Abstract
Rationale: Long-term survival after lung transplantation is limited by infectious complications and by bronchiolitis obliterans syndrome (BOS), a form of chronic rejection linked in part to microbial triggers.
Objectives: To define microbial populations in the respiratory tract of transplant patients comprehensively using unbiased high-density sequencing.
Methods: Lung was sampled by bronchoalveolar lavage (BAL) and upper respiratory tract by oropharyngeal wash (OW). Bacterial 16S rDNA and fungal internal transcribed spacer sequencing was used to profile organisms present. Outlier analysis plots defining taxa enriched in lung relative to OW were used to identify bacteria enriched in lung against a background of oropharyngeal carryover.
Measurements and Main Results: Lung transplant recipients had higher bacterial burden in BAL than control subjects, frequent appearance of dominant organisms, greater distance between communities in BAL and OW indicating more distinct populations, and decreased respiratory tract microbial richness and diversity. Fungal populations were typically dominated by Candida in both sites or by Aspergillus in BAL but not OW. 16S outlier analysis identified lung-enriched taxa indicating bacteria replicating in the lower respiratory tract. In some cases this confirmed respiratory cultures but in others revealed enrichment by anaerobic organisms or mixed outgrowth of upper respiratory flora and provided quantitative data on relative abundances of bacteria found by culture.
Conclusions: Respiratory tract microbial communities in lung transplant recipients differ in structure and composition from healthy subjects. Outlier analysis can identify specific bacteria replicating in lung. These findings provide novel approaches to address the relationship between microbial communities and transplant outcome and aid in assessing lung infections.
Keywords: microbiome, 16S, ITS, bronchiolitis obliterans syndrome
At a Glance Commentary
Scientific Knowledge on the Subject
Two of the major factors limiting long-term survival after lung transplantation are pulmonary infections and bronchiolitis obliterans syndrome, which is also linked to microbial triggers in the lung. Molecular analysis of microbial marker genes now enables comprehensive and quantitative identification of entire bacterial and fungal populations in a body site. Despite the importance of microbial factors in transplant outcome, these approaches have not been applied to define the respiratory microbiome in lung transplant recipients.
What This Study Adds to the Field
Bacterial and fungal communities in airways of lung transplant recipients differ from those of healthy people in structure, composition and dominant organisms, and higher overall bacterial burden and show greater differences between lower and upper respiratory tract organisms, indicating microbial outgrowth in transplant lungs. The specific bacteria replicating in transplant lungs were identified, independently of culture, by defining lung-enriched organisms in bronchoalveolar lavage against a background of upper respiratory tract flora. These approaches can be used to better understand how lung microbiota influence long-term transplant outcome.
The outcome for lung transplantation is much worse than other solid organ transplants, with only 53 and 30% survival at 5 and 10 years, respectively (1). Infection and primary graft dysfunction are the principal causes of death early after transplantation, whereas the major causes of later death are infection and bronchiolitis obliterans syndrome (BOS) (1). Lung shares with all organ transplants an increased susceptibility to infection resulting from immunosuppressive therapy, but additional factors make it uniquely susceptible. These include direct communication with the external environment and microbe-rich upper respiratory tract (URT), defective lower respiratory tract (LRT) mechanical defense due to vagal denervation cough reflex defects and abnormal mucocilliary clearance at anastamotic sites, and a high propensity for microaspiration (2, 3). Microbial triggers are also linked to BOS, the clinical manifestation of chronic rejection, based on epidemiological association with specific bacteria, fungi, or viruses detected in lung (4–8), linkage with genetic polymorphisms in innate microbial sensors (9–12), and responsiveness to azithromycin (13–15).
Traditional respiratory culture methods focus on individual organisms rather than whole populations, are only semiquantitative, and typically exclude organisms normally present in the upper respiratory tract due to the confounding issue of URT admixture in lung-derived specimens. Culture-independent molecular approaches can identify entire microbial populations present and can be highly quantitative. These methods are starting to be applied to the respiratory tract (16–19). Despite the unique susceptibility of lung transplants to microbial invasion and its importance in outcome, the lung transplant microbiome has not been studied by deep sequencing.
We used bacterial 16S rRNA gene and fungal internal transcribed spacer (ITS) sequencing to define the microbial populations in lung allografts from transplant recipients sampled by bronchoalveolar lavage (BAL). Because a principal challenge to LRT sampling by bronchoscopy is instrument carryover from the upper tract, lung microbiota in BAL were compared with URT populations sampled by oropharyngeal wash (OW). We report here that the transplant recipients’ lung microbial communities were notably different from those found in healthy control subjects in composition and structure, were less closely related to matched URT samples, and showed frequent lung-enrichment of specific bacteria. These methods help to define allograft colonization, may aid in the diagnosis of lung infections, and can provide insight into microbial contributors to BOS.
Methods
Subjects and Sample Collection
Lung transplant recipients undergoing clinical bronchoscopy provided a 10-ml saline oropharyngeal wash/gargle (OW) (20) followed by transoral bronchoscopy using standard clinical procedures. BAL was performed using 60 to 120 ml of saline with the bronchoscope wedged in the right middle lobe for bilateral or single-right transplants or in the left lingula for single-left transplants. BAL fluid was submitted for bacterial and fungal respiratory culture, and an aliquot was used for microbiome evaluation. To control for microbial DNA that might be present in instruments or reagents, 10 ml of saline was washed through the bronchoscope immediately before each procedure.
Nontransplant control subjects included healthy volunteers who underwent research bronchoscopy using a two-scope procedure previously reported (20). In that protocol, one bronchoscope was used to anesthetize the glottis and provide URT samples, and a second clean scope was used to sample the LRT, to minimize and precisely define potential carryover from the URT by a bronchoscope entering the LRT. Their BAL first return was used to best approximate clinical (single-scope) conditions (20). Another group of healthy volunteers underwent research bronchoscopy using a standard single-scope procedure identical to clinical bronchoscopy. Samples were also collected from two nontransplant subjects undergoing diagnostic bronchoscopy for evaluation of sarcoidosis and a lung nodule found to be adenocarcinoma.
All subjects gave written informed consent under Investigational Review Board–approved protocols.
DNA Extraction and 16S rDNA Quantification
For 16S analysis, genomic DNA was isolated from unfractionated OW and BAL using the PowerSoil DNA isolation kit (MoBio, Carlsbad, CA) as previously described (20). For ITS analysis, samples were additionally incubated at 95°C for 10 minutes before beadbeating. Bacterial 16S copy number was quantified by real-time PCR (21).
