Abstract
Objectives
To investigate the development of mutational resistance to antibiotics in staphylococcal biofilms.
Methods
Mutation frequencies to resistance against mupirocin and rifampicin were determined for planktonic cultures and for biofilms generated using either a novel static biofilm model or by continuous flow. DNA microarray analysis was performed to detect differences in transcriptional profiles between planktonic and biofilm cultures.
Results
The mutability of biofilm cultures increased up to 60-fold and 4-fold for S. aureus and S. epidermidis, respectively, compared with planktonic cultures. Incorporation of antioxidants into S. aureus biofilms reduced mutation frequencies, indicating that increased oxidative stress underlies the heightened mutability. Transcriptional profiling of early biofilm cultures revealed up-regulation of the superoxide dismutase gene, sodA, also suggestive of enhanced oxidative stress in these cultures. The addition of catalase to biofilms of S. aureus SH1000 reduced mutation frequencies, a finding which implicated hydrogen peroxide in increased biofilm mutability. However, catalase had no effect on biofilm mutability in S. aureus UAMS-1, suggesting that there is more than one mechanism by which the mutability of staphylococci may increase during the biofilm mode of growth.
Conclusion
Our findings suggest that biofilms represent an enriched source of mutational resistance to antibiotics in the staphylococci.
Introduction
In the natural environment bacteria frequently associate with surfaces [1]. This adherent mode of growth, usually associated with extracellular matrix production, is known as a biofilm. The biofilm growth state is also adopted by members of the human microbiome and by bacterial pathogens during a variety of acute and chronic infections [2], [3]. Biofilm-associated infections are notoriously difficult to eradicate due to the refractory nature of organisms in this growth state which are able to resist killing by the majority of antibiotics and the immune system [4], [5], [6], [7], [8]. Furthermore, there is limited but growing evidence to suggest that biofilms might facilitate the emergence of antibiotic resistance. Enhanced rates of conjugation have been reported in biofilms of enterococci and Pseudomonas spp [9], [10] and increased mutation frequencies to antibiotic resistance have been detected in biofilms of Pseudomonas aeruginosa and Streptococcus pneumoniae [11], [12].
Staphylococci cause an array of human infections which involve a biofilm component, including osteomyelitis, endocarditis, and infections associated with indwelling medical devices [13], [14], [15], [16]. It is unknown whether the staphylococcal biofilm might facilitate the emergence of resistance to antibacterial agents. Therefore we examined whether staphylococci become more mutable during the biofilm mode of growth compared with planktonic culture. The majority of established biofilm models are low throughput in nature, as with most flow systems [11], [17], whilst those that are high throughput (e.g. those generated using microtitre plate models) yield biofilms with comparatively low cell densities [18]. Therefore for this study we developed and validated a new static biofilm model to overcome the limitations of the existing approaches. The biofilms generated using our method achieved a yield of cells sufficient for mutation frequency determinations, and enabled biofilm culture for prolonged periods to permit investigation of the mutability of biofilms of differing maturity.
Results
Mutability of Staphylococcal Biofilms
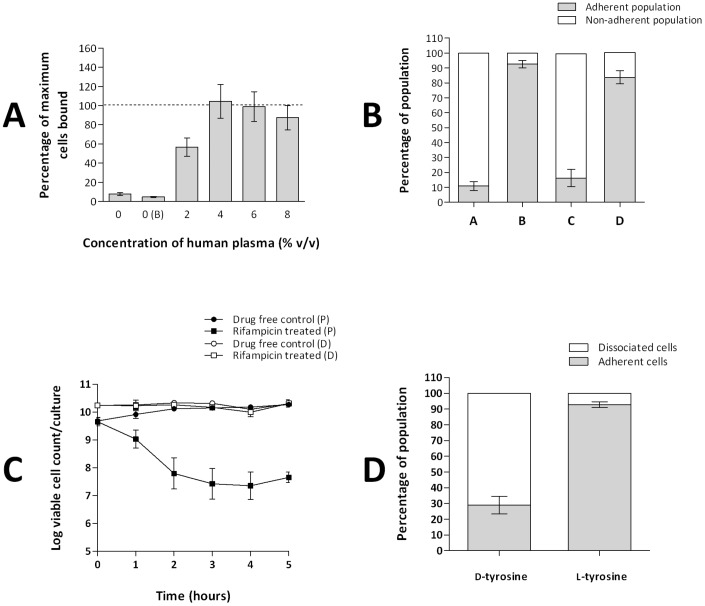
The limitations of existing biofilm models for performing mutation frequency determinations led us to develop a novel cellulose disk static (CDS) biofilm model better suited for this purpose. Although static biofilm models utilising membrane disks have previously been described [19], [20], they have to our knowledge not been widely used for culture of staphylococcal biofilms, nor have they been validated for this purpose. Extensive validation of the CDS model was performed to ensure that biofilms were indeed formed, including demonstration of cell adherence which was reversed in the presence of d-amino acids [21], [22] (Figure 1A, 1B and 1D), confirmation that bacteria associated with the filters exhibited recalcitrance to antibiotic-mediated killing (Figure 1C) and visualisation of biofilms by confocal and atomic force microscopy (data not shown). A description of the methods and results of the validation experiments can be found in Information S1.
Figure 1. Development and validation of the cellulose disk static biofilm model.
