria: (1) a minimum twofold upregulation under steady state iron starvation versus iron sufficiency in the wild type (genes induced by iron starvation); and (2) a minimum twofold downregulation during steady state iron-starved growth in the ΔhapX mutant relative to the wild type (genes whose induction by iron starvation depends on HapX). The screen identified a set of 88 HapX-dependent iron starvation–induced genes (see Supplemental Data Set 2 online), which was significantly enriched in predicted secreted proteins (22% compared with 11% of the total predicted genes in F. oxysporum). Some of the genes encode known or potential virulence factors, such as secreted aspartyl proteases, cell wall–degrading enzymes, PTH11-like integral membrane proteins, or the secreted in xylem (SIX3) protein (Table 2). Interestingly, the HapX-regulated gene set also includes two hAT family transposases (FOXG_14471 and FOXG_15009) and a reverse transcriptase (FOXG_12552) (see Supplemental Data Set 2 online).
Table 2. Putative Virulence-Related Genes Induced by Iron Starvation in a HapX-Dependent Manner.
| Gene | Gene Product | Wild Type ± Fe, log2 | Wild Type/hapX − Fe, log2 | SignalP | THMM |
|---|---|---|---|---|---|
| FOXG_04016 | Hypersensitive response–inducing protein | −3.54 | 1.09 | x | |
| FOXG_10677 | PTH11-like integral membrane protein | −2.30 | 1.64 | x | |
| FOXG_16469 | PTH11-like integral membrane protein | −1.93 | 1.07 | x | x |
| FOXG_15833 | Aspartyl protease | −1.49 | 1.50 | x | x |
| FOXG_09759 | Endo-1,3-beta-glucanase | −1.49 | 1.04 | x | |
| FOXG_11325 | Aspartyl protease | −1.20 | 1.02 | x | |
| FOXG_10097 | Aspartyl protease | −1.17 | 1.25 | x | |
| FOXG_10728 | Pectate lyase | −1.08 | 1.23 | x | |
| FOXG_09366 | ABC transporter | −1.08 | 1.19 | x | |
| FOXG_16838 | Endoglucanase | −1.06 | 1.19 | x | |
| FOXG_16398 | SIX3 | −1.05 | 1.19 |
Putative virulence-related genes of F. oxysporum are shown that fulfill the following criteria in microarray-based transcriptional profiling: (1) more than twofold upregulated in the wild type under steady state iron starvation versus iron sufficiency (Wild Type ± Fe) and (2) more than twofold downregulated during steady state iron-starved growth in ΔhapX compared with the wild type (Wild Type/hapx − Fe). Genes are ranked based on the level of downregulation in the Wild Type ± Fe condition. Genes predicted to encode a signal peptide and/or transmembrane domain are marked with an x in the columns SignalP and THMM, respectively.
HapX Governs Virulence of F. oxysporum on Tomato Plants and Immunodepressed Mice
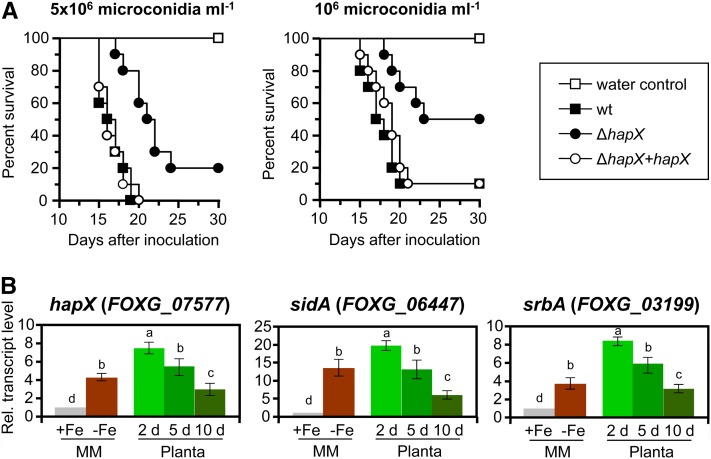
When tomato roots were inoculated with microconidial suspensions of the different fungal strains (5 × 106 or 106 microconidia ⋅ mL−1), development of vascular wilt symptoms and mortality was significantly lower (P = 0.0001 and P = 0.0059, respectively) in plants infected with the ΔhapX mutant than in those infected with the wild type or the complemented strain (Figure 7A). The hapX, sidA, and srbA genes were sharply upregulated in F. oxysporum during the early stages of tomato root infection, consistent with an extreme iron starvation response, and then gradually decreased during the later stages of the disease (Figure 7B).
Figure 7.
HapX Governs Virulence of F. oxysporum on Tomato Plants.
(A) Groups of 10 tomato plants (cv Monika) were inoculated by dipping roots into a suspension of 5 × 106 or 106 freshly obtained microconidia mL−1 of the indicated fungal strains. Percentage survival was plotted for 30 d. Data shown are from one representative experiment. wt, wild type.
(B) Quantitative real-time RT-PCR analysis was performed with total RNA obtained from the wild-type strain grown on MM with or without iron as described in Figure 2 or from inoculated tomato roots 2, 5, and 10 DAI. Transcript levels are expressed relative to those in iron-replete conditions (+Fe). Bars represent se from three independent experiments with three technical replicates each. Values with the same letter are not significantly different according to the Mann-Whitney test (P = 0.05).
[See online article for color version of this figure.]
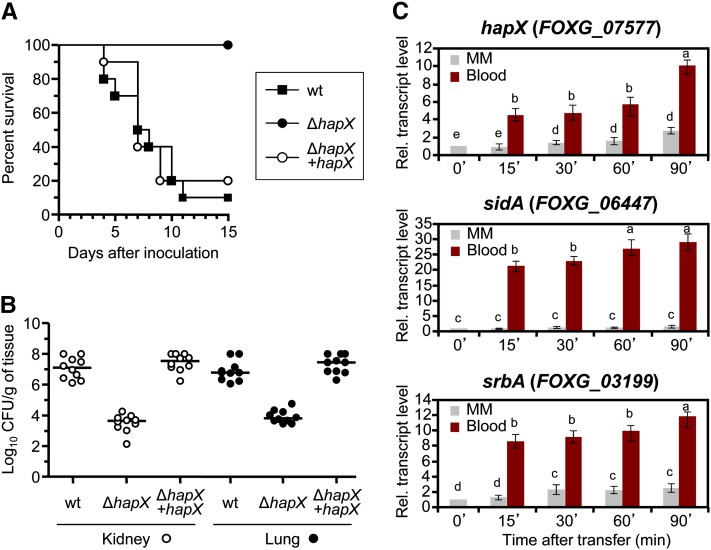
Besides causing vascular wilt on tomato plants, F. o. lycopersici strain 4287 can infect and kill immunodepressed mice (Ortoneda et al., 2004). We found that mortality rates in mice inoculated with the ΔhapX mutant were significantly lower (P < 0.0003) than in those inoculated with the wild type or the ΔhapX+hapX strain. All of the mice infected with the ΔhapX mutant survived the experiment, whereas the wild type and the complemented strain consistently caused death in 80 to 90% of the animals (Figure 8A). Fungal tissue burden in lungs and kidneys of surviving mice inoculated with the ΔhapX mutant was significantly lower (P < 0.0001) than in animals inoculated with the wild type or ΔhapX+hapX (Figure 8B). We also noted that transfer of F. oxysporum from iron-depleted minimal medium (MM) to human blood triggered a rapid transcriptional upregulation of the hapX, sidA, and srbA genes (Figure 8C). These results establish a key role for HapX in reprogramming of iron-dependent gene expression during infectious growth of F. oxysporum on plant and mammalian hosts.
Figure 8.
HapX Is Essential for Dissemination and Virulence of F. oxysporum in Immunodepressed Mice.
(A) Groups of 10 immunodepressed Oncins France 1 male mice were inoculated with 2 × 107 microconidia of the indicated strains by lateral tail vein injection. Percentage survival was plotted for 15 d. Data shown are from one representative experiment. wt, wild type.
(B) Ten randomly chosen surviving mice from each treatment were sacrificed 7 DAI, and homogenates obtained from the indicated organs were quantitatively cultured on PDA medium.
(C) Quantitative real-time RT-PCR analysis was performed with total RNA obtained from the wild-type strain germinated for 20 h (16 h at 28°C + 4 h at 37°C) in iron-depleted MM and then transferred for the indicated time periods (min) to fresh MM or whole human blood. Transcript levels are expressed relative to those in MM at time 0. Bars represent se from three independent experiments with three technical replicates each. Values with the same letter are not significantly different according to the Mann-Whitney test (P = 0.05).
