Abstract
How do molecular motors convert chemical energy to mechanical work? Helicases and nucleic acids offer simple motor systems for extensive biochemical and biophysical analyses. Atomic resolution structures of UvrD-like helicases complexed with DNA in the presence of AMPPNP, ADP·Pi, and Pi reveal several salient points that aid understanding mechano-chemical coupling. Each ATPase cycle causes two motor-domains to rotationally close and open. At a minimum, two motor-track contact points of alternating tight and loose attachment convert domain rotations to uni-directional movement. A motor is “in gear” for action only when fully in contact with its track and, if applicable, working against a load. The orientation of domain rotation relative to the track determines whether the movement is linear, spiral or circular. Motors powered by ATPases likely deliver each power stroke in two parts, before and after ATP hydrolysis. Implications of these findings for analyzing hexameric helicase, F1F0 ATPase and kinesin are discussed.
Keywords: motor-track-load, mechano-chemical coupling, ATPase, linear, spiral, circular, step size, inchworm, hand-over-hand
INTRODUCTION
All living organisms depend on motor proteins to move, transport cargos, divide, and do work. All motor proteins require electrochemical energy to overcome random thermal diffusion and produce directional motion (12). The most well known and best studied of motorized processes include actin-myosin based muscle contraction (29, 41–43, 57, 73), flagella-propelled bacteria or sperm cell movement (44, 97), microtubule-kinesin facilitated cargo transport (30, 38), membrane transporters moving molecules against concentration gradients (37, 66, 78, 83), and various helicases, which can separate double helices, alter DNA and RNA structures, and translocate, package or remodel genomic DNA (34, 80, 81, 91, 93). All motor proteins depend on chemical or electrical energy to perform mechanical work. For example, the rotational movement of bacterial flagellae is driven by proton or sodium ion gradients (52, 60, 63). In addition, electrochemical gradients of H+ or Na+ ions are used by F1F0 ATP synthase to make the universal high-energy molecule ATP (48, 105) (Fig. 1). Energy stored in ATP molecules is then released through hydrolysis and converted to mechanical work by many motor proteins. This review will use DNA helicase as an example to examine how chemical energy and mechanical work are exchanged and how the direction, form and step size of movement are controlled.
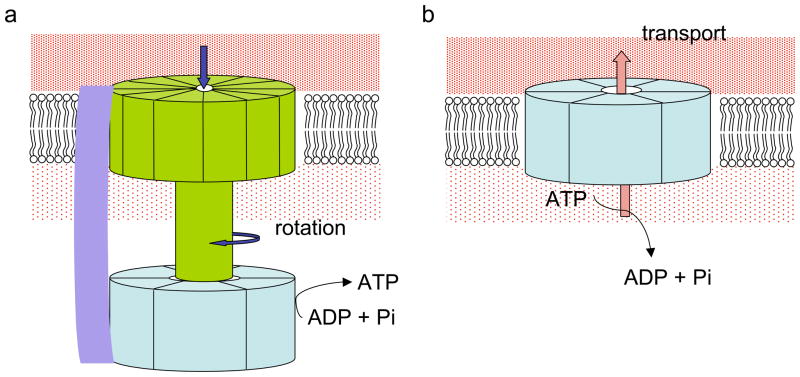
Figure 1.
(a) A schematic diagram of F1F0 ATP synthase using ion gradients to generate torque, which in turn powers ATP synthesis. (b) A diagram of using energy released from ATP hydrolysis to transport ions against its concentration gradients.
MOTOR, TRACK AND LOAD
Each motor system contains at a minimum a motor and a track. For example, muscle contraction powered by actinomyosin, microtubule-based cargo transport, translocation of helicases along RNA or DNA, rotation of F1 ATPase, and ion transport against concentration gradients across a membrane (29, 48, 53, 66, 68, 79, 85, 93, 105). The presence of a track molecule greatly stimulates the ATPase activity of its linked motor protein (31, 36). Molecular tracks also influence the path and form of movement. When embedded in a membrane, a motor protein may rotate within the plane of membrane or transport substrates across the membrane. Along a protein or nucleic acid filament, motors may move linearly or in a spiral. Since macromolecules are chiral and protein and nucleic acid polymers are polar, these tracks also help to determine the direction of motor movement. Without a track to define the path, form and direction, movement of motors would be random and futile. In addition to the track, most motor systems have a “work load” and encounter resistance, for example cargos in the microtubule-kinesin system, a concentration gradient across membrane, and the finite volume of a bacteriophage head for packaging DNA.
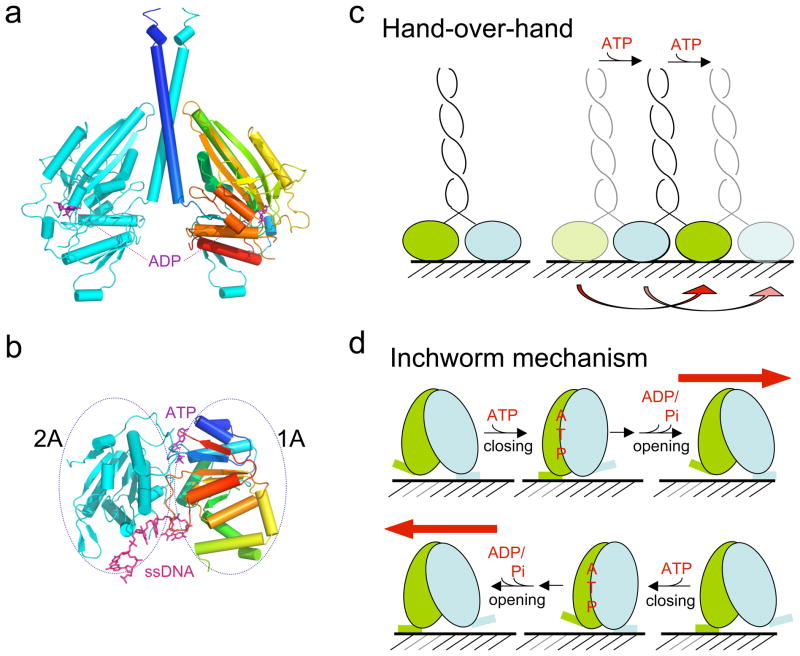
The overwhelming majority of motors are ATPases and use ATP as the energy source. All motor ATPases contain a Walker A or P-loop motif (32, 93, 100), and structurally can be separated into two large classes, myosin/kinesin-like (51, 82) and RecA-like (93, 100) (Fig. 2a–b). Kinesins and myosins are able to hydrolyze ATP in the monomeric form, but dimerization increases the turnover rate of ATPase (33, 35, 59, 70, 77). Typical kinesins are dimeric and are proposed to move processively by a hand-overhand mechanism (30, 38, 112) (Fig. 2c). In contrast, many RecA-like motor proteins require two juxtaposed RecA-like domains to form one ATP-binding site (1, 93). Two RecA-like motor domains may belong to one polypeptide chain in monomeric motors (98) or to adjacent protomers in a multimeric protein (25, 89). So far, RecA-like motors have been shown to move like an inchworm (104, 108) (Fig. 2d). The difference between hand-over-hand and inchworm mechanisms will be discussed at the end of this review.
