Abstract
Nanocarriers can fulfill essential functions in the stabilization and delivery of drugs: they prevent solubility issues and degradation, reduce side effects and modify the pharmacokinetic profile. However, particle based pharmaceuticals are very complex and thus challenging to scale up. As formulation routines account for a large fraction of production costs, reducing complexity in the process of assembly, loading and functionalization of nanoparticles is very desirable. Unlike existing approaches with similar goals, our protocol is designed to minimize usage of material and time. Prerequisite to this elegant one-step procedure is the controlled phase-separation of a hydrophobic peptide to nanoparticles, inducing concurrent cargo-entrapment and association of a protein corona. We demonstrate the process by assembling Flutax-2 containing peptide nanoparticles functionalized with transferrin. Cellular uptake of the particles and cargo release depend on specific particle-cell interactions via transferrin receptor. These data indicate corona-mediated delivery of membrane impermeable cargo in vitro by a particulate delivery system entirely composed of amino acids.
Keywords: Peptide, Nanoparticle, Drug Delivery, Targeting, Corona
1. Introduction
Nanocarriers [1] fulfill essential functions in the stabilization and delivery of drugs: they improve solubility [2] and in-vivo stability [3], reduce side effects and influence the kinetic drug release profile [4]. A targeted drug delivery system is minimally composed of carrier (matrix), cargo and ligand. The ligand binds to a membrane receptor and homes the system to a specified tissue. The matrix material is usually composed of lipids [5], polymers, silica, or metals [6] and assembled into vesicles, micelles or particles [7]. Particle properties that determine in vivo applicability include size, surface charge, and dispersibility, mainly governed by the hydrophobic effect [8]. An alternative to the classical carrier materials, colloidal delivery systems are emerging of which the matrix consists exclusively of peptides [9]. Such a matrix is desirable as it is derived fully synthetic (animal-free), yet can be degraded into single amino acids. Peptide synthesis yields purities up to 98 %, avoiding molecular polydispersity and thus batch-dependent variation of physicochemical properties. Additionally, the molecular properties of peptides can be easily fine-tuned by the exchange of single amino acids. However, the design of peptide particles has been exceedingly difficult, mainly due to the low solubility of short hydrophobic peptides in organic solvents.
In previous work we presented CD3ac, an uncharged peptide composed of the 10 amino acids [9]
as a matrix material with the ability to form particles in the nano- and micrometer size range. CD3ac is insoluble in aqueous solution but can be readily dissolved in a weakly apolar environment. Solubility in organic solvents originates in CD3ac′ unusual secondary structure induced by the sequence of alternating D- and L-amino acids. This property is essential as it allows for straightforward synthesis and purification by standard protocols applied in peptide research [10, 11]. Particles can be conveniently assembled from dissolved CD3ac by addition of water: an emulsion spontaneously forms as the ternary mixture (CD3ac, organic solvent, H2O) is brought into the two-phase region (CD3ac, H2O). The emulsification process resembles the ouzo effect [12], however, CD3ac droplets harden to solid particles as the organic solvent is removed. We showed previously that neutral as well as charged aromatic molecules efficiently migrate into the dispersed phase and get trapped during particle formation [9] (Movie S1). These results encouraged us to employ CD3ac for the targeted delivery of a payload into cultured cells. The efficiency of the actual particle assembly protocol prompted us to achieve targeting and delivery by similarly minimized experimental effort. Our drug delivery system consists of the CD3ac peptide matrix, entrapped cargo (Flutax-2) and a corona of transferrin labeled with Alexa Fluor 568 (Tfn-AF568). The assembly procedure takes a single step of 15 minutes and includes the encapsulation of Flutax-2 as well as the spontaneous formation of a Tfn-AF568 corona. Application to cell cultures does not require purification. The system allows for straightforward adjustment of particle size and entraps Flutax-2 at very high densities. Resulting nanoparticles bind specifically to transferrin receptor (TfR) and display cellular internalization by a change of fluorescent properties. Membrane impermeable Flutax-2 is delivered to the cytosol as detected by fluorescence measurements. Established delivery systems with similar performance tend to rely on much higher complexity [13] in design and production. Thus, our approach provides strong incentives for a practical application in the industrial formulation of particulate drug delivery systems.