[See online article for color version of this figure.]
HapX Mediates Iron Competition of F. oxysporum against Root-Colonizing Pseudomonads
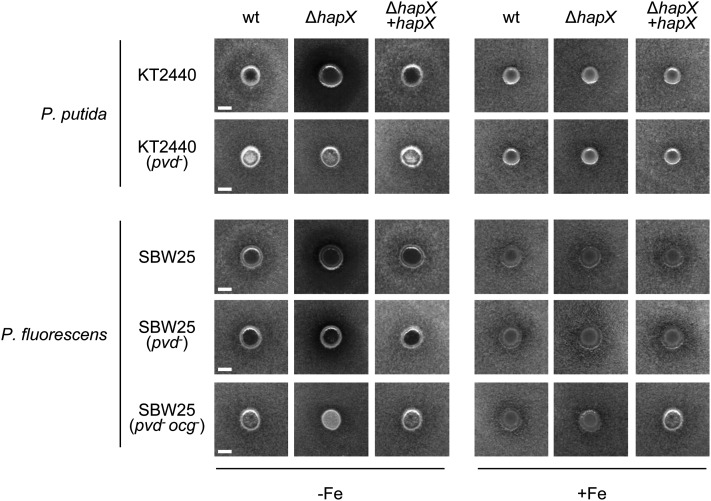
F. oxysporum competes for limited iron with other rhizosphere-inhabiting microorganisms, such as fluorescent pseudomonads (Scher and Baker, 1982; Simeoni et al., 1987). We tested the role of HapX in the interaction of F. oxysporum with two root-colonizing Pseudomonas isolates: Pseudomonas putida KT2440 producing the siderophore Pvd (Matthijs et al., 2009) and Pseudomonas fluorescens SBW25 producing both Pvd and Ocg (Matthijs et al., 2008). In vitro, both Pseudomonas spp displayed an antagonistic effect against F. oxysporum, visible as a halo of mycelial growth inhibition around the bacterial colony (Figure 9). The antagonistic effect was specific for iron-depleted conditions and was dependent on siderophore production by Pseudomonas spp, because it was abolished in the P. putida pvd− and P. fluorescens pvd− ocg− mutants. This finding strongly suggests that growth inhibition of F. oxysporum is linked to competition for iron. Further supporting this idea, the antagonistic effect of the Pseudomonas spp wild-type strains was exacerbated against the F. oxysporum ΔhapX mutant, but this effect was not detected in the P. putida pvd− and P. fluorescens pvd− ocg− mutants (Figure 9). Analogous results were obtained using a different in vitro antagonism assay (see Supplemental Figures 5 and 6 online).
Figure 9.
HapX Is Required for Efficient Iron Competition of F. oxysporum with Fluorescent Pseudomonads.
Growth of the indicated F. oxysporum strains was determined on solid MM in the presence of the rhizosphere-colonizing bacterial strains P. putida KT2440 (wild-type [wt] or pvd−, strain lacking the siderophore Pvd) or P. fluorescens SBW25 (wild-type, pvd−, or pvd− ocg− lacking the siderophores Pvd and Ocg). F. oxysporum microconidia were evenly spread on top of Glc casamino acids medium with or without iron. Bacterial strains were point-inoculated on the same day, and cultures were incubated for 4 d at 28°C. Mycelial growth is visible as a gray background, whereas growth inhibition appears as a dark halo surrounding the bacterial colony.
Bars = 5 mm.
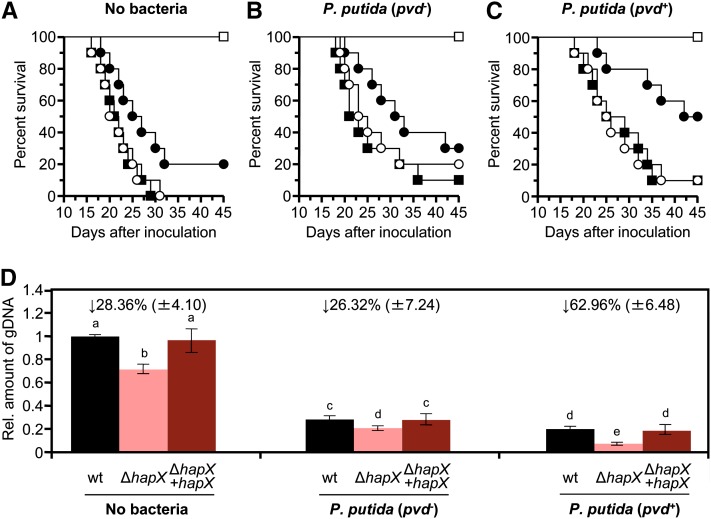
We next tested the role of HapX during plant infection by F. oxysporum in the presence of the rhizosphere-colonizing strain P. putida KT2440. Coinoculation was performed by dipping tomato roots for 2 h in a suspension of 109 bacterial cells ⋅ mL−1, followed by normal inoculation with F. oxysporum microconidia. As found in previous experiments (Figure 7), mortality rates of tomato plants inoculated with the ΔhapX mutant were significantly lower than those of plants inoculated with the wild type or ΔhapX+hapX strains (Figures 10A to 10C; see Supplemental Table 2 online). Coinoculation of tomato roots with P. putida KT2440 resulted in a significant delay in plant mortality caused by the different F. oxysporum strains, confirming the previously reported biocontrol activity of this bacterial isolate. Strikingly, the attenuation in virulence of the ΔhapX mutant was exacerbated by coinoculation with the P. putida KT2440 wild-type strain but not with the pvd− mutant (cf. Figures 10A to 10C). This strongly suggests that the inhibitory effect of P. putida KT2440 against F. oxysporum ΔhapX is partially caused by competition for iron in the tomato rhizosphere.
Figure 10.
Loss of HapX Increases Biocontrol Activity of Siderophore-Producing P. putida against F. oxysporum on Tomato Plants.
(A) Groups of 10 tomato plants (cv Monika) were dip-inoculated with a microconidial suspension of the following fungal strains as described in Figure 7A: wild-type, filled squares; ΔhapX, filled circles; ΔhapX+hapX, empty circles; water control, empty squares.
(B) and (C) Before fungal inoculation, roots were submerged for 2 h in a suspension of 109 cells mL−1 of the indicated P. putida strain. The severity of disease symptoms was monitored periodically and percentage survival plotted for 45 d. Data shown are from one representative experiment.
(D) Quantitative real-time PCR was used to measure the relative amount of fungal DNA in total genomic DNA extracted from tomato roots at 7 DAI with the indicated microorganisms. Amplification levels are expressed relative to those obtained from plants infected with the F. oxysporum wild-type (wt) strain. Numbers above columns indicate the percentage reduction of fungal biomass in the ΔhapX strain relative to the wild-type strain in the same condition. Bars represent se from three independent experiments with three technical replicates each.
[See online article for color version of this figure.]
Tomato plants infected with the ΔhapX mutant contained significantly less fungal biomass than those infected with the wild type or the complemented strain, as determined by real-time quantitative PCR of total DNA extracted from tomato roots at 7 d after inoculation (DAI) (Figure 10D). Concomitant with the reduction in disease severity, we noted a decrease of F. oxysporum biomass in plants coinoculated with P. putida KT2440. Strikingly, the relative decrease of fungal biomass in plants inoculated with the F. oxysporum ΔhapX mutant was 2.5 times stronger after coinoculation with the P. putida wild-type strain compared with the pvd− mutant (Figure 10D). Collectively, these results show that HapX functions in iron competition of F. oxysporum against siderophore-producing pseudomonads and that this role has a direct effect on the ability of the fungus to proliferate in the rhizosphere and to cause disease on tomato plants.
DISCUSSION
Iron is essential for virtually every organism. Although iron is abundant on earth, its availability is limited because of oxidation to insoluble forms by atmospheric oxygen. For this reason, and also because of its toxicity when present in excess, organisms have developed efficient strategies for iron homeostasis. In fungi, iron starvation increases expression of genes required for siderophore-mediated iron uptake (Mei et al., 1993; Haas et al., 2003; Oide et al., 2006; Greenshields et al., 2007; Schrettl et al., 2007). Meanwhile, iron-consuming pathways are rapidly downregulated to optimize the use of the limited iron resource (Oberegger et al., 2002; Hortschansky et al., 2007; Schrettl et al., 2010; Hsu et al., 2011).
