In legume plants, cytokinins are necessary and sufficient for symbiotic nodule organogenesis, allowing them to fix atmospheric nitrogen. Biochemical and reverse genetic approaches identified two transcription factors from the GRAS (NSP2) and bHLH families as direct targets of cytokinin signaling pathways in legume roots. These transcription factors act at the convergence of phytohormonal and nodulation symbiotic cues.
Abstract
Cytokinin regulates many aspects of plant development, and in legume crops, this phytohormone is necessary and sufficient for symbiotic nodule organogenesis, allowing them to fix atmospheric nitrogen. To identify direct links between cytokinins and nodule organogenesis, we determined a consensus sequence bound in vitro by a transcription factor (TF) acting in cytokinin signaling, the nodule-enhanced Medicago truncatula Mt RR1 response regulator (RR). Among genes rapidly regulated by cytokinins and containing this so-called RR binding site (RRBS) in their promoters, we found the nodulation-related Type-A RR Mt RR4 and the Nodulation Signaling Pathway 2 (NSP2) TF. Site-directed mutagenesis revealed that RRBS cis-elements in the RR4 and NSP2 promoters are essential for expression during nodule development and for cytokinin induction. Furthermore, a microRNA targeting NSP2 (miR171 h) is also rapidly induced by cytokinins and then shows an expression pattern anticorrelated with NSP2. Other primary targets regulated by cytokinins depending on the Cytokinin Response1 (CRE1) receptor were a cytokinin oxidase/dehydrogenase (CKX1) and a basic Helix-Loop-Helix TF (bHLH476). RNA interference constructs as well as insertion of a Tnt1 retrotransposon in the bHLH gene led to reduced nodulation. Hence, we identified two TFs, NSP2 and bHLH476, as direct cytokinin targets acting at the convergence of phytohormonal and symbiotic cues.
INTRODUCTION
Plant hormones play crucial roles in regulating organogenesis, and cytokinins have been shown to affect cell proliferation, elongation, differentiation, or senescence depending on the developmental context. In Arabidopsis thaliana, cytokinins are perceived through an extracellular Cyclase/Histidine Kinase-Associated Sensing Extracellular (CHASE) domain present in Cytokinin Response1/Authentic Histidine Kinase4 (CRE1/AHK4) transmembrane receptors as well as two other related sensors (AHK2 and AHK3; Werner and Schmülling, 2009). Downstream of these cytokinin receptors, a Histidyl-Aspartyl multistep phosphorelay is initiated, leading to the activation of Type-B response regulators (RRs). These plant Myb-related transcription factors (TFs; Stracke et al., 2001) can regulate cytokinin primary response genes, such as the Type-A RRs (Brandstatter and Kieber, 1998; D’Agostino et al., 2000; Sakai et al., 2000, 2001; Hwang and Sheen, 2001). DNA binding sites were identified for several Arabidopsis Type-B RRs (ARR1, ARR2, ARR10, and ARR11), but only a short consensus RR binding sequence was identified [5′-(A/G)GAT(T/C)-3′] (Sakai et al., 2000; Lohrmann et al., 2001; Hosoda et al., 2002; Imamura et al., 2003), preventing the reliable identification of transcripts directly regulated by these TFs. In addition, it was suggested that an extended sequence is likely required to allow specific interaction of each RR in a promoter context, notably to discriminate between various GOLDEN2/ARRs/PSR1/PHR1 (GARP)-type TFs (Taniguchi et al., 2007). In parallel, transcriptomic analyses performed in Arabidopsis revealed a wide variety of genes responding rapidly to exogenous cytokinins in seedlings of the wild type, transgenics, or mutants affected in cytokinin level or sensitivity (Hoth et al., 2003; Rashotte et al., 2003; Kiba et al., 2004; Brenner et al., 2005; Kiba et al., 2005; Rashotte et al., 2006; Lee et al., 2007; Taniguchi et al., 2007). Taniguchi et al. (2007) notably identified, based on the combined analysis of an inducible Type-B RR (ARR1) protein lacking its phosphoreceiver domain (ΔDDK), an arr1 mutant, and cycloheximide treatments, 17 genes regulated by cytokinins independently of de novo protein synthesis. ARR1 interaction with the promoter of one of these putative targets, coding for IAA3/SHY2, was validated by chromatin immunoprecipitation (ChIP), and this direct regulation of the auxin response by cytokinins was shown to control the balance between cell proliferation and differentiation at the root meristem transition zone (Dello Ioio et al., 2008).
Arabidopsis roots do not develop symbiotic interactions, whereas legume plants have the ability to interact with nitrogen-fixing bacteria under nitrogen starvation conditions. In response to a specific bacterial microsymbiont (e.g., Sinorhizobium meliloti for the model legume Medicago truncatula), a new organ is formed on host legume roots (the nitrogen-fixing nodules), and cytokinins are crucial for this organogenesis (Frugier et al., 2008). Systematic RNA interference (RNAi) targeting of the different M. truncatula cytokinin receptors revealed that the Cytokinin Response1 (CRE1) His kinase regulates nodule formation (Gonzalez-Rizzo et al., 2006). Similarly, the Lotus japonicus hyperinfected1 (hit1) mutant carrying a loss-of-function mutation in the L. japonicus Histidine Kinase1 (LHK1) gene (functionally related to CRE1) showed a strongly reduced nodulation (Murray et al., 2007). Interestingly, the L. japonicus spontaneous nodule formation2 (snf2) gain-of-function mutation affecting the same LHK1 receptor led to the formation of nodules in the absence of rhizobia (Tirichine et al., 2007). This result indicates that cytokinins are necessary and sufficient to activate cortical cell divisions and nodule organogenesis.
To search for genes directly regulated by the cytokinin cue and acting in legume root nodule development, we combined a transcriptomic analysis of M. truncatula root response to cytokinins with a systematic evolution of ligands by exponential enrichment (SELEX) approach for the nodule-associated RR Mt RR1 (Gonzalez-Rizzo et al., 2006). This allowed us to determine a large 12-bp RR binding site (RRBS), which was retrieved in promoters of several cytokinin-regulated genes, including two genes linked to nodulation: RR4, a Type-A RR (Plet et al., 2011), and the NSP2 TF (Kaló et al., 2005; Heckmann et al., 2006). These RRBSs were essential for cytokinin induction and symbiotic expression of these genes and allowed us to further identify direct targets of cytokinins acting in legume roots, including a new basic Helix-Loop-Helix (bHLH) TF, bHLH476. Downregulating bHLH476 expression, using either RNAi constructs or an insertional mutant, resulted in a reduced nodulation. Therefore, NSP2 and bHLH476 are two direct targets of cytokinins acting positively in symbiotic nodulation.
RESULTS
Cytokinin Target Genes and Functional Pathways Acting in M. truncatula Root Apices
To gain knowledge about cytokinin primary response genes in legume roots and nodules, we first developed a transcriptomic approach focused on the root apical region containing the zone required for rhizobial symbiotic interactions. This allowed us to identify transcripts regulated by a short-term exogenous cytokinin treatment (10−7 M of benzyl amino purine [BAP] for 1 h; see Supplemental Data Set 1 online) in this root region. The most significant differentially expressed functional categories were the flavonoid (see Supplemental Figures 1A and 1B online) and gibberellin (GA) metabolisms (see Supplemental Figure 1C online; P < 0.001, Fisher’s exact test, shown in Supplemental Table 1 online). Two enzymatic families were also very significantly affected (P < 0.002), the UDP glucosyl and glucuronyl transferases and the cytochrome P450 families (see Supplemental Figure 1D online), with several members related to flavonoid or cytokinin metabolism (i.e., zeatin O-glycosyltransferases and CYP735-A2 cytochrome P450 enzymes). Finally, a significant enrichment was observed for two families of TFs: bHLH and APETALA2/Ethylene Response Factor/Ethylene-Responsive Element Binding Protein (AP2/ERF/EREBP). The latter family was previously linked to cytokinin responses through the identification of cytokinin response factors (CRFs) (Rashotte et al., 2006). A BLASTX analysis revealed, however, that none of the identified AP2/ERF TFs regulated by cytokinins in M. truncatula roots were closely related to CRFs.
Overall, this transcriptomic analysis allowed us to identify genes rapidly regulated by cytokinins in legume roots, highlighting the enrichment for transcripts related to cytokinin, GA, and flavonoid metabolisms as well as for bHLH and AP2/ERF TFs.
Identification of a RRBS cis-Element Revealed New Candidate Cytokinin Primary Response Genes
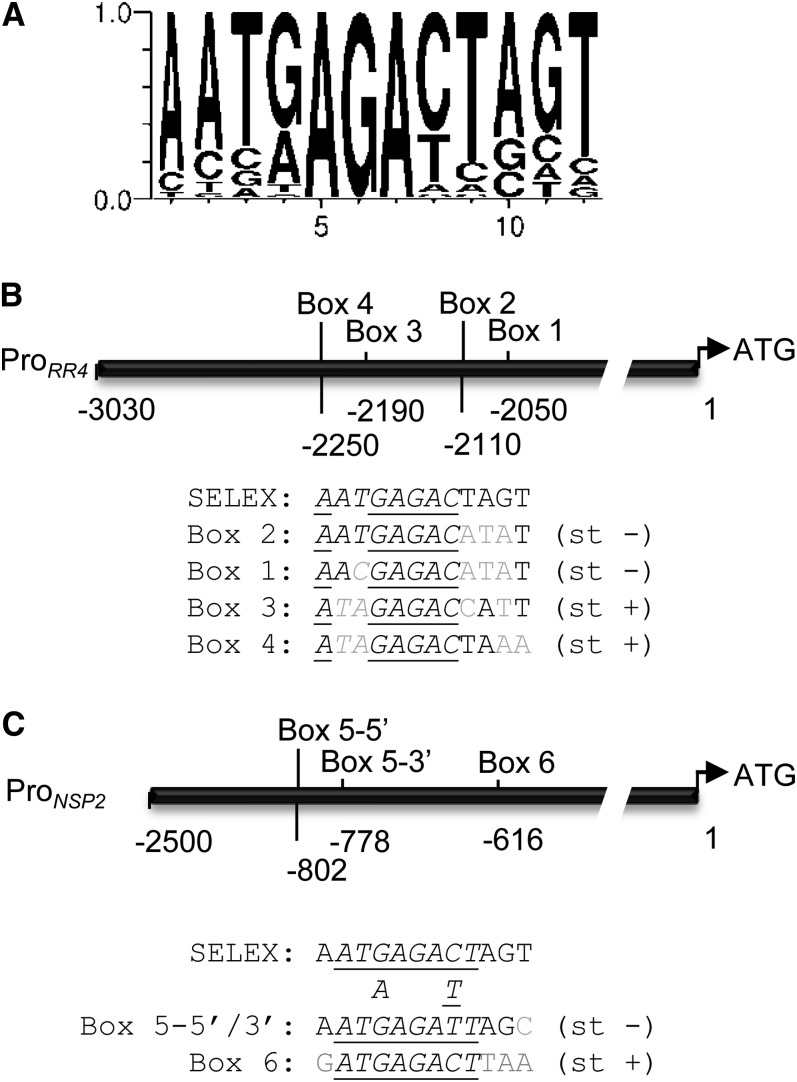
To identify direct molecular links between cytokinin regulation of gene expression and nodule organogenesis, we searched for targets of Mt RR1, a RR that is highly expressed during nodulation (Gonzalez-Rizzo et al., 2006) and that is able to interfere with root cytokinin sensitivity when expressed in Arabidopsis (see Supplemental Figure 2 online). To determine its DNA binding site, a region containing the RR1 DNA binding domain (BD) was expressed in Escherichia coli as a fusion with glutathione-S-transferase (GST) and purified by affinity chromatography. Seven rounds of PCR-assisted binding site selection (SELEX) were performed, and because no additional improvement was detected in an eighth round, 38 individual clones were randomly selected for sequencing. Every clone contained an AGA (or TCT) core, used as a reference to align all the sequences (see Supplemental Figure 3 online). Eighteen clones were identical, and the others were remarkably similar around the core sequence, leading to the following 12-bp consensus, referred to as the Mt RR1 RRBS consensus: 5′-AAT(G/A)AGA(C/T)TAGT-3′ [or the reverse complement 5′-ACTA(G/A)TCT(C/T)ATT-3′] (Figure 1A).