2. Materials & Methods
2.1. Stock solutions
Synthesis and purification of CD3ac was described in detail previously [9]. Briefly, the peptide was synthesized on solid phase using Fmoc protection group chemistry and purified on C18 reverse phase (RP) chromatography material applying a gradient of acetonitrile and water. Purity was determined by peak integration of RP-HPLC elution profiles at A280 and exceeds 95%. CD3ac stock solutions were prepared by dissolving the peptide in EtOH:H2O (1:1 v/v). The concentration was determined by absorption (Thermo Scientific Nanodrop 2000) at 280 nm in a mixture of EtOH:H2O:DMSO 1:1:2 considering ε280 = 21780. The peptide concentration was adjusted to 742 μM with EtOH:H2O (1:1 v/v), and aliquots of 200 μL were stored at −80 °C until further use. Tfn-AF568 (Invitrogen, T-23365) was dissolved to 500 μg/mL and stored at +4 C. Flutax-2 (Invitrogen, P22310) was dissolved to 40 μM in H2O:EtOH (1:1) and stored at −80 C.
2.2. Particle assembly, loading and corona formation
PNPs were assembled by mixing stock solutions of CD3ac, Tfn-AF568 and Flutax-2 to yield final concentrations of 123 μM CD3ac, 1.6 μM Flutax-2 and 10 μg/mL Tfn-AF568 in H2O:EtOH (1:1, v/v). Emulsification was induced by a first dilution step (1:1, H2O) followed by an equilibration period of 15 minutes before the ethanol content was further reduced to 12.5 % by the second dilution step (1:1, H2O). 50 μL aliquots of the resulting suspension were applied to 24-well sitting drop crystallization plates (Hampton Research, Cryschem) and counter-evaporated 3 times against 1 mL H2O during six hours.
2.3. Cultured Cell Experiments
CHO cells originate from the interdepartmental stock of Cell and Systems Biology at Harvard Medical School. TRVb cells were obtained from D. Cureton (Kirchhausen laboratory, Infectious Disease Institute, Boston, USA). Both cell lines were grown in F12:DMEM 1:1 (Cellgro, 10-090) plus 10% fetal bovine serum (Gibco). 2×104 cells in 0.5 mL media were seeded on cover glasses (VWR, 89015-724) in 24 well plates (Falcon, 353047) and incubated for 16 hours. Cells were washed 1x with PBS and incubated for an additional 30 min in Ham’s F12 medium (Cellgro, 10-080) before 50 μL NP-solution (as prepared above) in 250 μL F12 was applied. The concentration of CD3ac used to incubate cell thus corresponds to 8.3 μg/mL, ignoring the weight of associated Flutax-2 and Tfn-AF568. In competition assays cells were pre-incubated for 30 min in F12 medium containing 17 μM Tfn (Sigma, T1283) before a solution of 50 μL PNP in 250 μL F12 containing 17 μM Tfn was added. Samples were fixed with 3 % paraformaldehyde (Sigma, P6148) in PBS, mounted on glass slides using fluorescent mounting medium (Dako, S3023) and analyzed within 24 hours.
2.4. Fluorimetry
Fluorescence experiments were carried out on a BMG FLUOstar Omega plate reader on black 384 well-plates (MP100-1, Matrical). Dilution series of Tfn-AF568 and Flutax-2 were measured in H2O:DMSO:FBS 6:3:1 (V:V:V) and the data points fitted linearly. Resulting parameters are given in the table below:
To determine the encapsulation efficiencies of Flutax-2, PNPs were assembled applying the procedure described above in the presence of a fixed concentration of Tfn-AF568 (10 μg/mL) and various amounts of Flutax-2 (1.6, 4, 8, 12.5 and 16 μM). After assembly in crystallization plates (see above) PNP samples were normalized with H2O to 100 μL and centrifuged for 1 hour at 16,000 g before 80 μL were separated from the pellet fraction. Both fractions were normalized to 133.3 μL in H2O:DMSO:FBS 6:3:1 (v:v:v) before 120 μL were applied to the well-plate and the fluorescence intensity was measured.
2.5. TEM
PNP samples were prepared as described above. 5 μL of PNPs suspended in H2O were applied to a carbon film coated copper grid (400 square mesh, Electron Microscopy Sciences) and dried. The sample was stained with 10 μL 1 % uranyl acetate during one minute. Excess stain was removed with a filter paper and subsequently applied to a Tecnai G2 Spirit BioTWIN.