When initiating this study, we hypothesized that a soilborne pathogen, such as F. oxysporum, must face iron limitation both during saprophytic and pathogenic stages of its life cycle. In natural soils, total soluble Fe3+ represents as little as ∼10−10 M at equilibrium with soil iron (Simeoni et al., 1987), resulting in competition for iron among soil-inhabiting microorganisms (Haas and Défago, 2005). The availability of iron may be even more limited in mammalian or plant hosts, both of which have efficient mechanisms to sequester iron from invading microorganisms (Jurkevitch et al., 1993; Skaar, 2010). In support of this idea, loss of siderophores decreases virulence in microbial pathogens of humans (Ratledge and Dover, 2000; Schrettl et al., 2004; Schrettl et al., 2007; Cornelis, 2010) and plants (Mei et al., 1993; Schrettl et al., 2004; Oide et al., 2006; Greenshields et al., 2007; Schrettl et al., 2007). Here we find that the bZIP protein HapX, a conserved regulator of fungal iron homeostasis, is required for adaptation of F. oxysporum to iron-limiting conditions. Loss of HapX affects multiple aspects of the fungal life cycle, including saprophytic growth, competition with other microorganisms, as well as virulence on plant and mammalian hosts.
HapX Mediates Adaptation to Iron Starvation by Downregulating Iron-Consuming Pathways
Our data suggest that HapX is necessary for efficient growth of F. oxysporum under iron-limiting conditions but not under iron sufficiency. This result is similar to those reported in A. nidulans, A. fumigatus, C. neoformans, and C. albicans (Hortschansky et al., 2007; Jung et al., 2010; Schrettl et al., 2010; Hsu et al., 2011). The specific role during iron starvation suggests a key function of HapX in iron acquisition and/or use. However, the F. oxysporum ΔhapX mutant is not affected in iron uptake. This result is in line with a previous study in the human pathogen C. albicans (Hsu et al., 2011). By contrast, global RNA expression profiling revealed that HapX is essential for iron starvation–triggered downregulation of genes from different iron-dependent pathways. Thus, a key role of HapX in iron homeostasis of F. oxysporum is to shut down iron-consuming processes, such as respiration, amino acid metabolism, the citric acid cycle, or heme biosynthesis, when iron becomes limiting. Genome-wide transcriptional analyses in S. pombe, A. fumigatus, C. neoformans, and C. albicans also suggested that HapX orthologs are required for downregulation of iron-consuming genes during iron starvation (Mercier et al., 2006; Puig et al., 2008; Jung et al., 2010; Schrettl et al., 2010; Chen et al., 2011). This hypothesis is further corroborated by our finding that the F. oxysporum ΔhapX mutant has dramatically increased levels of PpIX as a consequence of heme pathway deregulation. Collectively, these results establish a conserved function of HapX in repression of iron-dependent pathways and highlight the essential role of this transcription factor in fungal adaptation to iron starvation conditions.
HapX Is a Regulator of Siderophore Biosynthesis
F. oxysporum ΔhapX mutants show an increase in transcript levels of siderophore biosynthetic genes under iron limitation. Accordingly, levels of the intracellular siderophores ferricrocin and ferrichrome C during iron limitation are much higher in the ΔhapX mutant than in the wild type. By contrast, loss of HapX in Aspergillus caused a reduction in triacetylfusarinine C (TAFC), an extracellular siderophore derived by N2-acetylation of FsC (Hortschansky et al., 2007; Schrettl et al., 2010). We detected only FsC, but not TAFC, in F. oxysporum culture supernatants. This result was unexpected, because TAFC has been reported both in Aspergillus spp and Fusarium graminearum (Oide et al., 2006; Schrettl et al., 2007; Blatzer et al., 2011b; Yasmin et al., 2012). The F. oxysporum genome encodes orthologs of all the known enzymes involved in TAFC biosynthesis, including SidG, which catalyzes N2-acetylation of FsC (Figure 4A; see Supplemental Table 1 online). Expression of sidG was detected by quantitative real-time RT-PCR (Figure 3B); therefore, we speculate that the F. oxysporum SidG homolog may lack sufficient enzymatic activity for TAFC production under the conditions tested.
Ferricrocin is the major intracellular siderophore in most Ascomycetes analyzed so far (Haas et al., 2008). The F. oxysporum genome encodes two NRPSs with similarity to ferrichrome-type NRPSs, such as A. fumigatus SidC, suggesting that at least two of the three intracellular siderophores detected in this study are synthesized by the same NRPS. Loss of HapX resulted in a coordinated increase of the ferricrocin and ferrichrome C contents, suggesting that the two siderophores may be synthesized by the same NRPS. The structure of ferrichrome C closely resembles that of ferricrocin, with the exception of an Ala replacing a Ser. The cellular content of ferricrocin is ∼15 times higher than that of ferrichrome C; therefore, it is feasible that ferrichrome C is synthesized by the ferricrocin-specific NRPS through relaxed specificity of the Ser-specific adenylation domain (Haas et al., 2008).
HapX is required for downregulation of sreA, encoding a repressor of siderophore biosynthesis, during iron-limiting conditions (this study; Hortschansky et al., 2007; Schrettl et al., 2010). HapX and SreA are interconnected by a negative feedback loop in A. nidulans, A. fumigatus, and S. pombe (Mercier et al., 2006; Hortschansky et al., 2007; Schrettl et al., 2010). The ΔhapX mutant was thus expected to display constitutive repression of siderophore genes and a reduction of the intra- and extracellular pool of siderophores during iron starvation. Instead, transcription of sidA, sidC, sidD, sidG, and mirB as well as intracellular siderophore levels were increased in the ΔhapX mutant during iron limitation. In A. nidulans, transcript levels of sidC during iron limitation were also increased in the ΔhapX mutant (Hortschansky et al., 2007). By contrast, siderophore biosynthetic genes in the A. fumigatus ΔhapX mutant were downregulated during iron starvation (Schrettl et al., 2010). How siderophore biosynthesis remains activated in the ΔhapX mutant while the sreA repressor gene is upregulated remains to be determined. It has been suggested that both SreA and HapX may posttranslationally sense iron (Haas et al., 1999; Hortschansky et al., 2007). In S. pombe, HapX and SreA orthologs interact with the monothiol glutaredoxin Grx3 for iron regulation (Mercier and Labbé, 2009; Jbel et al., 2011; Kim et al., 2011). Our data further support the idea that the SreA repressor is only functional under iron sufficiency. According to this model, sreA derepression in the ΔhapX mutant during iron limitation does not result in reduced siderophore production, because SreA is present in an inactive state. Further studies are needed to confirm this hypothesis.
HapX Contributes to Iron Competition of F. oxysporum against Soil- and Rhizosphere-Inhabiting Pseudomonads
Vascular wilts caused by F. oxysporum f spp are among the most common plant diseases in agriculture (Dean et al., 2012). Chemical control of the pathogen is largely unviable, because of regulatory restrictions and its high persistence in the soil (Agrios, 1997). Biological control measures, such as disease-suppressive soils, have been investigated for more than 70 years (Hornby, 1979; Alabouvette et al., 2009). The genus Pseudomonas is among the most extensively studied biocontrol agents (Alabouvette et al., 1979; Kloepper et al., 1980; Haas and Défago, 2005) and includes aggressive rhizosphere-colonizing strains that can increase plant growth and improve plant health (Haas and Défago, 2005; Weller, 2007; Cornelis, 2010). Known biocontrol mechanisms include the production of antibiotics, hydrogen cyanide, lytic exoenzymes (Thomashow and Weller, 1996), cyclic lipopeptides (Raaijmakers et al., 2006), competition for nutrient niches (Kamilova et al., 2005), siderophore-mediated iron competition (Thomashow and Weller, 1996), and induced systemic resistance (De Vleesschauwer and Höfte, 2009). The role of siderophores in antagonism of fluorescent pseudomonads against plant pathogenic fungi is well established (Haas and Défago, 2005; Weller, 2007; Cornelis, 2010), although several aspects, such as the presence of low-affinity siderophores besides the high-affinity siderophore Pvd, are not completely understood (Cornelis and Matthijs, 2002).
