Abstract
Rationale: DNA methylation is an important epigenetic mechanism, which often occurs in response to environmental stimuli and is crucial in regulating gene expression. It is likely that epigenetic alterations contribute to pathogenesis in idiopathic pulmonary fibrosis (IPF).
Objectives: To determine the DNA methylation changes in IPF and their effects on gene expression.
Methods: Total DNA methylation and DNA methyltransferase expression were compared in IPF and normal control lung tissues. IPF and normal tissues were subjected to comparative analysis of genome-wide DNA methylation and RNA expression using DNA hybridization to the Illumina HumanMethylation27 BeadChip and RNA hybridization to Illumina HumanHT-12 BeadChip. Functional analyses of differentially expressed and differentially methylated genes were done. Selected genes were validated at DNA, RNA, and protein levels.
Measurements and Main Results: DNA methylation status was altered in IPF. IPF samples demonstrated higher DNA methyltransferase expression without observed alterations in global DNA methylation. Genome-wide differences in DNA methylation status and RNA expression were demonstrated by array hybridization. Among the genes whose DNA methylation status and RNA expression were both significantly altered, 16 genes were hypermethylated in DNA associated with decreased mRNA expression or vice versa. We validated CLDN5, ZNF467, TP53INP1, and DDAH1 genes at the level of DNA methylation status, RNA, and protein-level expression.
Conclusions: Changes in DNA methylation correspond to altered mRNA expression of a number of genes, some with known and others with previously uncharacterized roles in IPF, suggesting that DNA methylation is important in the pathogenesis of IPF.
Keywords: idiopathic pulmonary fibrosis, DNA methylation, gene expression, microarray
At a Glance Commentary
Scientific Knowledge on the Subject
Idiopathic pulmonary fibrosis (IPF) is an often fatal disease, and epigenetic mechanisms have been implicated in the pathogenesis of IPF.
What This Study Adds to the Field
A comparative analysis of genome-wide DNA methylation combined with gene expression patterns from IPF and normal lung suggests that DNA methylation status and gene expression profiles are altered in IPF. The observation that gene expression of some of the critical genes in IPF is inversely related to DNA methylation of these genes suggests DNA methylation has a mechanistic role in IPF. An understanding of these epigenetic alterations may provide new insights into IPF pathogenesis.
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive, and usually fatal pulmonary disorder with no effective therapy, the prevalence of which has increased (1, 2). Prognosis in IPF is inversely correlated to the presence of fibroblastic foci, characterized by abnormal myofibroblasts (3). The changed phenotype of these fibrotic cells is due to the altered gene expression in these cells (4). These myofibroblasts are characterized not only by increased expression of α-smooth muscle actin and other muscle proteins, but also by decreased expression or function of regulatory genes, such as THY1 (5). Some of these genes have been characterized as “IPF suppressor genes,” for example, THY1 (6), CAV1 (7), and PTEN (8). Knockout mouse models of these IPF suppressor genes demonstrate more lung fibrosis, and restoring expression of these genes ameliorates the fibrosis (5, 7, 8). DNA hypermethylation may be an important mechanism contributing to down-regulation of these IPF suppressor genes (9). We previously reported that hypermethylation epigenetically decreases THY1 in IPF fibroblasts and in fibroblastic foci (10). On the other hand, hypomethylation of oncogene promoters can result in increased expression (11). Profiling DNA methylation changes may improve our understanding of the pathogenesis of IPF and uncover novel therapeutic targets.
Gene expression profiles are dependent on changes in the epigenome, including DNA methylation, histone modifications, and noncoding RNA regulation. DNA methylation usually occurs at clusters of CpG dinucleotides called CpG islands, generally in promoter regions, often associated with the transcriptional inactivation of the affected gene (12). CpG islands in gene promoters are mostly methylation-free in normal somatic tissues (13). DNA methylation is important in the fine-tuning of chromatin structure and histone modifications to regulate gene expression at various stages of differentiation and development (14). DNA methylation contributes to the pathogenesis of noncancerous diseases, such as cardiovascular disease, neurological and psychiatric disorders, and in pulmonary fibrosis (10, 15, 16). However, studies examining DNA methylation and gene expression on a global scale in IPF are lacking. In this article, we attempt to bridge this knowledge gap by studying genome-wide methylation profiles in IPF and normal lungs to identify differentially methylated genes that may play a role in the pathogenesis of IPF. By overlaying DNA methylation array data with RNA expression array data from lung tissue samples of IPF and normal control tissues, we explored possible relationships between the altered DNA methylation profile and corresponding changes in gene expression. Some of the results of these studies have been previously reported in the form of an abstract (17).
Methods
See the online supplement for details on methods.
Study Population
Lung tissue samples for microarray analysis, validation, and other studies in this project were obtained from the University of Alabama at Birmingham (UAB, Birmingham, AL) Tissue Procurement Facility or from the Lung Tissue Research Consortium (LTRC, Bethesda, MD). These included deidentified lung tissue from 12 patients with IPF (mostly late stage, severe) and 7 normal control subjects (see Table E1 in the online supplement). The diagnosis of IPF was based on American Thoracic Society and European Respiratory Society definitions (18). All cases were clinically and histologically reviewed by expert pulmonologists and pathologists, respectively. The study was approved by the UAB Institutional Review Board.
DNA and RNA Preparation and Related Assays
Genomic DNA and total RNA were extracted from whole lung tissue and prepared for microarrays. A portion of DNA was bisulfite modified and used for methylation-specific PCR (MSP) (10). Real-time reverse transcription-polymerase chain reaction (RT-PCR) or PCR was then performed (19). All primers are listed in Table E2. Quantitative DNA methylation assays were performed with a OneStep qMethyl kit (Zymo Research, Irvine, CA) with some modifications.
DNA Methylation Microarray and RNA Expression Microarray
DNA methylation and gene expression analysis were performed with the HumanMethylation27 BeadChip and iScan system or HumanHT-12 BeadChip and iScan system, respectively (Illumina Inc., San Diego, CA).
Immunohistochemistry
Sections from paraffin-embedded human IPF or normal lung tissues were used for immunohistochemistry with the following antibodies: anti-DNMT3a (Sigma, St. Louis, MO), anti-CLDN5 (cat. no. LS-C118405; Lifespan Biosciences, Seattle, WA), and anti-TP53INP1 (cat. no. LS-C48323; Lifespan Biosciences).