2.6. Microscopy
Fixed cells were analyzed on a Nikon Ti inverted microscope equipped with 60x Plan Apo NA 1.4 objective lens. DAPI fluorescence was excited with a 360/40 filter and collected with a 460/50 emission filter. Oregon Green fluorescence was excited with a 360/40 and collected with a 480/40 emission filter. AF568 fluorescence was excited with a 545/30 and collected with a 620/60 emission filter. Images were acquired with a Hamamatsu ORCA R2 cooled CCD camera controlled with MetaMorph 7 software. Gamma, brightness, and contrast were adjusted on displayed images (identically for compared image sets) using ImageJ software. z-Series optical sections were collected with a step size of 0.25 microns ranging from the glass slide to the highest detectable PNP using a Prior Proscan II focus motor. Samples observed after 1 and 6 hours of PNP-incubation are (merged) maximum stack-projections of AF568 and Oregon Green channels. The samples observed after 24 hours were obtained by average-projection of Oregon Green fluorescence and maximum-projection of AF-568 stacks. We used the average projection of Oregon Green to quantify differences in Flutax-2 fluorescence in the cytosol. Cell perimeters were segmented manually in DIC images. Fluorescence point maxima were extracted by ImageJ (v. 1.43u) using a noise tolerance of 50 in the public class MaximumFinder.
3. Results
3.1. Spatial Arrangement of Cargo and Corona
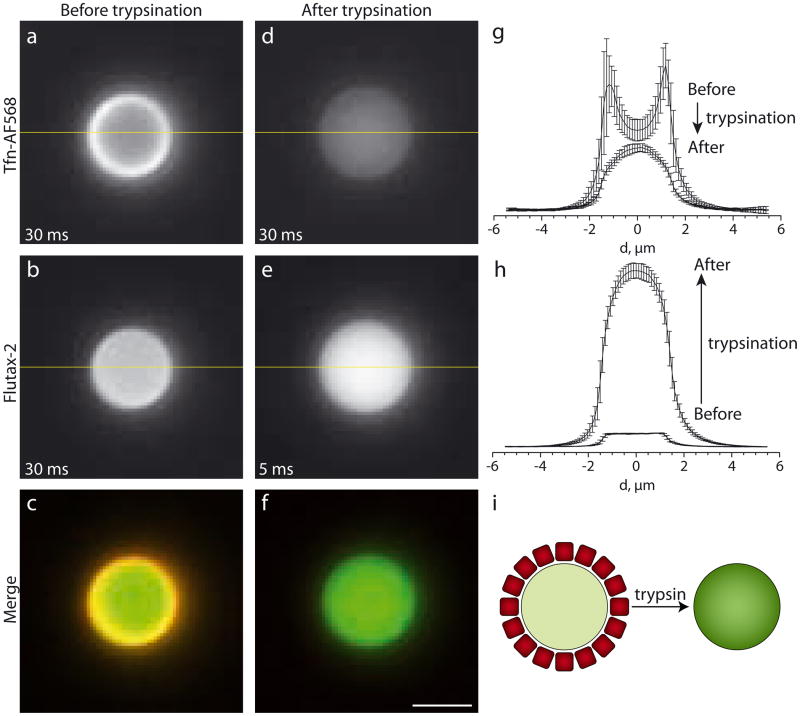
To evaluate the spatial arrangement of reagents in self-assembled CD3ac particles we dissolved peptides along with Flutax-2 and TfnAF568 in 50% EtOH and reduced the EtOH content in steps (see Materials and Methods). Fig. 1 (Fig. S1) show fluorescence images of the resulting particles. We designed them large enough in diameter (ca. 3 μm) to distinguish the distribution of fluorescence in the core and at the surface by conventional light microscopy. Tfn-AF568 shows a bright ring of fluorescence at the particle periphery whereas Flutax-2 fluorescence is equally distributed throughout the particle (Fig. 1a – c). We measured the entrapment efficiency by determining the partition coefficient of Flutax-2 between peptide particles and water (Fig. S2 and S3). We found a coefficient of 5.25, i.e. that under applied experimental conditions more than 80 % of co-dissolved Flutax-2 escapes the aqueous phase and gets entrapped in particles, a remarkably high value for a water soluble compound. Proteins are often surface-active and adsorb onto solid-liquid interfaces. Similarly, particles in contact with protein solutions get covered with a layer of proteins referred to as ‘protein corona’ [14]. Therefore, we hypothesized that the pronounced rim of red fluorescence represents a corona consisting of Tfn-AF568. To test this, we incubated particles for 6 hours in 50 μg/mL trypsin. As expected, the rim disappears while the spatial distribution of the Flutax-2 cargo remains unaltered (Fig. 1d – f). Quantification of gray-level profiles revealed that the intensity of the Tfn-AF568 rim is reduced 3-fold (Fig. 1g). At the same time, removal of the Tfn-AF568 corona resulted in an increase of green fluorescence up to a factor 13 (Fig. 1h). Excitation of Flutax-2 caused emission of Tfn-AF568 leading us to conclude that spectral overlap of the fluorophores leads to emission-absorption effects. To corroborate this interpretation we performed additional control experiments to exclude that trypsin changes the fluorescence intensity of Flutax-2 in solution or in peptide particles lacking a Tfn-AF568 corona (data not shown). Together, these experiments suggest that self-assembly of CD3ac, Flutax-2 and Tfn-AF568 leads to the formation of particles with entrapped Flutax-2 and a corona of surface-adsorbed Tfn-AF568. Trypsination of the particles results in proteolytic degradation of Tfn-Af568 followed by surface desorption of the fragments (Fig. 1i). Removal of the protein corona leads to decreased red and increased green fluorescence.
