Abstract
Rationale: Elevated long pentraxin-3 (PTX3) levels are associated with the development of primary graft dysfunction (PGD) after lung transplantation. Abnormalities in innate immunity, mediated by PTX3 release, may play a role in PGD pathogenesis.
Objectives: Our goal was to test whether variants in the gene encoding PTX3 are risk factors for PGD.
Methods: We performed a candidate gene association study in recipients from the multicenter, prospective Lung Transplant Outcomes Group cohort enrolled between July 2002 and July 2009. The primary outcome was International Society for Heart and Lung Transplantation grade 3 PGD within 72 hours of transplantation. Targeted genotyping of 10 haplotype-tagging PTX3 single-nucleotide polymorphisms (SNPs) was performed in lung transplant recipients. The association between PGD and each SNP was evaluated by logistic regression, adjusting for pretransplantation lung disease, cardiopulmonary bypass use, and population stratification. The association between SNPs and plasma PTX3 levels was tested across genotypes in a subset of recipients with idiopathic pulmonary fibrosis.
Measurements and Main Results: Six hundred fifty-four lung transplant recipients were included. The incidence of PGD was 29%. Two linked 5′ region variants, rs2120243 and rs2305619, were associated with PGD (odds ratio, 1.5; 95% confidence interval, 1.1 to 1.9; P = 0.006 and odds ratio, 1.4; 95% confidence interval, 1.1 to 1.9; P = 0.007, respectively). The minor allele of rs2305619 was significantly associated with higher plasma PTX3 levels measured pretransplantation (P = 0.014) and at 24 hours (P = 0.047) after transplantation in patients with idiopathic pulmonary fibrosis.
Conclusions: Genetic variants of PTX3 are associated with PGD after lung transplantation, and are associated with increased PTX3 plasma levels.
Keywords: primary graft dysfunction, single-nucleotide polymorphism, long pentraxin 3, lung transplantation
At a Glance Commentary
Scientific Knowledge on the Subject
There is expanding evidence of a central role for innate immune dysregulation in the development of primary graft dysfunction (PGD) after lung transplantation, and innate immunity is a target for evolutionary selection from infectious sources. Long pentraxin-3 (PTX3) is an innate immune mediator that has been shown to be a plasma marker in PGD, but the genetics of this relationship are incompletely understood.
What This Study Adds to the Field
We performed a candidate gene association study of long PTX3, an innate immune mediator, and PGD in the Lung Transplant Outcomes Group, a multicenter cohort of lung transplant recipients. We identified a significant association between polymorphisms in PTX3 and PGD, and functional evaluation revealed that PGD-associated PTX3 variants regulate plasma PTX3 levels.
Primary graft dysfunction (PGD), with an incidence of 10–30%, is a major cause of early morbidity and mortality after lung transplantation (1–4). Ischemia–reperfusion injury is a significant contributor to the development of PGD (5, 6). Furthermore, ischemia–reperfusion injury is also known to result in activation and propagation of the innate immune response (7–11).
Innate immune responses appear to be regulated by germline-encoded receptors, and their polymorphic variants influence response to infection and inflammatory conditions (12). Long pentraxin-3 (PTX3) is a phylogenetically conserved mediator of the innate immune response (13). We previously demonstrated that plasma PTX3 levels after lung transplantation were significantly associated with PGD, with the strongest relationship seen in transplant recipients with idiopathic pulmonary fibrosis (IPF) (14).
PTX3 is produced at sites of inflammation or injury by dendritic cells and other antigen-presenting cells as a result of IL-1 and Toll-like receptor-4 (TLR4) signaling pathways (13, 15–18). PTX3 blood levels are elevated under inflammatory and ischemic conditions, including myocardial infarction, acute lung injury, and sepsis (19–24). The PTX3 promoter responds to tumor necrosis factor-α and IL-1β stimulation and contains binding sites for nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) (25). On the basis of our prior findings of an association of plasma PTX3 levels with PGD, we performed a candidate gene analysis of PTX3 with the hypothesis that polymorphisms in PTX3 in lung transplant recipients are significantly associated with the development of severe PGD. On the basis of the previously identified association of elevated PTX3 plasma protein levels in IPF recipients with PGD, we further evaluated the association of PTX3 polymorphisms in IPF recipients with PGD. Some of the results of these studies have been previously reported in the form of an abstract (26).
Methods
Please see the online supplement for further details on PGD grading, genotyping method, measurement of PTX3 concentration, and statistical analysis, including power calculations.
Study Design and Subject Selection
Study subjects were enrolled from the multicenter Lung Transplant Outcomes Group cohort and patient-level data were collected prospectively as previously described (3, 4, 14, 27–29). This study was approved by the institutional review boards at each site and subjects provided consent for enrollment.
Phase 1 was a candidate gene association study evaluating the association of PTX3 single nucleotide polymorphisms (SNPs) and the risk of PGD after lung transplantation. Patients enrolled consecutively in the Lung Transplant Outcomes Group from July 2002 through July 2009 were included for analysis.
Phase 2 was a functional assessment of the SNPs identified in phase 1. Plasma PTX3 concentrations were measured in a subset of subjects with IPF (14). On the basis of a previously identified association of PTX3 plasma concentration in IPF recipients with PGD, this analysis was performed on the overall cohort and a subgroup limited to transplant recipients with IPF.