Bacterial 16S rDNA and Fungal ITS Gene Amplification and Sequence Analysis
16S rDNA genes were amplified using barcoded V1V2 primers, and amplicons were purified, pooled, and 454-pyrosequenced as described (22). Using the QIIME pipeline (23), reads were denoised, clustered into 97% similar operational taxonomic units (OTUs), aligned to full-length 16S rDNA sequences, assigned taxonomy with RDP Classifier, and used to generate de novo phylogenetic trees with FastTree2 (24). Sequences were removed if unalignable or identified as chimeras (25). ITS genes were amplified using barcoded ITS1F/ITS2 primers (26). Amplicons were purified, pooled, and 454-pyrosequenced, and sequences were denoised, clustered into OTUs at 95.2% sequence identity, and assigned taxonomy with the BROCC software pipeline as described (S. Dollive, unpublished observations). Additional details are provided in the online supplement.
Statistical Analysis
UniFrac (27) was used to measure β diversity as described by Lozupone and colleagues (28) and visualized using principal coordinate analysis of the distance matrix. We tested for differences in UniFrac distance, within versus between groups, by comparing the T statistic against 10,000 random permutations (28). For Procrustes analysis, the goodness of fit (M2 value) was measured by summing over the residuals, and significance was assessed by the Monte-Carlo label permutation method (29, 30). The Wilcoxon rank-sum test was used to compare the number of OTUs in healthy and transplant samples, rarefied to 3,000 sequence reads per sample. OTUs enriched in BAL versus OW were identified by computing the maximum likelihood parameters for a Dirichlet-multinomial distribution of OTU counts and then using the marginal form of this distribution to perform a one-sided test of OTU counts in the BAL sample (31, 32) (E.S. Charlson, unpublished observations).
Results
Lower Respiratory Tract Sampling from Lung Transplant Recipients
Paired BAL and OW were obtained from 21 lung transplant subjects (Tables 1 and 2 and Table E1 in the online supplement). Seven subjects were transplanted for COPD or emphysema due to α-1 antitrypsin deficiency. Seven subjects were transplanted for interstitial diseases including IPF, collagen-vascular associated interstitial lung disease (ILD), or sarcoidosis. One subject was transplanted for congenital heart disease with pulmonary hypertension, and six subjects were transplanted for suppurative lung diseases, mainly cystic fibrosis (CF). Most had bilateral transplants (81%), and two with severe pulmonary hypertension also received heart transplants. All bronchoscopies were routine surveillance procedures performed at regularly scheduled intervals during the first 15 months after transplant, except for Tx33, who had nonresolving clinical pneumonia despite treatment for culture-positive Pseudomonas aeruginosa, and Tx38, who was evaluated for declining FEV1 9.5 years after transplantation (late BOS). Two subjects also had early BOS identified by decreased FEV1 (Tx25, Tx40). All subjects were on immunosuppressives and antimicrobials typical of posttransplant management (Table 1). Nontransplant control subjects included healthy volunteers who underwent research bronchoscopy using procedures identical to clinical bronchoscopy or using a previously described two-bronchoscope procedure (20) and two subjects without infection or immunosuppression undergoing clinical diagnostic bronchoscopy.
TABLE 1.
CLINICAL FEATURES AND MICROBIOLOGICAL FINDINGS ASSOCIATED WITH 23 BAL SAMPLES FROM 21 LUNG TRANSPLANT RECIPIENTS STUDIED*
| Subject | Sample | Pretransplant Disease | Transplant Type | Months after Transplant | Bronch indication/status | Immunosuppression | Antimicrobials | BAL Bacterial Culture | BAL Fungal Culture |
| Tx24 | 24 | IPF | S-L | 2 | Routine | MMF, Pred, Tac | TMP/S, Valcyc, Nystatin | Staphylococcus aureus | None |
| Tx25 | 25 | A1AT | B | 6 | Routine/early BOS | MMF, Pred, Tac | Azithro, TMP/S, Valgan, Nystatin | None | None |
| Tx26 | 26 | CF | B | 9 | Routine | Pred, Tac | TMP/S, Valcyc | Pseudomonas aeruginosa | None |
| Tx30 | 30 | IPF | S-L | 15 | Routine | Aza, Pred, Tac | TMP/S, Valgan | None | None |
| Tx31 | 31 | CHD, PHTN | B-H | 5 | Routine | MMF, Pred, Tac | TMP/S, Valcyc | None | None |
| Tx32 | 32 | CF | B | 2 | Routine | Aza, Pred, Tac | TMP/S, Valgan | Achromobacter, P. aeruginosa | None |
| Tx33 | 33 | CF | B | 9 | Nonresolving PNA | Aza, Pred, Tac | Levofloxacin, Meropenem, Valcyc, Tobra | P. aeruginosa | None |
| Tx34 | 34 | A1AT | B | 4 | Routine | MMF, Pred, Tac | TMP/S, Valgan, Vori | None | None |
| Tx35 | 35 | PCD | B | 12 | Routine | MMF, Pred, Tac | TMP/S | Mycobacterium avium-intracellulare | Candida albicans |
| Tx36 | 36A | IPF | S-L | 2 | Routine | MMF, Pred, Tac | TMP/S, Valgan | P. aeruginosa | Aspergillus flavus, C. albicans |
| 36B | 4 | Routine | MMF, Pred, Tac | TMP/S, Valgan, Vori, Nystatin | Klebsiella oxytoca | None | |||
| Tx38 | 38 | CF | B | 114 | Late BOS | Pred, TAC | Azithro, TMP/S, Ciprofloxacin | None | C. albicans |
| Tx39 | 39A | COPD | B | 2 | Routine | Aza, Pred, Tac | TMP/S, Valgan | None | C. albicans |
| 39B | 4 | Routine | Aza, Pred, Tac | TMP/S, Valgan | None | C. albicans | |||
| Tx40 | 40 | CF | B | 18 | Routine/early BOS | MMF, CSA, Pred, pulse Methylpred | Azithro, TMP/S | P. aeruginosa | None |
| Tx41 | 41 | Sarc, PHTN | B-H | 1 | Routine | MMF, Pred, Tac | TMP/S, Valcyc, Nystatin | S. aureus | C. albicans |
| Tx42 | 42 | COPD | S-L | 3 | Routine | MMF, Pred, Tac | Atovaquone, Valgan | None | C. albicans |
| Tx43 | 43 | COPD | B | 5 | Routine | Aza, Pred, Tac | TMP/S, Valcyc, Vori | None | None |
| Tx44 | 44 | Sarc, PHTN | B | 12 | Routine | MMF, Pred, Tac | TMP/S | Burkholderia cepacia | Aspergillus fumigatus, Paecilomyces lilacinus |
| Tx45 | 45 | ILD | B | 4 | Routine | MMF, Pred, Tac | TMP/S, Nystatin | M. avium-intracellulare | C. albicans |
| Tx46 | 46 | COPD | B | 4 | Routine | Aza, Pred, Tac | Valgan | P. aeruginosa | None |
| Tx47 | 47 | COPD | B | 2 | Routine | Aza, Pred, Tac | TMP/S, Valgan, Nystatin | Enterobacter cloacae | C. albicans |
| Tx49 | 49 | ILD | B | 2 | Routine | MMF, Pred, Tac | TMP/S, Valgan | None | C. albicans |
Definition of abbreviations: A1AT = α-1 antitrypsin deficiency; Aza = azathioprine; Azithro = azithromycin; B = bilateral; BOS = bronchiolitis obliterans syndrome; CF = cystic fibrosis; CHD = congenital heart disease; CSA = cyclosporin A; H = heart transplant; ILD = interstitial lung disease associated with collagen vascular disease; IPF = idiopathic pulmonary fibrosis; MMF, mycophenolate mofetil; Nystatin = nystatin oral swish; PCD = primary cilliary dyskinesia; PHTN = pulmonary hypertension; PNA = pneumonia; Pred = prednisone; Sarc = sarcoid; S-L, single left; Tac = tacrolimus; TMP/S = trimethoprim/sulfamethoxazole; Valcyc = valcyclovir; Valgan = valgancyclovir; Vori = voriconazole;
BAL samples were obtained at the indicated times after transplant for routine postsurveillance bronchoscopy, except for Tx33 and Tx38 as noted.