Fig. 1.
3.2. Size Modulation
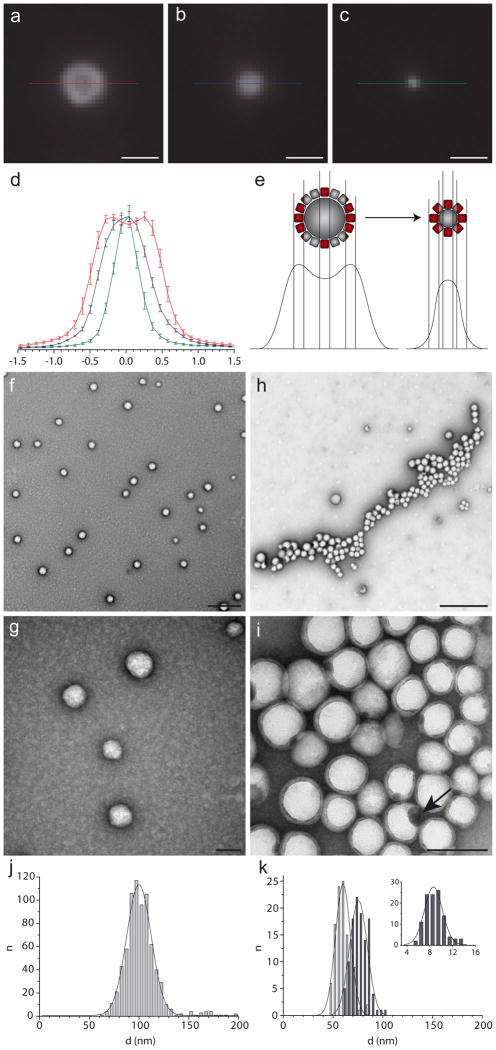
Particles for drug delivery are preferably between 8 and 200 nm in diameter as this size range is least likely to be cleared by kidney and liver [8]. Also, receptor mediated endocytosis, a possible mechanism for the uptake of targeted drug-containing particles, is size-dependent and most efficient for objects smaller than ca. 150 nm [15]. In order to reduce the size of CD3ac particles, we dissolved lower peptide concentrations prior to emulsification: Fig. 2a – c shows fluorescence microscopy images of peptide particles prepared from 492, 246 and 123 μM CD3ac, assembled in the presence of 10 μg/mL Tfn-AF568. The resulting size differences are summarized in Fig. 2d by intensity profiles of particle-associated fluorescence. The characteristic ring, still visible at 492 μM, cannot be observed on smaller particles due to the diffraction limited resolution of optical imaging, although light microscopy confirms the presence of Tfn-AF568 on particles smaller than 300 nm (Fig. 2e). To confirm corona formation of Tfn-AF568 on nanoparticles (d < 100 nm), we applied transmission electron microscopy (TEM). For the sake of brevity, we will use as an acronym for peptide nanoparticles self-assembled in the presence of cargo and corona. Fig. 2f – i show TEM images of PNPFlutax–2 (Fig. 2f, g) and (Fig. 2h, i). Both samples were stained with uranyl acetate, setting apart bright particles and dark background. PNPFlutax–2 were seen in large numbers, evenly distributed on the carbon film (Fig. 2f). By contrast, only few particles could be spotted in the sample: they pool together to clusters (Fig. 2h), reflecting the process of de-wetting and residual water evaporation during sample preparation and thus differential affinity to the hydrophobic carbon support. Higher magnification (Fig. 2i) reveals a rim of intermediate contrast on the nanoparticle surface. Its average thickness of 9.85 nm (st. dev. = 2.1, n = 99) is in agreement with the expected protein diameter for transferrin [16]. Although both samples were prepared by the same protocol, the average diameter of PNPFlutax–2 (100 nm, Fig. 2j) is twice that of (51 nm excluding corona, Fig. 2k). The average size of peptide particles does not only depend on the peptide concentration but also on the presence of surface active molecules which stabilize the emulsion early in the process of phase separation [12]. In conclusion, these electron microscopic analyses establish that peptide particle diameters can be controlled down to a few ten nanometers. Together with the fluorescence microscopy images of Fig. 1, they confirm the presence of a Tfn-AF568 corona on PNPs.