Western Blot
Antibodies used for immunoblotting were anti–α-smooth muscle actin (Biocarta US, San Diego, CA), anti-ZNF467 (cat. no. AP5309a; Abgent, San Diego, CA), anti-DDAH1 (cat. no. AP2898b; Abgent), anti-CLDN5, and anti-TP53INP1. Anti–β-tubulin antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used for loading control.
Data and Statistical Analysis
Statistical analysis for DNA and RNA arrays was done with the Illumina Genome Studio version 2011 analysis package, and pathway analysis was performed with Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA).
Initially liberal criteria were used in order not to miss true positive signals in the screening process, due to the relatively small sample size. A |DiffScore| of ≥13 (equivalent to P < 0.05) for DNA methylation array, a P value less than 0.05, and a fold change greater than 2 for RNA expression array were used. Genes chosen for further validation were those that fit these criteria and had an inverse relationship between DNA methylation and RNA expression. More stringent criteria were used to select differentially methylated and expressed genes for network analysis. Specifically, the false discovery rates (FDR) were calculated by the method of Benjamini and Hochberg (20) with correction for multiple testing. Genes with an FDR ≤ 0.05 were selected. For the four genes validated by additional experiments, P values were obtained by one-way analysis of variance or Kruskal-Wallis analysis of variance on ranks. The more stringent threshold of P < 0.0125 (= 0.05/4) according to the Bonferroni correction for multiple testing was used to claim statistical significance.
Data visualization and clustering were performed with GeneSpring GX version 11.5.1 software (Agilent Technologies, Santa Clara, CA).
Results
Alteration of DNA Methyltransferase Expression in IPF Lung Tissues
Global DNA methylation was similar in IPF and normal lung (Figure E1). Quantitative RT-PCR with whole lung extract demonstrated that patients with IPF had higher DNA methyltransferase (DNMT) 3a and DNMT3b expression, but DNMT1 expression was not different (Figure 1). Patients with IPF showed increased immunohistochemical staining for DNMT3a compared with normal control subjects (Figure 1D and Figure E4). Staining was primarily nuclear and seen in multiple cell types. In IPF, staining was most intense in hyperplastic epithelium overlying fibroblastic foci, but in general was not increased in myofibroblasts within the foci. In some samples, staining was most intense in a layer of cells between the outermost epithelial layer and the myofibroblasts (data not shown). Cells in this location have previously shown to express LAM5γ2 (laminin-5, γ2 chain), keratin-5, and heat shock protein 27 (HSP27) in what have been termed “sandwich foci” (21). In normal lung, DNMT3a staining was occasionally seen in bronchial epithelium (data not shown). These data suggest that de novo DNA methyltransferases are up-regulated in IPF, and the pattern of expression is altered. This could be responsible in part for differences in altered methylation patterns in specific cell types.
Figure 1.
Expression of DNA methyltransferases (DNMTs) in idiopathic pulmonary fibrosis (IPF) and normal lung (Norm) tissues. (A–C) Real-time reverse transcription-polymerase chain reaction (RT-PCR) results for DNMT3a (A; P = 0.015), DNMT3b (B; P = 0.01), and DNMT1 (C; P = 0.113) expression in IPF lung tissues (solid columns) compared with normal lung tissues (open columns); measurements are normalized to 18S. Experiments were performed in triplicate. Results are averages of at least three independent experiments, each experiment including at least three samples from each group. *P < 0.017 versus normal by analysis of variance. Columns and error bars represent means ± SD. (D) Hematoxylin and eosin (H&E) staining (top) and immunohistochemical staining for DNMT3a (bottom) on serial sections from a normal subject and patients with idiopathic pulmonary fibrosis (IPF). Fibroblastic foci are designated by asterisks. Arrows indicate intense staining in abnormal hyperplastic epithelium overlying fibroblastic foci. Staining shown is representative of all samples examined. In normal lung, DNMT3a staining was sometimes seen in bronchial epithelium (data not shown). (Additional immunohistochemistry of DNMT3a can be seen in Figure 4E.)
DNA Methylation Microarray Demonstrated Altered DNA Methylation Pattern in IPF Lung Tissues
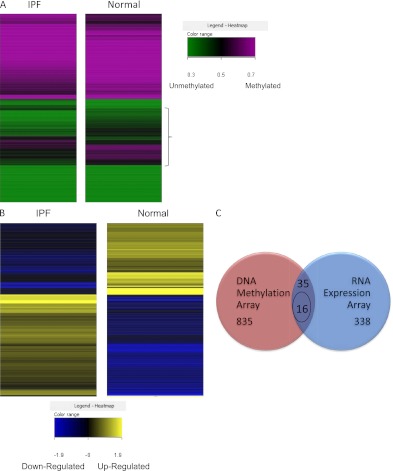
Genome-wide methylation profiles were compared between IPF and normal control groups. Using an equivalent of P value less than 0.05 (|DiffScore| ≥ 13), a total of 870 genes were identified as differentially methylated between IPF and normal groups out of more than 14,000 genes on the chip (Figure 2A). Four hundred and six genes were found to have lower, whereas 464 genes had higher, DNA methylation in IPF lung tissues compared with normal control lung tissues. Some of the identified differentially methylated genes are known to have roles in IPF, but many have not been previously associated with IPF (Table E7). For example, matrix metalloproteinase-7 (MMP7), which is known to have higher expression in patients with IPF (22), demonstrated lower DNA methylation in the array. Some differentially methylated genes have not been reported in IPF, but in other fibrotic diseases (Table E4). More stringent criteria were used for screening the differentially methylated genes for network analysis. Differentially methylated genes were grouped by Ingenuity Pathway Analysis (IPA) into several relevant functional categories, including cellular assembly and organization, cellular compromise, and small-molecule biochemistry, as shown in Table 1 and Table E3.
Figure 2.