Figure 1.
RRBSs in the Promoter of the RR4 and NSP2 Nodulation-Related Genes.
(A) Consensus sequence bound by the RR1 BD identified by SELEX. Numbers on the x and y axes, respectively, represent nucleotide positions and the frequency of occurrence (shown by letter size normalized from 0 to 1) of each nucleotide for each position (WebLogo; Crooks et al., 2004).
(B) and (C) Schematic diagram of RR4 (B) and NSP2 (C) promoter regions. The RRBS cis-element variants (boxes 1 to 6) are indicated. Alignment to the major SELEX consensus is shown below each promoter diagram. Gray indicates nonconserved nucleotides, underlined indicates conserved nucleotides between all boxes of each promoter, italics indicates 8-bp RRBS core, and st + or str − indicates DNA strand of sequences shown.
By combining these data, we developed a bioinformatic workflow to identify, among genes rapidly regulated by cytokinins in legume roots, those having a conserved RRBS core of at least 8 bp in their promoters. Comparison between 2.5-kb promoters of a similar number of randomly chosen cytokinin- and non–cytokinin-regulated genes (n = 150) revealed enrichment for multiple RRBS consensus cores in promoters of cytokinin-regulated genes (4% versus 1.5%). Promoters having more than two potential RRBS cis-elements corresponded to 13 genes upregulated by cytokinins, including RR4 and two TF-encoding genes from the NAM and GRAS families (Table 1; see Supplemental Data Set 2 online). RR4 is a Type-A RR previously characterized as a cytokinin primary response gene acting early in Medicago nodule organogenesis (Vernié et al., 2008; Plet et al., 2011). Analysis of a ∼3-kb region upstream of its ATG revealed four 12-bp regions highly similar to the RRBS consensus identified by SELEX, present between 1.9 to 2.2 kb before the predicted RR4 start codon and arranged in two tandems (Figure 1B, boxes 1 to 4). Six nucleotide positions identical among all four boxes corresponded to the consensus sequence identified by SELEX, and two to three nucleotides were additionally conserved on the different boxes (Figure 1B). Overall, each box showed a 66 to 75% identity (i.e., 8 to 9 bp out of 12) to the RRBS consensus. Strikingly, the GRAS TF identified was the NSP2 gene, which is crucial for symbiotic nodulation (Kaló et al., 2005; Heckmann et al., 2006). This promoter contains three RRBS cis-elements, a tandem of two identical boxes and another RRBS variant (Figure 1C, boxes 5 and 6). Overall, identity of eight to 11 bases was found between these boxes and the 12-bp RRBS.
Table 1. Selected Candidate Cytokinin-Regulated Genes with RRBS cis-Elements in Their Promoters.
| Genomic ID | Box 2 | Box 5 | Total No. of 8-bp Core RRBS | TIGR Database ID | FC BAP versus Control | Adjusted P-Value | Description |
|---|---|---|---|---|---|---|---|
| Medtr5g037580.1 | 1 | 0 | 4 | TC103991 | 11.12 | 3E−05 | RR Mt RR4 |
| Medtr3g097800.1 | 0 | 2 | 3 | TC98097 | 3.98 | 6E−04 | GRAS TF (Mt NSP2) |
| Medtr1g135030.1 | 1 | 0 | 2 | TC98397 | 3.46 | 0.004 | LOB TF |
| Medtr5g014710.1 | 0 | 1 | 2 | TC95476 | 1.92 | 0.027 | bHLH TF (Mt bHLH476) |
| Medtr2g116970.1 | 1 | 0 | 2 | TC109615 | 1.9 | 0.001 | GRAS TF |
| Medtr5g069060.1 | 1 | 1 | 3 | TC103195 | 1.76 | 5E−04 | Pathogenesis-related and ERF TF |
| Medtr4g097530.1 | 1 | 0 | 2 | AW684462 | 1.73 | 2E−04 | bZIP TF |
| Medtr7g005040.1 | 0 | 0 | 4 | TC95048 | 1.6 | 6E−04 | NAM TF |
| Medtr4g135740.1 | 0 | 2 | 2 | TC95247 | 1.78 | 0.035 | GST |
| Medtr1g019530.1 | 0 | 1 | 2 | TC95407 | 1.63 | 0.007 | Cytokinin oxidase (Mt CKX1) |
List of all genes encoding TFs induced in response to cytokinins (10−7 M of BAP for 1 h) in root apices (FC > 1.5, adjusted P < 0.05) and whose promoters (2.5 kb) contain at least three 8-bp core RRBSs or at least two RRBSs of which one is a box 2 or a box 5. Homologs of candidate cytokinin primary response genes previously reported in Arabidopsis (Tanigushi et al., 2007) are also indicated at the end of the table. Included for each gene: a genomic accession number, the number of box 2, box 5, and the total number of RRBS present in their promoter, a TC accession number (TIGR database), the FC and adjusted P-value, and a functional annotation.
A second screen was then performed with the RRBS core from each of the two nodulation-related genes that was the most closely related to the SELEX consensus (Figures 1B and 1C; see Supplemental Data Set 2 online, boxes 2 and 5 highlighted in green and yellow, respectively). Three additional TFs encoding genes from GRAS, LOB, and bZIP families were identified among cytokinin-regulated genes having at least one box 2 core and an additional RRBS core sequence (Table 1). Similarly, promoters from a bHLH TF and two homologs of potential Arabidopsis cytokinin primary response genes (Taniguchi et al., 2007) had a box 5 core associated with an additional RRBS core (Table 1).
Altogether, these different analyses led to the identification of 56 candidate cytokinin primary response genes, among which only two were previously reported in Arabidopsis. Seven TF-encoding genes upregulated by cytokinins were notably identified, and one, very interestingly, corresponded to NSP2, a known regulator of symbiotic nodulation.
Expression of RR4 and NSP2 Symbiotic Genes in Nodule Primordia and Cytokinin-Treated Roots Depends on RRBS cis-Elements
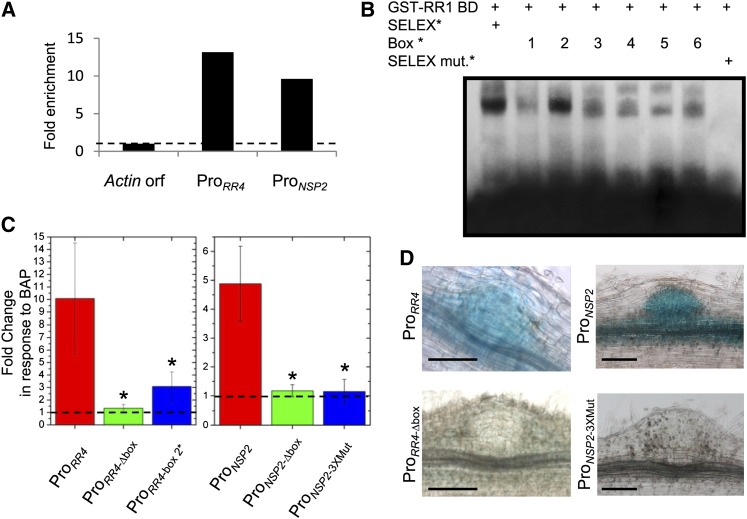
We then focused our interest on the two genes previously linked to legume symbiotic nodulation, RR4 and NSP2. ChIP followed by real-time PCR (ChIP-qPCR) was used first to validate in vivo the direct interaction of these promoters with RR1 (Figure 2A). In addition, electrophoretic mobility shift assays (EMSAs) showed that RR1 was able to recognize in vitro the six RRBS sequence variants present in these promoters (boxes 1 to 6, Figure 2B). When the central GAGA/TCTC core of the SELEX consensus was replaced by a CAGT/ACTG sequence, RR1 binding was abolished, indicating a specific in vitro interaction. Interestingly, the main sequence obtained by SELEX as well as the box most alike from the RR4 promoter (box 2) showed the best binding compared with the other boxes (Figure 2B). This result was further validated by competition EMSA performed with the different boxes (see Supplemental Figure 4 online).
Figure 2.
RRBS cis-Elements in the RR4 and NSP2 Promoters Are Required for Cytokinin Regulation and Expression in Nodule Primordia.
(A) ChIP-qPCR of RR1 binding to RR4 and NSP2 promoters. The Actin11 ORF was used as a negative control, and IP values were normalized for each gene against the input genomic DNA. Ratios with Actin11 were calculated to visualize RR1 binding fold enrichment (n > 30 independent transgenic roots/construct).
(B) EMSA analysis of the RR1 binding to different 12-bp oligonucleotides labeled with [α-32P] dATP (indicated by asterisk): the major SELEX consensus, boxes 1 to 4 (RR4 promoter), boxes 5 and 6 (NSP2 promoter), and a SELEX consensus where the central GAGA core was replaced by a ACTG sequence (SELEX mut.).
(C) Fluorometric quantification of GUS activity in roots expressing ProRR4:GUS (left) or ProNSP2:GUS (right) fusions after treatment with 10−7 M of BAP for 3 h. Error bars represent sd (n > 20 independent transgenic roots). A Kruskal-Wallis test was performed to assess significant differences (α < 0.05).
(D) Histochemical staining of GUS activity in nodule primordia expressing ProRR4:GUS (left) or ProNSP2:GUS (right) fusions. In [(C) and (D)], ProRR4-Δbox:GUS and ProNSP2-3XMut:GUS fusions were also used (see Methods for details).