Fig. 2.
3.3. Binding Specificity
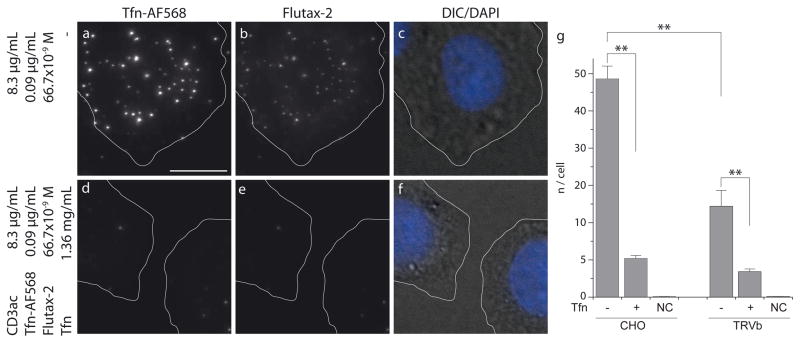
Selective binding to TfR depends on the functionality of the protein corona: its function can be compromised by protein denaturation, steric hindrance (crowding) or unfavorable orientation relative to the PNPs’ surface. Similarly, accumulation of PNPs on cell surfaces is not necessarily caused by specific corona-receptor interactions. Electrostatic (Coulomb) and electrodynamic (Van der Waals) forces can contribute to unspecific association [17–19]. In order to test functionality of the corona in mediating specific surface-receptor mediated binding we correlated the number of cell surface-associated PNPs to the density of available TfR using two independent experimental protocols: a) We competed PNP binding by Tfn in solution; and b) we compared PNP binding between TfR-expressing Chinese hamster ovary (CHO) cells and TRVb cells, which are derived from Chinese hamster ovary tissue that lacks endogenous TfR but expresses a low abundance of TfR2 [20]. Fig. 3 and S5 show microscopy images of CHO cells incubated for one hour with . We observed a significant accumulation of PNPs within the projected cell perimeter (Fig. 3a – d) which could be blocked by incubating CHO cells with 17 μM unlabeled Tfn (Fig. 3e – g, S5). This confirms that PNP interactions with the cell surface depend on freely valent TfR. Fig. 3h shows that the lower TfR density in TRVb leads to a significantly reduced association rate of . Incubation of TRVb with 17 μM unlabeled Tfn blocks binding of suggesting that in these cells interact mostly via the low-abundant receptor TfR2. Application of excess Tfn might not only compete with the PNPs for TfR but may also exchange fluorescent Tfn-AF568 in the particle corona with non-fluorescent Tfn. To test this possibility we compared the fluorescence intensity distribution of PNPTfn–AF568 incubated at 37°C in the presence and absence of 17 μM Tfn after 24 hours and could only detect insignificant differences (Fig. S4). This suggests that the rate of TfR-mediated binding of PNPs to the cell surface is much faster than the protein exchange on the PNP surface. Together with the results presented in Fig. 3 we conclude that PNPs bind specifically to TfR via the Tfn-AF568 corona.
Fig. 3.