A. Effect of human plasma on the adherence of S. aureus SH1000 to cellulose disks. On the x-axis, 0 represents no human plasma or buffer and 0(B) is buffer only. Data indicate the proportion of adherent cells, in 48 hr disk cultures, as a percentage of the maximum achieved at 10% (v/v) human plasma (dashed line) (1.8×1010 cfu/disk). Data from three experimental replicates. Error bars indicate standard error. B. Determination of the proportion of adherent and planktonic cells present in S. aureus SH1000 cellulose disk cultures. (A) Cultures grown for 48 hrs in the absence of human plasma. (B) Cultures grown for 48 hrs with human plasma (4% v/v) added prior to inoculation. (C) Cultures grown for 144 hrs with human plasma (4% v/v) added prior to inoculation. (D) Cultures grown for 144 hrs with human plasma (4% v/v) added prior to inoculation and every 48 hrs. Data from three experimental replicates. Error bars indicate standard error. C. Time-kill curves of S. aureus SH1000 exponential phase planktonic (P) and 48 hr cellulose disk (D) cultures exposed to 0.25 mg/L rifampicin. Data from three experimental replicates. Error bars indicate standard deviations. D. The dissociation of adherent cells of S. aureus SH1000 from cellulose disk cultures in the presence of d-tyrosine (100 µM) and l-tyrosine (100 µM). Data from three experimental replicates. Error bars indicate standard error. Methodology and additional information regarding validation of the cellulose disk model can be found in Supporting Information S1.
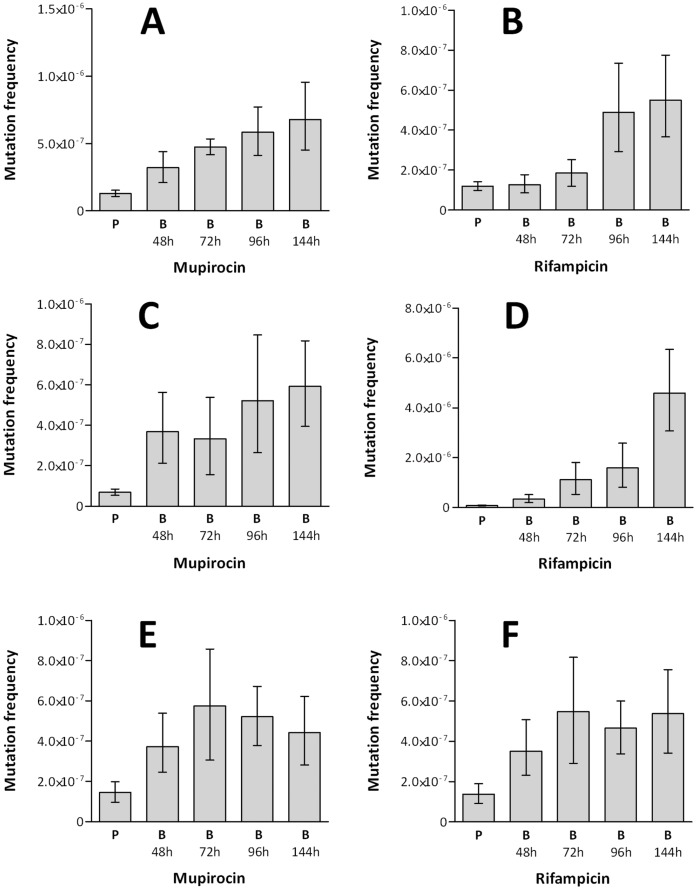
Mutation frequencies were determined for S. aureus SH1000 grown in planktonic cultures in MHB for 18 hrs, and for CDS biofilms of various maturation states, from 48 to 144 hrs. Compared with planktonic cultures, maximum mutation frequency increases of 4.1-fold and 4.6-fold were observed in the 96 hr biofilm cultures, using rifampicin and mupirocin selection, respectively (Figure 2A and 2B). To confirm that this increase in mutation frequency was not the result of using a different growth media in the biofilm cultures (BHA) compared with planktonic cultures (MHB), or as a consequence of the extended periods of incubation to which the biofilms were subjected, we also determined the mutation frequency of planktonic cultures of S. aureus SH1000 incubated for 96 hrs in BHB; no significant difference in mutability was observed for these cultures compared with planktonic cultures grown for 18 hrs in MHB (data not shown).
Figure 2. Mutation frequencies of static staphylococcal biofilms.
Mutation frequencies of planktonic and biofilm cultures (grown using the cellulose disk biofilm model) of S. aureus SH1000 (A & B), S. aureus UAMS-1 (C & D) and S. epidermidis RP62A (E & F), determined by selection of spontaneous mutants resistant to mupirocin (A, C & E) or rifampicin (B, D & F). P indicates planktonic cultures and 48 h, 72 h, 96 h and 144 h indicate biofilm (B) incubation time. Data from six experimental replicates. Error bars indicate 95% confidence intervals.
To examine whether increased mutability is associated with the biofilm mode of growth in other strains of S. aureus, equivalent experiments were performed with the prolific biofilm-forming strain, UAMS-1 [23]. CDS biofilms of UAMS-1 exhibited an even greater increase in mutability over planktonic cultures compared to SH1000; maximum mutability was observed in biofilms grown for 144 hr, at which point increases in mutation frequency to rifampicin and mupirocin resistance increased 59.5-fold and 8.6-fold, respectively (Figure 2C and 2D). Increased mutability in biofilms could also be demonstrated in staphylococci other than S. aureus. Mutation frequency determinations for planktonic and CDS biofilm cultures of S. epidermidis RP62A revealed maximal mutability after 72 hrs of biofilm growth, at which point mutation frequencies to rifampicin and mupirocin resistance were 4.0-fold greater than observed with planktonic cultures (Figure 2E and 2F).