Bars in (D) = 50 μm.
Using promoter-β-glucuronidase (GUS) fusions and site-directed mutagenesis, the significance of the RR4 promoter box 2, which is most closely related to the SELEX consensus and efficiently bound in vitro by RR1, was analyzed in vivo (Figure 2C, left, ProRR4-box 2*). This cis-element was essential to activate a GUS transcriptional fusion in response to cytokinins, indicating that the box 2 RRBS is a bona fide cis-element of the RR4 primary response gene. RR1 in vitro binding efficiency was similar for the two RRBS variants identified in the NSP2 promoter (boxes 5 and 6, Figure 2B); therefore, a simultaneous mutagenesis of the three cis-elements was performed, revealing their requirement for cytokinin induction (Figure 2C, right, ProNSP2-3Xmut). Finally, because both cre1 and nsp2 mutants are defective in early nodule organogenesis (Kalo et al., 2005; Plet et al., 2011), we analyzed the ProRR4 and ProNSP2:GUS patterns in nodule primordia (Figure 2D). ProNSP2 activity was abolished when the three RRBSs identified were mutagenized, similarly to a ProRR4 lacking the four RRBSs, suggesting that NSP2 expression in nodules depends on these cis-elements.
Overall, these results indicate that RR4 and NSP2 are direct targets of the RR1-dependent cytokinin signaling pathway and that RRBS cis-elements are crucial to regulate their expression in response to cytokinins and during early nodule development.
The MIR171 h Gene Has an Opposite Expression Pattern to Its NSP2 Target in Response to Cytokinins
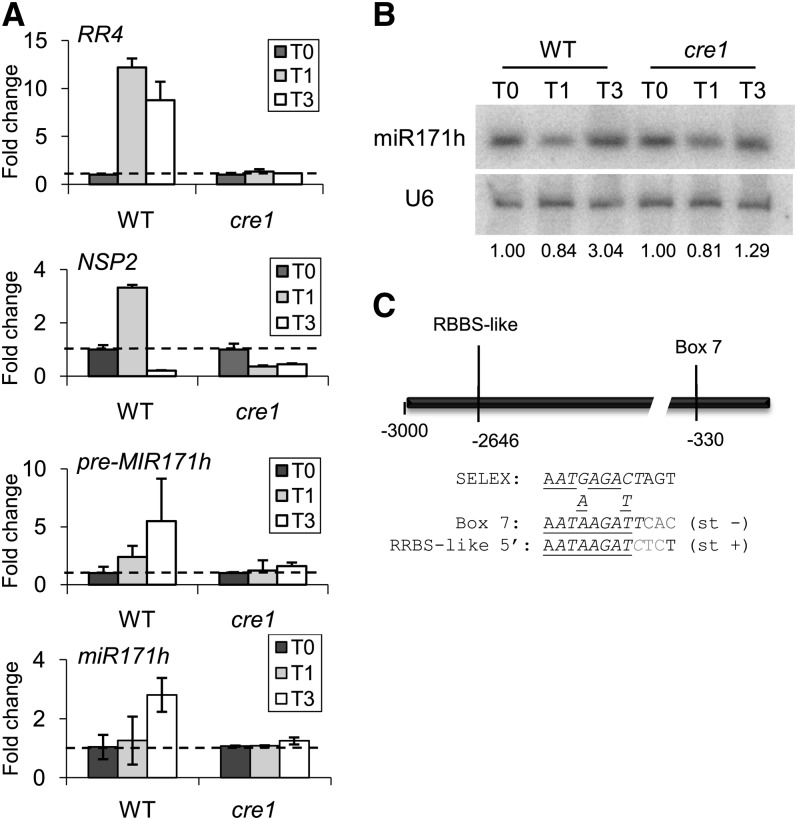
In contrast with RR4, the NSP2 expression pattern in response to cytokinins is very dynamic, with a transient induction followed by a rapid downregulation (Plet et al., 2011) (Figure 3A). Recently, a specific isoform of the miR171 microRNA (miRNA) was shown to cleave NSP2 transcripts efficiently, suggesting a posttranscriptional regulation of this TF (Devers et al., 2011). To explore whether the dynamic regulation of NSP2 in response to cytokinins could be linked to miR171 h, we analyzed its expression after a short-term cytokinin treatment. The MIR171 h precursor was continuously induced after 1- and 3-h treatments with 10−7 M of BAP, depending on the CRE1 pathway (Figure 3A), and displayed an opposite trend to NSP2 expression 3 h after cytokinin application. Accordingly, a CRE1-dependent accumulation of mature miR171 h was observed in response to a 3-h treatment with 10−7 M of BAP (Figures 3A and 3B). Although MIRNA genes were not present on microarrays and therefore could not be considered in the bioinformatic screen, a manual inspection of the MIR171 h promoter revealed the presence of a RRBS core sequence in the proximal region of the promoter as well as an additional RRBS core–like sequence (Figure 3C), indicating that expression of this miRNA isoform may also be directly regulated by cytokinin. These results suggest that cytokinin regulation of NSP2 is posttranscriptionally mediated by miR171 h.
Figure 3.
Cytokinins Regulate the miR171 h miRNA Targeting NSP2.
(A) Real-time RT-PCR analysis of RR4, NSP2, MIR171 h precursor (pre-MIR171 h), and mature miR171 h expression in roots of the wild type (WT) or of a CRE1 cytokinin receptor mutant (cre1-1) treated for 1 or 3 h with 10−7 M of BAP. Error bars represent sd of two technical replicates, and one representative biological replicate out of four is shown (n > 10 independent transgenic roots/condition).
(B) RNA gel blot analysis to detect miR171 h mature small RNA in response to 10−7 M of BAP for 1 or 3 h in the wild type or the cre1 mutant. The U6 RNA was used as a loading control, and calculated ratios normalized against the nontreated condition are indicated below the blots (n > 10 independent transgenic roots/condition).
(C) Schematic diagram of the MIR171 h promoter. A RRBS cis-element (box 7) is shown as well as an additional RRBS-like box (box 7 out of the 8-bp SELEX-core consensus). Alignment to the major SELEX consensus is shown below the promoter diagram. Gray indicates nonconserved nucleotides, underlined indicates conserved nucleotides between all boxes of each promoter, italics indicates 8-bp RRBS core, and st + or st − indicates DNA strand of sequences shown.
The bHLH476 TF Is a Direct Target of the CRE1 Pathway Positively Acting in Nodulation
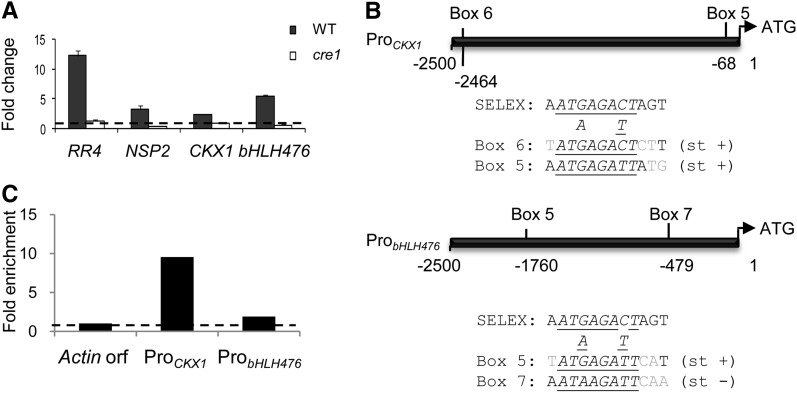
To identify new cytokinin primary target genes potentially acting in nodulation, we then tested whether cytokinin regulation of selected genes (shown in Table 1) was impaired in the cre1-1 mutant, which is essential for nodule organogenesis (Plet et al., 2011). In addition to RR4 and NSP2 genes, only transcripts encoding a bHLH TF (bHLH476) and a CKX (CKX1) were identified as CRE1-dependent (Figure 4A). The bHLH476 and CKX1 genes both contained a box 5 sequence, as in the case of NSP2, and an additional RRBS core variant (boxes 7 or 6, respectively; Figure 4B). ChIP-qPCR revealed that these two promoters were recognized in vivo by RR1 in M. truncatula roots (Figure 4C).
Figure 4.
CRE1-Dependent Regulation of Primary Cytokinin Response Genes.
(A) Real-time RT-PCR analysis of selected candidate cytokinin primary response genes in roots of the wild type (WT) or of a CRE1 cytokinin receptor mutant (cre1-1) treated for 1 h with 10−7 M of BAP. Genes tested are RR4, NSP2, CKX1, and bHLH476. The nontreated condition was set to 1 (dotted line) to visualize FCs. Error bars represent sd of two technical replicates, and one representative biological replicate out of four is shown (n > 10 independent transgenic roots/condition).
(B) Schematic diagram of the CKX1 and bHLH476 promoters. The different RRBS cis-elements variants (boxes 5 to 7) are indicated. Alignment to the major SELEX consensus is shown below each promoter diagram. Gray indicates nonconserved nucleotides, underlined indicates conserved nucleotides between all boxes of each promoter, italics indicates 8-bp RRBS core, and st + or st − indicates DNA strand of sequences shown.
(C) ChIP-qPCR of RR1 binding to CKX1 and bHLH476 promoters. Actin11 ORF was used as a negative control, and IP values were normalized for each gene against the input genomic DNA. Ratios with Actin11 were calculated to visualize RR1 binding fold enrichment (n > 30 independent transgenic roots/construct).