TABLE 2.
TRANSPLANT SUBGROUPS USED FOR ANALYSIS
| Nonsuppurative (n = 15) | |
| Age, yr (mean ± SD) | 57.5 ± 7.2 |
| Single/bilateral | 4/11 |
| Heart (n) | 2 |
| Suppurative (n = 6) | |
| Age, yr (mean ± SD) | 35.0 ± 11.3 |
| Single/bilateral | 0/6 |
| Heart (n) | 0 |
| ILD/IPF (n = 7) | |
| IPF (n) | 3 |
| Other ILD (n) | 4 |
| Age, yr (mean ± SD) | 60.7 ± 6.2 |
| Single/bilateral | 3/4 |
| Heart (n) | 1 |
| COPD/emphysema (n = 7) | |
| COPD (n) | 5 |
| A1AT deficiency (n) | 2 |
| Age, yr (mean ± SD) | 54.3 ± 7.0 |
| Single/bilateral | 1/6 |
| Heart (n) | 0 |
| Suppurative (n = 6) | |
| Cystic fibrosis (n) | 5 |
| Primary cilliary dyskinesia (n) | 1 |
| Age, yr (mean ± SD) | 35.0 ± 11.3 |
| Single/bilateral | 0/6 |
| Heart (n) | 0 |
Definition of abbreviations: A1AT = α-1 antitrypsin deficiency; COPD = chronic obstructive pulmonary disease; ILD = interstitial lung disease; IPF = idiopathic pulmonary fibrosis.
The top set of data shows group characteristics of subjects transplanted for nonsuppurative and suppurative indications. The lower data set shows underlying disease for the major subgroups of transplant subjects. One subject with congenital heart disease and pulmonary hypertension is included in the nonsuppurative group but not included in further subgroup analysis.
Bacterial Quantification in Lung Transplant Recipient Airways
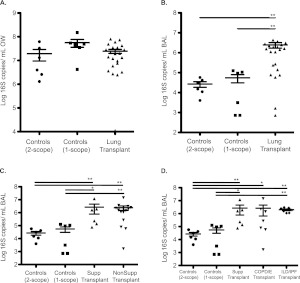
Bronchoscope prewashes and control samples demonstrated levels of 16S DNA close to or below the lower limit of qPCR quantification (LOQ: 725 copies/ml; data not shown) as we previously described (20). OW showed high numbers of 16S copies, and values were similar between subject groups (Figure 1A). In contrast, BAL of lung transplant subjects had a mean copy number that was 44-fold higher than control subjects sampled by single-scope bronchoscopy and 91-fold higher than BAL first return from healthy subjects sampled with a two-scope protocol (Figure 1B). Bacterial 16S DNA levels in BAL did not differ based on underlying indication for transplant whether grouped by suppurative versus nonsuppurative or among the nonsuppurative subgroups (Figures 1C and 1D).
Figure 1.
Quantification of bacterial 16S rDNA gene copies in oropharyngeal wash (OW) and bronchoalveolar lavage (BAL) from control and lung transplant subjects. 16S copy number in oropharyngeal (A) and BAL (B) samples of control and transplant subjects. Copy number was also examined in BAL samples of major transplant subgroups (C, D) as described in Table 2. The y axis indicates the 16S rRNA gene copy number by quantitative PCR. Each sample was analyzed in triplicate. *P < 0.05; **P < 0.01 (Mann-Whitney test).
Characterization of Bacterial Lineages in the Transplanted Lung by 454 Pyrosequencing
We surveyed bacterial lineages using deep sequencing of 16S rRNA genes (Figure 2). Bronchoscope prewash and control specimens, which had low copy numbers, showed bacterial lineages typical of water and skin organisms (see Figure E1 in the online supplement) as previously described (20).
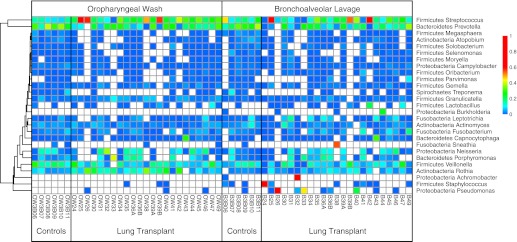
Figure 2.
Relative abundance of bacterial taxa derived from pyrosequencing data. Each column represents an individual sample. Sample type and subject group is indicated at the bottom of each group of columns. Each row corresponds to specific bacterial taxa, with their proportional representation in the given sample indicated by the color code at right. Bacterial operational taxonomic units (OTUs) were collected into genera, so some rows represent multiple OTUs. OTUs not assigned at the genera level and genera with less than 1,000 total reads are omitted. Hierarchical clustering groups the rows to emphasize lineages with similar abundance patterns.
Oropharyngeal samples contained high abundances of taxa characteristic of the oral cavity, including Streptococcus, Prevotella, Veillonella, Porphyromonas, Neisseria, and Rothia (22, 33, 34). Most transplant recipients had similar oropharyngeal flora as healthy subjects, except for four subjects whose OW were dominated by Streptococcus spp.