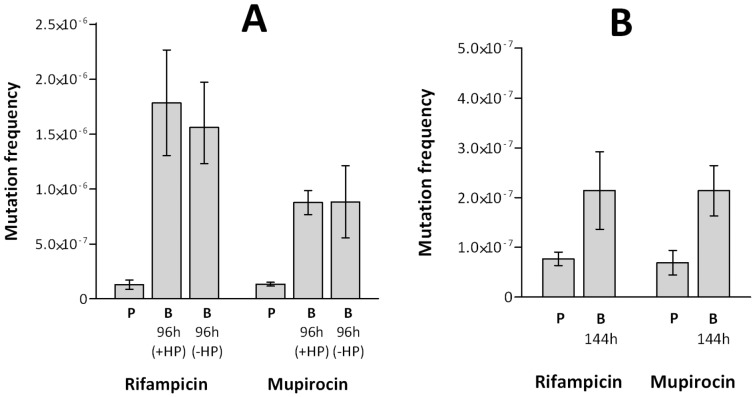
To ensure that the observed increases in mutability were neither unique to static biofilms formed in the absence of shear forces, nor an artefact of the CDS biofilm model, mutation frequencies to antibiotic resistance were also determined for biofilms of S. aureus SH1000 and UAMS-1 cultured using the Sorbarod flow system. Furthermore, to exclude the possibility of an impact on mutability by the human plasma used to prime the substratum for maximum biofilm yield in the CDS biofilm model, these experiments were conducted both in the presence and absence of human plasma. Sorbarod biofilms were grown for 96 hrs (SH1000) and 144 hrs (UAMS-1), since the greatest increases in mutability were observed at these time-points in the CDS model. S. aureus SH1000 Sorbarod biofilms exhibited 14.4-fold and 6.4-fold increases in mutation frequency to rifampicin and mupirocin resistance, respectively, compared with planktonic cultures (Figure 3A). Mutation frequency increases of 2.2-fold (rifampicin) and a 3.1-fold (mupirocin), compared with planktonic cultures, were also observed in S. aureus UAMS-1 Sorbarod biofilms (Figure 3B). Biofilms grown in the presence or absence of plasma exhibited no significant differences in mutation frequency (Figure 3A).
Figure 3. Mutation frequencies of S. aureus biofilms grown under constant flow.
Mutation frequencies of planktonic and biofilm cultures (grown in the Sorbarod model) of S. aureus SH1000 (A) and S. aureus UAMS-1 (B) determined by rifampicin and mupirocin selection. P indicates planktonic cultures and 96 h and 144 h indicate biofilm (B) incubation time. Biofilm cultures were incubated in the presence (+HP) and absence (−HP) of 4% (v/v) human plasma. Data are based on three experimental replicates. Error bars indicate 95% confidence intervals.
Oxidative Stress Contributes to Increased Biofilm Mutability
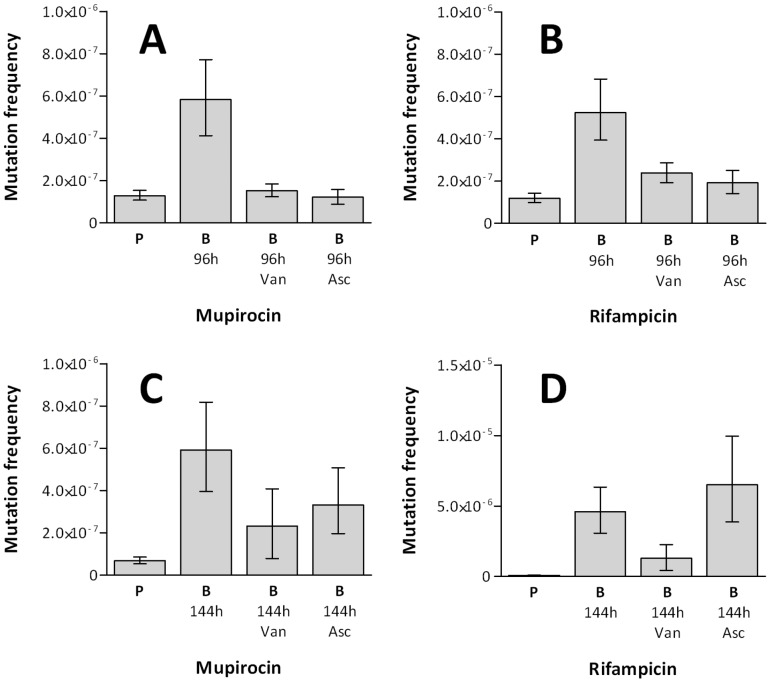
Previous studies have suggested that oxidative stress in P. aeruginosa and S. pneumoniae biofilms might prompt phenotypic and genotypic variation [11], [12], [24]. To investigate whether oxidative stress has a role in increasing mutability of staphylococcal biofilms, we examined the impact on mutation frequencies to antibiotic resistance of incorporating the antioxidants vanillin and ascorbic acid into biofilm cultures at ¼×MIC. CDS biofilms of S. aureus SH1000 demonstrated significant reductions in mutation frequency in the presence of either antioxidant compound (Figure 4A and 4B); in some cases the frequency of mutation decreased to that seen with planktonic cultures. Similarly, biofilms of S. aureus UAMS-1 demonstrated reduced mutation frequencies to mupirocin resistance for both antioxidants tested; however, only vanillin reduced the mutation frequency to rifampicin resistance (Figure 4C and 4D). By contrast, the addition of antioxidants to planktonic cultures of SH1000 or UAMS-1 did not reduce the basal mutation frequency to antibiotic resistance (data not shown).
Figure 4. The effect of antioxidants on S. aureus biofilm mutability.
Mutation frequencies of S. aureus SH1000 (A & B) and UAMS-1 (C & D) planktonic (P) and biofilm (B) cultures in the presence of antioxidants vanillin (Van) and ascorbic acid (Asc). Mutation frequencies were determined using mupirocin (A & C) and rifampicin (B & D) selection. Data from six experimental replicates. Error bars indicate 95% confidence intervals.