(A) Heatmap of DNA methylation array: methylated/unmethylated genes in idiopathic pulmonary fibrosis (IPF) and normal control samples. Each row represents a gene with its average methylation status of IPF samples or normal samples. A gradient color scale ranging between green (unmethylated) and purple (methylated) is included. Brackets indicate genes with the most significant differences in methylation status. (B) Heatmap of gene expression array in IPF and normal samples. Each row represents a gene with its average expression of IPF samples or normal samples. A gradient color scale ranging between blue (down-regulated in RNA expression) and gold (up-regulated in RNA expression) is shown. (C) Venn diagram of overlapped genes from DNA methylation array and RNA gene expression array data sets. Thirty-five genes in both data sets have a |DiffScore| of at least 13, which is equivalent to a P value less than 0.05, and are more than twofold changed in the RNA array, and 16 of these genes have inverse DNA methylation and RNA expression (i.e., DNA hypermethylation with RNA down-regulation, or DNA hypomethylation with RNA up-regulation). Entity List 1 (red): IPF filtered on expression (0.0–100.0)th percentile in the raw data (translated from IPF methylation), 870 entities (835 + 35). Entity List 2 (blue): t test unpaired [IPF] versus [normal] p ≤ 0.05, fold change ≥ 2.0, 373 entities (338 + 35).
TABLE 1.
NETWORKS ASSOCIATED WITH DIFFERENTIALLY METHYLATED GENES IN IDIOPATHIC PULMONARY FIBROSIS VERSUS NORMAL TISSUE
| Score | Focus Molecules* | Top Functions |
| 46 | 19 | Humoral immune response, protein synthesis, infectious disease |
| 28 | 13 | Energy production, lipid metabolism, small-molecule biochemistry |
| 20 | 10 | Cellular assembly and organization, cellular compromise, nervous system development and function |
| 3 | 1 | Molecular transport, protein synthesis, protein trafficking |
| 3 | 1 | DNA replication, recombination, and repair; gene expression; genetic disorder |
| 3 | 1 | Cellular growth and proliferation, dermatological diseases and conditions, genetic disorder |
| 2 | 1 | Protein trafficking, connective tissue disorders, immunological disease |
| 2 | 1 | DNA replication, recombination, and repair; digestive system development and function; drug metabolism |
For a list of molecules, see Table E3. The genes used in the network analysis were selected on the basis of a false positive discovery rate (FDR) ≤ 0.05 (see the online supplement for detail).
RNA Expression Array Analysis Demonstrates Differential Expression of Many Genes with Differential Methylation
Next, we examined transcriptional profiles using the HumanHT-12 BeadChip and iScan system, also from Illumina, Inc. Of the more than 30,000 genes on the chip, there were 373 genes with a P value less than 0.05 and fold change greater than 2 between IPF and normal control lung (Figure 2B). The top-ranked overexpressed genes in IPF by RNA expression array were associated with connective tissue disorders, such as collagens and genes involved in matrix remodeling and fibrogenesis, for example, SPARC (secreted protein acidic and rich in cysteine) (23) and MMP7 (22), previously reported in IPF. There were also genes involved in inflammatory response and cell signaling, for instance, the members of the interleukin family (Tables E5 and E8). Other top networks identified by IPA (using RNA with ≥2-fold change, P < 0.05) included cell–cell signaling, interaction, and cellular movement (Table 2 and Table E5). The highly ranked differentially expressed genes known to be related with IPF or other fibrotic diseases are listed in Table E6.
TABLE 2.
NETWORKS ASSOCIATED WITH DIFFERENTIALLY EXPRESSED GENES IN IDIOPATHIC PULMONARY FIBROSIS VERSUS NORMAL TISSUE
| Score | Focus Molecules* | Top Functions |
| 42 | 25 | Connective tissue disorders, genetic disorder, dermatological diseases and conditions |
| 36 | 23 | Inflammatory response, gene expression, cell-to-cell signaling and interaction |
| 35 | 22 | Cellular compromise, inflammatory response, cellular function and maintenance |
| 33 | 22 | Cell-to-cell signaling and interaction, cellular movement, immune cell trafficking |
| 31 | 20 | Hematological disease, cancer, reproductive system disease |
| 29 | 19 | Inflammatory response, cellular movement, hematopoiesis |
| 28 | 19 | Inflammatory response, cardiovascular disease, lipid metabolism |
| 23 | 17 | Gene expression, cell death, cell cycle |
| 23 | 16 | Embryonic development, tissue development, nervous system development and function |
| 21 | 17 | Cell death, liver necrosis/cell death, organ morphology |
| 21 | 15 | Cell morphology, carbohydrate metabolism, developmental disorder |
| 20 | 16 | Cardiovascular disease, cell death, gastrointestinal disease |
| 20 | 15 | Cell-to-cell signaling and interaction, cellular assembly and organization, cellular development |
| 19 | 15 | Cell cycle, lipid metabolism, nervous system development and function |
| 19 | 15 | Antigen presentation, cellular movement, hematological system development and function |
For a list of molecules, see Table E5. The genes used in the network analysis were selected on the basis of a false positive discovery rate (FDR). Because the genes with a fold change greater than 2 and a P value not greater than 0.05 had an FDR not exceeding 0.061, therefore all genes with a fold change greater than 2 and a P value not greater than 0.05 were used in the network analysis (see the online supplement for detail).
DNA methylation is important in the regulation of gene expression. There were only 35 genes identified with a fold change of 2 or more, a P value less than 0.05 for IPF versus normal, which also had significant differences in methylation on DNA methylation arrays (Figure 2C) by unpaired t test. Sixteen of these genes showed inverse DNA methylation and gene expression, that is, the genes with DNA hypermethylation demonstrated transcriptional down-regulation, or vice versa (Figure 3). Eight of the 16 genes have been reported to be associated with lung fibrosis; the direction of expression change of all but one (catalase) (24) is consistent with published data.
Figure 3.
Graphic representation of the 16 genes that were different in both data sets by P value. The columns represent fold changes in mRNA expression (idiopathic pulmonary fibrosis [IPF] vs. normal); the solid line represents the Δβ of DNA methylation status for IPF (vs. normal). *Genes previously reported to be involved in IPF: CLDN5 (PubMed ID [PMID] 22003091), HP (PMID 17044913), TP53INP1 (PMID 16166619, array data), DDAH1 (PMID 21677199), COL3A1 (PMID 15133032), MMP7 (PMID 18447576), and CTSK (PMID 15161653); †gene reported to be down-regulated in IPF: CAT (PMID 21190578).