BAL from healthy subjects shared a similar overall bacterial profile as OW samples, as we previously reported (20). Transplant subjects’ BAL largely shared bacterial profiles with OW samples, but a subset was clearly dominated by specific lineages that were absent in OW or detected at much lower abundances. Two BAL samples were dominated by P. aeruginosa (B26 and B33; both transplanted for CF) and revealed P. aeruginosa on culture. A high abundance of Staphylococcus aureus was seen in one BAL (B24; transplanted for IPF), also detected by culture. Similarly, one BAL showed dominant Achromobacter (B32; transplanted for CF), also identified by culture. In contrast, BAL from the subject with late-onset BOS (B38) was dominated by the unusual anaerobic lineage Sneathia (35), which was not identified by culture. Three BAL samples showed high levels of Streptococcus spp. (B25, B39B, B42; transplanted for COPD or α-1 antitrypsin deficiency) without any corresponding cultured organism, but Streptococcus spp. were also found at high or moderate abundance in OW from these subjects.
Characterization of Fungal Lineages in the Transplanted Lung and Matched OW
Fungi were analyzed by ITS gene sequencing (Figure 3; Figure E2). Although precise qPCR quantification is precluded by high length variation of the ITS1 gene, we used the amount of ITS PCR product as an estimate for the relative abundance of fungi (at top of the heat map).
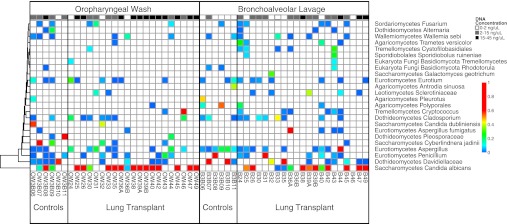
Figure 3.
Relative abundance of fungal taxa derived from pyrosequencing data. Fungal taxa identified in bronchoalveolar lavage and oropharyngeal wash samples are shown as described for bacterial lineages in Fig. 2. Fungal operational taxonomic units (OTUs) were collected into classes, so some rows represent multiple OTUs. OTUs not assigned at the class level and classes with less than 250 total reads are omitted. Along the top is the concentration of ITS DNA post-PCR for each sample, keyed by gray-scale at top right.
OW of healthy subjects revealed varying ITS amplification levels and a distribution similar to that recently reported in oral samples (26), including environmental agents as well as Candida and Asperillgus (Figure 3). One healthy subject had relatively high ITS amplification in OW (3B08), comprised mainly of Candida albicans. OW of transplant subjects revealed some samples with scant fungal sequences and others with robust ITS amplification. Of the transplant subjects with high OW fungal amplification, all but one were dominated by Candida (Figure 3). Some transplant subjects with intermediate or low levels of OW fungal sequences had high relative abundances of Candida, whereas others revealed no dominant lineage or environmental fungi. One transplant OW with an intermediate level of ITS amplification was dominated by Cryptococcus (OW46).
Healthy volunteers’ BAL yielded scant fungal amplification. Although Candida was identified in several healthy subjects’ OW, it was absent in their BAL. The few fungal reads present in healthy BAL were comprised largely of environmental agents such as Davidiellaceae and Cladosporium, also present in prewash control samples (Figure E2), as well as low abundances of Aspergillus.
BAL from lung transplant recipients showed a markedly different pattern. Four yielded high levels of ITS amplification; three of these were dominated by Candida (B38, B47, and B49), and one (B44) was dominated by Aspergillus. In each case, the corresponding fungus was found by culture. Several subjects with intermediate ITS levels revealed high abundance of Candida (B41, B25, B26, B39, B39B, and B42), and one was dominated by Aspergillus (B36, also found by culture). When Candida was seen in BAL, it was also always found at high abundance in OW, whereas for subjects with a high abundance of Aspergillus in BAL, this fungus was absent or at markedly lower abundance in OW. Cryptococcus was present in six transplant patients’ BAL but at relatively low abundance or in samples with low total ITS read numbers. Cryptococcus was not seen in BAL from the transplant subject who had this fungus at high abundance in OW (Tx46).
Community Richness, Completeness of Microbial Identification, and Species Diversity
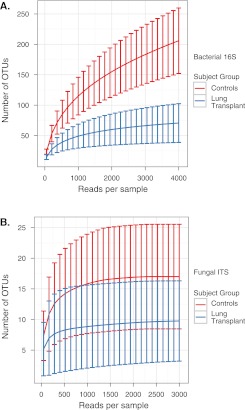
To determine whether transplant and healthy subjects’ microbial communities differed in species richness, we calculated the number of observed OTUs at various sequencing depths for bacteria and fungi in OW and BAL specimens. Rarefaction analysis of bacterial oropharyngeal OTUs (Figure 4A) indicated that richness was lower in transplant than healthy OW (P < 0.05, Wilcoxon rank sum) and that sampling was near saturation in transplant subjects but not in healthy subjects. Richness was lowest in the subjects transplanted for suppurative lung disease and intermediate in those transplanted for nonsuppurative disease (P < 0.05, Wilcoxon rank sum) but was not different between the COPD/emphysema and ILD/IPF groups. Oropharyngeal fungal microbiota from healthy and transplant subjects showed flat rarefaction curves at high sequencing depth, indicating that further sequencing would yield few additional fungal OTUs (Figure 4B), although transplant patients had fewer fungal lineages identified than healthy subjects (P < 0.05, Wilcoxon rank sum). We also saw higher richness in BAL of healthy subjects compared with transplant subjects (data not shown), although URT carryover contributing to BAL precludes definitive estimates of richness in lung.
Figure 4.
Rarefaction analysis of bacterial and fungal operational taxonomic units (OTUs) in the oropharynx of healthy and lung transplant subjects. The y axis denotes the number of OTUs detected by pyrosequencing of bacterial 16S genes (A) or fungal internal transcribed spacer genes (B) at the corresponding sequencing depths shown along the x axis. The subject group is indicated by the color key on the right. Error bars denote the standard deviation within subject groups.
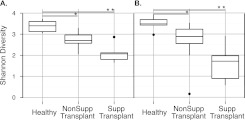
We then assessed bacterial community diversity using the Shannon Diversity Index (Figure 5). Transplant subjects had lower diversity in OW and BAL than healthy subjects (P < 0.05 Wilcoxon rank sum). Subjects transplanted for suppurative lung disease had the lowest community diversity, consistent with outgrowth of particular bacteria. Other transplant subjects were intermediate between the healthy and suppurative groups (P < 0.05, Wilcoxon rank sum).
Figure 5.
Alpha diversity of bacterial communities in healthy and transplant subjects. Diversity was calculated using the Shannon Index for oropharyngeal wash (A) and bronchoalveolar lavage (B). *P < 0.05; **P < 0.01 (Wilcoxon rank sum).