In P. aeruginosa biofilms, increased oxidative stress appears to result in part from down-regulation of enzymes that participate in the cellular defence to reactive oxygen species (ROS). To examine whether a reduction in the ability of sessile staphylococci to detoxify ROS could explain increased mutability of the staphylococcal biofilm, we performed transcriptome analysis by DNA microarray to look for de-regulation of genes implicated in ROS protection/detoxification in 48 and 144 hr biofilms of SH1000 compared to planktonic cultures (Table 1). Transcriptional profiles of 48 hr biofilm cultures showed numerous similarities with previously published expression data for S. aureus biofilms (Table 1) [25], including differential expression of SAOUHSC_01216 (sucC), SAOUHSC_01418 (odhA) and SAOUHSC_02350 (atpB). We noted up-regulation of at least two genes whose products are involved in protection against oxidative stress, namely sodA (SAOUHSC_01653), encoding superoxide dismutase, and qoxA (SAOUHSC_01002), encoding quinol oxidase subunit [26], [27]. Enhanced expression of sodA, following 48 hr biofilm growth, was confirmed using qRT-PCR (1.91-fold), although no differential expression of qoxA expression was detected (data not shown). Increased expression of antioxidant enzymes, such as SodA, in the biofilm further corroborates the idea that sessile staphylococci are subject to increased levels of oxidative stress, but suggests that increased oxidative stress is not the result of a reduced ability of staphylococci to detoxify ROS. We also performed transcriptional profiling of S. aureus biofilms grown for 144 hrs; to our knowledge, no transcriptional analyses are available in the published literature for S. aureus biofilms grown for longer than 48 hrs. However, the transcriptional profile of 144 hr biofilms exhibited little differential gene expression compared with planktonic cultures (Table 1).
Table 1. Differentially regulated genes in S. aureus SH1000 biofilms, grown for 48 hrs and 144 hrs, compared with planktonic cultures. ORFs encoding hypothetical proteins not shown.
| ORF | Gene | Function | Fold change |
| 48 hr Biofilms/Planktonic | |||
| SAOUHSC_00502 | ctsR | Transcriptional regulator CtsR | 2.20 up |
| SAOUHSC_00526 | Putative ribosomal protein L7Ae-like | 2.30 up | |
| SAOUHSC_00894 | rocD2 | Ornithine aminotransferase 2 | 2.61 up |
| SAOUHSC_00895 | Glutamate dehydrogenase, NAD-specific, putative | 2.36 up | |
| SAOUHSC_01002 | qoxA | Probable quinol oxidase subunit 2 | 2.00 up |
| SAOUHSC_01042 | Dihydrolipoamide S-acetyltransferase component of pyruvate dehydrogenase complex E2, putative | 2.17 up | |
| SAOUHSC_01103 | Succinate dehydrogenase cytochrome b-558 subunit, putative | 2.16 up | |
| SAOUHSC_01216 | sucC | Succinyl-CoA synthetase subunit beta | 2.44 up |
| SAOUHSC_01389 | pstS | Phosphate-binding protein PstS | 2.24 up |
| SAOUHSC_01407 | Putative XpaC protein | 2.04 up | |
| SAOUHSC_01408 | TelA-like protein | 2.32 up | |
| SAOUHSC_01418 | odhA | 2-oxoglutarate dehydrogenase E1 component | 2.02 up |
| SAOUHSC_01462 | gpsB | Cell cycle protein gpsB | 2.37 up |
| SAOUHSC_01477 | Putative Zn dependent protease | 2.38 up | |
| SAOUHSC_01653 | sodA | Superoxide dismutase [Mn] 1 | 2.77 up |
| SAOUHSC_01683 | dnaK | Chaperone protein DnaK | 2.47 up |
| SAOUHSC_01684 | grpE | Protein GrpE | 3.15 up |
| SAOUHSC_01685 | hrcA | Heat-inducible transcription repressor HrcA | 2.84 up |
| SAOUHSC_01720 | Putative Holliday junction resolvase | 2.00 up | |
| SAOUHSC_01801 | icd | Isocitrate dehydrogenase | 2.26 up |
| SAOUHSC_01880 | Transposase domain protein | 3.32 down | |
| SAOUHSC_02261 | agrB | Accessory gene regulator protein B | 2.76 up |
| SAOUHSC_02262 | agrD | Accessory gene regulator protein D | 3.56 up |
| SAOUHSC_02264 | agrC | Accessory gene regulator protein C | 2.62 up |
| SAOUHSC_02350 | atpB | ATP synthase subunit A | 2.03 up |
| SAOUHSC_02488 | rpmJ | 50S ribosomal protein L36 | 2.23 up |
| 144 hr Biofilms/Planktonic | |||
| SAOUHSC_01683 | dnaK | Chaperone protein DnaK | 2.17 up |
| SAOUHSC_01684 | grpE | Protein GrpE | 2.47 up |
| SAOUHSC_01685 | hrcA | Heat-inducible transcription repressor HrcA | 2.43 up |
| SAOUHSC_02260 | hld | Delta hemolysin | 3.55 down |
| SAOUHSC_02466 | Truncated MHC class II analog protein | 2.34 down | |
To further examine a potential role for antioxidant enzymes in the biofilm mutability of S. aureus, mutation frequencies for planktonic and biofilm cultures of the antioxidant enzyme-deficient strains S. aureus MHK11AM (SH1000 lacking the superoxide dismutases, SodA and SodM) and KC043 (SH1000 lacking the peroxidases, KatA and AhpC) were determined using rifampicin selection and compared with the wild-type. Although loss of these enzymes resulted in a further increase in biofilm mutability compared with S. aureus SH1000 (MHK11AM –2.2-fold, KC043–8.3-fold), a comparable increase in mutability was also observed in planktonic culture (MHK11AM –2.9-fold, KC043–10.8-fold). Since strains lacking antioxidant enzymes exhibited the same increased mutability in biofilms over planktonic culture as the wild-type strain, it appears that these enzymes are not involved in the phenomenon of biofilm mutability.