We provide several lines of evidence supporting a role of extracellular siderophores in the antagonism of Pseudomonas against F. oxysporum, both in vitro and on the tomato rhizosphere. First, the strongest in vitro growth inhibition effect with two bacterial strains was detected against the F. oxysporum ΔhapX mutant under iron-limiting conditions. Second, the in vitro antagonistic effect was strictly dependent on the production of bacterial siderophores. Third, coinoculation with the rhizosphere-colonizing P. putida strain KT2440 resulted in a marked decrease in F. oxysporum infection on tomato plants and a significant delay in the development of vascular wilt symptoms. It is important to note that part of the protective effect of P. putida was independent of siderophore production, because it was present both in the bacterial wild-type strain and the pvd− mutant. This suggests the presence of additional modes of action previously reported for Pseudomonas spp, such as antibiosis or competition for nutrients and space in the rhizosphere (Weinberg, 1986; Azegami et al., 1988; Gill and Warren, 1988). Interestingly, plants infected with the F. oxysporum ΔhapX mutant showed an additional decrease in fungal biomass and in severity of wilt symptoms, which was specifically associated with the P. putida wild-type strain, but not with the pvd− mutant. Thus, HapX is required by F. oxysporum for iron competition with fluorescent pseudomonads producing high-affinity siderophores, such as Pvd. Interestingly, the secondary siderophore Ocg, produced by P. fluorescens SBW25, also displayed growth-inhibiting activity toward the F. oxysporum ΔhapX mutant. Our results reveal a previously unrecognized role of HapX in rhizosphere competence of F. oxysporum, which may be of general relevance for other root-colonizing fungi.
Conserved Role of HapX in Virulence on Plants and Mammals
Iron uptake and metabolism is required for virulence of both bacterial (Expert, 1999; Taguchi et al., 2010) and fungal plant pathogens (Eichhorn et al., 2006; Oide et al., 2006; Greenshields et al., 2007). Although our data suggest that HapX is not required for in vitro iron uptake by F. oxysporum, we find that the ΔhapX mutants are significantly attenuated in their capacity to cause vascular wilt symptoms and mortality in tomato plants, thus providing evidence for a role of HapX in virulence of a plant pathogen. Biomass of the ΔhapX mutant in tomato roots was reduced; therefore, we conclude that HapX contributes to survival and proliferation of the fungus on the plant host. Interestingly, F. oxysporum strains lacking hapX were also unable to efficiently colonize and kill immunodepressed mice, confirming previous reports in the human pathogens C. neoformans, A. fumigatus, and C. albicans (Jung et al., 2010; Schrettl et al., 2010; Hsu et al., 2011). HapX is the first virulence determinant for which an essential role during plant and animal infection has been demonstrated in the same fungal pathogen. Previous studies in F. oxysporum identified either factors that are required for pathogenicity on tomato but not on mice, including the Fusarium Mitogen-activated protein Kinase1 (Fmk1), the small G protein Rho1, and the glucanosyltransferase Gas1 (Di Pietro et al., 2001; Caracuel et al., 2005; Martínez-Rocha et al., 2008), or vice versa, such as the pH response factor PacC, the light response factor White Collar-1 (Wc-1), or the secreted Fusarium Pathogenesis Related-1 (PR-1)–like protein Fpr1 (Caracuel et al., 2003; Ruiz-Roldán et al., 2008; Prados-Rosales et al., 2012). Our results suggest that HapX functions in two alternative genetic programs associated with infectious fungal growth on plants and mammals, most likely involving transcriptional reprogramming under severe iron limitation encountered in both types of hosts (Jurkevitch et al., 1993; Loper and Henkels, 1997; Weinberg, 1999; Weiss, 2002).
Deregulation of genes required for metabolic adaptation to iron deficiency may account for the impaired ability of the F. oxysporum ΔhapX mutant to proliferate in the host, as observed in the in vitro condition. However, our data suggest an additional role for HapX in the transcriptional activation of virulence-related genes. Iron starvation–induced genes, such as srbA, sidA, or hapX itself, are dramatically upregulated during growth of F. oxysporum in tomato roots or human blood. Microarray analysis identified an entire set of genes whose expression is activated by iron starvation in a HapX-dependent manner. A significant fraction of these genes encodes predicted secreted proteins that have been linked to fungal virulence, including cell wall–degrading enzymes, such as pectate lyase or endoglucanase (Di Pietro et al., 2009), aspartyl proteases (Naglik et al., 2003), or the SIX3 protein (van der Does et al., 2008). Additional virulence-related genes activated by HapX include those encoding integral membrane proteins, such as PTH11-like receptors (De Zwaan et al., 1999) or ABC transporters (Urban et al., 1999; Coleman and Mylonakis, 2009). Unexpectedly, we found that genes associated with mobile genetic elements, such as hAT family transposases and reverse transcriptase, are also induced by iron starvation via HapX. Collectively, these results suggest a role for HapX in the activation of iron starvation–induced virulence factors during fungal growth in the host. Further characterization of these potential virulence targets will provide new insights into the conserved role of the HapX-mediated iron response in fungal pathogenicity on plants and mammals.
METHODS
Fungal Strains and Culture Conditions
Fusarium oxysporum f sp lycopersici race 2 wild-type isolate 4287 (FGSC 9935) was used in all experiments. All fungal strains were stored as microconidial suspensions at −80°C with 30% glycerol. For extraction of genomic DNA and microconidia production, cultures were grown in potato dextrose broth at 28°C with shaking at 170 rpm (Di Pietro and Roncero, 1998). For analysis of gene expression, freshly obtained microconidia were germinated for 16 h in iron-depleted MM (Puhalla, 1968); mycelia were harvested by filtration, washed three times in sterile double-distilled water (ddH2O), and transferred for 10 h to fresh MM with or without 50 μM of Fe2(SO4)3. For analysis of gene expression in human blood, wild-type microconidia were germinated for 16 h at 28°C in iron-depleted MM containing 25 mM of sodium glutamate and 20 mM of HEPES, pH 7.4, transferred for 4 h to 37°C, and then transferred for different time periods to fresh MM or to heparinized human whole blood (Dunn Labortechnik GmbH) at 37°C. For analysis of in planta gene expression, wild-type microconidia were inoculated in iron-depleted MM in the presence of tomato (Solanum lycopersicum) roots as previously described (López-Berges et al., 2010), and the roots with the adhered mycelium were frozen after 2 d for RNA extraction. For later stages of infection, tomato plants inoculated with wild-type conidia were planted in vermiculite (see below), and roots were obtained 5 or 10 DAI and washed carefully before RNA extraction.
For intracellular iron determination, freshly obtained microconidia were germinated for 24 h in iron-depleted MM. Mycelia were harvested by filtration, washed three times in sterile ddH2O, and transferred for 1 h to fresh MM with or without 50 μM of Fe2(SO4)3. For analysis of colony growth, 2 × 104 fresh microconidia were spotted onto potato dextrose agar (PDA) (Scharlau) or MM with or without 50 μM of Fe2(SO4)3 and/or 0.2 mM of BPS. Plates were incubated at 28°C for the indicated time periods. All experiments included two replicate plates and were performed at least three times with similar results. For quantification of siderophores and PpIX, strains were grown at 28°C in Aspergillus MM (according to Pontecorvo et al. [1953]) containing 1% Glc as the carbon source and 20 mM of Gln as the nitrogen source for 5 d. Iron-replete media contained 30 μM of FeSO4. For iron-depleted conditions, iron was omitted.
Bacterial Strains
Two Pseudomonas isolates were used: Pseudomonas putida KT2440 producing the siderophore Pvd and its pvd− mutant (5A12) (Matthijs et al., 2009), and Pseudomonas fluorescens SBW25 producing both Pvd and Ocg and its pvd− mutant (SBW25) (Moon et al., 2008). For construction of the pvd− ocg− double mutant (15F3), the suicide plasmid pUT harboring the transposon mini-Tn5phoA3 (gentamycin-resistant) (de Chial et al., 2003) was used to generate transposon insertions in the chromosome of P. fluorescens SBW25::pvdL. Mid-log phase cultures of Escherichia coli SM10 (λpir), the host of pUT-mini-Tn5phoA3, were mixed with strain SBW25::pvdL in a 1:1 ratio. P. fluorescens SBW25::pvdL was kept at 45°C for 20 min just before mixing both strains to inactivate its restriction system. After overnight incubation on Luria-Bertani medium at 26°C, transposon insertions were selected on CAA supplemented with 100 μg mL−1 of gentamycin and 25 μg mL−1 of chloramphenicol. To avoid counterselection of mutants affected in iron uptake and metabolism, transposon-mutagenized pvdL recipients were selected on CAA amended with 50 μM of FeCl3. A bank of 2000 transconjugants was screened for mutants with loss of siderophore production as detected by the CAS assay (Schwyn and Neilands, 1987). Chromosomal DNA of 15F3 was isolated using the Gentra Puregene Genomic DNA Purification Kit (Qiagen), digested with SalI (Fermentas) and self-ligated, and the DNA flanking the mini-Tn5phoA3 was isolated and sequenced.