Validation of Selected Genes: ZNF467 and CLDN5 Are Down-regulated, whereas TP53INP1 and DDAH1 Are Up-regulated in IPF
We selected 4 genes (ZNF467, CLDN5, TP53INP1, and DDAH1) from the 16 genes that differed between IPF and control tissues by both DNA and RNA arrays, and which have never or only recently been reported as having a role in IPF.
The DNA methylation status was first analyzed by MSP, in which most samples demonstrated both M and U bands (data not shown), indicating a mixture of methylated and unmethylated DNA copies within these selected genes. The most striking difference was MSP for ZNF467, in which most IPF samples showed an M band, whereas normal control samples have none or only a faint M band (Figure 4B, part b). This indicates that IPF samples have more methylated ZNF467 than normal at the specific CpG sites of the primers. We confirmed the differences in DNA methylation using the OneStep qMethyl kit, which uses real-time PCR to quantify locus-specific DNA methylation (25, 26). CLDN5 and ZNF467 demonstrated more methylated DNA loci, whereas TP53INP1 and DDAH1 had fewer than the normal control sample (Figures 4B, 5B, 6B, and 7B). We then confirmed the RNA expression by real-time RT-PCR or at the protein level by Western blot. Compared with normal samples, IPF samples had lower expression of CLDN5 and ZNF467, but higher expression of TP53INP1 and DDAH1 (Figures 4A, 5A, 6A, and 7A). The protein expression by either Western blots or immunohistochemistry confirmed these findings in our samples (Figures 4C and 4D, 5C and 5D, 6C and 6D, and 7C and 7D; and Figure 8).
Figure 4.
ZNF467 is down-regulated in idiopathic pulmonary fibrosis (IPF) lung tissues. (A) mRNA expression of ZNF467 was determined by real-time reverse transcription-polymerase chain reaction (RT-PCR) in IPF (solid column) and normal (open column) samples, normalized to 18S. P ≤ 0.001. (B) a: Relative DNA methylation level by quantitative PCR of ZNF467 amplicon after digestion with methylation-sensitive restriction enzymes against its undigested control in IPF (solid column) and normal (open column) samples. P = 0.001. b: Methylation-specific PCR (MSP) with IPF or normal control DNA samples, using primers specifically designed for methylated (M) and unmethylated (U) sequence. (C) Representative Western blot for ZNF467 and α-smooth muscle actin (α-SMA) in IPF and normal lung tissues. β-Tubulin served as loading control. (D) Bar graph of ZNF467 (P = 0.005) (black column for IPF, open column for normal) and α-SMA (P = 0.003) (dark gray column for IPF, light gray column for normal) Western blots. Results are averages of at least three independent experiments of at least three samples of each group. *P < 0.0125 compared with normal control group. Columns and error bars represent mean ± SD.
Figure 5.
CLDN5 is down-regulated in idiopathic pulmonary fibrosis (IPF) lung tissues. (A) mRNA expression of CLDN5 was determined by real-time reverse transcription-polymerase chain reaction (RT-PCR) in IPF (solid column) and normal (open column) samples, normalized to 18S. P ≤ 0.001. (B) Relative DNA methylation level by quantitative PCR of CLDN5 amplicon after digestion with methylation-sensitive restriction enzymes against its undigested control in IPF (solid column) and normal (open column) samples. P ≤ 0.001. (C) Representative Western blot for CLDN5 and α-smooth muscle actin (α-SMA) in IPF and normal lung tissues. β-Tubulin served as loading control. (D) Bar graph of CLDN5 (P ≤ 0.001) (black column for IPF, open column for normal) and α-SMA (P ≤ 0.001) (dark gray column for IPF, light gray column for normal) Western blots. Results are averages of at least three independent experiments of at least three samples of each group. *P < 0.0125 compared with normal control group. Columns and error bars represent mean ± SD.
Figure 6.
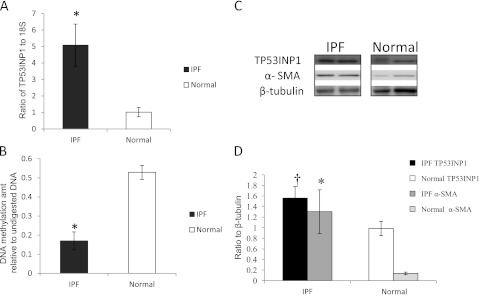
TP53INP1 is up-regulated in idiopathic pulmonary fibrosis (IPF) lung tissues. (A) mRNA expression of TP53INP1 was determined by real-time reverse transcription-polymerase chain reaction (RT-PCR) in IPF (solid column) and normal (open column) samples, normalized to 18S. P = 0.006. (B) Relative DNA methylation level by quantitative PCR of TP53INP1 amplicon after digestion with methylation-sensitive restriction enzymes against its undigested control in IPF (solid column) and normal (open column) samples. P ≤ 0.001. (C) Representative Western blot for TP53INP1 and α-smooth muscle actin (α-SMA) in IPF and normal lung tissues. β-Tubulin served as loading control. (D) Bar graph of TP53INP1 (P = 0.018) (black column for IPF, open column for normal) and α-SMA (P = 0.008) (dark gray column for IPF, light gray column for normal) Western blots. Results are averages of at least three independent experiments of at least three samples of each group. *P < 0.0125, †P > 0.0125 but < 0.05, compared with normal control group. Columns and error bars represent mean ± SD.
Figure 7.