Investigating a Role for Hydrogen Peroxide in Staphylococcal Biofilm Mutability
S. pneumoniae exhibits increased mutability during the biofilm mode of growth [12]. This phenomenon has been attributed to the activity of the pyruvate oxidase, SpxB [12], an enzyme that catalyzes the formation of acetyl phosphate from pyruvate, yielding the ROS hydrogen peroxide as a by-product. Since S. aureus encodes a pyruvate oxidase (CidC) with 33% identity to SpxB, we investigated the possibility that endogenous hydrogen peroxide may also be involved in staphylococcal biofilm mutability. Addition of catalase to biofilms of SH1000 reduced mutation frequencies (Figure 5), indicating that hydrogen peroxide indeed drives enhanced mutability in this strain during the biofilm mode of growth. In contrast, catalase did not significantly reduce the mutability of UAMS-1 biofilms (data not shown). To assess the role of the CidC pyruvate oxidase in SH1000 biofilm mutability, the mutation frequencies of SH1000ΔcidC biofilms were determined. However, disruption of cidC in SH1000 caused no significant reduction in the mutability of biofilms (Figure 5).
Figure 5. The effect of catalase and cidC inactivation on S. aureus biofilm mutability.
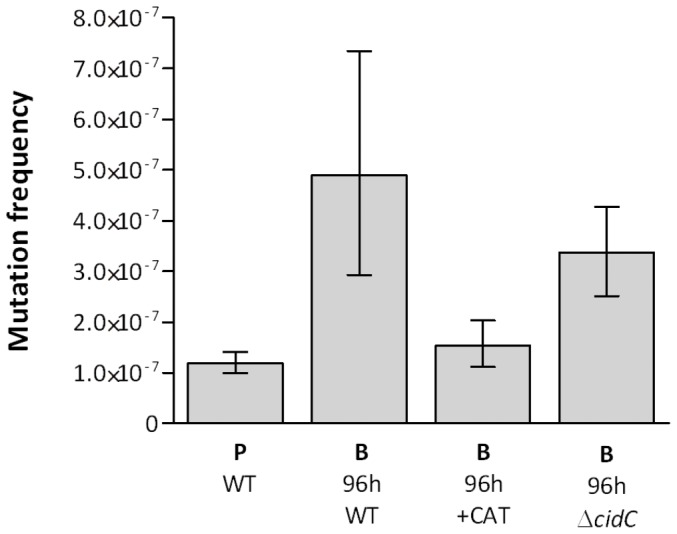
Mutation frequencies of S. aureus SH1000 planktonic (P) and 96 h biofilm (B) cultures, determined by rifampicin selection. WT indicates wild-type (S. aureus SH1000), +CAT indicates wild-type supplemented with catalase (4 U/ml) and ΔcidC indicates a CidC-deficient mutant of S. aureus SH1000. Data from six experimental replicates. Error bars indicate 95% confidence intervals.
Discussion
Mutation plays an important role in the development of antibiotic resistance in S. aureus and other staphylococci [28]. Single point mutations in the chromosome can cause clinically-significant levels of resistance to a variety of antistaphylococcal agents, including rifampicin, mupirocin, fusidic acid and fluoroquinolones [28], whilst the accumulation of mutations at multiple loci can lead to resistance to glycopeptides and daptomycin [29], [30]. Any factor that elevates the basal mutation frequency will serve to accelerate the emergence of antibiotic resistance; we show here that growth as a biofilm appears to represent one such factor for the staphylococci.
Across a series of experiments using different staphylococcal species and strains, selecting agents and biofilm models, we consistently observed enhanced mutability in biofilm cultures versus planktonic cultures, with increases in mutation frequency ranging between ∼2-fold and ∼60-fold. At the upper end of this range, the mutation frequencies are comparable to those exhibited by hypermutator strains of S. aureus defective for DNA repair [31]. Although a number of variables appear to affect the level of mutability observed in staphylococcal biofilms, biofilm maturity seems particularly important; the greatest increases in mutability were associated with biofilms that had been established for ≥96 hrs (Figure 2).
Although the increases in mutation frequencies of staphylococcal biofilm populations were in most cases relatively modest, this phenomenon may nonetheless have considerable impact on the development of antibiotic resistance in S. aureus and other staphylococci. This is particularly the case since staphylococcal infections frequently include a biofilm component [13], [14], [15], [32]. Furthermore, since biofilm cells are protected from antibiotic-mediated killing, bacteria within the biofilm will benefit not only from an elevated mutation frequency to antibiotic resistance, but will also experience extended periods of drug selection. Staphylococcal biofilms may therefore represent an important site for the development of antibiotic resistant mutants, which could then become dispersed from the biofilm during the dissemination phase of biofilm growth [33].
Loss of the increased mutability upon incorporation of antioxidants into biofilms indicates that staphylococci in biofilms are more mutable because of oxidative stress. This idea is supported by the transcriptional analysis of 48 hr biofilms, which revealed up-regulation of genes encoding antioxidant enzymes, such as SodA, compared with planktonic cultures. Furthermore, it is likely that this oxidative stress is generated within the staphylococcal cell, as sodA expression is responsive to endogenous, rather than exogenous, superoxide [34], [35]. Consistent with this hypothesis it has also been suggested that P. aeruginosa is subject to endogenous oxidative stress during biofilm growth [24].
Enhanced mutability in S. pneumoniae biofilms has been attributed to production of hydrogen peroxide by the pyruvate oxidase, SpxB. Although biofilm mutability in S. aureus SH1000 was also attributed to the presence of hydrogen peroxide, this was not the result of the staphylococcal pyruvate oxidase, CidC, suggesting that another metabolic source of hydrogen peroxide probably accounts for the increase in mutation frequency in this strain. The inability of catalase to affect the mutability of S. aureus UAMS-1 biofilms suggests that there is more than one mechanism by which the mutability of staphylococci may increase during the biofilm mode of growth.
In conclusion, we have shown that staphylococcal biofilms exhibit an increase in basal mutation frequency as a consequence of increased oxidative stress, a phenomenon which will accelerate the emergence of antibiotic resistance in this genus.