PGD Grading
PGD was defined as any episode of grade 3 PGD developing within 72 hours of allograft reperfusion, determined according to International Society for Heart and Lung Transplantation criteria, with grade 3 indicated by the presence of diffuse alveolar infiltrates in the allograft, a PaO2/FiO2 (fraction of inspired oxygen) ratio less than 200, and the exclusion of secondary causes (27, 30, 31). As a sensitivity analysis, grade 3 PGD present 48 or 72 hours after transplantation was used as an alternative PGD definition.
Genotyping Strategy
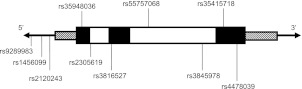
SNPs were selected to capture at least 50% of the variation in PTX3 at a minor allele frequency (MAF) greater than 5%, with additional SNPs selected to include known coding variants (Figure 1). Genotyping of 10 PTX3-tagging SNPs selected from HapMap and 1000 Genomes was performed with a combination of TaqMan polymerase chain reaction and the IBC chip (Illumina, San Diego, CA), a gene-centric array designed to assay SNPs in candidate genes affecting vascular, pulmonary, and metabolic phenotypes with a robust set of ancestry informative markers, filtered for the PTX3 gene (32–34).
Figure 1.
Gene structure of pentraxin-3 (PTX3), with approximate positions of single-nucleotide polymorphisms (SNPs) included for analysis. Exons are depicted as solid rectangles, whereas introns are open rectangles, and 5′ and 3′ untranslated regions (UTRs) are hatched.
Measurement of PTX3 Concentration
Plasma PTX3 concentrations were determined with a sandwich ELISA (Alexis Biochemicals, Lausen, Switzerland) in a previously measured subset of the patients with IPF (14).
Statistical Analysis
Subject characteristics were compared using t tests, Wilcoxon rank sum, or Kruskal-Wallis testing as appropriate. An odds ratio (OR) for PGD was calculated according to genotype for the seven SNPs with a minor allele frequency greater than 5%, with significance determined by χ2 test, assuming an additive model of genetic risk in PLINK (35). Genetically inferred ancestry was determined by principal components analysis and the approximately 1,800 ancestry informative markers (AIMs) on the IBC chip (36). To account for population stratification and potential clinical confounders, analyses included adjustment for key clinical covariates (cardiopulmonary bypass, predisposing lung disease) and two principal components derived from AIMs, using logistic regression. A Bonferroni-adjusted P < 7.1 × 10−3 was considered significant to account for the seven SNPs tested. We also performed analyses in the IPF subgroup and performed sensitivity analyses using altered outcome definitions. Haplotype analyses were performed with Haploview and PLINK (35, 37), using the confidence intervals method (38). Plasma PTX3 levels were analyzed across SNP genotypes, using the nonparametric test for trend. Except where noted, statistical analyses were performed with Stata 11.2 software (STATA Corp., College Station, TX).
Results
Patient Characteristics
Overall, 654 patients were included for analysis, with an incidence of PGD of 29% (189 of 654). The patient characteristics of the study subjects with and without PGD are described in Table 1. Subjects with PGD had a significantly higher usage of cardiopulmonary bypass compared with those without PGD (52 vs. 29%; P < 0.001). Age and sex were not significantly different between subjects with or without PGD.
TABLE 1.
SUBJECT CHARACTERISTICS
| Covariate | PGD (n = 189) | Non-PGD (n = 465) | P Value |
| Recipient variables | |||
| Age, mean (95% CI) | 52 (50, 54) | 52 (51, 54) | 0.6 |
| Male sex, n (%) | 97 (51) | 240 (52) | 0.7 |
| Pulmonary diagnosis, n (%) | <0.001 | ||
| COPD | 59 (31) | 209 (45) | |
| IPF | 73 (39) | 143 (31) | |
| CF | 14 (7) | 71(15) | |
| Other | 43 (23) | 42 (9) | |
| Race, n (%) | 0.004 | ||
| Caucasian | 147 (78) | 406 (87) | |
| African American | 31 (16) | 30 (6) | |
| Other | 11 (6) | 29 (6) | |
| Operative variables | |||
| Cardiopulmonary bypass use, yes, n (%) | 99 (52) | 133 (29) | <0.001 |
| Pulmonary arterial systolic pressure, mean (95% CI) | 34.6 (30.2, 39.0) | 30.4 (27.9, 33.0) | 0.1 |
Definition of abbreviations: CF = cystic fibrosis; CI = confidence interval; COPD = chronic obstructive pulmonary disease; IPF = idiopathic pulmonary fibrosis; PGD = primary graft dysfunction.
Percentages may not equal 100% because of rounding.
SNP Associations with PGD
After adjustment for pretransplantation diagnosis, genetically inferred ancestry using population stratification, and cardiopulmonary bypass use, two SNPs met our prespecified level of significance (Table 2). One SNP, rs2120243, resides in the 5′ promoter region and the minor allele is significantly associated with PGD (OR, 1.5; 95% confidence interval [CI], 1.1 to 1.9; P = 0.006). Variant rs2305619 is in the first intron and the minor allele at this position is also associated with PGD (OR, 1.4; 95% CI, 1.1 to 1.9; P = 0.007). The minor alleles at both SNP positions were common in the transplant population, with MAFs greater than 0.45.
TABLE 2.