Global Relationship between Lung and URT Bacterial Communities in Transplant and Healthy Subjects
We next investigated the relationship of lung and URT communities, following the hypothesis that due to the growth of microbes in transplanted lung, BAL would be more different from URT samples in transplant subjects than in healthy subjects. For this comparison we used UniFrac, which measures the similarity of bacterial communities based on the extent to which members of the communities are phylogenetically related (27). UniFrac distances were calculated for all pairs of samples, and then relationships among communities were plotted using principal coordinate analysis (PCoA).
UniFrac analysis showed that each subject’s BAL was more closely related to their own OW than to the BAL of different individuals (P < 0.05 for abundance-weighted and unweighted UniFrac). An additional nonphylogenetic measure of shared OTUs, the Jaccard Index (36, 37), also revealed that BAL from each individual was most tightly linked to their own OW (P < 0.05 binary/abundance Jaccard), consistent with BAL communities originating largely in the URT (20).
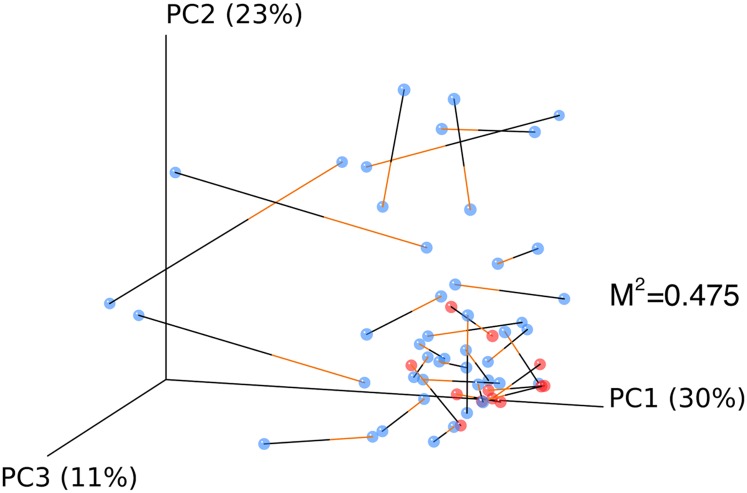
We then quantified the resemblance between lung and URT communities. The BAL and OW weighted UniFrac data were plotted using PCoA, and then the two PCoA plots were aligned using Procrustes (29, 30), which rotates and scales two matrices to maximize overlap (Figure 6). For healthy subjects, BAL and OW formed a tight cluster, separate from transplant subjects (P < 0.05 UniFrac and Jaccard), with small distances between communities from the same individual. In contrast, transplant subjects’ BAL and OW microbiota were often divergent and separated by greater distances. Thus, healthy subjects show close similarity between their BAL and OW microbiota, whereas distances are greater for lung transplant subjects, consistent with outgrowth of bacterial lineages in transplanted lungs.
Figure 6.
Relationship between bronchoalveolar lavage (BAL) and oropharyngeal bacterial communities within individuals. Weighted UniFrac distances were calculated between all pairs of samples within BAL or oropharyngeal wash (OW), and then each sample type was plotted separately in 3D space by principal coordinate analysis. The two plots (BAL and OW) were then transformed by Procrustes analysis to achieve maximum alignment. Each point corresponds to a bacterial community, with transplant subjects’ communities shown in blue, healthy subjects’ communities shown in red, and the two communities from each subject connected by a bar. The orange end of each bar connects to the OW sample data; the black end connects to the BAL sample data from the same individual. If BAL and OW plots are similar, then the relative distance between connected points (residuals) will be small. The overall similarity is summarized by the M2 value, and statistical goodness of fit is measured by a Monte Carlo label permutation approach (10,000 iterations).
We then determined whether the relationship between URT and BAL communities differed based on underlying indications for transplant. When BAL-OW distances within individuals were compared across groups, there was a trend for greater BAL-OW distances in the suppurative transplant group compared with all nonsuppurative or COPD/emphysema and ILD/IPF groups (Figure E3), suggesting that the greater BAL-OW distance among transplant subjects was driven mainly by the subgroup of subjects transplanted for suppurative diseases.
Comparing BAL and Oropharyngeal Bacterial Lineages to Identify Taxa Enriched in Lung
All bacterial and fungal organisms found by BAL culture were identified via 16S and ITS sequencing, except for Mycobacterium avium-intracellulare, which was found by culture in two subjects (Tx35, Tx45) but not by 16S sequencing (Tables 1 and E1). Respiratory pathogens identified by culture were sometimes the most abundant lineage identified by sequencing (B24, B26, B32, B33, B40, B44), but in many cases they were exceeded in abundance by taxa typical of upper respiratory tract flora (B36, B41, B46, B47, B50). Thus, potentially significant pathogens in the lung often represent a minority population against the background of oropharyngeal organisms in BAL. Furthermore, lung infection can result from bacteria that are normal inhabitants of the URT.
Therefore, to identify sequences that represent organisms genuinely replicating in lung, we developed a single-sided outlier test to compare each taxon’s abundance in BAL with its abundance in OW. This strategy assumes that microbes replicating in the LRT will be enriched in BAL compared with OW, whereas URT taxa present in BAL due to passive admixture such as bronchoscopic carryover or aspiration will be represented at a relative abundance no greater than that in OW. The model also accounts for uncertainty due to stochastics of sampling (E.S. Charlson, unpublished observations).
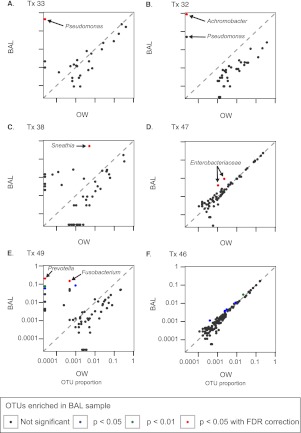
Figure 7 shows six outlier plots selected from among the 21 subjects to exemplify different patterns revealed by this analysis (full set in Figure E4). Each OTU is represented by a dot, with its relative abundance in BAL shown on the y axis and in OW on the x axis, and significant enrichment in BAL indicated by color-coding.
Figure 7.
Identification of lung-enriched bacteria using single-sided outlier plots to compare taxa abundances in bronchoalveolar lavage (BAL) and oropharyngeal wash (OW) samples. Six representative outlier plots are shown (A–F). Each dot represents an operational taxonomic unit (OTU), with its abundance in OW plotted on the x axis and in BAL on the y axis. Taxa found in equal proportions in lung and oropharynx are close to the diagonal line of identity from lower left to upper right, whereas those enriched in BAL are above the diagonal and to the left. Taxa enriched in BAL compared with OW at a level that reaches statistical significance (P < 0.05 after correction for false discovery rate) are highlighted in red.