Figure 2.
(a) Structure of a kinesin dimer (PDB: 2NCD) shown in ribbon diagrams. One subunit is shown in cyan and the other in a blue-to-red gradient from the N- to C-terminus. The bound ADPs are shown as purple sticks. (b) A ribbon diagram of the two RecA-like domains, 1A and 2A of UvrD (PDB: 2IS6). Domain 1A contains the Walker A and B motifs and is shown in blue-to-red gradient. (c) The hand-over-hand mechanism. (d) The inchworm mechanism. The same ATPase-driven domain rotation and the same two motor-track contacts may enable movement in opposite directions by switching the tight contact to loose and loose to tight with regard to the ATPase cycle. Figures 2a–b and 4 were made using PyMol (www.pymol.org).
METHODS AND LIMITS OF STUDYING MOTORS
Motion and motor proteins have been studied by an assortment of physiological, biochemical and biophysical techniques, from electrophysiology (74), light and electron microscopy (41, 65, 110), kinetic measurement of ATP hydrolysis (31, 47, 58), crystallography (39, 102), to more recent force and fluorescence single molecule microscopy (76, 86, 88, 101). These approaches complement one other and have revealed diverse architectures, organizations and mechanisms for motorized movement. For years the step size of a motor protein was a central question, which generated much debate and stimulated research of multiple approaches. Using kinetic, microscopic and crystallographic approaches, the myosin/kinesin and helicase fields have now come to general agreements. For example, the step size is ~80Å for kinesins (40, 99) and 1 bp (~ 3.4Å) for helicases per ATP hydrolyzed (14, 28, 53, 55, 71, 93).
The unresolved question is the mechanism for converting chemical energy to mechanical work. The details of mechano-chemical coupling are of sub-nano scale in time and space and require higher resolution analyses than the step sizes. To fully understand the energy conversion, one needs to examine a complete motor system, which includes the motor, track, and load if applicable.
Each available biophysical method usually focuses on one aspect of a multi-faceted process and suffers from various technical limitations. Crystallography offers the highest spatial resolution, but it is limited to materials that can be crystallized and provides only snapshots of low-energy states in a dynamic process. Electron microscopy is particularly powerful in imaging large complexes (3, 102). It complements crystallography well because of no requirement of crystals. But it suffers from relatively low resolution. Kinetic studies reveal the rate and amplitude of a chemical process, but only limited parameters of a complex process can be determined and are subject to model-bias in data fitting (46). Force and fluorescence-based single-molecule microscopies provide real-time observations of molecular motion. Yet microscopic observations are often indirect (via fluorescence label or force readout) and short of atomic resolution. For example, single-molecule approaches missed the mechano-chemical coupling and actual helicase step-size since the microscopic approaches detect kinetic pausing and cannot visualize the chemical reaction and mechanical movement simultaneously (18, 23, 55, 71).
Biochemical and biophysical analyses require ample amounts of homogeneous and well-behaving motor proteins, track molecules, and appropriate loads. Conformational heterogeneity of motor proteins resulting from their intrinsic dynamic nature may hamper biophysical studies, particularly in bulk solution where multiple species are recorded as an ensemble. The requirement of assembling a multi-component motor system, e.g. polymeric nature of actomyosin and microtubule or electrochemical gradients across a membrane, imposes further limits on high-resolution studies. Muscle contraction and cargo-bearing transport have so far been out of the reach of crystallization. How a proton gradient or external rotary force alters the kinetic and dynamic properties of F1F0 ATP synthase has also not been visualized at atomic resolution.
HELICASES AS A MODEL SYSTEM
Helicases, which translocate along and separate RNA or DNA double helices, have been successfully studied by a variety of biochemical and biophysical techniques (17, 55, 93). The success is largely due to the fact that pure and active motor proteins are relatively easy to produce, and the track molecules (DNA and RNA) can be chemically synthesized or generated in cell culture in large quantities. Moreover, the track molecules also serve as the “work loads” of helicases since the structures or macromolecular interactions of DNA or RNA are subjected to energy-dependent changes by helicases. Due to this simplicity, helicase translocation coupled with nucleic acid unwinding is one of the best-characterized motor systems.
Helicases are RecA-like ATPases. They exhibit increased ATPase activity in the presence of appropriate RNA or DNA track molecules. In addition to Walker A and B motifs, helicases contain distinctive sequence signatures and are divided into six superfamilies (SF), I to VI (93). The smallest functional units are monomers for SFI and SFII helicases, and cylindrical hexamers or dodecamers for the remaining four SFs. A monomeric helicase contains two RecA-like domains, one of which has the Walker A and B motifs for ATP hydrolysis and the other of which contributes to ATP binding but not catalysis (Fig. 2b). An ATP molecule is bound at the interface between two RecA-like domains (13, 49, 53, 90, 92, 95, 98). In contrast, each subunit of a hexameric helicase contains one RecA-like domain, and up to six ATP molecules are bound at the subunit interfaces between adjacent RecA domains (6, 11, 25, 27, 69, 94, 96).
Not all helicases necessarily unwind RNA or DNA double helices as the name may imply. Some helicases transolcate along single or double stranded RNA or DNA and remove whatever “road blocks” or associated molecules may exist along the way (5, 103). Chromatin remodeling ATPases (80) and the Holliday junction migration motor protein RuvB (107) are examples of motor proteins that translocate along dsDNA without making ssDNA. In addition, the helicases that seemingly unwind a double helix may separate two strands by translocating along one strand and displacing the other without actually untwisting the helix ((22, 45, 53) and references therein). Therefore, a helicase may be considered loaded when it unwinds double helices or translocates along a single strand.
UvrD and SFI helicases
UvrD helicase, also known as helicase II in E. coli (as the second helicase discovered) belongs to SFI (36, 84). It contains two RecA-like motor domains and uses ATP to unwind DNA duplexes in the 3′to 5′direction (53, 62). UvrD and its homologous SFI helicases Rep (50) and PcrA (98, 104) are among the most extensively studied and best-understood motors on nucleic acids. Extensive examination of these motors when bound to DNA substrate and undergoing ATP turnover by kinetic (2, 16, 20, 21, 26, 67), single-molecule microscopic (18, 71, 72), and particularly crystallographic studies (50, 53, 98, 104) reveals several key elements of mechano-chemical coupling, which may be generally applicable to other helicases and motor proteins.
In addition to the two RecA-like motor domains (1A and 2A), these SFI helicases have two DNA-interacting domains (1B and 2B) inserted in domains 1A and 2A, respectively (Fig. 3). A 40-residue unstructured C-terminal region of UvrD, which modifies the helicase activity and renders the protein polydispersive in vitro, was removed for crystallographic studies (15, 53, 64). The crystal structure of a Rep – ssDNA complex was the first helicase-DNA structure determined (50). Crystal structures of PcrA-DNA and PcrA-DNA-ATP analog complexes provided the first glimpse of the inchworm-like motion of helicases translocating along ssDNA in atomic detail (104). The full interactions between this family of helicase and a ss-ds DNA junction were eventually revealed by the crystal structures of UvrD-DNA complexes in three states: ATP free, ATP-analog (AMPPNP) bound, and an ATP hydrolysis transition state (53). These structures provide snapshots of ATP-driven directional translocation of UvrD along the ds- and ss-DNA and the atomic details of mechano-chemical coupling.