SINGLE-NUCLEOTIDE POLYMORPHISM ANALYSIS FOR ASSOCIATION WITH PRIMARY GRAFT DYSFUNCTION
| rs Number | Minor Allele | Risk Allele | MAF Affected | MAF Unaffected | OR (95% CI) | P Value | Location |
| rs9289983 | G | A | 0.41 | 0.50 | 0.8 (0.6, 1.0) | 0.04 | 5′ upstream |
| rs1456099 | A | T | 0.43 | 0.51 | 0.8 (0.6, 1.0) | 0.05 | 5′ upstream |
| rs2120243 | A | A | 0.49 | 0.42 | 1.5 (1.1, 1.9) | 0.006 | 5′ upstream |
| rs35948036 | — | — | 0.009 | 0.009 | — | — | First exon synonymous |
| rs2305619 | T | T | 0.54 | 0.46 | 1.4 (1.1, 1.9) | 0.007 | Intron |
| rs3816527 | G | G | 0.43 | 0.40 | 1.3 (1.0, 1.6) | 0.1 | Second exon nonsynonymous |
| rs55757068 | C | T | 0.06 | 0.07 | 0.7 (0.4, 1.3) | 0.1 | Intron |
| rs3845978 | C | T | 0.06 | 0.06 | 0.7 (0.4, 1.3) | 0.3 | Intron |
| rs35415718 | — | — | 0.02 | 0.01 | — | — | Third exon nonsynonymous |
| rs4478039 | — | — | 0.00 | 0.00 | — | — | Third exon nonsynonymous |
Definition of abbreviations: CI = confidence interval; MAF = minor allele frequency; OR = odds ratio.
OR and P values are based on an additive model. P for significance = 7.1 × 10−3, based on the testing of seven single-nucleotide polymorphisms.
Analysis is corrected for first two principal components derived from ancestry informative markers, cardiopulmonary bypass use, and preoperative lung disease.
Sensitivity Analysis for SNP Associations with Alternative PGD Definitions
When using grade 3 PGD present 48 hours after transplantation as an alternative outcome definition, 91 subjects met the criteria for PGD. The association between the minor alleles at the identified risk SNPs, rs2120243 (OR, 1.5; 95% CI, 1.04 to 2.0; P = 0.03) and rs2305619 (OR, 1.6, 95% CI, 1.1, 2.2, P = 0.01), and PGD is attenuated, based on the Bonferroni-corrected P value cutoff, in the setting of a smaller sample size but remains of similar magnitude to the primary outcome. Similarly, when using grade 3 PGD present 72 hours after transplantation as a secondary PGD definition (n = 78), the direction and magnitude of effect appeared similar to the overall population, although the result was similarly not statistically significant with the smaller sample size (for rs2120243: OR, 1.6; 95% CI, 1.1 to 2.3; P = 0.01; for rs2305619: OR, 1.6; 95% CI, 1.1 to 2.3; P = 0.01). The associations between the PTX3 SNPs and PGD at both alternative time points are significant when using a P < 0.05 cutoff.
Haplotype Analysis
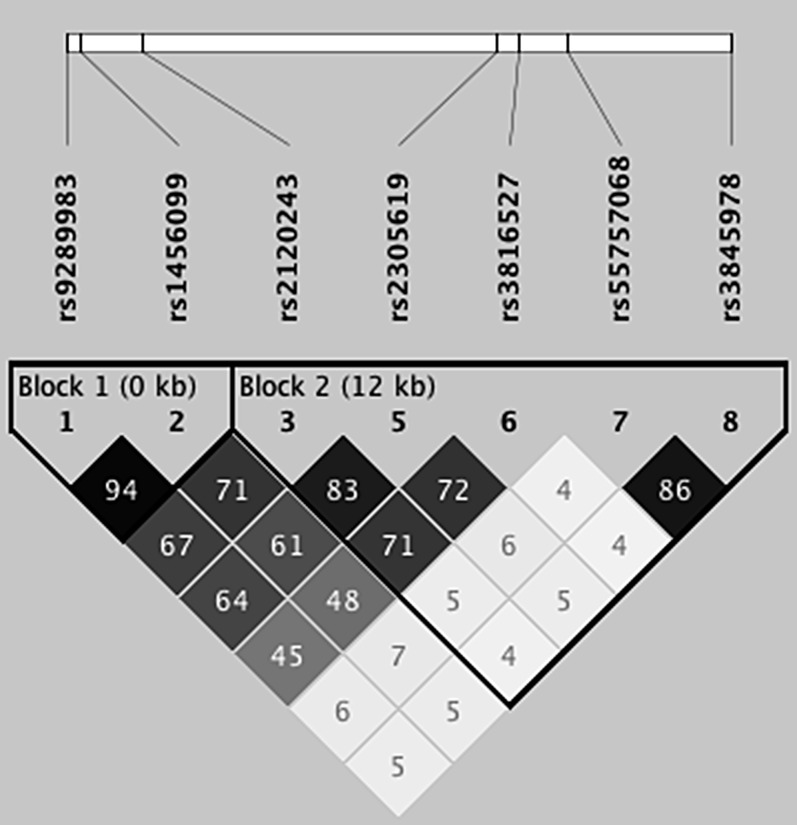
With two SNPs in PTX3 demonstrating significant association with PGD, we defined the linkage disequilibrium (LD) between markers and investigated whether PTX3 haplotypes, or combinations of alleles at different loci, were also associated with PGD. Two haplotype blocks were defined in our population for tested variants with MAF equal to or greater than 0.05 (Figure 2). Haplotype 1 blocks GA and AT and haplotype 2 blocks CCTCC and ATGCC were associated with PGD. Block 2 CCTCC (P = 0.02) and ATGCC (P = 0.03), defined by the 2 PGD-associated SNPs, demonstrated the strongest association (Table 3). There was significant linkage across the SNPs tested, with rs2120243 and rs2305619 demonstrating tight LD (r2 = 0.83). Furthermore, both markers from block 1 displayed moderate LD with block 2. Block 1 haplotypes were linked to the PGD-associated block 2 haplotypes; 73% of patients with haplotype block 1 AT had haplotype block 2 CCTCC while 98% of patients with haplotype 1 block GA had haplotype block 2 ATGCC.