3.4. Cellular Internalization
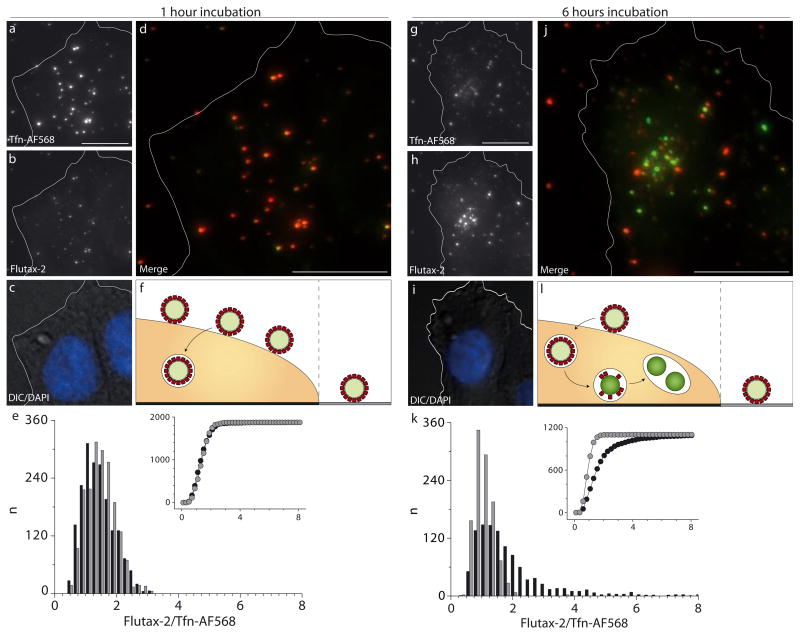
Although we established that bind specifically to cells via interactions with TfR, it is not self-evident that the particles will internalize. Tfn could dissociate from the PNP corona before internalization takes place, and the size difference between single Tfn proteins and a PNP may affect cellular uptake by endocytosis. Distinction between associated and internalized PNPs is not straightforward due to the flat shape of surface-adherent cells and a limited z-resolution of light microscopy. We showed above that removal of Tfn-AF568 from the particle surface can be detected by a pronounced shift of green to red fluorescence ratio (G/R). In the experiment shown in Fig. 4 and Fig. S6, respectively, we take advantage of this property to distinguish between PNPs associated and internalized to cells. We fixed and imaged CHO cells after 1 hour (Fig. 4a – e) and after 6 hours (Fig. 4g – k) of incubation with . After 1 hour the G/R distribution displayed a tight peak around 1.5, both for PNPs within (Fig. 4e, black bars) and outside (Fig. 4e, gray bars) the cell perimeter. After 6 hours, the population of PNPs within the cell perimeter displayed a significant shift towards higher G/R values, while the G/R distribution of PNPs outside the cell perimeter remained confined around 1.5 (Fig. 4k). We interpret this data as follows: After incubation for 1 hour, most PNPs have not yet reached a lysosomal compartment and those which have been internalized still have an intact corona containing Tfn-AF568. After six hours, the majority of PNPs has been transported into lysosomes and their protein corona proteolytically digested. The smaller degradation products dissociate from the particle surface due to weaker Van der Waals forces [21]. In analogy to the clear increase in G/R observed by removal of the corona by trypsin (Fig 1i) the proteolytic digestion of the PNP corona in lysosomes yields an increase of the G/R values of internalized PNPs. This interpretation is corroborated by the unchanged G/R values of PNPs seen on the glass surface. Of note because of the relatively slow digestion kinetics, changes in G/R can only be used as a qualitative indicator of internalized PNPs. Nevertheless, this was sufficient for the purpose of demonstrating that PNPs with a Tfn-AF568 corona enter cells, most likely by clathrin-mediated endocytosis via TfR [22].
Fig. 4.
3.5. Cargo Delivery
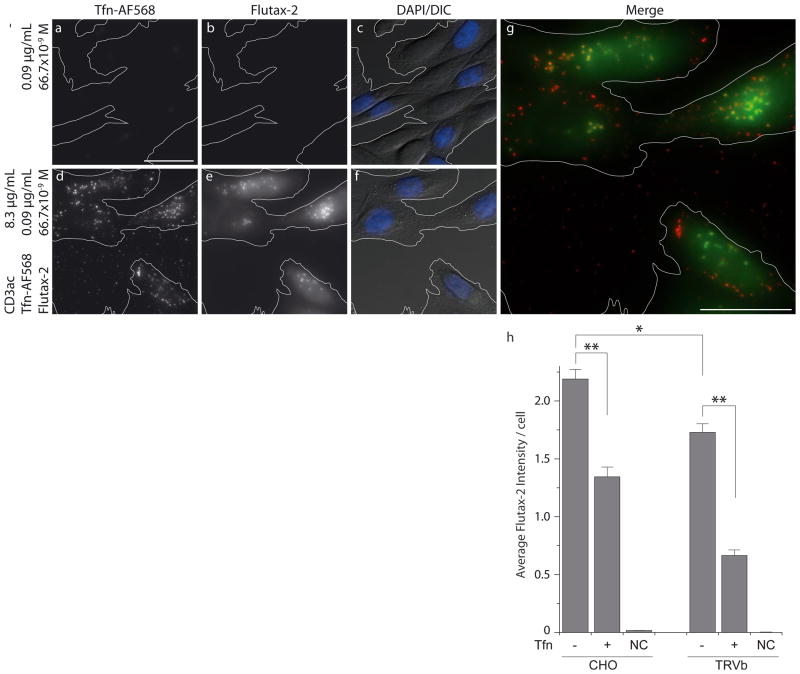
To test whether binding and internalization of PNPs would result in the selective import of small molecule cargo, we analyzed the release of encapsulated Flutax-2 24 hours post addition of (Fig. 5, S7). Flutax-2 is an Oregon Green (OG) modified derivative of paclitaxel [23], a mitotic inhibitor applied in cancer therapy [24]. Unlike its unlabeled form, Flutax-2 is charged and water-soluble at the applied concentration and does consequently not permeate cell membranes. Thus, it was an ideal model compound to investigate efficiency and specificity of small molecule delivery by PNPs to the cytosol. We incubated CHO cells for 24 hours with 0.67 μM Flutax-2, either dissolved in media (Fig. 5a – c) or entrapped in (Fig. 5d – g, Fig. S7). We averaged Flutax-2 emission in the cytosol to quantify the amount of delivered compound. Direct permeation of dissolved Flutax-2 through cell membranes could not be detected as the resulting fluorescence did not exceed the level of autofluorescence (Fig. 5b, Fig. S7). On the other hand, incubation with resulted in a strong diffuse green fluorescence signal (Fig. 5e and g), suggesting the delivery of Flutax-2 to the cytosol. The delivery was significantly reduced by competition of PNP-cell interactions by 17 μM dissolved and unlabeled Tfn in cell culture medium (Fig. 5h). Also, the overall rate of delivery was significantly lower for TRVb cells, which express only TfR2.