Tx33 (Figure 7A) underwent bronchoscopy because of nonresolving clinical pneumonia despite treatment for culture-positive P. aeruginosa. Molecular analysis revealed that P. aeruginosa was the only OTU significantly enriched compared with OW and represented more than 50% of all BAL sequences, confirming the etiology and the absence of other causes for infection.
Tx32 (Figure 7B) grew Achromobacter and P. aeruginosa in BAL culture. The outlier plot showed highly significant enrichment of Achromobacter, whereas Pseudomonas was above the line of identity but did not reach statistical significance. Achromobacter comprised more than 80% of bacteria in BAL and was 17-fold more abundant than Pseudomonas, indicating the relative quantity of each organism in lung.
Tx38 (Figure 7C) had late-onset BOS. Outlier analysis revealed high-level BAL enrichment of the anaerobic bacterium Sneathia, which represented more than 50% of all sequences in BAL but less than 0.5% in the oropharynx and indicates robust lung growth. No respiratory pathogens were identified in BAL culture, which would not detect anaerobes such as Sneathia.
Tx47 (Figure 7D) yielded Enterobacter cloacae in BAL culture. The outlier plot revealed clustering of many taxa along the line of identity, suggesting abundant oropharyngeal admixture with BAL. However, even in the background of high carryover, two Enterobacteriaceae taxa are significantly enriched in BAL relative to OW, confirming outgrowth in lung.
Tx49 (Figure 7E) was negative for respiratory pathogens by culture. However, outlier analysis revealed significant enrichment in BAL of several taxa normally found in the URT (Prevotella, Fusobacterium), suggesting outgrowth of mixed oropharyngeal flora not detected by standard respiratory culture.
Tx46 (Figure 7F) BAL culture revealed P. aeruginosa. However, outlier analysis revealed no taxa enriched in lung compared with URT. P. aeruginosa was present in BAL but represented only 0.01% of sequences and had a similar abundance in OW. This result suggests detection by culture in BAL likely from bronchoscopic carryover or passive aspiration without replication and enrichment in the lung.
Additional examples of clinical utility (Figure E4) include robust single-taxa lung enrichment (Tx24, Tx26, Tx44), lung outgrowth of mixed URT/anaerobic bacteria or other organisms not found on culture (Tx25, Tx41), and lack of outliers indicating absence of significant lung-enriched taxa (Tx30, Tx31).
Figure E4 shows a concise clinical report developed to summarize molecular microbiome findings, including the outlier analysis, culture, and clinical data for each subject. Statistical and software tools for report generation are embedded in a “reproducible ” format, which allows repeated analysis over user-specified data sets (E. S. Charlson, unpublished observations).
Discussion
Approximately 24% of lung transplant recipients die from posttransplant infections, most of which are bacterial or fungal pulmonary infections (1, 38, 39). The major cause of later death is BOS (1, 39), a form of chronic rejection that is not well understood but is linked to microbial triggers within the allograft (4–12, 40). Here we applied broad-range 16S bacterial and ITS fungal rRNA gene profiling to define microbial populations in the lungs of 21 transplant recipients. Transplant subjects showed marked differences from healthy subjects in airway microbial burden, community composition and structure, lower richness, and greater distances between lung and upper respiratory communities. We then used a novel approach to visualize taxa enriched in bronchoscopic specimens relative to oropharyngeal samples, allowing identification of specific bacteria replicating in allografts of individual subjects.
The richness and diversity of airway communities were reduced in transplant subjects compared with control subjects. Although richness and diversity was most reduced in subjects with CF, it was also reduced in subjects transplanted for other indications (Figure 5). Decreased airway community richness and diversity in CF has been correlated with deteriorating lung function (41, 42) but has not been investigated in relation to outcome from lung transplant either among subjects with CF or subjects with nonsuppurative lung disease.
Although molecular profiling can identify all taxa present and can quantify abundances, the source of those sequences is critical in determining their significance (43). By comparing the relative abundance of each bacterial taxon in BAL and oropharyngeal samples, we can identify enrichment in lung (E.S. Charlson, unpublished observations). Enrichment in BAL compared with the URT suggests replication within the lung and thus can distinguish such organisms from those entering BAL through bronchoscopic carryover or passive aspiration. A large proportion of transplant recipients had specific lung-enriched bacteria. Enriched taxa often confirmed organisms found by culture but also identified bacteria not amenable to culture, such as unusual anaerobes or normal oropharyngeal residents. Molecular profiling also determined proportional abundances of organisms, adding quantitative information to culture results. In some cases we showed that bacteria identified by culture were equally represented in BAL and URT, providing evidence against authentic LRT replication.
Transplant subjects had markedly higher levels of bacterial 16S DNA in BAL than controls subjects. This finding was true regardless of underlying indication for transplant, with no difference between subjects transplanted for COPD/emphysema, interstitial lung disease, or CF/suppurative lung disease. Although taxa present in BAL at abundances similar to the URT may result from bronchoscopic carryover, they may also reflect bacteria genuinely present in lung due to aspiration without replication and enrichment. The high 16S DNA levels seen even in subjects without outliers (or where outliers reflect a minority of BAL sequences) is consistent with such a source, which may result from increased propensity for aspiration or inability to clear aspirated bacteria effectively due to cough and mucocilliary clearance defects (3, 44, 45) Passively aspirated or even dead bacteria can be potent immunological activators, and aspiration has been implicated in BOS (46).
BOS is associated with polymorphisms in genes that regulate innate microbial responses (9–12), specific bacteria, fungi, or viruses in the LRT (4–8) and may be more responsive to the antibiotic azithromycin than to intensified immunosuppression (13–15). It is hypothesized that aberrant microbial populations in the lung, among other factors, trigger inflammatory pathways, resulting in graft injury (40). Our study was not designed to address BOS specifically. However, one subject had late-onset BOS, and 16S sequencing revealed high-level lung enrichment of the anaerobic lineage Sneathia. This organism is a component of the vaginal flora and a rare cause of obstetrical infections (35, 47–49) but has not previously been reported in the respiratory tract and would not be identified by standard culture. Two subjects with early BOS also had taxa enriched in lung that were not detected by culture (Tx25, Tx40). Longitudinal studies using broad-range profiling are needed to determine whether overall microbial burden or specific lung microbiome features, either early after transplant or at the time of BOS, have a role in late graft failure.