Figure 2.
Linkage disequilibrium (LD) structure of PTX3 in the study population. LD values and shading represent R2 values, with higher numbers and darker shading reflecting a higher degree of LD. Each single-nucleotide polymorphism in the haplotype is listed by its reference sequence (rs) number.
TABLE 3.
HAPLOTYPE ANALYSIS FOR ASSOCIATION WITH PRIMARY GRAFT DYSFUNCTION
| Haplotype | Frequency in PGD | Frequency in Non-PGD | OR | P Value |
| Block 1 | ||||
| GA | 0.59 | 0.49 | 1.3 | 0.048 |
| AT | 0.47 | 0.50 | 0.8 | 0.04 |
| GT | 0.01 | 0.01 | 1.1 | 0.8 |
| Block 2 | ||||
| CCTCC | 0.46 | 0.49 | 0.7 | 0.02 |
| ATGCC | 0.38 | 0.36 | 1.3 | 0.03 |
| CCTTT | 0.05 | 0.05 | 0.7 | 0.3 |
| ATTCC | 0.05 | 0.04 | 1.8 | 0.05 |
| CTGCC | 0.02 | 0.03 | 0.7 | 0.5 |
| CTTCC | 0.02 | 0.02 | 1.4 | 0.4 |
Definition of abbreviations: OR = odds ratio; PGD = primary graft dysfunction.
ORs for PGD for individual haplotypes are based on logistic regression controlling for population stratification, using principal component analysis, use of cardiopulmonary bypass, and preoperative pulmonary diagnosis. The alleles encoded by the identified risk single-nucleotide polymorphisms, rs2120243 and rs2305619, are the first two alleles in block 2; the risk alleles are A and T, respectively.
SNP Associations with PGD in IPF Subgroup
The results of the SNP-association study among patients with a preoperative diagnosis of IPF are presented in Table E1 (in the online supplement). When the analysis was restricted to subjects with IPF only, the direction and magnitude of effect appeared similar to the overall population, though the result was not statistically significant with the smaller sample size.
Functional Evaluation
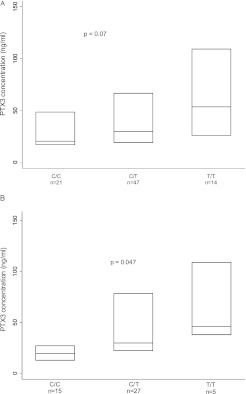
PTX3 SNP genotypes and plasma PTX3 levels were obtained from 82 subjects in the overall cohort, comprising subjects with varying preoperative diagnoses. Of these, 23 subjects (28%) had PGD and 59 (72%) did not have PGD. PTX3 plasma levels 24 hours after transplantation were higher with the presence of the risk allele at rs2305619, but did not achieve statistical significance (median [PTX3] C/C, 20.3 ng/ml; C/T, 29.6 ng/ml; T/T, 53.5 ng/ml) (P = 0.07) (Figure 3A).
Figure 3.
Box plot of plasma pentraxin-3 (PTX3) concentration 24 hours after transplantation stratified genotype at rs2305619 locus. (A) Eighty-two patients with PTX3 genotyping and plasma concentration measurements. (B) Forty-seven patents with a preoperative diagnosis of idiopathic pulmonary fibrosis (IPF) with PTX3 genotyping and plasma concentration measurements. Each horizontal line indicates median concentration. The upper and lower limits of each box indicate the interquartile range. The P value reported is from nonparametric test for trend.
Our prior work demonstrated a wider variability in plasma PTX3 levels among patients with IPF compared with those with chronic obstructive pulmonary disease. Of the 47 recipients with IPF who had both PTX3 genotyping and plasma PTX3 levels determined, 13 subjects (28%) had PGD and 34 (72%) did not have PGD. Circulating PTX3 plasma levels 24 hours after transplantation (median [PTX3] C/C, 19.5 ng/ml; C/T, 29.8 ng/ml; T/T, 46.0 ng/ml; P = 0.047) were significantly different by genotype for SNP rs2305619 (Figure 3B). PTX3 plasma levels 24 hours after transplantation were likewise higher with the presence of the risk allele at rs2120243, but did not achieve statistical significance (median [PTX3] C/C, 22.0 ng/ml; C/A, 29.2 ng/ml; A/A, 73.3 ng/ml) (P = 0.3) (Figure E1). Pretransplantation PTX3 levels were significantly associated with the genotype for SNP rs2305619 (median [PTX3] C/C, 1.7 ng/ml; C/T, 1.9 ng/ml; T/T, 7.9 ng/ml) (P = 0.014) (Figure E2).
Discussion
In a large diverse cohort of lung transplant recipients, we demonstrated two SNPs in PTX3 that were significantly associated with PGD after transplantation. The haplotypes defined by PGD-associated SNPs were also significantly associated with PGD after lung transplantation. Variation in one SNP, rs2305619, was also associated with plasma PTX3 concentration at baseline and 24 hours after transplantation in lung transplant recipients with IPF. We used a carefully defined phenotype for PGD that is widely accepted in the literature (27, 30, 39). Elevated plasma levels of the protein product of the PTX3 gene have been associated with PGD (14). The high-risk SNPs identified in PTX3 are in strong LD and are associated with a significant difference in PTX3 protein plasma levels, suggesting that PGD-associated SNPs or their LD partners may be functional. Our results are consonant with previous studies demonstrating associations between genetic variation in other innate immune genes with acute cellular rejection and bronchiolitis obliterans syndrome after lung transplantation (40–44).