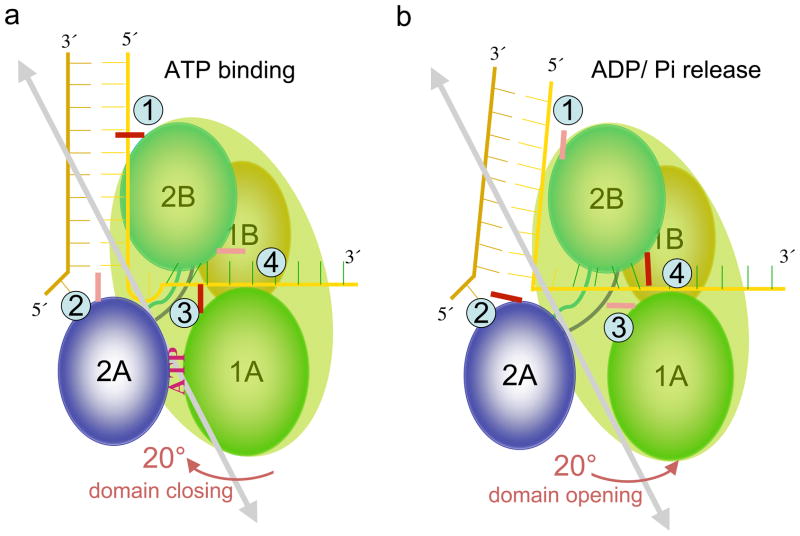
Figure 3.
Diagram of four domains (1A, 1B, 2A, 2B) in UvrD. The ds and ss DNA are roughly orthogonal to each other when bound to UvrD. The 1A, 1B and 2B domain form a rigid body when UvrD is in full contact with ds- and ss-DNA. There are four critical contact points between UvrD and DNA, labeled as 1, 2, 3 and 4. (a) The 20° domain closing induced by ATP binding, and (b) the reverse of domain opening by ADP/Pi release around the grey double-arrowed axis. The tight contacts are indicated in deep red and loose contacts are indicated in pink.
DOMAIN ROTATION AND INCHWORM MECHANISM
Binding of ATP between two RecA-like domains of UvrD or PcrA induces a ~20° rotation that closes the two domains; ATP hydrolysis and subsequent release of ADP and Pi result in a reverse 20° rotation and domain opening (Fig. 3) (53, 104). Interestingly the ~ 20° closing and opening motions driven by the ATP binding and ADP and Pi release have also been observed with structurally unrelated ATP-dependent chaperones (54). This ATPase-driven back-and-forth domain rotation of helicases is converted to mechanical movement of the motor along nucleic acid and therefore can be viewed as the engine of SFI helicases. Conversion of rotational motions to various forms of translocation is mediated by the 1B and 2B domains, which bridge the engine and the nucleic acid track.
The “inchworm” mechanism (104, 108) has two essential components: a rotational movement (closing and opening) of the engine (analogous to arching and extending the body of an inchworm), and two contacting points with alternating tight and loose interactions with a track (analogous to the front and hind legs of an inchworm) (Fig. 2d). Because the two alternating contact points are tightly coupled to the domain rotations, the outcome of domain closing and opening is uni-directional translocation and the distance traveled is the sum of the opening and closing rotations projected onto the track (Fig. 2d). The two components, the rotation axis for domain closing-and-opening and the track of translocation (which is equivalent to the line connecting the two contact points) have to be near orthogonal, so that the rotational movement of a motor core can be converted to linear translocation.
Direction of movement is determined by how the tight and loose contacts are coupled to the ATPase cycle. The contact point that is loose when the motor domains open (extend) and tight when the domains close (contract) is the head or front of movement (Fig. 2d). The other contact point, which has opposite tight and loose grip of the track relative to the domain rotation, is the tail and back of movement. The direction of movement will be reversed if the two contact points switch their tight and loose hold onto the track in coordination with the domain closing and opening (Fig. 2d).
The inchworm-like movement powered by domain rotations is exemplified in atomic detail by UvrD helicases.
HELICASE-DNA INTERACTIONS
UvrD, Rep and PcrA translocate along ssDNA in the 3′ to 5′ direction (20, 26, 72, 108). They bind ssDNA strongly and dsDNA weakly (7, 62, 72, 108). Stimulation of ATPase activity of these helicases is proportional to DNA binding, the tighter the DNA binding, the higher the ATPase activity. UvrD binds dsDNA with a 3′ single-stranded overhang tightest and also exhibits the highest ATPase activity in the presence of 3′-ssDNA overhang (53).
Full heliase-DNA contact for the motor to be “in gear”
The four domains in UvrD, Rep and PcrA helicases are open and flexible in the absence of a DNA track (98). Even when bound to ssDNA, Rep helicase still adopts two conformations. A half is “open” as in the DNA-free state and the other half is “closed” (50). When PcrA was bound to a DNA duplex with a 3′ single strand overhang, the four protein domains became stabilized in the “closed” conformation (104) (Fig. 3). However, because protein-dsDNA interactions were weak and somewhat disordered, each of the four domains moved in an uncoordinated fashion between the ATP-free and ATP-bound states (104). In contrast, in the UvrD-DNA complexes where both ds and ss portions of DNA are properly associated with UvrD, the four domains form two rigid bodies (Fig. 3). Domains 1A, 1B and 2B rotate in unison relative to domain 2A in response to ATP binding (53). Therefore the ATPase-powered domain rotation between two RecA-like domains, 1A and 2A, can be propagated throughout the helicase and to the bound DNA. Full contacts between the helicase and DNA, which is between the motor, track and load, appear to be essential to set the motor protein “in gear” so that the ATP-driven domain rotations are efficiently converted to DNA translocation instead of being dissipated.
Orthogonal ds- and ss-DNA
When bound to UvrD, 16 bps of duplex and 5 single-stranded nucleotides are in contact with the helicase and well defined by electron density (53). The duplex and single-stranded portions of DNA are roughly orthogonal to each other (Fig. 3). The orthogonal orientation allows the ds and ss DNA to move separately. Domains 1B and 2B form extensive interactions with dsDNA on one side, and 5 nucleotides of the ssDNA are sandwiched between these DNA-binding domains and the RecA-like domains 1A and 2A. Most of protein-DNA interactions are with the phosphosugar backbones. Interactions with DNA bases are often via base stacking and non-sequence specific. The direction of ssDNA is approximately perpendicular to the ATP-induced domain-rotation axis, and the dsDNA helical axis is at ~ 30° angle to the domain-rotation axis (Fig. 3).