Fig. 5.
4. Discussion
We assembled a targeted drug delivery system consisting of peptide matrix, Flutax-2 as cargo and Tfn-AF568 as a specific cell surface receptor ligand. All three components self-assemble to loaded and functionalized particles by application of a one-step-procedure. The simplicity of system and formation protocol originates in the concerted interaction of all involved components: CD3ac is not only matrix material, but supersedes encapsulation routines due to its high affinity to small aromatic molecules. The process of cargo uptake most likely resembles a two-phase liquid extraction where Flutax-2 escapes the aqueous phase and accumulates in peptide droplets, probably due to high affinity between delocalized ring systems of tryptophanes and Flutax-2. Additionally, the peptide’s solubility in mild organic solvents allows for concurrent dissolution and self-assembly of all involved components. The presence of Tfn-AF568 during emulsification of CD3ac results in the formation of a protein corona, targeting PNPs against TfR. Additionally, the presence of the protein allows for straightforward adjustment of particle size due to its surface activity and thus early stabilization of the peptide emulsion. Upon internalization of PNPs into lysosomal compartments, proteolytic digestion on a time scale of a few hours removes the corona and is followed by the release of cargo into the cytosol on a time scale of days.
Currently, we do not know the exact mechanisms of PNP endocytosis and cargo release from the lysosomal compartment. However, PNP binding to TfR and size range of the particles suggests uptake via clathrin-mediated endocytosis. We hypothesize that cargo release goes back to the proteolytic degradation of PNPs in the lysosome. The structure of charged CD3ac degradation products is likely to penetrate lipid membranes and might lead to the disruption of lysosomes [25]. Future, detailed cell biological analyses will be necessary to unveil these aspects.
5. Conclusion
Here, we focused on the global characterization of PNP’s performance as the first targeted drug delivery system based entirely on amino acids. Our work was inspired by the notion to create a nanoparticulate material with reduced preparation effort but complex functionality. This was achieved by the self-assembly of three molecular components to a particle whose function is greater than summed functions of its parts.
Supplementary Material
Table 1.
| Tfn-AF568 | Flutax-2 | |||
|---|---|---|---|---|
| Value | Std Error | Value | Std Error | |
| Intercept | 4501.69 | 234.76 | 4809.00 | 85.56 |
| Slope | 1.011×106 | 5750.49 | 5.573×1010 | 2.515×108 |
| R2 | 0.99977 | 0.99986 | ||
Acknowledgments
We thank the Nikon Imaging Center at Harvard Medical School for help with light microscopy.
This work was supported by fellowships from the Novartis Foundation and the Swiss National Science Foundation (C.D. and C.J.B.) and by the grant NIH R01 GM090317 (G.D.)