Nucleic Acid Manipulations
Total RNA and genomic DNA was extracted from F. oxysporum mycelia following previously reported protocols (Raeder and Broda, 1985; Chomczynski and Sacchi, 1987). The quality and quantity of extracted nucleic acids were determined by running aliquots in ethidium bromide–stained agarose gels and by spectrophotometric analysis in a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies), respectively. Routine nucleic acid manipulations were performed as described in standard protocols (Sambrook and Russell, 2001). DNA and protein sequence databases were searched using the BLAST algorithm (Altschul et al., 1990).
Targeted Gene Knockout
PCR reactions were routinely performed with the High Fidelity Template PCR system (Roche Diagnostics) using a MJ Mini Bio-Rad personal thermal cycler (see Supplemental Table 3 online for a complete list of primer sequences used in the study). All fungal transformations and purification of the transformants by monoconidial isolation were performed as described (Di Pietro and Roncero, 1998). Targeted replacement of the entire coding region of the F. oxysporum hapX gene with the hygromycin-resistance cassette (Punt et al., 1987) was performed using the double-joint PCR method (Yu et al., 2004) (see Supplemental Figure 2A online). DNA fragments flanking the hapX coding region were amplified from genomic DNA of F. oxysporum with primer pairs hapX-1 + hapX-M13-1 and hapX-2 + hapX-M13-2, respectively, and PCR fused with the hygromycin-resistance cassette using primers hapX-1n + hapX-2n. The obtained ΔhapX allele was used to transform protoplasts of the F. oxysporum wild-type strain to hygromycin resistance. Transformants showing homologous insertion of the construct were detected by PCR of genomic DNA with primer pair hapX-3 + hph-2 and by DNA gel blot analysis (see Supplemental Figures 2B and 2C online).
To generate a construct for complementation of the ΔhapX deletion mutant, a 5286-bp fragment spanning from 2322 bp upstream of the wild-type F. oxysporum hapX translation initiation codon to 1156 bp downstream of the translation termination codon was amplified by PCR with primer pair hapX-3 + hapX-2. The amplified fragment was used to cotransform protoplasts of the hapX deletion mutant with the phleomycin B–resistance gene under control of the Aspergillus nidulans gpdA promoter and trpC terminator, amplified with primers gpdA-15b + trpC-8b from the plasmid pAN8-1 (Mattern et al., 1988). Three out of eight phleomycin-resistant cotransformants were selected for their wild-type growth phenotype on solid MM without iron and were analyzed for the presence of a functional hapX allele by PCR with gene-specific primer pair hapX-4 + hapX-5 (see Supplemental Figure 2D online). We concluded that these transformants, designated ΔhapX+hapX, had integrated an intact copy of the F. oxysporum hapX gene into the genome.
Sequence Alignments, Phylogenetic Analysis, and Accession Numbers
HapX orthologs were identified by BLASTP searches in the Fusarium Comparative Genome database of the Broad Institute (http://www.broadinstitute.org/annotation/genome/fusarium_graminearum/MultiHome.html) or the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) websites, using the Aspergillus fumigatus HapX protein as bait. Full-length sequences were aligned with ClustalW (Thompson et al., 1994) and manually inspected. The SidC phylogenetic tree was built by maximum likelihood from a ClustalW alignment with PhyML version 4.0 using both parsimony and distance analysis (neighbor joining) with 1000 bootstrap replicates and represented as a phylogram with Dendroscope v1.2.3 (Guindon and Gascuel, 2003). Putative SidC orthologs in each fungal genome were identified by BLASTP searches on the Broad Institute or the National Center for Biotechnology Information websites, using the A. fumigatus SidC protein as bait.
Intracellular Iron Quantification
Intracellular iron concentration was measured using the BPS-based colorimetric assay (Tamarit et al., 2006; Hsu et al., 2011), with modifications. Briefly, mycelia were harvested by filtration, washed three times with ddH2O, and resuspended in 500 μL of 3% nitric acid. Suspensions were boiled for 2 h and centrifuged to discard cell debris. A total of 400 μL of supernatant was mixed with 160 μL of sodium ascorbate, 320 μL of BPS, and 126 μL of ammonium acetate, and reactions were incubated at room temperature for 5 min. OD535 of the BPS-Fe complex was measured with a SmartSpec Plus spectrophotometer (Bio-Rad). To eliminate the nonspecific absorbance, OD680 was subtracted from OD535. Intracellular iron concentration was expressed relative to that of the wild-type strain grown under iron-depleted conditions.
Quantitative Real-Time RT-PCR Analysis
Total RNA was treated with DNase I (Fermentas) and reverse-transcribed into first-strand cDNA with ribonuclease inhibitor RNasin Plus RNase inhibitor (Promega) and M-MLV reverse transcriptase (Invitrogen) using a poly-dT antisense primer. Gene-specific primers (see Supplemental Table 3 online) were designed to flank an intron if possible. Quantitative RT-PCR products were obtained using iQ SYBR Green Supermix (Bio-Rad) and an iCycler iQ real-time PCR System (Bio-Rad). Transcript levels were calculated by comparative Δcycle threshold (Livak and Schmittgen, 2001; Pfaffl, 2001) and normalized to act1. Expression values are presented as values relative to the expression in the wild-type strain under iron-replete conditions.
Transcriptional Profiling
A custom-made F. oxysporum microarray chip in the 4 × 44 K format (Agilent Technologies) containing 45,220 distinct 60-mer probes representing a total of 17,781 genes was used in this study. Three different probes were used for genes larger than 885 bp, and two were used for those smaller than 885 bp. RNA was prepared from the wild type or the ΔhapX strain grown in the presence or absence of Fe2(SO4)3 as described above. In total, three arrays were analyzed, representing three biological replicates. Microarray services were performed by the company Bioarray SL, an Agilent Certified Service Provider. Quality of RNA was analyzed using Nanodrop (Thermo Fisher Scientific) and Bioanalyzer 2100 (Agilent Technologies). RNA labeling with Cy3, array hybridization, and scanning were performed following Agilent's One-Color Microarray-Based Gene Expression Analysis protocol (http://www.chem.agilent.com/Library/usermanuals/Public/G4140-90040_GeneExpression_One-color_v6.5.pdf). Expression of a given gene was calculated based on the averaged processed data of the different probes corresponding to each gene.
Data background subtraction was performed using the “normexp” method with an offset value of 10. Interarray normalization was done using the “quantiles” method, which was implemented in R and included in the Bioconductor package (http://www.r-project.org/; Workman et al., 2002). Statistical analysis was conducted using the Bioconductor packages Limma, Marray, affy, PCAmethods, and EMA. The unpaired t test assuming equal variances was used for statistical comparison between the different data sets. A false discovery rate of 0.05 was considered significant.
Fluorescence Microscopy
Red autofluorescence of PpIX was visualized as described (Hortschansky et al., 2007) using an Imager M2 Zeiss Axioplan fluorescence microscope with a dsRed2 filter (excitation/emission at 546/590 nm) and a digital Photometrics Evolve camera for documentation.
Analysis of Fungal Siderophores and PpIX
Analysis of siderophores and PpIX was performed by CAS assay, reverse-phase HPLC, and high-resolution electrospray ionization MS as previously described (Oide et al., 2006). Siderophore analysis with reverse-phase HPLC was performed after saturation with FeSO4.
In Vitro Antagonism Assays with Pseudomonas spp
For the in vitro determination of F. oxysporum–Pseudomonas antagonism, 2.5 × 106 fresh fungal microconidia were spread onto Glc casamino acids solid medium (Cornelis et al., 1992) with or without 50 μM of Fe2(SO4)3 using a 1% agar-water top solution. A total of 5 μL of a bacterial overnight culture grown in Luria-Bertani medium and washed three times with ddH2O was spotted immediately on the surface of the plate, and plates were incubated for 4 d at 28°C. Alternatively, bacterial strains were point-inoculated with a toothpick on 25-mM-Gln MM plates with or without 50 μM of Fe2(SO4)3 and incubated at 28°C. After 24 h, 5 × 104 freshly obtained fungal microconidia were spotted at a distance of 20 mm, and cultures were incubated for an additional 5 d.
Plant Infection Assays
Tomato root inoculation assays were performed as described (Di Pietro and Roncero, 1998). Briefly, 2-week-old seedlings of tomato cv Monika were inoculated with F. oxysporum strains by immersing the roots in a microconidial suspension, planted in vermiculite, and maintained in a growth chamber. For bacterial/fungal coinoculation assays, seedlings were first dip-inoculated into a suspension of P. putida strains for 2 h and then infected with F. oxysporum as described above (Vitullo et al., 2011). The severity of disease symptoms and percentage survival were recorded each day for 30 to 45 d. Ten plants were used for each treatment. Virulence experiments were performed at least three times with similar results. Survival was estimated by the Kaplan-Meier method and compared among groups using the log-rank test. Data were analyzed with the software GraphPad Prism 4.