Four alternating tight-loose contacts
Four rather than two critical protein-DNA contact points are observed between UvrD and DNA (53). The first is between the 2B domain and the dsDNA ~10 bp upstream from the ds-ss junction. The second is between the ds-ss junction and domains 2A and 2B. The third is between the middle of ssDNA and the motif III (domain 1A) in the ATP-binding pocket near the γ-phosphate of ATP. The fourth occurs where the ssDNA exits UvrD. Domains 2B, 1B and 1A are all involved in forming this ssDNA exit gateway. An α-helix from domain 2B moves in coordination with the ATP/ADP binding state to close and open this gateway, thus regulating the ssDNA movement. Among the four contact points, the first two contact the dsDNA portion and the second, third and fourth contact the ssDNA in the direction of 5′ to 3′ (Fig. 3).
Comparison of the crystal structures of the ATP free, AMPPNP bound, and ATP hydrolysis intermediate states of UvrD reveals how these four contact points alternate tight and loose interactions. During the ATP-binding induced domain closing, the first and third contact points remain unchanged (therefore tight), while at the second and the fourth contacting points protein structures change and DNA and protein undergo relative movement. At the ds-ss junction (2nd contact), one base pair undergoes an unwinding movement and separates. The newly unpaired nucleotide bulges out (Fig. 3a). The ssDNA exit gateway (4th contact) becomes open and one nucleotide passes through it and leaves UvrD. During ADP and Pi release and domain closing, however, contact points 1 and 3 become loose and protein-DNA interactions change, while contacts 2 and 4 are held fast. The ssDNA exit gateway becomes closed. When domains 1A, 1B and 2B rotate away from domain 2A, ssDNA moves with these protein domains by 3.4Å, and the bulged-out nucleotide gets straightened. The loose contact 1 allows the protein to move along the dsDNA helix and re-establish the contact with the next base pair (Fig. 3b). The contact point 2 at the ds-ss junction is tight and prevents backward slippage and the newly separated base from re-pairing with its partner.
Such tight and loose alternations are evident in the crystal structures. When UvrD or PcrA was co-crystallized with DNA without ATP analogs, the exit gateway is always closed (53, 104), confirming that the protein-DNA interaction is tight. But when UvrD was crystallized with DNA and an ATP analog or an ATP hydrolysis intermediate, the gateway can be either open or closed (53), indicating that the DNA-protein interaction is flexible and thereby loose. A similar alternation of tight and loose protein-DNA interaction was observed at the first contact point between domain 2B and the DNA duplex. In the three crystal forms of UvrD-DNA complexes without ATP, the contact invariably occurs 10 bps from the ds-ss DNA junction. But in the 8 crystal structures of UvrD-DNA-ATP analog complexes, this contact point occurs 8, 9 or 10 bps from the ds-ss junction, apparently influenced by crystal lattice contacts, but centering at 9 bps (53).
The importance of these contacts is further supported by mutagenesis studies. Mutations of any one of the contacts in PcrA and UvrD impede the mechano-chemical coupling and diminish the efficiency, directionality and regularity of translocation by these helicases (19, 53). The seemingly enhanced helicase activity of Rep helicase by removing a dsDNA binding domain (2B) (10, 16) is likely due to the fact that 2B domain is not engaged in dsDNA binding without the necessary cofactor DBP (108). Since domain 2B limits ssDNA translocation by regulating the exit gateway (Fig. 3), its presence in Rep helicase without the benefit of dsDNA binding can explain the reduced ssDNA translocation (10, 16).
Direction of movement
Coupled with the ATP-driven domain rotation, these four alternating tight and loose protein-DNA contacts lead to an overall movement of UvrD in the 3′ to 5′ direction along the ssDNA overhang and unwinding of one base pair per ATPase cycle. As predicted by the inchworm mechanism, the head or front of movement is contact 1 because it is tight during domain closing (ATP binding) and loose during domain opening (ADP and Pi release), and the tail or back is contact 4 because it has the opposite tight and loose interactions between UvrD and DNA. To reverse the direction of translocation, a helicase may maintain the same protein-DNA interactions. By simply switching the tight-loose interactions at each contact point with regard to ATP binding and ADP/Pi release, it will travel along DNA in the opposite direction (Fig. 2d). This is indeed the case as observed with two 5′ —> 3′ helicases, RecD, also of SFI (87, 92).
FORM OF TRANSLOCATION
Linear movement along ssDNA
Linear translocation of a DNA helicase along ssDNA was first observed in atomic detail in the PcrA-DNA crystal structures with and without a bound ATP analog (104). The authors found that two contact points (equivalent to contacts 2 and 3 in UvrD-DNA complex) defined the direction and step size of the translocation along ssDNA. As shown in the case of UvrD, the second, third and fourth contacts along ssDNA form a line that is perpendicular to the rotational axis between the RecA-like motor domains. These contacts are loose-tight-loose in the ATP binding step and become tight-loose-tight during the ADP and Pi release step. The orientation of these contacts and the alternation of tight and loose interactions result in a linear single nucleotide translocation during each ATPase cycle.
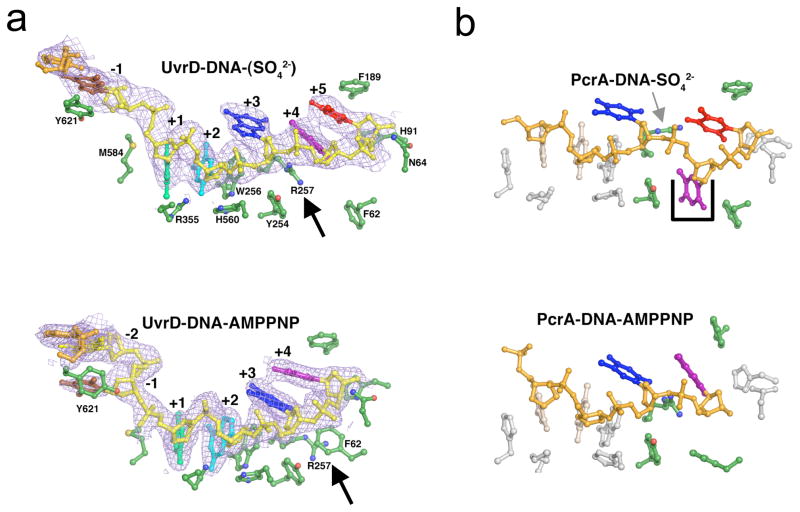
Interestingly, these contact points with ssDNA are not strictly conserved among UvrD-like helicases. In the absence of ATP, 5 single-stranded nucleotides are bound by the UvrD, PcrA and Rep helicases between the ds-ss junction and the ssDNA exit gateway. PcrA appears to secure its hold on ssDNA at contact 3 during ATP binding with a unique nucleotide “trap hole” that pockets the base of the flipped out 4th nucleotide, counting from the ds-ss junction (Fig. 4). After PcrA is bound to an ATP analog (AMPPNP), the ‘trap hole” is filled by a protein side chain and the nucleotide is released, thus allowing subsequent translocation of one nucleotide. The “trap hole” and nucleotide flipping in ssDNA have not been observed in NS3, Rep, UvrD, Hel328, VASA and eIF-4A complexed with either DNA or RNA (4, 8, 13, 49, 50, 53, 56, 90). In UvrD, an Arg sidechain instead of the “trap hole” at the third helicase-DNA contact point stops ssDNA from translocating in the 5′ to 3′ direction during ATP binding. After ATP is bound this Arg alters its rotamer conformation and moves out of the ssDNA pathway (Fig. 4). Therefore the contact point becomes loose and allows ssDNA to translocate in the 3′ to 5′ direction during ADP/Pi release.