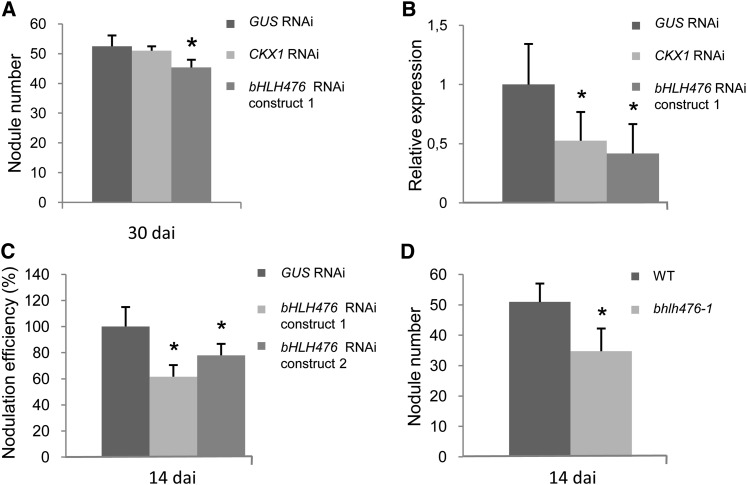
To determine the role of these genes in nodulation, bHLH476 and CKX1 expression was first analyzed using in situ hybridization. Whereas the expression level of the bHLH TF was beyond the detection threshold (in accord with the low expression levels detected using real-time RT-PCR; ∼29 to 30 cycles), the CKX1 gene was expressed in the different nodule zones (apical meristem, rhizobial infection/cell elongation zone, and nitrogen fixing zone; see Supplemental Figure 5 online). We then generated transgenic roots expressing RNAi constructs to silence these genes and tested their ability to nodulate under greenhouse optimal conditions. Only bHLH476 RNAi roots revealed a significant decrease in nodule number and density (Figure 5A; see Supplemental Figure 6A online), despite the fact that a similar level of downregulation was obtained for both transcripts with the respective RNAi constructs (Figure 5B) and that no close homolog of this TF could be detected as expressed in roots and nodules. This phenotype was further confirmed in vitro at an earlier nodulation stage and by using a second independent RNAi construct downregulating bHLH476 expression (Figure 5C). In addition, a Tnt1 insertional mutant predicted to generate a protein truncated in the middle of the bHLH domain (see Supplemental Figure 7 online) also showed a significantly reduced nodulation efficiency (Figure 5D; see Supplemental Figure 6B online). Microscopic analysis of bhlh476 mutant roots infected with a S. meliloti strain expressing a ProHemA:LACZ reporter did not reveal any detectable defect in root hairs or infection thread progression (see Supplemental Figure 8 online). Hence, we identified a new bHLH TF as a direct target of the CRE1-dependent cytokinin signaling pathway positively regulating nodulation.
Figure 5.
The bHLH476 Cytokinin Primary Response Transcription Factor Positively Regulates Symbiotic Nodulation.
(A) Quantification of the number of nodules formed under greenhouse conditions 30 DAI with S. meliloti on roots expressing RNAi constructs targeting CKX1, bHLH476, or GUS (as control), respectively.
(B) Real-time RT-PCR analysis of CKX1 and bHLH476 expression in roots expressing RNAi constructs targeting each gene or GUS (as control), respectively. The GUS RNAi control was set to 1 (dotted line). Error bars represent sd (n = 5 independent transgenic roots/construct), and a Kruskal-Wallis test was performed to assess significant differences (α < 0.05).
(C) Quantification of the nodulation efficiency under in vitro conditions (14 DAI) of roots expressing RNAi constructs targeting bHLH476 (two independent RNAi constructs) or GUS (as control), respectively.
(D) Quantification of the number of nodules formed under greenhouse conditions (14 DAI) on roots of a Tnt1 insertional mutant affecting the bHLH476 gene. WT, wild type.
In [(A), (C), and (D)], error bars represent confidence interval (α = 0.05; n > 30 independent transgenic roots/genotype), and a Kruskal-Wallis (A) or Mann-Whitney [(C) and (D)] test was performed to assess significant differences (α < 0.05).
DISCUSSION
In this study, we have identified primary targets of a cytokinin-related RR using a transcriptomic analysis focused on M. truncatula root apices. Most previous studies aiming to assess transcriptome changes in response to cytokinins were performed in whole seedlings, and to our knowledge, such targeted analysis in roots had not been previously done in any plant. The most significantly enriched functional pathways regulated by cytokinins in Medicago roots were linked to GAs and flavonoids. Previous transcriptomic analyses in Arabidopsis revealed a connection with GA metabolism and response. Indeed, two GA biosynthesis genes (a GA20 oxidase gene and the 3β-hydroxylase gene GA4) were shown to be rapidly downregulated by cytokinins, whereas two genes encoding negative regulators of GA signal transduction (GAI and RGA) were upregulated (Brenner et al., 2005; Kiba et al., 2005). Our analysis further suggests that in M. truncatula root apices, several genes encoding GA-related oxidases and β-hydroxylases are upregulated after a short-term cytokinin treatment. GAs and cytokinins were shown to have an antagonistic action in Arabidopsis shoot and root apical meristems (Greenboim-Wainberg et al., 2005; Alabadí et al., 2009). In addition, GA regulates root growth in Arabidopsis as well as nodule formation in legumes (Ferguson et al., 2005; Greenboim-Wainberg et al., 2005; Achard et al., 2009; Maekawa et al., 2009; Ubeda-Tomás et al., 2009). By contrast, connections between cytokinin action and flavonoid metabolism are poorly documented. In legumes, spatial expression of a gene encoding chalcone synthase is modulated by cytokinins in white clover (Trifolium repens) roots (Mathesius et al., 2000). Our transcriptomic data revealed that many genes related to metabolic pathways leading to the biosynthesis of different types of flavonoids were upregulated in Medicago roots after a short-term cytokinin application, including 10 genes linked to isoflavonoid pathways, four linked to chalcone/flavone pathways, three linked to flavanones, and three linked to flavonols (based on the P < 0.05 threshold). This result strongly points to a role of cytokinins in activating flavonoid pathways in roots, which may not have been previously observed in Arabidopsis, either because whole seedlings were used or, alternatively, because this crosstalk may be legume-specific. Indeed, flavonoids have a critical role in early stages of the legume symbiotic interaction with rhizobia in relation to the activation of bacterial Nod factor production and to the initiation of nodule organogenesis through the regulation of polar auxin transport (Wasson et al., 2006; Oldroyd and Downie, 2008).
Regarding genes encoding TFs, a significant enrichment for bHLH (discussed below) and AP2/ERF families was found. Previous transcriptomic analyses in Arabidopsis seedlings revealed that, in addition to the CRF subfamily, a few Arabidopsis AP2/ERF TFs were upregulated by cytokinins (Hoth et al., 2003; Rashotte et al., 2003; Brenner et al., 2005; Kiba et al., 2005). However, these genes are not closely related to the ones identified in Medicago roots. This result may indicate that legumes show differential regulation of TF subfamilies in response to cytokinins. In addition, the CRF requirement in cytokinin signaling may be different between seedlings and root apices.
The main goal of this study was to identify a set of cytokinin-regulated genes that could be direct primary targets of RR TFs acting in legume roots and nodules. Among several others, we selected the RR gene having the highest expression level in roots and nodules (Gonzalez-Rizzo et al., 2006; M. truncatula Gene Expression Atlas [http://mtgea.noble.org/v2/]). In Medicago roots, RR1 silencing using two different RNAi constructs targeting a specific or a conserved region (Mt RR1-MYB domain RNAi) and Mt RR1 overexpression did not reveal any significant cytokinin-sensitivity phenotype (Gonzalez-Rizzo et al., 2006; see Supplemental Figure 2 online). As observed in Arabidopsis, functional redundancy and compensation effects between different RR genes are likely to prevent the detection of cytokinin-sensitivity phenotypes (Mason et al., 2005). To circumvent these limitations, we ectopically expressed Mt RR1 in Arabidopsis. The altered root cytokinin sensitivity observed suggests that Mt RR1 is involved in cytokinin signaling, even though, in contrast with the positive role of Type-B RRs described in Arabidopsis (Mason et al., 2005; Ishida et al., 2008), reduced cytokinin sensitivity was observed (see Supplemental Figure 2 online). This may be because of the ectopic expression in a heterologous context or alternatively may reveal a noncanonical function. Interestingly, a study analyzing the role of Type A RR genes showing opposite regulations in response to rhizobial Nod factors suggested their paradoxical role in M. truncatula root and nodule development. An RNAi construct targeting these RRs indeed negatively affected both lateral root and nodule formation (Op den Camp et al., 2011), in contrast with other studies affecting either CKX gene expression in Lotus japonicus or Mt CRE1 (Lohar et al., 2004; Gonzalez-Rizzo et al., 2006), which revealed opposite functions of cytokinins in these organogeneses. Overall, these results suggest diverging roles for different members of either Type-B or Type-A RR families in nodulation. Further work is required to determine in each family which individual RRs act downstream of the CRE1 pathway to regulate, either positively or negatively, cytokinin response and nodule organogenesis.
In contrast with the short 5-bp–consensus RR binding sequence identified in Arabidopsis, our study identified a larger 12-bp cis-element consensus, which allowed the application of reliable bioinformatic searches for candidate cytokinin primary response genes. Based on a criteria of RRBS cis-element enrichment in promoters of cytokinin-regulated genes, we retrieved the expected Type-A RR Mt RR4 and two homologs of genes previously reported in Arabidopsis (CKX1 and GST; Taniguchi et al., 2007), indicating a certain conservation between these two plant species. Interestingly, both RR4 and CKX1 are proposed to mediate negative feedback on the cytokinin pathway by affecting signaling or the hormonal pool, respectively. A systematic search for homologs of previously reported Arabidopsis cytokinin primary response genes (Taniguchi et al., 2007) in the transcriptomic data revealed six additional homologs containing at least one RRBS 8-bp core sequence in their promoter and upregulated by cytokinins in Medicago root apices (see Supplemental Data Set 2 online). Among those, an AUX-IAA protein closely related to SHY2/IAA3 acting in Arabidopsis root meristems (Dello Ioio et al., 2008) contained three RRBS core sequences. The higher RR1 binding efficiency to box 2 than to box 5 (Figure 2B) suggests that the cis-element is better recognized when a C is at the eighth position of the AATGAGA(T/C)TAGT sequence. Further work is therefore needed to decipher precisely which RR binds to each RRBS variant and to determine the relevance of the number and location of RRBS cis-element combinations, because tandem organizations of RRBS were retrieved in the two promoters analyzed.
Among the candidate cytokinin primary response genes identified, NSP2 encodes one of the most upstream TFs previously described as acting in early nodulation, both in cortical and epidermal cells (Oldroyd and Downie, 2008; Madsen et al., 2010; Figure 6). RRBS cis-elements present in the NSP2 promoter are required in planta both for cytokinin regulation and expression in nodule primordia. This strongly suggests that a direct regulation of NSP2 expression by the CRE1-dependent signaling pathway occurs during early nodule organogenesis, where both CRE1 and NSP2 act (Kaló et al., 2005; Plet et al., 2011). Unfortunately, we were not able to complement the nsp2 mutant using a 2.5-kb ProNSP2:NSP2 construct, preventing us from testing the functional relevance of cytokinin regulation through the RRBS in nodulation. In addition, our results indicate that the tight and dynamic regulation of NSP2 expression (upregulated after a 1-h cytokinin treatment and repressed at later time points; Gonzalez-Rizzo et al., 2006; Maekawa et al., 2009; Plet et al., 2011) may involve posttranscriptional regulation by the miR171 h miRNA. Whereas several miRNAs were shown to be regulated by auxin and/or to target the auxin signaling pathway, this is the first cytokinin-regulated miRNA (Liu and Chen, 2009). Other TFs linked to nodulation and acting downstream of the CRE1 cytokinin receptor are Mt ERF Required for Nodulation1 (ERN1; Middleton et al., 2007) and Nodule Inception (NIN; Schauser et al., 1999; Madsen et al., 2010; Plet et al., 2011). Despite the fact that no enrichment for RRBS cis-elements was found in these promoters, certain cores similar to the SELEX consensus could be detected (see Supplemental Figure 9 online). We can speculate that cytokinin regulation of NIN and ERN1 may be also dependent on Type-B RRs.