Genetic variation in PTX3 is likely important in the development of PGD and may lead to functional differences in innate immune activity. Genetic variation in PTX3 has previously been associated with altered susceptibility to Pseudomonas aeruginosa colonization in patients with cystic fibrosis and altered pulmonary tuberculosis risk in West Africans (45, 46). Our identification of a PTX3–PGD association provides evidence to support the significance of altered innate immune activation in the development of PGD. We have previously demonstrated that elevated PTX3 concentrations after transplantation were associated with PGD in IPF transplant recipients (14). We have extended our understanding of the association between innate immunity and PGD by demonstrating the association of genetic variation in PTX3 with PGD and also by highlighting the association of the genetic variation with changes in plasma PTX3 concentrations. SNP level variation in PTX3 may explain some of the variance in post-transplantation PTX3 plasma concentrations, although the results are preliminary.
The region of PTX3 demonstrating association with PGD spanned the 5′ promoter region (rs2120243) and the first intron (rs2305619). Variation at these sites may alter transcriptional regulation, which might explain the variation in circulating PTX3 plasma levels observed in subjects with IPF. This is consistent with the observation that the PTX3 promoter increases gene transcription in response to the proinflammatory mediators tumor necrosis factor-α and IL-1β and binding of the transcription factor NF-κB (25). Alternatively, variation in the 5′ and first intronic region might result in abnormal splicing, or altered translational dynamics in the setting of acute ischemia–reperfusion. Alternatively spliced isoforms of PTX3 have not been reported to date. It is equally possible that these SNPs merely tag an untyped functional variant. Our genotyping was designed to test the reported coding variants in PTX3, but three of four known coding SNPs were rare in our population. The only common exonic SNP, rs3816527, displays moderate LD with the PGD-associated SNPs but did not itself demonstrate association with the PGD phenotype, perhaps because of low MAF. Given the high LD observed across PTX3, it may be necessary to pursue further sequencing and in vitro promoter assays, including fine mapping and mutagenesis studies, to more fully assess functionality.
Small-molecule and monoclonal antibody innate immune modulators are under investigation in models of acute lung injury, lentiviral infection, and inflammatory disorders (20, 47, 48). Eritoran, a small-molecule antagonist of TLR4, an upstream mediator of PTX3 production, is currently under investigation for treating severe sepsis (49). The potential role of therapies aimed at mitigating or preventing PGD through modification of host innate immune responses might be the subject of future trials. Proper modulation of PTX3 levels and activity are likely key to normal function of the innate immune responses to infection and inflammation, which may be particularly relevant for an injury response such as PGD. In PTX3 knockout mice, PTX3 deficiency was associated with more severe LPS-induced experimental lung injury whereas in wild-type mice, PTX3 overexpression was strongly associated with LPS-induced lung injury (19, 20). Differences in the timing and magnitude of PTX3 generation and release may explain some of these differences. Given the evidence for a potential role played by transcriptional regulation in the association between genetic variation in PTX3 and altered susceptibility to PGD, genotype may be an important consideration in future trial design for innate immune modulators.
We have previously demonstrated that post-transplantation biomarker profiles differ across pretransplantation diagnostic categories and have hypothesized that cellular pathways leading to PGD may be dependent on pretransplantation diagnosis (4, 14). On the basis of the observed correlation of genetic variation in PTX3 and PTX3 plasma concentrations in patients with IPF, there may be a genetic underpinning to these differences. There is also expanding evidence for the role played by innate immune mediators, including CXCL17, TLRs, and surfactant proteins, in the pathogenesis of interstitial lung diseases (50–54). The role of PTX3 and genetic variation in innate immune genes in the development of pulmonary fibrosis and the interplay between fibrotic lung disease and post-transplantation PGD are areas of future study.
There are several limitations to our study. The PTX3 SNP–PGD association has yet to be confirmed in a replication cohort. Supporting the veracity of the PTX3–PGD association are the previous association of PTX3 plasma levels with PGD, the single-gene design of our genetic association study, and the regulation of PTX3 plasma level by genotype. We focused on the relationship between PTX3 genotype and circulating PTX3 among subjects with IPF because subjects with IPF demonstrate the most variability in post-transplantation PTX3 concentrations. It will be important to assess the association between PTX3 polymorphisms and plasma protein levels in patients with other preoperative diagnoses as well as in the alveolar compartment.
As this was a candidate gene association study, we evaluated the influence of variation in PTX3 alone. There may be important variation in other innate immune genes, including TLRs, that impact the risk of developing the PGD phenotype, and different innate immunity loci may interact to modify PGD risk. Furthermore, it may be that PGD is a phenotype influenced by both donor and recipient genotype. Our analysis focused on allograft recipients and was essentially blinded to the potential influence of donor innate immune responses. Further work to elucidate the loci or environmental stimuli regulating the expression of PTX3 after transplantation is warranted.
In summary, we identified an association between polymorphisms in PTX3 and risk of PGD after lung transplantation. Furthermore, we identified a correlation between a PGD-associated PTX3 SNP and pre- and post-transplantation PTX3 plasma concentrations in patients with IPF. Improved mechanistic understanding of the genetic and clinical risk factors for PGD may allow improved prognostication pretransplantation and may aid in the development of personalized post-transplantation therapy tailored to an individual recipient’s risk.