Figure 4.
Different ways of forming tight and loose contact with ssDNA by homologous UvrD and PcrA. (a) In UvrD-DNA complexes, each nucleotide from +1 to +5 position counting from the ds-ss junction is color-coded and highlighted by the electron densities. The Arg (R257) (indicated by the black arrow head) stacks with DNA bases (+4 and +5) in the ATP-free state and holds ssDNA in place during ATP binding. R257 together with F62 change their rotamer conformations when UvrD is bound to an ATP analogue and allows ssDNA translocation during ADP and Pi release. (b) In PcrA-DNA complexes, the last three nucleotides corresponding to +3, +4 and +5 in UvrD are colored accordingly. The trap-hole that stops ssDNA translocation during ATP binding is shown in a black box. Interestingly, PcrA does contain the equivalent of R257 (indicated by the grey arrow).
Spiral movement around dsDNA
In contrast to the orthogonal orientation of ssDNA relative to the domain rotation axis and linear translocation as an outcome, the ds-DNA helical axis is at a ~30° angle to the motor domain rotation axis in the UvrD-DNA complex (Fig. 3). Although the motor domains undergo the same closing-and-opening rotations, the orientation of the two contact points between UvrD and dsDNA are along the DNA helical axis and slightly inclined to the motor core rotation, thus resulting in a spiral movement (consisting of both rotational and translational components) of the helicase along dsDNA (Fig. 3). This spiral movement of UvrD was likened to a ratchet “wrench” in repeated ATPase cycles (53). Fundamentally, it is a derivative of inchworm mechanism.
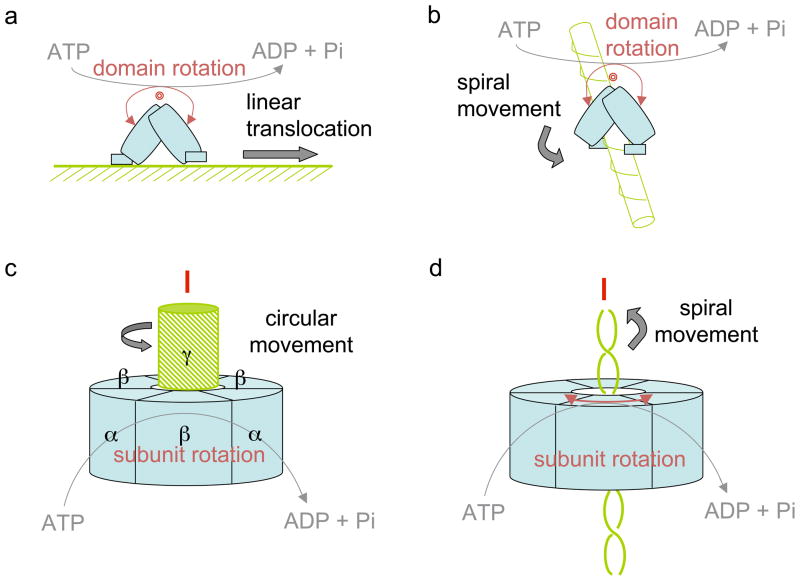
Since molecular movement is based on the ATPase-driven domain rotation, how does domain rotation lead to linear, circular or spiral movement? The inchworm mechanism may seemingly imply a linear form of translocation. But the UvrD helicase demonstrates that spiral movement can result from the same motor domain rotations. Theoretically, the inter-domain closing-and-opening rotations can result in linear, circular or spiral movement depending on the orientation of contact points relative to the domain rotational axis. If the direction of a track (the line connecting the two contacting points) is perpendicular to the inter-domain rotation axis, the movement will be linear. If the track is parallel or collinear to the rotation axis, the movement will be circular. If the two are at an angle, the resulting movement will be a spiral (Fig. 5).
Figure 5.
Linear, spiral and circular movement. (a) Linear movement by monomeric motor in an inchworm fashion. (b) Same monomeric motor can move along a helix in the spiral form. (c) Cylindrical motors often conduct circular (rotary) movement. (d) They can also move in a spiral form, e.g. around the helical backbone of nucleic acid, which may appear to be linear translocation.
COMPARISON WITH OTHER MOTORS
Non-linear movement by rotary motors
Many motor proteins are cylindrical with multiple subunits related by a rotational symmetry including, for example, F1F0 ATP synthase (48, 105) and hexameric or dodecameric helicases of superfamily III to VI (6, 11, 25, 27, 69, 94, 96). Each subunit contains one RecA-like domain, and ATP molecules are usually bound at the subunit interface. ATP binding and release of ADP and Pi lead to the relative rotation of adjacent subunits towards and away from each other (1). Naturally, movement powered by the cylindrical motors is often rotational as observed with the F1F0 ATPase (as the reversal of ATP synthesis) (76). This is because the ATPase-powered subunit rotation is concentric with the protein symmetry and the direction of the track (central stalk, γ subunit) (Fig. 5c).
The relative orientation of a hexameric helicase and its track molecule, nucleic acid threading through the center of the circular motor (22, 25) (Fig. 5d), is analogous to that between the γ and α3β3 subunits of F1 ATPase. Yet ATPase-powered subunit rotation leads to translocation along rather than merely rotation around ss or dsDNA in the center. Because of a co-linear component in the relationship between the nucleic acid helical axis and subunit-rotation axis, translocation of helicase along nucleic acid cannot be linear. The seemingly linear translocation most likely results from a spiral translocation of the helicase around the ss or ds DNA or RNA backbone. As demonstrated by UvrD, spiral movement is possible if the contact points between protein and nucleic acid are at an angle to the inter-subunit rotational axis. This can be readily achieved since protein motors interact with the helical backbone of nucleic acid (25).
Two parts of a power stroke
Each ATPase cycle can be divided into two halves separated by the chemical reaction (ATP hydrolysis): ATP binding and ADP/Pi release. The ATP hydrolysis reaction is reversible and ATP can reform (9). Only after release of ADP, Pi or both is the energy stored in the phosphodiester bond extracted. In the UvrD case, each half of the ATPase cycle is associated with a motor-domain rotation, closing and opening; and both do work. A similar division of labor before and after ATP hydrolysis has been observed with the ATP powered rotation of F1F0 ATPase (as a reversal of ATP synthesis) (75, 109) (see discussion later). ATP binding alone is associated with a large change of free energy, which can be used to do work. But only after release of hydrolysis products ADP or Pi does the work performed become permanent and irreversible. In the case of UvrD, during the first half of the ATPase cycle ATP binding is coupled to domain closing and leads to unwinding of 1 bp. In the second half of the ATPase cycle, ADP and Pi release is coupled with domain opening and leads to ssDNA translocation by 1 nt. Neither of these two elements can exist without the other, and together they achieve translocation and unwinding of 1 bp.