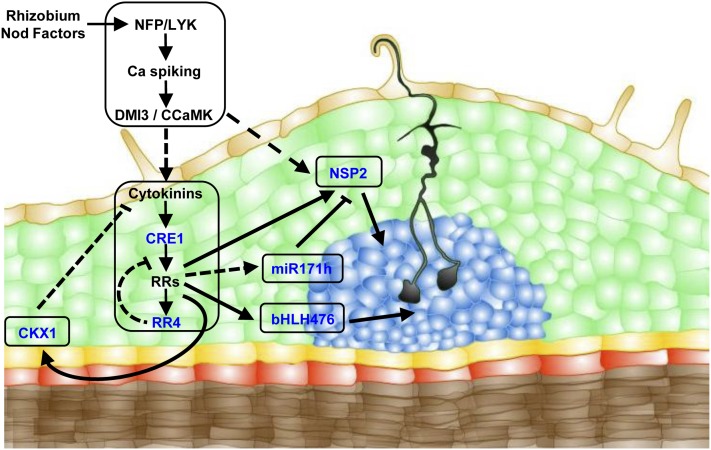
Figure 6.
Downstream Cytokinin Signaling Events in Symbiotic Nodulation.
Schematic representation of a M. truncatula symbiotic nodule primordium, including selected known components of the Rhizobium Nod factor signaling pathway (Nod Factor Perception/Lys-M Kinase receptor [NFP/LYK], Does Not Make Infections/Calcium Calmodulin Protein Kinase [DMI3/CCaMK], Nod factor Signaling Pathway2 [NSP2]) and of the cytokinin signaling pathway (Cytokinin Response1 [CRE1], Response Regulators [RRs]). New genes identified in this study as acting downstream of the CRE1/RR cytokinin pathway are also indicated: the miR171 h isoform targeting NSP2 and the new direct cytokinin targets bHLH476 and CKX1. Proven interactions are indicated by full lines, and putative interactions by dotted lines.
Orange indicates epidermis, green indicates cortex, yellow indicates endodermis, red indicates pericycle, brown indicates stele, and blue indicates nodule primordium.
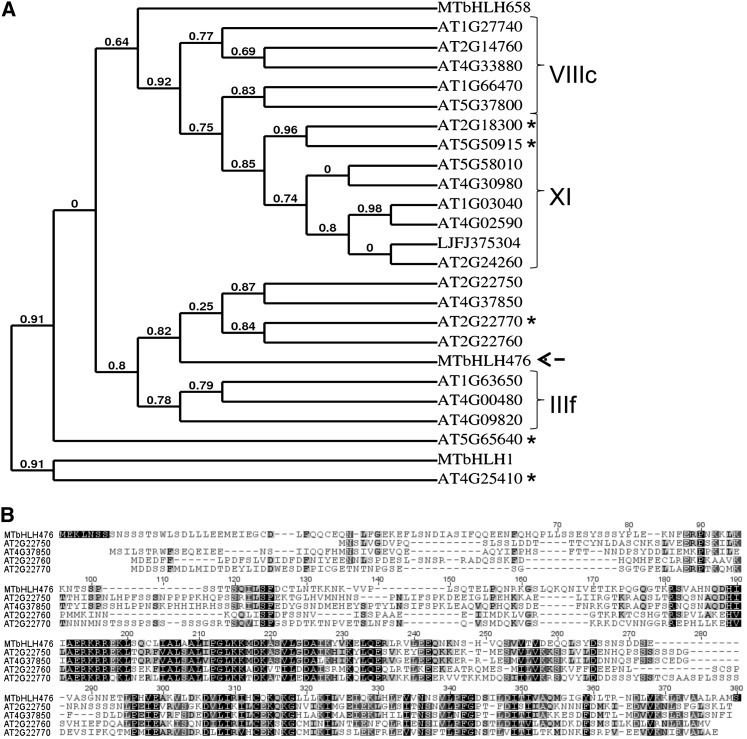
Our study additionally identified a new regulatory gene acting directly downstream of the CRE1 cytokinin pathway and positively regulating symbiotic nodulation: the bHLH476 TF (Figure 6). Previous transcriptomic analyses performed in Arabidopsis seedlings revealed that at least five bHLH TFs were rapidly upregulated by cytokinins (At2g18300, At2g22770, At4g25410, At5g50915, At5g65640; Hoth et al., 2003; Brenner et al., 2005; Kiba et al., 2005; Lee et al., 2007). Apart from its bHLH DNA BD, the Medicago bHLH476 TF is not closely related to any of these genes or to any of the bHLH TFs functionally characterized in legumes (Figure 7). Accordingly, in contrast with root hair less1/slippery mutants (rhl1; Karas et al., 2009), we did not detect any root hair phenotype in bhlh476 mutants or in plants carrying RNAi constructs targeting this gene. Furthermore, infection thread progression was similar to the wild type. Future experiments, including overexpression studies, will help to determine whether bHLH476 is involved in nodule organogenesis. Hence, the bHLH476 cytokinin primary target may have been specifically recruited in legume roots at the convergence of cytokinin and nodulation cues.
Figure 7.
bHLH476 Is Not Closely Related to any Known Cytokinin Response and Nodulation Genes.
(A) Phylogenetic tree of bHLH TFs functionally characterized in legumes (LjFJ375304/ROOTHAIRLESS1 [RHL1]/SLIPPERY [Karas et al., 2009]; MtbHLH1 [Godiard et al., 2011]; MtbHLH658 [Zahaf et al., 2012]), previously linked to Arabidopsis cytokinin response, or related to MtbHLH476 (arrow). Complete proteins were aligned at http://www.phylogeny.fr/ using MUSCLE 3.7, PhyML 3.0 phylogenetic analysis, and TreeDyn (PHYLIP package) displays. Numbers indicate the branch support values as the likelihood a posteriori. Asterisks indicate the cytokinin-responsive bHLH genes identified in Arabidopsis. Groups and subgroups of bHLH proteins are indicated at the right, according to Karas et al. (2009). Sequences are available from The Arabidopsis Information Resource (http://www.Arabidopsis.org/) and from GenBank (http://www.ncbi.nlm.nih.gov/) or TIGR (http://compbio.dfci.harvard.edu/tgi/plant.html) databases for legume species. The alignment is available in Supplemental Data Set 4 online.
(B) Protein alignment (using ClustalW) of MtbHLH476 with its closer Arabidopsis homologs: At2g22750, At2g22760, At2g22770, and At4g37850. Black boxes indicate identity between the five sequences, and gray boxes indicate identity in at least three sequences.
Overall, using a combination of transcriptomic, biochemical, and molecular approaches, we unveiled new transcriptional and posttranscriptional networks acting in symbiotic nodule organogenesis downstream of the CRE1 signaling pathway. Among the novel primary targets of cytokinin action identified, the NSP2 and bHLH476 TFs were linked to nodulation, suggesting their recruitment in legumes into specific symbiotic functions. The identification of extended RRBS cis-elements also opens global perspectives to develop artificial reporters to monitor specifically cytokinin responses in plants.
METHODS
Biological Material
Medicago truncatula cv Jemalong A17 wild type and the cre1-1 mutant allele (Plet et al., 2011) were used in this study, as well as Arabidopsis thaliana (Colombia-0 ecotype). In addition, a M. truncatula mutant derived from the T00007H line of the Jemalong A17 Tnt1 collection was identified as having an insertion located at nucleotide 664 (from the ATG) in the bHLH476 gene (bhlh476-1 allele; see Supplemental Figure 7 and Supplemental Data Set 3 online for genotyping primers). Seeds were sterilized for 20 min in bleach (12% [v/v] sodium hypochlorite). After washing with sterilized water, seeds were sown on 1% agar plates and stored for 2 d at 4°C before incubating overnight at 24°C in the dark to ensure uniform germination. Germinated seedlings were transferred to square plates containing appropriate medium (see below) and grown vertically in chambers at 24°C under long-day conditions (16 h light at 150 μE light intensity/8 h dark).
For nodulation experiments, germinated seeds were grown in vitro on low-nitrogen medium (Fahraeus medium without nitrogen; Truchet et al., 1985). Roots were inoculated with 10 mL of the Sinorhizobium meliloti strain 1021 suspension (OD600 = 0.05) per plate for 1 h. A derivative S. meliloti strain 2011 (GMI6390, pMH682) (GMI6526; Ardourel et al., 1994) carrying the pXLGD4 plasmid containing a ProHemA:LACZ transcriptional fusion was additionally used.
Hormone Treatments
Fifteen germinated seedlings were placed on a grid in a Magenta box with 30 mL of low-nitrogen liquid medium (“i”; Blondon, 1964) and grown in a shaking incubator (125 rpm) at 24°C under long-day conditions (16 h light/8 h dark). After 5 d, seedlings were treated with or without 10−7 M of BAP (Sigma-Aldrich) and maintained under the same growth conditions for various incubation times (0, 1, and 3 h). In parallel, mock experiments were performed, which consisted of collecting nontreated roots across the kinetic (0, 1, and 3 h). No significant variation in expression of any of the genes tested was observed in three independent replicates. Roots were collected at the indicated time points and immediately frozen in liquid nitrogen for RNA extraction. In all cases, four independent biological experiments were performed (n > 10).
Cloning, Expression, and Purification of Recombinant Protein
A region of Mt RR1 (JQ647414, GenBank database [http://www.ncbi.nlm.nih.gov]) containing the DNA BD coding sequence (RR1 BD) was selected (residues 69 to 229; see Supplemental Figure 10 online), PCR-amplified, and inserted in frame into the BamHI and EcoRI sites of the expression vector pGEX-3X (Smith and Johnson, 1988) to generate a GST fusion. Amplifications were performed using oligonucleotides RR1-BD1 (5′-CGCGGATCCCAATTTCCCACGGTGCCC-3′) and RR1-BD2 (5′-CACGAATTCGTCTGGATATCCTTGCTG-3′), where sequences underlined correspond to EcoR1 and BamH1 restriction sites, respectively. Escherichia coli cells, strain JM109 (GE Healthcare), were transformed with the Mt RR1 BD pGEX-3X fusions and then grown and induced for 3 h as described in Palena et al. (1998). However, the incubation after induction with isopropyl-β-d-1-thiogalactopyranoside (Sigma-Aldrich) was performed at 28°C. Recombinant proteins were purified using the GST-tag (GE Healthcare; Kaelin et al., 1992), and stored at −80°C in 5% glycerol.