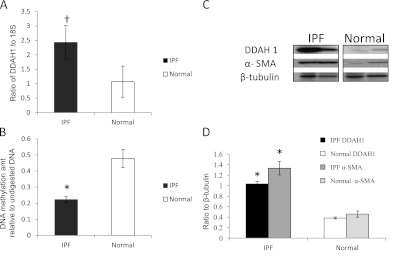
DDAH1 is up-regulated in idiopathic pulmonary fibrosis (IPF) lung tissues. (A) mRNA expression of DDAH1 was determined by real-time reverse transcription-polymerase chain reaction (RT-PCR) in IPF (solid column) and normal (open column) samples, normalized to 18S. P = 0.029. (B) Relative DNA methylation level by quantitative PCR of DDAH1 amplicon after digestion with methylation-sensitive restriction enzymes against its undigested control in IPF (solid column) and normal (open column) samples. P = 0.002. (C) Representative Western blot for DDAH1 and α-smooth muscle actin (α-SMA) in IPF and normal lung tissues. β-Tubulin served as loading control. (D) Bar graph of DDAH1 (P ≤ 0.001) (black column for IPF, open column for normal) and α-SMA (P ≤ 0.001) (dark gray column for IPF, light gray column for normal) Western blots. Results are averages of at least three independent experiments of at least three samples of each group. *P < 0.0125, †P > 0.0125 but < 0.05, compared with normal control group. Columns and error bars represent mean ± SD.
Figure 8.
Immunohistochemistry of TP53INP1 and CLDN5 in idiopathic pulmonary fibrosis (IPF) tissues and normal lung tissues. Shown are hematoxylin and eosin (H&E) staining (left) and immunohistochemical staining for TP53INP1 and CLDN5 (middle and right) on serial sections from a normal subject (top) and two patients with IPF (bottom). A fibroblastic focus is designated by an asterisk. Arrow indicates faint staining of CLDN5 in epithelium overlying fibroblastic foci. Staining shown is representative of all samples examined. Insets indicate region outlined by dashed lines in top right panel in Figure E4.
Figure 8 shows characteristic staining for CLDN5 and TP53INP1 on IPF and normal lung tissues. TP53INP1 shows increased staining in IPF, especially in the cell layer immediately beneath the epithelium covering fibroblastic foci, a pattern similar to staining for DNMT3a (Figure 1D and Figure E4). Normal tissues had little staining for TP53INP1. CLDN5, however, was present in endothelial cells and in some interstitial cells in normal lung tissue, but was decreased in IPF, especially in areas with fibroblastic foci. Occasional faint staining for CLDN5 was seen in epithelium overlying fibroblastic foci.
Interestingly, many of the top-ranked genes from the DNA methylation and RNA arrays could be organized into a single network by IPA (Figure E5). Some of the molecules involved in this network are either known to be associated with pulmonary fibrosis, such as MMP7 (22), HP (27), or have been validated in this study.
Discussion
This is the first study to examine the DNA methylation changes and altered expression of genes on a genome-wide level in IPF and normal lung tissues. The DNA methylation changes at promoter regions of specific genes may change cell phenotypes and contribute to the pathogenesis of IPF (10, 16).
In this study, we compared global DNA methylation levels, and the changes in related DNA methylation enzymes. DNA is methylated by DNA methyltransferases, including DNMT1, DNMT3a, and DNMT3b (28). DNMT3a and DNMT3b are de novo methyltransferases, whereas DNMT1 maintains established methylation patterns (28). We did not observe differences in global DNA methylation between IPF and normal lung (Figure E1), but found higher DNMT3a and DNMT3b expression levels in IPF. Immunohistochemistry for DNMT3a demonstrated increased staining in IPF, most prominent in hyperplastic epithelium overlying fibroblastic foci and in some cases in cells previously identified as arising from bronchiolar basal cells (21), suggesting that there are active methylation changes within specific anatomic regions, where cells are undergoing phenotypic alteration. This is consistent with our previous finding of THY1 silencing associated with DNA hypermethylation in myofibroblasts within fibroblastic foci (10).
Even though the relationships among DNMT expression, global methylation, and methylation of specific genes are not always straightforward (29), we examined the DNA methylation status in IPF, as there should be additional genes with altered expression caused by changes in DNA methylation. Although we found multiple genes for which DNA methylation is inversely correlated with RNA expression, the overall correlation of methylation and expression failed to achieve statistical significance because of the small sample size, heterogeneous nature of IPF, and the incomplete overlaps of the genes in these two arrays (see online supplement data). There were other genes for which DNA methylation status differed significantly between IPF and normal samples, but did not correlate with gene expression. This is not surprising, as DNA methylation is only one of the many epigenetic mechanisms that regulate gene expression (30). There are also examples of genes for which hypermethylation at certain CpG islands is associated with increased expression (31).
We chose to validate several genes not previously associated with IPF at the time the studies were performed. ZNF467 is a zinc finger protein, a transcription factor that enhances nuclear retention and trans-activation of STAT3 (signal transducer and activator of transcription-3) (32). STAT3 trans-activation has been demonstrated to be altered in IPF (33). Studies have demonstrated that ZNF467 may also trans-activate a peroxisome proliferator-activated receptor (PPAR) response element and recruit a histone deacetylase complex (34). The hypermethylation of ZNF467 can result in lowered ZNF467 expression in IPF, which could potentially decrease the activation of PPARγ (34). PPARγ ligands are potential antifibrotic agents (35, 36). Histone deacetylases are important in regulation of the myofibroblast phenotype (37). Thus, there is significant biological plausibility for a role for ZNF467 in the pathogenesis of IPF.
The other down-regulated gene in this study is CLDN5, a transmembrane protein belonging to the claudin family that is strongly expressed in normal lung endothelium and is a major contributor to tight junction formation in these cells (38). A report, published while this manuscript was in preparation, demonstrated that claudin-5 is decreased in bleomycin-induced pulmonary fibrosis (39), which indicated that decreased CLDN5 expression may be involved in epithelial–mesenchymal transition (EMT). EMT often involves promoter methylation to turn off epithelial genes (40, 41). The hypermethylation of claudin-5 in IPF lung tissues could be an indicator of EMT occurring in specific cell populations. We found only faint staining of CLDN5 in epithelium overlying fibroblastic foci and in small vessels adjacent to fibroblastic foci, in contrast to more diffuse expression in normal lung (Figure 8).