For UvrD, the two parts of an ATPase cycle also separate the linear translocation along ssDNA and the spiral movement along dsDNA. The division ensures the attachment of motor to its track, and a minimum of two contact points are maintained at any given time. It also reinforces the direction and step size of UvrD movement and prevents any slippage on ss or dsDNA. The energy released from each ATP hydrolysis, 7–8 kcal per mole, clearly exceeds the free energy in a base pair (106). The excess may be rationalized as being spent on assuring the direction, form and step size of motion.
It is hard to imagine that a two-part ATPase cycle and division of labor is unique only to UvrD-like helicases and the F1F0 ATPase. A single power stroke is often used to describe the movement of myosin V during Pi release (24, 39) because attachment of the motor to the track has been shown to be tight only in the ADP-bound form. Yet it is possible that when fully loaded and in gear, myosin has other attachment sites and delivers a power stroke during ATP binding. As exemplified in DNA unwinding by UvrD, there must be more than one critical contact point between a motor and its track to ensure the direction and step size. Even when one or two contacts are loose and weak, a power stroke can still be delivered.
Step size of F1F0 ATPase
F1F0 ATP synthase consists of a membrane-embedded F0 rotor empowered by ion gradients and the F1 ATP synthesis center (105). Without F0 or ion gradients, F1F0 ATP synthase becomes an ATPase. The minimal F1 made of 3 αβ and 1 γ subunits (α3β3γ) can hydrolyze ATP and rotates the central γ subunit (76) (Fig. 1a, 5c). The β subunits contain the ATPase activity, while the α subunits merely bind ATPs (1). Each ATP hydrolyzed by a β subunit results in a 120° rotation of the γ subunit relative to the surrounding α3β3, so that three ATPs hydrolyzed by the circular F1 ATPase lead to a full-circle rotation of γ. Each 120° rotation is further divided as 80°–90° during ATP binding and 30°–40° during Pi release as determined by fluorescence-based single molecule microscopy (75, 109). Yet 80° is a large number. It surpasses any recorded domain or subunit rotation driven by an ATPase in a single step. It is larger than ~ 60° span of each α or β subunit and much larger than the ~20° domain rotation observed with UvrD-like helicases. An 80° rotation entails that the γ subunit has to skip one subunit each time an ATP is bound (Fig. 5c). One wonders if the α subunits, which bind but don’t hydrolyze ATP, may undergo simple ATP binding and release powered by the ATPase activity of an adjacent β subunit, and thereby undergo similar conformational changes as the β subunits and contribute to the overall γ subunit rotation. The 120° rotation per ATP hydrolyzed may be conducted by both α and β subunits in four sub-steps, which result from two ATP binding, one ATP release, and ADP and Pi release steps. By this mechanism, an average of 30° rotation sub-steps can be correlated to the nucleotide binding- and release-induced protein conformational changes and is much closer to the 20° rotation observed with UvrD-like helicases.
Inchworm vs. hand-over-hand
Both inchworm and hand-over-hand mechanisms are based on a domain or subunit rotation powered by ATPase. Monomeric helicases have been shown in atomic detail to translocate by the inchworm mechanism whether linearly or spirally (53, 93). The dimeric kinesin and myosin V molecules have been shown to move by a hand-overhand mechanism (111, 112). The feature that has been used to differentiate the two mechanisms is whether the two subunits of the dimeric motor alternatively take the lead position. The hand-over-hand mechanism predicts that they alternate, whereas the inchworm mechanism predicts that one subunit maintains the lead position (111, 112). The microscopic observations clearly support the hand-over-hand mechanism. However, the critical question that remains to be addressed is the number of contact points between each motor subunit and track and how they alternate tight and loose interactions according to the ATP, ADP and nucleotide-free states. The UvrD-like helicases require at least two contacts, but the kinesin/myosin motors appear to have one contact by each ATPase subunit. Physically a single contact point between a motor protein and track with alternating tight and loose interaction cannot track the direction and step size of movement. However, myosin and kinesin have two motor subunits, and they may alternately have tight and loose interactions with the track. The control of directional movement then rests on the tight coupling of the two motor subunits. Interestingly, the proposed coiled-coil link between two kinesin subunits is distal to the microtubule- and ATP-binding sites (Fig. 2a). Moreover, the linker (known as neck linker) between the motor head domain and coiled coil appears flexible (61). It will be interesting to uncover the atomic details of myosin and kinesin motors that are loaded and in full contact with their respective track, how they move with defined direction in large steps, and what the fundamental differences between inchworm and hand-over-hand mechanisms turn out to be.
SUMMARY POINTS.
A motor system includes not only a motor but also a track and a load.
The heart of all molecular engines is likely relative domain or subunit rotations powered by ATP hydrolysis or ion gradients.
Domain rotations are then converted to linear, circular or spiral movements depending on the orientation of the domain/subunit rotational axis relative to the contacts with the track. A monomeric or dimeric motor may move in a linear or spiral form, like UvrD, kinesin and myosin V. A cylindrical motor can move in a circular or spiral path, but not linearly, e.g. F0F1 ATP synthase and hexameric helicases.
The Inchworm mechanism requires at a minimum two contact points between a motor and its track and that these two contact points alternate tight and loose interactions in coordination with the ATP, ADP and nucleotide free states of the motor.
Mechano-chemical coupling requires a motor to be in full contact with its track and load if applicable.
Motors powered by ATPases likely deliver each power stroke in two parts, coordinated with ATP binding and ADP/Pi release in the case of helicases. In the cases of kinesin and myosin where ADP release is relatively slow, the two parts may be ATP binding and Pi release.
FUTURE ISSUES.
Details of mechano-chemical coupling in many motor systems are not clear. The following is only a short list.
What is the rotational step size of F1 ATPase? Do the α subunits undergo ATP binding and release cycles and contribute to γ subunit rotation?
For the hexameric helicases, the only available helicase-DNA complex structure, papilomavirus E1 (25), resembles the apo-protein structure (89). This observation raises the question of whether ATP merely stabilizes a pre-existing thermally fluctuating state or that key ATP-induced conformational states are yet missing.
Even with the UvrD-like helicases, the structure of helicase-DNA complex after Pi release and with ADP bound is still missing, and the effect of Pi release is unknown.
Finally, how do the two myosin and kinesin motor heads communicate? How do these motor proteins take large steps without falling off and getting lost from their tracks?
Acknowledgments
I apologize to all the investigators who could not be appropriately cited owing to space limitations. I thank Drs. D. J. Leahy and R. Craigie and Mr. M. Gregory for reviewing the manuscript.