PCR-Assisted Binding Site Selection
To select DNA molecules specifically bound by the purified recombinant RR1 BD, the random oligonucleotide selection technique (SELEX; Oliphant et al., 1989) was applied, using procedures described by Blackwell and Weintraub (1990). A 32P-labeled (30,000 cpm) 51-mer double-stranded oligonucleotide containing a 12-bp random central core [5′-GATGAAGCTTCCTGGACAAT(12N)GCAGTCACTGAAGAATTCT-3′] was incubated with purified GST-Mt RR1 BD as described above. Bound DNA molecules were separated by EMSA and eluted from gel slices with 0.5 mL of 0.5 M NH4Ac, 10 mM MgCl2, 1 mM EDTA, and 0.1% (w/v) SDS. The selected DNA molecules were amplified using oligonucleotides R1 (5′-GATGAAGCTTCCTGGACAAT-3′) and R2 (5′-CAGAATTCTTCAGTGACTGC-3′). Amplification reactions were performed as follows: 18 cycles of 1 min at 94°C, 1 min at 53°C, and 1 min at 72°C. The number of cycles was decreased to 12 after the fourth round. After purification through polyacrylamide gels, the amplified molecules were subjected to new cycles of binding, elution, and amplification. Enrichment in sequences bound specifically by Mt RR1 BD was monitored by binding and competition analysis in EMSA. After seven rounds of selection, the population of oligonucleotides was cloned into the pCR 2.1-TOPO vector (Invitrogen). Thirty-eight randomly picked clones were sequenced.
DNA Binding Assays
For EMSA performed with synthetic probes, aliquots of the purified proteins were incubated with double-stranded DNA (0.3 to 0.6 ng, 30,000 cpm, labeled with [α-32P] dATP by filling in the 3′-ends using the Klenow fragment of DNA polymerase) generated by hybridization of the complementary synthetic oligonucleotides 5′-AATTCACATATAATGAGACTAGTTGAG-3′ and 5′-GATCCTCAACTAGTCTCATTATATGTG-3′ or derivatives with modifications within the binding sequence (underlined) as described in the Results and Supplemental Data Set 3 online. Binding reactions (20 μL) containing 20 mM of HEPES (pH 7.5), 50 mM of KCl, 2 mM of MgCl2, 0.5 mM of EDTA, 1.0 mM of DTT, 0.5% Triton X-100, 22 ng/μL of BSA, 1 μg of poly(dI-dC), and 10% glycerol were incubated for 15 min at room temperature, supplemented with 2.5% Ficoll, and immediately loaded onto a running gel (5% acrylamide, 0.08% bis-acrylamide in 0.5× TBE plus 2.5% glycerol; 1× TBE is 90 mM of Tris-borate, pH 8.3, and 2 mM of EDTA). The gel was run in 0.5× TBE at 20 mA for 2 h and dried before autoradiography. When competition assays were performed, 100-fold unlabeled double-stranded oligonucleotides were included in the binding reaction mix and incubated for 10 min before the addition of the selected labeled oligonucleotide. For competition EMSAs, fresh DTT was added to avoid the formation of the double band typically caused by GST oligomerization.
In Situ Hybridization, Real-time RT-PCR, ChIP, and RNA Gel Blot Analysis
In situ hybridizations were performed as described in Bustos-Sanmamed et al. (2012) on nodules 21 d after S. meliloti inoculation using an Intavis InsituPro automat (http://www.intavis.com/en/). Antisense RNA probes corresponding to Mt bHLH476 and Mt CKX1 were generated as well as an Mt CKX1 sense probe as negative control (primers shown in Supplemental Data Set 3 online).
For real-time RT-PCR, total RNA was extracted from frozen roots using the RNeasy plant mini kit (Qiagen). First-strand cDNA was synthesized from 1.5 μg of total RNA using the Superscript II First-Strand Synthesis System (Invitrogen). Primer design was performed using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/). Primer combinations showing a minimum amplification efficiency of 90% were used in real-time RT-PCR experiments (see Supplemental Data Set 3 online). Real-time RT-PCR reactions were performed using the LightCycler480 SYBR Green I Master Kit on a LightCycler480 apparatus according to manufacturer's instructions (Roche). Cycling conditions were as follows: 95°C for 10 min, 40 cycles at 95°C for 5 s, 60°C for 5 s, and 72°C for 15 s. PCR amplification specificity was verified using a dissociation curve (55 to 95°C), as well as systematic sequencing of PCR amplicons. A negative control without cDNA template was always included for each primer combination. Technical replicates (on two independent syntheses of cDNA derived from the same RNA sample) and four independent biological experiments (n > 10) were performed in all cases. Ratios were done with constitutive controls for gene expression to normalize the data between different biological conditions. Mt ACTIN, Mt RBP1, and Mt H3L were selected using Genorm software (http://medgen.ugent.be/∼jvdesomp/genorm/; Vandesompele et al., 2002) as reference genes for experiments involving hormonal treatments in wild-type and cre1 mutant roots (primers shown in Supplemental Data Set 3 online). Mt RBP1 was chosen to calculate ratios, and the value of the experimental control condition was set to 1 as a reference to determine relative expression or induction factors.
For ChIP experiments, roots expressing the Mt RR1-hemagglutinin (HA) fusion were used (see below). Purification of root nuclei expressing this construct or an empty vector and HA immunoprecipitation (IP) were done as in Ariel et al. (2010). The different promoters analyzed, as well as the Actin11 open reading frame (ORF, used as negative control, same primers as for RT-PCR), were amplified by real-time PCR from the input genomic DNA (i.e., before IP) and after IP (primers indicated in Supplemental Data Set 3 online; cycling conditions as for RT-PCR). For each gene, the input data were used to normalize results obtained from the IP extracts and the Actin11 ORF as a calibrator. Two independent biological experiments (each based on a pool of n > 30 independent transgenic roots per construct) were performed.
Small RNA gel blots were performed following the protocol described in Boualem et al. (2006), using the 5′-GAGTGATATTGATTCGGCTCG-3′ probe to detect the specific miR171 h isoform. The U6 RNA, used as loading control, was detected with the following probe: 5′-GCAGGGGCCATGCTAATCTTCTCTGTATCGT-3′. The expression profile of the miR171 h isoform was assayed in parallel using stem–loop real-time RT-PCR, as described by Varkonyi-Gasic et al. (2007), using the primer miR171 h-RT 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAGTGA-3′ and primers indicated in Supplemental Data Set 3 online for subsequent real-time PCR.
Hybridization of Microarrays, Data Processing, Statistical Analyses, MapMan Display, and Bioinformatic Workflow
For microarray analysis of the root apices’ response to cytokinins, germinated seedlings were transferred to perlite:sand (3:1) pots without bottoms on a grid and were grown until roots passed through the grid (as described in Gruber et al., 2009). The liquid plant growth media SN/2 (Soluplant 18.6.26; Duclos International) in which the emerging root apices were immersed was replaced by fresh medium with or without 10−7 M of BAP for 1 h. Four biological replicates for each condition (treated and nontreated) were performed (using at least nine plants per replicate). Root apices (1 cm) were harvested, immediately frozen in liquid nitrogen, and stored at −80°C for RNA extraction.
RNAs were extracted as described above for RT-PCR. RNA quality was checked using an Agilent 2100 Bioanalyzer (Agilent). A total of 2 μg of each sample were used to synthesize Cy5/Cy3-labeled cDNA using the Amino Allyl Message Amp II aRNA Amplification kit (Ambion) according to the manufacturer’s instructions. Samples were balanced with respect to dyes: for each experimental condition, the four independent biological samples were labeled with both the Cy5 and Cy3 dyes. This ensures that the effects of interest are not biased by genes interacting with dyes. Cy5/Cy3-labeled cDNA were hybridized with the 70-mer Mt16K+ oligonucleotide microarrays (http://www.ebi.ac.uk/microarray-as/ae/; accession number A-MEXP-138, ArrayExpress database) for 16 h at 60°C in a rotating oven (6 rpm) using an Agilent hybridization chamber system. The hybridized slides were washed in 1× SSC and 0.2% SDS for 10 min at 50°C, 0.1× SSC for 10 min at room temperature, 0.05× SSC for 5 min at room temperature twice, and water at room temperature for 1 min. All washes were done in the dark with gentle agitation.
Microarray slides were scanned with a GenePix two-laser scanner (Axon Instruments), and the resulting images were analyzed with the GenePix 6.0 software (Molecular Devices). Data transformations and normalization, performed with the MAnGO R script (version 1.0; Marisa et al. 2007), consisted of a local background correction, omitting flagged spots, and then an intensity-dependent print-tip loess normalization and a scale between array normalization (Yang et al. 2002). Minimum Information About a Microarray Experiment (MIAME)–compliant data were deposited in the ArrayExpress database (http://www.ebi.ac.uk/microarray-as/ae/) under the accession number E-MEXP-1442. Differential analysis was based on an empirical Bayes moderated t test adjusted with the false discovery rate (Benjamini and Hochberg, 1995) multiple test correction. Differentially expressed genes were selected based on adjusted P-values, and a background threshold was defined based on a mean intensity of the two channels (A-mean) > 7 on a log2 scale (threshold based on negative controls available in the microarray).
Data were visualized using the MapMan software (http://gabi.rzpd.de/projects/MapMan/; Thimm et al., 2004; Tellström et al., 2007), following instructions provided on the website. To predict functional categories showing an enrichment in the differentially regulated genes, we used│fold change (FC)│> 1.5 and adjusted P < 0.001 thresholds to maximize the relevance of the analysis. MapMan bins were used to perform a Fisher’s exact test (P > 0.05), and categories with more than 10 elements and for which at least three were differentially regulated were retained. To scan promoter regions for the RRBS cores, the REMORA Workflow GetAndScanPromoterRegionsFromAListOfMedicagoIDsToUniqueIDs was used (http://www.legoo.org/; Carrere and Gouzy, 2006). Cytokinin-regulated genes were defined based on less stringent criteria (│FC│> 1.5 and adjusted P < 0.05) to allow the identification of the maximum number of promoter regions. The following parameters were set in the workflow: coverage used to define genomic regions corresponding to The Institute for Genomic Research (TIGR) tentative consensus (TC) accession numbers, 90%; promoter length, 2500 bp (defined based on our analysis of RRBS in the Mt RR4 promoter; see Results). PatScan strings were based on box sequences indicated in Supplemental Data Set 2 online followed by [0,0,0], meaning that only perfect matches were allowed.
Cloning, Site-Directed Mutagenesis, and Agrobacterium spp Transformation
For Mt RR4 transcriptional fusion, a 3030-bp sequence upstream of the RR4 start codon was amplified by PCR using a Pfx polymerase (Invitrogen) and primers RR4pro-F and RR4pro-R (as described in Plet et al. [2011]) (see Supplemental Data Set 3 online). A 1133-bp proximal promoter region (referred to as Δbox) was also amplified with primers prox-RR4-F and RR4pro-R (see Supplemental Data Set 3 online). ProRR4 box 2 was mutagenized using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) using the primers RR4-box2-F and RR4-box2-R (see Supplemental Data Set 3 online).