In planta quantification of fungal biomass was performed as described (Pareja-Jaime et al., 2010), with modifications. Briefly, total genomic DNA was extracted from infected tomato roots at 7 DAI. Quantitative real-time PCR was performed, and relative amounts of fungal genomic DNA were calculated by comparative cycle threshold of the fungus-specific six1 gene (Rep et al., 2004) normalized to the tomato gadph gene.
Animal Infection Assays
Mice were cared for in accordance with the principles outlined by the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (http://conventions.coe.int/Treaty/en/Treaties/Html/123.htm). Experimental conditions were approved by the Animal Welfare Committee of the Faculty of Medicine, Universitat Rovira i Virgili.
Infection assays with immunodepressed mice were performed as described (Ortoneda et al., 2004). Briefly, groups of 10 Oncins France 1 male mice (Charles River, Criffa S.A.) were immunosuppressed with a single intraperitoneal 200 mg kg−1 dose of cyclophosphamide (Laboratorios Funk S.A.) and with a single intravenous 150 mg kg−1 dose of 5-fluorouracil (Fluoro-uracil, Roche S.A.) and infected by injecting 0.2 mL of an inoculum of 108 conidia mL−1 of sterile saline into a lateral vein of the tail. Survival was recorded each day for 15 d. Infection experiments with each individual strain were performed at least three times. Survival was estimated by the Kaplan-Meier method and compared among groups using the log-rank test. To determine fungal tissue burden, randomly chosen surviving mice were sacrificed 7 DAI. Kidneys and lungs were aseptically removed, weighed, and homogenized in sterile saline, and 10-fold serial dilutions were spread onto PDA. Plates were incubated at 28°C, colonies were counted after 3 d, and the number of colony forming units per gram of organ was calculated. Fungal colony counts were converted to log10 and compared using the analysis of variance test. Data were analyzed with the software GraphPad Prism 4.
Accession Numbers
Microarray data are deposited in the Gene Expression Omnibus database (approved GEO Series GSE39325) and can be accessed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39325. Sequence data can be found in the GenBank/EMBL database or in the Fusarium Comparative Genome database under the following accession numbers: HapX, FOXG_07577; CycA, FOXG_12815; AcoA, FOXG_03713; LysF, FOXG_12892; HemA, FOXG_02151; SreA, FOXG_06266; SidA, FOXG_06447; SidC, FOXG_06448; SidD, FOXG_09785; SidG, FOXG_00516; MirB, FOXG_00517; SrbA, FOXG_03199; Act1, FOXG_01569; Six1, FOXG_16418; Gapdh, M64114; pAN7-1 (PgpdA-hygr-TtrpC), Z32698; pAN8-1 (PgpdA-phleor-TtrpC), Z32751.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignment of Fungal Orthologs of the bZIP Protein HapX.
Supplemental Figure 2. Targeted Disruption of the F. oxysporum hapX Gene.
Supplemental Figure 3. The Genus Fusarium Contains an Extra Ortholog of the NRPS SidC.
Supplemental Figure 4. CCAAT Sequences in the Promoter Regions of HapX-Regulated Genes.
Supplemental Figure 5. P. putida–F. oxysporum Antagonism Assay.
Supplemental Figure 6. P. fluorescens–F. oxysporum Antagonism Assay.
Supplemental Table 1. Predicted Orthologs of Siderophore Biosynthetic and Regulatory Genes of A. fumigatus in F. oxysporum.
Supplemental Table 2. Statistical Significance (P Values) of Tomato Plant Survival Curves in Coinoculation Experiments of F. oxysporum and P. putida.
Supplemental Table 3. Primer Sequences Used in This Study.
Supplemental Data Set 1. Genes Derepressed in ΔhapX under Iron Starvation.
Supplemental Data Set 2. Genes Induced by Iron Starvation in a HapX-Dependent Manner.
Acknowledgments
We thank Esther Martínez Aguilera for valuable technical assistance. This research was supported by the following grants: BIO2010-15505 from Ministerio de Ciencia e Innovación (MICINN), European Research Area (ERA)-NET/PathoGenoMics project TRANSPAT (BIO2008-04479-E from MICINN), EUI2009-03942 from MICINN/Plant KBBE, BIO-3847 from Junta de Andalucia to A.D.P., Marie Curie Initial Training Network ARIADNE (FP7-PEOPLE-ITN-237936) to A.D.P., and ERA-NET/PathoGenoMics project TRANSPAT (FWF I282-B09 from Austrian Science Foundation) to H.H. M.S.L.-B. received a PhD fellowship from MICINN.
AUTHOR CONTRIBUTIONS
M.S.L.-B., J.C., D.T., L.S., S.M., C.J., J.G., H.H., and A.D.P. designed the research. M.S.L.-B., J.C., D.T., L.S., S.M., and C.J. performed the research. J.C., S.M., P.C., J.G., H.H., and A.D.P. contributed reagents/materials/analysis tools. M.S.L.-B., J.C., D.T., L.S., S.M., C.J., P.C., J.G., H.H., and A.D.P. analyzed data. M.S.L.-B., D.T., S.M., P.C., H.H., and A.D.P. wrote the article.
Glossary
- Pvd
pyoverdine
- Ocg
ornicorrugatin
- BPS
bathophenanthrolinedisulfonic acid
- CAS
chrome azurol S
- HPLC
high-performance liquid chromatography
- MS
mass spectroscopy
- TCA
trichloroacetic acid
- PpIX
protoporphyrin IX
- DAI
days after inoculation
- TAFC
triacetylfusarinine C
- NRPS
nonribosomal peptide synthetase
- MM
minimal medium
- ddH2O
double-distilled water
- PDA
potato dextrose agar
- MICINN
Ministerio de Ciencia e Innovación
- FsC
fusarinine C
References
- Agrios G.N. (1997). Plant Pathology. (San Diego, CA: Academic Press) [Google Scholar]
- Alabouvette C., Olivain C., Migheli Q., Steinberg C. (2009). Microbiological control of soil-borne phytopathogenic fungi with special emphasis on wilt-inducing Fusarium oxysporum. New Phytol. 184: 529–544 [DOI] [PubMed] [Google Scholar]
- Alabouvette C., Rousel F., Louvet J. (1979). Characteristics of Fusarium wilt: Suppressive soils and prospects for their utilization in biological control. In Soil-Borne Plant Pathogens, B. Schippers and W. Gums, eds (New York: Academic Press), pp. 165–182 [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Armstrong G.M., Armstrong J.K. (1981). Formae speciales and races of Fusarium oxysporum causing wilt diseases. In Fusarium: Diseases, Biology and Taxonomy, R. Cook, ed (University Park, PA: Penn State University Press), pp. 391–399 [Google Scholar]
- Azegami K., Nishiyama K., Kato H. (1988). Effect of iron limitation on “Pseudomonas plantarii” growth and tropolone and protein production. Appl. Environ. Microbiol. 54: 844–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. (1968). Mechanisms of biological control of soil-borne pathogens. Annu. Rev. Phytopathol. 6: 263–294 [Google Scholar]
- Blatzer M., Barker B.M., Willger S.D., Beckmann N., Blosser S.J., Cornish E.J., Mazurie A., Grahl N., Haas H., Cramer R.A. (2011a). SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus. PLoS Genet. 7: e1002374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatzer M., Schrettl M., Sarg B., Lindner H.H., Pfaller K., Haas H. (2011b). SidL, an Aspergillus fumigatus transacetylase involved in biosynthesis of the siderophores ferricrocin and hydroxyferricrocin. Appl. Environ. Microbiol. 77: 4959–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracuel Z., Martínez-Rocha A.L., Di Pietro A., Madrid M.P., Roncero M.I.G. (2005). Fusarium oxysporum gas1 encodes a putative beta-1,3-glucanosyltransferase required for virulence on tomato plants. Mol. Plant Microbe Interact. 18: 1140–1147 [DOI] [PubMed] [Google Scholar]
- Caracuel Z., Roncero M.I.G., Espeso E.A., González-Verdejo C.I., García-Maceira F.I., Di Pietro A. (2003). The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum. Mol. Microbiol. 48: 765–779 [DOI] [PubMed] [Google Scholar]
- Chen C., Pande K., French S.D., Tuch B.B., Noble S.M. (2011). An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 10: 118–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Coleman J.J., Mylonakis E. (2009). Efflux in fungi: La pièce de résistance. PLoS Pathog. 5: e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis P. (2010). Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 86: 1637–1645 [DOI] [PubMed] [Google Scholar]
- Cornelis P., Anjaiah V., Koedam N., Delfosse P., Jacques P., Thonart P., Neirinckx L. (1992). Stability, frequency and multiplicity of transposon insertions in the pyoverdine region in the chromosomes of different fluorescent pseudomonads. J. Gen. Microbiol. 138: 1337–1343 [DOI] [PubMed] [Google Scholar]
- Cornelis P., Matthijs S. (2002). Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: Not only pyoverdines. Environ. Microbiol. 4: 787–798 [DOI] [PubMed] [Google Scholar]