Appendix. Supplementary Material
Supplementary data associated with this article can be found, in the online version, at doi:
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotechnology. 2007;2:751–60. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 2.Knop K, Hoogenboom R, Fischer D, Schubert US. Poly(ethylene glycol) in drug delivery: pros and cons as well as potential alternatives. Angew Chem Int Ed Engl. 2010;49:6288–308. doi: 10.1002/anie.200902672. [DOI] [PubMed] [Google Scholar]
- 3.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–38. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen TM, Cullis PR. Drug delivery systems: entering the mainstream. Science. 2004;303:1818–22. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- 5.Multifunctional nanocarriers. Adv Drug Deliv. 2006;58:1532–55. doi: 10.1016/j.addr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Stark WJ. Nanoparticles in biological systems. Angew Chem Int Ed Engl. 2011;50:1242–58. doi: 10.1002/anie.200906684. [DOI] [PubMed] [Google Scholar]
- 7.Soussan E, Cassel S, Blanzat M, Rico-Lattes I. Drug delivery by soft matter: matrix and vesicular carriers. Angew Chem Int Ed Engl. 2009;48:274–88. doi: 10.1002/anie.200802453. [DOI] [PubMed] [Google Scholar]
- 8.Nel AE, Mädler L, Velegol D, Xia T, Hoek EM, Somasundaran P, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8:543–57. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 9.Dittrich C, Meier W. Solid Peptide Nanoparticles: Structural Characterization and Quantification of Cargo Encapsulation. Macromolecular Bioscience. 2010;10:1406–15. doi: 10.1002/mabi.201000221. [DOI] [PubMed] [Google Scholar]
- 10.Merrifield RB. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J Am. 1963;85:2149–54. [Google Scholar]
- 11.Nilsson BL, Soellner MB, Raines RT. Chemical synthesis of proteins. Annu Rev Bioph Biom. 2005;34:91–118. doi: 10.1146/annurev.biophys.34.040204.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sitnikova NL, Sprik R, Wegdam G, Eiser E. Spontaneously formed trans-anethol/water/alcohol emulsions: mechanism of formation and stability. Langmuir. 2005;21:7083–9. doi: 10.1021/la046816l. [DOI] [PubMed] [Google Scholar]
- 13.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Baldelli BF, et al. Physical-Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. J Am Chem Soc. 2011;133:2525–34. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, et al. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 15.MacGillivray RTA, Moore SA, Chen J, Anderson BF, Baker H, Luo Y, et al. Two High-Resolution Crystal Structures of the Recombinant N-Lobe of Human Transferrin Reveal a Structural Change Implicated in Iron Release. Biochemistry. 1998;37:7919–28. doi: 10.1021/bi980355j. [DOI] [PubMed] [Google Scholar]
- 16.Min Y, Akbulut M, Kristiansen K, Golan Y, Israelachvili J. The role of interparticle and external forces in nanoparticle assembly. Nat Mater. 2008;7:527–38. doi: 10.1038/nmat2206. [DOI] [PubMed] [Google Scholar]
- 17.Fleck CC, Netz RR. Electrostatic colloid-membrane binding. Europhys Lett. 2004;67:314–20. [Google Scholar]
- 18.Dagastine RR, Manica R, Carnie SL, Chan DY, Stevens GW, Grieser F. Dynamic forces between two deformable oil droplets in water. Science. 2006;313:210–3. doi: 10.1126/science.1125527. [DOI] [PubMed] [Google Scholar]
- 19.McGraw TE, Greenfield L, Maxfield FR. Functional expression of the human transferrin receptor cDNA in Chinese hamster ovary cells deficient in endogenous transferrin receptor. J Cell Biol. 1987;105:207–14. doi: 10.1083/jcb.105.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parhi P, Golas A, Barnthip N, Noh H, Vogler EA. Volumetric interpretation of protein adsorption: Capacity scaling with adsorbate molecular weight and adsorbent surface energy. Biomaterials. 2009;30:6814–24. doi: 10.1016/j.biomaterials.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97:329–39. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lillo MP, Cañadas O, Dale RE, Acuña AU. Location and properties of the taxol binding center in microtubules: a picosecond laser study with fluorescent taxoids. Biochemist. 2002;41:12436–49. doi: 10.1021/bi0261793. [DOI] [PubMed] [Google Scholar]
- 23.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Can. 2004;4:253–65. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 24.Stewart KM, Horton KL, Kelley SO. Cell-penetrating peptides as delivery vehicles for biology and medicine. Org Biomol Chem. 2008;6:2242–55. doi: 10.1039/b719950c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.