Our study has several limitations. (1) To not “over-call” lung-enriched taxa, we used a 95% confidence threshold (E.S. Charlson, unpublished observations). There may be organisms authentically replicating in the lung that do not reach statistical significance for enrichment, although they are likely to be present at low abundance and/or with modest fold-increase over OW. (2) Although OW is a clinically tractable way to sample the URT, there may be subtle differences in outliers identified when BAL is compared with the most accurate representation of potential carryover, direct peri-glottic specimens obtained with high stringency two-scope sampling (20; E.S. Charlson, unpublished observations). (3) Although most pathogens found by culture were detected by molecular analysis, two subjects had M. avium-intracellulare infections on culture that were not found by 16S sequencing. Although MAI is occasionally isolated as a laboratory contaminant, this result suggests that mycobacterial detection may be a limitation of 16S rDNA surveys with the primers used (50). (4) A relatively small number of subjects was included in each transplant group here, which should be expanded to larger numbers of subjects and serial specimens.
In summary, aberrant microbial communities and specific lung-enriched taxa are common in lung after transplantation. Future studies are needed to determine whether particular community structures or individual microorganisms identified by microbiome analysis are associated with late graft failure and BOS and whether broad-range 16S bacterial or ITS fungal profiling added to standard culture improves diagnosis and management of lung colonization and infection in transplant and other lung diseases.
Supplementary Material
Acknowledgments
The authors thank the research subjects who volunteered for this study and the clinicians who assisted with specimen collection; W. Russell and D. Frame for critical study assistance; the Penn Center for AIDS Research (P30-AI045008) for technical support; J. Christie for valuable discussion and insights; and members of the Bushman and Collman laboratories for helpful discussions.
Footnotes
This work was supported in part by the Lung HIV Microbiome Project Grant U01 HL098957 from the National Institutes of Health (R.G.C. and F.D.B.), by grant T32 AI060516 (E.S.C.), and grant K12 HL090021 (J.M.D), and by grants from the Penn Genome Frontiers Institute and the Pennsylvania Department of Health; the Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusion. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions: E.S.C, J.M.D., F.D.B., and R.G.C. designed the research; J.M.D., A.R.H., A.S.F., and A.Y. collected the samples; E.S.C. performed research; E.S.C., K.B., F.D.B., and R.G.C. analyzed data; and E.S.C., F.D.B. and R.G.C. wrote the paper.
This article has an online supplement, which is accessible from this issue's table of content at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201204-0693OC on July 12, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Christie JD, Edwards LB, Kucheryavaya AY, Benden C, Dobbels F, Kirk R, Rahmel AO, Stehlik J, Hertz MI. The registry of the international society for heart and lung transplantation: twenty-eighth adult lung and heart-lung transplant report–2011. J Heart Lung Transplant 2011;30:1104–1122 [DOI] [PubMed] [Google Scholar]
- 2.Robertson AG, Griffin SM, Murphy DM, Pearson JP, Forrest IA, Dark JH, Corris PA, Ward C. Targeting allograft injury and inflammation in the management of post-lung transplant bronchiolitis obliterans syndrome. Am J Transplant 2009;9:1272–1278 [DOI] [PubMed] [Google Scholar]
- 3.Atkins BZ, Trachtenberg MS, Prince-Petersen R, Vess G, Bush EL, Balsara KR, Lin SS, Davis RD., Jr Assessing oropharyngeal dysphagia after lung transplantation: altered swallowing mechanisms and increased morbidity. J Heart Lung Transplant 2007;26:1144–1148 [DOI] [PubMed] [Google Scholar]
- 4.Botha P, Archer L, Anderson RL, Lordan J, Dark JH, Corris PA, Gould K, Fisher AJ. Pseudomonas aeruginosa colonization of the allograft after lung transplantation and the risk of bronchiolitis obliterans syndrome. Transplantation 2008;85:771–774 [DOI] [PubMed] [Google Scholar]
- 5.Khalifah AP, Hachem RR, Chakinala MM, Schechtman KB, Patterson GA, Schuster DP, Mohanakumar T, Trulock EP, Walter MJ. Respiratory viral infections are a distinct risk for bronchiolitis obliterans syndrome and death. Am J Respir Crit Care Med 2004;170:181–187 [DOI] [PubMed] [Google Scholar]
- 6.Weigt SS, Elashoff RM, Huang C, Ardehali A, Gregson AL, Kubak B, Fishbein MC, Saggar R, Keane MP, Lynch JP, III, et al. Aspergillus colonization of the lung allograft is a risk factor for bronchiolitis obliterans syndrome. Am J Transplant 2009;9:1903–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotsimbos TC, Snell GI, Levvey B, Spelman DW, Fuller AJ, Wesselingh SL, Williams TJ, Ostergaard L. Chlamydia pneumoniae serology in donors and recipients and the risk of bronchiolitis obliterans syndrome after lung transplantation. Transplantation 2005;79:269–275 [DOI] [PubMed] [Google Scholar]
- 8.Heng D, Sharples LD, McNeil K, Stewart S, Wreghitt T, Wallwork J. Bronchiolitis obliterans syndrome: incidence, natural history, prognosis, and risk factors. J Heart Lung Transplant 1998;17:1255–1263 [PubMed] [Google Scholar]
- 9.Munster JM, van der Bij W, Breukink MB, van der Steege G, Zuurman MW, Hepkema BG, Verschuuren EA, van Son WJ, Seelen MA. Association between donor MBL promoter haplotype and graft survival and the development of BOS after lung transplantation. Transplantation 2008;86:1857–1863 [DOI] [PubMed] [Google Scholar]
- 10.Palmer SM, Burch LH, Trindade AJ, Davis RD, Herczyk WF, Reinsmoen NL, Schwartz DA. Innate immunity influences long-term outcomes after human lung transplant. Am J Respir Crit Care Med 2005;171:780–785 [DOI] [PubMed] [Google Scholar]
- 11.Palmer SM, Klimecki W, Yu L, Reinsmoen NL, Snyder LD, Ganous TM, Burch L, Schwartz DA. Genetic regulation of rejection and survival following human lung transplantation by the innate immune receptor CD14. Am J Transplant 2007;7:693–699 [DOI] [PubMed] [Google Scholar]
- 12.Kastelijn EA, van Moorsel CH, Rijkers GT, Ruven HJ, Karthaus V, Kwakkel-van Erp JM, van de Graaf EA, Zanen P, van Kessel DA, Grutters JC, et al. Polymorphisms in innate immunity genes associated with development of bronchiolitis obliterans after lung transplantation. J Heart Lung Transplant 2010;29:665–671 [DOI] [PubMed] [Google Scholar]