List of important abbreviations and acronyms
- ATP
adenosine triphosphate
- AMPPNP
adenylyl-imidodiphosphate
- PDB
protein data bank
- ds
double stranded
- ss
single stranded
References
- 1.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–8. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 2.Ali JA, Lohman TM. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–80. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 3.Amos LA, van den Ent F, Lowe J. Structural/functional homology between the bacterial and eukaryotic cytoskeletons. Curr Opin Cell Biol. 2004;16:24–31. doi: 10.1016/j.ceb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–72. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 5.Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell. 2009;35:105–15. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318:459–63. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- 7.Bird LE, Brannigan JA, Subramanya HS, Wigley DB. Characterisation of Bacillus stearothermophilus PcrA helicase: evidence against an active rolling mechanism. Nucleic Acids Res. 1998;26:2686–93. doi: 10.1093/nar/26.11.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–25. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Boyer PD, Stokes BO, Wolcott RG, Degani C. Coupling of “high-energy” phosphate bonds to energy transductions. Fed Proc. 1975;34:1711–7. [PubMed] [Google Scholar]
- 10.Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci U S A. 2005;102:10076–81. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, et al. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci U S A. 2008;105:20191–6. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustamante C, Keller D, Oster G. The physics of molecular motors. Acc Chem Res. 2001;34:412–20. doi: 10.1021/ar0001719. [DOI] [PubMed] [Google Scholar]
- 13.Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–52. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 14.Byrd AK, Raney KD. Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry. 2005;44:12990–7. doi: 10.1021/bi050703z. [DOI] [PubMed] [Google Scholar]
- 15.Centore RC, Leeson MC, Sandler SJ. UvrD303, a hyperhelicase mutant that antagonizes RecA-dependent SOS expression by a mechanism that depends on its C terminus. J Bacteriol. 2009;191:1429–38. doi: 10.1128/JB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng W, Brendza KM, Gauss GH, Korolev S, Waksman G, Lohman TM. The 2B domain of the Escherichia coli Rep protein is not required for DNA helicase activity. Proc Natl Acad Sci U S A. 2002;99:16006–11. doi: 10.1073/pnas.242479399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: Structures and properties of isolated helicases. Q Rev Biophys. 2002;35:431–78. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- 18.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci U S A. 2004;101:6439–44. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dillingham MS, Soultanas P, Wigley DB. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 1999;27:3310–7. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–12. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 21.Dillingham MS, Wigley DB, Webb MR. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–51. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- 22.Donmez I, Patel SS. Mechanisms of a ring shaped helicase. Nucleic Acids Res. 2006;34:4216–24. doi: 10.1093/nar/gkl508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr, et al. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–8. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenberg E, Hill TL. Muscle contraction and free energy transduction in biological systems. Science. 1985;227:999–1006. doi: 10.1126/science.3156404. [DOI] [PubMed] [Google Scholar]
- 25.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–5. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 26.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119:47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Galletto R, Jezewska MJ, Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: quantitative analysis of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J Mol Biol. 2004;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 29.Geeves MA, Holmes KC. The molecular mechanism of muscle contraction. Adv Protein Chem. 2005;71:161–93. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- 30.Gennerich A, Vale RD. Walking the walk: how kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hackney DD. Kinesin ATPase: rate-limiting ADP release. Proc Natl Acad Sci U S A. 1988;85:6314–8. doi: 10.1073/pnas.85.17.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hackney DD. Kinesin and myosin ATPases: variations on a theme. Philos Trans R Soc Lond B Biol Sci. 1992;336:13–7. doi: 10.1098/rstb.1992.0038. discussion 17–8. [DOI] [PubMed] [Google Scholar]
- 33.Hackney DD. Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc Natl Acad Sci U S A. 1994;91:6865–9. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci. 2007;64:2306–22. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hertzer KM, Ems-McClung SC, Kline-Smith SL, Lipkin TG, Gilbert SP, Walczak CE. Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK. Mol Biol Cell. 2006;17:700–10. doi: 10.1091/mbc.E05-08-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hickson ID, Arthur HM, Bramhill D, Emmerson PT. The E. coli uvrD gene product is DNA helicase II. Mol Gen Genet. 1983;190:265–70. doi: 10.1007/BF00330649. [DOI] [PubMed] [Google Scholar]
- 37.Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 38.Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88:1089–118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 39.Houdusse A, Sweeney HL. Myosin motors: missing structures and hidden springs. Curr Opin Struct Biol. 2001;11:182–94. doi: 10.1016/s0959-440x(00)00188-3. [DOI] [PubMed] [Google Scholar]
- 40.Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–8. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 41.Huxley AF, Niedergerke R. Structural changes in muscle during contraction; interference microscopy of living muscle fibres. Nature. 1954;173:971–3. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- 42.Huxley AF, Simmons RM. Proposed mechanism of force generation in striated muscle. Nature. 1971;233:533–8. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- 43.Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–65. [PubMed] [Google Scholar]
- 44.Inaba K. Molecular architecture of the sperm flagella: molecules for motility and signaling. Zoolog Sci. 2003;20:1043–56. doi: 10.2108/zsj.20.1043. [DOI] [PubMed] [Google Scholar]
- 45.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson KA. Advances in transient-state kinetics. Curr Opin Biotechnol. 1998;9:87–9. doi: 10.1016/s0958-1669(98)80089-x. [DOI] [PubMed] [Google Scholar]
- 47.Johnson KA, Gilbert SP. Pathway of the microtubule-kinesin ATPase. Biophys J. 1995;68:173S–76S. discussion 76S–79S. [PMC free article] [PubMed] [Google Scholar]
- 48.Junge W, Sielaff H, Engelbrecht S. Torque generation and elastic power transmission in the rotary F(O)F(1)-ATPase. Nature. 2009;459:364–70. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- 49.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, et al. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 50.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–47. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 51.Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–5. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larsen SH, Adler J, Gargus JJ, Hogg RW. Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc Natl Acad Sci U S A. 1974;71:1239–43. doi: 10.1073/pnas.71.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–60. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–20. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 56.Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. Embo J. 2008;27:3209–19. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lymn RW, Taylor EW. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 1971;10:4617–24. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- 58.Ma YZ, Taylor EW. Kinetic mechanism of kinesin motor domain. Biochemistry. 1995;34:13233–41. doi: 10.1021/bi00040a039. [DOI] [PubMed] [Google Scholar]