For Mt NSP2 transcriptional fusion, a 2.5-kb sequence upstream of the NSP2 ATG start codon was similarly amplified using primers NSP2-pro-F and NSP2-pro-R (see Supplemental Data Set 3 online). The region between −616 bp and −802 bp was replaced by a BamH1 restriction site by PCR amplification (Δbox construct; primers NSP2-del-F and NSP2-del-R; see Supplemental Data Set 3 online) and subcloned in a pBluescript-SK− vector (Addgene).
The three RRBSs in the NSP2 promoter were successively mutagenized using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) to replace the GAGA central core by an ACTG sequence, leading to the ProNSP2-3XMut construct. This was achieved using the following primers for each box: mutB5-5′F/mut B5-5′R, mutB5-3′F/mut B5-3′R, and mut B6 F/mut B6 R (see Supplemental Data Set 3 online).
The different promoter constructs were finally cloned using Gateway technology from the pTopo-Entry vector (Invitrogen) into the pkGWFS7 vector (http://www.psb.ugent.be/gateway/index.php) carrying a green fluorescent protein (GFP)-GUS fusion downstream of the cloning recombination site.
The RNAi constructs targeting either the Mt RR1 conserved MYB domain (Mt RR1-MYB domain) or the Mt bHLH476 gene were cloned in the pFRN vector (Gonzalez-Rizzo et al., 2006) using primers indicated in Supplemental Data Set 3 online. The pFRN GUS RNAi control is described in Gonzalez-Rizzo et al. (2006). The 35S-CaMV:RR1-HA construct was cloned in-frame with a C-terminal HA tag and was transferred into a pMF plant expression vector (Merchan et al., 2007; primers indicated in Supplemental Data Set 3 online). The empty vector, containing a 35S:GUS cassette, was used as control.
The resulting constructs were introduced into Agrobacterium rhizogenes ARqua1 (streptomycin-resistant derivative strain of A4T; Quandt et al., 1993) and were used for M. truncatula root transformation. The transgenic roots were obtained after kanamycin (25 mg/L) selection for 2 weeks as described by Boisson-Dernier et al. (2001). Composite plants were then transferred onto growth papers (Mega International) on Fahraeus medium without nitrogen (Truchet et al., 1985) for 4 to 6 d and were further used to test cytokinin or nodulation responses (as described before). In addition, the 35S-CaMV:RR1-HA construct was introduced in Arabidopsis ecotype Columbia-0 using the A. tumefaciens floral dipping method, and several independent transformed lines were selected on kanamycin (50 mg/L).
Fluorometric Assays, Histochemical Staining, and Microscopic Analyses
To analyze expression of the GUS reporter in response to a short-term cytokinin treatment, a fluorometric assay was performed to quantify the induction FC in response to BAP treatment. Composite plants generated as described above were transferred to Fahraeus liquid medium and grown for another 3 d. Plants were individually identified, a part of each root (∼0.5 mg) was frozen in liquid nitrogen, and samples were stored at −80°C. After 2 d, the same plants were treated for 3 h with 10−7 M of BAP, and a new sample of each root was frozen with liquid nitrogen. Specific GUS activity in protein extracts was measured using the fluorogenic substrate 4-methylumbelliferyl β-d-glucuronide (Sigma-Aldrich) essentially as described by Welchen and Gonzalez (2005). Total protein extracts were prepared by grinding root tissues in extraction buffer (50 mM sodium phosphate, pH 7.0, 10 mM EDTA, 10 mM β-mercaptoethanol) containing 0.1% (w/v) SDS and 1% Triton X-100, followed by centrifugation at 13,000g for 10 min. GUS activity was measured with 1 mM of 4-methylumbelliferyl β-d-glucuronide and 20% methanol. The results were expressed as a ratio between the values obtained after and before the treatment for each individual root (n > 20).
To analyze expression of the GUS reporter during nodulation, composite plants were inoculated with the S. meliloti strain 1021 (as described before). Three independent biological experiments were performed (n = 25). Histochemical stainings were performed as previously described (Pichon et al., 1992); samples were incubated in darkness for up to 16 h at 37°C. Roots and nodules infected by the ProHemA:LACZ strain were used for β-galactosidase staining (overnight at 30°C), as described in Ardourel et al. (1994). In both cases, after staining, roots were observed 3 to 5 d after inoculation (DAI) with S. meliloti in bright field using a Nikon AZ100 microscope equipped with a DFC 300 camera.
Phylogenetic Analysis
The phylogenetic reconstruction was performed using the default parameters of the PHYLIP package (http://www.phylogeny.fr/). Sequences were aligned with MUSCLE (v3.7; Edgar, 2004) configured for highest accuracy (MUSCLE with default settings), and the Gblocks program (v0.91b) was used to eliminate poorly aligned positions and divergent regions. The alignment is available in Supplemental Data Set 4 online. The phylogenetic tree was reconstructed using the maximum likelihood method implemented in the PhyML program (v3.0 aLRT). The WAG substitution model was selected, assuming an estimated proportion of invariant sites (of 0.073) and four γ-distributed rate categories to account for rate heterogeneity across sites. The γ shape parameter was estimated directly from the data (γ = 3.959). Reliability for internal branch was assessed using the aLRT test (SH-Like). Graphical representation and editing of the phylogenetic tree were performed with TreeDyn (v198.3). Numbers indicate the branch support values as the likelihood a posteriori. Sequences are available from The Arabidopsis Information Resource (http://www.Arabidopsis.org/) and from GenBank (http://www.ncbi.nlm.nih.gov/) or TIGR (http://compbio.dfci.harvard.edu/tgi/plant.html) for legume species.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Mt RR1, JQ647414. The 70-mer Mt16K+ oligonucleotide microarrays are deposited in the ArrayExpress database (http://www.ebi.ac.uk/microarray-as/ae/) under accession number A-MEXP-138, and microarray experiment–compliant data of the transcriptomic experiment are deposited under the accession number E-MEXP-1442.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Metabolic Processes, Hormonal Pathways, and Large Enzymatic Families Regulated in Response to a Short-Term Cytokinin Treatment in Root Apices.
Supplemental Figure 2. Mt RR1 Overexpression in Arabidopsis Affect Root Sensitivity to Cytokinins.
Supplemental Figure 3. Raw Sequences Cloned after Seven Rounds of Binding Site Selection Using SELEX.
Supplemental Figure 4. Competition EMSA Comparing the in Vitro Binding Preference of GST-RR1 BD for Alternative RRBS.
Supplemental Figure 5. Expression Pattern of Mt CKX1 in Nodules.
Supplemental Figure 6. Nodule Density of bHLH476 RNAi Roots.
Supplemental Figure 7. Nucleotidic and Protein Sequence of the bHLH476 TF and Location of the Tnt1 Insertion.
Supplemental Figure 8. Root Hair and Rhizobium Infection Phenotype of bhlh476 Mutants.
Supplemental Figure 9. Schematic Diagrams of Promoter Regions of ERN1 and NIN Cytokinin Nodulation-Related Genes.
Supplemental Figure 10. Protein Sequence of the Mt RR1 RR.
Supplemental Table 1. Differentially Expressed Functional Categories in Response to Cytokinins (10−7 M of BAP for 1 h) in Root Apices Based on MapMan Bins.
Supplemental Data Set 1. Detailed List of Differentially Expressed Genes in Response to Cytokinins (10−7 M of BAP for 1 h) in Root Apices.
Supplemental Data Set 2. Detailed List of Candidate Cytokinin-Regulated Genes with RRBS Boxes in Their Promoters.
Supplemental Data Set 3. List of Primers.
Supplemental Data Set 4. Text File of the Alignment Used to Generate the Phylogenetic Tree Shown in Figure 7.
Acknowledgments
We thank Hervé Delacroix and Sandrine Imbeaud (Gif/Orsay DNA MicroArray Platform, Centre National de la Recherche Scientifique, Gif-sur-Yvette, France) for statistical analyses of the microarray data, Marion Verdenaud (Laboratoire des Interactions Plantes Micro-organismes, Castanet-Tolosan, France) for her involvement in setting up the bioinformatic workflow, and Elena Carvajal (Tétraèdre, France) for artwork. We also thank the anonymous reviewers for their constructive comments. Microscopy was done on the Imagif platform (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France). This project was done in the frame of the French Agence Nationale de la Recherche project LEGUROOT, which also funded M.B.-H. The Tnt1 mutant collection was generated in the frame of the GRAIN LEGUME (GLIP-EEC-FP6) project. Work in Argentina was supported by Agencia Nacional de Promoción Científica y Tecnológica (PICT 2005 38103 and PICT 2007 37000/022). R.L.C. is a member of Consejo Nacional de Investigaciones Científicas y Técnicas, and F.A. was a fellow of the same institution. We thank the ECOS-Sud program A07B03, the Argentinean Ministry of Education, the French Embassy, and the Banco Santa Fe Foundation for providing short-term fellowships to F.A. J.P. was the recipient of a doctoral grant from the Ministère de la Recherche et de la Technologie (France), and S.B. was supported by a GRAIN LEGUME (GLIP-EEC-FP6) fellowship.
AUTHOR CONTRIBUTIONS
F.F. designed the research; F.A., M.B.-H., C.L., E.H., M.B., J.P., M.M, S.B., and R.L.C. performed research; S.C. and M.C. contributed new computational tools; F.A., M.B.-H., C.L., E.H., J.P., M.M, S.B., J.L.I., R.L.C., and F.F. analyzed data; F.A., M.B.-H., M.C., and F.F. wrote the article.