TP53INP1 and DDAH1 were the genes we confirmed as increased in IPF at the RNA or protein level. TP53INP1 is a p53-inducible cell stress response protein, a major mediator of p53 antioxidant function (42). In certain cancers, TP53INP1 is overexpressed and correlates with poor prognosis (43, 44). In a study in prostate cancer, antisense oligonucleotides down-regulated TP53INP1, which can inhibit cell proliferation and induce apoptosis (45). TP53INP1 was also reported to be increased in a previous IPF microarray analysis (46). DNA hypomethylation may contribute to the up-regulation of TP53INP1 expression in IPF. Whether the increased TP53INP1 has similar effects in IPF as in prostate cancer is under further investigation. The other up-regulated gene, DDAH1, belongs to the dimethylarginine dimethylaminohydrolase (DDAH) gene family. Increased DDAH could decrease endogenous inhibitors of nitric oxide synthase (NOS) activity, which could result in increased NOS in IPF (47). As we were completing our validation of DDAH1, a study was published describing slightly increased expression of DDAH1 and higher expression of DDAH2 in IPF (48). The study demonstrated that DDAH activity contributes to the pathogenesis of lung fibrosis through the modulation of endogenous NOS inhibitors. In their animal model, DDAH1-overexpressing mice had greater pulmonary fibrosis compared with wild-type mice. We did not examine DDAH2 expression, but in our IPF samples, DDAH1 was increased significantly at the protein level. Further functional and mechanistic studies of the regulation of TP53INP1, DDAH1, ZNF467, and CLDN5 in IPF, as well as how gain or loss of DNA methylation affects RNA/protein expression, are ongoing in our laboratories.
Our studies have some important limitations, including individual variations, and the temporally heterogeneous nature of IPF. Different stages of the disease can have different phenotypes. Even in the same patient, different cells could be at different stages. Specific gene DNA methylation status has been related to stages of cancer progression and outcomes (49, 50). For example, AIM1 gene promoter methylation in specific regions is higher in more advanced melanoma disease stages (51). Previously we demonstrated hypermethylation of the THY1 promoter in fibroblasts within fibroblastic foci, which represent “active” disease, but not in adjacent epithelium, and not in areas of dense, “established” fibrosis. This finding suggests that methylation is controlled temporally and spatially in the lung in IPF. Description of changes at different stages of IPF and within specific cell types must await novel techniques that are still in development. Another limitation of our study was the small sample size, which required us to use less stringent criteria during the screening process, in order not to miss true positive data. In addition, although we have shown an inverse correlation between DNA methylation and gene expression for certain genes, this does not imply that this is the mechanism by which these genes are regulated, or that the gene expression changes occur subsequent to the methylation changes. To prove that the DNA methylation changes drive the changes in gene expression would require mechanistic studies in animal models or cell culture targeting these genes, and such inhibition/overexpression experiments are not possible in human studies. We are currently designing studies evaluating how gain or loss of DNA methylation affects gene expression of the identified genes in animal models. Finally, the lung tissue samples from the normal control subjects are from a slightly younger cohort, as it is a challenge to obtain an exactly age-matched normal cohort. It is possible that some of the changes we observed may in part be due to aging. Nevertheless, as IPF is a disorder seen in more advanced ages, it is possible that age-related changes in methylation may be contributory to IPF.
Different studies use different array platforms resulting in differing RNA array data (46, 52). We used whole lung tissue, which contains signals from multiple cell types and regions with differing histopathology. We confirmed the presence of fibroblastic foci within tissue immediately adjacent to tissue used for array analysis, but this is still an important limitation. However, this approach confirms that many molecules known to be altered in IPF could be epigenetically regulated, and identifies novel molecules and pathways that can be confirmed at the individual gene and cellular level, as we have done previously for THY1 (10). Also, this study makes a stronger case for exploring alteration of DNA methylation as a novel therapeutic approach in IPF.
Inconsistency between single-gene DNA methylation status and array data has also been encountered by investigators in lung cancer research (53). We previously reported that THY1 is hypermethylated in IPF fibrotic foci and hypomethylated with increased gene expression in the overlying epithelium, but the whole lung array data indicated THY1 has lower methylation in IPF samples (P > 0.05), and increased expression in RNA array (P < 0.05; a greater than twofold change). Thus, it is critically important that findings from array approaches be validated at the tissue level.
Epigenetic regulation in fibrotic diseases is a relatively new area. Although this is the first study to apply a whole-genome approach to DNA methylation in IPF, a great deal remains to be explored. There are other important DNA methylation–related proteins, such as methyl-CpG binding domain proteins, which have been reported to act together with histone-modifying enzymes to regulate gene expression (30, 54). However, microarray approaches can provide valuable information about methylation patterns in IPF. Using a validation process, novel genes of clinical interest can be identified. By combining gene expression and DNA methylation status, we can generate more reliable and accurate biomarkers for IPF. Also, it could be helpful by measurement of epigenetic alterations to select specific patients who may benefit from epigenetic therapy. Overall, understanding the DNA methylation profile in idiopathic pulmonary fibrosis will not only yield new insights into the pathogenesis of this devastating disease but will also help improve diagnosis and treatment of patients with IPF.
Supplementary Material
Footnotes
Supported by National Institutes of Health grant R03HL097006 (Y.Y.S.), R01HL092906 (N.A.), R01HL067967 (V.J.T.), R01HL082818 (J.S.H.), and AHA09SDG2260095 (Y.Y.S.).
Author Contributions: J.S.H. and Y.Y.S. designed the experiments; B.H., X.Z., and H.L. performed the experiments; D.C., M.B., and K.Z. prepared and analyzed the array data; Y.Y.S., N.A., V.J.T., and J.S.H. analyzed and interpreted the data; Y.Y.S. and J.S.H. drafted the manuscript; and Y.Y.S., N.A., and J.S.H. revised the manuscript.