Materials and Methods
Planktonic Growth of Bacterial Strains and Antibiotic Susceptibility Determinations
Strains used in this study are listed in Table 2. Antibiotic susceptibility testing was performed by agar dilution following the guidelines of the British Society for Antimicrobial Chemotherapy (BSAC), but used Mueller-Hinton broth and agar (MHB and MHA) in place of Iso-Sensitest medium [36]. All chemicals were from Sigma-Aldrich (Dorset, UK), with the exception of mupirocin and vanillin which were from USP Reference Standards (Rockville, USA) and MP Biomedicals (Illkirch, France), respectively. Planktonic cultures were grown for 18 hrs at 37°C with shaking in MHB or brain-heart infusion broth (BHB). Where required, antioxidants were added to cultures at ¼×MIC.
Table 2. Bacterial strains used in this study.
| Strain | Comments | MIC (µg/ml) | MIC (mg/ml) | Source | ||
| RIF | MUP | VAN | ASC | |||
| S. aureus SH1000 | Standard laboratory strain | 0.008 | 0.125 | 4 | 0.125 | [46], [47] |
| S. aureus UAMS-1 | Biofilm proficient strain | 0.016 | 0.125 | 4 | 0.125 | [23] |
| S. epidermidis RP62A | Biofilm proficient strain (ATCC35984) | 0.016 | 0.125 | nd | nd | ATCC |
| S. aureus KC043 | KatA/AhpC defective derivative of SH10000 | 0.032 | nd | nd | nd | [48] |
| S. aureus MHK11AM | SodA/SodM defective derivative of SH1000 | 0.032 | nd | nd | nd | [35] |
| S. aureus USA300ΔcidC | CidC defective derivative of USA300 (strain NE564) | nd | nd | nd | nd | NARSA |
| S. aureus SH1000ΔcidC | CidC defective derivative of SH1000 | 0.008 | nd | nd | nd | This study |
RIF – Rifampicin, MUP – Mupirocin, VAN – Vanillin, ASC – Ascorbic acid. nd – Not determined. NARSA – Network on Antimicrobial Resistance in Staphylococcus aureus.
Cellulose Disk Static (CDS) Biofilm Model
Mixed cellulose-ester membrane filter disks (25 mm diameter, 0.22 µm pore size; Millipore, Billerica, USA) were used as a substratum for the formation of staphylococcal biofilms. To promote biofilm formation, disks were immersed in 4% v/v normal pooled human plasma (Sera Laboratories International, Bolney, UK) diluted in 0.05 M carbonate buffer [37] and incubated at 4°C overnight prior to use. Plasma-soaked disks were immersed in a saturated culture of stationary phase bacteria, placed on brain heart infusion agar (BHA), and incubated at 37°C. To harvest cells from S. aureus biofilms for mutation frequency determinations, loosely-associated cells were first removed from the disks by gentle agitation in sterile saline, and the disks then incubated for 30 mins at 37°C in the presence of cellulase (1 mg/ml in 0.05 M citrate buffer pH 5.0) [38]. In the case of S. epidermidis biofilms, cells were liberated from the disks by incubation in phosphate-buffered saline pH 7.4 containing 625 µM sodium metaperiodate, 3.125 mM sodium acetate for 60 mins at 37°C [39]. In both cases, detached cells were then washed in sterile saline, harvested by centrifugation for 10 mins at 5000×g, and resuspended in sterile saline before proceeding to mutation frequency determinations as described below. Where required, biofilm cultures were supplemented with antioxidants (ascorbic acid, vanillin and catalase) by incorporation into BHA and by addition drop-wise directly to the cell population. Ascorbic acid and vanillin were added at ¼×MIC (0.03 mg/ml and 1 mg/ml, respectively); catalase was added at 4 U/ml. Methodology regarding the development and validation of this biofilm model can be found in Information S1.
Sorbarod (Constant Flow) Biofilm Model
Inoculation, incubation and harvesting of biofilms on compacted cellulose (Sorbarod) filters were performed essentially as previously described [11]. To promote biofilm formation, Sorboarod filters were pre-treated with 1 ml of 4% (v/v) human plasma for 16 hrs at 4°C. Cells were harvested by vigorously vortexing the filters, followed by sonication for 15 mins.
Mutation Frequency Determinations
Mutation frequencies were determined essentially as described by O’Neill et al [40]. Bacterial suspensions containing ∼109 cfu/ml were spread onto selective MHA containing 4×MIC (Table 2) of rifampicin or mupirocin to recover spontaneous antibiotic-resistant mutants. Culture dilutions containing ∼102 cfu/ml were spread onto non-selective MHA to determine viable cell numbers. Agar plates were incubated for 48–72 hrs at 37°C, and mutation frequencies expressed as the number of antibiotic-resistant mutants recovered as a proportion of the total cell count. Errors were calculated as 95% confidence intervals using Fieller’s Theorem [41]. Mutation frequencies were determined for six experimental replicates, each consisting of three biological replicates.