Supplementary Material
Footnotes
Supported by NIH grants R01 HL087115, R01 HL081619, R01 HL096845, and K24 HL103844.
Author Contributions: Conception and design: J.M.D., N.J.M., R.F., S.M.K., R.J.S., S.M.P., L.B.W., J.D.C.; acquisition of data: R.F., D.J.L., J.C.L., E.C., V.N.L., S.B., M.C., E.D., J.S., K.W., J.O., A.W., D.W., S.A., P.D.S., J.A.B., D.W., L.B.W., S.M.P., J.D.C.; analysis and interpretation of data: J.M.D., N.J.M., M.R., S.M.K., S.M.P., L.B.W., J.D.C.; drafting or revising the manuscript for important intellectual content: J.M.D., N.J.M., R.F., M.R., S.M.K., D.J.L., J.C.L., E.C., R.J.S., V.N.L., S.B., M.C., E.D., J.S., K.W., J.O., A.W., D.W., S.A., P.D.S., J.A.B., D.W., L.B.W., S.M.P., J.D.C.; final approval of the version to be published: J.M.D., N.J.M., R.F., M.R., S.M.K., D.J.L., J.C.L., E.C., R.J.S., V.N.L., S.B., M.C., E.D., J.S., K.W., J.O., A.W., D.W., S.A., P.D.S., J.A.B., D.W., L.B.W., S.M.P., J.D.C.
The participating centers for this study were as follows: University of Pennsylvania, Columbia University, University of Alabama at Birmingham, Vanderbilt University, Stanford University, Johns Hopkins University, University of Michigan, Duke University, University of Pittsburgh, and University of Chicago.
This article has an online supplement, which is available from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201204-0692OC on July 19, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Arcasoy SM, Fisher A, Hachem RR, Scavuzzo M, Ware LB. Report of the ISHLT Working Group on Primary Lung Graft Dysfunction. V. Predictors and outcomes. J Heart Lung Transplant 2005;24:1483–1488 [DOI] [PubMed] [Google Scholar]
- 2.Christie JD, Kotloff RM, Ahya VN, Tino G, Pochettino A, Gaughan C, DeMissie E, Kimmel SE. The effect of primary graft dysfunction on survival after lung transplantation. Am J Respir Crit Care Med 2005;171:1312–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christie JD, Kotloff RM, Pochettino A, Arcasoy SM, Rosengard BR, Landis JR, Kimmel SE. Clinical risk factors for primary graft failure following lung transplantation. Chest 2003;124:1232–1241 [DOI] [PubMed] [Google Scholar]
- 4.Diamond JM, Kawut SM, Lederer DJ, Ahya VN, Kohl B, Sonett J, Palmer SM, Crespo M, Wille K, Lama VN, et al. Elevated plasma Clara cell secretory protein concentration is associated with high-grade primary graft dysfunction. Am J Transplant 2011;11:561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diamond JM, Christie JD. The contribution of airway and lung tissue ischemia to primary graft dysfunction. Curr Opin Organ Transplant 2010;15:552–557 [DOI] [PubMed] [Google Scholar]
- 6.Moreno I, Vicente R, Ramos F, Vicente JL, Barbera M. Determination of interleukin-6 in lung transplantation: association with primary graft dysfunction. Transplant Proc 2007;39:2425–2426 [DOI] [PubMed] [Google Scholar]
- 7.Frantz S, Kobzik L, Kim YD, Fukazawa R, Medzhitov R, Lee RT, Kelly RA. Toll4 (TLR4) expression in cardiac myocytes in normal and failing myocardium. J Clin Invest 1999;104:271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oyama J, Blais C, Jr, Liu X, Pu M, Kobzik L, Kelly RA, Bourcier T. Reduced myocardial ischemia–reperfusion injury in Toll-like receptor 4–deficient mice. Circulation 2004;109:784–789 [DOI] [PubMed] [Google Scholar]
- 9.Riad A, Jager S, Sobirey M, Escher F, Yaulema-Riss A, Westermann D, Karatas A, Heimesaat MM, Bereswill S, Dragun D, et al. Toll-like receptor-4 modulates survival by induction of left ventricular remodeling after myocardial infarction in mice. J Immunol 2008;180:6954–6961 [DOI] [PubMed] [Google Scholar]
- 10.Vakeva AP, Agah A, Rollins SA, Matis LA, Li L, Stahl GL. Myocardial infarction and apoptosis after myocardial ischemia and reperfusion: role of the terminal complement components and inhibition by anti-C5 therapy. Circulation 1998;97:2259–2267 [DOI] [PubMed] [Google Scholar]
- 11.Yasojima K, Kilgore KS, Washington RA, Lucchesi BR, McGeer PL. Complement gene expression by rabbit heart: upregulation by ischemia and reperfusion. Circ Res 1998;82:1224–1230 [DOI] [PubMed] [Google Scholar]
- 12.Sorensen TI, Nielsen GG, Andersen PK, Teasdale TW. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med 1988;318:727–732 [DOI] [PubMed] [Google Scholar]
- 13.Garlanda C, Bottazzi B, Bastone A, Mantovani A. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu Rev Immunol 2005;23:337–366 [DOI] [PubMed] [Google Scholar]
- 14.Diamond JM, Lederer DJ, Kawut SM, Lee J, Ahya VN, Bellamy S, Palmer SM, Lama VN, Bhorade S, Crespo M, et al. Elevated plasma long pentraxin-3 levels and primary graft dysfunction after lung transplantation for idiopathic pulmonary fibrosis. Am J Transplant 2011;11:2517–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breviario F, d’Aniello EM, Golay J, Peri G, Bottazzi B, Bairoch A, Saccone S, Marzella R, Predazzi V, Rocchi M, et al. Interleukin-1–inducible genes in endothelial cells: cloning of a new gene related to C-reactive protein and serum amyloid P component. J Biol Chem 1992;267:22190–22197 [PubMed] [Google Scholar]