This article has an online supplement, which is available from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201201-0077OC on June 14, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Hardie WD, Hagood JS, Dave V, Perl AK, Whitsett JA, Korfhagen TR, Glasser S. Signaling pathways in the epithelial origins of pulmonary fibrosis. Cell Cycle 2010;9:2769–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olson AL, Swigris JJ, Lezotte DC, Norris JM, Wilson CG, Brown KK. Mortality from pulmonary fibrosis increased in the United States from 1992 to 2003. Am J Respir Crit Care Med 2007;176:277–284 [DOI] [PubMed] [Google Scholar]
- 3.Pardo A, Selman M. Idiopathic pulmonary fibrosis: new insights in its pathogenesis. Int J Biochem Cell Biol 2002;34:1534–1538 [DOI] [PubMed] [Google Scholar]
- 4.Phan SH. Fibroblast phenotypes in pulmonary fibrosis. Am J Respir Cell Mol Biol 2003;29:S87–S92 [PubMed] [Google Scholar]
- 5.Hagood JS, Prabhakaran P, Kumbla P, Salazar L, MacEwen MW, Barker TH, Ortiz LA, Schoeb T, Siegal GP, Alexander CB, et al. Loss of fibroblast Thy-1 expression correlates with lung fibrogenesis. Am J Pathol 2005;167:365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders YY, Kumbla P, Hagood JS. Enhanced myofibroblastic differentiation and survival in Thy-1(–) lung fibroblasts. Am J Respir Cell Mol Biol 2007;36:226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang XM, Zhang Y, Kim HP, Zhou Z, Feghali-Bostwick CA, Liu F, Ifedigbo E, Xu X, Oury TD, Kaminski N, et al. Caveolin-1: a critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. J Exp Med 2006;203:2895–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White ES, Atrasz RG, Hu B, Phan SH, Stambolic V, Mak TW, Hogaboam CM, Flaherty KR, Martinez FJ, Kontos CD, et al. Negative regulation of myofibroblast differentiation by PTEN (phosphatase and tensin homolog deleted on chromosome 10). Am J Respir Crit Care Med 2006;173:112–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lung HL, Bangarusamy DK, Xie D, Cheung AK, Cheng Y, Kumaran MK, Miller L, Liu ET, Guan XY, Sham JS, et al. THY1 is a candidate tumour suppressor gene with decreased expression in metastatic nasopharyngeal carcinoma. Oncogene 2005;24:6525–6532 [DOI] [PubMed] [Google Scholar]
- 10.Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, Hagood JS. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol 2008;39:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007;128:683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003;349:2042–2054 [DOI] [PubMed] [Google Scholar]
- 13.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA 1993;90:11995–11999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skinner MK. Role of epigenetics in developmental biology and transgenerational inheritance. Birth Defects Res C Embryo Today 2011;93:51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin-Subero JI, Esteller M. Profiling epigenetic alterations in disease. Adv Exp Med Biol 2011;711:162–177 [DOI] [PubMed] [Google Scholar]
- 16.Huang SK, Fisher AS, Scruggs AM, White ES, Hogaboam CM, Richardson BC, Peters-Golden M. Hypermethylation of PTGER2 confers prostaglandin E2 resistance in fibrotic fibroblasts from humans and mice. Am J Pathol 2010;177:2245–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanders YY, Halloran B, Hagood JS. Epigenetic changes suggest genomic instability in IPF patient samples compared to controls. Abstract presented in part at the American Thoracic Society International Conference, May 13–18, 2011, Denver, CO.
- 18. American Thoracic Society, European Respiratory Society. Idiopathic pulmonary fibrosis: diagnosis and treatment [international consensus statement]. Am J Respir Crit Care Med2000;161:646–664. [DOI] [PubMed]
- 19.Sanders YY, Tollefsbol TO, Varisco BM, Hagood JS. Epigenetic regulation of Thy-1 by histone deacetylase inhibitor in rat lung fibroblasts. Am J Respir Cell Mol Biol 2011;45:16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, B 1995;57:289–300 [Google Scholar]
- 21.Chilosi M, Zamo A, Doglioni C, Reghellin D, Lestani M, Montagna L, Pedron S, Ennas MG, Cancellieri A, Murer B, et al. Migratory marker expression in fibroblast foci of idiopathic pulmonary fibrosis. Respir Res 2006;7:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosas IO, Richards TJ, Konishi K, Zhang Y, Gibson K, Lokshin AE, Lindell KO, Cisneros J, Macdonald SD, Pardo A, et al. MMP1 and MMP7 as potential peripheral blood biomarkers in idiopathic pulmonary fibrosis. PLoS Med 2008;5:e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang W, Wei K, Jacobs SS, Upadhyay D, Weill D, Rosen GD. SPARC suppresses apoptosis of idiopathic pulmonary fibrosis fibroblasts through constitutive activation of β-catenin. J Biol Chem 2010;285:8196–8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odajima N, Betsuyaku T, Nagai K, Moriyama C, Wang DH, Takigawa T, Ogino K, Nishimura M. The role of catalase in pulmonary fibrosis. Respir Res 2010;11:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holemon H, Korshunova Y, Ordway JM, Bedell JA, Citek RW, Lakey N, Leon J, Finney M, McPherson JD, Jeddeloh JA. Methylscreen: DNA methylation density monitoring using quantitative PCR. Biotechniques 2007;43:683–693 [DOI] [PubMed] [Google Scholar]
- 26.Badal V, Menendez S, Coomber D, Lane DP. Regulation of the p14ARF promoter by DNA methylation. Cell Cycle 2008;7:112–119 [DOI] [PubMed] [Google Scholar]
- 27.Fietta A, Bardoni A, Salvini R, Passadore I, Morosini M, Cavagna L, Codullo V, Pozzi E, Meloni F, Montecucco C. Analysis of bronchoalveolar lavage fluid proteome from systemic sclerosis patients with or without functional, clinical and radiological signs of lung fibrosis. Arthritis Res Ther 2006;8:R160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 2005;74:481–514 [DOI] [PubMed] [Google Scholar]
- 29.Ehrlich M, Woods CB, Yu MC, Dubeau L, Yang F, Campan M, Weisenberger DJ, Long T, Youn B, Fiala ES, et al. Quantitative analysis of associations between DNA hypermethylation, hypomethylation, and DNMT RNA levels in ovarian tumors. Oncogene 2006;25:2636–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev 2005;15:490–495 [DOI] [PubMed] [Google Scholar]