- de Chial M., Ghysels B., Beatson S.A., Geoffroy V., Meyer J.M., Pattery T., Baysse C., Chablain P., Parsons Y.N., Winstanley C., Cordwell S.J., Cornelis P. (2003). Identification of type II and type III pyoverdine receptors from Pseudomonas aeruginosa. Microbiology 149: 821–831 [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D., Höfte M. (2009). Rhizobacteria-induced systemic resistance. Adv. Bot. Res. 51: 223–281 [Google Scholar]
- Dean R., Van Kan J.A., Pretorius Z.A., Hammond-Kosack K.E., Di Pietro A., Spanu P.D., Rudd J.J., Dickman M., Kahmann R., Ellis J., Foster G.D. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13: 414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan T.M., Carroll A.M., Valent B., Sweigard J.A. (1999). Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell 11: 2013–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro A., García-MacEira F.I., Méglecz E., Roncero M.I.G. (2001). A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39: 1140–1152 [PubMed] [Google Scholar]
- Di Pietro A., Roncero M.I.G. (1998). Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant Microbe Interact. 11: 91–98 [DOI] [PubMed] [Google Scholar]
- Di Pietro A., Roncero M.I.G., Ruiz-Roldán M.C. (2009). From tools of survival to weapons of destruction: Role of cell wall-degrading enzymes in plant infection. In The Mycota, Plant Relationships, 2nd ed, Vol. V, H. Deising, ed (Heidelberg, Berlin: Springer Verlag), pp. 181–200 [Google Scholar]
- Eichhorn H., Lessing F., Winterberg B., Schirawski J., Kämper J., Müller P., Kahmann R. (2006). A ferroxidation/permeation iron uptake system is required for virulence in Ustilago maydis. Plant Cell 18: 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery T. (1980). Malonichrome, a new iron chelate from Fusarium roseum. Biochim. Biophys. Acta 629: 382–390 [DOI] [PubMed] [Google Scholar]
- Expert D. (1999). Withholding and exchanging iron: Interactions between Erwinia spp. and their plant hosts. Annu. Rev. Phytopathol. 37: 307–334 [DOI] [PubMed] [Google Scholar]
- Gill P.R., Jr, Warren G.J. (1988). An iron-antagonized fungistatic agent that is not required for iron assimilation from a fluorescent rhizosphere pseudomonad. J. Bacteriol. 170: 163–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenshields D.L., Liu G., Feng J., Selvaraj G., Wei Y. (2007). The siderophore biosynthetic gene SID1, but not the ferroxidase gene FET3, is required for full Fusarium graminearum virulence. Mol. Plant Pathol. 8: 411–421 [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Haas D., Défago G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3: 307–319 [DOI] [PubMed] [Google Scholar]
- Haas H., Eisendle M., Turgeon B.G. (2008). Siderophores in fungal physiology and virulence. Annu. Rev. Phytopathol. 46: 149–187 [DOI] [PubMed] [Google Scholar]
- Haas H., Schoeser M., Lesuisse E., Ernst J.F., Parson W., Abt B., Winkelmann G., Oberegger H. (2003). Characterization of the Aspergillus nidulans transporters for the siderophores enterobactin and triacetylfusarinine C. Biochem. J. 371: 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H., Zadra I., Stöffler G., Angermayr K. (1999). The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J. Biol. Chem. 274: 4613–4619 [DOI] [PubMed] [Google Scholar]
- Halliwell B., Gutteridge J.M. (1984). Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby D. (1979). Take-all decline: A theorist's paradise. In Soil-Borne Plant Pathogens, B. Schippers and W. Gams, eds (New York: Academic Press), pp. 133–156 [Google Scholar]
- Hortschansky P., et al. (2007). Interaction of HapX with the CCAAT-binding complex—a novel mechanism of gene regulation by iron. EMBO J. 26: 3157–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.C., Yang C.Y., Lan C.Y. (2011). Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryot. Cell 10: 207–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jbel M., Mercier A., Labbé S. (2011). Grx4 monothiol glutaredoxin is required for iron limitation-dependent inhibition of Fep1. Eukaryot. Cell 10: 629–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W.H., Saikia S., Hu G., Wang J., Fung C.K., D’Souza C., White R., Kronstad J.W. (2010). HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog. 6: e1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkevitch E., Hadar Y., Chen Y., Chino M., Mori S. (1993). Indirect utilization of the phytosiderophore mugineic acid as an iron source to rhizosphere fluorescent Pseudomonas. Biometals 6: 119–123 [DOI] [PubMed] [Google Scholar]
- Kamilova F., Validov S., Azarova T., Mulders I., Lugtenberg B. (2005). Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environ. Microbiol. 7: 1809–1817 [DOI] [PubMed] [Google Scholar]
- Kim K.D., Kim H.J., Lee K.C., Roe J.H. (2011). Multi-domain CGFS-type glutaredoxin Grx4 regulates iron homeostasis via direct interaction with a repressor Fep1 in fission yeast. Biochem. Biophys. Res. Commun. 408: 609–614 [DOI] [PubMed] [Google Scholar]
- Kloepper J.W., Leong J., Teintze M., Schroth M.N. (1980). Pseudomonas siderophores: A mechanism explaining disease-suppressive soils. Curr. Microbiol. 4: 317–320 [Google Scholar]
- Lagopodi A.L., Ram A.F., Lamers G.E., Punt P.J., Van den Hondel C.A., Lugtenberg B.J., Bloemberg G.V. (2002). Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol. Plant Microbe Interact. 15: 172–179 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Loper J.E., Henkels M.D. (1997). Availability of iron to Pseudomonas fluorescens in rhizosphere and bulk soil evaluated with an ice nucleation reporter gene. Appl. Environ. Microbiol. 63: 99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Berges M.S., Rispail N., Prados-Rosales R.C., Di Pietro A. (2010). A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell 22: 2459–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L.J., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Rocha A.L., Roncero M.I., López-Ramirez A., Mariné M., Guarro J., Martínez-Cadena G., Di Pietro A. (2008). Rho1 has distinct functions in morphogenesis, cell wall biosynthesis and virulence of Fusarium oxysporum. Cell. Microbiol. 10: 1339–1351 [DOI] [PubMed] [Google Scholar]
- Mattern I.E., Punt P.J., van den Hondel C.A.M.J.J. (1988). A vector of Aspergillus transformation conferring phleomycin resistance. Fungal Genet. Newslett. 35: 25 [Google Scholar]
- Matthijs S., Budzikiewicz H., Schäfer M., Wathelet B., Cornelis P. (2008). Ornicorrugatin, a new siderophore from Pseudomonas fluorescens AF76. Z. Naturforsch., C, J. Biosci. 63: 8–12 [DOI] [PubMed] [Google Scholar]
- Matthijs S., Laus G., Meyer J.M., Abbaspour-Tehrani K., Schäfer M., Budzikiewicz H., Cornelis P. (2009). Siderophore-mediated iron acquisition in the entomopathogenic bacterium Pseudomonas entomophila L48 and its close relative Pseudomonas putida KT2440. Biometals 22: 951–964 [DOI] [PubMed] [Google Scholar]
- Mei B., Budde A.D., Leong S.A. (1993). sid1, a gene initiating siderophore biosynthesis in Ustilago maydis: Molecular characterization, regulation by iron, and role in phytopathogenicity. Proc. Natl. Acad. Sci. USA 90: 903–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado-Blanco J., van der Drift K.M., Olsson P.E., Thomas-Oates J.E., van Loon L.C., Bakker P.A. (2001). Analysis of the pmsCEAB gene cluster involved in biosynthesis of salicylic acid and the siderophore pseudomonine in the biocontrol strain Pseudomonas fluorescens WCS374. J. Bacteriol. 183: 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier A., Labbé S. (2009). Both Php4 function and subcellular localization are regulated by iron via a multistep mechanism involving the glutaredoxin Grx4 and the exportin Crm1. J. Biol. Chem. 284: 20249–20262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier A., Pelletier B., Labbé S. (2006). A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 5: 1866–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier A., Watt S., Bähler J., Labbé S. (2008). Key function for the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryot. Cell 7: 493–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon C.D., Zhang X.X., Matthijs S., Schäfer M., Budzikiewicz H., Rainey P.B. (2008). Genomic, genetic and structural analysis of pyoverdine-mediated iron acquisition in the plant growth-promoting bacterium Pseudomonas fluorescens SBW25. BMC Microbiol. 8: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.E., Emery T. (1976). Nα-Acetylfusarinines: Isolation, characterization, and properties. Biochem. 15: 2719–2723 [DOI] [PubMed] [Google Scholar]