- 16.Lee GW, Lee TH, Vilcek J. TSG-14, a tumor necrosis factor– and IL-1–inducible protein, is a novel member of the pentaxin family of acute phase proteins. J Immunol 1993;150:1804–1812 [PubMed] [Google Scholar]
- 17.Mantovani A, Garlanda C, Bottazzi B. Pentraxin 3, a non-redundant soluble pattern recognition receptor involved in innate immunity. Vaccine 2003;21:S43–S47 [DOI] [PubMed] [Google Scholar]
- 18.Ortega-Hernandez OD, Bassi N, Shoenfeld Y, Anaya JM. The long pentraxin 3 and its role in autoimmunity. Semin Arthritis Rheum 2009;39:38–54 [DOI] [PubMed] [Google Scholar]
- 19.Han B, Haitsma JJ, Zhang Y, Bai X, Rubacha M, Keshavjee S, Zhang H, Liu M. Long pentraxin PTX3 deficiency worsens LPS-induced acute lung injury. Intensive Care Med 2010;37:334–342 [DOI] [PubMed] [Google Scholar]
- 20.He X, Han B, Bai X, Zhang Y, Cypel M, Mura M, Keshavjee S, Liu M. Ptx3 as a potential biomarker of acute lung injury: supporting evidence from animal experimentation. Intensive Care Med 2010;36:356–364 [DOI] [PubMed] [Google Scholar]
- 21.He X, Han B, Liu M. Long pentraxin 3 in pulmonary infection and acute lung injury. Am J Physiol Lung Cell Mol Physiol 2007;292:L1039–L1049 [DOI] [PubMed] [Google Scholar]
- 22.Mauri T, Coppadoro A, Bellani G, Bombino M, Patroniti N, Peri G, Mantovani A, Pesenti A. Pentraxin 3 in acute respiratory distress syndrome: an early marker of severity. Crit Care Med 2008;36:2302–2308 [DOI] [PubMed] [Google Scholar]
- 23.Muller B, Peri G, Doni A, Torri V, Landmann R, Bottazzi B, Mantovani A. Circulating levels of the long pentraxin PTX3 correlate with severity of infection in critically ill patients. Crit Care Med 2001;29:1404–1407 [DOI] [PubMed] [Google Scholar]
- 24.Wagenaar JF, Goris MG, Gasem MH, Isbandrio B, Moalli F, Mantovani A, Boer KR, Hartskeerl RA, Garlanda C, van Gorp EC. Long pentraxin PTX3 is associated with mortality and disease severity in severe leptospirosis. J Infect 2009;58:425–432 [DOI] [PubMed] [Google Scholar]
- 25.Basile A, Sica A, d’Aniello E, Breviario F, Garrido G, Castellano M, Mantovani A, Introna M. Characterization of the promoter for the human long pentraxin PTX3: role of NF-κB in tumor necrosis factor-α and interleukin-1β regulation. J Biol Chem 1997;272:8172–8178 [DOI] [PubMed] [Google Scholar]
- 26.Diamond JM, Feng R, Meyer NJ, Lederer D, Lee J, Kawut S, Ahya V, Cantu E, Palmer S, Weinacker A, et al. PTX3 polymorphisms are associated with primary graft dysfunction after lung transplantation [abstract]. J Heart Lung Transplant 2011;30:S23 [Google Scholar]
- 27.Christie JD, Bellamy S, Ware LB, Lederer D, Hadjiliadis D, Lee J, Robinson N, Localio AR, Wille K, Lama V, et al. Construct validity of the definition of primary graft dysfunction after lung transplantation. J Heart Lung Transplant 2010;29:1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christie JD, Robinson N, Ware LB, Plotnick M, De Andrade J, Lama V, Milstone A, Orens J, Weinacker A, Demissie E, et al. Association of protein C and type 1 plasminogen activator inhibitor with primary graft dysfunction. Am J Respir Crit Care Med 2007;175:69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christie JD, Shah CV, Kawut SM, Mangalmurti N, Lederer DJ, Sonett JR, Ahya VN, Palmer SM, Wille K, Lama V, et al. Plasma levels of receptor for advanced glycation end products, blood transfusion, and risk of primary graft dysfunction. Am J Respir Crit Care Med 2009;180:1010–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christie JD, Carby M, Bag R, Corris P, Hertz M, Weill D. Report of the ISHLT Working Group on Primary Lung Graft Dysfunction. II. Definition: a consensus statement of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant 2005;24:1454–1459 [DOI] [PubMed] [Google Scholar]
- 31.Christie JD, Van Raemdonck D, de Perrot M, Barr M, Keshavjee S, Arcasoy S, Orens J. Report of the ISHLT Working Group on Primary Lung Graft Dysfunction. I. Introduction and methods. J Heart Lung Transplant 2005;24:1451–1453 [DOI] [PubMed] [Google Scholar]
- 32.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International HapMap Consortium. The International HapMap Project. Nature 2003;426:789–796 [DOI] [PubMed] [Google Scholar]
- 34.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, et al. Concept, design and implementation of a cardiovascular gene–centric 50K SNP array for large-scale genomic association studies. PLoS One 2008;3:e3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christie JD, Wurfel MM, Feng R, O’Keefe GE, Bradfield J, Ware LB, Christiani DC, Calfee CS, Cohen MJ, Matthay M, et al. Genome wide association identifies PPFIA1 as a candidate gene for acute lung injury risk following major trauma. PLoS One 2012;7:e28268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265 [DOI] [PubMed] [Google Scholar]