Transcriptional Profiling by DNA Microarray
Planktonic cultures of SH1000 were combined with two volumes of RNAprotect Bacteria Reagent (Qiagen, Crawley, UK), and the cells harvested by centrifugation at 5000×g. CDS biofilm cultures of SH1000 were immersed in RNAprotect, detached with buffered cellulase and then centrifuged at 5000×g to pellet cells. Cells were washed in RNAprotect and stored at −80°C until required. Pellets were thawed at room temperature, washed in 10 ml of TE buffer (pH 7.4), resuspended in 1 ml of TE buffer (pH 7.4) containing 200 µg lysostaphin/ml (Sigma-Alrich, Dorset, UK), and incubated at 37°C for 60–90 mins. Proteinase K (40 µg/ml) (Sigma-Alrich, Dorset, UK) was then added to the mixture, and incubated continued for 10 mins at room temperature. Total RNA was purified from these samples using the RNeasy Midi kit (Qiagen) following the manufacturer’s instructions, including an on-column DNase digestion using the RNase-free DNase kit (Qiagen). cDNA synthesis, cDNA labelling, hybridisation using 4×72K multiplex microarrays, and quantification of gene expression were performed by Roche Nimblegen (Madison, USA). Of the 2892 predicted protein coding open reading frames of the S. aureus NCTC8325 genome, 2887 were represented on the microarrays. Data were analysed using ArrayStar™ (DNASTAR, Madison, USA), and genes were considered differentially regulated if the average gene expression value showed ≥2-fold increase or decrease with a P value of ≤0.05 by student’s t test [42], [43]. Transcriptional profiling was performed for three biological replicates, and the data deposited at the NCBI Gene Expression Omnibus under Accession GSE35837.
To confirm the transcriptional profiling data, real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed. Synthesis of cDNA using total RNA (RT) and qPCR were performed using the Quantitect RT kit and Quantitect SYBR Green PCR kit (Qiagen), respectively, according to the manufacturer’s instructions. The relative quantities of gyrA (endogenous control) and sodA mRNA transcripts in planktonic and 48 hr biofilm cultures were determined by qPCR using primers GyrA_Fwd (5′-ATAGAAATGGTAAGATTGCGATT-3′), GyrA_Rev (5′-ACCTTTCACACCCGTTGC-3′), SodA_Fwd (5′-GCGCCAATGTAGTCAGGGCGTTTG-3′) and SodA_Rev (5′-TGCACGCTTTGGTTCAGGTTGGG-3′) by relative standard curve quantification. Data were analysed using MxPro Mx3005P software (Agilent, Wokingham, UK). Relative expression levels of sodA were calculated with respect to gyrA as the endogenous control. Data were regarded as significant if a P value = ≤0.05 was returned following a Student’s t-test.
Inactivation of the cidC Gene in S. aureus SH1000
The transposon-disrupted cidC gene from S. aureus USA300ΔcidC (NE564; Nebraska transposon mutant library [44]) was transduced into S. aureus SH1000 using Φ11 [45], to generate S. aureus SH1000ΔcidC. Successful transduction of this locus to SH1000 was confirmed by PCR.
Supporting Information
Methodology and additional information regarding optimisation and validation of the cellulose disk model.
(DOCX)
Acknowledgments
We thank Prof E. Ingham and Dr D. Thomas for assistance with qRT-PCR, Prof S. Foster for provision of strains KC043 and MHK11AM, and Dr A. Hedges for advice on statistical analysis.
Funding Statement
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC PhD studentship) and the British Society for Antimicrobial Chemotherapy (Grant no. GA849). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zobell CE (1943) The effect of solid surfaces upon bacterial activity. J Bacteriol 46: 39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Macfarlane S (2008) Microbial biofilm communities in the gastrointestinal tract. J Clin Gastroenterol 42: S142–143. [DOI] [PubMed] [Google Scholar]
- 3. Hall-Stoodley L, Costerton JW, Stoodley P (2004) Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol 2: 95–108. [DOI] [PubMed] [Google Scholar]
- 4. Jensen PO, Givskov M, Bjarnsholt T, Moser C (2010) The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol 59: 292–305. [DOI] [PubMed] [Google Scholar]
- 5. Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, et al. (2004) A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem 279: 54881–54886. [DOI] [PubMed] [Google Scholar]
- 6. Stewart PS, Costerton JW (2001) Antibiotic resistance of bacteria in biofilms. Lancet 358: 135–138. [DOI] [PubMed] [Google Scholar]
- 7. Ashby MJ, Neale JE, Knott SJ, Critchley IA (1994) Effect of antibiotics on non-growing planktonic cells and biofilms of Escherichia coli . J Antimicrob Chemother 33: 443–452. [DOI] [PubMed] [Google Scholar]
- 8. Izano EA, Shah SM, Kaplan JB (2009) Intercellular adhesion and biocide resistance in nontypeable Haemophilus influenzae biofilms. Microb Pathog 46: 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cvitkovitch DG (2004) Genetic Exchange in Biofilms. In: Ghannoum M, O’Toole GA, editors. Microbial Biofilms. Washington DC: ASM Press. 192–205.