- Naglik J.R., Challacombe S.J., Hube B. (2003). Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67: 400–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucci M., Anaissie E. (2007). Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 20: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberegger H., Schoeser M., Zadra I., Schrettl M., Parson W., Haas H. (2002). Regulation of freA, acoA, lysF, and cycA expression by iron availability in Aspergillus nidulans. Appl. Environ. Microbiol. 68: 5769–5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oide S., Moeder W., Krasnoff S., Gibson D., Haas H., Yoshioka K., Turgeon B.G. (2006). NPS6, encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 18: 2836–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortoneda M., Guarro J., Madrid M.P., Caracuel Z., Roncero M.I., Mayayo E., Di Pietro A. (2004). Fusarium oxysporum as a multihost model for the genetic dissection of fungal virulence in plants and mammals. Infect. Immun. 72: 1760–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareja-Jaime Y., Martín-Urdíroz M., Roncero M.I., González-Reyes J.A., Roldán M. del C. (2010). Chitin synthase-deficient mutant of Fusarium oxysporum elicits tomato plant defence response and protects against wild-type infection. Mol. Plant Pathol. 11: 479–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Nadales E., Di Pietro A. (2011). The membrane mucin Msb2 regulates invasive growth and plant infection in Fusarium oxysporum. Plant Cell 23: 1171–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo G., Roper J.A., Hemmons L.M., MacDonald K.D., Bufton A.W. (1953). The genetics of Aspergillus nidulans. Adv. Genet. 5: 141–238 [DOI] [PubMed] [Google Scholar]
- Prados-Rosales R.C., Roldán-Rodríguez R., Serena C., López-Berges M.S., Guarro J., Martínez-del-Pozo A., Di Pietro A. (2012). A PR-1-like protein of Fusarium oxysporum functions in virulence on mammalian hosts. J. Biol. Chem. 287: 21970–21979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhalla J.E. (1968). Compatibility reactions on solid medium and interstrain inhibition in Ustilago maydis. Genetics 60: 461–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S., Vergara S.V., Thiele D.J. (2008). Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 7: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt P.J., Oliver R.P., Dingemanse M.A., Pouwels P.H., van den Hondel C.A. (1987). Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56: 117–124 [DOI] [PubMed] [Google Scholar]
- Raaijmakers J.M., de Bruijn I., de Kock M.J. (2006). Cyclic lipopeptide production by plant-associated Pseudomonas spp.: Diversity, activity, biosynthesis, and regulation. Mol. Plant Microbe Interact. 19: 699–710 [DOI] [PubMed] [Google Scholar]
- Raeder U., Broda P. (1985). Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1: 17–20 [Google Scholar]
- Ratledge C., Dover L.G. (2000). Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54: 881–941 [DOI] [PubMed] [Google Scholar]
- Ravel J., Cornelis P. (2003). Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 11: 195–200 [DOI] [PubMed] [Google Scholar]
- Rep M., van der Does H.C., Meijer M., van Wijk R., Houterman P.M., Dekker H.L., de Koster C.G., Cornelissen B.J. (2004). A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 53: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Ruiz-Roldán M.C., Garre V., Guarro J., Mariné M., Roncero M.I.G. (2008). Role of the white collar 1 photoreceptor in carotenogenesis, UV resistance, hydrophobicity, and virulence of Fusarium oxysporum. Eukaryot. Cell 7: 1227–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. (2001). Molecular Cloning: A Laboratory Manual, 3rd ed (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press) [Google Scholar]
- Scher F.M., Baker R. (1982). Effect of Pseudomonas putida and a synthetic iron chelator on induction of soil suppressiveness to Fusarium wilt pathogens. Phytopathology 72: 1567–1573 [Google Scholar]
- Schrettl M., Bignell E., Kragl C., Joechl C., Rogers T., Arst H.N., Jr, Haynes K., Haas H. (2004). Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 200: 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Bignell E., Kragl C., Sabiha Y., Loss O., Eisendle M., Wallner A., Arst H.N., Jr, Haynes K., Haas H. (2007). Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 3: 1195–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., et al. (2010). HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 6: e1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwyn B., Neilands J.B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160: 47–56 [DOI] [PubMed] [Google Scholar]
- Simeoni L.A., Lindsay W.L., Baker R. (1987). Critical iron level associated with biological control of Fusarium wilt. Phytopathology 77: 1057–1061 [Google Scholar]
- Skaar E.P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6: e1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Suzuki T., Inagaki Y., Toyoda K., Shiraishi T., Ichinose Y. (2010). The siderophore pyoverdine of Pseudomonas syringae pv. tabaci 6605 is an intrinsic virulence factor in host tobacco infection. J. Bacteriol. 192: 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamarit J., Irazusta V., Moreno-Cermeño A., Ros J. (2006). Colorimetric assay for the quantitation of iron in yeast. Anal. Biochem. 351: 149–151 [DOI] [PubMed] [Google Scholar]
- Tanaka A., Kato M., Nagase T., Kobayashi T., Tsukagoshi N. (2002). Isolation of genes encoding novel transcription factors which interact with the Hap complex from Aspergillus species. Biochim. Biophys. Acta 1576: 176–182 [DOI] [PubMed] [Google Scholar]
- Thomashow L.S., Weller D.M. (1996). Current concepts in the use of introduced bacteria for biological disease control: Mechanisms and antifungal metabolites. In Plant Microbe Interactions, Vol. 1, G. Stacey and N. Keen, eds (London: Chapman and Hall), pp. 187–236 [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban M., Bhargava T., Hamer J.E. (1999). An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 18: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Does H.C., Lievens B., Claes L., Houterman P.M., Cornelissen B.J., Rep M. (2008). The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ. Microbiol. 10: 1475–1485 [DOI] [PubMed] [Google Scholar]
- Vitullo D., Di Pietro A., Romano A., Lanzotti V., Lima G. (2011). Role of new bacterial surfactins in the antifungal interaction between Bacillus amyloliquefaciens and Fusarium oxysporum. Plant Pathol. 61: 689–699 [Google Scholar]
- Weinberg E.D. (1986). Regulation of secondary metabolism by trace metals. In Cell Metabolism: Growth and Environment, T.A.V. Subramarian, ed (Boca Raton, FL: Chemical Rubber Corporation Press), pp. 151–160 [Google Scholar]
- Weinberg E.D. (1999). The role of iron in protozoan and fungal infectious diseases. J. Eukaryot. Microbiol. 46: 231–238 [DOI] [PubMed] [Google Scholar]
- Weiss G. (2002). Iron and immunity: A double-edged sword. Eur. J. Clin. Invest. 32(Suppl 1): 70–78 [DOI] [PubMed] [Google Scholar]
- Weller D.M. (2007). Pseudomonas biocontrol agents of soilborne pathogens: Looking back over 30 years. Phytopathology 97: 250–256 [DOI] [PubMed] [Google Scholar]
- Workman C., Jensen L.J., Jarmer H., Berka R., Gautier L., Nielser H.B., Saxild H.H., Nielsen C., Brunak S., Knudsen S. (2002). A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 3: research0048.1–research0048.16 [DOI] [PMC free article] [PubMed]
- Yasmin S., Alcazar-Fuoli L., Gründlinger M., Puempel T., Cairns T., Blatzer M., Lopez J.F., Grimalt J.O., Bignell E., Haas H. (2012). Mevalonate governs interdependency of ergosterol and siderophore biosyntheses in the fungal pathogen Aspergillus fumigatus. Proc. Natl. Acad. Sci. USA 109: E497–E504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.H., Hamari Z., Han K.H., Seo J.A., Reyes-Domínguez Y., Scazzocchio C. (2004). Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41: 973–981 [DOI] [PubMed] [Google Scholar]