- 38.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science 2002;296:2225–2229 [DOI] [PubMed] [Google Scholar]
- 39.Cypel M, Yeung JC, Liu M, Anraku M, Chen F, Karolak W, Sato M, Laratta J, Azad S, Madonik M, et al. Normothermic ex vivo lung perfusion in clinical lung transplantation. N Engl J Med 2011;364:1431–1440 [DOI] [PubMed] [Google Scholar]
- 40.Kastelijn EA, van Moorsel CH, Rijkers GT, Ruven HJ, Karthaus V, Kwakkel-van Erp JM, van de Graaf EA, Zanen P, van Kessel DA, Grutters JC, et al. Polymorphisms in innate immunity genes associated with development of bronchiolitis obliterans after lung transplantation. J Heart Lung Transplant 2010;29:665–671 [DOI] [PubMed] [Google Scholar]
- 41.Munster JM, van der Bij W, Breukink MB, van der Steege G, Zuurman MW, Hepkema BG, Verschuuren EA, van Son WJ, Seelen MA. Association between donor MBL promoter haplotype and graft survival and the development of BOS after lung transplantation. Transplantation 2008;86:1857–1863 [DOI] [PubMed] [Google Scholar]
- 42.Palmer SM, Burch LH, Davis RD, Herczyk WF, Howell DN, Reinsmoen NL, Schwartz DA. The role of innate immunity in acute allograft rejection after lung transplantation. Am J Respir Crit Care Med 2003;168:628–632 [DOI] [PubMed] [Google Scholar]
- 43.Palmer SM, Burch LH, Trindade AJ, Davis RD, Herczyk WF, Reinsmoen NL, Schwartz DA. Innate immunity influences long-term outcomes after human lung transplant. Am J Respir Crit Care Med 2005;171:780–785 [DOI] [PubMed] [Google Scholar]
- 44.Palmer SM, Klimecki W, Yu L, Reinsmoen NL, Snyder LD, Ganous TM, Burch L, Schwartz DA. Genetic regulation of rejection and survival following human lung transplantation by the innate immune receptor CD14. Am J Transplant 2007;7:693–699 [DOI] [PubMed] [Google Scholar]
- 45.Chiarini M, Sabelli C, Melotti P, Garlanda C, Savoldi G, Mazza C, Padoan R, Plebani A, Mantovani A, Notarangelo LD, et al. PTX3 genetic variations affect the risk of Pseudomonas aeruginosa airway colonization in cystic fibrosis patients. Genes Immun 2010;11:665–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olesen R, Wejse C, Velez DR, Bisseye C, Sodemann M, Aaby P, Rabna P, Worwui A, Chapman H, Diatta M, et al. DC-SIGN (CD209), pentraxin 3 and vitamin D receptor gene variants associate with pulmonary tuberculosis risk in West Africans. Genes Immun 2007;8:456–467 [DOI] [PubMed] [Google Scholar]
- 47.Ansari AA, Reimann KA, Mayne AE, Takahashi Y, Stephenson ST, Wang R, Wang X, Li J, Price AA, Little DM, et al. Blocking of α4β7 gut-homing integrin during acute infection leads to decreased plasma and gastrointestinal tissue viral loads in simian immunodeficiency virus–infected rhesus macaques. J Immunol 2011;186:1044–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee KH, Liu YJ, Biswas A, Ogawa C, Kobayashi KS. A novel aminosaccharide compound blocks immune responses by Toll-like receptors and nucleotide-binding domain, leucine-rich repeat proteins. J Biol Chem 2011;286:5727–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimamoto A, Chong AJ, Yada M, Shomura S, Takayama H, Fleisig AJ, Agnew ML, Hampton CR, Rothnie CL, Spring DJ, et al. Inhibition of Toll-like receptor 4 with eritoran attenuates myocardial ischemia–reperfusion injury. Circulation 2006;114:I270–I274 [DOI] [PubMed] [Google Scholar]
- 50.Barreiro LB, Ben-Ali M, Quach H, Laval G, Patin E, Pickrell JK, Bouchier C, Tichit M, Neyrolles O, Gicquel B, et al. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet 2009;5:e1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferwerda B, McCall MB, Alonso S, Giamarellos-Bourboulis EJ, Mouktaroudi M, Izagirre N, Syafruddin D, Kibiki G, Cristea T, Hijmans A, et al. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc Natl Acad Sci USA 2007;104:16645–16650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma X, Liu Y, Gowen BB, Graviss EA, Clark AG, Musser JM. Full-exon resequencing reveals Toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS One 2007;2:e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plantinga TS, Ioana M, Alonso S, Izagirre N, Hervella M, Joosten LA, van der Meer JW, de la Rua C, Netea MG. The evolutionary history of TLR4 polymorphisms in Europe. J Innate Immun 2012;4:168–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vasseur E, Patin E, Laval G, Pajon S, Fornarino S, Crouau-Roy B, Quintana-Murci L. The selective footprints of viral pressures at the human RIG-I–like receptor family. Hum Mol Genet 2011;20:4462–4474 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.