- 31.Xue Q, Zhou YF, Zhu SN, Bulun SE. Hypermethylation of the CpG island spanning from exon II to intron III is associated with steroidogenic factor 1 expression in stromal cells of endometriosis. Reprod Sci 2011;18:1080–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakayama K, Kim KW, Miyajima A. A novel nuclear zinc finger protein EZI enhances nuclear retention and transactivation of STAT3. EMBO J 2002;21:6174–6184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pechkovsky DV, Prêle CM, Wong J, Hogaboam CM, McAnulty RJ, Laurent GJ, Zhang SS, Selman M, Mutsaers SE, Knight DA. STAT3-mediated signaling dysregulates lung fibroblast–myofibroblast activation and differentiation in UIP/IPF. Am J Pathol 2012;180:1398–1412 [DOI] [PubMed] [Google Scholar]
- 34.Quach JM, Walker EC, Allan E, Solano M, Yokoyama A, Kato S, Sims NA, Gillespie MT, Martin TJ. Zinc finger protein 467 is a novel regulator of osteoblast and adipocyte commitment. J Biol Chem 2011;286:4186–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakatos HF, Thatcher TH, Kottmann RM, Garcia TM, Phipps RP, Sime PJ. doi: 10.1155/2007/71323. The role of PPARs in lung fibrosis. PPAR Res 2007;2007:71323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milam JE, Keshamouni VG, Phan SH, Hu B, Gangireddy SR, Hogaboam CM, Standiford TJ, Thannickal VJ, Reddy RC. PPAR-γ agonists inhibit profibrotic phenotypes in human lung fibroblasts and bleomycin-induced pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2008;294:L891–L901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo W, Shan B, Klingsberg RC, Qin X, Lasky JA. Abrogation of TGF-β1–induced fibroblast–myofibroblast differentiation by histone deacetylase inhibition. Am J Physiol Lung Cell Mol Physiol 2009;297:L864–L870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaarteenaho-Wiik R, Soini Y. Claudin-1, -2, -3, -4, -5, and -7 in usual interstitial pneumonia and sarcoidosis. J Histochem Cytochem 2009;57:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohta H, Chiba S, Ebina M, Furuse M, Nukiwa T. Altered expression of tight junction molecules in alveolar septa in lung injury and fibrosis. Am J Physiol Lung Cell Mol Physiol 2011;14:14. [DOI] [PubMed] [Google Scholar]
- 40.Williams CS, Zhang B, Smith JJ, Jayagopal A, Barrett CW, Pino C, Russ P, Presley SH, Peng D, Rosenblatt DO, et al. BVES regulates EMT in human corneal and colon cancer cells and is silenced via promoter methylation in human colorectal carcinoma. J Clin Invest 2011;121:4056–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ke XS, Qu Y, Cheng Y, Li WC, Rotter V, Oyan AM, Kalland KH. Global profiling of histone and DNA methylation reveals epigenetic-based regulation of gene expression during epithelial to mesenchymal transition in prostate cells. BMC Genomics 2010;11:669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cano CE, Gommeaux J, Pietri S, Culcasi M, Garcia S, Seux M, Barelier S, Vasseur S, Spoto RP, Pebusque MJ, et al. Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer Res 2009;69:219–226 [DOI] [PubMed] [Google Scholar]
- 43.Giusiano S, Garcia S, Andrieu C, Dusetti NJ, Bastide C, Gleave M, Taranger-Charpin C, Iovanna JL, Rocchi P. TP53INP1 overexpression in prostate cancer correlates with poor prognostic factors and is predictive of biological cancer relapse. Prostate 2012;72:117–128 [DOI] [PubMed] [Google Scholar]
- 44.Taieb D, Giusiano S, Sebag F, Marcy M, de Micco C, Palazzo FF, Dusetti NJ, Iovanna JL, Henry JF, Garcia S, et al. Tumor protein p53-induced nuclear protein (TP53INP1) expression in medullary thyroid carcinoma: a molecular guide to the optimal extent of surgery? World J Surg 2010;34:830–835 [DOI] [PubMed] [Google Scholar]
- 45.Giusiano S, Baylot V, Andrieu C, Fazli L, Gleave M, Iovanna JL, Taranger-Charpin C, Garcia S, Rocchi P. TP53INP1 as new therapeutic target in castration-resistant prostate cancer. Prostate 2011;27:22477. [DOI] [PubMed] [Google Scholar]
- 46.Selman M, Pardo A, Barrera L, Estrada A, Watson SR, Wilson K, Aziz N, Kaminski N, Zlotnik A. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am J Respir Crit Care Med 2006;173:188–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saleh D, Barnes PJ, Giaid A. Increased production of the potent oxidant peroxynitrite in the lungs of patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 1997;155:1763–1769 [DOI] [PubMed] [Google Scholar]
- 48.Pullamsetti SS, Savai R, Dumitrascu R, Dahal BK, Wilhelm J, Konigshoff M, Zakrzewicz D, Ghofrani HA, Weissmann N, Eickelberg O, et al. The role of dimethylarginine dimethylaminohydrolase in idiopathic pulmonary fibrosis. Sci Transl Med 2011;3:87ra53. [DOI] [PubMed] [Google Scholar]
- 49.Ramos EA, Grochoski M, Braun-Prado K, Seniski GG, Cavalli IJ, Ribeiro EM, Camargo AA, Costa FF, Klassen G. Epigenetic changes of CXCR4 and its ligand CXCL12 as prognostic factors for sporadic breast cancer. PLoS One 2011;6:e29461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka M, Chang P, Li Y, Li D, Overman M, Maru DM, Sethi S, Phillips J, Bland GL, Abbruzzese JL, et al. Association of CHFR promoter methylation with disease recurrence in locally advanced colon cancer. Clin Cancer Res 2011;17:4531–4540 [DOI] [PubMed] [Google Scholar]
- 51.Hoshimoto S, Kuo CT, Chong KK, Takeshima TL, Takei Y, Li MW, Huang SK, Sim MS, Morton DL, Hoon DS. AIM1 and LINE-1 epigenetic aberrations in tumor and serum relate to melanoma progression and disease outcome. J Invest Dermatol 2012;132:1689–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konishi K, Gibson KF, Lindell KO, Richards TJ, Zhang Y, Dhir R, Bisceglia M, Gilbert S, Yousem SA, Song JW, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2009;180:167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heller G, Zielinski CC, Zochbauer-Muller S. Lung cancer: from single-gene methylation to methylome profiling. Cancer Metastasis Rev 2010;29:95–107 [DOI] [PubMed] [Google Scholar]
- 54.Ikegami K, Ohgane J, Tanaka S, Yagi S, Shiota K. Interplay between DNA methylation, histone modification and chromatin remodeling in stem cells and during development. Int J Dev Biol 2009;53:203–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.