- 59.Ma YZ, Taylor EW. Interacting head mechanism of microtubule-kinesin ATPase. J Biol Chem. 1997;272:724–30. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- 60.Manson MD, Tedesco P, Berg HC, Harold FM, Van der Drift C. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci U S A. 1977;74:3060–4. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marx A, Muller J, Mandelkow EM, Hoenger A, Mandelkow E. Interaction of kinesin motors, microtubules, and MAPs. J Muscle Res Cell Motil. 2006;27:125–37. doi: 10.1007/s10974-005-9051-4. [DOI] [PubMed] [Google Scholar]
- 62.Matson SW, George JW. DNA helicase II of Escherichia coli. Characterization of the single-stranded DNA-dependent NTPase and helicase activities. J Biol Chem. 1987;262:2066–76. [PubMed] [Google Scholar]
- 63.Matsura S, Shioi J, Imae Y. Motility in Bacillus subtilis driven by an artificial protonmotive force. FEBS Lett. 1977;82:187–90. doi: 10.1016/0014-5793(77)80581-4. [DOI] [PubMed] [Google Scholar]
- 64.Mechanic LE, Latta ME, Matson SW. A region near the C-terminal end of Escherichia coli DNA helicase II is required for single-stranded DNA binding. J Bacteriol. 1999;181:2519–26. doi: 10.1128/jb.181.8.2519-2526.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Milligan RA. Protein-protein interactions in the rigor actomyosin complex. Proc Natl Acad Sci U S A. 1996;93:21–6. doi: 10.1073/pnas.93.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moller JV, Nissen P, Sorensen TL, le Maire M. Transport mechanism of the sarcoplasmic reticulum Ca2+-ATPase pump. Curr Opin Struct Biol. 2005;15:387–93. doi: 10.1016/j.sbi.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 67.Moore KJ, Lohman TM. Kinetic mechanism of adenine nucleotide binding to and hydrolysis by the Escherichia coli Rep monomer. 1. Use of fluorescent nucleotide analogues. Biochemistry. 1994;33:14550–64. doi: 10.1021/bi00252a023. [DOI] [PubMed] [Google Scholar]
- 68.Morth JP, Pedersen BP, Toustrup-Jensen MS, Sorensen TL, Petersen J, et al. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–9. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 69.Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135:623–34. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moyer ML, Gilbert SP, Johnson KA. Pathway of ATP hydrolysis by monomeric and dimeric kinesin. Biochemistry. 1998;37:800–13. doi: 10.1021/bi9711184. [DOI] [PubMed] [Google Scholar]
- 71.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–6. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–5. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 73.Nath S. Molecular mechanisms of energy transduction in cells: engineering applications and biological implications. Adv Biochem Eng Biotechnol. 2003;85:125–80. doi: 10.1007/3-540-36466-8_5. [DOI] [PubMed] [Google Scholar]
- 74.Newman D. New Theory of Contraction of Striated Muscle, and Demonstration of the Composition of the Broad Dark Bands. J Anat Physiol. 1879;13:549–76. [PMC free article] [PubMed] [Google Scholar]
- 75.Nishizaka T, Oiwa K, Noji H, Kimura S, Muneyuki E, et al. Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat Struct Mol Biol. 2004;11:142–8. doi: 10.1038/nsmb721. [DOI] [PubMed] [Google Scholar]
- 76.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 77.Okada Y, Higuchi H, Hirokawa N. Processivity of the single-headed kinesin KIF1A through biased binding to tubulin. Nature. 2003;424:574–7. doi: 10.1038/nature01804. [DOI] [PubMed] [Google Scholar]
- 78.Olesen C, Picard M, Winther AM, Gyrup C, Morth JP, et al. The structural basis of calcium transport by the calcium pump. Nature. 2007;450:1036–42. doi: 10.1038/nature06418. [DOI] [PubMed] [Google Scholar]
- 79.Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–4. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- 80.Racki LR, Narlikar GJ. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr Opin Genet Dev. 2008;18:137–44. doi: 10.1016/j.gde.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–81. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 82.Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, et al. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–8. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 83.Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat Rev Mol Cell Biol. 2009;10:218–27. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Richet E, Moreau J, Kohiyama M. DNA-dependent ATPases from Escherichia coli K12. Cold Spring Harb Symp Quant Biol 43 Pt. 1979;1:345–8. doi: 10.1101/sqb.1979.043.01.041. [DOI] [PubMed] [Google Scholar]
- 85.Ross JL, Ali MY, Warshaw DM. Cargo transport: molecular motors navigate a complex cytoskeleton. Curr Opin Cell Biol. 2008;20:41–7. doi: 10.1016/j.ceb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sabbert D, Engelbrecht S, Junge W. Intersubunit rotation in active F-ATPase. Nature. 1996;381:623–5. doi: 10.1038/381623a0. [DOI] [PubMed] [Google Scholar]
- 87.Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell. 2009;137:849–59. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 88.Saleh OA, Allemand JF, Croquette V, Bensimon D. Single-molecule manipulation measurements of DNA transport proteins. Chemphyschem. 2005;6:813–8. doi: 10.1002/cphc.200400635. [DOI] [PubMed] [Google Scholar]
- 89.Sanders CM, Kovalevskiy OV, Sizov D, Lebedev AA, Isupov MN, Antson AA. Papillomavirus E1 helicase assembly maintains an asymmetric state in the absence of DNA and nucleotide cofactors. Nucleic Acids Res. 2007;35:6451–7. doi: 10.1093/nar/gkm705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 91.Sherratt DJ, Soballe B, Barre FX, Filipe S, Lau I, et al. Recombination and chromosome segregation. Philos Trans R Soc Lond B Biol Sci. 2004;359:61–9. doi: 10.1098/rstb.2003.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–93. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 93.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 94.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 95.Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 96.Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–46. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 97.Sowa Y, Berry RM. Bacterial flagellar motor. Q Rev Biophys. 2008;41:103–32. doi: 10.1017/S0033583508004691. [DOI] [PubMed] [Google Scholar]
- 98.Subramanya HS, Bird LE, Brannigan JA, Wigley DB. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–83. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 99.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–7. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 100.Tucker PA, Sallai L. The AAA+ superfamily--a myriad of motions. Curr Opin Struct Biol. 2007;17:641–52. doi: 10.1016/j.sbi.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 101.Vale RD. Microscopes for fluorimeters: the era of single molecule measurements. Cell. 2008;135:779–85. doi: 10.1016/j.cell.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 103.Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, et al. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. Embo J. 2005;24:180–9. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 105.von Ballmoos C, Wiedenmann A, Dimroth P. Essentials for ATP synthesis by F1F0 ATP synthases. Annu Rev Biochem. 2009;78:649–72. doi: 10.1146/annurev.biochem.78.081307.104803. [DOI] [PubMed] [Google Scholar]
- 106.von Hippel PH, Delagoutte E. Macromolecular complexes that unwind nucleic acids. Bioessays. 2003;25:1168–77. doi: 10.1002/bies.10369. [DOI] [PubMed] [Google Scholar]
- 107.Yamada K, Ariyoshi M, Morikawa K. Three-dimensional structural views of branch migration and resolution in DNA homologous recombination. Curr Opin Struct Biol. 2004;14:130–7. doi: 10.1016/j.sbi.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 108.Yarranton GT, Gefter ML. Enzyme-catalyzed DNA unwinding: studies on Escherichia coli rep protein. Proc Natl Acad Sci U S A. 1979;76:1658–62. doi: 10.1073/pnas.76.4.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yasuda R, Noji H, Yoshida M, Kinosita K, Jr, Itoh H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 2001;410:898–904. doi: 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 110.Yeo GF. On the Normal Duration and Significance of the “Latent Period of Excitation” in Muscle-contraction. J Physiol. 1888;9:396–453. doi: 10.1113/jphysiol.1888.sp000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–5. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 112.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-overhand. Science. 2004;303:676–8. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]