Glossary
- TF
transcription factor
- RRBS
response regulator binding site
- RR
response regulator
- ChIP
chromatin immunoprecipitation
- RNAi
RNA interference
- SELEX
systematic evolution of ligands by exponential enrichment
- BAP
benzyl amino purine
- CRF
cytokinin response factor
- BD
binding domain
- ChIP-qPCR
chromatin immunoprecipitation followed by real-time PCR
- EMSA
electrophoretic mobility shift assay
- GUS
β-glucuronidase
- miRNA
microRNA
- GA
gibberellin
- IP
immunoprecipitation
- FC
fold change
- HA
hemagglutinin
- DAI
days after inoculation
- TIGR
The Institute for Genomic Research
- TC
tentative consensus
- ORF
open reading frame
References
- Achard P., Gusti A., Cheminant S., Alioua M., Dhondt S., Coppens F., Beemster G.T., Genschik P. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19: 1188–1193 [DOI] [PubMed] [Google Scholar]
- Alabadí D., Blázquez M.A., Carbonell J., Ferrándiz C., Pérez-Amador M.A. (2009). Instructive roles for hormones in plant development. Int. J. Dev. Biol. 53: 1597–1608 [DOI] [PubMed] [Google Scholar]
- Ardourel M., Demont N., Debellé F., Maillet F., de Billy F., Promé J.C., Dénarié J., Truchet G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F., Diet A., Verdenaud M., Gruber V., Frugier F., Chan R., Crespi M. (2010). Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell 22: 2171–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57: 289–300 [Google Scholar]
- Blackwell T.K., Weintraub H. (1990). Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 250: 1104–1110 [DOI] [PubMed] [Google Scholar]
- Blondon F. (1964). Contribution à l’étude du développement de graminées fourragères: Ray-grass et dactyle. Rev. Gen. Bot. 71: 293–381 [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Brandstatter I., Kieber J.J. (1998). Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner W.G., Romanov G.A., Köllmer I., Bürkle L., Schmülling T. (2005). Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Bustos-Sanmamed P., Laffont C., Frugier F., Lelandais-Brière C., Crespi M. (2012). Analyzing small and long RNAs in plant development using non-radioactive in situ hybridization. In Plant Organogenesis: Methods and Protocols (Methods in Molecular Biology), Vol. 959, I. De Smet, ed (New York: Humana Press), in press. [DOI] [PubMed] [Google Scholar]
- Carrere S., Gouzy J. (2006). REMORA: A pilot in the ocean of BioMoby web-services. Bioinformatics 22: 900–901 [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino I.B., Deruère J., Kieber J.J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M.T., Aoyama T., Costantino P., Sabatini S. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Devers E.A., Branscheid A., May P., Krajinski F. (2011). Stars and symbiosis: microRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 156: 1990–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B.J., Ross J.J., Reid J.B. (2005). Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol. 138: 2396–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugier F., Kosuta S., Murray J.D., Crespi M., Szczyglowski K. (2008). Cytokinin: Secret agent of symbiosis. Trends Plant Sci. 13: 115–120 [DOI] [PubMed] [Google Scholar]
- Godiard L., Lepage A., Moreau S., Laporte D., Verdenaud M., Timmers T., Gamas P. (2011). MtbHLH1, a bHLH transcription factor involved in Medicago truncatula nodule vascular patterning and nodule to plant metabolic exchanges. New Phytol. 191: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S., Crespi M., Frugier F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y., Maymon I., Borochov R., Alvarez J., Olszewski N., Ori N., Eshed Y., Weiss D. (2005). Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber V., Blanchet S., Diet A., Zahaf O., Boualem A., Kakar K., Alunni B., Udvardi M., Frugier F., Crespi M. (2009). Identification of transcription factors involved in root apex responses to salt stress in Medicago truncatula. Mol. Genet. Genomics 281: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann A.B., Lombardo F., Miwa H., Perry J.A., Bunnewell S., Parniske M., Wang T.L., Downie J.A. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142: 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda K., Imamura A., Katoh E., Hatta T., Tachiki M., Yamada H., Mizuno T., Yamazaki T. (2002). Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14: 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S., Ikeda Y., Morgante M., Wang X., Zuo J., Hanafey M.K., Gaasterland T., Tingey S.V., Chua N.H. (2003). Monitoring genome-wide changes in gene expression in response to endogenous cytokinin reveals targets in Arabidopsis thaliana. FEBS Lett. 554: 373–380 [DOI] [PubMed] [Google Scholar]
- Hwang I., Sheen J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Imamura A., Kiba T., Tajima Y., Yamashino T., Mizuno T. (2003). In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 44: 122–131 [DOI] [PubMed] [Google Scholar]
- Ishida K., Yamashino T., Yokoyama A., Mizuno T. (2008). Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 49: 47–57 [DOI] [PubMed] [Google Scholar]
- Kaelin W.G., Jr, et al. (1992). Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell 70: 351–364 [DOI] [PubMed] [Google Scholar]
- Kaló P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Karas B., Amyot L., Johansen C., Sato S., Tabata S., Kawaguchi M., Szczyglowski K. (2009). Conservation of lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol. 151: 1175–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Aoki K., Sakakibara H., Mizuno T. (2004). Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol. 45: 1063–1077 [DOI] [PubMed] [Google Scholar]
- Kiba T., Naitou T., Koizumi N., Yamashino T., Sakakibara H., Mizuno T. (2005). Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His->Asp phosphorelay circuitry. Plant Cell Physiol. 46: 339–355 [DOI] [PubMed] [Google Scholar]
- Lee D.J., Park J.Y., Ku S.J., Ha Y.M., Kim S., Kim M.D., Oh M.H., Kim J. (2007). Genome-wide expression profiling of ARABIDOPSIS RESPONSE REGULATOR 7(ARR7) overexpression in cytokinin response. Mol. Genet. Genomics 277: 115–137 [DOI] [PubMed] [Google Scholar]
- Liu Q., Chen Y.Q. (2009). Insights into the mechanism of plant development: Interactions of miRNAs pathway with phytohormone response. Biochem. Biophys. Res. Commun. 384: 1–5 [DOI] [PubMed] [Google Scholar]
- Lohar D.P., Schaff J.E., Laskey J.G., Kieber J.J., Bilyeu K.D., Bird D.M. (2004). Cytokinins play opposite roles in lateral root formation, and nematode and Rhizobial symbioses. Plant J. 38: 203–214 [DOI] [PubMed] [Google Scholar]
- Lohrmann J., Sweere U., Zabaleta E., Bäurle I., Keitel C., Kozma-Bognar L., Brennicke A., Schäfer E., Kudla J., Harter K. (2001). The response regulator ARR2: A pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol. Genet. Genomics 265: 2–13 [DOI] [PubMed] [Google Scholar]
- Madsen L.H., Tirichine L., Jurkiewicz A., Sullivan J.T., Heckmann A.B., Bek A.S., Ronson C.W., James E.K., Stougaard J. (April 12, 2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus Nature Com. (online), doi/10.1038/ncomms1009.10.1038/ncomms1009 [DOI] [PMC free article] [PubMed]
- Maekawa T., Maekawa-Yoshikawa M., Takeda N., Imaizumi-Anraku H., Murooka Y., Hayashi M. (2009). Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 58: 183–194 [DOI] [PubMed] [Google Scholar]
- Marisa L., Ichanté J.L., Reymond N., Aggerbeck L., Delacroix H., Mucchielli-Giorgi M.H. (2007). MAnGO: An interactive R-based tool for two-colour microarray analysis. Bioinformatics 23: 2339–2341 [DOI] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U., Charon C., Rolfe B.G., Kondorosi A., Crespi M. (2000). Temporal and spatial order of events during the induction of cortical cell divisions in white clover by Rhizobium leguminosarum bv. trifolii inoculation or localized cytokinin addition. Mol. Plant Microbe Interact. 13: 617–628 [DOI] [PubMed] [Google Scholar]
- Merchan F., de Lorenzo L., Rizzo S.G., Niebel A., Manyani H., Frugier F., Sousa C., Crespi M. (2007). Identification of regulatory pathways involved in the reacquisition of root growth after salt stress in Medicago truncatula. Plant J. 51: 1–17 [DOI] [PubMed] [Google Scholar]
- Middleton P.H., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.D., Karas B.J., Sato S., Tabata S., Amyot L., Szczyglowski K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104 [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Oliphant A.R., Brandl C.J., Struhl K. (1989). Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: Analysis of yeast GCN4 protein. Mol. Cell. Biol. 9: 2944–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op den Camp R.H., De Mita S., Lillo A., Cao Q., Limpens E., Bisseling T., Geurts R. (2011). A phylogenetic strategy based on a legume-specific whole genome duplication yields symbiotic cytokinin type-A response regulators. Plant Physiol. 157: 2013–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palena C.M., Gonzalez D.H., Guelman S.A., Chan R.L. (1998). Expression of sunflower homeodomain containing proteins in Escherichia coli: Purification and functional studies. Protein Expr. Purif. 13: 97–103 [DOI] [PubMed] [Google Scholar]
- Pichon M., Journet E.P., Dedieu A., de Billy F., Truchet G., Barker D.G. (1992). Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell 4: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J., Wasson A., Ariel F., Le Signor C., Baker D., Mathesius U., Crespi M., Frugier F. (2011). MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 65: 622–633 [DOI] [PubMed] [Google Scholar]
- Quandt H.J., Pühler A., Broer I. (1993). Transgenic root nodules of Vicia hirsuta: A fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol. Plant Microbe Interact. 6: 699–706 [Google Scholar]
- Rashotte A.M., Carson S.D.B., To J.P.C., Kieber J.J. (2003). Expression profiling of cytokinin action in Arabidopsis. Plant Physiol. 132: 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte A.M., Mason M.G., Hutchison C.E., Ferreira F.J., Schaller G.E., Kieber J.J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 103: 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Aoyama T., Oka A. (2000). Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 24: 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H., Honma T., Aoyama T., Sato S., Kato T., Tabata S., Oka A. (2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Smith D.B., Johnson K.S. (1988). Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67: 31–40 [DOI] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Taniguchi M., Sasaki N., Tsuge T., Aoyama T., Oka A. (2007). ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol. 48: 263–277 [DOI] [PubMed] [Google Scholar]
- Tellström V., Usadel B., Thimm O., Stitt M., Küster H., Niehaus K. (2007). The lipopolysaccharide of Sinorhizobium meliloti suppresses defense-associated gene expression in cell cultures of the host plant Medicago truncatula. Plant Physiol. 143: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tirichine L., Sandal N., Madsen L.H., Radutoiu S., Albrektsen A.S., Sato S., Asamizu E., Tabata S., Stougaard J. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107 [DOI] [PubMed] [Google Scholar]
- Truchet G., Debellé F., Vasse J., Terzaghi B., Garnerone A.M., Rosenberg C., Batut J., Maillet F., Dénarié J. (1985). Identification of a Rhizobium meliloti pSym2011 region controlling the host specificity of root hair curling and nodulation. J. Bacteriol. 164: 1200–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Federici F., Casimiro I., Beemster G.T., Bhalerao R., Swarup R., Doerner P., Haseloff J., Bennett M.J. (2009). Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19: 1194–1199 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed]
- Varkonyi-Gasic E., Wu R., Wood M., Walton E.F., Hellens R.P. (2007). Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernié T., Moreau S., de Billy F., Plet J., Combier J.P., Rogers C., Oldroyd G., Frugier F., Niebel A., Gamas P. (2008). EFD Is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell 20: 2696–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson A.P., Pellerone F.I., Mathesius U. (2006). Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E., Gonzalez D.H. (2005). Differential expression of the Arabidopsis cytochrome c genes Cytc-1 and Cytc-2. Evidence for the involvement of TCP-domain protein-binding elements in anther- and meristem-specific expression of the Cytc-1 gene. Plant Physiol. 139: 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Schmülling T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P. (2002). Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahaf O., et al. (2012). Comparative transcriptomic analysis of salt adaptation in roots of contrasting Medicago truncatula genotypes. Mol. Plant 5: 1068–1081 [DOI] [PubMed] [Google Scholar]