- 13.Gerhardt SG, McDyer JF, Girgis RE, Conte JV, Yang SC, Orens JB. Maintenance azithromycin therapy for bronchiolitis obliterans syndrome: results of a pilot study. Am J Respir Crit Care Med 2003;168:121–125 [DOI] [PubMed] [Google Scholar]
- 14.Vos R, Vanaudenaerde BM, Verleden SE, De Vleeschauwer SI, Willems-Widyastuti A, Van Raemdonck DE, Schoonis A, Nawrot TS, Dupont LJ, Verleden GM. A randomised controlled trial of azithromycin to prevent chronic rejection after lung transplantation. Eur Respir J 2011;37:164–172 [DOI] [PubMed] [Google Scholar]
- 15.Jain R, Hachem RR, Morrell MR, Trulock EP, Chakinala MM, Yusen RD, Huang HJ, Mohanakumar T, Patterson GA, Walter MJ. Azithromycin is associated with increased survival in lung transplant recipients with bronchiolitis obliterans syndrome. J Heart Lung Transplant 2010;29:531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE 2011;6:e16384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flanagan JL, Brodie EL, Weng L, Lynch SV, Garcia O, Brown R, Hugenholtz P, DeSantis TZ, Andersen GL, Wiener-Kronish JP, et al. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J Clin Microbiol 2007;45:1954–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, Kaess H, Deterding RR, Accurso FJ, Pace NR. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA 2007;104:20529–20533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010;5:e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 2011;184:957–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol 2010;3:148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, Hwang J, Bushman FD, Collman RG. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS ONE 2010;5:e15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. Qiime allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price MN, Dehal PS, Arkin AP. Fasttree 2: approximately maximum-likelihood trees for large alignments. PLoS ONE 2010;5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, et al. Chimeric 16s rRNA sequence formation and detection in sanger and 454-pyrosequenced pcr amplicons. Genome Res 2011;21:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 2010;6:e1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 2005;71:8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 2007;73:1576–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gower JC. Generalized Procrustes analysis. Psychometrika 1975;40:33–51 [Google Scholar]
- 30.Hurley JR, Cattell RB. The Procrustes program: producing direct rotation to test a hypothesized factor structure. Behav Sci 1962;7:258–262 [Google Scholar]
- 31.Weir BS, Hill WG. Estimating f-statistics. Annu Rev Genet 2002;36:721–750 [DOI] [PubMed] [Google Scholar]
- 32.Ng KWTG, Tang ML. Dirichlet and related distributions. Chichester, UK: Wiley; 2011 [Google Scholar]
- 33.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science 2009;326:1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio 2010;1 doi: 10.1128/mBio.00129-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins MD, Hoyles L, Tornqvist E, von Essen R, Falsen E. Characterization of some strains from human clinical sources which resemble “Leptotrichia sanguinegens”: description of sneathia sanguinegens sp. Nov., gen. Nov. Syst Appl Microbiol 2001;24:358–361 [DOI] [PubMed] [Google Scholar]
- 36.Chao A, Chazdon RL, Colwell RK, Shen T-J. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett 2005;8:148–159 [Google Scholar]
- 37.Jaccard P. The distribution of the flora in the alpine zone. New Phytol 1912;11:37–50 [Google Scholar]
- 38.Valentine VG, Bonvillain RW, Gupta MR, Lombard GA, LaPlace SG, Dhillon GS, Wang G. Infections in lung allograft recipients: ganciclovir era. J Heart Lung Transplant 2008;27:528–535 [DOI] [PubMed] [Google Scholar]
- 39.Christie JD, Edwards LB, Kucheryavaya AY, Aurora P, Dobbels F, Kirk R, Rahmel AO, Stehlik J, Hertz MI. The registry of the international society for heart and lung transplantation: twenty-seventh official adult lung and heart-lung transplant report–2010. J Heart Lung Transplant 2010;29:1104–1118 [DOI] [PubMed] [Google Scholar]
- 40.Nakajima T, Palchevsky V, Perkins DL, Belperio JA, Finn PW. Lung transplantation: infection, inflammation, and the microbiome. Semin Immunopathol 2011;33:135–156 [DOI] [PubMed] [Google Scholar]
- 41.Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, Karaoz U, Andersen GL, Brown R, Fujimura KE, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS ONE 2010;5:e11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klepac-Ceraj V, Lemon KP, Martin TR, Allgaier M, Kembel SW, Knapp AA, Lory S, Brodie EL, Lynch SV, Bohannan BJ, et al. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ Microbiol 2010;12:1293–1303 [DOI] [PubMed] [Google Scholar]
- 43.Bartlett JG, Alexander J, Mayhew J, Sullivan-Sigler N, Gorbach SL. Should fiberoptic bronchoscopy aspirates be cultured? Am Rev Respir Dis 1976;114:73–78 [DOI] [PubMed] [Google Scholar]
- 44.Herve P, Silbert D, Cerrina J, Simonneau G, Dartevelle P. Impairment of bronchial mucociliary clearance in long-term survivors of heart/lung and double-lung transplantation. The Paris-Sud lung transplant group. Chest 1993;103:59–63 [DOI] [PubMed] [Google Scholar]
- 45.Higenbottam T, Jackson M, Woolman P, Lowry R, Wallwork J. The cough response to ultrasonically nebulized distilled water in heart-lung transplantation patients. Am Rev Respir Dis 1989;140:58–61 [DOI] [PubMed] [Google Scholar]
- 46.D'Ovidio F, Mura M, Tsang M, Waddell TK, Hutcheon MA, Singer LG, Hadjiliadis D, Chaparro C, Gutierrez C, Pierre A, et al. Bile acid aspiration and the development of bronchiolitis obliterans after lung transplantation. J Thorac Cardiovasc Surg 2005;129:1144–1152 [DOI] [PubMed] [Google Scholar]
- 47.Ling Z, Kong J, Liu F, Zhu H, Chen X, Wang Y, Li L, Nelson KE, Xia Y, Xiang C. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genomics 2010;11:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA 2011;108:4680–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Martino SJ, Mahoudeau I, Brettes JP, Piemont Y, Monteil H, Jaulhac B. Peripartum bacteremias due to leptotrichia amnionii and sneathia sanguinegens, rare causes of fever during and after delivery. J Clin Microbiol 2004;42:5940–5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bogner JR, Rusch-Gerdes S, Mertenskotter T, Loch O, Emminger C, Baumgarten R, Brockmeyer NH, Brockhaus W, Jablonowski H, Stoehr A, et al. Patterns of Mycobacterium avium culture and PCR positivity in immunodeficient HIV-infected patients: progression from localized to systematic disease, German AIDS Study Group (GASG/IDKF). Scand J Infect Dis 1997;29:579–584 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.