- 10. Ehlers LJ, Bouwer EJ (1999) RP4 plasmid transfer among species of Pseudomonas in a biofilm reactor. Water Science and Technology 39: 163–171. [Google Scholar]
- 11. Driffield K, Miller K, Bostock JM, O’Neill AJ, Chopra I (2008) Increased mutability of Pseudomonas aeruginosa in biofilms. J Antimicrob Chemother 61: 1053–1056. [DOI] [PubMed] [Google Scholar]
- 12. Allegrucci M, Sauer K (2008) Formation of Streptococcus pneumoniae non-phase-variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J Bacteriol 190: 6330–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akiyama H, Hamada T, Huh WK, Yamasaki O, Oono T, et al. (2003) Confocal laser scanning microscopic observation of glycocalyx production by Staphylococcus aureus in skin lesions of bullous impetigo, atopic dermatitis and pemphigus foliaceus. Br J Dermatol 148: 526–532. [DOI] [PubMed] [Google Scholar]
- 14. Brady RA, Leid JG, Calhoun JH, Costerton JW, Shirtliff ME (2008) Osteomyelitis and the role of biofilms in chronic infection. FEMS Immunol Med Microbiol 52: 13–22. [DOI] [PubMed] [Google Scholar]
- 15. Heimberger TS, Duma RJ (1989) Infections of prosthetic heart valves and cardiac pacemakers. Infect Dis Clin North Am 3: 221–245. [PubMed] [Google Scholar]
- 16. Schierle CF, De la Garza M, Mustoe TA, Galiano RD (2009) Staphylococcal biofilms impair wound healing by delaying reepithelialization in a murine cutaneous wound model. Wound Repair Regen 17: 354–359. [DOI] [PubMed] [Google Scholar]
- 17. Herles S, Olsen S, Afflitto J, Gaffar A (1994) Chemostat flow cell system: an in vitro model for the evaluation of antiplaque agents. J Dent Res 73: 1748–1755. [DOI] [PubMed] [Google Scholar]
- 18. Ceri H, Olson ME, Stremick C, Read RR, Morck D, et al. (1999) The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37: 1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Anderl JN, Franklin MJ, Stewart PS (2000) Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother 44: 1818–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walters MC III, Roe F, Bugnicourt A, Franklin MJ, Stewart PS (2003) Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother 47: 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, et al. (2010) D-amino acids trigger biofilm disassembly. Science 328: 627–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hochbaum AI, Kolodkin-Gal I, Foulston L, Kolter R, Aizenberg J, et al. (2011) Inhibitory effects of D-amino acids on Staphylococcus aureus biofilm development. J Bacteriol 193: 5616–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, et al. (1995) Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun 63: 3373–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boles BR, Singh PK (2008) Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci U S A 105: 12503–12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Resch A, Rosenstein R, Nerz C, Gotz F (2005) Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol 71: 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Jubelirer S, Garcia Costas AM, Frigaard NU, Bryant DA (2009) Multiple antioxidant proteins protect Chlorobaculum tepidum against oxygen and reactive oxygen species. Arch Microbiol 191: 853–867. [DOI] [PubMed] [Google Scholar]
- 27. Karavolos MH, Horsburgh MJ, Ingham E, Foster SJ (2003) Role and regulation of the superoxide dismutases of Staphylococcus aureus . Microbiology 149: 2749–2758. [DOI] [PubMed] [Google Scholar]
- 28. Woodford N, Ellington MJ (2007) The emergence of antibiotic resistance by mutation. Clin Microbiol Infect 13: 5–18. [DOI] [PubMed] [Google Scholar]
- 29. Kato Y, Suzuki T, Ida T, Maebashi K (2010) Genetic changes associated with glycopeptide resistance in Staphylococcus aureus: predominance of amino acid substitutions in YvqF/VraSR. J Antimicrob Chemother 65: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, et al. (2011) Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med 365: 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O’Neill AJ, Chopra I (2002) Insertional inactivation of mutS in Staphylococcus aureus reveals potential for elevated mutation frequencies, although the prevalence of mutators in clinical isolates is low. J Antimicrob Chemother 50: 161–169. [DOI] [PubMed] [Google Scholar]
- 32. Donlan RM (2001) Biofilms and device-associated infections. Emerg Infect Dis 7: 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaplan JB (2010) Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res 89: 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clements MO, Watson SP, Foster SJ (1999) Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J Bacteriol 181: 3898–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karavolos MH, Horsburgh MJ, Ingham E, Foster SJ (2003) Role and regulation of the superoxide dismutases of Staphylococcus aureus . Microbiology 149: 2749–2758. [DOI] [PubMed] [Google Scholar]
- 36.BSAC (1991) A guide to sensitivity testing. Report of the Working Party on Antibiotic Sensitivity Testing of the British Society for Antimicrobial Chemotherapy. J Antimicrob Chemother 27 Suppl D: 1–50. [PubMed]
- 37. Beenken KE, Blevins JS, Smeltzer MS (2003) Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun 71: 4206–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cescutti P, Toffanin R, Fett WF, Osman SF, Pollesello P, et al. (1998) Structural investigation of the exopolysaccharide produced by Pseudomonas flavescens strain B62 - degradation by a fungal cellulase and isolation of the oligosaccharide repeating unit. Eur J Biochem 251: 971–979. [DOI] [PubMed] [Google Scholar]
- 39. Kogan G, Sadovskaya I, Chaignon P, Chokr A, Jabbouri S (2006) Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol Lett 255: 11–16. [DOI] [PubMed] [Google Scholar]
- 40. O’Neill AJ, Cove JH, Chopra I (2001) Mutation frequencies for resistance to fusidic acid and rifampicin in Staphylococcus aureus . J Antimicrob Chemother 47: 647–650. [DOI] [PubMed] [Google Scholar]
- 41. Cumming G (2009) Inference by eye: reading the overlap of independent confidence intervals. Stat Med 28: 205–220. [DOI] [PubMed] [Google Scholar]
- 42. Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, et al. (2004) Global Gene Expression in Staphylococcus aureus Biofilms. J Bacteriol 186: 4665–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sauer K, Cullen MC, Rickard AH, Zeef LA, Davies DG, et al. (2004) Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol 186: 7312–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bae T, Glass EM, Schneewind O, Missiakas D (2008) Generating a collection of insertion mutations in the Staphylococcus aureus genome using bursa aurealis . Methods Mol Biol 416: 103–116. [DOI] [PubMed] [Google Scholar]
- 45.Foster TJ (1998) Molecular Genetic Analysis of Staphylococcal Virulence. In: Peter Williams JK, George S, editors. Methods in Microbiology: Academic Press. 433–454.
- 46. Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, et al. (2002) σB modulates virulence determinant expression and stress resistance: Characterization of a functional rsbU strain derived from Staphylococcus aureus 8325–4. J Bacteriol 184: 5457–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. O’Neill AJ (2010) Staphylococcus aureus SH1000 and 8325–4: comparative genome sequences of key laboratory strains in staphylococcal research. Lett Appl Microbiol 51: 358–361. [DOI] [PubMed] [Google Scholar]
- 48. Cosgrove K, Coutts G, Jonsson IM, Tarkowski A, Kokai-Kun JF, et al. (2007) Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus . J Bacteriol 189: 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methodology and additional information regarding optimisation and validation of the cellulose disk model.
(DOCX)